ABSTRACT
Bradyrhizobium encompasses a variety of bacteria that can live in symbiotic and endophytic associations with leguminous and nonleguminous plants, such as rice. Therefore, it can be expected that rice endophytic bradyrhizobia can be applied in the rice-legume crop rotation system. Some endophytic bradyrhizobial strains were isolated from rice (Oryza sativa L.) tissues. The rice biomass could be enhanced when supplementing bradyrhizobial strain inoculation with KNO3, NH4NO3, or urea, especially in Bradyrhizobium sp. strain SUTN9-2. In contrast, the strains which suppressed rice growth were photosynthetic bradyrhizobia and were found to produce nitric oxide (NO) in the rice root. The expression of genes involved in NO production was conducted using a quantitative reverse transcription-PCR (qRT-PCR) technique. The nirK gene expression level in Bradyrhizobium sp. strain SUT-PR48 with nitrate was higher than that of the norB gene. In contrast, the inoculation of SUTN9-2 resulted in a lower expression of the nirK gene than that of the norB gene. These results suggest that SUT-PR48 may accumulate NO more than SUTN9-2 does. Furthermore, the nifH expression of SUTN9-2 was induced in treatment without nitrogen supplementation in an endophytic association with rice. The indole-3-acetic acid (IAA) and 1-amino-cyclopropane-1-carboxylic acid (ACC) deaminase produced in planta by SUTN9-2 were also detected. Enumeration of rice endophytic bradyrhizobia from rice tissues revealed that SUTN9-2 persisted in rice tissues until rice-harvesting season. The mung bean (Vigna radiata) can be nodulated after rice stubbles were decomposed. Therefore, it is possible that rice stubbles can be used as an inoculum in the rice-legume crop rotation system under both low- and high-organic-matter soil conditions.
IMPORTANCE This study shows that some rice endophytic bradyrhizobia could produce IAA and ACC deaminase and have a nitrogen fixation ability during symbiosis inside rice tissues. These characteristics may play an important role in rice growth promotion by endophytic bradyrhizobia. However, the NO-producing strains should be of concern due to a possible deleterious effect of NO on rice growth. In addition, this study reports the application of endophytic bradyrhizobia in rice stubbles, and the rice stubbles were used directly as an inoculum for a leguminous plant (mung bean). The degradation of rice stubbles leads to an increased number of SUTN9-2 in the soil and may result in increased mung bean nodulation. Therefore, the persistence of endophytic bradyrhizobia in rice tissues can be developed to use rice stubbles as an inoculum for mung bean in a rice-legume crop rotation system.
KEYWORDS: rice endophytic bradyrhizobia, mung bean, rice-legume crop rotation
INTRODUCTION
Nitrogen-fixing root nodule formation is the result of a symbiotic relationship between rhizobia and their legume host plants. Rhizobia are able to infect the specific leguminous host roots and form nodules through complex interactions between plants and microbes (1). It is now well established that in addition to symbiotic association with legumes, rhizobia may occur as an endophyte (is colonized in intercellular spaces) of the root in nonlegumes, such as rice (Oryza sativa L. [2, 3], Oryza breviligulata [4], or O. sativa L. cv. Pelde [5]), wheat (Triticum aestivum) (6), sugarcane (Saccharum officinarum), and maize (Zea mays) (7), and some rhizobia can promote plant growth and productivity (2–7). The genus Bradyrhizobium also encompasses a variety of bacteria that can live in symbiotic and endophytic associations with legumes and nonlegumes (3).
In fact, legumes are suitable rotational crops with rice and can be planted before or after rice-harvesting season (8). For example, precultivation of rice with mung bean crop (Vigna radiata) significantly increases rice dry weight and also provides the advantage of marketable mung bean grain (8). For the inoculum carrier, peat that is exhaustible natural resources has been commonly used as a carrier for rhizobia (9). However, in cases where peat is not available, other carriers have also been utilized, such as s liquid carrier, plant residues, and wastewater sludge, but these resources are exhaustible and have some limitations, such as their unstable availability, sterilization method, and shelf-life (10). Therefore, if we can select endophytic bradyrhizobia that can nodulate mung bean and establish themselves in rice tissues, it is possible that rice stubbles can be used as an inoculum in field-grown legumes to reduce the use of bacterial inoculum. In this study, endophytic bradyrhizobia with the ability to establish symbiosis with mung bean and that have the potential to promote rice growth were selected based on the response of bacteria to different types of N chemical fertilizers and the plant growth-promoting characteristics.
RESULTS
Characterization of rice endophytic bradyrhizobia and their symbiotic properties with mung bean.
Among bradyrhizobial strains, Bradyrhizobium sp. strain SUT-PR9 produced the greatest amount of indole-3-acetic acid (IAA), followed by Bradyrhizobium sp. strains SUT-R74 and SUT-PR48 (29.44, 10.04, and 5.41 mg · mg of protein−1, respectively) at a free-living stage. On the other hand, the lowest production of IAA was found in Bradyrhizobium sp. strain SUT-R55 (0.44 mg · mg of protein−1). SUT-PR9 also had the highest 1-amino-cyclopropane-1-carboxylic acid (ACC) deaminase activity (5.34 μmol · h−1 · mg of protein−1), and the lowest activity was detected in SUT-PR48 (2.18 μmol · h−1 · mg of protein−1). All strains in free-living form were also assessed for their nitrogenase activity. The highest nitrogenase activity was observed in SUT-R74 (0.93 nmol · h−1 · mg of protein−1). However, nitrogenase activity was not detected in Bradyrhizobium sp. strain SUT-R3, ORS285, or PRC008 (Table 1).
TABLE 1.
Plant growth promotion characteristics of the rice endophytic bradyrhizobial strainsa
| Bacterial strain | mg of IAA · mg of protein−1 | μmol alpha ketobutyrate · h−1 · mg of protein−1 | nmol ARA · h−1 · mg of protein−1 under free-living conditions |
|---|---|---|---|
| SUT-R3 | 1.89 ± 1.25 B | 4.29 ± 0.57 AB | 0 ± 0.00 D |
| SUT-PR9 | 29.44 ± 15.29 A | 5.34 ± 0.26 A | 0.01 ± 0.00 D |
| SUT-PR48 | 5.41 ± 2.20 B | 2.18 ± 0.10 D | 0.11 ± 0.07 CD |
| SUT-R55 | 0.44 ± 0.00 B | 3.29 ± 0.29 BCD | 0.08 ± 0.06 CD |
| SUT-PR64 | 2.76 ± 1.25 B | 3.73 ± 0.52 BC | 0.43 ± 0.12 B |
| SUT-R74 | 10.04 ± 1.38 B | 3.56 ± 0.46 BC | 0.93 ± 0.24 A |
| SUTN9-2 | 0.75 ± 0.36 B | 3.48 ± 0.10 BC | 0.23 ± 0.11 C |
| ORS285 | 4.79 ± 2.60 B | 2.63 ± 0.87 CD | 0 ± 0.00 D |
| PRC008 | 0.52 ± 0.16 B | 2.55 ± 0.62 CD | 0 ± 0.00 D |
Different letters in the same column indicate significant differences between treatments (P ≤ 0.05) (n = 3).
Under symbiotic conditions with mung bean, strain SUTN9-2 had the highest nitrogenase activity, while the nitrogenase activities of SUT-R3, SUT-PR9, SUT-PR48, SUT-PR64, and SUT-R74 were not significantly different from that of the control. The highest number and dry weights of nodules were produced by commercial strain PRC008, followed by SUTN9-2 and SUT-R55, while SUT-PR9, SUT-PR48, SUT-PR64, and SUT-R74 did not form nodules after 1 month of inoculation. Maximum plant dry weight was obtained from the treatment inoculated with commercial strain PRC008. Bradyrhizobial strain SUTN9-2 did not produce a significantly different plant dry weight from that of PRC008. In addition, strain PRC008 was not a rice endophyte (data not shown). The results suggested that SUTN9-2 is more effective than other rice endophytic bradyrhizobial strains (Table 2).
TABLE 2.
Effects of rice endophytic bradyrhizobia on growth, nodulation, and nitrogen fixation on mung bean (cv. SUT4)a
| Bacterial strain | ARA (nmol ethylene · h−1 · nodule dry weight−1) | No. of nodules per plant | Nodule (mg [dry wt] · plant−1) | Plant (g [dry wt] · plant−1) |
|---|---|---|---|---|
| Control | 0.00 ± 0.00 D | 0.00 ± 0.00 C | 0.00 ± 0.00 C | 0.42 ± 0.02 CD |
| SUT-R3 | 7.07 ± 0.15 D | 6.00 ± 1.53 C | 2.63 ± 2.01 BC | 0.42 ± 0.07 CD |
| SUT-PR9 | 0.00 ± 0.00 D | 0.00 ± 0.00 C | 0.00 ± 0.00 C | 0.58 ± 0.16 BC |
| SUT-PR48 | 0.00 ± 0.00 D | 0.00 ± 0.00 C | 0.00 ± 0.00 C | 0.32 ± 0.10 D |
| SUT-R55 | 44.88 ± 9.99 B | 16.00 ± 0.57 B | 8.66 ± 0.50 BC | 0.62 ± 0.19 BC |
| SUT-PR64 | 0.00 ± 0.00 D | 0.00 ± 0.00 C | 0.00 ± 0.00 C | 0.45 ± 0.07 CD |
| SUT-R74 | 0.00 ± 0.00 D | 0.00 ± 0.00 C | 0.00 ± 0.00 C | 0.40 ± 0.07 CD |
| SUTN9-2 | 81.11 ± 3.11 A | 17.00 ± 4.50 B | 12.13 ± 2.73 B | 0.80 ± 0.10 AB |
| ORS285 | 0.39 ± 0.01 D | 5.00 ± 2.00 C | 5.00 ± 2.00 BC | 0.43 ± 0.04 CD |
| PRC008 | 17.50 ± 1.33 C | 60.00 ± 14.90 A | 49.83 ± 17.40 A | 0.94 ± 0.19 A |
Different letters in the same column indicate significant differences between treatments (P ≤ 0.05) (n = 3).
