Abstract
In the face of the current extinction crisis and severely limited conservation resources, safeguarding the tree of life is increasingly recognized as a high priority. We conducted a first systematic global assessment of the conservation of phylogenetic diversity (PD) that uses realistic area targets and highlights the key areas for conservation of the mammalian tree of life. Our approach offers a substantially more effective conservation solution than one focused on species. In many locations, priorities for PD differ substantially from those of a species-based approach that ignores evolutionary relationships. This discrepancy increases rapidly as the amount of land available for conservation declines, as does the relative benefit for mammal conservation (for the same area protected). This benefit is equivalent to an additional 5900 Myr of distinct mammalian evolution captured simply through a better informed choice of priority areas. Our study uses area targets for PD to generate more realistic conservation scenarios, and tests the impact of phylogenetic uncertainty when selecting areas to represent diversity across a phylogeny. It demonstrates the opportunity of using rapidly growing phylogenetic information in conservation planning and the readiness for a new generation of conservation planning applications that explicitly consider the heritage of the tree of life's biodiversity.
Keywords: mammals, phylogenetic diversity, conservation planning
1. Background
In the face of the current extinction crisis [1] and severely limited conservation resources, safeguarding the tree of life is increasingly recognized as a high priority [2–5]. This has led to a widespread focus on measures informed by evolutionary relationships, such as phylogenetic diversity (PD; [6]). These measures are used for many purposes now, but were originally devised to tackle questions of triage or conservation priority, based on a recognition that named taxa such as species are not equal in the amount of life's diversity that would be lost if a given taxon became extinct. PD counts the phylogenetic branch lengths joining a set of species (or other biota) to the root of their phylogenetic tree, and thus indicates their collective contribution to the tree of life, typically in terms of amounts of neutral genetic variation, or the amount of time for which a given lineage has existed separately. The reasons for conserving PD are largely the same as for other approaches to biodiversity conservation: actual and potential utility to humanity such as ecosystem services, as well as an intrinsic obligation to reduce the rate of anthropogenic decline of life's diversity. Whereas species counts represent the number of taxonomic units, PD quantifies the amount of difference represented by these units. PD has been associated with ecosystem productivity [7] and plant medicinal uses [8], maximizing the persistence of variants with potential to thrive under new environmental conditions, and protecting diversity that is not yet known to be important [9].
A renewed focus on evolutionary relationships in conservation is due, in part, to advances in both phylogenetic and spatial data over the past decade. These advances have radically improved our ability to estimate phylogenies, to quantify PD, to assess how patterns of relatedness and PD vary across geography [9–13] and to identify places where much PD is restricted to small areas [13–17]. This work is crucial in providing an inventory of the world's diversity and a better understanding of the processes responsible for its spatial variation.
These areas of research stand distinct from systematic conservation planning (SCP) which is a widely used technique to identify areas or actions that, in combination, maximize conservation benefit while minimizing the overall cost [18,19]. Spatial SCP accounts for the size and overlap of conservation features when finding the sets of areas that can meet conservation goals most efficiently. Apart from being able to include the real or opportunity costs of conservation (such as land value, management costs and foregone economic uses), SCP can also respond to other processes, such as ecosystem connectivity or threats, to contribute to complex real-world conservation challenges such as in forest management [20], and freshwater [21] and marine environments [22].
Various studies have demonstrated the utility and implementation of these area-selection approaches for PD (including [8,23–27]). However, despite continuing growth in spatial and phylogenetic biodiversity data [10,28,29] and ongoing research interest [30], the broader application of phylogenies in spatial SCP has remained surprisingly limited. Some evidence from simulations [31] and empirical work [32] suggested that the areas selected by species-based conservation also perform well for PD conservation, especially when trees are balanced (i.e. with similar numbers of species in clades of similar age), or evolutionarily distinct species are concentrated in species-rich areas [31]. Even where the use of PD was shown to make a large difference to the choice of areas [8], the actual conservation gain found from this strategy was small (electronic supplementary material figure S3C in [8]). However, the question of whether species-based conservation is an effective surrogate for PD has not been tested in a more realistic scenario—for example, including proper conservation targets—and mounting evidence of diversification and geographical distribution of clades [10,33] suggests that such cases, in which phylogenetic information offers little conservation improvement, should be uncommon.
The choice of conservation targets is known to be important in SCP [34], and we suggest that the target strategy may be central in understanding previous findings of limited conservation benefit of using PD over species in SCP. The single-occurrence approach to phylogenetic SCP, found in most previous studies, considers a species or branch is to be sufficiently protected by reserving one geographical location where it occurs. Ecologically sound conservation requires setting meaningful area targets larger than a single occurrence [35]. While recent studies have used targets, to assess PD conservation [36,37] in existing protected areas (PAs), area targets have not been evaluated in conservation planning, but could support better conservation prioritization and enable us to re-assess the benefits of phylogenetic SCP.
Two developments make it timely to re-assess the role that phylogeny-informed conservation planning could play. First, the greatly improved extent, resolution and availability of phylogenetic [10,28,38] and spatial information now allow for meaningful, phylogenetically informed global conservation assessments for major clades such as mammals and birds [4,39]. Second, earlier assessments have usually relied on custom-built in-house research code [8,26,32], but recent studies [27,40,41] have opened the way to the use of PD with mainstream SCP software packages which are designed to support far more complex and realistic conservation scenarios. These include area targets, as well as integrating biodiversity information with factors such as cost and spatial configuration, to provide spatial conservation solutions in real landscapes. Importantly, they are also supported by a global user community of land-use planners and researchers.
