ABSTRACT
Patients with community-onset (CO) methicillin-resistant Staphylococcus aureus (MRSA) infections contribute to MRSA contamination of the home environment and may be reexposed to MRSA strains from this reservoir. This study evaluates One Health risk factors, which focus on the relationship between humans, animals, and the environment, for the increased prevalence of multiple antimicrobial-resistant MRSA isolates in the home environment. During a trial of patients with CO-MRSA infection, MRSA was isolated from the household environment at the baseline and 3 months later, following randomization of patients and household members to mupirocin-based decolonization therapy or an education control group. Up to two environmental MRSA isolates collected at each visit were tested. MRSA isolates were identified in 68% (65/95) of homes at the baseline (n = 104 isolates) and 51% (33/65) of homes 3 months later (n = 56 isolates). The rates of multidrug resistance (MDR) were 61% among isolates collected at the baseline and 55% among isolates collected at the visit 3 months later. At the baseline, 100% (14/14) of MRSA isolates from rural homes were MDR. While antimicrobial use by humans or pets was associated with an increased risk for the isolation of MDR MRSA from the environment, clindamycin use was not associated with an increased risk for the isolation of MDR MRSA. Incident low-level mupirocin-resistant MRSA strains were isolated at 3 months from 2 (5%) of 39 homes that were randomized to mupirocin treatment but none of the control homes. Among patients recently treated for a CO-MRSA infection, MRSA and MDR MRSA were common contaminants in the home environment. This study contributes to evidence that occupant use of antimicrobial drugs, except for clindamycin, is associated with MDR MRSA in the home environmental reservoir. (This study has been registered at ClinicalTrials.gov under registration no. NCT00966446.)
IMPORTANCE MRSA is a common bacterial agent implicated in skin and soft tissue infections (SSTIs) in both community and health care settings. Patients with CO-MRSA infections contribute to environmental MRSA contamination in these settings and may be reexposed to MRSA strains from these reservoirs. People interact with natural and built environments; therefore, understanding the relationships between humans and animals as well as the characteristics of environmental reservoirs is important to advance strategies to combat antimicrobial resistance. Household interactions may influence the frequency and duration of exposure, which in turn may impact the duration of MRSA colonization or the probability for recurrent colonization and infection. Therefore, MRSA contamination of the home environment may contribute to human and animal recolonization and decolonization treatment failure. The aim of this study was to evaluate One Health risk factors that may be amenable to intervention and may influence the recovery of MDR and mupirocin resistance in CO-MRSA isolates.
KEYWORDS: MRSA, Staphylococcus aureus, multidrug resistance, mupirocin, environment, household, One Health
INTRODUCTION
Staphylococcus aureus, including methicillin-resistant S. aureus (MRSA) strains, is one of the most common bacterial agents implicated in skin and soft tissue infections (SSTIs) in both community and health care settings (1–3). MRSA nasal colonization has been shown to increase the risk for the development of clinical infection (4). Antimicrobial-resistant pathogens, which include MRSA, have human costs in morbidity and mortality, and they have been estimated to contribute to excess health care costs (5). Understanding One Health risk factors, encompassing the relationships between humans and animals as well as the characteristics of environmental reservoirs, is important to advance strategies to combat antimicrobial resistance (AMR) (6).
People interact with the natural and built environments, which may serve as reservoirs for both pathogens and antimicrobial resistance determinants (7). In the community, households are increasingly recognized as critical to cycles of recolonization and infection of patients without hospital-associated risk factors (8). Household interactions may influence the frequency and duration of exposure, which in turn may impact the duration of MRSA colonization or the probability for recurrent colonization and infection (9). Patients with MRSA SSTIs and their colonized household members contribute to MRSA contamination of the home and then may be reexposed, including to multidrug-resistant (MDR) (10) and mupirocin-resistant (Mupr) MRSA strains, from this reservoir. Household occupants include domestic pets, which have been implicated in household MRSA transmission (11–13). Transmission involving humans and/or pets can occur directly or indirectly through the environment (8). Therefore, MRSA contamination of the home environment may contribute to human and animal recolonization and decolonization treatment failure.
Mupirocin is an important antimicrobial drug that is typically used in humans for MRSA decolonization (14). However, MDR and Mupr in MRSA isolates limit treatment and decolonization options (14–16). Therefore, the aim of this study was to evaluate One Health risk factors that may be amenable to intervention and may influence the recovery of MDR isolates, defined by the Sentry Antimicrobial Surveillance Program to be isolates nonsusceptible to four or more classes of antimicrobials (10), and Mupr isolates from among community-onset MRSA (CO-MRSA) isolates. We hypothesized that the primary risk factors that could drive MDR resistance and Mupr in CO-MRSA isolates were antimicrobial drug use in humans and animals and household decolonization treatment and that secondary risk factors could include sample location, home location, household size, the presence of domestic pets, evidence of unwanted pests, season, and reported use of disinfectants. We tested this hypothesis with MRSA isolates collected from surfaces in the homes of people recently diagnosed with a MRSA SSTI before and after these households were randomized to a mupirocin-based decolonization treatment or an education control group.
(Portions of this work were previously presented at two American Society for Microbiology conferences [17, 18].)
RESULTS
Identification and characterization of MRSA isolates.
Environmental sites in 95 homes of patients diagnosed with a community-onset MRSA infection were sampled. Sampling was repeated in 65 homes 3 months after the baseline visit (referred to here as the 3-month visit) and following randomization to decolonization therapy or an education control group for people. MRSA isolates were identified in 68% (65/95) of the homes at the baseline and 51% (33/65) of the homes 3 months later. At the baseline, 104 isolates were identified as MRSA. At the 3-month visit, 56 isolates were identified as MRSA. In each home, 0 to 2 MRSA isolates were identified per visit. Table S1 in the supplemental material displays the results of spa typing and shows that 91 of the 160 (57%) isolates were spa type t008. Isolates of spa type t008 were not more likely than other spa types to be MDR (adjusted odds ratio [aOR], 0.64; 95% confidence interval [CI], 0.27, 1.51; P = 0.31), accounting for clustering within the home and accounting for the visit.
Subset analysis of household surfaces.
We conducted a PCR evaluation of the 196 presumptive coagulase-positive staphylococci (CPS) identified from 308 environmental samples in a subset of 25 homes: 25 of 95 (26%) homes at the enrollment (baseline) visit and 14 of 65 (22%) of the same homes sampled again at the 3-month visit. As previously described, this subset was identified a priori as the first 20 homes of individuals enrolled from the four urban hospitals and the first 5 homes of individuals enrolled from the rural hospital (19). Repository surfaces were contaminated with MRSA more often than frequently touched sites (aOR, 1.51; 95% CI, 0.93, 2.44; P = 0.09), and contamination rates were lower at the 3-month visit than at the enrollment visit (aOR, 0.38; 95% CI, 0.14, 1.06; P = 0.06), but neither of these two estimates achieved statistical significance. A self-report of site-specific recent cleaning or laundering (on the same day as the visit or within the prior 3 days) did not impact either CPS or MRSA recovery from surfaces in the subset and did not impact CPS recovery from the surfaces of all homes considered (data not shown).
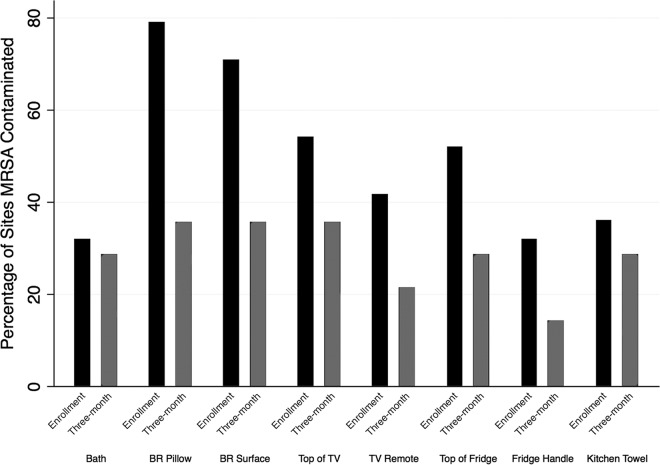
Subset data demonstrated that no house was misclassified as MRSA negative on the basis of isolate selection. Figure 1 shows the results from these homes according to the site sampled. In this subset, sites in the bedroom of the index participant were more often contaminated with MRSA than sites in the common room (aOR, 2.74; 95% CI, 1.78, 4.25; P < 0.001). Recent cleaning or laundering was not associated with a reduction in the rate of recovery of MRSA.
