Abstract
The VCP-Ufd1-Npl4 complex regulates proteasomal processing within cells by delivering ubiquitinated proteins to the proteasome for degradation. Mutations in VCP are associated with two neurodegenerative diseases, amyotrophic lateral sclerosis (ALS) and inclusion body myopathy with Paget's disease of the bone and frontotemporal dementia (IBMPFD), and extensive study has revealed crucial functions of VCP within neurons. By contrast, little is known about the functions of Npl4 or Ufd1 in vivo. Using neuronal-specific knockdown of Npl4 or Ufd1 in Drosophila melanogaster, we infer that Npl4 contributes to microtubule organization within developing motor neurons. Moreover, Npl4 RNAi flies present with neurodegenerative phenotypes including progressive locomotor deficits, reduced lifespan and increased accumulation of TAR DNA-binding protein-43 homolog (TBPH). Knockdown, but not overexpression, of TBPH also exacerbates Npl4 RNAi-associated adult-onset neurodegenerative phenotypes. In contrast, we find that neuronal knockdown of Ufd1 has little effect on neuromuscular junction (NMJ) organization, TBPH accumulation or adult behaviour. These findings suggest the differing neuronal functions of Npl4 and Ufd1 in vivo.
Keywords: Proteasome, TDP-43 accumulation, Neurodegeneration, Drosophila
Abbreviations: ALS, amyotrophic lateral sclerosis; IBMPDF, inclusion body myopathy with Paget's disease of the bone and frontotemporal dementia; NMJ, neuromuscular junction; Npl4, nuclear protein localization homolog 4; TBPH, TAR DNA-binding protein-43 homolog; Ufd1, ubiquitin fusion degradation 1; UPS, ubiquitin proteasome system; VCP, valosin-containing protein
1. Introduction
The ubiquitin proteasome system (UPS) is the primary protein processing system of the cell which ensures that misfolded, ubiquitin-tagged proteins are translocated to the proteasome where they are degraded. Central to this process is the action of the highly conserved chaperone complex VCP-Ufd1-Npl4, made up of the hexameric VCP (valosin-containing protein) bound to Ufd1 (ubiquitin fusion degradation 1) and Npl4 (nuclear protein localization homolog 4). Misfolded proteins are identified and polyubiquitinated by the action of ubiquitin activating (E1), conjugating (E2), and ligase (E3) enzymes (Pickart, 2001). Subsequently, the VCP-Ufd1-Npl4 complex binds to the ubiquitin-tagged proteins, extracts them out and away from non-ubiquitinated partners, and delivers them to the proteasome for processing (Ye et al., 2001). All components of the VCP-Ufd1-Npl4 complex are highly conserved across species and loss of any one of these proteins impairs UPS function, triggering accumulation of ubiquitinated proteins in yeast, Drosophila and mammalian systems (Dai and Li, 2001, Ye et al., 2001, Wojcik et al., 2004, Beskow et al., 2009).
Of the proteins in the VCP-Ufd1-Npl4 complex, VCP has been extensively studied and has been shown to have multifaceted roles in various cellular pathways. Specificity of VCP function is determined by its association with different protein co-factors such that VCP binding to p37 functions in Golgi/ER biogenesis (Uchiyama et al., 2006), VCP binding to p47 functions in membrane fusion (Roy et al., 2000), VCP binding to UBXD1 functions in endosomal sorting (Ritz et al., 2011), and VCP binding to Npl4-Ufd1 regulates UPS functions (Ye et al., 2003). Within post-mitotic neurons, regulation of the UPS by VCP is critically important. Mutations in VCP are causative of two fatal proteinopathies: amyotrophic lateral sclerosis (ALS) and inclusion body myopathy with Paget's disease of the bone and frontotemporal dementia (IBMPFD) (Watts et al., 2004, Johnson et al., 2010, Abramzon et al., 2012). Both of these degenerative diseases are characterised by cytoplasmic aggregates containing ubiquitinated and disease-causing mutant proteins within neurons and glia, most frequently the RNA-binding protein TAR-DNA binding protein 43 (TDP-43) (Neumann et al., 2006). Moreover, neurodegeneration caused by pathogenic VCP mutations is exacerbated by cytoplasmic mislocalization of TDP-43 in Drosophila models of IMBPFD (Ritson et al., 2010). Together, these results highlight an important role for VCP in neuronal maintenance in vivo.
In contrast, relatively little is known about the functions of the VCP co-factors Npl4 and Ufd1, and specific roles for these proteins in neuronal function and dysfunction have not been investigated. Npl4 and Ufd1 undergo bipartite binding to VCP, via the UBXL domain on Npl4 and the SHP1 domain on Ufd1, to extract ubiquitinated polypeptides for degradation by the proteasome (Ye et al., 2001, Bruderer et al., 2004, Hanzelmann and Schindelin, 2016). The substrates of VCP-Ufd1-Npl4-mediated UPS include transcription factors (Zhang et al., 2015), cell cycle regulators (Cao et al., 2003), mitochondrial targets for the clearance of damaged mitochondria (Kim et al., 2013) and aggregate prone proteins (Ballar et al., 2011). Though little work has investigated independent roles for Ufd1 and Npl4, some in vitro studies have revealed different requirements for Ufd1 and Npl4 in the UPS degradation of substrates of certain ubiquitin E3 ligases (Ballar et al., 2006, Ballar et al., 2011, Cao et al., 2007). For example, in yeast, the action of Npl4, but not Ufd1, is required to degrade the primary pathogenic form of the cystic fibrosis transmembrane conductance regulator (CFTR) ΔF508 (Ballar et al., 2011). Interestingly, CFTRΔF508 forms cytoplasmic aggregates in the affected tissues of cystic fibrosis patients, resembling TDP-43 aggregates in ALS and IMBPFD patients, though it is not yet known how this contributes to disease pathogenesis (Du et al., 2015).
In this study, we set out to better understand the role of the VCP co-factors Npl4 and Ufd1 in neuronal function in vivo. We have generated Npl4 and Ufd1 reduced expression Drosophila by targeted knockdown of these genes. We find that both Ufd1 and Npl4 are required for neuronal organization and function, though knockdown of Npl4 results in much more severe phenotypes. Furthermore, knockdown of Npl4, but not Ufd1, is associated with increased accumulation of TBPH, the Drosophila homolog of TDP-43, and modulates loss of TBPH-associated neurodegenerative phenotypes. These findings suggest a specific role for Npl4-dependent proteasomal function in neurons and provide a novel model to decipher the pathogenic effect of proteasomal inhibition within neurons in vivo.
2. Results
2.1. Npl4 and Ufd1 have roles in the development and maintenance of neuronal function
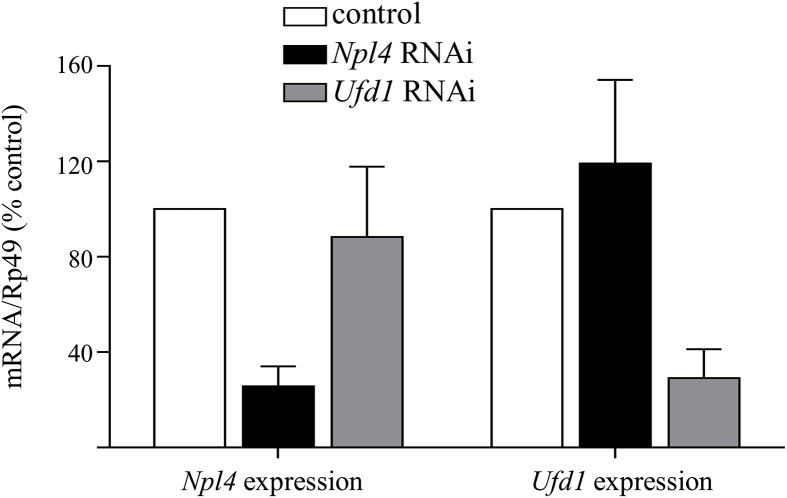
All three components of the VCP-Ufd1-Npl4 complex have well conserved Drosophila homologs: Ter94, Ufd1-like and Npl4, that share 85%, 56% and 48% protein sequence identity with human sequences, respectively. Drosophila lines containing Ufd1 and Npl4 RNAi insertions are available from the Vienna Drosophila RNAi centre (Dietzl et al., 2007; www.VDRC.at). Expression of Npl4 RNAi or Ufd1 RNAi under the control of the ubiquitous driver da-GAL4 results in similarly robust knockdown of gene expression (Figs. 1A and S1) and lethality during late larval or pupal stages of development. This is consistent with the essential functions of the VCP-Ufd1-Npl4 complex during development, with loss of Ufd1 or Npl4 causing developmental lethality (Mouysset et al., 2006). The targeted knockdown is specific for each gene such that Npl4 RNAi has no significant effect on Ufd1 expression and vice versa. Furthermore, neither Npl4 nor Ufd1 RNAi lines resulted in altered VCP expression levels. RNAi lines from the KK library were used throughout this study as the Npl4 RNAi and Ufd1 RNAi lines from the GD library failed to strongly modify expression of the target genes, with Drosophila surviving all stages of development. The 60100 w1118 control stock, which is the genetic background for all KK RNAi lines, was used as the control line throughout this study.