In this study, we build on these advances to provide global, target-area-based SCP that addresses PD, conservation targets and phylogenetic uncertainty. We use a straightforward method to include phylogenetic information in readily accessible spatial planning and decision-support tools [42,43] focusing here on two tools, Marxan [42] and Prioritizr [44], which each allow the setting of area targets for the protection of any feature of conservation value, and explicitly define the conservation objectives being sought. We aim to provide a straightforward and general approach to phylogenetically informed spatial conservation planning for target-based area prioritization, supported by scripts which prepare phylogenetic data for Marxan.
Instead of the traditional use of species as conservation features, we use branches on a phylogeny [27,40,45] and their geographical distribution. Although the phylogeny is still occasionally represented by phylogenetic groups as conservation features [46], branches of a phylogeny represent an easily interpretable and understood conservation metric (millions of years of evolutionary history). Faith [47] argues that branch lengths represent features which capture known and unknown traits which will contribute to human utility (option value [48]) and future evolution. Asmyhr et al. [40] demonstrated the application of Marxan to phylogenetic SCP, using a model system with a very limited number of sites and clades. We use a similar approach, but at a global scale, to optimize across thousands of areas and phylogenetic branches, and to set species and clade level targets for area to be conserved. This supports more reliable and efficient real-world conservation solutions by allowing for ecologically or expert-driven minimum areas. While our target approach is generic, conservation planning at a finer resolution could use detailed information about species biology for targets that are more responsive to the requirements of individual species.
Here, we apply phylogenetically informed SCP with real area targets to 4778 terrestrial mammal species. We (i) identify priority areas for the efficient safeguarding of mammalian evolutionary diversity and (ii) compare PD-based conservation planning with a traditional species-focused approach while (iii) using meaningful area targets and (iv) accounting for phylogenetic uncertainty. We aim to make these approaches readily available to conservation practitioners, with the expectation that phylogenetic approaches to conservation planning could become commonplace alongside those focused on species.
2. Material and methods
(a). Spatial and phylogenetic data for land mammals
We extracted expert range maps for the world's terrestrial mammal species [49] over an equal area grid (Behrman projection) of 110 km (1° at the equator, area 1.2 × 104 km2), with each grid cell representing a planning unit (PU). For each PU, we recorded the proportion of its area intersecting the species range. Timing and ancestral relationships in the mammal radiation were represented at species level by a 5020 species supertree [28]. Taxonomic alignment resulted in a dataset of 4778 terrestrial species with spatial and phylogenetic data, across 12 146 PUs [14]. To account for phylogenetic uncertainty due to unresolved nodes on the tree, we drew 100 trees at random from the distribution of fully resolved trees generated by Kuhn et al. [50]. Repeating the conservation analysis over trees resolved by this technique provides a more realistic branch length distribution and offers an avenue to make phylogenetic uncertainty explicit in the results. In future work, this sampling could be done from a formal Bayesian posterior distribution.
(b). Phylogenetic systematic conservation planning
Our phylogenetic approach to SCP allows the conservation planning software designed for features such as species or ecosystems, to select areas to optimize the representation of PD, given other constraints. Our method extends recent approaches [27,40,45] in four important ways: (i) scaling its operation to a global scale across thousands of areas and phylogenetic tips; (ii) explorating the use of area targets in phylogenetic SCP; (iii) incorporating phylogenetic uncertainty in phylogenetic SCP; and (iv) importantly for further practical application, providing code that prepares phylogenetic and spatial data for use in the standard SCP software.
In this approach, summarized in electronic supplementary material, figure S1, the conservation features are branches on a phylogeny. Each branch, at any level on the tree, has a geographical distribution defined by the union of the ranges of the tree tips descending from that branch. Or, for the case of abundances, a branch could be represented by the sum of populations or individuals at the tips. Alternatively with species distribution models, the probability of occurrence of each internal branch would be calculated from the probabilities for its descendent tips [27,51,52].
Each branch is then assigned a weight proportional to its length, indicating the relative importance of meeting the conservation target for that branch. Simply providing the conservation planning algorithm, with the geographical distribution of each branch and its length-based weighting, enables the software to seek a solution which maximizes PD. Thus, specialized conservation planning software is not needed to handle the hierarchical relationship between phylogenetic branches, because it is implicit in the nested geographical ranges of terminal taxa and their ancestral branches. Because each branch is weighted in proportion to its length, there is no risk that an internal branch will be given greater (or lesser) priority due to the number of its descendent lineages.
These steps to prepare the input data (electronic supplementary material, figure S1) should be generally applicable to support phylogenetic SCP in standard conservation planning tools which are not ‘aware’ of the hierarchical structure of the phylogeny:
(1) Create a sites-by-branches matrix, which extends a standard sites-by-species matrix by adding a column for each internal branch (representing a group of species or other tips). A branch occurs in a PU if any of its descendant tips occur there. Here, we used the proportion of each PU occupied by a species as a measure of occurrence. Where multiple descendants of a branch occur in the same PU, we summed the areas occupied within the PU, up to the total area of the PU.
(2) Assign a representation target to each branch, based on the geographical range of the branch and the rules for targets discussed below.
(3) Assign a weighting to each terminal and internal branch, proportional to its length. This weight—conservation feature penalty factor (CFPF)—adjusts the penalty for falling short of the representation target for a branch in proportion to its length and to the area of the shortfall. Longer branches have higher penalties and are thus more important.
We captured phylogenetic uncertainty by preparing multiple versions of these input data, one for each of the 100 alternative mammal trees sampled from a Bayesian posterior distribution, as well as an equivalent species-only conservation scenario for comparison. To ensure, for subsequent comparison, that the PD and species conservation scenarios placed equivalent weight on meeting representation targets, we set the CFPF for each species in the species scenario to equal the mean CFPF value for terminal taxa in the PD scenario.