Effects of different nitrogen sources on rice growth promotion when inoculated with rice endophytic bradyrhizobia.
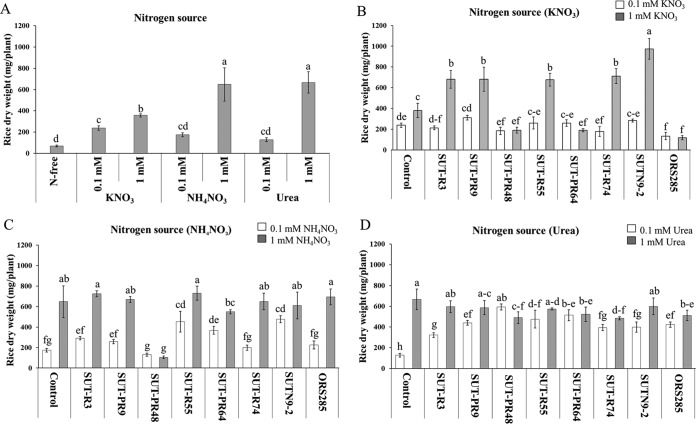
The nitrogen sources for plants at 0.1 and 1 mM KNO3, urea, and NH4NO3 were applied into N-free Hoagland's medium (11). The uninoculated control with a nitrogen source from 1 mM urea and NH4NO3 produced a significantly different plant dry weight compared to with that of the KNO3 and N-free treatment (Fig. 1A). Among eight bradyrhizobial strains, only the inoculation of nonphotosynthetic Bradyrhizobium (SUT-R3, SUT-PR9, SUT-R55, SUT-R74, and SUTN9-2) with 1 mM KNO3 (Fig. 1B) and NH4NO3 obviously increased the rice dry weight (Fig. 1C). All bradyrhizobial inoculations significantly increased rice biomass with 0.1 mM urea supplementation (Fig. 1D). In contrast, the inoculation of photosynthetic Bradyrhizobium (SUT-PR48, SUT-PR64, and ORS285) with 0.1 and 1 mM urea showed significantly higher total plant dry weight than that with KNO3, NH4NO3, and N-free treatment (Fig. 1B to D). To understand the mechanism of endophytic bradyrhizobia on rice growth promotion and suppression, SUTN9-2 (representing the mung bean-nodulating and growth-promoting rice strain) and SUT-PR48 (representing nonnodulating mung bean and growth-suppressing rice strain) were selected for further experiments.
FIG 1.
Effect of different nitrogen sources and rice endophytic bradyrhizobial strains on rice growth; uninoculated control (A), KNO3 (B), NH3NO3 (C), and urea (D) supplementations. Within each N supplementation, means labeled with different letters are statistically different at a P value of ≤0.05 (n = 3).
Localization of endophytic bradyrhizobia in rice.
To confirm that bradyrhizobia are rice endophytes, strains SUTN9-2 and SUT-PR48 in rice roots were investigated at 3 and 7 days after inoculation (DAI) by using scanning electron microscopy (SEM). Rod-shaped bacterial cells were observed and were mostly located in groups or distributed so as to be covering the root surface (Fig. S1A and B). Such damage of the epidermal surface on heavily colonized areas suggests an active invasion mechanism probably associated with a high-density bacterial population. Fig. S1A and B show that bradyrhizobia invaded the inner tissues through the epidermal cells, eventually migrating to the cortex cells after 3 days of inoculation (Fig. S1C and D). At 7 days, bradyrhizobia were observed entering the intercellular and intracellular spaces (Fig. S1E and F). Strains SUTN9-2 and SUT-PR48 showed a similar invasion at 3 and 7 days (Fig. S1A to F). In this experiment, we demonstrated that Bradyrhizobium spp. can invade rice roots, spreading rapidly and systematically through the plant tissues.
NO production according to different nitrogen sources and bradyrhizobial strains.
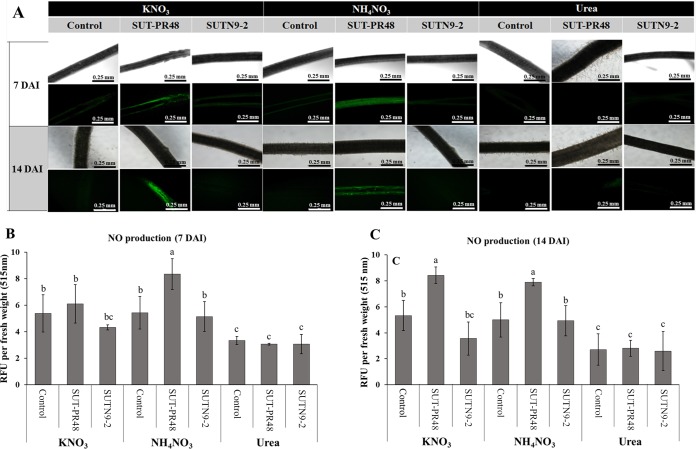
Since some bradyrhizobia are known to have denitrification activity, it is possible that the intermediate nitric oxide (NO) gas, which is toxic to plant cells, accumulates in plant cells and inhibits plant growth. The NO produced by plant cells without bacterial inoculation was not significantly different from those in N-supplied (KNO3, HH4NO3, and urea) and N-free nutrient solutions (Fig. S3A). Thus, the NO production in rice root inoculated with different bradyrhizobia under supplementation of different N sources was determined. Distinct fluorescence indicating NO production was detected when the rice roots were inoculated with bradyrhizobial strains SUT-PR48 and SUTN9-2 in the treatments supplied with KNO3 and NH4NO3 (Fig. 2). In contrast, fluorescence was not observed when those rice plants were inoculated with SUT-PR48 and SUTN9-2 in the treatment supplied with urea (Fig. 2B and C). Fluorescence was found at 14 DAI with photosynthetic bradyrhizobial strain SUT-PR48 in both treatments of KNO3 and NH4NO3 amendments (Fig. 2C). However, some fluorescence was also observed in nonphotosynthetic Bradyrhizobium SUT-R3 (see Fig. S2 in the supplemental material). For NO production in free-living cells, NO production of SUT-PR48 was significantly higher than that of SUTN9-2 in both the KNO3 and NH4NO3 supplementations (Fig. S3B). It seems that high-level NO production from strain SUT-PR48 (Fig. 2 and S3B) correlated with the significant suppression of rice growth (Fig. 1B and C).
FIG 2.
(A) Effect of different nitrogen sources and some rice endophytic bradyrhizobial strains, SUT-PR48 and SUTN9-2, on nitric oxide production detected by DAF-FM DA solution under confocal laser scanning microscopy. (B and C) NO production in rice roots: quantification of NO produced in rice roots (O. sativa PT1) at 1 week (B) and 2 weeks (C) after inoculation. Relative fluorescence unit (RFU) values per rice root fresh weight at 515 nm were estimated. Means labeled with different letters are statistically different at a P value of ≤0.05 (n = 3).
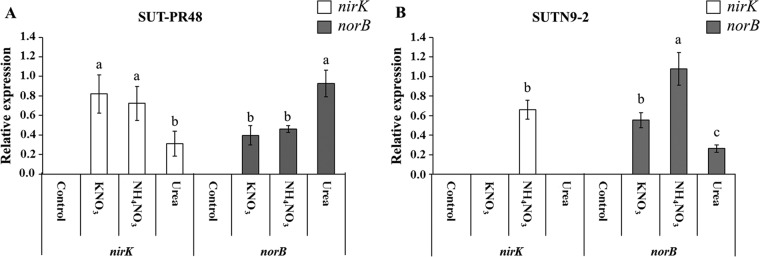
Relative expression of genes involved in nitric oxide production of endophytic bradyrhizobia SUT-PR48 and SUTN9-2 in rice roots.
The endophytic bradyrhizobium-related genes were selected from their mode of nitric oxide production, including the Cu-containing nitrite reductase gene (nirK) and nitric oxide reductase gene (norB). The expression levels of the nirK and norB genes of SUT-PR48 and SUTN9-2 are presented in Fig. 3. The inoculation of SUT-PR48 with KNO3 demonstrated the highest nirK gene expression level (0.82-fold), followed by that with inoculation with NH4NO3 (0.69-fold). However, the nirK gene expression level of SUT-PR48 was significantly the lowest when treated with urea (0.31-fold). In contrast, the highest relative expression level of norB was detected in the inoculation of SUT-PR48 with urea (0.93-fold), followed by that with inoculation with NH4NO3 (0.46-fold) and KNO3 (0.39-fold), respectively (Fig. 3A).
FIG 3.
Relative expression of nirK gene and norB gene of Bradyrhizobium sp. SUT-PR48 (A) and SUTN9-2 (B) in rice roots at 7 DAI in response to different nitrogen sources. The housekeeping gene atpD was used as an endogenous control. Letters indicate the N-amended treatments. Control, uninoculated control; KNO3, KNO3 supplementation; NH4NO3, NH4NO3 supplementation; urea, urea supplementation. The statistical analysis was separately calculated between nirK and norB genes. Means labeled with different letters are statistically different at a P value of ≤0.05 (n = 3).
The expression levels of the nirK and norB genes of SUTN9-2 are depicted in Fig. 3B. The nirK gene expression level of SUTN9-2 was not detected when KNO3 or urea was used as a source of nitrogen. However, the inoculation of SUTN9-2 with NH4NO3 induced the nirK expression level by 0.66-fold. In the case of the norB gene, the expression level in NH4NO3 (1.08-fold) was also significantly higher than that of the other nitrogen sources, KNO3 (0.55-fold) and urea (0.26-fold), respectively.