FIG 1.
Percentage of sites contaminated with MRSA at the enrollment visit (baseline) and the 3-month visit. Samples were collected from eight standardized locations in the common room, kitchen, and bedroom (BR) of each household.
Multidrug resistance in environmental MRSA isolates.
Sixty-two percent (64/104) of MRSA isolates were classified as MDR at the baseline, and 55% (31/56) were classified as MDR at the 3-month visit. All 160 isolates were positive for the mecA gene, which confers beta-lactam resistance. The rates of susceptibility to other antimicrobial drugs, determined by disk diffusion testing, were as follows: erythromycin, 13%; ciprofloxacin, 48%; clindamycin, 74%; gentamicin, 74%; tetracycline, 90%; trimethoprim-sulfamethoxazole, 93%; linezolid, 97%; chloramphenicol, 97%; quinupristin-dalfopristin (Synercid), 99%; and vancomycin 100%.
Risk factors for home contamination with MDR MRSA.
At the baseline visit, rural home location and small household size (2 or fewer people) were significant household or environmental predictors of MDR among MRSA isolates (Table 1); 100% (14/14) of the MRSA isolates from rural homes were multidrug resistant (P = 0.001), and 10 (71%) of these were of spa type t008. When all 160 isolates collected at both visits from all households were evaluated, 91% (20/22) of isolates from rural households were found to be MDR MRSA. Also at the baseline visit, the use of any antimicrobial drugs (other than clindamycin) by a human or domestic pet occupant was associated with the detection of MDR MRSA isolates. Clindamycin use was associated with the detection of MRSA isolates that were not MDR (Table 2). Index patient age was moderately negatively correlated with household size (P < 0.003); only household size was included in the model. Because isolation from a rural household completely predicted MDR, this variable was excluded from subsequent logistic regression models. The antimicrobial use characteristics of rural households differed from those of nonrural households (more use of trimethoprim-sulfamethoxazole than clindamycin); the isolate-level comparison is shown in Table S2.
TABLE 1.
Household and environmental risk factorsa
| Factor | Baseline visit |
Three-month visit |
||||||
|---|---|---|---|---|---|---|---|---|
| No. (%) of isolates |
P valueb | No. (%) of isolates |
P valueb | |||||
| Total | Sentry MDR (n = 64) | Not MDR (n = 40) | Total | Sentry MDR (n = 31) | Not MDR (n = 25) | |||
| Room location | ||||||||
| Common room | 51 | 28 (54.90) | 23 (45.10) | 28 | 15 (53.57) | 13 (46.43) | ||
| Bedroom | 53 | 36 (67.92) | 17 (32.08) | 0.17 | 28 | 16 (57.14) | 12 (42.86) | 0.79 |
| Location | ||||||||
| Rural | 14 | 14 (100.00) | 0 (0.00) | 8 | 6 (75.00) | 2 (25.00) | ||
| Nonrural | 90 | 50 (55.56) | 40 (44.44) | 0.001*** | 48 | 25 (52.08) | 23 (47.92) | 0.24 |
| House type | ||||||||
| Attached | 63 | 34 (53.97) | 29 (46.03) | Ref | 30 | 14 (46.67) | 16 (53.33) | Ref |
| Single family | 29 | 21 (72.41) | 8 (27.59) | 0.10 | 21 | 14 (66.67) | 7 (33.33) | 0.16 |
| Apartment | 12 | 9 (75.00) | 3 (25.00) | 0.19 | 5 | 3 (60.00) | 2 (40.00) | 0.58 |
| HH size (no. of people) | ||||||||
| 1–2 | 26 | 20 (76.92) | 6 (23.08) | 0.04* | 11 | 7 (63.64) | 4 (36.36) | 0.58 |
| 3 | 19 | 10 (52.63) | 9 (47.37) | 0.92 | 11 | 6 (54.55) | 5 (45.45) | 0.97 |
| 4 | 16 | 12 (75.00) | 4 (25.00) | 0.11 | 8 | 4 (50.00) | 4 (50.00) | 0.85 |
| ≥5 | 43 | 22 (51.16) | 21 (48.84) | Ref | 26 | 14 (53.85) | 12 (46.15) | Ref |
| Site | ||||||||
| Touched site | 69 | 45 (65.22) | 24 (34.78) | 36 | 20 (55.56) | 16 (44.44) | ||
| Repository site | 35 | 19 (54.29) | 16 (45.71) | 0.28 | 20 | 11 (55.00) | 9 (45.00) | 0.97 |
| HH petsc | 2.48 (0–14) | 1.55 (0–10) | 0.10 | 2.97 (0–10) | 1.36 (0–3) | 0.04* | ||
| Unwanted pests | ||||||||
| Yes | 89 | 53 (59.55) | 36 (40.45) | 47 | 23 (48.94) | 24 (51.06) | ||
| No | 15 | 11 (73.33) | 4 (26.67) | 0.32 | 9 | 8 (88.89) | 1 (11.11) | 0.05* |
| Season | ||||||||
| Summer | 24 | 15 (62.50) | 9 (37.50) | Ref | 22 | 9 (40.91) | 13 (59.09) | Ref |
| Fall | 16 | 12 (75.00) | 4 (25.00) | 0.41 | 18 | 13 (72.22) | 5 (27.78) | 0.05 |
| Winter | 26 | 17 (65.38) | 9 (34.62) | 0.83 | 11 | 7 (63.64) | 4 (36.36) | 0.22 |
| Spring | 38 | 20 (52.63) | 18 (47.37) | 0.45 | 5 | 2 (40.00) | 3 (60.00) | 0.97 |
| EPA cleaner | ||||||||
| Yes | 65 | 39 (60.00) | 26 (40.00) | 37 | 24 (64.86) | 13 (35.14) | ||
| No | 39 | 25 (64.10) | 14 (35.90) | 0.68 | 19 | 7 (36.84) | 12 (63.16) | 0.05* |
Sentry MDR, Sentry Antimicrobial Surveillance Program definition of MDR; HH, household; Ref, reference group.
P values for all covariates except rural were obtained using logistic regression. The P value for rural was obtained using chi-square analysis. Significance, indicated in bold, is as follows: *, P ≤ 0.05; ***, P ≤ 0.001.
Data represent the mean (range) number of isolates.
TABLE 2.
Household occupant antimicrobial usea
| Factor | Baseline visit |
Three-month visit |
||||||
|---|---|---|---|---|---|---|---|---|
| No. (%) of isolates |
P valueb | No. (%) of isolates |
P valueb | |||||
| Total | Sentry MDR (n = 64) | Not MDR (n = 40) | Total | Sentry MDR (n = 31) | Not MDR (n = 25) | |||
| Any HH Abx use | ||||||||
| Human or pet use | 96 | 63 (65.62) | 33 (34.38) | 17 | 10 (58.82) | 7 (41.18) | ||
| No use | 8 | 1 (12.50) | 7 (87.50) | 0.02* | 39 | 21 (53.85) | 18 (46.15) | 0.73 |
| Individual Abx use | ||||||||
| Clinda | ||||||||
| Yes | 41 | 18 (43.90) | 23 (56.10) | 3 | 1 (33.33) | 2 (66.67) | ||
| No | 63 | 46 (73.02) | 17 (26.98) | 0.003** | 53 | 30 (56.60) | 23 (43.40) | 0.45 |
| Sulfa | ||||||||
| Yes | 52 | 36 (69.23) | 16 (30.77) | 4 | 2 (50.00) | 2 (50.00) | ||
| No | 52 | 28 (53.85) | 24 (46.15) | 0.11 | 52 | 29 (55.77) | 23 (44.23) | 0.82 |
| Mup | ||||||||
| Yes | 12 | 6 (50.00) | 6 (50.00) | 5 | 3 (60.00) | 2 (40.00) | ||
| No | 92 | 58 (63.04) | 34 (36.96) | 0.39 | 51 | 28 (54.90) | 23 (45.10) | 0.83 |
| Decolonization | ||||||||
| Yes | 30 | 17 (56.67) | 13 (43.33) | |||||
| No | 26 | 14 (53.85) | 12 (46.15) | 0.83 | ||||
Sentry MDR, Sentry Antimicrobial Surveillance Program definition of MDR; HH, household; Abx, antimicrobial; Clinda, clindamycin; Sulfa, trimethoprim-sulfamethoxazole; Mup, mupirocin.
P values for all covariates were obtained using logistic regression. Significance, indicated in bold, is as follows: *, P ≤ 0.05; **, P ≤ 0.01.
