ABSTRACT
The genomes of many bacteria that participate in nitrogen cycling through the process of nitrification contain putative genes associated with acyl-homoserine lactone (AHL) quorum sensing (QS). AHL QS or bacterial cell-cell signaling is a method of bacterial communication and gene regulation and may be involved in nitrogen oxide fluxes or other important phenotypes in nitrifying bacteria. Here, we carried out a broad survey of AHL production in nitrifying bacteria in three steps. First, we analyzed the evolutionary history of AHL synthase and AHL receptor homologs in sequenced genomes and metagenomes of nitrifying bacteria to identify AHL synthase homologs in ammonia-oxidizing bacteria (AOB) of the genus Nitrosospira and nitrite-oxidizing bacteria (NOB) of the genera Nitrococcus, Nitrobacter, and Nitrospira. Next, we screened cultures of both AOB and NOB with uncharacterized AHL synthase genes and AHL synthase-negative nitrifiers by a bioassay. Our results suggest that an AHL synthase gene is required for, but does not guarantee, cell density-dependent AHL production under the conditions tested. Finally, we utilized mass spectrometry to identify the AHLs produced by the AOB Nitrosospira multiformis and Nitrosospira briensis and the NOB Nitrobacter vulgaris and Nitrospira moscoviensis as N-decanoyl-l-homoserine lactone (C10-HSL), N-3-hydroxy-tetradecanoyl-l-homoserine lactone (3-OH-C14-HSL), a monounsaturated AHL (C10:1-HSL), and N-octanoyl-l-homoserine lactone (C8-HSL), respectively. Our survey expands the list of AHL-producing nitrifiers to include a representative of Nitrospira lineage II and suggests that AHL production is widespread in nitrifying bacteria.
IMPORTANCE Nitrification, the aerobic oxidation of ammonia to nitrate via nitrite by nitrifying microorganisms, plays an important role in environmental nitrogen cycling from agricultural fertilization to wastewater treatment. The genomes of many nitrifying bacteria contain genes associated with bacterial cell-cell signaling or quorum sensing (QS). QS is a method of bacterial communication and gene regulation that is well studied in bacterial pathogens, but less is known about QS in environmental systems. Our previous work suggested that QS might be involved in the regulation of nitrogen oxide gas production during nitrite metabolism. This study characterized putative QS signals produced by different genera and species of nitrifiers. Our work lays the foundation for future experiments investigating communication between nitrifying bacteria, the purpose of QS in these microorganisms, and the manipulation of QS during nitrification.
KEYWORDS: Nitrobacter, Nitrosospira, Nitrospira, acyl-homoserine lactone, ammonia oxidation, nitrification, nitrite oxidation, quorum sensing
INTRODUCTION
Nitrification, the aerobic sequential oxidation of ammonia (NH3) to nitrite (NO2−) and then to nitrate (NO3−), has a profound impact on environmental nitrogen (N) cycling (1). In particular, nitrification contributes to N availability for plants, N oxide gas emissions, and NO2− and NO3− leaching that leads to eutrophication (1, 2). Both the rate of nitrification and the coupling of NH3 oxidation to NO2− oxidation may influence the above-mentioned processes. During nitrification, high NH3 oxidation rates are associated with the increased production of nitric oxide (NO) and the greenhouse gas nitrous oxide (N2O), collectively called N oxide gases (3–5). The transient accumulation of NO2− is also associated with N oxide emissions and potential toxic effects (6–9). Finally, the accumulation of NO3− often leads to leaching and eutrophication (2). A greater understanding of the nitrification process is important for efficient fertilization of agricultural crops, wastewater treatment, and control of N oxide emissions.
Diverse microorganisms participate in nitrification. NH3 oxidation is generally carried out by NH3-oxidizing bacteria (AOB) and NH3-oxidizing archaea (AOA), while NO2−-oxidizing bacteria (NOB) carry out nitrite oxidation (1, 10–13). In addition, complete ammonia oxidation to nitrate (comammox) was discovered in Nitrospira lineage II, a subset of the NOB (14, 15). Genome sequencing of many nitrifying bacteria revealed genes associated with bacterial cell-cell signaling or quorum sensing (QS), via N-acyl-homoserine lactone (AHL) chemical signals (also referred to as autoinducers) (14–21). The role of AHLs in nitrifying bacteria is largely unknown, but a quorum-quenching transcriptome-sequencing (mRNA-Seq) study on the NOB Nitrobacter winogradskyi suggested that AHL QS regulates N oxide fluxes during the metabolism of NO2− (22).