(c). Targets for representative conservation
We set targets using a bounded percentage strategy, similar to previous global studies [53,54], to prioritize the representation of narrowly distributed species and lineages. A target of 25% of the geographical range of each species and internal branch was set, constrained to a minimum target of 1 PU (1.2 × 104 km2) to avoid range losses for endemic biota, and a maximum of 25 PU (3.1 × 105 km2) to ensure that widespread species and clades are protected, but do not monopolize scarce conservation resources. Setting a maximum target for very large ranges may be relevant for species SCP, but far more so for phylogenetic SCP in which conservation features include many deep branches with widespread descendent clades. We calculated ranges based on the proportion of each PU occupied, so a target of one PU could be met by several PU partly occupied by the species or lineage. We did not consider the difference between cost of areas for conservation explicitly, but made cost proportional to the land area of each PU, which varied only when a PU was partly covered by sea.
An example of the data formats, and the R scripts to generate them from a phylogeny and species ranges, are available at github.com/danrosauer/phylospatial.
(d). Optimized selection of areas
We ran Marxan (version 2.4.3; [42]) on a computing cluster to select sets of PUs which best met the targets for representation of terrestrial mammal PD. Marxan generates multiple results for each analysis, giving a distribution of potential solutions through repeated independent searches of the solution space. Marxan ran separately for the following scenarios: (i) 10 repeats for each of the 100 alternative trees for the PD scenario and (ii) 10 repeats for each of the 100 alternative trees for the species scenario. In the species scenario, the trees did not affect the choice of areas, but were used to evaluate its effectiveness at capturing PD. For each set of 1000 runs, irreplaceability [55] was calculated as the proportion of times that a given PU was selected. We compared irreplaceability for each PU between the PD and species scenarios.
Phylogenetic approaches to conservation are needed (and were conceived) for situations where it is not possible to meet conservation goals for all species [5]. If conservation resources were available to meet targets for all species, then PD (at species level and above) would also be protected without any need to consider it explicitly. It is precisely when not all species targets can be met, and we must prioritize which elements of biodiversity to protect, that a difference is expected between species and PD-based conservation planning. This is also representative of real-world constraints in which sufficient areas to be managed for conservation are rarely available in the required locations. We thus, limited the total available for conservation in each scenario with limits, defined as a percentage of total global land, from 2% to 17%. A clear indication of the most irreplaceable areas under the most limited conservation scenarios may also be of practical use to target improved management. We therefore, examined a wide range in area available for spatial conservation planning and how this affects the efficacy of a PD-informed approach. Conveniently, the R package Prioritizr [44] is able to solve problems using the input files formatted for Marxan, but it was far slower than Marxan for our analyses, limiting its use across multiple trees. We chose Prioritizr for the explicit comparison of optimization performance, because it specifies the level of optimality which was achieved, but used the Marxan results to represent phylogenetic uncertainty across multiple trees, and to map irreplaceability. For each of the area limits, the PD optimization was compared with its matched species run, to calculate the proportional change in PD captured due to the use of phylogenetic information, as (PDPD/PDspecies) − 1.
(e). Model evaluation
To evaluate the effect of novel components of this method, we performed five further analyses, described fully in the supplementary methods. First, we partitioned the variation between results to determine how much of the variation was due to phylogenetic uncertainty. Second, we explored how the difference in conservation outcomes between species and phylogenetic SCP might affect particular species, by evaluating the outcome for evolutionarily distinct species identified through the EDGE project (http://edgeofexistence.org; [56]). Third, we compared the irreplaceability of each PU with the proportion of that unit which is currently in PAs. Fourth, we compared our targets, which are applied to each branch (representing a species or clade), with targets placed on species only, in which each branch is treated as protected if any one of its descendent species meets its reservation target. Finally, to test the sensitivity of our results to the choice of area targets, we compared them with a single-occurrence strategy used in previous studies [8,31,32] by re-running the SCP analyses with a target of one occurrence of each branch.
3. Results and discussion
(a). Effectiveness of phylogenetically informed systematic conservation planning
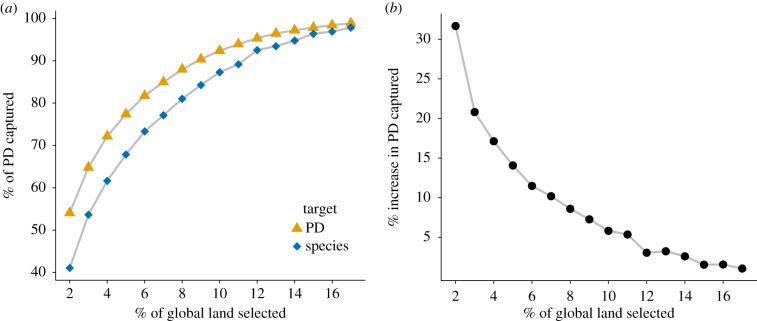
The benefits of phylogenetically informed conservation planning for retaining the tree of life strongly increase as the area available for conservation declines (figure 1b). The proportional gain due to use of PD was greatest for the smallest conservation scenario that we tested (2% of global land), in which phylogenetically informed SCP captured 31.7% more global mammal PD than the species scenario within the same total area, a substantial benefit across the whole mammal radiation. As expected, the proportion of overall PD conserved at first decreases slowly, then strongly as available area decreases (figure 1a). While a carefully chosen 17% of land could safeguard 98.9% of PD, this drops to just 54.1% for 2% of land. But it is the difference between the PD and species sceanarios that is most striking. An additional 5943 Myr of distinct mammal evolutionary history could be conserved (in an optimally selected 2% of global land) simply by a better informed choice of areas. For comparison, the same increase in a species conservation strategy would be equivalent to meeting the conservation targets for an additional 621 mammal species without increasing the area protected. The size of this effect is specific to the reservation targets used, and while we expect this result to be typical of other large groups, this must be determined empirically. It is also likely that for smaller clades the benefit of using PD in conservation would be more variable, with even larger benefits in some cases. A prudent approach before major conservation investment is thus to assess the size of the conservation benefit of using phylogenetic information when identifying priority areas, as the cost of doing so is small compared with the potential for improved conservation outcomes.