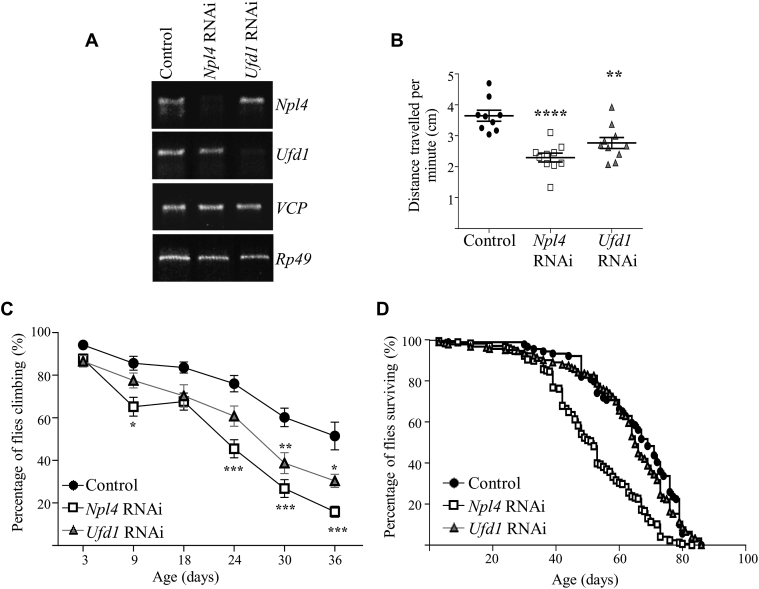
Fig. 1.
Neuronal knockdown of Drosophila Npl4 or Ufd1 causes progressive degeneration. A: Targeted expression of RNAi lines successfully knocked down Npl4 or Ufd1 expression as evidenced by PCR amplification of Npl4, Ufd1 and Rp49 cDNA from progeny of da-GAL4 crossed to either 60100 w1118 (control), Npl4 RNAi or Ufd1 RNAi flies. B and C: Neuronal-specific loss of Npl4 or Ufd1 (generated using nSyb-GAL4) caused locomotor deficits in larvae (B, values significantly different from control were determined by one-way ANOVA and Tukey's post-tests; ****P < 0.0001, **P < 0.01; n = 9–10 experiments; 10 larvae per experiment) and adult flies (C, values significantly different from control were determined by two-way ANOVA and Bonferroni's post-tests; *, P < 0.05; **, P < 0.01; ***, P < 0.005; n = 11–19 experiments; 10 flies per experiment) compared to controls. D: Neuronal-specific loss of Npl4, but not Ufd1, caused decreased lifespan compared to controls (P < 0.0001 by log-rank test; n = 90–180 flies).
To study whether Npl4 or Ufd1 has neuronal-specific functions, we investigated the effect of neuronal-specific knockdown of these genes by crossing RNAi lines to the pan-neuronal driver nSyb-GAL4. Two Drosophila lines with neuronal loss of Npl4 or Ufd1 are viable and appear healthy but has discernible locomotor deficits compared to controls at the larval L3 stage, with a more severe locomotor deficit detectable in Npl4 RNAi larvae than Ufd1 RNAi larvae (Fig. 1B). In adult Drosophila, neuronal knockdown of Npl4 or Ufd1 causes an early onset progressive decline in locomotor activity relative to controls, from 9 to 24 days post-eclosion in Npl4 RNAi flies and from 30 days post-eclosion in Ufd1 RNAi flies (Fig. 1C). Lifespan assays of these flies reveal that Npl4 RNAi flies have significantly impaired survival compared to controls, reflecting a degenerative phenotype resulting from neuronal loss of Npl4 (Fig. 1D; Tables S1 and S2). In contrast, survival in Ufd1 RNAi flies is unchanged from that of controls.
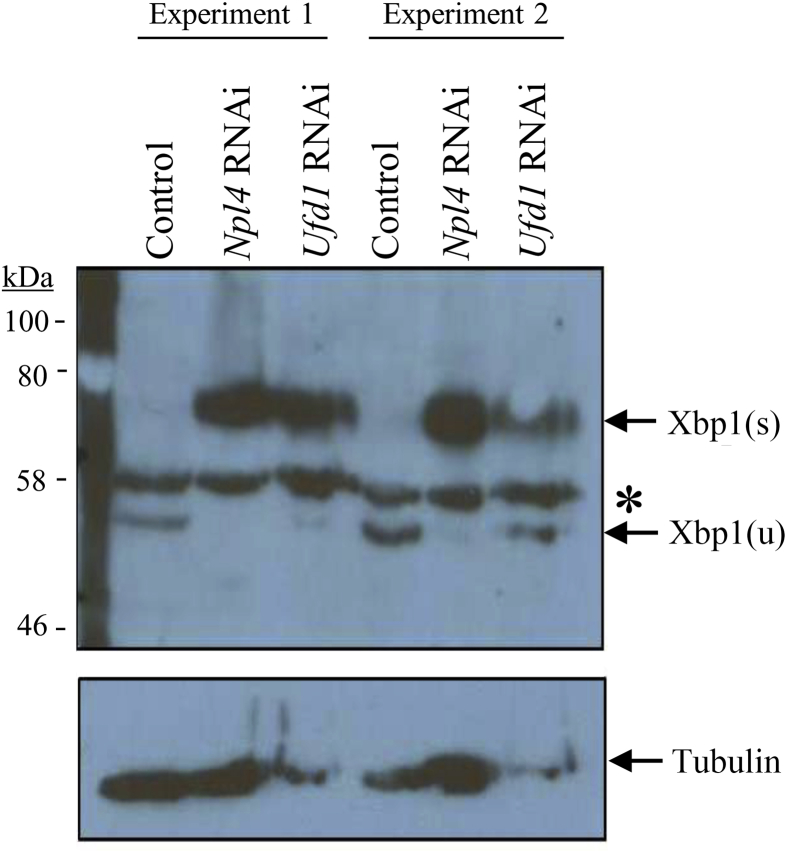
VCP interacts with Npl4 and Ufd1 to mediate the proteasomal clearance of ubiquitinated proteins as part of the UPS. Loss of either cofactor results in a failure of this clearance and an accumulation of ubiquitinated proteins, which is not seen upon loss of other VCP cofactors (Wojcik et al., 2004). Accumulation of unfolded or misfolded proteins in the endoplasmic reticulum (ER) activates the ER stress response, a major branch of which is the IRE1/Xbp1 pathway (Sone et al., 2013). In this pathway, Xbp1 mRNA is spliced by activated IRE1 resulting in the translation of the active transcription factor Xbp1(s) that induces the expression of downstream target genes including quality control proteins (Yamamoto et al., 2007). In the absence of ER stress, Xbp1 mRNA is not spliced and the resulting Xbp1(u) cannot function to activate transcription of these genes. The presence of Xbp1(s) can therefore be used as a read-out of increased ER stress. To determine whether there was an upregulation of ER stress in response to loss of Npl4 or Ufd1, we used an antibody which detects both Xbp1(s) and Xbp1(u). Loss of either Npl4 or Ufd1 results in expression of activated Xbp1(s) which is not detected in control larvae (Fig. 2). Together, these results demonstrate that knockdown of Npl4 or Ufd1 induces ER stress and causes progressive neurodegeneration.