Expression of genes involved in nitrogen fixation of endophytic Bradyrhizobium SUTN9-2 in rice tissues.
In a filtrate of rice tissues, the SUTN9-2 nifH-GUS-labeled strain was separated from the rice root and leaf sheath at 14, 28, 70, and 84 DAI. The lowest nifH gene expression of SUTN9-2 was produced in the leaf sheath (0.035 ± 0.009 Miller units) and root (0.23 ± 0.05 Miller units) at 14 DAI in an N-free solution. The highest nifH gene expression in a root was detected at 28 DAI (109.16 ± 30.04 Miller units). However, the nifH gene expression in a root was clearly decreased at 70 and 84 DAI, respectively. In contrast, the highest nifH gene expression in a leaf sheath was detected at 70 DAI (213.97 ± 14.55 Miller units) but slightly decreased at 84 DAI (Fig. 4).
FIG 4.
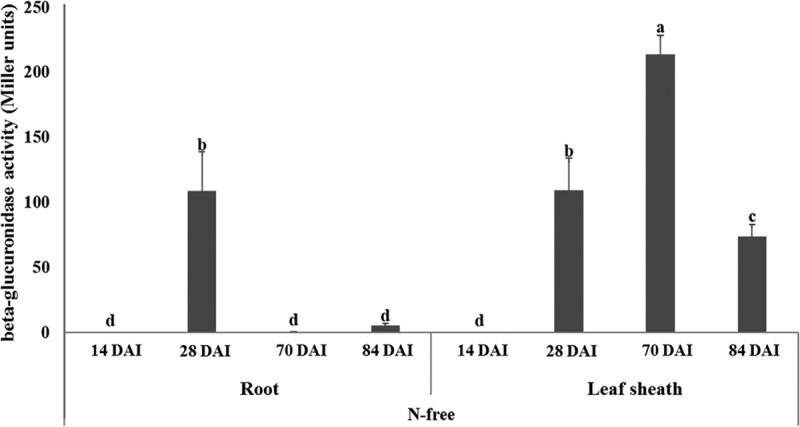
Expression of nifH gene of Bradyrhizobium sp. SUTN9-2 nifH-GUS fusion labeled strain in rice leaf sheaths and roots at 14, 28, 70, and 84 DAI in response to N-free solution compared to uninoculated control. Means labeled with different letters are statistically different at a P value of ≤0.05 (n = 3).
IAA and ACC deaminase production by Bradyrhizobium sp. SUTN9-2 in rice tissues.
SUTN9-2 produced a greater amount of IAA than the uninoculated control. The IAA production by SUTN9-2 in the rice root and leaf sheath increased from 26.46 ± 3.75 to 37.04 ± 2.80 and 92.42 ± 1.01 to 131.17 ± 7.77 mg of IAA · g (dry weight) of plant−1, respectively, compared with the uninoculated control. SUTN9-2 was also able to produce ACC deaminase, and the ACC deaminase activity of SUTN9-2 in the rice root and leaf sheath was significantly higher than that of the uninoculated control (Table 3).
TABLE 3.
IAA and ACC deaminase production by Bradyrhizobium sp. SUTN9-2 in rice tissuesa
| Treatment | mg of IAA · g−1 (dry wt) of plant | ACC deaminase (μmol alpha ketobutyrate · g−1 [dry wt] of plant) |
|---|---|---|
| Control (without SUTN9-2) | ||
| Leaf sheath | 92.42 ± 1.01 B | 368.98 ± 6.99 B |
| Root | 26.46 ± 3.75 D | 79.63 ± 8.63 D |
| SUTN9-2 | ||
| Leaf sheath | 131.17 ± 7.77 A | 414.81 ± 13.45 A |
| Root | 37.04 ± 2.80 C | 210.00 ± 8.38 C |
Different letters in the same column indicate significant differences between treatments (P ≤ 0.05) (n = 3).
Enumeration of rice SUTN9-2 in rice tissues.
The population densities and the persistence of SUTN9-2 (SUTN9-2GUS) in rice tissues were determined by plant most probable number (MPN) count. The population size of SUTN9-2 was found in both root and leaf sheath tissues, and the population densities varied from 104 to 106 (MPN/g of inoculant/g [fresh weight] of rice). The population densities of the leaf sheath and root significantly decreased after 14 and 21 DAI, respectively. However, no significant differences in population densities were observed in leaf sheath (at 14 to 28 DAI) or root (at 7, 14, and 28 DAI) tissues. In addition, SUTN9-2 still persisted in rice tissues from 7 to 28 DAI (Fig. 5).
FIG 5.
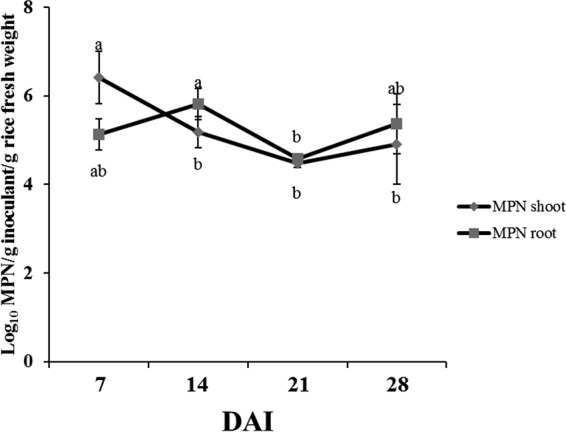
Enumeration of rice endophytic Bradyrhizobium sp. SUTN9-2 in rice tissues at different times using mung bean most probable number (MPN). Means labeled with different letters are statistically different at a P value of ≤0.05 (n = 3).
Investigation of the persistence of SUTN9-2 in the rice plant under pot trial conditions.
The rice inoculated with of SUTN9-2 (SUTN9-2GUS) planted in both soil containing low nutrients and high organic matter (paddy soil) showed no significant differences between the number of plants per hill and panicles per hill compared to the uninoculated control (Fig. S5). The persistence of SUTN9-2 in rice grown in pots under greenhouse conditions was evaluated by plate count of the CFU. The numbers of the blue colonies were counted to display the SUTN9-2 population densities in different rice tissues. The results revealed that the population density of SUTN9-2 was determined in all tissues of rice and was similar in the low-organic-matter soil and the high-organic-matter soil (paddy soil). The highest population densities were observed in leaf sheaths, and the other plant tissues (stubbles and roots) had slightly lower population densities (103 to 104 CFU/g [fresh weight]). However, the population densities were not significantly different in the tissues. On the other hand, the lowest population densities were observed in seed and leaf tissues (101 to 102 CFU/g [fresh weight]) (Fig. S6).
Investigation of the nodulation of SUTN9-2 from rice stubbles in mung bean.
At 1 week after rice stubbles were incorporated into soil, mung bean nodulation from rice stubbles was not observed. However, the nodulation was observed at 2 weeks with around 3 to 4 nodules (data not shown). An increase in the number of mung bean nodules was obtained at 5 weeks after rice stubbles were incorporated into the low-organic-matter soil and high-organic-matter soil, with nodule numbers around 60 and 35 nodules, respectively. No significant differences in the nodule dry weight, plant dry weight, and acetylene reduction activity were observed between low- and high-organic-matter soils, and the activity was significantly higher than that of the uninoculated control (Table 4). In addition, the population of SUTN9-2 (SUTN9-2GUS) in the soil was also determined by the plant infection (MPN) method, and the nodules formed by SUTN9-2 were confirmed by β-glucuronidase (GUS) staining. The results revealed that a small amount of SUTN9-2 remained in the soil (8 MPN/g of stubble/g of soil [dry weight]) before the step of rice incorporation, and then the population densities of SUTN9-2 increased at 1 week after incorporating the rice stubbles into the soil (50 MPN/g of stubble/g of soil [dry weight]) (data not shown).
TABLE 4.
Nodulation of SUTN9-2 in mung bean 5 weeks after incorporated rice stubbles into the soila
| Soil type | ARA (nmol ethylene · h−1 · nodule dry weight−1) | No. of nodules per plant | Nodule (mg [dry wt] · plant−1) | Plant (g [dry wt] · plant−1) |
|---|---|---|---|---|
| Control (without SUT9-2) | 0.00 ± 0.00 B | 0.00 ± 0.00 C | 0.00 ± 0.00 B | 0.13 ± 0.006 B |
| Low-organic-matter soil | 26.30 ± 6.20 A | 60.00 ± 5.00 A | 0.58 ± 0.16 A | 0.26 ± 0.07 A |
| High-organic-matter soil | 25.90 ± 5.17 A | 35.00 ± 11.00 B | 0.70 ± 0.35 A | 0.22 ± 0.04 A |
Different letters in the same column indicate significant differences between treatments (P ≤ 0.05) (n = 3).
DISCUSSION
Investigation of the influence of N sources on rice growth demonstrated that rice growth was differently affected by the supplementation of urea, NH4NO3, and KNO3 (Fig. 1A). Jang et al. (12) reported that rice (O. sativa L.) growth was also higher in ammonium-based N fertilizers than in nitrate-based fertilizers. In rice, the application of NH4+ is preferred over NO3− as a nitrogen source because NH4+ metabolism requires less energy than that of NO3− (12, 13), since the absorption of NH4+ occurs faster than the absorption of NO3− (14). While NH4+ can be assimilated directly into amino acid, NO3− must first be reduced into NO2− and then NH4+ via nitrate reductase and nitrite reductase (15). Thus, NH4+ is the main form of N available to rice. In contrast, NO3− is the dominant form of nitrogen for plant uptake in aerobic soil. Fageria et al. (16) reported that plants supplemented with equal proportions of NH4+ and NO3− grew as well as those supplemented with any single amount of an N form. In addition, plants can also absorb both forms of N equally, and the N form absorbed is mainly determined by what form is abundant and assessable at any given time (17, 18).