Figure 1.
(a) Percentage of total mammal PD captured when optimizing for PD (triangles) and for species (diamonds). Almost 54% of our PD conservation targets could be achieved in just 2% of global land by optimizing for PD explicitly, increasing with total area protected to 99% in 17% of all land. (b) Increase in PD captured by optimizing for PD rather than for species, which is the proportional difference between the results in (a). The improvement is tiny for large land areas, but the benefit of using PD grows rapidly as available land reduces, to almost 32% more PD captured for 2% of land conserved. (Online version in colour.)
Our study found a far larger benefit of phylogenetic SCP than the previous studies [8,31,32]. We propose that this could be due to our use of more realistic targets related to range size, compared with the single area occurrences in previous comparative studies. With single-occurrence targets, the benefit of using PD compared with species optimization was far lower than for range-based targets and close to zero when greater than 9% of land was reserved (electronic supplementary material, figure S2). In other words, when we adopted a simple, single-occurrence strategy, we found far less benefit of using phylogenetic SCP, with results closer to previous studies using that strategy. This strengthens our conclusion that under more realistic conservation targets, phylogenetic SCP can offer an important advantage.
Land and funds to implement successful protection strategies are limited. While current PAs cover 12.9% of non-Antarctic global land, the resources and land available, in practice, to specifically target for conservation of known portions of the tree of life may be far more limited than the size of existing conservation areas, or current agreements, would suggest. There are strong biases in the location of existing reserves [57], and even designated PAs are often not managed effectively for conservation, so a clear indication of the very most irreplaceable areas under the most limited conservation scenarios may be of practical use to target improved management and identify new foci for globally important conservation.
(b). Key areas for global mammal conservation
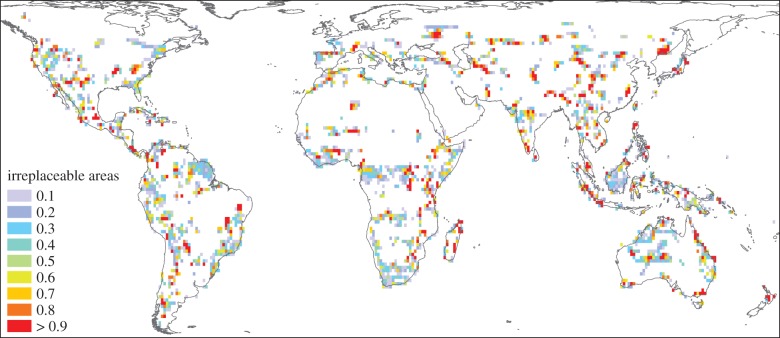
The key areas for safeguarding the mammalian tree of life, given 10% of the land surface, are shown in figure 2. A total of 2.7% of global land was found to be highly irreplaceable (red in figure 2) for conserving the greatest possible portion of mammal PD within the set limit of global land area, and thus critically important for ensuring the future of the world's diverse mammal lineages. These areas are widely scattered, as one would expect when the objective is to sample as widely as possible from mammalian evolutionary diversity which is itself structured spatially by biogeographic history and environmental gradients [58]. Variation among areas in frequency of selection by Marxan reflects flexibility in the choice of areas to meet these conservation objectives efficiently and uncertainty in the phylogenetic relationships. Moderate irreplaceability, as found in South Africa, Borneo and the Guyana Plateau in South America (figure 2), does not indicate that these areas are of low importance, but rather there is choice within such regions about which particular areas to include. With a lower area available for planned conservation (e.g. 2.5%, electronic supplementary material, figure S3), there is far less flexibility, and a majority of areas chosen were highly irreplaceable (1.5% of global land). Some of the highlighted areas in figure 2 are already well managed for conservation, so not all areas would require major investment to protect diversity. However, all the high-ranking areas are crucial to the future of the mammalian tree of life, and should be evaluated further for inclusion (or retention) in a protected area or for active conservation management.
Figure 2.
Priority areas for conserving PD across 4778 terrestrial mammal species. Colours indicate the frequency with which each area was selected in 1000 alternative reserve solutions, when constrained to protecting 10% of global land area. Red areas were selected with frequency greater than 90%. For interactive map see https://mol.org/patterns/facets.
(c). Phylogenetically informed prioritization identifies important areas that species conservation planning misses
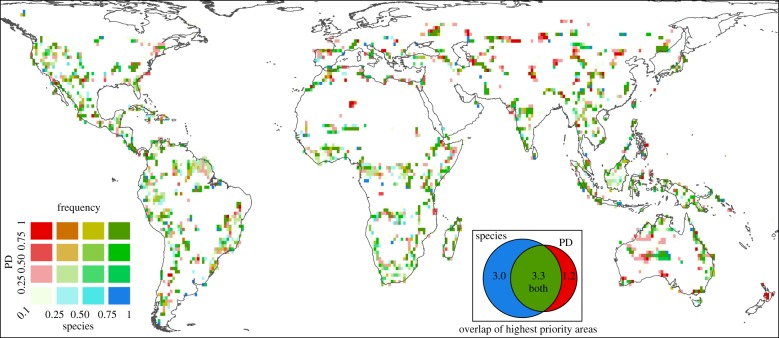
Phylogenetic information had a large effect on spatial conservation priorities, with important differences in the areas chosen compared with a traditional, species-based prioritization that is agnostic of evolutionary relationships. High-priority areas for efficient conservation of PD, which were overlooked in the species analysis (red in figure 3; electronic supplementary material, figure S4), were found across all continents, but particularly in parts of Central Asia, Australia, Spain and North Africa. A smaller range of areas (blue) were far less important for PD than species conservation. Areas of agreement, in the top quartile for species and PD (green), covered 3.3% of global land, with 3.0% ranking highly for species only and 1.2% for PD only. From a theoretical viewpoint, one would expect correspondence between the priorities for PD conservation (figure 2), and centres of phylogenetic endemism for mammals [14] where much evolutionary diversity is restricted to small areas. Furthermore, areas with endemic species on deep branches (palaeoendemism) would tend to be far more important for PD than species conservation, and such areas (figure 3a in reference 14) are significantly associated with long-term isolation by sea (continuous through Late Pleistocene glacial cycles), while this is not the case for species endemism.