Tables 3 and 4 show the results from unadjusted and adjusted logistic regression models at the baseline and at the follow-up visit, respectively, and demonstrate that household size, pet ownership, and antimicrobial use were associated with MDR MRSA in the adjusted models. Of note, clindamycin use by either humans or animals was not associated with a risk of MDR, while trimethoprim-sulfamethoxazole use was associated with a higher risk for the detection of home environmental MDR MRSA.
TABLE 3.
Unadjusted and adjusted logistic regression models at the baseline visita
| Outcome variable | Unadjusted model |
Adjusted model Ab |
Adjusted model Bc |
|||
|---|---|---|---|---|---|---|
| ORd (95% CI) | P valuee | OR (95% CI) | P valuee | OR (95% CI) | P valuee | |
| Bedroom | 1.74 (0.91–3.33) | 0.09 | 2.94 (0.89–9.74) | 0.08 | 1.81 (0.67–4.87) | 0.24 |
| Touched site | 1.58 (0.65–3.81) | 0.31 | 0.61 (0.15–2.54) | 0.49 | 1.34 (0.41–4.43) | 0.62 |
| HH size (no. of people) | ||||||
| 1–2 | 3.18 (0.88–11.48) | 0.08 | 7.06 (1.06–47.14) | 0.04* | 3.88 (0.69–21.87) | 0.12 |
| 3 | 1.06 (0.30–3.71) | 0.93 | 1.23 (0.31–4.85) | 0.76 | 0.45 (0.10–2.02) | 0.30 |
| 4 | 2.86 (0.50–16.36) | 0.23 | 10.86 (1.11–106.65) | 0.04* | 1.88 (0.26–13.50) | 0.53 |
| ≥5 | Ref | Ref | Ref | Ref | Ref | Ref |
| House type | ||||||
| Attached | Ref | Ref | Ref | Ref | Ref | Ref |
| Single family | 2.23 (0.70–7.20) | 0.17 | 1.95 (0.40–9.59) | 0.41 | 1.59 (0.37–6.77) | 0.52 |
| Apartment | 2.56 (0.41–15.90) | 0.31 | 4.68 (0.55–39.79) | 0.16 | 2.08 (0.20–21.85) | 0.54 |
| Domestic pets | 1.18 (0.94–1.48) | 0.16 | 1.32 (0.98–1.77) | 0.07 | 1.31 (1.04–1.65) | 0.02* |
| Season | ||||||
| Summer | Ref | Ref | Ref | Ref | Ref | Ref |
| Fall | 1.80 (0.39–8.36) | 0.45 | 2.20 (0.32–15.20) | 0.42 | 1.42 (0.23–8.93) | 0.70 |
| Winter | 1.13 (0.39–8.36) | 0.45 | 3.23 (0.54–19.26) | 0.19 | 4.14 (0.60–28.73) | 0.15 |
| Spring | 0.67 (0.17–2.58) | 0.55 | 1.93 (0.29–12.91) | 0.49 | 1.68 (0.26–10.91) | 0.58 |
| Unwanted pests | 0.54 (0.10–3.00) | 0.47 | 1.29 (0.14–11.67) | 0.82 | 0.63 (0.10–4.05) | 0.62 |
| No EPA cleaner | 1.19 (0.46–3.10) | 0.72 | 1.59 (0.54–4.70) | 0.40 | 0.75 (0.22–2.57) | 0.65 |
| Decolonization | ||||||
| Any HH Abx use | ||||||
| No use | Ref | Ref | Ref | Ref | ||
| Human or pet use | 13.36 (1.36–131.15) | 0.03* | 70.35 (1.33–3718.66) | 0.04* | ||
| Individual Abx use | ||||||
| Clinda | 0.29 (0.11–0.80) | 0.02* | 0.27 (0.08–0.90) | 0.03* | ||
| Sulfa | 1.93 (0.73–5.07) | 0.18 | 3.06 (0.98–9.56) | 0.05* | ||
| Mup | 0.59 (1.02–2.84) | 0.46 | 0.76 (0.06–9.84) | 0.83 | ||
Data are for 104 isolates. HH, household; Abx, antimicrobial; Clinda, clindamycin; Sulfa, trimethoprim-sulfamethoxazole; Mup, mupirocin; Ref, reference group.
Model A evaluates any use of antimicrobial drugs by human or domestic animal occupants.
Model B evaluates mupirocin, clindamycin, and trimethoprim-sulfamethoxazole use as individual variables.
ORs for adjusted models used logistic regression and took all listed covariates into account. Rural was not included in the models due to a zero stratum (cf. Table 1).
P values for all covariates were obtained using logistic regression. Significance, indicated in bold, is as follows: *, P ≤ 0.05.
TABLE 4.
Unadjusted and adjusted logistic regression models at the follow-up visita
| Outcome variable | Unadjusted |
Adjusted model Ab |
Adjusted model Bc |
|||
|---|---|---|---|---|---|---|
| ORd (95% CI) | P valuee | OR (95% CI) | P valuee | OR (95% CI) | P valuee | |
| Bedroom | 1.16 (0.51–2.61) | 0.72 | 1.02 (0.05–21.03) | 0.99 | 0.67 (0.04–10.80) | 0.77 |
| Touched site | 1.02 (0.41–2.56) | 0.96 | 1.56 (0.07–33.36) | 0.77 | 3.37 (0.22–50.95) | 0.37 |
| HH size (no. of people) | ||||||
| 1–2 | 1.50 (0.24–9.41) | 0.66 | 8.34 (0.61–113.21) | 0.12 | 9.72 (0.27–352.32) | 0.21 |
| 3 | 1.03 (0.16–6.78) | 0.98 | 0.18 (0.01–3.57) | 0.25 | 0.11 (0.001–12.00) | 0.35 |
| 4 | 0.86 (0.16–4.45) | 0.85 | 1.34 (0.07–26.44) | 0.84 | 1.21 (0.06–22.94) | 0.90 |
| ≥5 | Ref | Ref | Ref | Ref | Ref | Ref |
| House type | ||||||
| Attached | Ref | Ref | Ref | Ref | Ref | Ref |
| Single family | 2.29 (0.54–9.72) | 0.25 | 3.28 (0.50–21.30) | 0.21 | 9.61 (0.53–174.06) | 0.12 |
| Apartment | 1.71 (0.17–17.63) | 0.64 | 0.47 (0.03–8.33) | 0.60 | 0.18 (0.006–5.26) | 0.31 |
| Domestic pets | 1.36 (1.02–1.81) | 0.04* | 1.39 (0.80–2.39) | 0.23 | 1.37 (0.89–2.10) | 0.14 |
| Season | ||||||
| Summer | Ref | Ref | Ref | Ref | Ref | Ref |
| Fall | 3.76 (0.70–20.13) | 0.12 | 6.83 (0.67–69.80) | 0.10 | 14.67 (1.16–184.96) | 0.04* |
| Winter | 2.53 (0.44–14.62) | 0.29 | 0.39 (0.01–24.4) | 0.65 | 0.11 (0.001–8.71) | 0.31 |
| Spring | 0.96 (0.06–16.17) | 0.98 | 11.99 (0.36–395.78) | 0.16 | 35.83 (0.95–1351.37) | 0.05* |
| Unwanted pests | 0.12 (0.01–1.04) | 0.05* | 0.13 (0.01–1.37) | 0.09 | 0.21 (0.01–6.04) | 0.35 |
| No EPA cleaner | 0.32 (0.07–1.38) | 0.12 | 0.03 (0.001–0.75) | 0.03* | 0.01 (0.0002–0.34) | 0.01* |
| Decolonization | 1.12 (0.29–4.32) | 0.86 | 4.70 (0.39–56.63) | 0.21 | 8.26 (0.29–237.35) | 0.21 |
| Any HH Abx use | ||||||
| No use | Ref | Ref | Ref | Ref | Ref | Ref |
| Human or pet use | 1.22 (0.28–5.34) | 0.78 | 0.34 (0.01–7.86) | 0.49 | ||
| Individual Abx use | ||||||
| Clinda | 0.38 (0.08–1.95) | 0.24 | 0.01 (0.0001–0.46) | 0.02* | ||
| Sulfa | 0.79 (0.04–16.01) | 0.88 | 0.15 (0.001–25.07) | 0.45 | ||
| Mup | 1.23 (0.18–8.22) | 0.82 | ||||
Data are for 56 isolates. HH, household; Abx, antimicrobial; Clinda, clindamycin; Sulfa, trimethoprim-sulfamethoxazole; Mup, mupirocin; Ref, reference group.
Model A evaluates any use of antimicrobial drugs by human or domestic animal occupants.