Bacterial QS is a widespread process that uses diffusible chemical signals to coordinate gene expression in response to cell density, diffusion dynamics, and spatial distribution (23–25). QS through the production of AHLs, the best-studied chemical signal or autoinducer, can control a variety of different cooperative and stress-associated phenotypes, such as exoenzyme secretion, exopolysaccharide production, biofilm formation, luminescence, conjugation, and adaptation to starvation (25–27). AHL QS generally employs a LuxI homolog autoinducer synthase and a LuxR homolog signal receptor/transcription factor (24). The continuous basal expression of the autoinducer synthase produces AHLs that initially diffuse or are transported out of the cell. When a critical concentration is reached, AHLs make their way back into the cell and are bound by the signal receptor. The receptor/transcription factor activates a variety of genes, generally including the autoinducer synthase. This process often creates a feed-forward loop that coordinates gene expression in whole populations of cells (25).
AHL production has been demonstrated for only a few nitrifying bacteria. In the AOB Nitrosomonas europaea, AHLs were detected in the culture supernatant, but its genome lacks any known AHL synthase or AHL receptor homologs (28, 29). The genome of the AOB Nitrosospira multiformis contains putative QS genes, and AHLs were previously identified by heterologous expression, but that study was unable to detect AHLs in pure cultures (17, 30). The NOB Nb. winogradskyi was shown to produce AHLs and express a QS-controlled phenotype in previous studies (22, 31). A subsequent study identified the same AHLs in Nb. winogradskyi and observed lower concentrations of AHLs during mixotrophic growth (32). In addition, a metagenomic clone of an AHL synthase potentially from the phylum Nitrospirae, which includes NOB and comammox bacteria, produced AHLs (33). Due to the previous discrepancies between genomic and phenotype evidence, some confusion remains about which genes are responsible for AHL production and how ubiquitous the production of AHLs is in nitrifying bacteria.
In this study, we investigated putative AHL QS genes and AHL production in diverse nitrifying bacteria through genomic analyses and screening of pure cultures. We thoroughly screened genomic databases for AHL autoinducer synthase LuxI homologs and autoinducer receptor/transcription factor LuxR homologs in nitrifying bacteria and analyzed the evolutionary history of LuxI and LuxR homologs in nitrifiers. Next, we addressed the discrepancy between genomic and phenotypic evidence by screening pure cultures of nitrifying bacteria both with and without putative QS genes. Finally, we identified AHLs produced by species of the genera Nitrosospira, Nitrobacter, and Nitrospira by ultraperformance liquid chromatography-mass spectrometry (UPLC-MS).
RESULTS
AHL QS genes form distinct clades in AOB, NOB, and comammox bacteria.
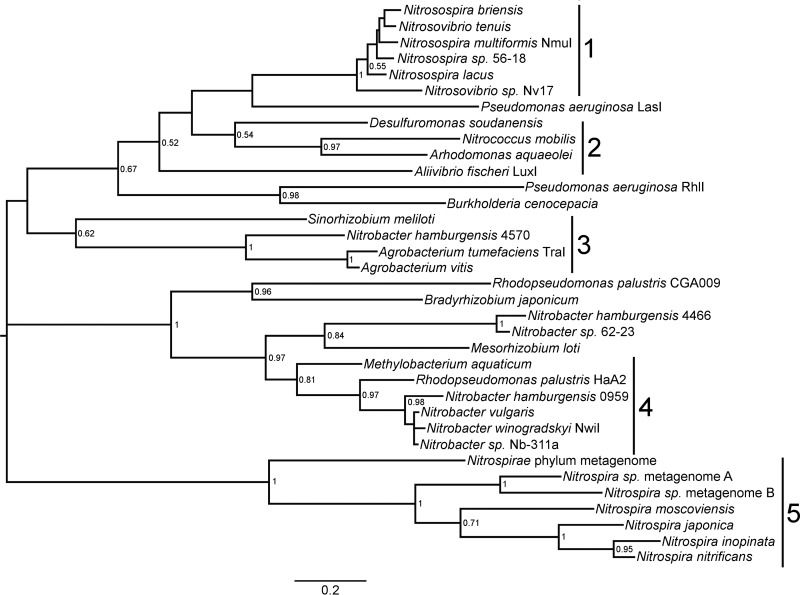