Figure 3.
Comparing irreplaceability for mammal conservation by PD and species, constrained to 10% of global land area. Red = highly irreplaceable for PD only, blue = high irreplaceability for species only. Intermediate colours are graded such that deepest green = high irreplaceability in the PD and species-based prioritizations. Red thus indicates important areas for PD conservation missed in species-based analysis, while dark green indicates agreement between methods on high importance. The Venn diagram shows the overlap of areas selected with a frequency of 0.75 or more for species, PD or both, as a percentage of all land area.
The difference in areas chosen to efficiently preserve mammal species or PD also becomes more stark as available land declines. For example, with 2.5% of global land available for protection (electronic supplementary material, figure S4), the majority of the areas identified for PD conservation in western Russia, Australia, Madagascar, South Africa and northwest United States did not match areas selected for species conservation. If more limited (and arguably more achievable) conservation options are considered, or if the pool of potential areas for conservation is smaller in the future, the choice between species- and PD-based priorities will matter greatly, and the use of phylogenetic information will be more important to protect the tree of life.
These differences in locations also show implications of the difference in priorities between the PD- and species-based targets in practice, with PD favouring evolutionarily distinct species with fewer close relatives. While both methods performed similarly across targets for all species (in 10% of global land, species prioritization met 61% and PD 58%), for the top 10 species in our study on the list of evolutionarily distinct and globally endangered species (EDGE; [56]), on average 41% of conservation targets were met in the species scenario, increasing to 73% with choices informed by PD. These effects were consistent across the range of total area for conservation (electronic supplementary material, figure S5). This result shows the PD conservation strategy performing as it should: it gives higher priority to species that represent more unique evolutionary diversity.
(d). Phylogenetic uncertainty
Phylogenetic uncertainty is a typical element of studies based on evolutionary relationships, but has not been applied in phylogenetic SCP. A recent study looked at the impact of phylogenetic uncertainty in conservation prioritization and concluded that it played ‘a very minor role in the prioritization outcome’ [59]. The study's method was similar to phylogenetic SCP, but merely prioritized higher PD areas for conservation, rather than seeking a complementary representation of this diversity. This is analogous to simply selecting areas of high species richness, rather than complementary areas which together represent the region's species.
We found that phylogenetic uncertainty affects the frequency with which areas are selected (figure 2) and that the solutions for each tree tend to be similar in the areas selected, clustering together in ordination space (electronic supplementary material, figures S6 and S7). In other words, variation between solutions is greater between trees (phylogenetic uncertainty) than within trees (model optimization uncertainty), as confirmed by MANOVA (F = 368, d.f. = 19, p < 10−15). By running the prioritization across a Bayesian posterior distribution of trees, this approach reflects phylogenetic uncertainty in the solution. Our finding highlights the limitations of planning for conservation from just a single best tree, and shows how further sequencing-based or bioinformatic improvements to the tree of life may have direct relevance for an efficient and cost-effective spatial conservation prioritization.
(e). Branch reservation targets
The focus on area targets for PD in conservation planning in this study is novel (but see [41]), and one aspect deserves further discussion. Targets for internal branches are, in effect, conservation goals for the clade descended from each branch, and their rationale is similar to species-level targets. This logic of scoring solutions based on their ability to protect the range of internal branches has been increasingly applied in recent studies [27,39,41], though largely without targets. The alternative, sometimes used [36,37], involves counting areas protected only for the tips and treating internal branches as protected if any one of their descendants is protected. The tip-targets approach to PD conservation is based on a reasonable premise that a lineage persists if even one of its descendant species persists. This logic is sound only if meeting reservation targets can guarantee the persistence of a given species, and thus of its lineage, but this is rarely the case in conservation planning. Achieving targets to reserve part of the range of a species can only improve the probability—but not guarantee—that a species or clade persists. We, therefore, argue that a branch should not be considered protected on the basis of a single descendent tip meeting its target; instead, the protection required for any part of the phylogeny should take account of how widespread its extant descendants are, with more protection required for more widespread clades, to enhance their likelihood of persistence.
The tip-targets approach may not lead to good conservation outcomes in cases where a large clade is treated as protected if a single, narrowly distributed species meets its reservation target (electronic supplementary material, figure S9). For example, if we consider a branch that has as its descendants a large cosmopolitan clade (e.g. the Rodentia), should we count the PD of that branch as adequately protected if a single endemic species on a small island was well conserved? In many cases, the targets for internal branches will be easily met while meeting the targets for their descendent species, but if the logic is right, then the method will handle the trade-offs for which the PD metric was devised.
One reason targets are needed is that we often do not know how likely a species is to persist, especially for large, global groups. When there is more detailed knowledge of probability of persistence under different conservation options, this target-based method could be customized, for example using probabilistic PD [51]. Despite the conceptual difference, for a large conservation estate the performance of the two target strategies was similar, with optimization under each strategy performing well for the other. However, as the available land declined, the two approaches diverged. The same solutions (sets of selected areas) were rated as more effective for the tip-only PD targets than for the more demanding branch targets. With optimization for branch targets (in 2% land area) up to 11% more PD was deemed captured based on the tip targets, while under the tip-target strategy, the difference peaked at 30%, allowing much conservation, defined by branch targets, to be missed.