The rice dry weights in the treatments of KNO3 and NH4NO3 after inoculation with nonphotosynthetic bradyrhizobia were higher than those in the treatments of urea. On the other hand, the rice dry weights from inoculation with photosynthetic bradyrhizobia were lower in the treatments of KNO3 and NH4NO3 than in urea (Fig. 1). Bradyrhizobium has the capability of denitrification in the dissimilatory reduction of nitrate (NO3−) to N2 via the gaseous intermediates nitric oxide (NO) and nitrous oxide (N2O) (19). NO is an inorganic free radical that can become very toxic for the plant cells (20–22). The results suggested that some endophytic bradyrhizobial strains may produce nitric oxide, resulting in rice growth suppression (Fig. 2). Therefore, to select the endophytic bacteria as a biofertilizer inoculum, the NO production from bacterial strains must first be considered.
The production of NO by bradyrhizobia has been previously reported, especially in Bradyrhizobium japonicum (15, 23, 24). Our results revealed that NO in the rice root was produced from the treatments of NH4NO3 and KNO3 inoculated with photosynthetic bradyrhizobium (SUT-PR48) (Fig. 2). This result implied that some photosynthetic bradyrhizobia accumulated NO using NO3− from NH4NO3 and KNO3 via denitrification. On the other hand, an associated function of denitrification is the detoxification of cytotoxic compounds, such as NO2− and NO produced as intermediates during denitrification reactions (18). This function was found in some strains (SUT-R3, SUT-R55, SUT-R74, and SUTN9-2) because NO was not observed from the treatments of NH4NO3 and KNO3 inoculated with nonphotosynthetic bradyrhizobia, especially strain SUTN9-2 (Fig. 2). Thus, it can be assumed that inoculation with bradyrhizobia producing NO in the rice roots may also inhibit rice growth. In addition, NO production was detected only in rice roots inoculated with SUT-PR48 (Fig. 2). In this study, although the fluorescent signals representing NO production were detected in free-living cells (Fig. S3), it could be the artifact from the N supplementation, since the control medium (without bacterial cells) produced a high signal level (approximately 5 relative fluorescence units [RFU] per optical density [OD]) (Fig. S3B). The decrease in fluorescence signal in medium containing bacterial cells may be because a certain amount of the N source was consumed by bacteria, resulting in that signal reduction (approximately 0.5 RFU per OD) (Fig. S3B) compared with the control medium. In addition, since this experiment was performed under aerobic conditions, denitrification in bradyrhizobia may not have occurred. The verification of NO production in free-living bradyrhizobia remains to be further examined under anaerobic conditions.
Several studies have clearly shown that the production of NO occurred in early stages of rhizobium-legume symbiosis (25–28). Thus, the nirK genes of Bradyrhizobium were found to be essential for denitrification; then, norB catalyzes the two-electron reduction of NO to the greenhouse gas N2O in the detoxification process (25). The results of this study revealed that the nirK gene expression was activated by nitrogen source containing nitrate, especially in Bradyrhizobium sp. SUT-PR48 (Fig. 3). In contrast, nitrate did not activate the expression of the norB gene, which is required for NO detoxification and led to the accumulation of NO in rice plants. This may be the reason why SUT-PR48 showed a suppression in rice growth when nitrate was used as a nitrogen source. However, NO accumulation in the treatment of NH4NO3 was significantly higher than that of KNO3 (Fig. 2A), whereas nirK expression of NH4NO3 treatment was not significantly different from that with KNO3 supplementation (Fig. 3A). It seems that the amount of transcripts does not always correspond to NO activity levels. Perhaps, plant root tissues also contribute to NO production (and to NO reduction as well) (29). Likewise, bradyrhizobial genes other than nirK and norB may contribute to NO production and detoxification in the oxygen-limited host environment (30, 31).
In accordance with our results in SUTN9-2, nirK gene expression was detected only in the treatment of NH4NO3. It is possible that the NH4+ from this source of fertilizer could be assimilated more easily than NO3−. The remaining NO3− may be accumulated at high levels and may induce the expression of nirK when rice is supplemented with NH4NO3 (Fig. 3B). However, the norB gene of SUTN9-2 showed a higher expression level than the nirK gene. This result implied that SUTN9-2 may perform the NO detoxifications which contribute to the rice growth promotion.
In this study, the physiological characteristics of rice endophytic bradyrhizobia, such as IAA and ACC production (Table 1), were also investigated. The strains which promoted rice growth had the capability to produce IAA and ACC deaminase. Thus, these characteristics may affect plant growth promotion. For the nitrogenase activity (determined by acetylene reduction assay [ARA]), bradyrhizobial strains are known to fix nitrogen under free-living conditions (32). This feature may provide bradyrhizobia an advantage to survive under oligotrophic conditions. However, this experiment aimed to explore the possibility of plant growth promotion characteristics derived from those plant growth-promoting rhizobial (PGPR) characteristics which may facilitate rice growth promotion. Although it could not be concluded that the low nitrogen fixation activity detected from rice endophytic bradyrhizobial strains would support their efficiency in plant growth promotion, SUTN9-2 in rice tissues showed the ability to produce both IAA and ACC deaminase at 1 month after inoculation. Bhattacharjee et al. (33) found that Rhizobium leguminosarum bv. trifolii SN10 was able to synthesize ACC deaminase and IAA and could enhance the growth of various varieties of rice grown in the subcontinent. Therefore, we can speculate that SUTN9-2 cells or IAA and ACC deaminase produced by SUTN9-2 may account for the increasing rice growth promotion. Nitrogen accumulation in rice may be caused by biological nitrogen fixation of endophytic bradyrhizobia. This reaction occurs by the activity of the nitrogenase enzyme and the gene that encodes the nitrogenase structural component is nifH. During nitrogen fixation, the nifH gene was expressed in the treatment without nitrogen source (N-free) at 14, 28, 70, and 84 DAI (Fig. 4 and S4). This indicated that this gene was induced under the treatment without a nitrogen source. Similarly, Terakado-Tonooka et al. (34) suggested that bradyrhizobia colonize and express the nifH gene not only in the root nodules of leguminous plants but also in sweet potatoes as diazotrophic endophytes. However, it has been observed that R. leguminosarum bv. trifolii does not fix nitrogen in association with rice (2, 35). Thus, the nitrogen fixation ability of endophytic bacteria is also dependent on the variety of rice and diazotrophic endophytes. Thus, the nitrogen fixation of SUTN9-2 may be suspected as one of the factors involved in rice growth promotion.
Chi et al. (36) examined the persistence of viable populations of endophytic rhizobia within rice plants and found that rhizobia inoculated into the rhizosphere of rice were recovered from within surface-sterilized leaf sheaths, leaves, and roots. In addition, R. leguminosarum bv. trifolii utilizes a dynamic infection process that permits them to migrate endophytically upward into the stem base, leaf base, leaf sheaths, and some leaves of rice (37). However, our results also revealed that SUTN9-2 persisted in rice tissues until rice-harvesting season (Fig. S6). In addition, endophytic rhizobia persisted in rice throughout the vegetative and into the reproductive phases of development (36). These results indicated that it is possible to use endophytic bradyrhizobia in the rice-legume cropping system.
The maximum viable number of rhizobia per seed of mung bean was 107 to 108 rhizobia/seed (38). Perkins (39) found that increasing the inoculum level above 100 rhizobia/seed did not increase the nodule number. However, smaller amounts of inoculum resulted in abundant nodulation on the lateral roots of soybeans grown in the growth chamber, but smaller amounts of inoculum failed to produce good nodulation in the field (40). The results of this study indicated that small and large amounts of SUTN9-2 were released from rice stubbles harvested after 1 week and 1 month, respectively. This implied that the nodulation of mung bean using rice stubbles as an inoculum may be affected by the degradation of rice stubbles which release SUTN9-2 into the soil. Therefore, the persistence of endophytic bradyrhizobia in rice tissue can be developed for using rice stubbles as the inoculum for mung bean in the rice-legume crop rotation system. Further investigation of rice stubble inoculum will be performed under field conditions to compare it with the normal inoculum to assess the ability of nitrogen fixation in legume plants as well as frequency of normal inoculum application.
MATERIALS AND METHODS
Plants and bacterial strains.
Rice (Oryza sativa L.) cv. Pathum Thani 1 and mung bean (Vigna radiata L.) cv. SUT4 were used in this study. The bradyrhizobial strains, including photosynthetic and nonphotosynthetic bradyrhizobial strains (PB and non-PB strains, respectively) are listed in Table 5. Bradyrhizobial strains were cultured and maintained on yeast extract-mannitol (YEM) medium (41). Escherichia coli strains were grown in Luria-Bertani (LB) medium at 37°C. When required, the media were supplemented with the appropriate antibiotics at the following concentrations: kanamycin, 100 μg/ml; nalidixic acid, 25 μg/ml; and cefotaxime, 20 μg/ml.
TABLE 5.
