ABSTRACT
Glyphosate is the most widely used herbicide worldwide and a critical tool for weed control in no-till cropping systems. However, there are concerns about the nontarget impacts of long-term glyphosate use on soil microbial communities. We investigated the impacts of repeated glyphosate treatments on bacterial communities in the soil and rhizosphere of wheat in soils with and without long-term history of glyphosate use. We cycled wheat in the greenhouse using soils from 4 paired fields under no-till (20+-year history of glyphosate) or no history of use. At each cycle, we terminated plants with glyphosate (2× the field rate) or by removing the crowns, and soil and rhizosphere bacterial communities were characterized. Location, cropping history, year, and proximity to the roots had much stronger effects on bacterial communities than did glyphosate, which only explained 2 to 5% of the variation. Less than 1% of all taxa were impacted by glyphosate, more in soils with a long history of use, and more increased than decreased in relative abundance. Glyphosate had minimal impacts on soil and rhizosphere bacteria of wheat, although dying roots after glyphosate application may provide a “greenbridge” favoring some copiotrophic taxa.
IMPORTANCE Glyphosate (Roundup) is the most widely used herbicide in the world and the foundation of Roundup Ready soybeans, corn, and the no-till cropping system. However, there have been recent concerns about nontarget impacts of glyphosate on soil microbes. Using next-generation sequencing methods and glyphosate treatments of wheat plants, we described the bacterial communities in the soil and rhizosphere of wheat grown in Pacific Northwest soils across multiple years, different locations, and soils with different histories of glyphosate use. The effects of glyphosate were subtle and much less than those of drivers such as location and cropping systems. Only a small percentage of the bacterial groups were influenced by glyphosate, and most of those were stimulated, probably because of the dying roots. This study provides important information for the future of this important tool for no-till systems and the environmental benefits of reducing soil erosion and fossil fuel inputs.
KEYWORDS: glyphosate, Roundup, soil microbiome, rhizosphere, tillage, wheat, rhizosphere-inhabiting microbes
INTRODUCTION
Glyphosate [N-(phosphonomethyl)glycine] is the most widely used herbicide worldwide, with 6.1 billion kg applied in the last decade (1). The popularity of glyphosate is due to its effectiveness at low concentrations (∼0.84 kg acid equivalents/ha), low mammalian toxicity, short environmental half-life (∼3 to 90 days, 44 to 60 days on average), and low activity/mobility in soil, making it one of the most environmentally friendly herbicides available (2, 3). Glyphosate is considered an essential tool for weed management in no-till cropping systems throughout the United States, which are key for reducing soil erosion and fuel use and increasing soil quality, organic matter, and water retention (4–7). However, despite the minimal environmental impact of glyphosate and its role in sustainable cropping systems, there are persistent concerns about potential nontarget effects of glyphosate on plant-beneficial microbial communities (3, 8–11).
Glyphosate specifically acts on the enzyme 5-enolpyruvylshikimic acid-3-phosphate synthase (EPSPS; EC 2.5.1.19) and inhibits the production of aromatic amino acids, ultimately resulting in plant death. While many bacteria and fungi also contain glyphosate-sensitive EPSPS enzymes and are inhibited by glyphosate (12), others are able to metabolize it, and microbial activity is the primary route of glyphosate degradation. Two pathways of glyphosate catabolism by bacteria have been characterized, which are widespread across bacterial lineages (13–15). However, although glyphosate metabolizers are found commonly in soils, even those with no history of glyphosate use, relatively few taxa are known to metabolize glyphosate as a source of P, C, or N, and many may be nonculturable (13, 16, 17). Because some taxa are glyphosate sensitive, whereas others are insensitive or may benefit from it as a nutrient source, glyphosate is likely to have negative, neutral, or positive effects on soil microbial populations and to modify the microbiomes of crop plants. Consequently, it is not surprising that studies on the effects of glyphosate on microbial populations have yielded conflicting evidence, suggesting detrimental effects (9–11, 18–20), minimal or no effects (21–27), or stimulatory effects (23, 28–31) on bacterial abundance, activity, and diversity.
The inconsistent findings regarding glyphosate effects on soil microbiota may be due to complex interactions between glyphosate and soil characteristics (2). Soil minerology and chemistry influence the binding of glyphosate to soil particles and its leaching and accessibility to microorganisms. Although glyphosate binds tightly to Fe- and Al-oxides, sorption decreases at high pH. Furthermore, P competes with glyphosate for binding sites, which may be blocked by soil organic matter, suppressing glyphosate binding (2). However, even sorbed glyphosate might be available to microbial degradation (32). Additionally, it has been suggested that microbial populations in soils with a history of glyphosate exposure have adapted and can better tolerate (i.e., are less responsive) or more rapidly break down (i.e., are more responsive to) the herbicide (24, 25, 29, 33).
Plant proximity also may influence the effects of glyphosate on microbial communities. Since plants select specific rhizosphere microbial consortia via root exudates (34), they may recruit a consistent suite of rhizobacteria regardless of small changes in soil communities due to glyphosate. On the contrary, because unbound glyphosate residues or degradation products (e.g., aminomethylphosphonic acid [AMPA]) may alter plant growth, defense, or root exudation (9, 35, 36), the buildup of residues in soil may impact the quantity or composition of resources available in the rhizosphere, with subsequent impacts on rhizosphere communities. However, the likelihood of detecting subtle changes in microbiome composition has been limited by low-resolution techniques, and the potential impacts of glyphosate on soil and plant-associated microbial communities remain a controversial issue of great concern to growers and the general public alike.
To address these concerns, we investigated the following questions: (i) do repeated applications of glyphosate impact the composition or diversity of soil and rhizosphere bacteria? (ii) Does the response of soil bacterial communities to glyphosate application depend on the history of glyphosate use in the field? (iii) Does the presence of a host plant influence the effects of glyphosate on soil and the plant-associated bacterial community?
RESULTS
In this study, we sampled four farms distributed across a wide precipitation gradient in the spring of 2 years. At each farm, we sampled locations with and without a history of glyphosate application, and there were four replicates taken from each location. Each sample was split into two pots and seeded with wheat. One pot was treated with glyphosate, and the other was not. Wheat was grown for 4 growth cycles, and bulk soil and rhizosphere samples were taken at the end of the first and fourth cycles, corresponding to three rounds of termination with glyphosate. We obtained 9,065,781 sequences binned into 19,686 operational taxonomic units (OTUs) at 97% similarity (2,946,020 sequences in 18,787 OTUs after rarefaction; see Fig. S1 in the supplemental material). Bacterial communities tended to be dominated by Proteobacteria (33.3% ± 9.6%), Actinobacteria (19.3% ± 8.3%), Bacteroidetes (14.3% ± 9.6%), and Acidobacteria (9.0% ± 3.8%) (Fig. S2). Abundant genera included Kaistobacter (0.01% ± 0.007%), Flavobacterium (0.009% ± 0.023%), Bradyrhizobium (0.006% ± 0.004%), Acidovorax (0.006% ± 0.014%), Devosia (0.006% ± 0.004%), and Chitinophaga (0.005% ± 0.013%) (Fig. S3). Because bacterial communities differed between sampling years (Fig. S4), communities from each year were analyzed independently.
Effects of glyphosate on bacterial community structure and diversity.