4. Conclusion
We find that a phylogenetically informed spatial conservation prioritization using phylogenetic branches as conservation units is substantially a more effective way of conserving the mammalian tree of life than a prioritization focused on species alone. The benefit of using phylogenetic information in conservation planning increases sharply as the area available for optimally placed conservation reserves declines, with up to 32% more PD included. We identify those areas that are likely to be essential for future persistence of evolutionary diversity in mammals and importantly show that areas differ to an important degree from the priorities identified when evolutionary relationships are ignored. Given the cost-effectiveness of including phylogenetic information in conservation planning, the potential to capture up to an additional 5943 Myr of distinct mammalian evolutionary history, simply through better choice of areas, is a strong argument for the use of phylogenetic information in conservation.
Supplementary Material
Acknowledgements
We thank members of the ‘PD Mafia’ working group for discussions about methods, Jeffrey Hanson for help with prioritizr software, and three anonymous reviewers for their helpful comments.
Data accessibility
Data used in this study have all been previously published, but to support replication, the data files as formatted for this study are deposited in Dryad: http://dx.doi.org/10.5061/dryad.rc416 [60].
Authors' contributions
D.F.R. carried out the data preparation and analysis, developed the scripts to support PD conservation planning and prepared the initial draft of the manuscript. S.L. analysed variance between reserve solutions. All authors contributed to the study design, writing, design of figures and gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
D.F.R. was supported by an Australian Research Council (ARC) DECRA Fellowship DE160100035, and an ARC Laureate Fellowship to Craig Moritz. L.J.P. was supported by a Marie Curie Fellowship (CLEF) AMD-659422-1, and S.L. by ARC DECRA Fellowship DE130100565. W.J. acknowledges support from NSF grants DBI-1262600 and DEB-1441737 and NASA Grant NNX11AP72G.
References
- 1.Newbold T, et al. 2016. Has land use pushed terrestrial biodiversity beyond the planetary boundary? A global assessment. Science 353, 288–291. ( 10.1126/science.aaf2201) [DOI] [PubMed] [Google Scholar]
- 2.Mace GM, Gittleman JL, Purvis A. 2003. Preserving the tree of life. Science 300, 1707–1709. ( 10.1126/science.1085510) [DOI] [PubMed] [Google Scholar]
- 3.Rosauer DF, Mooers AO. 2013. Nurturing the use of evolutionary diversity in nature conservation. Trends Ecol. Evol. 28, 322–323. ( 10.1016/j.tree.2013.01.014) [DOI] [PubMed] [Google Scholar]
- 4.Jetz W, Thomas GH, Joy JB, Redding DW, Hartmann K, Mooers AO. 2014. Global distribution and conservation of evolutionary distinctness in birds. Curr. Biol. 24, 919–930. ( 10.1016/j.cub.2014.03.011) [DOI] [PubMed] [Google Scholar]
- 5.Vane-Wright RI, Humphries CJ, Williams PH. 1991. What to protect?—Systematics and the agony of choice. Biol. Conserv. 55, 235–254. ( 10.1016/0006-3207(91)90030-D) [DOI] [Google Scholar]
- 6.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. ( 10.1016/0006-3207(92)91201-3) [DOI] [Google Scholar]
- 7.Cadotte MW. 2013. Experimental evidence that evolutionarily diverse assemblages result in higher productivity. Proc. Natl Acad. Sci. USA 110, 8996–9000. ( 10.1073/pnas.1301685110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forest F, et al. 2007. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 445, 757–760. ( 10.1038/nature05587) [DOI] [PubMed] [Google Scholar]
- 9.Davies RG, Orme CDL, Webster AJ, Jones KE, Blackburn TM, Gaston KJ. 2007. Environmental predictors of global parrot (Aves: Psittaciformes) species richness and phylogenetic diversity. Glob. Ecol. Biogeog. 16, 220–233. ( 10.1111/j.1466-8238.2007.00282.x) [DOI] [Google Scholar]
- 10.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. 2012. The global diversity of birds in space and time. Nature 491, 444–448. ( 10.1038/nature11631) [DOI] [PubMed] [Google Scholar]
- 11.Thuiller W, Lavergne S, Roquet C, Boulangeat I, Lafourcade B, Araujo MB. 2011. Consequences of climate change on the tree of life in Europe. Nature 470, 531–534. ( 10.1038/nature09705) [DOI] [PubMed] [Google Scholar]
- 12.González-Orozco CE, et al. 2015. Assessing biodiversity and endemism using phylogenetic methods across multiple taxonomic groups. Ecol. Evol. 5, 5177–5192. ( 10.1002/ece3.1747) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Chen B, Liu C, Lai J, Zhang J, Ma K. 2011. Identifying hotspots of endemic woody seed plant diversity in China. Divers. Distrib. 18, 673–688. ( 10.1111/j.1472-4642.2011.00845.x) [DOI] [Google Scholar]
- 14.Rosauer DF, Jetz W. 2015. Phylogenetic endemism in terrestrial mammals. Glob. Ecol. Biogeogr. 24, 168–179. ( 10.1111/Geb.12237) [DOI] [Google Scholar]
- 15.Mishler BD, Knerr N, González-Orozco CE, Thornhill AH, Laffan SW, Miller JT. 2014. Phylogenetic measures of biodiversity and neo- and paleo-endemism in Australian Acacia. Nat. Commun. 5, 4473 ( 10.1038/ncomms5473) [DOI] [PubMed] [Google Scholar]
- 16.González-Orozco CE, Laffan SW, Miller JT. 2011. Spatial distribution of species richness and endemism of the genus Acacia in Australia. Aust. J. Bot. 59, 601–609. ( 10.1071/BT11112) [DOI] [Google Scholar]
- 17.Rosauer D, Laffan SW, Crisp MD, Donnellan SC, Cook LG. 2009. Phylogenetic endemism: a new approach for identifying geographical concentrations of evolutionary history. Mol. Ecol. 18, 4061–4072. ( 10.1111/j.1365-294X.2009.04311.x) [DOI] [PubMed] [Google Scholar]
- 18.Kirkpatrick JB. 1983. An iterative method for establishing priorities for the selection of nature reserves: an example from Tasmania. Biol. Conserv. 25, 127–134. ( 10.1016/0006-3207(83)90056-3) [DOI] [Google Scholar]