Bradyrhizobial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics and source of isolationa | Source or reference |
|---|---|---|
| Strains | ||
| PB strainsb | ||
| SUT-PR48 | Rice root isolate (rice with crop rotation) | 3 |
| SUT-PR64 | Rice root isolate (rice with crop rotation) | 3 |
| ORS285 | Aeschynomene afraspera root or stem nodule isolate | 63 |
| Non-PB strains | ||
| SUT-PR9 | Rice root isolate (rice with crop rotation) | 3 |
| SUT-R3 | Rice root isolate (rice with crop rotation) | 3 |
| SUT-R55 | Rice root isolate (rice with crop rotation) | 3 |
| SUT-R74 | Rice root isolate/paddy soil isolate (rice with crop rotation) | 3 |
| SUTN9-2 | Aeschynomene americana nodule isolate (paddy crop) | 62 |
| SUTN9-2GUS | SUTN9-2 marked with mTn5SSgusA20 (pCAM120); Smr Spr | 54 |
| SUTN9-2nifH-GUS | Transcriptional nifH::gus reporter construct (pVO155nifHpm9-2gus) | This work |
| PRC008 | Recommended for mung bean/non-rice endophyte | Department of Agriculture (DOA), Thailand |
| Escherichia coli S17-1 | pro recA RP4-2 (Tcs::Mu) (Kms::Tn7), Mob+ | 64 |
| Plasmids | ||
| pCAM120 | mTn5SSgusA20 in pUT/mini-Tn5 | S. Okazakic; 65 |
| pVO155nifHpm9-2gus | nifH::gus reporter construct into pVO155-npt2-cefo-npt2-gfp; Kmr Cefor | This work |
Smr, spectinomycin resistance; Spr, streptomycin resistance; Tcs, tetracycline susceptibility; Kms, kanamycin susceptibility; Kmr, kanamycin resistance; Cefor, cefotaxime resistance.
PB, photosynthetic Bradyrhizobium.
Department of International Environmental and Agricultural Science, Graduate School of Agriculture, Tokyo University of Agriculture and Technology, Tokyo, Japan.
Physiological characteristics of rice endophytic bradyrhizobia.
The physiological characteristics of selected bradyrhizobial strains on nitrogenase activity in free-living form, indole acetic acid (IAA) production, and 1-amino-cyclopropane-1-carboxylic acid (ACC) deaminase activity were assayed. Each isolate was grown in YEM medium (41) at 28°C with agitation (125 rpm) for 5 days prior to characterization, as follows.
(i) ARA.
The nitrogenase enzyme activities of rice endophytic bradyrhizobia in free-living form were investigated on the basis of the acetylene reduction assay (ARA) (4). The reactions were carried out in a 21-ml test tube containing a bacterial culture in LG medium (10 g of glucose, 0.41 g of KH2PO4, 0.52 g of K2HPO4, 0.2 g of CaCl2, 0.05 g of Na2SO4, 0.1 g of MgSO4·7H2O, 0.005 g of FeSO4·7H2O, 0.0025 g of Na2MoO4·2H2O per liter) (42) and incubated at 28 ± 2°C for 7 days. Ten percent (vol/vol) of gas phase in the headspace was replaced with acetylene and further incubated at 28 ± 2°C for 24 h. After incubation, the gas from the vessel was injected into a gas chromatograph (GC) with a flame ionization detector equipped with polyethylene (PE)-alumina packed column (50 m by 0.32 mm by 0.25 μm) (PerkinElmer, USA). The standard curve of ethylene was constructed using various concentrations of pure ethylene (43). The total protein concentrations of cell suspension were determined using Lowry's method (44).
(ii) IAA production assay.
The IAA production was colorimetrically determined according to the method of Costacurta et al. (45). The bradyrhizobial strains were grown in YEM broth medium supplemented with l-tryptophan (100 mg · liter−1) at 28°C. The supernatant of stationary-phase culture was obtained by centrifugation at 10,000 × g for 15 min. The amount of IAA produced per mg of protein was detected as described by Costacurta et al. (45). Pure IAA (Sigma, USA) was used as a standard.
(iii) ACC deaminase activity assay.
The bacterial cells were collected by centrifugation at 4,000 × g for 5 min and washed twice with minimal medium (46). Cell pellets were suspended in 15 ml of minimal medium supplemented with 1 mM ACC and further incubated at 28°C for 40 h with shaking at 125 rpm to induce ACC deaminase enzyme production. ACC deaminase activity was measured according to protocol of Tittabutr and coworkers (47).
Determination of rice endophytic bradyrhizobia on nodulation and growth promotion of mung bean.
Mung bean seeds were surface-sterilized (48) prior to being put into sterilized petri dishes containing wet sterilized tissue paper and kept at room temperature for 2 days. The germinated seeds were then transplanted into the Leonard's jar containing sterilized vermiculite and N-free solution under aseptic conditions. One milliliter of 5 × 108 CFU · ml−1 rice endophytic bradyrhizobial inoculum was applied to each seedling at 2 days after transplanting. Plants were grown under controlled environmental conditions of 28 ± 2°C on 16/8-h day/night cycle (full light, 639 microeinsteins [μE] · m−2 · S−1). After 28 DAI, plants were harvested, and the root part was used for analysis of nitrogenase activity by measurement of acetylene-reducing activity. After the ARA assay, nodules were detached from the roots, and the number of the nodules was scored. The plant and nodule dry weights were determined after drying at 70°C for 72 h.
Investigation of rice growth promotion by endophytic bradyrhizobia inoculation under different N sources.
The Leonard's jar was filled with 1:3 (wt/wt) sterilized sand and vermiculite. The N-free nutrient solution contained 7 mM CaSO4·2H2O, 17.8 mM Fe-EDTA, 1.0 mM K2SO4, 0.25 mM KH2PO4, 0.625 mM K2HPO4, and 2.0 mM MgSO4·7H2O, and micronutrients adjusted pH to 6.8 (49) were added into the Leonard's jar. The different N sources of 0.1 and 1 mM KNO3, urea, and NH4NO3 were added separately into the N-free solution and applied through a wick to provide nutrients to the plants. The whole apparatus was autoclaved (90 min at 121°C) prior to applying the rice seedlings. Surface-disinfected rice seeds were germinated on 0.85% agar of YEM medium for 1 day. Then, three replicates of germinated seeds were soaked overnight in broth containing various bacterial isolates (5 ml of 108 CFU · ml−1) and then transplanted into the Leonard's jar under aseptic conditions. This was conducted as three replicates per single bradyrhizobial isolate as well as the bradyrhizobial reference strains (SUTN9-2 and ORS285). Rice was grown under controlled environmental conditions of 28 ± 2°C on 16/8-h day/night cycle (full light, 639 μE · m−2 · S−1). The rice plants were harvested after 1 month of planting, and the dry weight was determined.
Investigation of rice endophytic bradyrhizobia inside plant tissues using SEM.
The roots from rice after inoculation with Bradyrhizobium strains SUT-PR48 and SUTN9-2 at 3 and 7 days of culture were fixed with 2.5% (vol/vol) glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.2) for 2 h and postfixed in 1% (wt/vol) osmium tetroxide in the same buffer for 2 h. The fixed roots were dehydrated in a graded ethanol series. Then, the samples were treated with CO2, mounted on an aluminum cylinder (stub), and covered with a steam of carbon and ionized gold (49, 50). The samples were examined under an SEM (JSM 7800F; JEOL Ltd., Tokyo, Japan).
Detection of NO production.
The rice plants inoculated with bradyrhizobial strains were grown in the test tube containing N-free nutrient solution with different N sources of 1 mM KNO3, urea, and NH4NO3. One- and two-week-old-plants were collected for diaminofluorescein-FM diacetate (DAF-FM DA) detection. A stock solution of 5 mM DAF-FM DA in dimethyl sulfoxide was diluted 500-fold (10 μM DAF-FM final concentration) in water before use (51). The rice roots were placed for 30 min on filter paper soaked with the DAF-FM DA solution, and the fluorescence images of the roots were then observed under confocal laser scanning microscopy (excitation, 488 nm; emission, 515 to 530 nm; Nikon Model Ni-E, Nikon Instech Co., Ltd., Tokyo, Japan).
Quantification of NO production by fluorescence spectrophotometry.
A stock solution of DAF-FM was diluted 1,000-fold in water before use. Detached rice roots at 7 and 14 DAI were soaked in the DAF-FM solution for 1 min. The relative fluorescence units (RFU) of the DAF-FM solution were measured using a microplate reader fluorescence spectrophotometer (Varioskan; Thermo Scientific, USA). The wavelengths for excitation and emission were 470 and 515 nm, respectively (52). To examine the NO production from free-living cells of strains SUT-PR48 and SUTN9-2, the protocol described by Fukudome and coworkers (53) was modified. Bradyrhizobial strains SUT-PR48 and SUTN9-2 were grown in YM medium at 30°C for 5 days. Bacterial cells were collected by centrifugation at 10,000 × g for 5 min and were incubated with different N sources of 1 mM KNO3, urea, and NH4NO3 in 1× phosphate-buffered saline (PBS) buffer (supplemented with 5 μM DAF-FM final concentration) for 16 h. The cell mixtures were centrifuged at 10,000 × g for 5 min. The supernatants were used to analyze NO production using a microplate reader fluorescence spectrophotometer (Varioskan; Thermo Scientific). The wavelengths for excitation and emission were 470 and 515 nm, respectively.
Total RNA extraction and qRT-PCR analysis.
The fresh rice roots were harvested at 7 days after inoculation with endophytic bradyrhizobial strains SUT-PR48 and SUTN9-2. Rice samples were sterilized with 70% ethanol for 30 s and 3% sodium hypochlorite, washed 5 to 6 times with sterilized water, immediately frozen in liquid nitrogen, and stored at −80°C for further total RNA extraction.
Total RNAs were directly isolated from plant samples using the RNeasy plant minikit (Qiagen, USA), according to the manufacturer's protocol. RNAs were treated with the DNase I to prevent contamination of genomic DNA and then converted to cDNA using iScript cDNA synthesis (Bio-Rad). The transcription levels were determined by real-time quantitative reverse transcription-PCR (qRT-PCR). The primers used for amplification (atpD, nirK, norB, and nifH) are listed in Table 6. qRT-PCR amplification was performed using QuantStudio 3 real-time PCR system mix (Applied Biosystems) with the following cycling conditions: an initial denaturation step at 95°C for 2 min; 35 cycles of 2 min at 95°C; 30 s at annealing temperatures of 53°C (atpD), 53°C (nirK), 54°C (norB), and 48°C (nifH), followed by a final 5-min extension at 72°C (Table 6). The relative gene expression was analyzed by comparative threshold cycle (CT) method (−ΔΔCT) that was normalized to the endogenous housekeeping gene (atpD). Three biological replicates were pooled and analyzed. At least three PCR amplifications were performed for each sample.