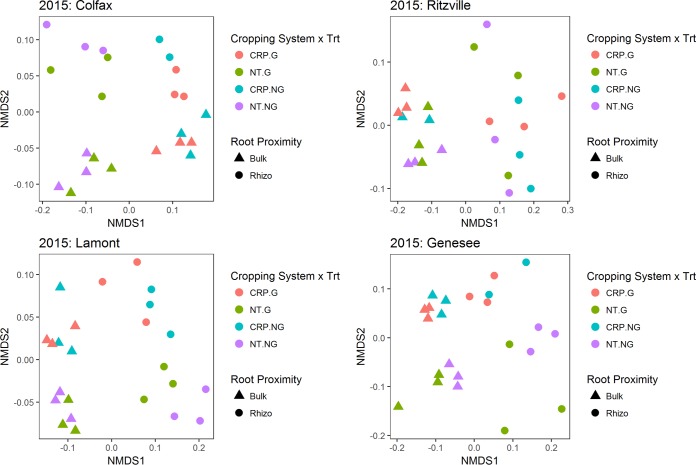
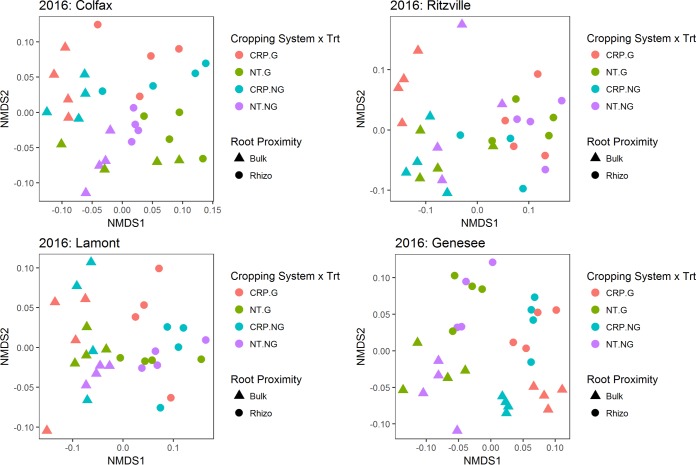
Within each location in 2015 and 2016, bacterial communities clustered primarily by cropping system and root proximity, with no clear or consistent grouping of glyphosate-treated soil or rhizosphere bacterial communities in soil from no-till (NT; history of glyphosate use) or Conservation Reserve Program (CRP; no history of glyphosate use) cropping systems (Fig. 1 and 2). However, in a few cases, bacterial communities from glyphosate-treated soil or rhizospheres formed clusters distinct from those of nontreated samples in the same location, cropping system, and root proximity. For example, in 2015, there was little overlap between glyphosate-treated and nontreated rhizosphere communities from Colfax soil, or in glyphosate-treated or nontreated rhizosphere communities in Lamont CRP soil (Fig. 1). Similarly, there was little overlap between glyphosate-treated and nontreated communities from Genesee bulk soil (Fig. 1). In 2016, glyphosate-treated bulk soil communities from CRP systems at Ritzville and Genesee clustered separately from nontreated communities (Fig. 2). Permutational multivariate analysis of variance (PERMANOVA) supported the small and inconsistent effect of glyphosate on bacterial communities, with glyphosate treatment explaining only ∼2 to 5% of the overall variation in the community structure at each site in 2015 and 2016 (Tables 1 and 2, respectively). Nonetheless, glyphosate treatment was a significant source of variation at the Genesee location in 2015, and at Colfax, Lamont, and Genesee in 2016 (Tables 1 and 2). In contrast to the weak and inconsistent impacts of glyphosate, cropping system and root proximity consistently explained significant portions of variation in bacterial communities (∼7% to 47% and ∼12% to 48% of the variation, respectively; Tables 1 and 2). Together, cropping system and root proximity explained ∼59% to 65% of the total variation in bacterial community structure at each location in 2015 and ∼37% to 50% in 2016, indicating that these two factors are primary drivers of bacterial communities within locations and that the effects on bacterial communities varied between years.
FIG 1.
NMDS plots of weighted UniFrac distances among bacterial communities for each location in 2015 (stress = Colfax, 0.07; Ritzville, 0.08; Lamont, 0.07; Genesee, 0.09). Trt, treatment; G, glyphosate; Rhizo, rhizosphere.
FIG 2.
NMDS plots of weighted UniFrac distances among bacterial communities for each location in 2016 (stress = Colfax, 0.17; Ritzville, 0.13; Lamont, 0.10; Genesee, 0.15).
TABLE 1.
PERMANOVA of impacts of cropping system, root proximity, and glyphosate on bacterial community structure at different locations in 2015
| Location | Factora | F value | r2 | P valueb |
|---|---|---|---|---|
| Colfax | CS | 19.92 | 0.467 | 0.001 |
| RP | 15.64 | 0.183 | 0.001 | |
| G | 1.39 | 0.016 | 0.236 | |
| CS × RP | 0.68 | 0.013 | 0.59 | |
| CS × G | 0.49 | 0.009 | 0.77 | |
| RP × G | 0.95 | 0.018 | 0.41 | |
| CS × RP × G | 0.88 | 0.017 | 0.46 | |
| Lamont | CS | 9.04 | 0.153 | 0.001 |
| RP | 26.9 | 0.455 | 0.001 | |
| G | 2.23 | 0.038 | 0.083 | |
| CS × RP | 1.6 | 0.027 | 0.132 | |
| CS × G | 0.083 | 0.014 | 0.446 | |
| RP × G | 1.42 | 0.024 | 0.181 | |
| CS × RP × G | 1.09 | 0.018 | 0.319 | |
| Ritzville | CS | 5.91 | 0.108 | 0.002 |
| RP | 26.33 | 0.48 | 0.001 | |
| G | 1.72 | 0.032 | 0.142 | |
| CS × RP | 0.054 | 2.83 | 0.049 | |
| CS × G | 0.017 | 0.087 | 0.443 | |
| RP × G | 0.025 | 1.3 | 0.229 | |
| CS × RP × G | 0.017 | 0.88 | 0.418 | |
| Genesee | CS | 14.73 | 0.267 | 0.001 |
| RP | 19.24 | 0.348 | 0.001 | |
| G | 2.87 | 0.052 | 0.025 | |
| CS × RP | 0.22 | 0.004 | 0.98 | |
| CS × G | 1.18 | 0.021 | 0.32 | |
| RP × G | 1.47 | 0.027 | 0.2 | |
| CS × RP × G | 0.58 | 0.01 | 0.71 |
CS, cropping system; RP, root proximity; G, glyphosate.
Values in bold represent factors with a significant effect (P < 0.05).
TABLE 2.
PERMANOVA of impacts of cropping system, root proximity, and glyphosate on bacterial community structure at different locations in 2016
| Location | Factora | F value | r2 | P valueb |
|---|---|---|---|---|
| Colfax | CS | 17.41 | 0.38 | 0.001 |
| RP | 11.08 | 0.12 | 0.001 | |
| G | 2.1 | 0.022 | 0.043 | |
| CS × RP | 0.955 | 0.022 | 0.46 | |
| CS × G | 1.52 | 0.035 | 0.12 | |
| RP × G | 0.899 | 0.02 | 0.52 | |
| CS × RP × G | 0.839 | 0.019 | 0.56 | |
| Lamont | CS | 3.27 | 0.066 | 0.006 |
| RP | 15.51 | 0.315 | 0.001 | |
| G | 2.3 | 0.047 | 0.038 | |
| CS × RP | 0.78 | 0.016 | 0.596 | |
| CS × G | 1.02 | 0.021 | 0.343 | |
| RP × G | 1.28 | 0.026 | 0.228 | |
| CS × RP × G | 1.03 | 0.021 | 0.342 | |
| Ritzville | CS | 5.01 | 0.108 | 0.002 |
| RP | 12.03 | 0.259 | 0.001 | |
| G | 1.48 | 0.032 | 0.134 | |
| CS × RP | 0.73 | 0.016 | 0.673 | |
| CS × G | 2 | 0.043 | 0.055 | |
| RP × G | 1.08 | 0.023 | 0.326 | |
| CS × RP × G | 0.97 | 0.021 | 0.379 | |
| Genesee | CS | 15.84 | 0.265 | 0.001 |
| RP | 12.64 | 0.211 | 0.001 | |
| G | 2.11 | 0.035 | 0.032 | |
| CS × RP | 0.713 | 0.012 | 0.65 | |
| CS × G | 1.36 | 0.023 | 0.21 | |
| RP × G | 1.75 | 0.029 | 0.082 | |
| CS × RP × G | 1.29 | 0.022 | 0.22 |
CS, cropping system; RP, root proximity; G, glyphosate.