Model B evaluates mupirocin, clindamycin, and trimethoprim-sulfamethoxazole use as individual variables.
ORs for adjusted models used logistic regression and took all listed covariates into account. Rural was not included in the models due to a zero stratum (cf. Table 1).
P values for all covariates were obtained using logistic regression. Significance, indicated in bold, is as follows: *, P ≤ 0.05.
At the 3-month visit, the presence of unwanted pests (cockroaches and rodents) and no Environmental Protection Agency (EPA)-listed cleaners, such as Lysol or Clorox bleach, were significant predictors of non-MDR MRSA at the household level (Tables 1, 2, 5, and 6). In the unadjusted models, each additional pet in the home was associated with a higher risk of contamination with MDR MRSA. Conversely, MRSA isolates from households with pest infestation (cockroaches and rodents) at the 3-month visit were more likely to not be MDR. This was also true for the baseline visit in the unadjusted models, but adjustment for covariates attenuated the effect. In both model A and model B, homes that did not use an EPA-listed cleaner were more likely to have MRSA isolates that were not MDR. Sensitivity analysis that excluded rural homes from the analysis did not strongly impact the direction or the significance of the estimates of an association for home environmental MDR MRSA. Tables 5 and 6 illustrate the results when, instead of an a priori approach, we performed data-driven forward stepwise selection of risk factors for the adjusted logistic regression models at the baseline and 3-month visits, respectively.
TABLE 5.
Adjusted regression model using stepwise selection at the baseline visita
| Outcome variable | Adjusted model Ab |
Adjusted model Bc |
||
|---|---|---|---|---|
| OR (95% CI) | P valued | OR (95% CI) | P valued | |
| Bedroom | 2.21 (0.93–5.26) | 0.07 | 2.07 (0.91–4.73) | 0.08 |
| HH size (no. of people) | ||||
| 1–2 | 5.06 (1.14–22.34) | 0.03* | 4.98 (1.00–24.87) | 0.05* |
| 3 | 1.20 (0.30–4.75) | 0.79 | 0.52 (0.13–2.13) | 0.36 |
| 4 | 7.20 (0.70–73.19) | 0.09 | 2.75 (0.46–16.62) | 0.26 |
| ≥5 | Ref | Ref | Ref | Ref |
| House type | ||||
| Attached | Ref | Ref | ||
| Single family | 1.31 (0.34–5.00) | 0.69 | ||
| Apartment | 6.29 (0.80–49.63) | 0.08 | ||
| Domestic pets | 1.23 (0.97–1.57) | 0.09 | 1.31 (1.08–1.60) | 0.01* |
| Season | ||||
| Summer | Ref | Ref | ||
| Fall | 1.41 (0.25–8.02) | 0.70 | ||
| Winter | 3.28 (0.61–17.62) | 0.16 | ||
| Spring | 1.48 (0.34–6.48) | 0.60 | ||
| HH Abx use | ||||
| No use | Ref | Ref | ||
| Human and pet use | 63.80 (1.50–2,718.77) | 0.03* | ||
| Individual Abx use | ||||
| Clinda | 0.27 (0.09–0.79) | 0.02* | ||
| Sulfa | 2.39 (0.74–7.73) | 0.14 | ||
Data are for 104 isolates. HH, household; Abx, antimicrobial; Clinda, clindamycin; Sulfa, trimethoprim-sulfamethoxazole; Ref, reference group.
Model A evaluates any use of antimicrobial drugs by human or domestic animal occupants.
Model B evaluates mupirocin, clindamycin, and trimethoprim-sulfamethoxazole use as individual variables.
Significance, indicated in bold, is as follows: *, P ≤ 0.05.
TABLE 6.
Adjusted regression model using stepwise selection at the follow-up visita
| Outcome variable | Adjusted model Ab |
Adjusted model Bc |
||
|---|---|---|---|---|
| OR (95% CI) | P valued | OR (95% CI) | P valued | |
| HH size (no. of people) | ||||
| 1–2 | 7.51 (0.87–64.67) | 0.07 | 8.80 (0.82–94.13) | 0.07 |
| 3 | 0.61 (0.07–5.11) | 0.64 | 0.85 (0.10–7.58) | 0.88 |
| 4 | 1.09 (0.08–14.84) | 0.95 | 1.03 (0.06–18.67) | 0.98 |
| ≥5 | Ref | Ref | Ref | Ref |
| Domestic pets | 1.37 (0.90–2.08) | 0.13 | 1.41 (0.88–2.27) | 0.15 |
| Season | ||||
| Summer | Ref | Ref | Ref | Ref |
| Fall | 7.40 (0.90–61.04) | 0.06 | 11.78 (0.92–150.75) | 0.06 |
| Winter | 1.21 (0.07–21.51) | 0.89 | 1.09 (0.04–29.51) | 0.96 |
| Spring | 5.40 (0.31–95.52) | 0.31 | 5.19 (0.31–87.11) | 0.24 |
| Unwanted pests | 0.10 (0.01–1.21) | 0.07 | 0.11 (0.01–1.58) | 0.10 |
| No EPA cleaner | 0.08 (0.01–0.62) | 0.02* | 0.05 (0.004–0.67) | 0.03* |
| HH Clinda use | 0.09 (0.01–0.92) | 0.04* | ||
Data are for 56 isolates. HH, household; Clinda, clindamycin; Ref, reference group.
Model A evaluates any use of antimicrobial drugs by human or domestic animal occupants.
Model B evaluates mupirocin, clindamycin, and trimethoprim-sulfamethoxazole use as individual variables.
Significance, indicated in bold, is as follows: *, P ≤ 0.05.
Sensitivity analysis for antibiotic use.
At the baseline, all homes with pet use of antimicrobials also reported the use of antimicrobial drugs by humans. Therefore, we conducted a sensitivity analysis comparing the 84 (80%) and 12 (12%) samples from homes with human-only antimicrobial use and both human and pet antimicrobial use, respectively, to the 8 samples (8%) from homes where no human or animal antimicrobial use was reported. We identified that samples from homes with human-only antimicrobial use were 68 times more likely to be multidrug resistant (P < 0.04) and that samples from homes with human and pet antimicrobial use were 116 times more likely to be multidrug resistant (P < 0.04) than samples from homes with no antimicrobial use. This suggests that pet antimicrobial use did not detract from and may have contributed to the risk of multidrug resistance in MRSA from the environmental reservoir.
Results of stepwise selection.
In model A, small households (those with 1 to 2 people) were 5 times more likely than the reference group of households with 5 or more people to be contaminated with MDR MRSA, and households with any human or pet use of antimicrobials were over 60 times more likely than households reporting no prior or current antimicrobial use to be contaminated with MDR MRSA. In model B, the presence of domestic pets was associated with a 1.25 times increased odds of home contamination with MDR MRSA, and either pet or human use of clindamycin was associated with protection against home contamination with MDR MRSA. At the 3-month visit, household size, the presence of domestic pets, the presence of unwanted pests (mice, cockroaches, etc.), season, and the use of EPA-listed cleaners were retained in both models. In model A, households that did not use an EPA-listed cleaner were associated with significant protection against home contamination with MDR MRSA. In model B, this was also true, and the use of clindamycin by either pet or human occupants in the household was again associated with protection against home contamination with MDR MRSA.
Mupirocin resistance.
At the baseline visit, 94/104 (90%) MRSA isolates were susceptible to mupirocin, and at the 3-month visit, 50/56 (89%) were susceptible. The household prevalence of mupirocin susceptibility by Etest assessment was 94% for isolates collected at both visits. All (n = 10) Mupr MRSA isolates collected at the baseline home visit were high-level Mupr, as tested by quantitative PCR (qPCR), in which Mupr was mediated through a genetic mechanism (mupA). Among 39 households randomized to mupirocin treatment, 2 (5%) had incident MRSA isolates with low-level Mupr (in which phenotypic resistance was determined by Etest and was found to not be mediated by mupA on the basis of qPCR testing) in the home environment at the 3-month visit; none of the 26 households randomized to the education control group were found to have Mupr MRSA at 3 months.
Risk factors for detection and development of mupirocin resistance.