TABLE 6.
Primers used in this study
| Target gene by type | Primer name | Gene description | Primer sequence (5′→3′) | Description of design (reference or accession no.) |
|---|---|---|---|---|
| Housekeeping | ||||
| atpD | atpDF-SUT-PR48 | ATP synthase subunit beta | TGTTGTCGACGAAGAACAGC | Designed from atpD of Bradyrhizobium sp. strain ORS285 (LT859959.1) |
| atpDR-SUT-PR48 | CACGAGTTCATCGAGTCCAA | |||
| atpDF-SUTN9-2 | TGTTGTCGACGAAGAACAGC | Designed from atpD of Bradyrhizobium sp. strain SUTN9-2 (LAXE00000000) | ||
| atpDR-SUTN9-2 | CACGAGTTCATCGAGTCCAA | |||
| Nitric oxide production | ||||
| norB | norB F-SUT-PR48 | Nitric oxide reductase | AAGACCACGGTGACCAACAT | Designed from norB of Bradyrhizobium sp. strain ORS285 (LT859959.1) |
| norB R-SUT-PR48 | CGATCGATACCGTTGAGCTT | |||
| norBF-SUTN9-2 | ACAGGAAGAAGATCGCAACG | Designed from norB of Bradyrhizobium sp. strain SUTN9-2 (LAXE00000000) | ||
| norBR-SUTN9-2 | GTGGCTGTGGTCGGTTATCT | |||
| nirK | nirK F-SUT-PR48 | Cu-containing nitrite reductase | TGCTGATCGTCCATTCTCAG | Designed from nirK of Bradyrhizobium sp. strain ORS285 (LT859959.1) |
| nirK R-SUT-PR48 | TGTGGGTGACGTAAGCGTAG | |||
| nirK F-SUTN9-2 | TTGAAGTTGCCCTTCTCGTC | Designed from nirK of Bradyrhizobium sp. strain SUTN9-2 (LAXE00000000) | ||
| nirK R-SUTN9-2 | GGCGTGTTCGTGTATCACTG | |||
| Nitrogen fixation | ||||
| nifH | nifH F | Dinitrogenase reductase | TACGGNAARGGSGGNATCGGCAA | Noisangiam et al. (62) |
| nifH R | AGCATGTCYTCSAGYTCNTCCA | |||
| nifH-GUS (transcriptional fusion) | ||||
| nifHpm.SUTN9-2.F | nifH promoter region | ACCTATGTCGACGTGCTGAGCTGACTGAGTGG | Designed from nifH promoter region of Bradyrhizobium sp. strain SUTN9-2 (LAXE00000000) | |
| nifHpm.SUTN9-2.R | GCGTCGTCTAGAACTCAGCCCTCACTCAGTGT | |||
| pVO.Bis.r | pVO155 plasmid | GCACAGCAATTGCCCGGCTTTCTTG | Designed from pVO155 plasmid (to confirm nifH-GUS integrated into SUTN9-2 chromosome) | |
| CefoM15-Fin.f | Cefotaxime gene | GCTATGGCACCACCAACGATATC | Designed from CefoM15 gene (to confirm nifH-GUS integrated into SUTN9-2 chromosome) | |
Rice endophytic bradyrhizobia nifH-GUS assay and nitrogen fixation assay.
To obtain nifH-GUS-labeled strain, the plasmid pVO155nifHpm9-2gus was constructed by amplification of the nifH promoter region from the Bradyrhizobium sp. SUTN9-2 genome (54). This plasmid, which is nonreplicative in Bradyrhizobium strains, is a derivative of plasmid pVO155 and carries the promoterless gusA gene (55). The resulting plasmid was introduced into E. coli S17-1 by electroporation (15 kV/cm, 100 Ω, and 25 μF) and was transferred into SUTN9-2 by triparental mating (56). Transconjugants were selected on YEM plates supplemented with 25 μg/ml nalidixic acid, 20 μg/ml cefotaxime, and 100 μg/ml kanamycin.
The rice plants inoculated with the SUTN9-2 nifH-GUS-labeled strain were grown in the test tube containing N-free nutrient and a 1 mM urea solution. The fresh rice roots and leaf sheaths were harvested at 2, 4, 10, and 12 weeks after inoculation. Rice samples were surface-sterilized and then macerated separately in a sterilized mortar and pestle with sterilized water. The SUTN9-2 nifH-GUS-labeled strain was separated from rice tissues by filtration using a 3-layer Miracloth and 8.0-μm membrane filter, respectively. Glucuronidase was assayed in a buffer consisting of 50 mM sodium phosphate (pH 7.0), 10 mM 2 mercaptoethanol, 0.1% Triton X-100, and 1 mM p-nitrophenyl β-d-glucuronide. Reactions occurred in a 1-ml volume mixture at 37°C and were terminated by the addition of 0.4 ml of 2.5 M 2-amino-2-methylpropanediol. p-Nitrophenol absorbance was measured at 415 nm (57).
The symbiotic abilities of bradyrhizobial strains were determined in Leonard's jars containing sterilized vermiculite, and 1 ml of bacterial strain SUTN9-2 equivalent to 107 cells was inoculated onto germinated O. sativa cv. PT1 seeds. Plants were harvested after 2, 4, 8, and 12 weeks, and five plants were used to analyze nitrogenase activity using the acetylene reduction assay (ARA) (3). The reactions were carried out in an 80-ml test tube. Five percent (vol/vol) of gas phase in the headspace was replaced with acetylene and further incubated at 28°C for 24 h. Ethylene production was measured by using a gas chromatograph (GC) with a flame ionization detector and equipped with a PE-alumina column (50 m by 0.32 mm by 0.25 μm; PerkinElmer, USA).
Production of IAA and ACC deaminase by Bradyrhizobium sp. SUTN9-2 in rice tissues.
The rice roots and leaf sheaths were harvested at 1 month after inoculation with endophytic bradyrhizobial strain SUTN9-2 and with an uninoculated control. Whole-rice samples were sterilized with 70% ethanol for 30 s and 3% sodium hypochlorite for 3 min, washed 5 to 6 times with sterilized water, and then macerated separately in liquid nitrogen with a sterilized mortar and pestle. Then, the supernatant was collected by centrifugation at 10,000 × g for 5 min. IAA and ACC deaminase production was determined as previously described (references 45 and 47, respectively).
Enumeration of endophytic bradyrhizobia.
The plant most probable number (MPN) method was used to enumerate the bacterial endophytic population (19). Rice was grown under controlled environmental conditions of 28 ± 2°C on 16/8-h day/night cycle (full light, 639 μE · m−2 · S−1) for 1, 2, 3, and 4 weeks. Then, the rice was surface-sterilized, and excised samples of roots and leaf sheaths were macerated with a sterilized mortar and pestle, diluted with saline solution (0.85% NaCl), and inoculated into plastic pouches using mung bean as a plant host for enumerating the population of endophytic bradyrhizobial nodulating mung bean.
Preparation of rice stubbles as bradyrhizobial inoculum for mung bean.
The experiment was conducted under pot trial conditions. The experimental units consisted of pots (size 25.5 by 22.5 cm) sterilized with a 3% sodium hypochlorite solution overnight and then washed by adding boiled water into pots before filling them with 5 kg of low-organic-matter soil (pH 6.8; electrical conductivity [EC], 408 μS/cm; % organic matter [%O.M.], 0.63%; phosphorus [P], 49.9 ppm; potassium [K], 141 ppm; calcium [Ca], 689 ppm) and high-organic-matter soil (paddy soil) (pH 7.65; EC, 1,066 μS/cm; %O.M., 3.57%; P, 86.1 ppm; K, 932 ppm; Ca, 3,001 ppm). Soils were partially sterilized by adding 3 liters of boiled water into pots containing soil before seeding in order to eliminate native (indigenous) bradyrhizobia. Rice seeds were surface disinfected by washing them in 95% ethanol for 30 s, hydrogen peroxide (10% [vol/vol]) for 10 min, sodium hypochlorite solution (3% [vol/vol]) for 3 min, and then 5 to 6 times with sterilized water. Seeds were germinated in the dark at 30°C for 2 days on plates containing 0.85% agar YEM medium. Germinated seeds were soaked overnight with a culture of SUTN9-2 (108 CFU · ml−1) containing the GUS reporter gene (SUTN9-2GUS) (Table 5). The experiment was conducted as five replicates of the following treatments: (i) control (without inoculation), (ii) low-organic-matter soil inoculated with SUTN9-2, and (iii) high-organic-matter soil collected from a paddy field inoculated with SUTN9-2. Rice plants were grown in the greenhouse until the seed maturation stage of rice. The number of plants per hill and the number of panicles per hill were evaluated.
Enumeration of rice endophytic bradyrhizobia from rice tissues.
To enumerate the endophytic bradyrhizobia in rice tissues, 1- to 4-week-old rice tissues were surface-sterilized as previously described, and excised sample seeds, leaves, stems, roots, and stubbles were macerated separately with a sterilized mortar and pestle and then diluted in saline solution prior to spreading on YEM plates containing streptomycin (200 μg · ml−1) and 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc) (10 mg · ml−1). After 5 days of incubation at 28°C, the numbers of the blue colonies were counted to display the SUTN9-2 population densities in different rice tissues.
Enumeration of rice endophytic bradyrhizobia from soil.