Values in bold represent factors with a significant effect (P < 0.05).
As with bacterial community structure, patterns in bacterial richness and diversity in response to glyphosate treatment were significant in only a few cases and were inconsistent among locations, cropping systems, root proximities, and sampling years (Fig. S5 and S6 and Tables S2 and S3). When a significant effect of glyphosate on bacterial diversity was found, diversity tended to be higher in glyphosate-treated communities. For example, in Colfax soils in 2015, glyphosate-treated communities tended to have higher Shannon and Simpson diversity than nontreated communities. Similar patterns were observed for Ritzville (for Simpson's diversity) and Lamont (for Shannon diversity) in 2015. However, in 2016, these patterns were not significant, and in some cases, such as for Colfax NT communities, diversity appeared to follow an opposite trend with glyphosate treatment from that in 2015.
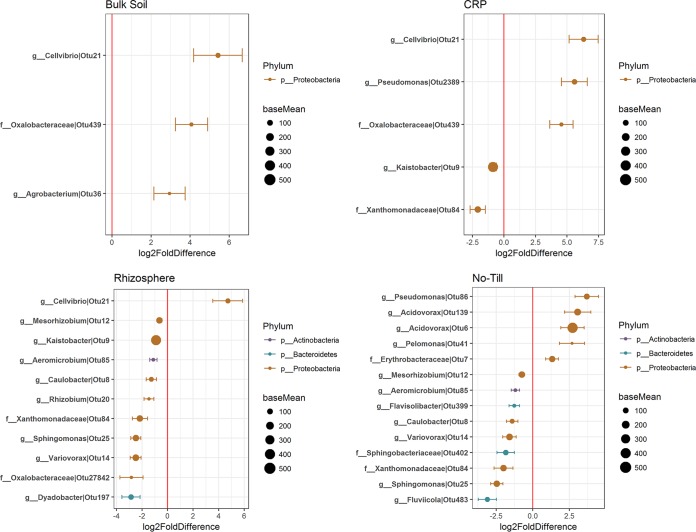
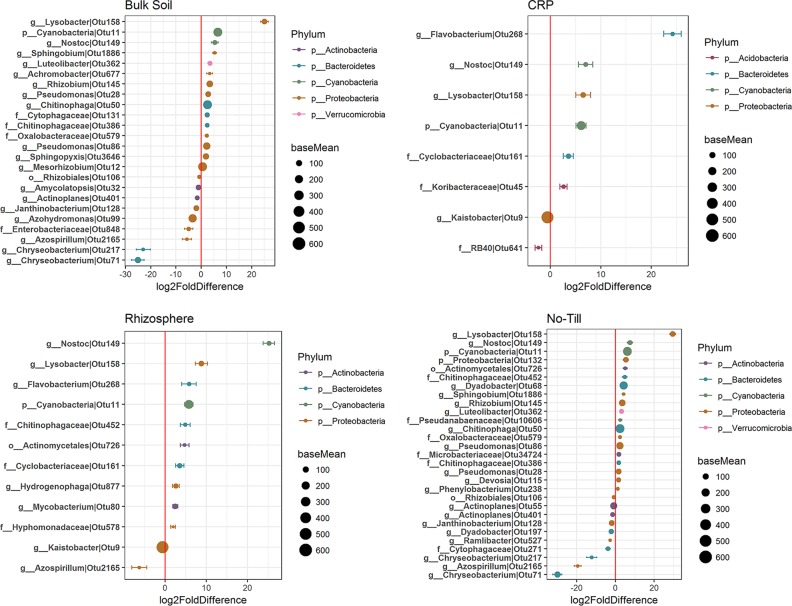
Despite the minor effects of glyphosate treatment on bacterial community composition and diversity as a whole, the relative abundances of a small number of taxa significantly increased or decreased in response to glyphosate treatment for each cropping system and root proximity in 2015 and 2016 (Fig. 3 and 4 and Table 3). Differentially abundant bacterial taxa (DAotus) affected by glyphosate represented only a small percentage of total OTUs (≤0.41% and ≤0.99% of OTUs detected in 2015 and 2016, respectively) and comprised a small fraction of the total community, generally accounting for <5% of the sequences (Table 3). The DAotus with the largest changes in relative abundance were typically of low relative abundance over all samples, and those DAotus with high relative abundances generally had small log2-fold changes with glyphosate treatment (Fig. 3 and 4). In general, there was a greater number of DAotus detected in 2016 than in 2015, and these typically comprised a larger percentage of the bacterial community (Fig. 3 and 4 and Table 3).
FIG 3.
Distribution of log2 fold changes in DAotus in 2015. Diagrams show the normalized sequence count (base mean) of DAotus in each soil type that were increased or decreased by glyphosate treatment, colored by the bacterial phyla to which they belong. Only DAotus with base means of >30 are presented. The red vertical line represents a zero-fold change, where OTUs to the right of the line (positive values) are increased in relative abundance with glyphosate, and those to the left of the line (negative values) are reduced in relative abundance with glyphosate. g, genus; f, family; p, phylum.
FIG 4.
Distribution of log2 fold changes in DAotus in 2016. Diagrams show the normalized sequence count (base mean) of DAotus in each soil type that were increased or decreased by glyphosate treatment, colored by the bacterial phyla to which they belong. Only DAotus with base means of >30 are presented. The red vertical line represents a zero-fold change, where OTUs to the right of the line (positive values) are increased in relative abundance with glyphosate, and those to the left of the line (negative values) are reduced in relative abundance with glyphosate.
TABLE 3.
Counts and percentages of DAotus increasing and decreasing within bacterial communities for each factor
| Factor | Responsea | 2015 |
2016 |
||
|---|---|---|---|---|---|
| Increase | Decrease | Increase | Decrease | ||
| Bulk | No. of DAotus | 10 | 1 | 45 | 23 |
| Mean ± SD % of sequences | 0.137 ± 0.29 | 0.013 ± 0.08 | 2.74 ± 3.5 | 1.14 ± 1.0 | |
| Rhizosphere | No. of DAotus | 3 | 19 | 38 | 16 |
| Mean ± SD % of sequences | 0.399 ± 0.98 | 5.77 ± 3.9 | 1.80 ± 2.6 | 1.60 ± 1.0 | |
| CRP | No. of DAotus | 9 | 5 | 23 | 9 |
| Mean ± SD % of sequences | 0.407 ± 1.1 | 2.23 ± 2.1 | 1.56 ± 2.8 | 1.57 ± 0.88 | |
| NT | No. of DAotus | 18 | 13 | 74 | 34 |
| Mean ± SD % of sequences | 2.04 ± 5.9 | 2.42 ± 2.6 | 3.82 ± 3.8 | 1.77 ± 1.7 | |
Percentages are based on DESeq2-normalized counts.
DAotus belonged to diverse phylogenetic groups but included all major phyla commonly found in soil or rhizosphere communities (Fig. 3 and 4). A large portion of DAotus belonged to the phyla Proteobacteria and Bacteroidetes. These two groups comprised 69 to 91% and 44 to 68% of DAotus in 2015 and 2016, respectively, while making up only ∼32% of all OTUs among samples. Members of these phyla showed both increases and decreases in response to glyphosate treatment in each location, even among OTUs belonging to the same genus (Fig. 3 and 4), suggesting little phylogenetic consistency in the specific responses of soil and rhizosphere communities to glyphosate and that the effects of glyphosate on relative abundances of soil bacteria are likely to be limited to a small number of specific OTUs or bacterial species.