In addition to associations between mupirocin use and the detection of MDR among MRSA isolates, we also evaluated the MRSA isolates for mupirocin resistance as an outcome due to the clinical and public health importance of this resistance phenotype. Due to small numbers, statistical adjustment was not performed. The presence of domestic pets in the home was associated with a 95% decreased odds of the detection of environmental Mupr MRSA at the baseline (odds ratio [OR], 0.05; 95% CI, 0.01, 0.54; P = 0.01). Domestic pets were not treated with mupirocin as part of the trial, and during interviews regarding pet-specific risk factors at each visit, no owners reported any use of mupirocin by pets prior to or during the study. Prior use of mupirocin by humans in the household before the trial was associated with a 7-fold increased odds of the detection of environmental Mupr MRSA at the baseline, but this did not achieve statistical significance (OR, 7.10; 95% CI, 0.90, 55.7; P = 0.06). Households with mupA-positive MRSA in the environment at the baseline were significantly more likely to have it present in the environment at the 3-month visit (OR, 20.00; 95% CI, 1.00, 403.6; P = 0.05). The home (n = 1) with persistent mupA-positive MRSA environmental contamination was associated with mupA-positive MRSA colonization in people and was associated with the failure of the index patient to successfully clear MRSA colonization during treatment.
DISCUSSION
In this study, we found that 68% (65/95) of homes of patients recently diagnosed with a CO-MRSA SSTI were contaminated with MRSA. The majority, 57% (91/160), of the MRSA strains identified were spa type t008, a dominant community strain in the United States that has been associated with household transmission (20). Among the MRSA-contaminated homes, we evaluated factors associated with multidrug and mupirocin resistance in the home environmental MRSA reservoir in the context of a randomized controlled trial of household-wide decolonization treatment. The majority, 59% (95/160), of the MRSA isolates that we characterized were multidrug resistant.
The literature on risk factors that may contribute to multidrug and/or mupirocin resistance in the environmental CO-MRSA reservoir is limited. One prior study characterized the antimicrobial susceptibility among S. aureus isolates collected from household inhabitants, environmental surfaces, and the pets of children with CO-MRSA, but that study did not evaluate the risk factors associated with the prevalence of MDR or mupirocin resistance in the home environmental MRSA reservoir (21). Although the overall rates of mupirocin resistance were low, our study demonstrated that the household environment can act as a reservoir of mupirocin-resistant MRSA, that the presence of Mupr MRSA in households has the potential to be associated with treatment failure due to recolonization or persistent colonization, and that the use of mupirocin in decolonization treatment has the potential to be associated with incident low-level mupirocin resistance in the home environmental MRSA reservoir.
We found that, prior to randomization and initiation of household-wide decolonization treatment, rural location, small household size, and any prior human or animal use of antimicrobials were associated with an increased odds for home MRSA isolates to be MDR. Small household size was associated with older index patients, who may have more comorbidities that increase their frequency of contact with the health care system and increase their antimicrobial use; however, the rate of prior antimicrobial use was high among both large and small households. The prior use of antimicrobial drugs has been associated with an increased risk for MRSA and drug resistance in previous studies of people (22, 23). After households were randomized to the use of twice-daily nasal mupirocin and two chlorhexidine body washes for all people in the home, two consistent protective effects emerged. MRSA-contaminated households that did not report the use of a cleaner on the EPA list of agents with known effective biocidal activity against MRSA and households reporting the use of clindamycin by at least one human or animal were associated with the presence of more susceptible (not MDR) MRSA isolates. Therefore, this study is the first, to our knowledge, to report that the use of nonbiocidal cleaning products and that the use of clindamycin in either humans or domestic animals is not associated with a risk of MDR in the home environmental MRSA reservoir. It is possible that cleaners with biocidal activity against MRSA exert selective pressure, contributing to expansion of the MDR MRSA reservoir if strains carry genes for disinfectant resistance, such as qacA (24). Prior studies have shown that certain disinfectants are less effective against biofilm-producing S. aureus strains; we did not test our environmental strains for biofilm production (25, 26). Further research needs to be done to replicate and elucidate the mechanism of this effect.
We identified that 68% MRSA isolates from the home environment were susceptible to clindamycin, which is consistent with the 66% rate of clindamycin susceptibility observed among colonizing MRSA isolates collected from people in a population representative of that of the United States from 2012 to 2014 (27). Morelli et al. determined that 90% of environmental S. aureus isolates were susceptible to clindamycin but did not report the distribution of susceptibility among MRSA versus methicillin-susceptible S. aureus (MSSA) isolates, precluding a direct comparison (21). The finding of a protective role for clindamycin use by humans or domestic animals in the household is consistent with a prior report of a study with this cohort that the use of clindamycin is associated with both the earlier clearance and the lower persistence of colonization among the index patients (28, 29). It is possible that these effects could be mediated, at least in part, by the home environmental MRSA reservoir. All clindamycin-resistant environmental MRSA isolates were also MDR, and at the 3-month visit, none of the homes reporting the use of clindamycin were contaminated with clindamycin-resistant MRSA isolates. It is also possible that environmental effects could be related to changes in carriage among human household members, although this is less likely, given that our analysis was limited to the MRSA environmental reservoir and excluded homes with no MRSA contamination. In contrast, the use of trimethoprim-sulfamethoxazole was not associated with either protection or an increased odds of detection of MDR strains in the home environmental MRSA reservoir. This suggests that although clindamycin and trimethoprim-sulfamethoxazole drugs have been found to be equivalently effective for the treatment of community MRSA SSTIs, they may exert different effects in terms of selective pressure (30–32).
Living in a rural household predicted MDR completely at the baseline visit. The risk of MDR because of rural residence is a novel finding, as rural residence has been considered a risk factor for human MRSA colonization, but rural households have not previously been considered a potential environmental reservoir for AMR (29). This finding may be due to multiple factors. In our study population, 42% of homes classified as rural reported being able to see or smell a farm. The agricultural use of antimicrobials is a potential source of selection pressure in rural communities that may contribute to an increased prevalence of MDR MRSA through both direct (occupational) and indirect (environmental) routes (33, 34). A difference in prescribing practices in rural areas was also observed in our study population. These practices may contribute to the differences in MDR patterns among MRSA isolates from rural homes. We observed that people and domestic pets in rural homes were more likely to report trimethoprim-sulfamethoxazole than clindamycin use than people and domestic pets in nonrural homes. However, the exclusion of rural homes from our analysis did not strongly impact the direction or significance of estimates of an association of clindamycin and trimethoprim-sulfamethoxazole use with the detection of home environmental MDR MRSA.
We found interesting effects according to the presence of domestic pets and unwanted pests. The presence of domestic pets was associated with an increased odds for the presence of MDR MRSA in the home environment, while the presence of unwanted pests was associated with protection against MDR MRSA strains at the 3-month visit, following randomization to the decolonization intervention in people. Domestic pets are known to be a potential reservoir for CO-MRSA, although the prevalence of MRSA carriage in pets in this study was low (35). While the adjusted models accounted for pet antimicrobial use, the models did not capture all other risk factors that could contribute to selection for drug-resistant strains, such as contact with veterinary health care settings. In contrast, unwanted pests are not a direct target for antimicrobial treatment. It is possible that more susceptible strains may influence the environmental reservoir and dilute the pool of MDR MRSA isolates; this effect could be magnified as humans undergo decolonization treatment (reducing their shedding into the home environmental MRSA reservoir). This hypothesis could also explain the finding that domestic pets were associated with protection against contamination of the home environmental MRSA reservoir with mupirocin-resistant strains. No owners reported that their pets had been treated with mupirocin in the year prior to or during the conduct of the study. Of concern, it is possible that if mupirocin use becomes more common in veterinary practice, this potential effect will diminish.
This study has several limitations. Due to a small sample size, the study may not have had a sufficient power to determine associations for some risk factors with a modest effect difference. In addition, only MRSA isolates were included in this analysis, which may have biased our assessment of the household resistome. However, the home environmental MRSA reservoir is clinically relevant, particularly in the context of consideration of the household unit as part of a therapeutic intervention. Although we evaluated only two MRSA isolates per household and selected isolates that were more likely to be resistant to methicillin, no households were misclassified as MRSA negative and our selection process was systematic. Finally, we did not evaluate biofilm formation or perform universal testing for disinfectant resistance phenotypes and genotypes as part of this analysis; these are targets for future research. Our study was strengthened by the inclusion of an inspection-based assessment of the household, in addition to the incorporation of pet-related antimicrobial use.
In conclusion, our study demonstrates a need to consider the home environmental MRSA reservoir to help prevent recurrent, multidrug-resistant MRSA infections in the community. The potential for mupirocin decolonization treatment to select for mupirocin-resistant strains that may enter this reservoir deserves further scrutiny. Future studies should also attempt to replicate the novel potential risk factors identified here, specifically, rural location and the use in the home of biocides that are known to have effectiveness against MRSA. Antimicrobial-resistant bacteria present a growing threat to public health nationally and globally, and it is increasingly important to consider the household environment as an important location for interventions.