Plant most probable number (MPN) count was carried out for enumeration of the bacterial number in soil from rice cultivation where the rice was harvested and harvested 1 week later. The soils were mixed with sterilized water (1:1 [wt/wt]). All visible roots were removed from each suspension. The plant MPN using mung bean as the plant host was calculated from the dry weight of the soil. The number of bradyrhizobia was estimated by using tenfold dilutions (41). Three parallel dilution series based on a statistical treatment of the counting methods were used for enumeration analyses (58).
Investigation of mung bean nodulation from rice stubbles containing endophytic bradyrhizobia.
After harvesting the rice inoculated with SUTN9-2, the remaining stubbles (∼40g/pot) were immediately incorporated into the soil in each pot. After 1 week, 3 mung bean seeds were planted in each pot for 3 weeks under greenhouse conditions. The nodules from mung bean roots were collected and stained by a GUS assay (59).
Statistical analysis.
Statistical analysis was performed with the SPSS 16.0 for Windows software (SPSS, Inc., Chicago, IL). The experimental data were statistically analyzed according to Steel and Torrie (60), and means were compared by Duncan's multiple range test (61).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Suranaree University of Technology (SUT) and by the Office of the Higher Education Commission under the NRU project of Thailand.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01488-17.
REFERENCES
- 1.Spaink HP. 2000. Root nodulation and infection factors produced by rhizobial bacteria. Annu Rev Microbiol 54:257–288. doi: 10.1146/annurev.micro.54.1.257. [DOI] [PubMed] [Google Scholar]
- 2.Biswas JC, Ladha JK, Dazzo FB, Yanni YG, Rolfe BG. 2000. Rhizobial inoculation influences seedling vigor and yield of rice. Agron J 92:880–886. doi: 10.2134/agronj2000.925880x. [DOI] [Google Scholar]
- 3.Piromyou P, Greetatorn T, Teamtisong K, Okubo T, Shinoda R, Nuntakij A, Tittabutr P, Boonkerd N, Minamisawa K, Teaumroong N. 2015. Preferential association of endophytic bradyrhizobia with different rice cultivars and its implications for rice endophyte evolution. Appl Environ Microbiol 81:3049–3061. doi: 10.1128/AEM.04253-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaintreuil C, Giraud E, Prin Y, Lorquin J, Bâ A, Gillis M, de Lajudie P, Dreyfus B. 2000. Photosynthetic bradyrhizobia are natural endophytes of the African wild rice Oryza breviligulata. Appl Environ Microbiol 66:5437–5447. doi: 10.1128/AEM.66.12.5437-5447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perrine FM, Prayitno J, Weinman JJ, Dazzo FB, Rolfe BG. 2001. Rhizobium plasmids are involved in the inhibition or stimulation of rice growth and development. Funct Plant Biol 28:923–937. doi: 10.1071/PP01046. [DOI] [Google Scholar]
- 6.Hilali A, Prevost D, Broughton W, Antoun H. 2001. Effects of inoculation with Rhizobium leguminosarum biovar trifolii on wheat cultivated in clover crop rotation agricultural soil in Morocco. Can J Microbiol 47:590–593. (In French.) [PubMed] [Google Scholar]
- 7.Bhattacharjee RB, Singh A, Mukhopadhyay S. 2008. Use of nitrogen-fixing bacteria as biofertiliser for non-legumes: prospects and challenges. Appl Microbiol Biotechnol 80:199–209. doi: 10.1007/s00253-008-1567-2. [DOI] [PubMed] [Google Scholar]
- 8.Polthanee A, Promkhambut A, Treloges V. 2012. Effect of pre-rice mungbean and cattle manure application on growth and yield of organic rice. Int J Environ Rural Dev 3:1–9. [Google Scholar]
- 9.Smith R. 1995. Inoculant formulations and applications to meet changing needs, p 653–657. In Tikhonovich IA, Provorov NA, Romanov VI, Newton WE (ed), Nitrogen fixation: fundamentals and applications. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 10.Ben Rebah F, Tyagi RD, Prevost D. 2002. Wastewater sludge as a substrate for growth and carrier for rhizobia: the effect of storage conditions on survival of Sinorhizobium meliloti. Bioresour Technol 83:145–151. doi: 10.1016/S0960-8524(01)00202-4. [DOI] [PubMed] [Google Scholar]
- 11.Hotter GS, Scott DB. 1991. Exopolysaccharide mutants of Rhizobium loti are fully effective on a determinate nodulating host but are ineffective on an indeterminate nodulating host. J Bacteriol 173:851–859. doi: 10.1128/jb.173.2.851-859.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang S-W, Hamayun M, Sohn W, Kang S-M, Choi K-I, Shin D-H, Lee I-J. 2008. Growth and gibberellins levels of two rice cultivars as influenced by different nitrogen containing compounds. J Crop Sci Biotech 11:223–226. [Google Scholar]
- 13.Jolivet E. 1987. Nitrate and ammonium nutrition in plants. Plant Physiol Biochem 25:805–812. [Google Scholar]
- 14.Gaudin R, Dupuy J. 1999. Ammoniacal nutrition of transplanted rice fertilized with large urea granules. Agron J 91:33–36. doi: 10.2134/agronj1999.00021962009100010006x. [DOI] [Google Scholar]
- 15.Meakin G, Jepson B, Richardson D, Bedmar E, Delgado M. 2006. The role of Bradyrhizobium japonicum nitric oxide reductase in nitric oxide detoxification in soya bean root nodules. Biochem Soc Trans 34:195–196. doi: 10.1042/BST0340195. [DOI] [PubMed] [Google Scholar]
- 16.Fageria NK, Baligar VC, Clark R. 2006. Physiology of crop production. CRC Press, Boca Raton, FL. [Google Scholar]
- 17.Mangel M, Stamps J. 2001. Trade-offs between growth and mortality and the maintenance of individual variation in growth. Evol Ecol Res 3:611–632. [Google Scholar]
- 18.Delgado MJ, Casella S, Bedmar EJ. 2006. Denitrification in rhizobia-legume symbiosis, p 83–93. In Bothe H, Ferguson SJ, Newton WE (ed), Biology of the nitrogen cycle. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 19.Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61:533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maxwell CA, Hartwig UA, Joseph CM, Phillips DA. 1989. A chalcone and two related flavonoids released from alfalfa roots induce nod genes of Rhizobium meliloti. Plant Physiol 91:842–847. doi: 10.1104/pp.91.3.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beligni MV, Lamattina L. 2000. Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 210:215–221. doi: 10.1007/PL00008128. [DOI] [PubMed] [Google Scholar]
- 22.Millar AH, Day DA. 1996. Nitric oxide inhibits the cytochrome oxidase but not the alternative oxidase of plant mitochondria. FEBS Lett 398:155–158. doi: 10.1016/S0014-5793(96)01230-6. [DOI] [PubMed] [Google Scholar]
- 23.Mesa S, Velasco L, Manzanera ME, Delgado MJ, Bedmar EJ. 2002. Characterization of the norCBQD genes, encoding nitric oxide reductase, in the nitrogen fixing bacterium Bradyrhizobium japonicum. Microbiology 148:3553–3560. doi: 10.1099/00221287-148-11-3553. [DOI] [PubMed] [Google Scholar]
- 24.Bedmar EJ, Robles EF, Delgado MJ. 2005. The complete denitrification pathway of the symbiotic, nitrogen-fixing bacterium Bradyrhizobium japonicum. Biochem Soc Trans 33:141–144. doi: 10.1042/BST0330141. [DOI] [PubMed] [Google Scholar]
- 25.Sánchez C, Cabrera JJ, Gates AJ, Bedmar EJ, Richardson DJ, Delgado MJ. 2011. Nitric oxide detoxification in the rhizobia–legume symbiosis. Biochem Soc Trans 39:184–188. doi: 10.1042/BST0390184. [DOI] [PubMed] [Google Scholar]
- 26.Hérouart D, Baudouin E, Frendo P, Harrison J, Santos R, Jamet A, Van de Sype G, Touati D, Puppo A. 2002. Reactive oxygen species, nitric oxide and glutathione: a key role in the establishment of the legume-Rhizobium symbiosis? Plant Physiol Biochem 40:619–624. doi: 10.1016/S0981-9428(02)01415-8. [DOI] [Google Scholar]
- 27.Meilhoc E, Boscari A, Bruand C, Puppo A, Brouquisse R. 2011. Nitric oxide in legume-Rhizobium symbiosis. Plant Sci 181:573–581. doi: 10.1016/j.plantsci.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Baudouin E, Pieuchot L, Engler G, Pauly N, Puppo A. 2006. Nitric oxide is formed in Medicago truncatula-Sinorhizobium meliloti functional nodules. Mol Plant Microbe Interact 19:970–975. doi: 10.1094/MPMI-19-0970. [DOI] [PubMed] [Google Scholar]
- 29.Rockel P, Strube F, Rockel A, Wildt J, Kaiser WM. 2002. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J Exp Bot 53:103–110. [PubMed] [Google Scholar]
- 30.Gomes CM, Giuffre A, Forte E, Vicente JB, Saraiva LM, Brunori M, Teixeira M. 2002. A novel type of nitric-oxide reductase Escherichia coli flavorubredoxin. J Biol Chem 277:25273–25276. doi: 10.1074/jbc.M203886200. [DOI] [PubMed] [Google Scholar]
- 31.Dalsing BL, Truchon AN, Gonzalez-Orta ET, Milling AS, Allen C. 2015. Ralstonia solanacearum uses inorganic nitrogen metabolism for virulence, ATP production, and detoxification in the oxygen-limited host xylem environment. mBio 6(2):e02471-14. doi: 10.1128/mBio.02471-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alazard D. 1990. Nitrogen fixation in pure culture by rhizobia isolated from stem nodules of tropical Aeschynomene species. FEMS Microbiol Lett 68:177–182. [Google Scholar]