- 19.Margules CR, Pressey RL. 2000. Systematic conservation planning. Nature 405, 243–253. ( 10.1038/35012251) [DOI] [PubMed] [Google Scholar]
- 20.Pressey RL. 1998. Algorithms, politics and timber: an example of the role of science in a public, political negotiation process over new conservation areas in production forests. In Ecology for everyone: communicating ecology to scientists, the public and the politicians (eds Wills R, Hobbs R), pp. 73–87. Sydney, Australia: Surrey Beatty and Sons. [Google Scholar]
- 21.Nel JL, et al. 2016. Knowledge co-production and boundary work to promote implementation of conservation plans. Conserv. Biol. 30, 176–188. ( 10.1111/cobi.12560) [DOI] [PubMed] [Google Scholar]
- 22.Fernandes L, et al. 2005. Establishing representative no-take areas in the Great Barrier Reef: large-scale implementation of theory on marine protected areas. Conserv. Biol. 19, 1733–1744. ( 10.1111/j.1523-1739.2005.00302.x) [DOI] [Google Scholar]
- 23.Rodrigues ASL, Gaston KJ. 2002. Maximising phylogenetic diversity in the selection of networks of conservation areas. Biol. Conserv. 105, 103–111. ( 10.1016/S0006-3207(01)00208-7) [DOI] [Google Scholar]
- 24.Faith DP, Walker PA. 1994. DIVERSITY: a software package for sampling phylogenetic and environmental diversity. Reference and user's guide. v.2.1. Canberra, Australia: CSIRO Division of Wildlife and Ecology. [Google Scholar]
- 25.Faith DP, Walker PA. 1996. Environmental diversity: on the best-possible use of surrogate data for assessing the relative biodiversity of sets of areas. Biodivers. Conserv. 5, 399–415. ( 10.1007/bf00056387) [DOI] [Google Scholar]
- 26.Billionnet A. 2013. Solution of the generalized Noah's Ark problem. Syst. Biol. 62, 147–156. ( 10.1093/sysbio/sys081) [DOI] [PubMed] [Google Scholar]
- 27.Pollock LJ, Rosauer DF, Thornhill AH, Kujala H, Crisp MD, Miller JT, McCarthy MA. 2015. Phylogenetic diversity meets conservation policy: small areas are key to preserving eucalypt lineages. Phil. Trans. R. Soc. B 370, 20140007 ( 10.1098/rstb.2014.0007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fritz SA, Bininda-Emonds ORP, Purvis A. 2009. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol. Lett. 12, 538–549. ( 10.1111/j.1461-0248.2009.01307.x) [DOI] [PubMed] [Google Scholar]
- 29.Pyron AR, Wiens JJ. 2011. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 61, 543–583. ( 10.1016/j.ympev.2011.06.012) [DOI] [PubMed] [Google Scholar]
- 30.Winter M, Devictor V, Schweiger O. 2013. Phylogenetic diversity and nature conservation: where are we? Trends Ecol. Evol. 28, 199–204. ( 10.1016/j.tree.2012.10.015) [DOI] [PubMed] [Google Scholar]
- 31.Rodrigues ASL, Brooks TM, Gaston KJ. 2005. Integrating phylogenetic diversity in the selection of priority areas for conservation: does it make a difference? In Phylogeny and conservation (eds Purvis A, Gittleman JL, Brooks T), pp. 101–119. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 32.Rodrigues AS, et al. 2011. Complete, accurate, mammalian phylogenies aid conservation planning but not much. Phil. Trans. R. Soc. B 366, 2652–2660. ( 10.1098/rstb.2011.0104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pigot AL, Owens IPF, Orme CDL. 2012. Speciation and extinction drive the appearance of directional range size evolution in phylogenies and the fossil record. PLoS Biol. 10, e1001260 ( 10.1371/journal.pbio.1001260) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carwardine J, Klein CJ, Wilson KA, Pressey RL, Possingham HP. 2009. Hitting the target and missing the point: target-based conservation planning in context. Conserv. Lett. 2, 4–11. ( 10.1111/j.1755-263X.2008.00042.x) [DOI] [Google Scholar]
- 35.Pressey RL, Cowling RM, Rouget M. 2003. Formulating conservation targets for biodiversity pattern and process in the cape floristic region, South Africa. Biol. Conserv. 112, 99–127. ( 10.1016/S0006-3207(02)00424-X) [DOI] [Google Scholar]
- 36.Guilhaumon F, et al. 2015. Representing taxonomic, phylogenetic and functional diversity: new challenges for Mediterranean marine-protected areas. Divers. Distrib. 21, 175–187. ( 10.1111/ddi.12280) [DOI] [Google Scholar]
- 37.Thuiller W, Maiorano L, Mazel F, Guilhaumon F, Ficetola GF, Lavergne S, Renaud J, Roquet C, Mouillot D. 2015. Conserving the functional and phylogenetic trees of life of European tetrapods. Phil. Trans. R. Soc. B 370, 20140005 ( 10.1098/rstb.2014.0005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pyron RA, Burbrink FT, Wiens JJ. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 13, 1–54. ( 10.1186/1471-2148-13-93) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pollock LJ, Thuiller W, Jetz W. 2017. Large conservation gains possible for global biodiversity facets. Nature 546, 141–144. ( 10.1038/nature22368) [DOI] [PubMed] [Google Scholar]
- 40.Asmyhr MG, Linke S, Hose G, Nipperess DA. 2014. Systematic conservation planning for groundwater ecosystems using phylogenetic diversity. PLoS ONE 9, e115132 ( 10.1371/journal.pone.0115132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carvalho SB, Velo-Antón G, Tarroso P, Portela AP, Barata M, Carranza S, Moritz C, Possingham HP. 2017. Spatial conservation prioritization of biodiversity spanning the evolutionary continuum. Nat. Ecol. Evol. 1, 0151 ( 10.1038/s41559-017-0151) [DOI] [PubMed] [Google Scholar]
- 42.Ball IR, Possingham HP, Watts M. 2009. Marxan and relatives: software for spatial conservation prioritisation. Spatial conservation prioritisation: quantitative methods and computational tools, pp. 185–195. Oxford, UK: Oxford University Press. [Google Scholar]
- 43.Moilanen A. 2007. Landscape zonation, benefit functions and target-based planning: unifying reserve selection strategies. Biol. Conserv. 134, 571–579. ( 10.1016/j.biocon.2006.09.008) [DOI] [Google Scholar]
- 44.Hanson JO, Schuster R, Morrell N, Srimas-Mackey M, Watts ME, Arcese P, Bennett J, Possingham HP. 2017. prioritizr: systematic conservation prioritization in R. R package. (version 1.0.1.0 ed).