MATERIALS AND METHODS
Household recruitment and questionnaire. (i) Study design.
Study participants were recruited as part of a randomized-controlled trial (RCT; ClinicalTrials.gov registration no. NCT00966446) at one of five participating institutions in the mid-Atlantic United States, which included two urban adult care hospitals, an adult community hospital, an urban children's hospital, and a rural adult and pediatric hospital. The conduct and results of the RCT have been described previously, and the main goal of this trial was to determine the impact of household-wide decolonization treatment on recurrent MRSA infection (28). Briefly, outpatients were recruited between January and December 2012 and were included on the basis of a laboratory-confirmed MRSA skin or soft tissue infection (SSTI). To be included in the study, a study subject (i.e., index patient), including adults and children, and all members of his or her household were required to agree to participate. Informed consent or assent was obtained from all index patients and household members. As part of the RCT protocol, all human household members were cluster randomized to a 1-week decolonization treatment (two arms) or an education control group (one arm). Participants in households randomized to treatment were assigned twice-daily nasal mupirocin for 7 days and a chlorhexidine body wash on the first and last day of mupirocin application; the treatment week was scheduled to occur 6 weeks after the baseline visit.
This analysis was limited to participants who consented to home environmental sampling for the Pets and Environmental Transmission of Staphylococci (PETS) study, parts of which have been described previously (19, 36, 37). Participants were administered verbal questionnaires at each visit. Trained personnel used an iFormBuilder (iFormBuilder, Herndon, VA) application for iPad (Apple, Cupertino, CA) to collect data by interview and inspection regarding household-related and pet-related characteristics. Study personnel also collected data on participant characteristics by interview and diary.
(ii) Rural versus urban classification.
A priori, all counties included in the catchment area for the RCT and the PETS study were determined to fall into an urban classification on the basis of the 2010 census and the Office of Management and Budget definition (38). As a result, study staff assigned households to a subjective category on the basis of the characteristics of the neighborhood and surrounding community. To capture potential indirect contact with livestock, study staff recorded the proximity of the house to livestock and crop agricultural areas and queried the heads of households regarding odors and other indications of agricultural influence.
(iii) Home inspection.
Study staff performed home inspections for unwanted pests (primarily flies, mice, and cockroaches) and other household characteristics at each of the home visits. Domestic pets were identified and sampled as previously described (19).
This study was approved by the institutional review boards and the animal care and use committees of the participating institutions (University of Pennsylvania, Johns Hopkins University).
(iv) Household sampling.
Autoclave-sterilized electrostatic cloths (Swiffer; Proctor & Gamble) were used to collect household surface dust samples for bacterial culture. Samples were collected from eight standardized locations in the common room, kitchen, and bedroom of each household, although the participants could decline sampling of the bedroom (two of the eight samples). Electrostatic cloths were used to sample typically a 30- by 30-cm area in the various rooms, as previously described (39). The samples were then placed in sterile stomacher bags for transport to the laboratory.
Bacterial culture.
Samples were subjected to a two-arm culture method. Arm 1 was optimized for the isolation of coagulase-positive methicillin-susceptible (MS) Staphylococcus spp., and arm 2 was optimized for the isolation for methicillin-resistant (MR) organisms as previously described (39, 40).
Bacterial culture method.
The electrostatic cloths were cultured as previously described (39, 40). Briefly, the cloths were enriched in 60 ml Mueller-Hinton broth supplemented with 6.5% NaCl, and then (for the MR enrichment arm only) 1 ml was subcultured to 9 ml tryptic soy broth supplemented with 2.5% NaCl, 3.5 mg/liter cefoxitin, and 10 mg/liter aztreonam. The broths were incubated at 37°C for 16 to 20 h, and then a 10-μl aliquot was plated onto BBL Columbia CNA blood agar; the plates were incubated at 37°C for 16 to 20 h. After incubation, presumptive staphylococcal colonies on CNA agar were subcultured to Baird-Parker (BP) agar. All probable coagulase-positive staphylococci (CPS) (based on the phenotype on BP agar) were archived to Microbank tubes (Pro-Lab Diagnostics, Canada) and held at −80°C. S. aureus ATCC 43300, S. pseudintermedius ATCC 49444, and S. schleiferi VHUP1939-05 were used as positive controls for culture and PCR.
Selection and molecular characterization of isolates.
Up to two isolates from each home visit were selected on the basis of the phenotype on blood agar by one member of the study team (M.F.D.) for species identification by PCR and antimicrobial susceptibility testing. Hemolytic, yellow-pigmented colonies (presumptive S. aureus isolates) from the arm of the protocol that was selective for methicillin resistance were chosen over nonhemolytic, nonpigmented colonies from the arm of the protocol that was nonselective for resistance, identical to a selection process used for animal isolates analyzed in the PETS study (19). To identify S. aureus, S. pseudintermedius, or S. schleiferi, a multiplex PCR assay that amplifies species-specific segments of the nuclease gene (nuc) was performed as previously described (41). Methicillin-resistant isolates (MRSA and MR S. pseudintermedius isolates) were determined by the presence of a universal mecA-mecC sequence, with ATCC 43300 and LGA251 used as mecA- and mecC-positive controls, respectively (42). Isolates confirmed to be MRSA were subjected to S. aureus protein A (spa) typing as previously described (43, 44).
Subset analysis.
To determine whether selection of only two isolates contributed to bias in assessment of the home MRSA status, all CPS isolates from the first 20 homes recruited from the urban enrollment centers and the first 5 homes recruited from the rural enrollment center were tested by PCR.
Antimicrobial susceptibility testing.
Testing for susceptibility to 10 antimicrobials (chloramphenicol, vancomycin, quinupristin-dalfopristin [Synercid], linezolid, tetracycline, gentamicin, trimethoprim-sulfamethoxazole, clindamycin, ciprofloxacin, and erythromycin) and erythromycin-induced resistance to clindamycin (D-test) was conducted for selected isolates prior to cryopreservation using disk diffusion methods following CLSI guidelines (45). The Sentry definition of nonsusceptibility to four or more classes of antimicrobials (resistance to methicillin by design plus resistance to antimicrobials in three additional classes) was used to define multidrug resistance (10). Mupirocin susceptibility was evaluated following cryopreservation using Etest (bioMérieux, France) and established real-time PCR methods (46).
Statistical analysis.
Statistical analysis was restricted to the MRSA isolates from each household. Data collected during home visits, which included the baseline visit and a 3-month follow-up visit, were analyzed using Stata (version 14) software (StataCorp, College Station, TX). Unadjusted and adjusted logistic regression to generate odds ratios (ORs) and adjusted ORs (aORs), respectively, was performed to estimate associations between the antimicrobial resistance of MRSA isolates and antimicrobial use and other household risk factors, including sample location, home location, household size, the presence of domestic pets, the presence of evidence of unwanted pests, season, and reported use of disinfectants. Survey-weighting techniques (SVY commands in Stata) were used to account for the clustering of multiple isolates within a household. To assess overall household antimicrobial use, model A represented models in which a variable for any use of antimicrobial drugs by human or domestic animal occupants was employed; model B was stratified instead on the basis of individual antimicrobial use and evaluated the target drug mupirocin and the two antimicrobials commonly used for the treatment of MRSA infection in humans: clindamycin and trimethoprim-sulfamethoxazole (30–32). These three variables were considered independently, and households could report the use of any combination. For the baseline visit, the antimicrobial drugs used by humans and domestic animals in the prior year were considered; for the second visit, only those used during the 3-month interval between the visits were considered.
The sites sampled were categorized into touched or repository sites. Touched sites were selected as surfaces more commonly touched in the household and included the handle of the refrigerator, the kitchen towel, the television remote, the bathroom faucet, and the bedroom pillow of the index participant. Repository sites were selected as areas that a patient was not likely to touch on a daily basis and included the top of the refrigerator, the top of the television, and the top of a wardrobe or other dusty surface in the bedroom. Pest variables were aggregated from self-reported data and home inspections. Season was defined as winter, spring, summer, and fall using cutoffs for the end date of each season of 20 March, 21 June, 22 September, and 21 December, respectively. Via interviews, the participants provided the names of all disinfectants typically used. These names were checked against Environmental Protection Agency (EPA)-registered products considered to be effective against MRSA and vancomycin-resistant Enterococcus faecalis or E. faecium (vancomycin-resistant enterococci [VRE]) (EPA, list H), and cleaners matching those on this list were considered EPA-listed household cleaners, e.g., Lysol and Clorox bleach. All EPA-listed cleaners were grouped into whether they were used in the household or not. For categorical variables, reference groups were assigned as the largest stratum.