- 33.Bhattacharjee RB, Jourand P, Chaintreuil C, Dreyfus B, Singh A, Mukhopadhyay SN. 2012. Indole acetic acid and ACC deaminase-producing Rhizobium leguminosarum bv. trifolii SN10 promote rice growth, and in the process undergo colonization and chemotaxis. Biol Fertil Soils 48:173–182. doi: 10.1007/s00374-011-0614-9. [DOI] [Google Scholar]
- 34.Terakado-Tonooka J, Ohwaki Y, Yamakawa H, Tanaka F, Yoneyama T, Fujihara S. 2008. Expressed nifH genes of endophytic bacteria detected in field-grown sweet potatoes (Ipomoea batatas L.). Microbes Environ 23:89–93. doi: 10.1264/jsme2.23.89. [DOI] [PubMed] [Google Scholar]
- 35.Yanni YG, Rizk RY, El-Fattah FKA, Squartini A, Corich V, Giacomini A, de Bruijn F, Rademaker J, Maya-Flores J, Ostrom P, Vega-Hernandez M, Hollingsworth RI, Martinez-Moline E, Mateos P, Velazquez E, Wopereis J, Triplett E, Umali-Garcia M, Anarna JA, Rolfe BG, Ladha JK, Hill J, Mujoo R, Ng PK, Dazzo FB. 2001. The beneficial plant growth-promoting association of Rhizobium leguminosarum bv. trifolii with rice roots. Aust J Plant Biol 28:845–870. doi: 10.1071/PP01069. [DOI] [Google Scholar]
- 36.Chi F, Shen S-H, Cheng H-P, Jing Y-X, Yanni YG, Dazzo FB. 2005. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl Environ Microbiol 71:7271–7278. doi: 10.1128/AEM.71.11.7271-7278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yanni YG, Rizk R, Corich V, Squartini A, Ninke K, Philip-Hollingsworth S, Orgambide G, De Bruijn F, Stoltzfus J, Buckley D. 1997. Natural endophytic association between Rhizobium leguminosarum bv. trifolii and rice roots and assessment of its potential to promote rice growth. Plant Soil 194:99–114. doi: 10.1023/A:1004269902246. [DOI] [Google Scholar]
- 38.Hafeez F, Idris M, Malik K. 1989. Growth and survival of cowpea bradyrhizobia in various carrier materials. Biol Fertil Soils 7:279–282. doi: 10.1007/BF00709662. [DOI] [Google Scholar]
- 39.Perkins AT. 1925. The effect of bacterial numbers on the nodulation of Virginia soybeans. J Agric Res 30:95–96. [Google Scholar]
- 40.Weaver RW. 1970. Populations of Rhizobium japonicum in Iowa soils and inoculum level needed for nodulation of Glycine max (L.) Merrill. Ph.D. dissertation, Iowa State University, Ames, IA. [Google Scholar]
- 41.Somasegaran P, Hoben HJ. 2012. Collecting nodules and isolating rhizobia, p 7–23. In Handbook for rhizobia: methods in legume-Rhizobium technology. Springer Science & Business Media, New York, NY. [Google Scholar]
- 42.Turner G, Gibson A. 1980. Measurement of nitrogen fixation by indirect means, p 111–138. In Bergersen FJ. (ed), Methods for evaluating biological nitrogen fixation. John Wiley and Sons, Chichester, United Kingdom. [Google Scholar]
- 43.Nuntagij A, Abe M, Uchiumi T, Seki Y, Boonkerd N, Higashi S. 1997. Characterization of Bradyrhizobium strains isolated from soybean cultivation in Thailand. J Gen Appl Microbiol 43:183–187. doi: 10.2323/jgam.43.183. [DOI] [PubMed] [Google Scholar]
- 44.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275. [PubMed] [Google Scholar]
- 45.Costacurta A, Mazzafera P, Rosato YB. 1998. Indole-3-acetic acid biosynthesis by Xanthomonas axonopodis pv. citri is increased in the presence of plant leaf extracts. FEMS Microbiol Lett 159:215–220. doi: 10.1111/j.1574-6968.1998.tb12863.x. [DOI] [Google Scholar]
- 46.Penrose DM, Glick BR. 2003. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant 118:10–15. doi: 10.1034/j.1399-3054.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- 47.Tittabutr P, Awaya JD, Li QX, Borthakur D. 2008. The cloned 1-aminocyclopropane-1-carboxylate (ACC) deaminase gene from Sinorhizobium sp. strain BL3 in Rhizobium sp. strain TAL1145 promotes nodulation and growth of Leucaena leucocephala. Syst Appl Microbiol 31:141–150. doi: 10.1016/j.syapm.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Shaharoona B, Arshad M, Zahir Z. 2006. Effect of plant growth promoting rhizobacteria containing ACC-deaminase on maize (Zea mays L.) growth under axenic conditions and on nodulation in mung bean (Vigna radiata L.). Lett Appl Microbiol 42:155–159. doi: 10.1111/j.1472-765X.2005.01827.x. [DOI] [PubMed] [Google Scholar]
- 49.Nowell J, Pawley J. 1979. Preparation of experimental animal tissue for SEM. Scan Electron Microsc II:1–19. [PubMed] [Google Scholar]
- 50.Bacilio-Jiménez M, Aguilar-Flores S, del Valle MV, Pérez A, Zepeda A, Zenteno E. 2001. Endophytic bacteria in rice seeds inhibit early colonization of roots by Azospirillum brasilense. Soil Biol Biochem 33:167–172. doi: 10.1016/S0038-0717(00)00126-7. [DOI] [Google Scholar]
- 51.Nagata M, Murakami E-i, Shimoda Y, Shimoda-Sasakura F, Kucho K-i, Suzuki A, Abe M, Higashi S, Uchiumi T. 2008. Expression of a class 1 hemoglobin gene and production of nitric oxide in response to symbiotic and pathogenic bacteria in Lotus japonicus. Mol Plant Microbe Interact 21:1175–1183. doi: 10.1094/MPMI-21-9-1175. [DOI] [PubMed] [Google Scholar]
- 52.Tominaga A, Nagata M, Futsuki K, Abe H, Uchiumi T, Abe M, Kucho K-i, Hashiguchi M, Akashi R, Hirsch AM, Arima S, Suzuki A. 2009. Enhanced nodulation and nitrogen fixation in the abscisic acid low-sensitive mutant enhanced nitrogen fixation1 of Lotus japonicus. Plant Physiol 151:1965–1976. doi: 10.1104/pp.109.142638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fukudome M, Calvo-Begueria L, Kado T, Osuki K-i, Rubio MC, Murakami E-i, Nagata M, Kucho K-i, Sandal N, Stougaard J. 2016. Hemoglobin LjGlb1-1 is involved in nodulation and regulates the level of nitric oxide in the Lotus japonicus–Mesorhizobium loti symbiosis. J Exp Bot 67:5275–5283. doi: 10.1093/jxb/erw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piromyou P, Songwattana P, Greetatorn T, Okubo T, Kakizaki KC, Prakamhang J, Tittabutr P, Boonkerd N, Teaumroong N, Minamisawa K. 2015. The type III secretion system (T3SS) is a determinant for rice-endophyte colonization by non-photosynthetic Bradyrhizobium. Microbes Environ 30:291–300. doi: 10.1264/jsme2.ME15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oke V, Long SR. 1999. Bacterial genes induced within the nodule during the Rhizobium–legume symbiosis. Mol Microbiol 32:837–849. doi: 10.1046/j.1365-2958.1999.01402.x. [DOI] [PubMed] [Google Scholar]
- 56.Tittabutr P, Sripakdi S, Boonkerd N, Tanthanuch W, Minamisawa K, Teaumroong N. 2015. Possible role of 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity of Sinorhizobium sp. BL3 on symbiosis with mung bean and determinate nodule senescence. Microbes Environ 30:310–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beliaeff B, Mary J-Y. 1993. The “most probable number” estimate and its confidence limits. Water Res 27:799–805. doi: 10.1016/0043-1354(93)90143-6. [DOI] [Google Scholar]
- 59.Manassila M, Nuntagij A, Kotepong S, Boonkerd N, Teaumroong N. 2007. Characterization and monitoring of selected rhizobial strains isolated from tree legumes in Thailand. Afr J Biotechnol 6:1393. [Google Scholar]
- 60.Steel RG, Torrie JH. 1980. Principle and procedures of statistic: a biometrical approach. McGraw-Hill, New York, NY. [Google Scholar]
- 61.Duncan DB. 1955. Multiple range and multiple F tests. Biometrics 11:1–42. doi: 10.2307/3001478. [DOI] [Google Scholar]
- 62.Noisangiam R, Teamtisong K, Tittabutr P, Boonkerd N, Toshiki U, Minamisawa K, Teaumroong N. 2012. Genetic diversity, symbiotic evolution, and proposed infection process of Bradyrhizobium strains isolated from root nodules of Aeschynomene americana L. in Thailand. Appl Environ Microbiol 78:6236–6250. doi: 10.1128/AEM.00897-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Molouba F, Lorquin J, Willems A, Hoste B, Giraud E, Dreyfus B, Gillis M, de Lajudie P, Masson-Boivin C. 1999. Photosynthetic bradyrhizobia from Aeschynomene spp. are specific to stem-nodulated species and form a separate 16S ribosomal DNA restriction fragment length polymorphism group. Appl Environ Microbiol 65:3084–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vinuesa P, Silva C, Werner D, Martínez-Romero E. 2005. Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol Phylogenet Evol 34:29–54. doi: 10.1016/j.ympev.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 65.Wilson KJ, Sessitsch A, Corbo JC, Giller KE, Akkermans AD, Jefferson RA. 1995. β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other Gram-negative bacteria. Microbiology 141:1691–1705. doi: 10.1099/13500872-141-7-1691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.