Fig. 2.
Knockdown of Ufd1 and Npl4 induces splicing of ER stress factor, Xbp1. Western blot analysis of protein samples purified from larval progeny of da-GAL4 crossed to either control, Npl4 RNAi or Ufd1 RNAi flies from two independent experiments is shown. Knockdown of Npl4 or Ufd1 resulted in the production of activated Xbp1(s) and the reduction of inactive Xbp1(u). The asterisk denotes a non-specific band. The nitrocellulose membrane was stripped and reprobed with antibody against α-tubulin.
2.2. Npl4 is required for normal microtubule and neuromuscular junction organization
Given the locomotor phenotypes evident in Npl4 RNAi Drosophila, and to a lesser extent in Ufd1 RNAi Drosophila, we investigated the effect of knockdown of Npl4 or Ufd1 on neuronal structure. Synaptic morphology was examined in control and neuronal loss of Npl4 or Ufd1 stage L3 larvae by staining neuromuscular junctions (NMJs) with anti-HRP antibodies, permitting visualization of the presynaptic membrane. Image analysis of Npl4 RNAi larvae revealed a significantly elongated NMJ morphology indicated by increased branch length at muscle 6/7 in abdominal segment A3 compared to controls (Fig. 3A and B). This phenotype may reflect defective neuronal pruning, and is reminiscent of the long peripheral neuronal dendritic arborisations evident in VCP mutants (Rumpf et al., 2014). In contrast, Ufd1 RNAi NMJ branch length was unchanged from control.
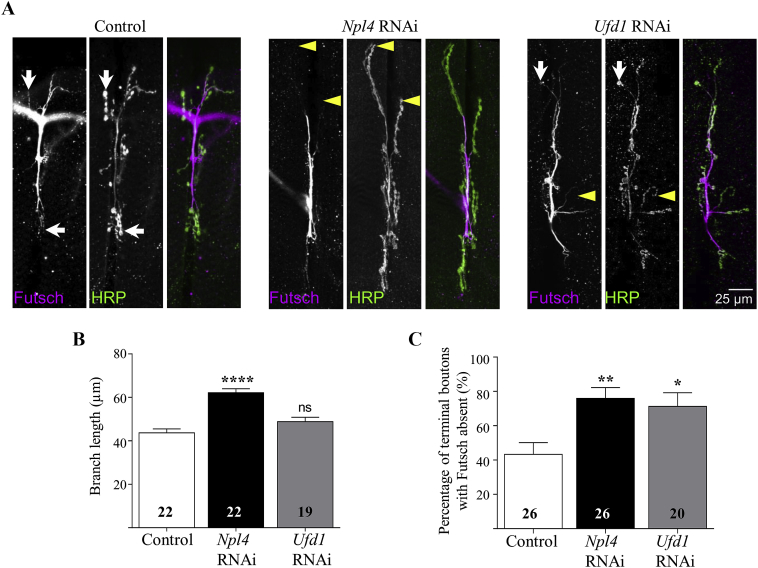
Fig. 3.
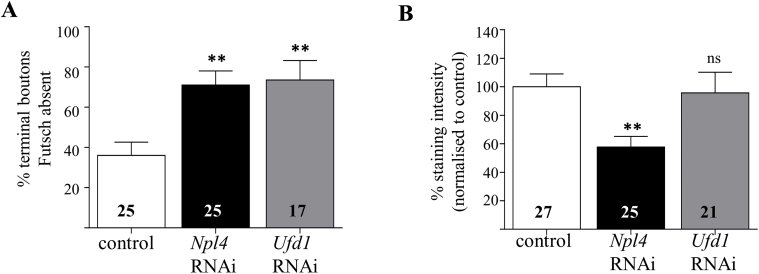
Neuromuscular junction organization in neuronal knockdown Npl4 or Ufd1 larvae. A: Confocal images showing NMJs on muscles 6/7 of L3 larvae stained for the microtubule marker Futsch (magenta) and the neuronal membrane marker HRP (green). Larvae are progeny of nSyb-GAL4 crossed to either control, Npl4 RNAi or Ufd1 RNAi flies. B: Quantification of NMJ branch length per NMJ. C: Graph represents percentage of branches in which the terminal boutons lack of Futsch staining. Knockdown of Npl4 or Ufd1 impaired microtubule organization as evidenced by the absence of Futsch staining in the most distal terminal boutons (yellow arrowheads in A) compared to controls (arrows in A). Data are expressed as mean ± SEM (n = 20–26 larvae from three independent experiments with n for each genotype indicated on graphs), and values significantly different from control were determined by one-way ANOVA and Dunnett's post-tests (*, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, P > 0.05).
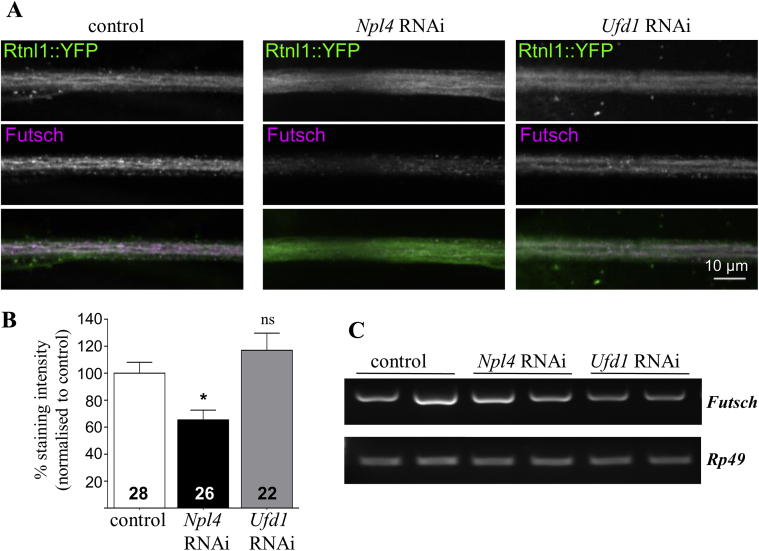
These NMJ morphological defects prompted us to investigate the organization of the neuronal cytoskeleton within presynaptic terminals by staining with antibodies to the microtubule (MT)-associated protein Futsch (the Drosophila homolog of vertebrate MAP1B) (Hummel et al., 2000). MT Futsch staining extends the length of neurons and is normally present in the distal terminal boutons of wild-type controls (Fig. 3A, arrows). However, Futsch staining is notably absent from the terminal boutons of Npl4 RNAi and many Ufd1 RNAi, and NMJ branches (Fig. 3A, arrowheads). Quantification reveals a significant increase in the proportion of branches in which the terminal bouton is lacking Futsch staining in Npl4 and Ufd1 RNAi NMJs at muscle 6/7 in anterior segment A3 (Fig. 3C). Futsch staining is also reduced within axon bundles of Npl4 RNAi neurons compared to controls (Fig. S2A and B). Comparable morphological and Futsch staining results were obtained when NMJs at muscle 6/7 and axon bundles in posterior segment A7 were analyzed (Fig. S3A and B), indicating that there is no length-dependent effect due to loss of Npl4 or Ufd1 from neurons as has been identified in other models of neurodegeneration (Fowler and O'Sullivan, 2016). Futsch mRNA expression levels were unaffected by loss of Npl4 or Ufd1, suggesting that the observed disruption is not caused by altered Futsch gene expression (Fig. S2C).