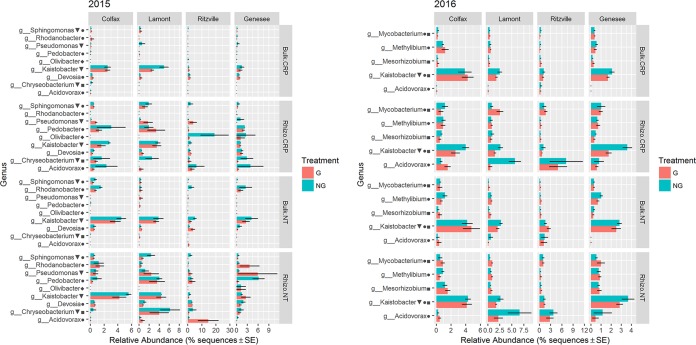
The relative abundances of a small number of genera representing more than 0.5% of the sequences in each year were significantly influenced by glyphosate (Fig. 5). Notably, multiple genera belonging to Sphingomonadaceae (Sphingomonas and Kaistobacter) and Sphingobacteriaceae (Pedobacter and Olivibacter) tended to be relatively less abundant in glyphosate treatments. However, only Chryseobacterium, Kaistobacter, and Sphingomonas in 2015 and “Candidatus Solibacter” and Kaistobacter in 2016 varied significantly with glyphosate after correction for false-discovery rates (FDR; P > 0.1).
FIG 5.
Bar plots of genera significantly influenced by glyphosate (triangle), a glyphosate × location interaction (circle), root proximity × glyphosate interaction (square), or cropping system × glyphosate interaction (diamond) with an uncorrected P value of <0.05 based on ANOVA of log10(1 + x)-transformed sequence counts. SE, standard error.
Adaptation of bacterial communities to long-term glyphosate use.
Adaptation of bacterial communities to long-term glyphosate applications should be reflected in differential responses of communities between cropping systems with a long history of glyphosate exposure (NT) and those with no known exposure (CRP). However, there were no consistent patterns in clustering of glyphosate-treated communities in nonmetric multidimensional scaling (NMDS) plots, and PERMANOVA detected no significant interactions between glyphosate treatment with either cropping system at any location in 2015 or 2016 (Fig. 1 and 2 and Tables 1 and 2). These patterns suggest that the major constituents of bacterial communities are unlikely to be adapted to glyphosate, even after many decades of use. Although not statistically significant, there were weak near-significant interactions between cropping system and glyphosate treatment for two locations in 2016 (Table 2), likely due to subtle glyphosate-induced shifts in CRP bulk soil communities (Fig. 2).
Consistent with patterns in bacterial community structure, there was no evidence that glyphosate use history significantly impacted the response of bacterial diversity to glyphosate treatment. There were no significant interactions between cropping history and glyphosate treatment in 2015 in a comparison of bacterial richness or diversity (Table S2). In 2016, bacterial communities in Colfax NT soils treated with glyphosate tended to be less diverse than those in nontreated soils (Fig. S6). However, the opposite pattern was observed in Ritzville soils in 2016, where communities from glyphosate-treated NT soils tended to be more diverse (Fig. S6). The lack of consistent patterns in diversity in response to glyphosate in NT and CRP systems across locations and years indicates that the diversity of bacterial communities is resilient in the face of glyphosate contamination, regardless of the history of glyphosate use in the field.
A larger proportion of DAotus responded to glyphosate in NT than in CRP soils (2015, n = 31 of 7,531 OTUs and 14 of 8,408 OTUs, respectively, χ2 = 7.63, P = 0.006; 2016, n = 108 of 10,869 OTUs and 32 of 11,888 OTUs, respectively, χ2 = 47.56, P < 0.0001). DAotus did not differ significantly in the direction of their responses between NT and CRP systems in either year (2015, χ2 = 0.004, P = 0.95; 2016, χ2 = 0.021, P = 0.89). Most DAotus in both cropping systems tended to increase in relative abundance with glyphosate treatment in each year (64% and 56% in 2015 and 72% and 69% in 2016 of DAotus for CRP and NT systems, respectively). Together, these data indicate that although relatively rare overall, a greater number of bacterial OTUs respond to glyphosate applications in NT systems than those from communities lacking a history of glyphosate exposure (CRP), but the proportions of OTUs with relative increases or decreases are similar between cropping histories (Table 3).
A small number of DAotus were shared among cropping systems (Fig. 3, 4, and 6). In 2015, only 2 OTU responded to glyphosate in both CRP and NT systems (OTU 84 [Xanthomonadaceae] decreased and OTU 20180 [Fluviicola] increased with glyphosate treatment [Fig. 6]). In 2016, 12 DAotus increased in relative abundance, whereas 2 decreased in both CRP and NT systems (Fig. 6). DAotus that increased in both cropping systems in 2016 predominantly included those belonging to the Bacteroidetes (OTUs 699, 1084, 3528, and 674), Cyanobacteria (OTUs 11 and 149), and Proteobacteria (OTUs 158 and 1391), whereas those with relative decreases in abundance included Chloroflexi (OTU 6578) and Fibrobacteres (OTU 24177) (Fig. 4). No DAotus were consistent between CRP and NT systems in either 2015 or 2016. Thus, although there were more DAotus in NT, the responses of most individual taxa were inconsistent among cropping systems and years.
FIG 6.
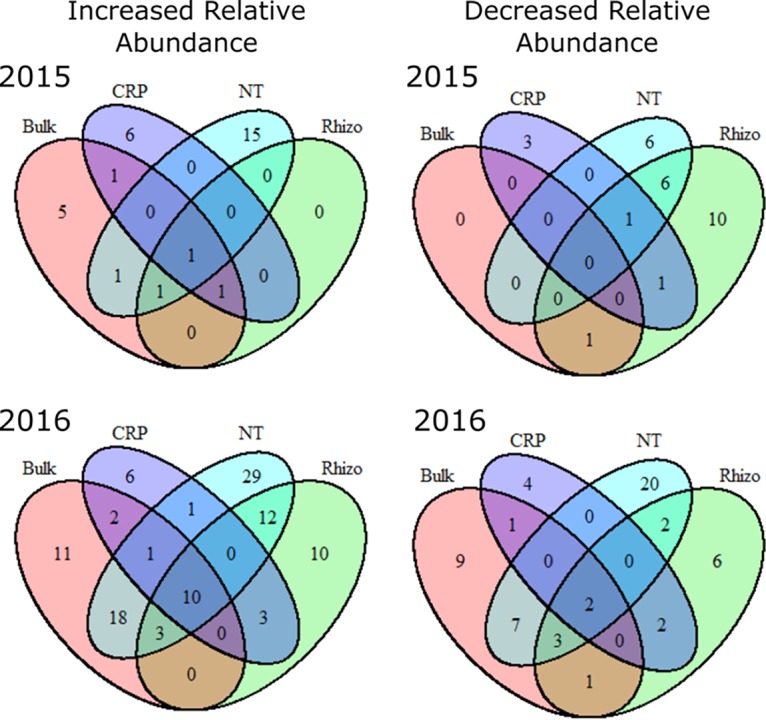
Venn diagrams showing overlap in DAotus increasing and decreasing in relative abundance in 2015 and 2016.
RP effects of glyphosate on bacterial communities.
Plant-dependent effects of glyphosate on microbiomes should be evident in differential responses of bacterial communities to glyphosate treatment in bulk versus rhizosphere soil. However, there were no significant interactions between root proximity (RP) and glyphosate treatment in 2015 or 2016 (Tables 1 and 2), and responses of bacterial communities to glyphosate were not rhizosphere or bulk soil specific (Fig. 1 and 2). In 2016, there was a near-significant interaction between root proximity and glyphosate treatment at the Genesee location (Table 2), likely due to a small effect of glyphosate treatment on bacterial communities in CRP bulk soil (Fig. 2). In total, there were no clear or consistent effects of glyphosate on either bulk soil or rhizosphere communities across locations, cropping systems, or years, suggesting that the potential impacts of glyphosate on microbiomes are not strongly influenced by proximity to a plant.
Similarly, in most cases, the response of bacterial diversity to glyphosate treatment did not depend on root proximity (Tables S2 and S3). Moreover, in the small number of cases where the RP × glyphosate interaction was significant, there were no consistent patterns among locations, years, root proximity, or diversity measures used. For example, richness and Shannon diversity tended to be higher only in Lamont rhizosphere soils in 2015, yet in 2016, bulk soil communities from the Ritzville or Genesee locations tended to be less rich or diverse, respectively (Fig. S5 and S6 and Tables S2 and S3).