In order to identify putative genes associated with AHL QS, Nb. winogradskyi LuxI/R homologs (NwiI/R) were used to perform a comprehensive BLAST search of nitrifier genomes in the KEGG and NCBI databases. This search identified both novel putative QS genes and previously characterized genes of the genera Nitrosospira, Nitrococcus, Nitrobacter, and Nitrospira (see Data Set S1 in the supplemental material). The identification of QS gene homologs in the genus Nitrococcus and in Nitrospira lineage II was particularly interesting, as AHL QS genes had not been reported previously for these genera in the literature. The amino acid sequences of the identified QS genes were aligned with MUSCLE, and MEGA was used to construct a phylogenetic tree of LuxI and LuxR homologs identified in nitrifying bacteria along with several well-known QS-proficient bacteria (Fig. 1, Fig. S1, and Data Set S1). Phylogenetic analysis of LuxI autoinducer synthase homolog sequences showed better overall bootstrap support than that for LuxR homolog sequences, so we focused our analysis on autoinducer synthase genes (Fig. 1 and Fig. S1). As previously shown for other QS-proficient bacteria, the evolutionary history of LuxI and LuxR homologs in nitrifiers loosely resembles the 16S rRNA phylogeny, suggesting an ancient origin of AHL QS genes in these bacteria (34).
FIG 1.
Phylogenetic tree of selected LuxI homologs from nitrifiers and other bacteria. The bar indicates the mean number of substitutions per site. Bootstrap values from 500 resamplings are shown only for nodes with values of 0.5 or higher. Numbered bars indicate clades 1 through 5, as described in Results. The protein name or gene number of the LuxI homolog follows the genus, species, and strain designation, if applicable.
The autoinducer synthase genes identified in nitrifying bacteria formed five distinct clades with important distinguishing features (Fig. 1). Clade 1 included members of the genera Nitrosospira and Nitrosovibrio and was the only clade that included AOB with putative AHL QS genes (Fig. 1). Clade 1 LuxI homologs showed on average 28 to 30% identity to Nb. winogradskyi NwiI. LuxI/R homologs were previously identified in Nss. multiformis but have not been characterized in other AOB (30). Clade 2 contained only one known nitrifying bacterium, the marine NOB Nitrococcus mobilis (Fig. 1). In our analysis, the Nc. mobilis autoinducer synthase gene grouped with autoinducer synthase genes from QS-proficient halophiles and sulfur-reducing bacteria (Arhodomonas aquaeolei and Desulfuromonas soudanensis, respectively). The Nc. mobilis LuxI homolog was 24% similar to NwiI and has not been characterized previously. Clade 3 contained LuxI homologs associated with the conjugation of the Ti plasmid commonly associated with Agrobacterium tumefaciens (35) (Fig. 1). The genome of Nitrobacter hamburgensis contains three plasmids possessing some conjugation genes similar to those on the Ti plasmid and one autoinducer synthase belonging to clade 3, showing 28% identity to NwiI (18). Clade 4 included all known Nitrobacter species, including Nb. winogradskyi, Nb. hamburgensis, and Nb. vulgaris, and also includes metabolically diverse members of the Rhizobiales, such as Rhodopseudomonas palustris (Fig. 1). Although the LuxI homologs from these NOB showed 86 to 100% identity to NwiI, two autoinducer synthase genes from a Nb. hamburgensis plasmid and a metagenome formed a small separate clade with only 39 to 44% identity. Finally, clade 5 contained LuxI homologs belonging to the phylum Nitrospirae (Fig. 1). Our search identified several autoinducer synthase genes belonging to Nitrospira lineage II NOB and the two recently identified complete-ammonia-oxidizing (comammox) bacteria (Fig. 1). These LuxI homologs showed 25 to 28% identity to NwiI, and AHL QS genes have not been reported previously for Nitrospira.
Production of AHLs by Nitrosospira, Nitrobacter, and Nitrospira is cell density dependent during batch culturing.