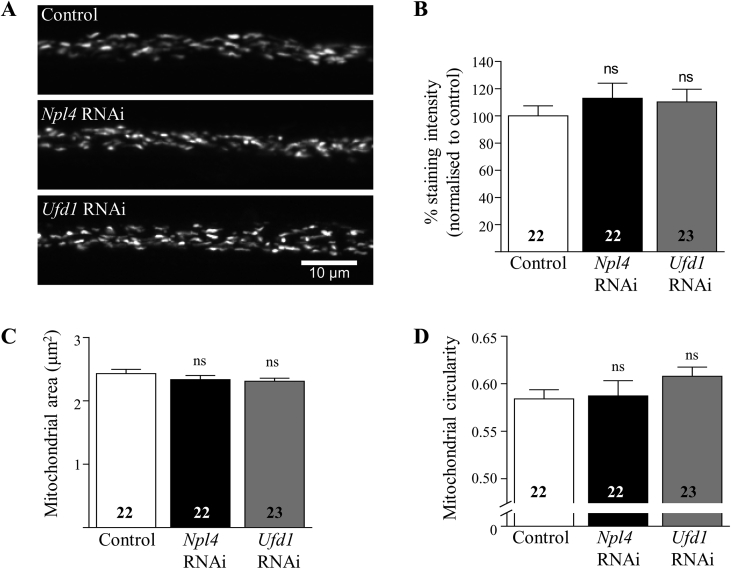
We investigated whether other intracellular compartments were disrupted by loss of Npl4 or Ufd1. Axonal staining of the smooth ER marker Rtnl1:YFP is unchanged in Npl4 RNAi or Ufd1 RNAi motor neurons compared to controls (Fig. S2A). Furthermore, analysis of mitochondrial intensity, area and circularity revealed no change in mitochondrial organization in the axons of these larvae (Fig. S4). These findings suggest that the disruption of MT organization in Npl4 RNAi motor neurons in vivo occurs independently of gross, widespread defects in intracellular compartments.
2.3. Knockdown of Npl4 increases neuronal mislocalization of TBPH
Disease-causing mutations in VCP cause cytoplasmic redistribution of the ALS-associated protein TDP-43 (TBPH in Drosophila) (Ritson et al., 2010). We therefore examined the effect of knockdown of Npl4 or Ufd1 on TBPH distribution in neurons. Venus-tagged TBPH (Wang et al., 2011) was expressed in neurons under the control of nSyb-GAL4 in control, Npl4 RNAi or Ufd1 RNAi larvae. Entire ventral nerve cords (VNCs) were imaged at 5 μm intervals and TBPH::Venus staining was quantified. In VNCs of control and Ufd1 RNAi larvae, a subset of neurons showed intense TBPH::Venus staining, identified as positively stained when analyzed in ImageJ. Knockdown of Npl4 caused a striking increase in the number of TBPH::Venus positive neurons (Fig. 4A and B). This suggests that loss of Npl4 renders neurons more susceptible to accumulation of this aggregate-prone protein, possibly through impaired proteasomal processing.
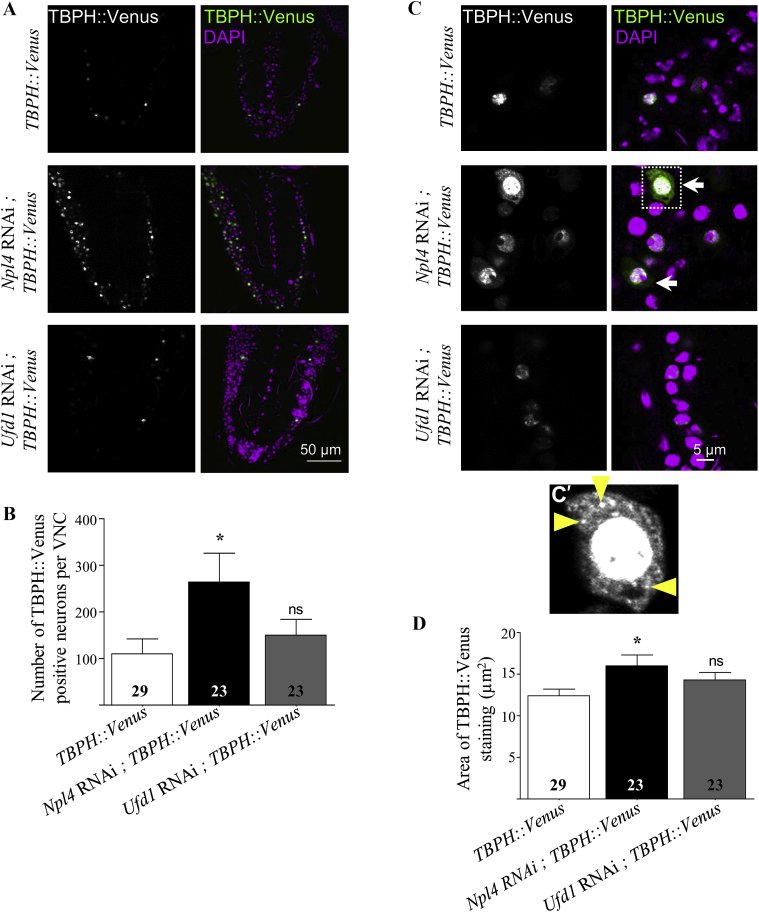
Fig. 4.
Neuronal knockdown of Npl4 increases TBPH accumulation. A: Confocal sections of larval ventral nerve cord (VNC) showing Venus-tagged TBPH (green) and DAPI (magenta) staining in control, Npl4 RNAi or Ufd1 RNAi neurons (generated using nSyb-GAL4). B: Graph shows quantification of the number of TBPH::Venus positive neurons throughout the VNC in each genotype. C: Knockdown of Npl4 caused increased mislocalization of TBPH to the cytoplasm compared to control (arrows). Inset (C′) shows cytoplasmic accumulations of TBPH detectable in a subset of Npl4 RNAi neurons (yellow arrowheads). D: Graph shows quantification of area of neuronal TBPH::Venus positive staining in each genotype. All data are expressed as mean ± SEM (n = 23–29 larvae from 5 independent experiments), and values significantly different from control were determined by one-way ANOVA and Dunnett's post-tests (*, P < 0.05; ns, P > 0.05).
TDP-43 mislocalization (to the cytoplasm from the nucleus) as well as cytoplasmic and nuclear aggregates of TDP-43 are common features in ALS/FTD models and in neurons from ALS/FTD patients (Mouysset et al., 2006, Neumann, 2009). TBPH::Venus expressed in control and Ufd1 RNAi neurons overlapped with DAPI staining and therefore localized to nuclei (Fig. 4C), consistent with previous findings (Alami et al., 2014). In contrast, loss of Npl4 caused increased localization of TBPH in the cytoplasm of neurons (Fig. 4C, arrows) with cytoplasmic aggregates of TBPH evident in a subset of Npl4 RNAi neurons (Fig. 4C′, arrowheads). Analysis of all TBPH::Venus-positive cells revealed that loss of Npl4 results in a larger area of staining of TBPH::Venus, confirming cytoplasmic mislocalization of TBPH (Fig. 4D).