The rhizosphere and bulk soil communities did not differ in the number of OTUs impacted by glyphosate (2015, 22 of 8,959 OTUs and 11 of 8,937 OTUs, respectively, χ2 = 3.06, P = 0.08; 2016, 54 of 12,784 OTUs and 68 of 12,316 OTUs, respectively, χ2 = 0.98, P = 0.32), supporting the finding that glyphosate impacts are not restricted to or buffered by the rhizosphere. However, in 2015, DAotus had contrasting responses between bulk and rhizosphere soil (χ2 = 15.25, P < 0.0001). Most DAotus in bulk soil (10/11 [91% of DAotus]) tended to increase in relative abundance with glyphosate treatment, whereas only a single DAotu decreased with glyphosate treatment (1/11 [9% of DAotus]) (Table 3). In contrast, in the rhizosphere, most DAotus decreased in relative abundance with glyphosate treatment (19/22 [86% of DAotus]), and only a few DAotus increased (3/22 [14% of DAotus]). However, there were no significant differences in DAotu responses in 2016 (χ2 = 0.09, P < 0.77), where 66% of DAotus in bulk soil increased (n = 45/68) and 34% decreased (23/68) (Table 3). In the rhizosphere in 2016, most DAotus (38/54 [70%]) increased in relative abundance rather than decreased (16/54 [30%]).
There was little consistency in the identities of DAotus between bulk and rhizosphere soils. Only 4 OTUs were influenced by glyphosate in both bulk and rhizosphere soil in 2015 (OTUs 20180 [Fluviicola spp.], 161 [Cyclobacteriaceae spp.], and 21 [Cellvibrio spp.] increased in relative abundance, whereas OTU 4045 [Bacteroidetes spp.] decreased) (Fig. 3). In 2016, 13 OTUs increased in relative abundance, while 6 decreased in both bulk and rhizosphere soil (Table 3). Those DAotus that increased in relative abundance included members of Bacteroidetes (OTUs 1084, 3528, 699, 674, and 2630), Proteobacteria (OTUs 1057, 158, 1391, and 1023), and Cyanobacteria (OTUs 149 and 11), whereas those with relative decreases included members of Gemmatimonadetes (OTU 1933), Chloroflexi (OTU 6576), Acidobacteria (OTU 6369), Fibrobacteres (OTU 24177), Cyanobacteria (OTU 453), and Proteobacteria (OTU 2165) (Fig. 4). No OTUs responded consistently in both bulk and rhizosphere soil in the two years.
DISCUSSION
Though glyphosate is a lynchpin of no-till practices and is widely considered an environmentally safe herbicide, there have been persistent questions regarding its potential long-term impacts on plant-associated microbes and plant health. In this work, the most detailed to date, to our knowledge, of glyphosate effects on wheat-associated bacterial communities across one of the most productive growing regions in the United States, we found that repeated glyphosate applications had minimal impacts on soil and rhizosphere bacterial community composition and diversity and that the small effects of glyphosate were inconsistent between locations, years, root proximities, and cropping systems. Thus, despite numerous claims of the detrimental effects of glyphosate, our results support the idea that field rate applications of glyphosate for preemergence weed control are unlikely to substantially impact soil or rhizosphere bacterial communities. This is likely due to the low application rates and volumes of glyphosate, its rapid binding to soil, and its strong adherence to the upper layers of the soil profile (<2 mm from surface) (3).
Although there may be transient effects of acute glyphosate exposure on soil microbial communities that we may not have detected after 6 weeks (21), the high diversity and interconnectedness of microbial ecosystems are likely to render communities resistant or resilient to small perturbations, such as low levels of glyphosate exposure. Many microbes may tolerate low concentrations of glyphosate (37, 38), survive in dormant states (e.g., spores), or inhabit protected microniches or deeper soil strata. Consequently, communities are likely to rapidly recover to a predisturbance state with little impact of glyphosate on microbial community functions.
Despite weak or insignificant effects of glyphosate on soil and rhizosphere microbial communities, a small number of taxa increased or decreased in relative abundance after glyphosate treatment. However, the specific DAotus detected were largely inconsistent among locations, years, and cropping systems. These varied patterns in the responses to glyphosate suggest that rather than a consistent direct positive or negative effect of glyphosate, other factors likely control the population dynamics of bacterial taxa. Although the affected DAotus represented common soil phyla, a disproportionate number belonged to Proteobacteria and Bacteroidetes, both of which are considered to be copiotrophic (39). Differences in resource availability, potentially as an indirect effect of glyphosate or its residues on plant root exudation or degradation dynamics, may generate distinct competitive outcomes among these taxa, resulting in small shifts in relative abundances. This is consistent with the finding that closely related taxa (belonging to the same genus), which likely share common life history strategies and niche preferences, often had differential responses to glyphosate, potentially indicative of competitive interactions.
Changes in root-derived resources with glyphosate treatment may generate a “greenbridge” effect, similar to that observed with root-rotting fungal pathogens (8, 40). The shikimic acid pathway, which is inhibited by glyphosate, is crucial for the formation of aromatic amino acids and plays a role in plant defense. With reduced defensive compounds, dying roots are more readily colonized by root pathogens and survive to infect subsequent crops. In the same way, dying roots may selectively be colonized by fast-growing copiotrophic bacteria which increase in abundance after repeated glyphosate applications. Thus, the changes in the relative abundances of copiotrophic taxa after glyphosate application likely are not due to the utilization of glyphosate as a nutrient source but rather to the resources provided by dying roots. Importantly, the taxa that responded to glyphosate exposure are those with a little-clear role in plant health, comprised only a small portion of the bacterial community, and are unlikely to play a major role in plant productivity in agroecosystems.
Consistent with the small impact of glyphosate on soil and rhizosphere bacterial communities as a whole, neither communities from environments with a long history of glyphosate use (NT) nor those with no history of glyphosate exposure (CRP) exhibited strong community-wide shifts after glyphosate applications. Thus, even after decades of glyphosate use, most bacterial taxa do not differ in their capacity to tolerate or utilize glyphosate as a nutrient source. This finding is consistent with the idea that glyphosate degradation is largely a cometabolic process that does not provide a significant fitness benefit to the organisms producing glyphosate-degrading enzymes (14). Moreover, most glyphosate degraders potentially benefit by accessing inorganic P or N only in settings where these nutrients are limiting (14, 41). Consequently, it is unlikely that glyphosate-degrading phenotypes will be under strong selection in agricultural systems where soil nutrient availability is maintained at high levels and glyphosate is applied at field rates.
Even though bacterial communities from NT or CRP systems did not show community-wide shifts in composition under glyphosate treatment, a significantly greater proportion of individual taxa increased or decreased in relative abundance in NT versus CRP in both sampling years. Though still a small portion of the bacterial community (<1% of OTUs in each cropping system), populations from cropping systems with a long history of glyphosate use were somewhat more likely to be affected by glyphosate treatment than those lacking such a history. This predisposition of some bacterial taxa with a history of glyphosate exposure to respond to additional glyphosate treatment may be due to higher densities of a few taxa that have adapted to better tolerate or grow in the presence of glyphosate or to the accumulation of inhibitory residues that reduce populations of less-tolerant taxa. However, the lack of consistency in the identities of glyphosate-responsive taxa among sampling years may suggest that a diversity of generalist copiotrophic bacteria are capable of multiplying on dying roots and that patterns in specific taxa among years will be contingent on other drivers of microbial communities, such as location/soil type or environmental conditions. Although glyphosate is expected to have little selective effect on microbes and adaptation of microbes may play a small role in glyphosate degradation rates, second-order effects of glyphosate mediated via plants may allow some taxa to predominate.