Stepwise selection.
Data-driven forward stepwise selection of risk factors for the adjusted logistic regression models at the baseline and 3-month visits was performed. In the baseline models, bedroom site, household size, the presence of domestic pets, and household antimicrobial use were retained in both model A and model B.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the study participants and to study personnel and students, particularly Julie Vallati, Amy Shelly, Jacqueleen Wise, Robyn Smith, Grace Ndicu, John Ndicu, Aimee Vasse, Danielle Searson, Elana Youssef, Haley Keller, and Krista Reynolds. John Groopman, David Sack, and Ellen Silbergeld provided laboratory and other resources. We thank Jesper Larsen for assistance with spa typing.
This research was supported by the Commonwealth Universal Research Enhancement (CURE) Program of the Pennsylvania Department of Health (to E.L.), the Johns Hopkins Center for a Livable Future (to M.F.D.), a Johns Hopkins Faculty Innovation grant (to M.F.D.), the Morris Animal Foundation (to M.F.D.), and the American College of Veterinary Dermatology/American Academy of Veterinary Dermatology (to D.O.M.). Investigators were supported by an NIAID K24 grant (AI080942 to E.L.), a postdoctoral fellowship on a NIEHS T32 grant (ES7141-29 to M.F.D.), and an ORIP K01 grant (K01OD019918 to M.F.D.). This research has been funded by a CDC cooperative agreement (FOA#CK11-001, Epicenters for the Prevention of Healthcare Associated Infections).
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01369-17.
REFERENCES
- 1.Low DE, Keller N, Barth A, Jones RN. 2001. Clinical prevalence, antimicrobial susceptibility, and geographic resistance patterns of enterococci: results from the Sentry Antimicrobial Surveillance Program, 1997-1999. Clin Infect Dis 32(Suppl 2):133–145. [DOI] [PubMed] [Google Scholar]
- 2.Salgado CD, Farr BM, Calfee DP. 2003. Community-acquired methicillin-resistant Staphylococcus aureus: a meta-analysis of prevalence and risk factors. Clin Infect Dis 36:131–139. doi: 10.1086/345436. [DOI] [PubMed] [Google Scholar]
- 3.Boyce JM, Cookson B, Christiansen K, Hori S, Vuopio-Varkila J, Kocagöz S, Öztop AY, Vandenbroucke-Grauls CM, Harbarth S, Pittet D. 2005. Meticillin-resistant Staphylococcus aureus. Lancet Infect Dis 5:653–663. doi: 10.1016/S1473-3099(05)70243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David MZ, Daum RS. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones RN. 1996. The emergent needs for basic research, education, and surveillance of antimicrobial resistance. Problems facing the report from the American Society for Microbiology Task Force on Antibiotic Resistance. Diagn Microbiol Infect Dis 25:153–161. [DOI] [PubMed] [Google Scholar]
- 6.Schwarz S, Enne VI, van Duijkeren E. 2016. 40 years of veterinary papers in JAC—what have we learnt? J Antimicrob Chemother 71:2681–2690. doi: 10.1093/jac/dkw363. [DOI] [PubMed] [Google Scholar]
- 7.Huijbers PMC, Blaak H, de Jong MCM, Graat EAM, Vandenbroucke-Grauls CMJE, de Roda Husman AM. 2015. Role of the environment in the transmission of antimicrobial resistance to humans: a review. Environ Sci Technol 49:11993–12004. doi: 10.1021/acs.est.5b02566. [DOI] [PubMed] [Google Scholar]
- 8.Davis MF, Iverson SA, Baron P, Vasse A, Silbergeld EK, Lautenbach E, Morris DO. 2012. Household transmission of meticillin-resistant Staphylococcus aureus and other staphylococci. Lancet Infect Dis 12:703–716. doi: 10.1016/S1473-3099(12)70156-1. [DOI] [PubMed] [Google Scholar]
- 9.Wang YC, Coxson P, Shen Y-M, Goldman L, Bibbins-Domingo K. 2012. A penny-per-ounce tax on sugar-sweetened beverages would cut health and cost burdens of diabetes. Health Aff (Millwood) 31:199–207. doi: 10.1377/hlthaff.2011.0410. [DOI] [PubMed] [Google Scholar]
- 10.Moet GJ, Jones RN, Biedenbach DJ, Stilwell MG, Fritsche TR. 2007. Contemporary causes of skin and soft tissue infections in North America, Latin America, and Europe: report from the SENTRY Antimicrobial Surveillance Program (1998-2004). Diagn Microbiol Infect Dis 57:7–13. doi: 10.1016/j.diagmicrobio.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Bramble M, Morris D, Tolomeo P, Lautenbach E. 2011. Potential role of pet animals in household transmission of methicillin-resistant Staphylococcus aureus: a narrative review. Vector Borne Zoonotic Dis 11:617–620. doi: 10.1089/vbz.2010.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faires MC, Tater KC, Weese JS. 2009. An investigation of methicillin-resistant Staphylococcus aureus colonization in people and pets in the same household with an infected person or infected pet. J Am Vet Med Assoc 235:540–543. doi: 10.2460/javma.235.5.540. [DOI] [PubMed] [Google Scholar]
- 13.Ferreira JP, Fowler VG, Correa MT, Lyman R, Ruffin F, Anderson KL. 2011. Transmission of methicillin-resistant Staphylococcus aureus between human and hamster. J Clin Microbiol 49:1679–1680. doi: 10.1128/JCM.02469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Septimus EJ, Schweizer ML. 2016. Decolonization in prevention of health care-associated infections. Clin Microbiol Rev 29:201–222. doi: 10.1128/CMR.00049-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel JB, Gorwitz RJ, Jernigan JA. 2009. Mupirocin resistance. Clin Infect Dis 49:935–941. doi: 10.1086/605495. [DOI] [PubMed] [Google Scholar]
- 16.Bathoorn E, Hetem DJ, Alphenaar J, Kusters JG, Bonten MJM. 2012. Emergence of high-level mupirocin resistance in coagulase-negative staphylococci associated with increased short-term mupirocin use. J Clin Microbiol 50:2947–2950. doi: 10.1128/JCM.00302-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shahbazian JH, Torrie A, Ferguson J, Baron P, Julian K, Nachamkin I, Rankin SC, Morris DO, Lautenbach E, Davis MF. 2015. Multidrug resistance in environmental methicillin-resistant Staphylococcus aureus (MRSA) collected from the homes of people diagnosed with a community-onset (CO-) MRSA infection, abstr 60 In Abstr 4th ASM Conf Antimicrobial Resistance in Zoonotic Bacteria and Foodborne Pathogens. American Society for Microbiology, Washington, DC. [Google Scholar]
- 18.Hahn PD, Shahbazian JH, Spicer K, Christ A, Ludwig S, Tolomeo P, Cluzet VC, Nachamkin I, Rankin SC, Morris DO, Lautenbach E, Davis MF. 2015. Mupirocin susceptibility in environmental methicillin-resistant Staphylococcus aureus from homes with and without pets in the context of a mupirocin-based randomized clinical trial, abstr 22 In Abstr 4th ASM-ESCMID Conf Methicillin-Resistant Staphylococci Anim Vet Public Health Implications. American Society for Microbiology, Washington, DC. [Google Scholar]
- 19.Iverson SA, Brazil AM, Ferguson JM, Nelson K, Lautenbach E, Rankin SC, Morris DO, Davis MF. 2015. Anatomical patterns of colonization of pets with staphylococcal species in homes of people with methicillin-resistant Staphylococcus aureus (MRSA) skin or soft tissue infection (SSTI). Vet Microbiol 176:202–208. doi: 10.1016/j.vetmic.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Miller LG, Eells SJ, Taylor AR, David MZ, Ortiz N, Zychowski D, Kumar N, Cruz D, Boyle Vavra S, Daum RS. 2012. Staphylococcus aureus colonization among household contacts of patients with skin infections: risk factors, strain discordance, and complex ecology. Clin Infect Dis 54:1523–1535. doi: 10.1093/cid/cis213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morelli JJ, Hogan PG, Sullivan ML, Muenks CE, Wang JW, Thompson RM, Burnham C-AD, Fritz SA. 2015. Antimicrobial susceptibility profiles of Staphylococcus aureus isolates recovered from humans, environmental surfaces, and companion animals in households of children with community-onset methicillin-resistant S. aureus infections. Antimicrob Agents Chemother 59:6634–6637. doi: 10.1128/AAC.01492-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgenstern M, Erichsen C, Hackl S, Mily J, Militz M, Friederichs J, Hungerer S, Bühren V, Moriarty TF, Post V, Richards RG, Kates SL. 2016. Antibiotic resistance of commensal Staphylococcus aureus and coagulase-negative staphylococci in an international cohort of surgeons: a prospective point-prevalence study. PLoS One 11:e0148437. doi: 10.1371/journal.pone.0148437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Bijnen EME, Paget J, de Lange-de Klerk ESM, den Heijer CDJ, Versporten A, Stobberingh EE, Goossens H, Schellevis FG, collaboration with the APRES Study Team . 