2.4. TBPH knockdown modulates Npl4 RNAi- and Ufd1 RNAi-associated neurodegenerative phenotypes
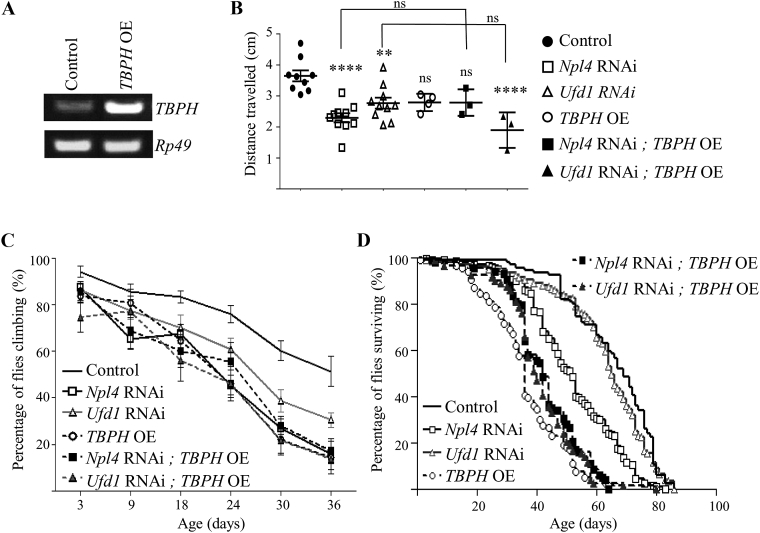
To determine whether this increased TPBH accumulation caused by expression of TPBH::Venus in Npl4 knockdown neurons has any physiological consequences, we looked for effects on the locomotor or survival deficits caused by loss of Npl4 or Ufd1 alone. Neuronal overexpression (OE) of TBPH (Fig. 5A) causes impaired locomotion in adult flies and reduction of lifespan consistent with previous findings (Fig. 5C and D; Tables S1‒S4) (Diaper et al., 2013). The combined effect of neuronal knockdown of Npl4 and OE of TBPH on adult locomotion and survival is intermediate to the effect of either Npl4 RNAi or TBPH OE alone (Fig. 5B–D; Table S1‒S4). Similarly, the combined effect of Ufd1 RNAi and TBPH OE on adult locomotion and survival is similar to the effect of TBPH OE alone (Fig. 5C and D; Table S1‒S4). This argues against an additive effect of TBPH OE in either Npl4 RNAi or Ufd1 RNAi Drosophila.
Fig. 5.
Overexpression of TBPH does not modify neurodegeneration associated with knockdown of Npl4 or Ufd1. A: PCR validation of TBPH overexpression (OE) in UAS-TBPH::Venus larvae compared to control larvae. B and C: Neuronal-specific OE of TBPH did not significantly modify locomotor deficits caused by knockdown of Npl4 or Ufd1 (generated using nSyb-GAL4) in either larvae (B, P values determined by one-way ANOVA and Tukey's post-tests; n = 3–10 experiments, 10 larvae from per experiment; ns P > 0.05, **P < 0.01, ****P < 0.0001) or adult flies (C, n = 8–18 experiments, 10 flies per experiment). D: Neuronal OE of TBPH did not significantly modify the lifespan of these flies (P > 0.05 by log-rank tests; n = 84–180 flies).
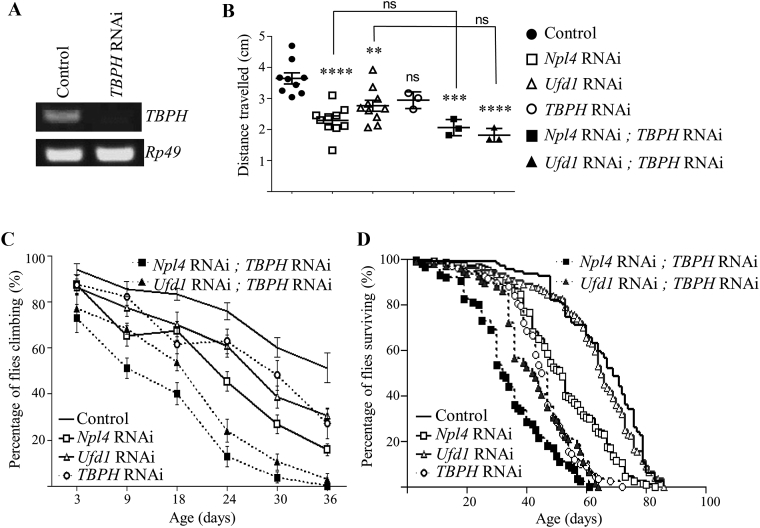
Previous reports have suggested that in Drosophila, TBPH aggregates may not themselves confer neurotoxicity but instead loss-of-function phenotypes may underlie TBPH-dysfunction in Drosophila (Diaper et al., 2013, Cragnaz et al., 2014). TBPH is required during development and few TBPH knockout Drosophila survive to adulthood (Feiguin et al., 2009, Diaper et al., 2013). We therefore investigated the effect of loss of TBPH on Npl4 RNAi and Ufd1 RNAi Drosophila using TBPH RNAi (Diaper et al., 2013) under the control of nSyb-GAL4. While neuronal TBPH RNAi (Fig. 6A) does not significantly disrupt larval locomotion (Fig. 6B), adult climbing and survival are reduced in TBPH RNAi adults (Fig. 6C and D; Table S1‒S4). In contrast to the findings with TBPH OE, TBPH RNAi exacerbates neurodegenerative phenotypes caused by knockdown of Npl4 and both adult climbing and survival were significantly reduced in Npl4 RNAi and TBPH RNAi double knockdown Drosophila compared to either Npl4 RNAi or TBPH RNAi alone (Fig. 6C and D; Tables S1 and S2). Adult locomotion, but not larval locomotion or survival, was also significantly impaired in Ufd1 RNAi and TBPH RNAi double knockdown Drosophila compared to either Ufd1 RNAi or TBPH RNAi alone (Fig. 6B–D; Table S1‒S4). These results suggest that in Drosophila, age-dependent neurodegeneration associated with Npl4 and Ufd1 knockdown is more sensitive to reduced TPBH expression than TBPH OE.
Fig. 6.
Knockdown of TBPH exacerbates neurodegeneration associated with knockdown of Npl4 or Ufd1. A: PCR validation of TBPH knockdown in TBPH RNAi larvae compared to control larvae. B and C: Locomotor deficits caused by neuronal specific loss of Npl4 or Ufd1 (generated using nSyb-GAL4) were exacerbated by TBPH knockdown in adult flies only. Graphs show quantification of locomotor scores in larvae (B, P values determined by one-way ANOVA and Tukey's post-tests; n = 3–10 experiments, 10 larvae per experiment; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, P > 0.05) or adult flies (C, P values determined by two-way ANOVA and Bonferroni's post-tests; n = 8–18 experiments, 10 flies per experiment). D: Loss of TBPH significantly shortened the lifespan of Npl4 knockdown flies compared to loss of either Npl4 or TBPH alone (P < 0.0001 by log-rank tests; n = 84–180 flies). Note that control, Npl4 RNAi and Ufd1 RNAi values are the same as those used in Fig. 5.
3. Discussion
The VCP-Ufd1-Npl4 complex is critically required in all cells to regulate protein processing and degradation via UPS. However, while VCP has been studied extensively in cell and animal systems, the co-factors Npl4 and Ufd1 remain relatively poorly examined (Roy et al., 2000, Ye et al., 2003, Watts et al., 2004, Uchiyama et al., 2006, Johnson et al., 2010, Ritson et al., 2010, Ritz et al., 2011, Abramzon et al., 2012, O'Sullivan et al., 2013). Here, we have generated Drosophila in which neuronal expression of Npl4 and Ufd1 has been knocked down to identify specific roles for these proteins in post-mitotic neurons in vivo.
An important finding of this study is that comparable knockdown of Npl4 and Ufd1 from Drosophila neurons causes overlapping yet distinct neurological consequences. In fact, we have found that knockdown of Npl4 results in more severe neurodevelopmental and neurodegenerative behavioural phenotypes than knockdown of Ufd1. These differences in neuronal phenotypes occur despite the similarly robust reduction of either Npl4 or Ufd1 gene expression in their respective knockdown lines and the induction of the ER stress response, as determined by Xbp1 splicing, in both lines. Instead, we have found evidence that these behavioural phenotypes may be brought about by defects in neuronal microtubule network formation and aberrant NMJ organization which are particularly evident in Npl4 knockdown Drosophila. Loss of VCP has recently been shown to regulate dendritic spine density in cultured mammalian neurons, via a p47-and protein synthesis-dependent pathway and knockdown of Ufd1 did not alter dendritic spine density (Shih and Hsueh, 2016), consistent with the findings presented here. Unfortunately, knockdown of Npl4 was not investigated, therefore it is not currently known whether Npl4 functions along with VCP in mammalian neuronal dendritic spine formation.