Because the presence of plants may be a crucial mediator of glyphosate-induced shifts in soil microbiomes, plants may buffer against the effects of glyphosate on microbial communities in bulk soil by selecting for specific plant-associated taxa in the rhizosphere. Alternatively, if residual glyphosate or its breakdown products modify plant physiology in ways that inhibit the production of defense compounds or increase root exudation, the presence of plants may intensify the effects of glyphosate on the microbiome through altered degradation of plant biomass. However, the community-wide response of bacteria to glyphosate treatment was weak and did not appear to differ consistently between bulk soil and rhizosphere environments. Thus, close contact of a bacterial community with plant roots is not likely to play a major role in mediating the effects of glyphosate on bacterial communities of wheat, suggesting that the buildup of some populations on dying roots is more likely an important route of glyphosate effects on soil communities. However, due to significant differences in the composition of bulk soil and rhizosphere communities, many of the taxa affected by glyphosate were specific to these regions.
Other recent studies have observed subtle or insignificant impacts of glyphosate on bacterial communities, though in a more limited number of soils or with little experimental replication (11, 14, 18, 26). For example, Newman et al. (11) described slight increases in the relative abundances of Proteobacteria and decreases in Acidobacteria, consistent with the concept of copiotrophic bacteria being favored over oligotrophs in the rhizospheres of glyphosate-treated plants (28). However, there are few investigations of how a history of glyphosate use impacts the microbial community response to additional herbicide applications. Existing work has documented both increases (24) and decreases (25) in microbial respiration in soils with a history of glyphosate exposure, potentially due to toxic effects of glyphosate on microbial populations or glyphosate metabolism by adapted taxa, but with little corresponding impact on bacterial community composition. In contrast to these, we identified a small but significantly greater portion of taxa that respond to glyphosate in soils with a long history of exposure than in unexposed soils.
This work demonstrates a minimal impact of glyphosate on bacterial communities in the soil and rhizosphere of a dryland wheat cropping system. The lack of a consistent cohort of OTUs that were significantly impacted by glyphosate indicates that it has no major impact on bacterial communities in soil or the rhizosphere of wheat. Rather, a small number of taxa may be influenced by an increase in biomass of dying roots in the soil.
MATERIALS AND METHODS
Field sampling and greenhouse assays.
Soil was collected from four farms spanning a rainfall gradient across eastern Washington. Fields were chosen that had a long history (>30 years) of glyphosate use as a result of long-term no-till (NT) cropping practices. A detailed analysis of the soils at these locations is presented in Table S4. At the Genesee location, which receives an average of 631 mm precipitation annually, the typical rotation is winter wheat/spring wheat/peas. At the Colfax location, which receives an average of 512 mm precipitation annually, the typical rotation is winter wheat/spring wheat/canola. At the Lamont location, which receives an average of 439 mm precipitation annually, the typical rotation is winter wheat/spring wheat or spring canola and then a fallow year. At the Ritzville location, which receives an average of 314 mm precipitation annually, the typical rotation is winter wheat followed by a year of fallow. At each location, nearby fields that had no known exposure to glyphosate and were in the Conservation Reserve Program (CRP) also were chosen for comparison. At Genesee, Colfax, and Lamont, CRP was a mixture of native and introduced grasses. At Ritzville, CRP was native sage and native grasses. Soil was collected from the top 30 cm of each field using a clean spade and transported to the greenhouse. Soil was potted into 130 cm by 130 cm by 150 cm black plastic pots, and wheat (Triticum aestivum L. cv. Louise) was planted (5 seeds/pot). Pots were arranged in a randomized block design, and plants were grown with regular watering and a single round of fertilization (Miracle Gro; Scotts, Inc., Maryville, OH) at each cycle. The greenhouse temperature was 23°C during the day and 18°C at night, with 16-h/day supplemental lighting with sodium vapor lamps. At the end of 6 weeks, two treatments were applied to paired pots. Plants were either killed with a glyphosate spray (2 times the recommended field rate, 1.68 kg/ha; RT3 with 0.25% nonionic surfactant [NIS] adjuvant and ammonium sulfate) or by manual clipping at the soil line. After plants were allowed to senesce in the greenhouse for 2 weeks, the remaining above-ground biomass was removed, and a subsequent cycle of wheat growth was initiated. After 4 such cycles (3 glyphosate sprays), bulk and rhizosphere soils were sampled from each pot. Bulk soil (∼1 g) was taken between wheat plants using a sterile cork borer (∼0.5 cm diameter). For rhizosphere samples, a single plant was removed from each replicate pot and shaken vigorously to remove loosely adhering soil from the root systems. Root systems were then transferred to a sterile 50-ml tube, submersed in 10 ml of H2O, vortexed for 1 min, and sonicated for 1 min to remove the tightly adhering rhizosphere soil. The first experiment was initiated in January 2015 and terminated in June 2015. The second experiment was initiated in November 2015 and terminated in April 2016.
DNA extraction and Illumina sequencing.
DNA was extracted from soil and rhizosphere samples using the Mo Bio PowerSoil kit (Mo Bio/Qiagen, Carlsbad, CA), according to the manufacturer's instructions. For rhizosphere samples, soil suspensions were centrifuged (16,000 × g, 1 min) to pellet rhizosphere soil, H2O was removed, and soil was suspended in the PowerSoil buffer. Samples were transferred to PowerSoil bead tubes and processed according to standard protocols. Homogenization of the samples was performed on a FastPrep bead beater (MP Biomedical, Santa Ana, CA) using the “soil” program. DNA was quantified on a NanoDrop spectrophotometer and submitted for amplification of the V1–V3 region of the 16S rRNA gene and paired-end (2 × 300 bp) sequencing on the Illumina MiSeq platform using version 3 chemistry. Samples from two locations in 2015 (Genesee and Ritzville) were submitted to the MR DNA lab (Shallowater, TX), whereas all other samples were submitted to the University of Minnesota Genomics Center (UMGC). Although the same region was amplified by each sequencing provider (V1–V3), there were differences in amplification protocols and primer designs among sequence providers, confounding the location effects in the 2015 experiment. PCR amplification at MR DNA used HotStartTaq Plus master mix (Qiagen, USA) and primers 27F (5′-AGRGTTTGATCMTGGCTCAG-3′) and 519R (5′-GWATTACCGCGGCKGCTG-3′). The thermocycling conditions consisted of 94°C for 3 min, followed by 28 cycles of 94°C for 30 s, 53°C for 40 s, and 72°C for 1 min, with a final elongation step at 72°C for 5 min. Samples were checked for successful amplification on a 2% agarose gel, pooled in equal proportions, purified with AMPure XP beads, and prepared for sequencing with the Illumina TruSeq DNA library preparation protocol. PCR amplification at the University of Minnesota UMGC used a dual-indexing approach, as described by Gohl et al. (42). Briefly, this consisted of a first round of PCR using template-specific primers MN_27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and MN_534R (5′-ATTACCGCGGCTGCTGG-3′) with Kapa HiFi HotStart polymerase. The PCR conditions consisted of an initial denaturing at 95°C for 5 min, followed by 25 cycles of 98°C for 20 s, 55°C for 15 s, and 72°C for 1 min, with a final extension at 72°C for 1 min. The products from the first PCR were diluted 1:100, and 5 μl was included in a second PCR using indexing primers. The second PCR consisted of an initial denaturation at 95°C for 5 min, 10 cycles of 98°C for 20 s, 55°C for 15 s, and 72°C for 1 min, with a final extension at 72°C for 5 min. The products were pooled, size selected, and spiked with 20% PhiX prior to sequencing with an Illumina MiSeq 600 cycle version 3 kit.
Sequence processing.