Pure cultures of nitrifiers with sequenced genomes and uncharacterized autoinducer synthase genes were selected from each clade to screen for AHL production (Table 1). We also chose to screen the AOB Nss. multiformis since a previous characterization study was unable to detect AHLs from pure cultures (30). Because previous research reported AHL production by the NH3 oxidizer Nm. europaea, whose genome does not contain known autoinducer synthase or receptor genes, we also evaluated several nitrifiers with sequenced genomes that do not contain LuxI/R homologs (28) (Table 1).
TABLE 1.
Bacterial strains and plasmids
| Genus and species | Type of bacteriuma | QSb | Strain designation | Reference(s) |
|---|---|---|---|---|
| Nitrosospira multiformis | AOB | +/+ | ATCC 25196T | 17 |
| Nitrosospira briensis | AOB | +/+ | C-128 | 20 |
| Nitrosomonas europaea | AOB | −/− | ATCC 19718 | 29 |
| Nitrosomonas eutropha | AOB | −/− | C91 | 62 |
| Nitrosococcus oceani | AOB | −/− | ATCC 19707 | 63 |
| Nitrobacter hamburgensis | NOB | +/− | X14 | 18, 64 |
| Nitrobacter vulgaris | NOB | +/+ | AB1 | 21, 65 |
| Nitrococcus mobilis | NOB | +/− | Nb−231 | 48 |
| Nitrospina gracilis | NOB | −/− | 3/211 | 48, 66 |
| Nitrospira defluvii | NOB | −/− | A17 | 40, 67 |
| Nitrospira moscoviensis | NOB | +/+ | M1 | 19, 41 |
| Agrobacterium tumefaciens | −/− | KYC55(pJZ372)(pJZ384)(pJZ410) | 36, 54 |
Ammonia-oxidizing bacterium (AOB) or nitrite-oxidizing bacterium (NOB).
Data indicate whether the species genome contains a putative AHL synthase (LuxI homolog) and a putative AHL receptor (LuxR homolog) and if AHLs were detected in pure cultures by a bioassay, respectively. For example, +/+ indicates that the genome contains QS genes and that AHLs were detected.
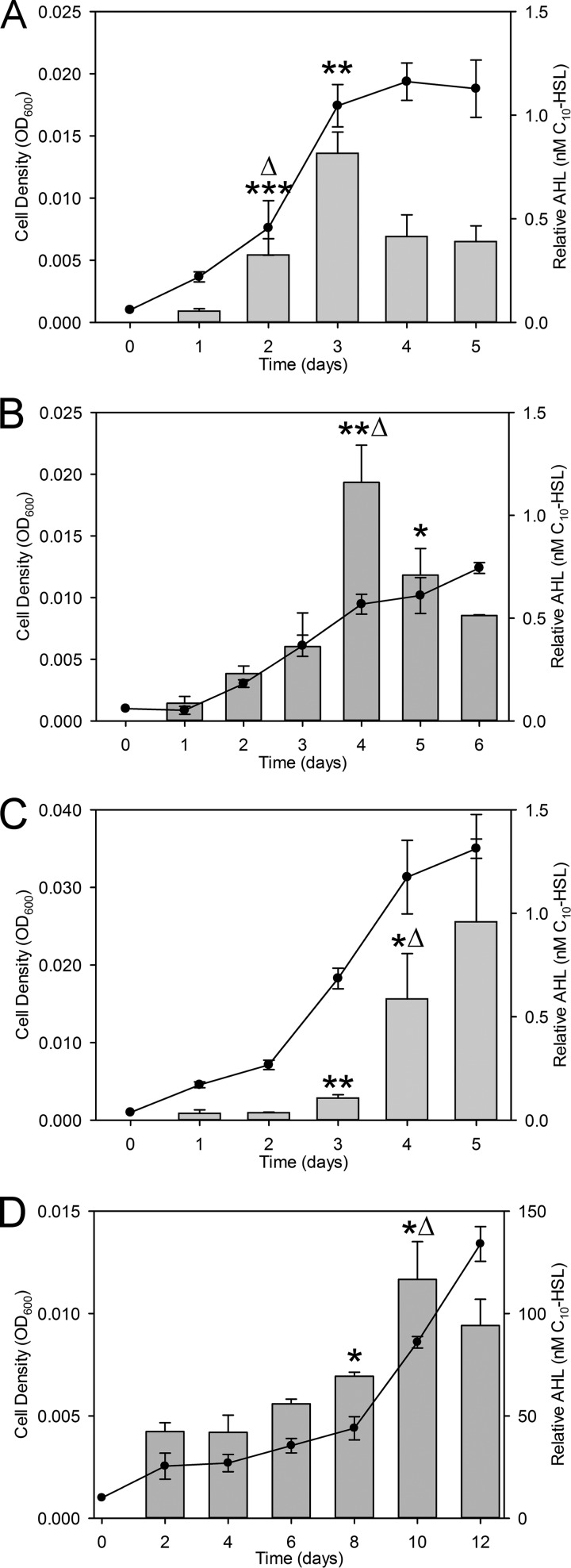
To initially screen nitrifying pure cultures for AHL production, we used a broad-range, ultrasensitive A. tumefaciens bioassay previously shown to detect a wide range of AHLs with diverse acyl chain lengths and saturation and oxidation states (36, 37). As shown in Table 1, AHLs were detected in pure cultures of the NH3 oxidizers Nss. multiformis and Nitrosospira briensis and the NO2− oxidizers Nb. vulgaris and Nitrospira moscoviensis (Fig. 2). Under the batch culture conditions tested, we were unable to detect AHLs from cultures of the NH3 oxidizers Nm. europaea, Nitrosomonas eutropha, and Nitrosococcus oceani and the NO2− oxidizers Nb. hamburgensis, Nc. mobilis, Nitrospina gracilis, and Nitrospira defluvii (Table 1). The results of this screen suggested that the presence of AHL synthase genes is required for the production of AHLs, yet the presence of putative AHL synthase and