Mutations in the gene encoding VCP cause cytoplasmic aggregation of TDP-43/TBPH in animal models of IMBPFD (Ritson et al., 2010) and in the muscles of IMBPFD patients (Weihl et al., 2008). We now provide evidence that reduced expression of the VCP co-factor Npl4 is sufficient to induce TBPH aggregation within a subset of neuronal cell bodies. These findings may be explained by a model whereby VCP-Npl4, and perhaps to a lesser extent VCP-Ufd1-Npl4, are required for the correct processing of TDP-43. While this function cannot be directly linked to the degenerative phenotypes observed in the Npl4 RNAi Drosophila, we have found that Npl4 knockdown exacerbates neurodegeneration associated with loss of TBPH. This finding contrasts with previous work showing that TDP-43 RNAi partially rescues neurotoxicity associated with overexpression of mutant forms of VCP, while overexpression of TDP-43 exacerbates neurotoxicity in the model (Ritson et al., 2010). Nonetheless, our results point to a role for Npl4 in neuronal maintenance which may be important when considering therapeutic strategies to increase the clearance of neurotoxic TDP-43 in IMBPFD or ALS (Azuma et al., 2014).
An important question that remains to be addressed is the mechanism by which Npl4 and Ufd1 have distinct functions in neurons. One possible explanation could be that functional VCP-Npl4 complexes form even in the absence of Ufd1. 3D electron cryomicroscopic reconstructions of the VCP-Ufd-Npl4 complex reveal that Npl4 mediates the interaction between Npl4/Ufd1 and VCP by binding to the N-domain of VCP, while binding sites on Ufd1 allow for higher affinity binding through the large degree of conformational flexibility in the VCP-Ufd1-Npl4 complex (Bebeacua et al., 2012). Therefore, VCP-Npl4 complexes may form in cells independent of Ufd1. A second, not mutually exclusive, explanation is that there exist distinct substrates for Npl4-and Ufd1-containing complexes. The E3 ubiquitin ligase gp78 has been shown to mediate degradation of its substrates via the VCP-Npl4 complex, independently of Ufd1 (Ballar et al., 2011). gp78 is highly involved in the degradation of neurodegenerative disease proteins and its substrates are known to include the aggregate-prone proteins SOD1, Ataxin-3, mutant Huntingtin and CFTRΔF508 (Yang et al., 2007, Yang et al., 2010, Ying et al., 2009, Ballar et al., 2011). Given the findings presented here, it is interesting to speculate that VCP-Npl4 complexes may function within neurons in vivo to regulate the degradation of aggregate-prone proteins such as TDP-43.
4. Materials and methods
4.1. Fly stocks
All fly stocks were maintained as described previously (Fowler and O'Sullivan, 2016). Briefly, flies were maintained at 25°C with 12 h:12 h light:dark and were transferred to fresh cornmeal media containing vials every 2–3 days. For knockdown experiments, the following fly lines were used: UAS-Npl4-RNAi line 109309 (construct KK102533); UAS-Ufd1-RNAi line 104713 (construct KK108134); the 60100 w1118 control stock (all obtained for the Vienna Drosophila RNAi Centre, www.vdrc.at) (Dietzl et al., 2007); UAS-TBPH::Venus (generated by Brian McCabe) (Wang et al., 2011); UAS-TBPH-RNAi (generated by Frank Hirth) (Diaper et al., 2013). UAS lines were crossed either to da-GAL4 (Perrin et al., 2003), nSyb-GAL4 (Bushey et al., 2009) or OK6-GAL4 (Aberle et al., 2002) as indicated in the text.
4.2. Behavioural assays
All behavioural assays were conducted on progeny of nSyb-Gal4 crossed to 60100 w118 (control) or RNAi flies. Larval locomotor abilities were tested by placing larvae on the centre square of a grid on a 9 cm Petri dish, marked with a 1 cm2 grid, containing 2% agarose (poured and set prior to testing) and counting the number of grid lines crossed in 1 min. 10 larvae per genotype per day were investigated. A minimum of 3 independent experiments, testing 2–3 vials of 10 male flies each, were used to examine adult locomotion. Flies were maintained at 25°C and assessed on designated days post-eclosion. The number of flies climbing to the top of a vertical glass vial (10 cm length, 2.5 cm diameter) over 15 s was determined. Survival assays consisted of taking a daily mortality score on male flies from climbing assays.
4.3. Semi-quantitative PCR
PCR was performed on cDNA produced from RNA isolated and purified from 10 larvae collected in TRIzol reagent (Invitrogen, Paisley, UK) as previously described (Fowler and O'Sullivan, 2016). Primers used were: Rp49-F: 5ʹ-CCGACCACGTTACAAGAACTCTC; Rp49-R: CGCTTCAAGGGACAGTATCTGA; Npl4-F: 5ʹ-GCGAACGTGATAGCAAGCTG-3ʹ; Npl4-R: 5ʹ-TACTCGCATCGTCCGTGATG-3ʹ; Ufd1-F: 5ʹ-TACTCGAGTTTGTGGCCGAC-3ʹ; Ufd1-R: 5ʹ-AGCACCTTCTTAACGACCGG-3ʹ; TPBH-F: AAGTACCGCAACCTGGACAC-3ʹ; TBPH-R: 5ʹ-CGGGATCAAGGAAGGTTACA-3ʹ; Futsch-F: 5ʹ-GCGAGTGCATCAACGGATTC-3ʹ Futsch-R: 5ʹ-GTTAAAGTCGCGCGAATCCC-3ʹ. PCR conditions were 95°C for 30 s, 60°C for 30 s and 72°C for 1 min, repeated 20 cycles for Rp49, 29 cycles for Npl4 and Ufd1, 26 cycles for TBPH and 35 cycles for Futsch. Quantification analysis was conducted on at least five different samples for each genotype. PCR products were run on ethidium bromide containing agarose gel for visualisation and band intensities were quantified using ImageJ (ImageJ; NIH, Bethesda, USA). Rp49 intensities were used as control for cDNA concentration in the PCR reaction.
4.4. Immunoblot test
Soluble protein lysate was purified from L3 stage larvae as described previously (O'Sullivan et al., 2013). Samples were run on 10% acrylamide separating gels and transferred to nitrocellulose membrane (GE Healthcare Life Sciences, Buckinghamshire, UK) by electrophoresis. After blocking in 5% non-fat milk in 10 mM Tris-HCL, 100 mM NaCl and 0.05% Tween-20 (TBS-T) for 1 h, the membrane was then subjected to overnight incubation with primary antibodies against Xbp-1 (M-186; Santa Cruz Biotechnology, Heidelberg, Germany) and α-tubulin (T9026; Sigma, Poole, UK) at 4°C. Following incubation with HRP (horseradish peroxidase)-coupled secondary antibody for 1 h at room temperature and chemiluminescent substrate (WesternBright ECL; Advansta, USA), the bands were visualised by autoradiography.
4.5. Histology and immunomicroscopy
Immunostaining of Drosophila larvae was carried out as described previously (Fowler and O'Sullivan, 2016). Briefly, third-instar larvae were dissected in chilled Ca+2-free HL3 solution (Stewart et al., 1994), fixed in 4% formaldehyde in PBS for 30 min, permeabilised in 0.1% Triton X-100 in PBS and incubated with primary antibodies against Futsch (22C10; Hybridoma) (Fujita et al., 1982) and HRP (P7899; Sigma). Following incubation with the appropriate secondary antibodies, fixed preparations were mounted in Vectashield containing the nuclear stain DAPI (Vector Laboratories; Peterborough, UK) and were viewed using an Olympus IX81 confocal head mounted on an Olympus Fluoview FV-1000 microscope. All images were acquired using a 60×/1.35NA objective and the FV10-ASW ver.04.01 software. We imaged NMJs at muscle 6/7 of segment A3. Z-stacks of whole VNCs were imaged at 5 μm intervals.
4.6. Image analysis
Mean branch length per NMJ was determined in ImageJ, with a branch defined as at least two boutons connected to the NMJ arbor. To quantify tubulin staining within terminal synaptic boutons at NMJs, Futsch staining was classified as present, i.e., detectable staining within terminal bouton stained with Dlg, or absent. Staining was categorized on images in a blinded fashion, without knowing which image corresponded to which sample, and then subsequently ‘un-blinded’.