Despite primer and amplification differences for some samples in 2015, all reads spanned the same 16S region. Sequences were trimmed to an identical region, leaving no terminal gaps, and were processed together. Forward and reverse reads were paired using PEAR (version 0.9.6) (43). Barcodes and primer sequences were removed with cutadapt (version 1.91) (44), and sequences with ambiguous bases or shorter than 350 bp were removed. Processed sequences were clustered following the UPARSE pipeline (45) using vsearch (46) for all steps, with the exception of OTU clustering, which used usearch (version 8.1) (45). Briefly, reads were quality filtered using a maximum expected error rate of 1 and dereplicated, and singletons were removed prior to OTU clustering at 97% similarity threshold using the cluster_otus command. Processed reads were then mapped to OTU clusters to generate an OTU abundance table. Taxonomy was assigned to OTU representative sequences (centroids) with the RDP naive Bayesian Classifier (47) using the Greengenes 13_8 reference (48) database and an 80% confidence threshold. OTUs were filtered to remove any nonbacterial OTUs or those classified as mitochondria or chloroplasts using QIIME scripts (version 1.9.1) (49). OTUs with a total sequence count of <20 were removed to reduce poor-quality OTUs, and the OTU table was rarefied to 13,391 sequences/sample prior to analysis. Further, UniFrac distances were calculated after OTUs were aligned to the Greengenes reference using PyNAST, and a phylogenetic tree was constructed using FastTree in QIIME. Unrarefied OTU tables were retained for differential abundance analysis with DESeq2 (version 1.12.4) (50).
Data analysis.
Nonmetric multidimensional scaling (NMDS) and PERMANOVA were performed using weighted UniFrac distances to assess the significance of location, year, cropping system, root proximity, and glyphosate on bacterial community structure using the metaMDS and adonis functions of the vegan package (version 2.4.1 [51]) in R (52). Richness and diversity metrics (Shannon's [H′] and inverse Simpson's [1/D]) were estimated from rarefied OTU tables and evaluated among factors using analysis of variance (ANOVA). DESeq2 was used to identify differentially abundant bacterial taxa (DAotus) that were impacted by glyphosate treatment. Briefly, unrarefied OTU tables were filtered to contain only OTUs with normalized counts of >5 and that were present in 3 or more samples. In each year, glyphosate DAotus were evaluated in rhizosphere and bulk soil after accounting for location and cropping system effects using the model: ∼location + cropping system + root proximity × glyphosate. Similarly, DAotus were evaluated for NT and CRP soils after accounting for location and root proximity using the model: ∼location + root proximity + cropping system × glyphosate. Contrasts were performed between glyphosate treatments, and OTUs with adjusted P values of <0.1 were considered to be differentially abundant. Pearson's chi-square tests were used to compare the number of DAotus in different root proximity or cropping system treatments. ANOVA on log10(x + 1)-transformed sequence counts was used to test for the main effects of glyphosate treatment and interactions with location, root proximity, and cropping system on abundant bacterial genera (those representing >0.5% of all sequences). Distributions of sequence counts of genera were tested for normality using a Shapiro-Wilk test and examined manually to ensure that they approximated normal distributions. Data from the 2 years were analyzed separately to isolate the effects of glyphosate (glyphosate treatment) on bacterial community structure in no-till (NT) and CRP cropping systems while controlling for the effects of sampling year (Fig. S4 and Table S1).
Supplementary Material
ACKNOWLEDGMENTS
D.C.S. was funded by an administrator-funded USDA-ARS Postdoctoral Research Associate Award. This research was funded by USDA-ARS and Regional Approaches to Climate Change-Pacific Northwest Agriculture (REACCH) award 2011-68002-30191 from the USDA National Institute for Food and Agriculture.
We thank the following growers for their collaboration: John Aeschliman, Ron Jirava, Tracy Eriksen, and Russ Zenner.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01354-17.
REFERENCES
- 1.Benbrook CM. 2016. Trends in glyphosate herbicide use in the United States and globally. Environ Sci Eur 28:3. doi: 10.1186/s12302-016-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borggaard OK, Gimsing AL. 2008. Fate of glyphosate in soil and the possibility of leaching to ground and surface waters: a review. Pest Manag Sci 64:441–456. doi: 10.1002/ps.1512. [DOI] [PubMed] [Google Scholar]
- 3.Duke SO, Lydon J, Koskinen WC, Moorman TB, Chaney RL, Hammerschmidt R. 2012. Glyphosate effects on plant mineral nutrition, crop rhizosphere microbiota, and plant disease in glyphosate-resistant crops. J Agric Food Chem 60:10375–10397. doi: 10.1021/jf302436u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huggins DR, Reganold JP. 2008. No-till: the quiet revolution. Sci Am 299:70–77. doi: 10.1038/scientificamerican0708-70. [DOI] [PubMed] [Google Scholar]
- 5.Williams JD, Wuest SB, Long DS. 2014. Soil and water conservation in the Pacific Northwest through no-tillage and intensified crop rotations. J Soil Water Conserv 69:495–504. doi: 10.2489/jswc.69.6.495. [DOI] [Google Scholar]
- 6.Lal R, Reicosky DC, Hanson JD. 2007. Evolution of the plow over 10,000 years and the rationale for no-till farming. Soil Tillage Res 93:1–12. doi: 10.1016/j.still.2006.11.004. [DOI] [Google Scholar]
- 7.Calegari A, Hargrove WL, Rheinheimer DDS, Ralisch R, Tessier D, de Tourdonnet S, de Fatima Guimarães M. 2008. Impact of long-term no-tillage and cropping system management on soil organic carbon in an Oxisol: a model for sustainability. Agron J 100:1013. doi: 10.2134/agronj2007.0121. [DOI] [Google Scholar]
- 8.Hammerschmidt R. 2017. How glyphosate affects plant disease development: it is more than enhanced susceptibility. Pest Manag Sci, in press. doi: 10.1002/ps.4521. [DOI] [PubMed] [Google Scholar]
- 9.Kremer RJ, Means NE. 2009. Glyphosate and glyphosate-resistant crop interactions with rhizosphere microorganisms. Eur J Agron 31:153–161. doi: 10.1016/j.eja.2009.06.004. [DOI] [Google Scholar]
- 10.Zobiole LHS, Kremer RJ, Oliveira RS Jr, Constantin J. 2011. Glyphosate affects micro-organisms in rhizospheres of glyphosate-resistant soybeans. J Appl Microbiol 110:118–127. doi: 10.1111/j.1365-2672.2010.04864.x. [DOI] [PubMed] [Google Scholar]
- 11.Newman MM, Hoilett N, Lorenz N, Dick RP, Liles MR, Ramsier C, Kloepper JW. 2016. Glyphosate effects on soil rhizosphere-associated bacterial communities. Sci Total Environ 543:155–160. doi: 10.1016/j.scitotenv.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Bentley R. 1990. The shikimate pathway—a metabolic tree with many branches. Crit Rev Biochem Mol Biol 25:307–384. doi: 10.3109/10409239009090615. [DOI] [PubMed] [Google Scholar]
- 13.Hove-Jensen B, Zechel DL, Jochimsen B. 2014. Utilization of glyphosate as phosphate source: biochemistry and genetics of bacterial carbon-phosphorus lyase. Microbiol Mol Biol Rev 78:176–197. doi: 10.1128/MMBR.00040-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh BK, Walker A. 2006. Microbial degradation of organophosphorus compounds. FEMS Microbiol Rev 30:428–471. doi: 10.1111/j.1574-6976.2006.00018.x. [DOI] [PubMed] [Google Scholar]