- 45.Strecker AL, Olden JD, Whittier JB, Paukert CP. 2011. Defining conservation priorities for freshwater fishes according to taxonomic, functional, and phylogenetic diversity. Ecol. Appl. 21, 3002–3013. ( 10.1890/11-0599.1) [DOI] [Google Scholar]
- 46.Brum FT, Graham CH, Costa GC, Hedges SB, Penone C, Radeloff VC, Rondinini C, Loyola R, Davidson AD. 2017. Global priorities for conservation across multiple dimensions of mammalian diversity. Proc. Natl Acad. Sci. USA 114, 7641–7646. ( 10.1073/pnas.1706461114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faith DP. 1992. Systematics and conservation—on predicting the feature diversity of subsets of taxa. Cladistics 8, 361–373. ( 10.1111/j.1096-0031.1992.tb00078.x) [DOI] [PubMed] [Google Scholar]
- 48.Faith DP. 1994. Phylogenetic pattern and the quantification of organismal biodiversity. Phil. Trans. R. Soc. Lond. B 345, 45–58. ( 10.1098/rstb.1994.0085) [DOI] [PubMed] [Google Scholar]
- 49.IUCN. 2010. IUCN Red List of Threatened Species—Mammal Range Polygons. (October 2010 ed, IUCN).
- 50.Kuhn TS, Mooers AØ, Thomas GH. 2011. A simple polytomy resolver for dated phylogenies. Methods Ecol. Evol. 2, 427–436. ( 10.1111/j.2041-210X.2011.00103.x) [DOI] [Google Scholar]
- 51.Faith DP. 2008. Threatened species and the potential loss of phylogenetic diversity: conservation scenarios based on estimated extinction probabilities and phylogenetic risk analysis. Conserv. Biol. 22, 1461–1470. ( 10.1111/j.1523-1739.2008.01068.x) [DOI] [PubMed] [Google Scholar]
- 52.Rosauer DF, et al. 2016. Phylogeography, hotspots and conservation priorities: an example from the Top End of Australia. Biol. Conserv. 204, 83–93. ( 10.1016/j.biocon.2016.05.002) [DOI] [Google Scholar]
- 53.Venter O, et al. 2014. Targeting global protected area expansion for imperilled biodiversity. PLoS Biol. 12, e1001891 ( 10.1371/journal.pbio.1001891) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodrigues ASL, et al. 2004. Global gap analysis: priority regions for expanding the global protected-area network. BioScience 54, 1092–1100. ( 10.1641/0006-3568(2004)054%5B1092:GGAPRF%5D2.0.CO;2) [DOI] [Google Scholar]
- 55.Pressey RL, Humphries CJ, Margules CR, Vane-Wright RI, Williams PH. 1993. Beyond opportunism: key principles for systematic reserve selection. Trends Ecol. Evol. 8, 124–128. ( 10.1016/0169-5347(93)90023-i) [DOI] [PubMed] [Google Scholar]
- 56.Collen B, Turvey ST, Waterman C, Meredith HMR, Kuhn TS, Baillie JEM, Isaac NJB. 2011. Investing in evolutionary history: implementing a phylogenetic approach for mammal conservation. Phil. Trans. R. Soc. B 366, 2611–2622. ( 10.1098/rstb.2011.0109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carwardine J, Wilson KA, Ceballos G, Ehrlich PR, Naidoo R, Iwamura T, Hajkowicz SA, Possingham HP. 2008. Cost-effective priorities for global mammal conservation. Proc. Natl Acad. Sci. USA 105, 11 446–11 450. ( 10.1073/pnas.0707157105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Penone C, Weinstein BG, Graham CH, Brooks TM, Rondinini C, Hedges SB, Davidson AD, Costa GC. 2016. Global mammal beta diversity shows parallel assemblage structure in similar but isolated environments. Proc. R. Soc. B 283, 20161028 ( 10.1098/rspb.2016.1028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arponen A, Zupan L. 2016. Representing hotspots of evolutionary history in systematic conservation planning for European mammals. In Biodiversity conservation and phylogenetic systematics: preserving our evolutionary heritage in an extinction crisis (eds Pellens R, Grandcolas P), pp. 265–286. Berlin, Germany: Springer. [Google Scholar]
- 60.Rosauer DF, Pollock LJ, Linke S, Jetz W. 2017. Data from: Phylogenetically informed spatial planning is required to conserve the mammalian tree of life. Dryad Digital Repository. ( 10.5061/dryad.rc4167) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in this study have all been previously published, but to support replication, the data files as formatted for this study are deposited in Dryad: http://dx.doi.org/10.5061/dryad.rc416 [60].