To measure the intensity of Futsch or mito:GFP staining within axons, the mean grey intensity for axonal regions of interest was quantified using Image J. For analysis of mitochondrial area and circularity, images were i) thresholded and converted to binary images, ii) the watershed function was used on these binarized images to separate adjoined mitochondria, and iii) the mitochondrial area calculated in ImageJ. Mitochondrial circularity was quantified using the Shape Descriptors option in the ImageJ/Analyze menu which uses equation 4 π x [Area]/[Perimeter]2 to generate a value between 0 and 1, where 1 is a perfect circle and as the value approaches 0 it becomes increasingly elongated.
For TBPH analysis in VNC, confocal sections were acquired throughout the VNC at 5 μm intervals. For analysis of TBPH-positive puncta, images were: i) thresholded and converted to binary images, ii) the watershed function was used on these binarized images to separate adjoined puncta and the number, and iii) the area of TBPH-positive puncta was quantified in ImageJ.
For all analyses, at least four independent experiments were performed, with at least five larvae per genotype in each experiment.
4.7. Statistical analysis
All data were exported to Prism (GraphPad Software, Inc.; La Jolla, CA, USA) for statistical analysis. Statistical significance was determined using one-way ANOVA and Dunnett's post-tests, except for locomotion and lifespan assays. Statistical significance for larval crawling assays was determined using one-way ANOVA and Tukey's post-tests. Statistical significance for adult locomotion assays was determined using two-way ANOVA and Bonferroni post-tests. Lifespan assays were analyzed using the log-rank test.
Acknowledgments
We thank F. Hirth, B. McCabe, the Developmental Studies Hybridoma Bank and the Bloomington and Vienna Drosophila Stock Centres for antibodies and stocks. D.J.B. was supported by a Wellcome Trust-NIH PhD studentship (200927/Z/16/Z) and a Wellcome Trust Vacation Studentship (WT200927).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.jgg.2017.06.003.
Supplementary data
The following are the supplementary data related to this article:
FIGS1.
FIGS2.
FIGS3.
FIGS4.
References
- Aberle H., Haghighi A.P., Fetter R.D., McCabe B.D., Magalhaes T.R., Goodman C.S. Wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron. 2002;33:545–558. doi: 10.1016/s0896-6273(02)00589-5. [DOI] [PubMed] [Google Scholar]
- Abramzon Y., Johnson J.O., Scholz S.W., Taylor J.P., Brunetti M., Calvo A., Mandrioli J., Benatar M., Mora G., Restagno G., Chio A., Traynor B.J. Valosin-containing protein (VCP) mutations in sporadic amyotrophic lateral sclerosis. Neurobiol. Aging. 2012;33:2231. doi: 10.1016/j.neurobiolaging.2012.04.005. e1-2231.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alami N.H., Smith R.B., Carrasco M.A., Williams L.A., Winborn C.S., Han S.S., Kiskinis E., Winborn B., Freibaum B.D., Kanagaraj A., Clare A.J., Badders N.M., Bilican B., Chaum E., Chandran S., Shaw C.E., Eggan K.C., Maniatis T., Taylor J.P. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–543. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y., Tokuda T., Shimamura M., Kyotani A., Sasayama H., Yoshida T., Mizuta I., Mizuno T., Nakagawa M., Fujikake N., Ueyama M., Nagai Y., Yamaguchi M. Identification of ter94, Drosophila VCP, as a strong modulator of motor neuron degeneration induced by knockdown of Caz, Drosophila FUS. Hum. Mol. Genet. 2014;23:3467–3480. doi: 10.1093/hmg/ddu055. [DOI] [PubMed] [Google Scholar]
- Ballar P., Pabuccuoglu A., Kose F.A. Different p97/VCP complexes function in retrotranslocation step of mammalian ER-associated degradation (ERAD) Int. J. Biochem. Cell Biol. 2011;43:613–621. doi: 10.1016/j.biocel.2010.12.021. [DOI] [PubMed] [Google Scholar]
- Ballar P., Shen Y., Yang H., Fang S. The role of a novel p97/valosin-containing protein-interacting motif of gp78 in endoplasmic reticulum-associated degradation. J. Biol. Chem. 2006;281:35359–35368. doi: 10.1074/jbc.M603355200. [DOI] [PubMed] [Google Scholar]
- Bebeacua C., Forster A., McKeown C., Meyer H.H., Zhang X., Freemont P.S. Distinct conformations of the protein complex p97-Ufd1-Npl4 revealed by electron cryomicroscopy. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1098–1103. doi: 10.1073/pnas.1114341109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beskow A., Grimberg K.B., Bott L.C., Salomons F.A., Dantuma N.P., Young P. A conserved unfoldase activity for the p97 AAA-ATPase in proteasomal degradation. J. Mol. Biol. 2009;394:732–746. doi: 10.1016/j.jmb.2009.09.050. [DOI] [PubMed] [Google Scholar]
- Bruderer R.M., Brasseur C., Meyer H.H. The AAA ATPase p97/VCP interacts with its alternative co-factors, Ufd1-Npl4 and p47, through a common bipartite binding mechanism. J. Biol. Chem. 2004;279:49609–49616. doi: 10.1074/jbc.M408695200. [DOI] [PubMed] [Google Scholar]
- Bushey D., Tononi G., Cirelli C. The Drosophila fragile X mental retardation gene regulates sleep need. J. Neurosci. 2009;29:1948–1961. doi: 10.1523/JNEUROSCI.4830-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Wang J., Qi W., Miao H.H., Wang J., Ge L., DeBose-Boyd R.A., Tang J.J., Li B.L., Song B.L. Ufd1 is a cofactor of gp78 and plays a key role in cholesterol metabolism by regulating the stability of HMG-CoA reductase. Cell Metab. 2007;6:115–128. doi: 10.1016/j.cmet.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Cao K., Nakajima R., Meyer H.H., Zheng Y. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell. 2003;115:355–367. doi: 10.1016/s0092-8674(03)00815-8. [DOI] [PubMed] [Google Scholar]
- Cragnaz L., Klima R., Skoko N., Budini M., Feiguin F., Baralle F.E. Aggregate formation prevents dTDP-43 neurotoxicity in the Drosophila melanogaster eye. Neurobiol. Dis. 2014;71:74–80. doi: 10.1016/j.nbd.2014.07.009. [DOI] [PubMed] [Google Scholar]
- Dai R.M., Li C.C. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat. Cell Biol. 2001;3:740–744. doi: 10.1038/35087056. [DOI] [PubMed] [Google Scholar]
- Diaper D.C., Adachi Y., Sutcliffe B., Humphrey D.M., Elliott C.J., Stepto A., Ludlow Z.N., Vanden Broeck L., Callaerts P., Dermaut B., Al-Chalabi A., Shaw C.E., Robinson I.M., Hirth F. Loss and gain of Drosophila TDP-43 impair synaptic efficacy and motor control leading to age-related neurodegeneration by loss-of-function phenotypes. Hum. Mol. Genet. 2013;22:1539–1557. doi: 10.1093/hmg/ddt005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., Couto A., Marra V., Keleman K., Dickson B.J. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Du K., Karp P.H., Ackerley C., Zabner J., Keshavjee S., Cutz E., Yeger H. Aggregates of mutant CFTR fragments in airway epithelial cells of CF lungs: new pathologic observations. J. Cyst. Fibros. 2015;14:182–193. doi: 10.1016/j.jcf.2014.09.012. [DOI] [PubMed] [Google Scholar]
- Feiguin F., Godena V.K., Romano G., D'Ambrogio A., Klima R., Baralle F.E. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–1592. doi: 10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Fowler P.C., O'Sullivan N.C. ER-shaping proteins are required for ER and mitochondrial network organization in motor neurons. Hum. Mol. Genet. 2016;25:2827–2837. doi: 10.1093/hmg/ddw139. [DOI] [PubMed] [Google Scholar]