2015. Antibiotic exposure and other risk factors for antimicrobial resistance in nasal commensal Staphylococcus aureus: an ecological study in 8 European countries. PLoS One 10:e0135094. doi: 10.1371/journal.pone.0135094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wassenaar T, Ussery D, Nielsen L, Ingmer H. 2015. Review and phylogenetic analysis of qac genes that reduce susceptibility to quaternary ammonium compounds in Staphylococcus species. Eur J Microbiol Immunol 5:44–61. doi: 10.1556/EuJMI-D-14-00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adukwu EC, Allen SC, Phillips CA. 2015. A comparison of the sensitivity of four Staphylococcus aureus isolates to two chlorine-based disinfectants and an eco-friendly commercially available cleaning agent. Int J Environ Health Res 25:115–125. doi: 10.1080/09603123.2014.903905. [DOI] [PubMed] [Google Scholar]
- 26.Almatroudi A, Gosbell IB, Hu H, Jensen SO, Espedido BA, Tahir S, Glasbey TO, Legge P, Whiteley G, Deva A, Vickery K. 2016. Staphylococcus aureus dry-surface biofilms are not killed by sodium hypochlorite: implications for infection control. J Hosp Infect 93:263–270. doi: 10.1016/j.jhin.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 27.Sader HS, Mendes RE, Jones RN, Flamm RK. 2016. Antimicrobial susceptibility patterns of community- and hospital-acquired methicillin-resistant Staphylococcus aureus from United States hospitals: results from the AWARE Ceftaroline Surveillance Program (2012-2014). Diagn Microbiol Infect Dis 86:76–79. doi: 10.1016/j.diagmicrobio.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 28.Cluzet VC, Gerber JS, Nachamkin I, Metlay JP, Zaoutis TE, Davis MF, Julian KG, Royer D, Linkin DR, Coffin SE, Margolis DJ, Hollander JE, Mistry RD, Gavin LJ, Tolomeo P, Wise JA, Wheeler MK, Bilker WB, Han X, Hu B, Fishman NO, Lautenbach E. 2015. Duration of colonization and determinants of earlier clearance of colonization with methicillin-resistant Staphylococcus aureus. Clin Infect Dis 60:1489–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cluzet VC, Gerber JS, Nachamkin I, Coffin SE, Davis MF, Julian KG, Zaoutis TE, Metlay JP, Linkin DR, Tolomeo P, Wise JA, Bilker WB, Hu B, Lautenbach E, CDC Prevention Epicenters Program . 2017. Factors associated with persistent colonisation with methicillin-resistant Staphylococcus aureus. Epidemiol Infect 145:1409–1417. doi: 10.1017/S0950268817000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller LG, Daum RS, Creech CB, Young D, Downing MD, Eells SJ, Pettibone S, Hoagland RJ, Chambers HF, DMID 07-0051 Team. 2015. Clindamycin versus trimethoprim-sulfamethoxazole for uncomplicated skin infections. N Engl J Med 372:1093–1103. doi: 10.1056/NEJMoa1403789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frei CR, Miller ML, Lewis JS, Lawson KA, Hunter JM, Oramasionwu CU, Talbert RL. 2010. Trimethoprim-sulfamethoxazole or clindamycin for community-associated MRSA (CA-MRSA) skin infections. J Am Board Fam Med 23:714–719. doi: 10.3122/jabfm.2010.06.090270. [DOI] [PubMed] [Google Scholar]
- 32.Holmes L, Ma C, Qiao H, Drabik C, Hurley C, Jones D, Judkiewicz S, Faden H. 2016. Trimethoprim-sulfamethoxazole therapy reduces failure and recurrence in methicillin-resistant Staphylococcus aureus skin abscesses after surgical drainage. J Pediatr 169:128–134.e1. doi: 10.1016/j.jpeds.2015.10.044. [DOI] [PubMed] [Google Scholar]
- 33.Love DC, Davis MF, Bassett A, Gunther A, Nachman KE. 2011. Dose imprecision and resistance: free-choice medicated feeds in industrial food animal production in the United States. Environ Health Perspect 119:279–283. doi: 10.1289/ehp.1002625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Köck R, Werner P, Friedrich AW, Fegeler C, Becker K, Prevalence of Multiresistant Microorganisms (PMM) Study Group . 2016. Persistence of nasal colonization with human pathogenic bacteria and associated antimicrobial resistance in the German general population. New Microbes New Infect 9:24–34. doi: 10.1016/j.nmni.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loeffler A, Lloyd DH. 2010. Companion animals: a reservoir for methicillin-resistant Staphylococcus aureus in the community? Epidemiol Infect 138:595–605. doi: 10.1017/S0950268809991476. [DOI] [PubMed] [Google Scholar]
- 36.Misic AM, Davis MF, Tyldsley AS, Hodkinson BP, Tolomeo P, Hu B, Nachamkin I, Lautenbach E, Morris DO, Grice EA. 2015. The shared microbiota of humans and companion animals as evaluated from Staphylococcus carriage sites. Microbiome 3:2. doi: 10.1186/s40168-014-0052-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis MF, Misic AM, Morris DO, Moss JT, Tolomeo P, Beiting DP, Nachamkin I, Lautenbach E, Rankin SC. 2015. Genome sequencing reveals strain dynamics of methicillin-resistant Staphylococcus aureus in the same household in the context of clinical disease in a person and a dog. Vet Microbiol 180:304–307. doi: 10.1016/j.vetmic.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cromartie J, Parker T. What is rural? Economic Research Service, U.S. Department of Agriculture, Washington, DC: https://www.ers.usda.gov/topics/rural-economy-population/rural-classifications/what-is-rural.aspx. [Google Scholar]
- 39.Davis MF, Baron P, Price LB, Williams DL, Jeyaseelan S, Hambleton IR, Diette GB, Breysse PN, McCormack MC. 2012. Dry collection and culture methods for recovery of methicillin-susceptible and methicillin-resistant Staphylococcus aureus strains from indoor home environments. Appl Environ Microbiol 78:2474–2476. doi: 10.1128/AEM.06886-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis MF, Hu B, Carroll KC, Bilker WB, Tolomeo P, Cluzet VC, Baron P, Ferguson JM, Morris DO, Rankin SC, Lautenbach E, Nachamkin I. 2016. Comparison of culture-based methods for identification of colonization with methicillin-resistant and methicillin-susceptible Staphylococcus aureus in the context of cocolonization. J Clin Microbiol 54:1907–1911. doi: 10.1128/JCM.00132-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sasaki T, Tsubakishita S, Tanaka Y, Sakusabe A, Ohtsuka M, Hirotaki S, Kawakami T, Fukata T, Hiramatsu K. 2010. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J Clin Microbiol 48:765–769. doi: 10.1128/JCM.01232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alvarez CA, Yomayusa N, Leal AL, Moreno J, Mendez-Alvarez S, Ibañez M, Vanegas N. 2010. Nosocomial infections caused by community-associated methicillin-resistant Staphylococcus aureus in Colombia. Am J Infect Control 38:315–318. doi: 10.1016/j.ajic.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 43.Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, Bost DA, Riehman M, Naidich S, Kreiswirth BN. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol 37:3556–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mellmann A, Weniger T, Berssenbrügge C, Rothgänger J, Sammeth M, Stoye J, Harmsen D. 2007. Based upon repeat pattern (BURP): an algorithm to characterize the long-term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC Microbiol 7:98. doi: 10.1186/1471-2180-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clinical and Laboratory Standards Institute. 2010. 2009 S. aureus CLSI breakpoint guide. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 46.McGann P, Milillo M, Kwak YI, Quintero R, Waterman PE, Lesho E. 2013. Rapid and simultaneous detection of the chlorhexidine and mupirocin resistance genes qacA/B and mupA in clinical isolates of methicillin-resistant Staphylococcus aureus. Diagn Microbiol Infect Dis 77:270–272. doi: 10.1016/j.diagmicrobio.2013.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.