- 15.Sviridov AV, Shushkova TV, Ermakova IT, Ivanova EV, Epiktetov DO, Leontievsky AA. 2015. Microbial degradation of glyphosate herbicides (review). Appl Biochem Microbiol 51:188–195. doi: 10.1134/S0003683815020209. [DOI] [PubMed] [Google Scholar]
- 16.Van Eerd LL, Hoagland RE, Zablotowicz RM, Hall JC. 2003. Pesticide metabolism in plants and microorganisms. Weed Sci 51:472–495. doi: 10.1614/0043-1745(2003)051[0472:PMIPAM]2.0.CO;2. [DOI] [Google Scholar]
- 17.Forlani G, Mangiagalli A, Nielsen E, Suardi C. 1999. Degradation of the phosphonate herbicide glyphosate in soil: evidence for a possible involvement of unculturable microorganisms. Soil Biol Biochem 31:991–997. doi: 10.1016/S0038-0717(99)00010-3. [DOI] [Google Scholar]
- 18.Newman MM, Lorenz N, Hoilett N, Lee NR, Dick RP, Liles MR, Ramsier C, Kloepper JW. 2016. Changes in rhizosphere bacterial gene expression following glyphosate treatment. Sci Total Environ 553:32–41. doi: 10.1016/j.scitotenv.2016.02.078. [DOI] [PubMed] [Google Scholar]
- 19.Druille M, García-Parisi PA, Golluscio RA, Cavagnaro FP, Omacini M. 2016. Repeated annual glyphosate applications may impair beneficial soil microorganisms in temperate grassland. Agric Ecosyst Environ 230:184–190. doi: 10.1016/j.agee.2016.06.011. [DOI] [Google Scholar]
- 20.Cherni AE, Trabelsi D, Chebil S, Barhoumi F, Rodríguez-Llorente ID, Zribi K. 2015. Effect of glyphosate on enzymatic activities, Rhizobiaceae and total bacterial communities in an agricultural Tunisian soil. Water Air Soil Pollut 226:145. doi: 10.1007/s11270-014-2263-8. [DOI] [Google Scholar]
- 21.Weaver MA, Krutz LJ, Zablotowicz RM, Reddy KN. 2007. Effects of glyphosate on soil microbial communities and its mineralization in a Mississippi soil. Pest Manag Sci 63:388–393. doi: 10.1002/ps.1351. [DOI] [PubMed] [Google Scholar]
- 22.Hart MM, Powell JR, Gulden RH, Dunfield KE, Pauls KP, Swanton CJ, Klironomos JN, Antunes PM, Koch AM, Trevors JT. 2009. Separating the effect of crop from herbicide on soil microbial communities in glyphosate-resistant corn. Pedobiologia 52:253–262. doi: 10.1016/j.pedobi.2008.10.005. [DOI] [Google Scholar]
- 23.Ratcliff AW, Busse MD, Shestak CJ. 2006. Changes in microbial community structure following herbicide (glyphosate) additions to forest soils. Appl Soil Ecol 34:114–124. doi: 10.1016/j.apsoil.2006.03.002. [DOI] [Google Scholar]
- 24.Lane M, Lorenz N, Saxena J, Ramsier C, Dick RP. 2012. Microbial activity, community structure and potassium dynamics in rhizosphere soil of soybean plants treated with glyphosate. Pedobiologia 55:153–159. doi: 10.1016/j.pedobi.2011.12.005. [DOI] [Google Scholar]
- 25.Zabaloy MC, Gómez E, Garland JL, Gómez MA. 2012. Assessment of microbial community function and structure in soil microcosms exposed to glyphosate. Appl Soil Ecol 61:333–339. doi: 10.1016/j.apsoil.2011.12.004. [DOI] [Google Scholar]
- 26.Barriuso J, Marín S, Mellado RP. 2011. Potential accumulative effect of the herbicide glyphosate on glyphosate-tolerant maize rhizobacterial communities over a three-year cultivation period. PLoS One 6:e27558. doi: 10.1371/journal.pone.0027558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zabaloy MC, Carné I, Viassolo R, Gómez MA, Gomez E. 2016. Soil ecotoxicity assessment of glyphosate use under field conditions: microbial activity and community structure of Eubacteria and ammonia-oxidising bacteria. Pest Manag Sci 72:684–691. doi: 10.1002/ps.4037. [DOI] [PubMed] [Google Scholar]
- 28.Imparato V, Santos SS, Johansen A, Geisen S, Winding A. 2016. Stimulation of bacteria and protists in rhizosphere of glyphosate-treated barley. Appl Soil Ecol 98:47–55. doi: 10.1016/j.apsoil.2015.09.007. [DOI] [Google Scholar]
- 29.Lancaster SH, Hollister EB, Senseman SA, Gentry TJ. 2010. Effects of repeated glyphosate applications on soil microbial community composition and the mineralization of glyphosate. Pest Manag Sci 66:59–64. doi: 10.1002/ps.1831. [DOI] [PubMed] [Google Scholar]
- 30.Mijangos I, Becerril JM, Albizu I, Epelde L, Garbisu C. 2009. Effects of glyphosate on rhizosphere soil microbial communities under two different plant compositions by cultivation-dependent and -independent methodologies. Soil Biol Biochem 41:505–513. doi: 10.1016/j.soilbio.2008.12.009. [DOI] [Google Scholar]
- 31.Haney RL, Senseman SA, Hons FM, Zuberer DA. 2000. Effect of glyphosate on soil microbial activity and biomass. Weed Sci 48:89–93. doi: 10.1614/0043-1745(2000)048[0089:EOGOSM]2.0.CO;2. [DOI] [Google Scholar]
- 32.Schnürer Y, Persson P, Nilsson M, Nordgren A, Giesler R. 2006. Effects of surface sorption on microbial degradation of glyphosate. Environ Sci Technol 40:4145–4150. doi: 10.1021/es0523744. [DOI] [PubMed] [Google Scholar]
- 33.Nye M, Hoilett N, Ramsier C, Renz P, Dick RP. 2014. Microbial community structure in soils amended with glyphosate-tolerant soybean residue. Appl Ecol Environ Sci 2:74–81. doi: 10.12691/aees-2-3-1. [DOI] [Google Scholar]
- 34.Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 35.Gomes MP, Smedbol E, Chalifour A, Henault-Ethier L, Labrecque M, Lepage L, Lucotte M, Juneau P. 2014. Alteration of plant physiology by glyphosate and its by-product aminomethylphosphonic acid: an overview. J Exp Bot 65:4691–4703. doi: 10.1093/jxb/eru269. [DOI] [PubMed] [Google Scholar]
- 36.Rosenbaum KK, Miller GL, Kremer RJ, Bradley KW. 2014. Interactions between glyphosate, Fusarium infection of common waterhemp (Amaranthus rudis), and soil microbial abundance and diversity in soil collections from Missouri. Weed Sci 62:71–82. doi: 10.1614/WS-D-13-00071.1. [DOI] [Google Scholar]
- 37.Shehata AA, Schrödl W, Aldin AA, Hafez HM, Krüger M. 2013. The effect of glyphosate on potential pathogens and beneficial members of poultry microbiota in vitro. Curr Microbiol 66:350–358. doi: 10.1007/s00284-012-0277-2. [DOI] [PubMed] [Google Scholar]
- 38.Pollegioni L, Schonbrunn E, Siehl D. 2011. Molecular basis of glyphosate resistance—different approaches through protein engineering. FEBS J 278:2753–2766. doi: 10.1111/j.1742-4658.2011.08214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fierer N, Bradford MA, Jackson RB. 2007. Toward and ecological classification of soil bacteria. Ecology 88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 40.Babiker EM, Hulbert SH, Schroeder KL, Paulitz TC. 2011. Optimum timing of preplant applications of glyphosate to manage Rhizoctonia root rot in barley. Plant Dis 95:304–310. doi: 10.1094/PDIS-05-10-0354. [DOI] [PubMed] [Google Scholar]
- 41.Kryuchkova YV, Burygin GL, Gogoleva NE, Gogolev YV, Chernyshova MP, Makarov OE, Fedorov EE, Turkovskaya OV. 2014. Isolation and characterization of a glyphosate-degrading rhizosphere strain, Enterobacter cloacae K7. Microbiol Res 169:99–105. doi: 10.1016/j.micres.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 42.Gohl DM, Vangay P, Garbe J, MacLean A, Hauge A, Becker A, Gould TJ, Clayton JB, Johnson TJ, Hunter R, Knights D, Beckman KB. 2016. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol 34:942–949. doi: 10.1038/nbt.3601. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 45.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 46.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oksanen J, Blanchette FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin D, O'Hara B, Simpson G, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2016. Vegan: community ecology package. https://cran.r-project.org/web/packages/vegan/index.html. [Google Scholar]
- 52.R Core Development Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.