- Fujita S.C., Zipursky S.L., Benzer S., Ferrus A., Shotwell S.L. Monoclonal antibodies against the Drosophila nervous system. Proc. Natl. Acad. Sci. U. S. A. 1982;79:7929–7933. doi: 10.1073/pnas.79.24.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelmann P., Schindelin H. Characterization of an additional binding surface on the p97 N-terminal domain involved in bipartite cofactor interactions. Structure. 2016;24:140–147. doi: 10.1016/j.str.2015.10.027. [DOI] [PubMed] [Google Scholar]
- Hummel T., Krukkert K., Roos J., Davis G., Klambt C. Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron. 2000;26:357–370. doi: 10.1016/s0896-6273(00)81169-1. [DOI] [PubMed] [Google Scholar]
- Johnson J.O., Mandrioli J., Benatar M., Abramzon Y., Van Deerlin V.M., Trojanowski J.Q., Gibbs J.R., Brunetti M., Gronka S., Wuu J., Ding J., McCluskey L., Martinez-Lage M., Falcone D., Hernandez D.G., Arepalli S., Chong S., Schymick J.C., Rothstein J., Landi F., Wang Y.D., Calvo A., Mora G., Sabatelli M., Monsurro M.R., Battistini S., Salvi F., Spataro R., Sola P., Borghero G., Consortium, I. Galassi G., Scholz S.W., Taylor J.P., Restagno G., Chio A., Traynor B.J. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.C., Tresse E., Kolaitis R.M., Molliex A., Thomas R.E., Alami N.H., Wang B., Joshi A., Smith R.B., Ritson G.P., Winborn B.J., Moore J., Lee J.Y., Yao T.P., Pallanck L., Kundu M., Taylor J.P. VCP is essential for mitochondrial quality control by PINK1/Parkin and this function is impaired by VCP mutations. Neuron. 2013;78:65–80. doi: 10.1016/j.neuron.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouysset J., Kahler C., Hoppe T. A conserved role of Caenorhabditis elegans CDC-48 in ER-associated protein degradation. J. Struct. Biol. 2006;156:41–49. doi: 10.1016/j.jsb.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Neumann M. Molecular neuropathology of TDP-43 proteinopathies. Int. J. Mol. Sci. 2009;10:232–246. doi: 10.3390/ijms10010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., McCluskey L.F., Miller B.L., Masliah E., Mackenzie I.R., Feldman H., Feiden W., Kretzschmar H.A., Trojanowski J.Q., Lee V.M. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- O'Sullivan N.C., Drager N., O'Kane C.J. Characterization of the Drosophila atlastin interactome reveals VCP as a functionally related interactor. J. Genet. Genomics. 2013;40:297–306. doi: 10.1016/j.jgg.2013.04.008. [DOI] [PubMed] [Google Scholar]
- Perrin L., Bloyer S., Ferraz C., Agrawal N., Sinha P., Dura J.M. The leucine zipper motif of the Drosophila AF10 homologue can inhibit PRE-mediated repression: implications for leukemogenic activity of human MLL-AF10 fusions. Mol. Cell. Biol. 2003;23:119–130. doi: 10.1128/MCB.23.1.119-130.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. Ubiquitin enters the new millennium. Mol. Cell. 2001;8:499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- Ritson G.P., Custer S.K., Freibaum B.D., Guinto J.B., Geffel D., Moore J., Tang W., Winton M.J., Neumann M., Trojanowski J.Q., Lee V.M., Forman M.S., Taylor J.P. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J. Neurosci. 2010;30:7729–7739. doi: 10.1523/JNEUROSCI.5894-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz D., Vuk M., Kirchner P., Bug M., Schutz S., Hayer A., Bremer S., Lusk C., Baloh R.H., Lee H., Glatter T., Gstaiger M., Aebersold R., Weihl C.C., Meyer H. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat. Cell Biol. 2011;13:1116–1123. doi: 10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy L., Bergeron J.J., Lavoie C., Hendriks R., Gushue J., Fazel A., Pelletier A., Morre D.J., Subramaniam V.N., Hong W., Paiement J. Role of p97 and syntaxin 5 in the assembly of transitional endoplasmic reticulum. Mol. Biol. Cell. 2000;11:2529–2542. doi: 10.1091/mbc.11.8.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpf S., Bagley J.A., Thompson-Peer K.L., Zhu S., Gorczyca D., Beckstead R.B., Jan L.Y., Jan Y.N. Drosophila Valosin-Containing Protein is required for dendrite pruning through a regulatory role in mRNA metabolism. Proc. Natl. Acad. Sci. U. S. A. 2014;111:7331–7336. doi: 10.1073/pnas.1406898111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih Y.T., Hsueh Y.P. VCP and ATL1 regulate endoplasmic reticulum and protein synthesis for dendritic spine formation. Nat. Commun. 2016;7:11020. doi: 10.1038/ncomms11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M., Zeng X., Larese J., Ryoo H.D. A modified UPR stress sensing system reveals a novel tissue distribution of IRE1/XBP1 activity during normal Drosophila development. Cell Stress Chaperones. 2013;18:307–319. doi: 10.1007/s12192-012-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart B.A., Atwood H.L., Renger J.J., Wang J., Wu C.F. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol. 1994;175:179–191. doi: 10.1007/BF00215114. [DOI] [PubMed] [Google Scholar]
- Uchiyama K., Totsukawa G., Puhka M., Kaneko Y., Jokitalo E., Dreveny I., Beuron F., Zhang X., Freemont P., Kondo H. p37 is a p97 adaptor required for Golgi and ER biogenesis in interphase and at the end of mitosis. Dev. Cell. 2006;11:803–816. doi: 10.1016/j.devcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Wang J.W., Brent J.R., Tomlinson A., Shneider N.A., McCabe B.D. The ALS-associated proteins FUS and TDP-43 function together to affect Drosophila locomotion and life span. J. Clin. Invest. 2011;121:4118–4126. doi: 10.1172/JCI57883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts G.D., Wymer J., Kovach M.J., Mehta S.G., Mumm S., Darvish D., Pestronk A., Whyte M.P., Kimonis V.E. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- Weihl C.C., Temiz P., Miller S.E., Watts G., Smith C., Forman M., Hanson P.I., Kimonis V., Pestronk A. TDP-43 accumulation in inclusion body myopathy muscle suggests a common pathogenic mechanism with frontotemporal dementia. J. Neurol. Neurosurg. Psychiatry. 2008;79:1186–1189. doi: 10.1136/jnnp.2007.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik C., Yano M., DeMartino G.N. RNA interference of valosin-containing protein (VCP/p97) reveals multiple cellular roles linked to ubiquitin/proteasome-dependent proteolysis. J. Cell Sci. 2004;117:281–292. doi: 10.1242/jcs.00841. [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Yang H., Liu C., Zhong Y., Luo S., Monteiro M.J., Fang S. Huntingtin interacts with the cue domain of gp78 and inhibits gp78 binding to ubiquitin and p97/VCP. PLoS One. 2010;5:e8905. doi: 10.1371/journal.pone.0008905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Zhong X., Ballar P., Luo S., Shen Y., Rubinsztein D.C., Monteiro M.J., Fang S. Ubiquitin ligase Hrd1 enhances the degradation and suppresses the toxicity of polyglutamine-expanded huntingtin. Exp. Cell Res. 2007;313:538–550. doi: 10.1016/j.yexcr.2006.10.031. [DOI] [PubMed] [Google Scholar]
- Ye Y., Meyer H.H., Rapoport T.A. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y., Meyer H.H., Rapoport T.A. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Z., Wang H., Fan H., Zhu X., Zhou J., Fei E., Wang G. Gp78, an ER associated E3, promotes SOD1 and ataxin-3 degradation. Hum. Mol. Genet. 2009;18:4268–4281. doi: 10.1093/hmg/ddp380. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wang Y., Li C., Shi Z., Hao Q., Wang W., Song X., Zhao Y., Jiao S., Zhou Z. The Transitional endoplasmic reticulum ATPase p97 regulates the alternative nuclear factor NF-kappaB signaling via partial degradation of the NF-kappaB subunit p100. J. Biol. Chem. 2015;290:19558–19568. doi: 10.1074/jbc.M114.630061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.