receptor genes does not guarantee the production of detectable AHLs under the applied conditions (Table 1). In all AHL-positive nitrifiers except Ns. moscoviensis, AHLs accumulated in batch cultures, and there were statistically significant increases as the optical density at 600 nm (OD600) approached 0.01 (Fig. 2A to C). Ns. moscoviensis showed a statistically significant increase in the AHL concentration at an extremely low OD600 of 0.005, and AHL concentrations also increased significantly at an OD600 of 0.01 (Fig. 2D). Since cell density, measured as the optical density, was often below an OD600 of 0.01, we also measured nitrite accumulation and consumption for AOB and NOB, which corresponded to cell density during batch culturing (Fig. 2; see also Fig. S2 in the supplemental material). In addition, there was a statistically significant increase in the amount of AHLs produced per cell, based on the OD600, on days 2, 4, 4, and 10 for Nm. multiformis, Nm. briensis, Nb. vulgaris, and Ns. moscoviensis, respectively (Fig. 2). A particularly interesting result was the accumulation of approximately 100-fold-higher relative concentrations of AHL in Ns. moscoviensis despite lower cell densities (Fig. 2D). However, all of our bioassay results were normalized to N-decanoyl-l-homoserine lactone (C10-HSL) standards. The enhanced detection of AHL in Ns. moscoviensis suggested the production of a different AHL that the bioassay was extremely sensitive to, as previously reported (36).
FIG 2.
AHL production during growth in batch culture. AHLs were detected by a bioassay during batch culture of the NH3 oxidizers Nss. multiformis (A) and Nss. briensis (B) and the NO2− oxidizers Nb. vulgaris (C) and Ns. moscoviensis (D). Circles indicate the cell density (OD600) (left y axis), and bars represent the relative AHL concentration detected (equivalent to nanomolar concentrations of C10-HSL) (right y axis) over time (days) (x axis). Relative AHL concentrations were quantified based on C10-HSL standards reported previously, but actual concentrations vary depending on the AHL being measured (22, 36). Asterisks indicate statistically significant changes in AHL concentrations compared to previous measurements (*, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 5e−4 [versus all previous measurements, as determined by a two-tailed t test]). Delta indicates a statistically significant increase in the amount of AHLs produced per OD600 compared to the amount measured the previous day (Δ, P ≤ 0.05). Values are the means of data from three independent biological replicates. Error bars indicate the standard deviations of the means.
Identification of AHLs by UPLC-MS.
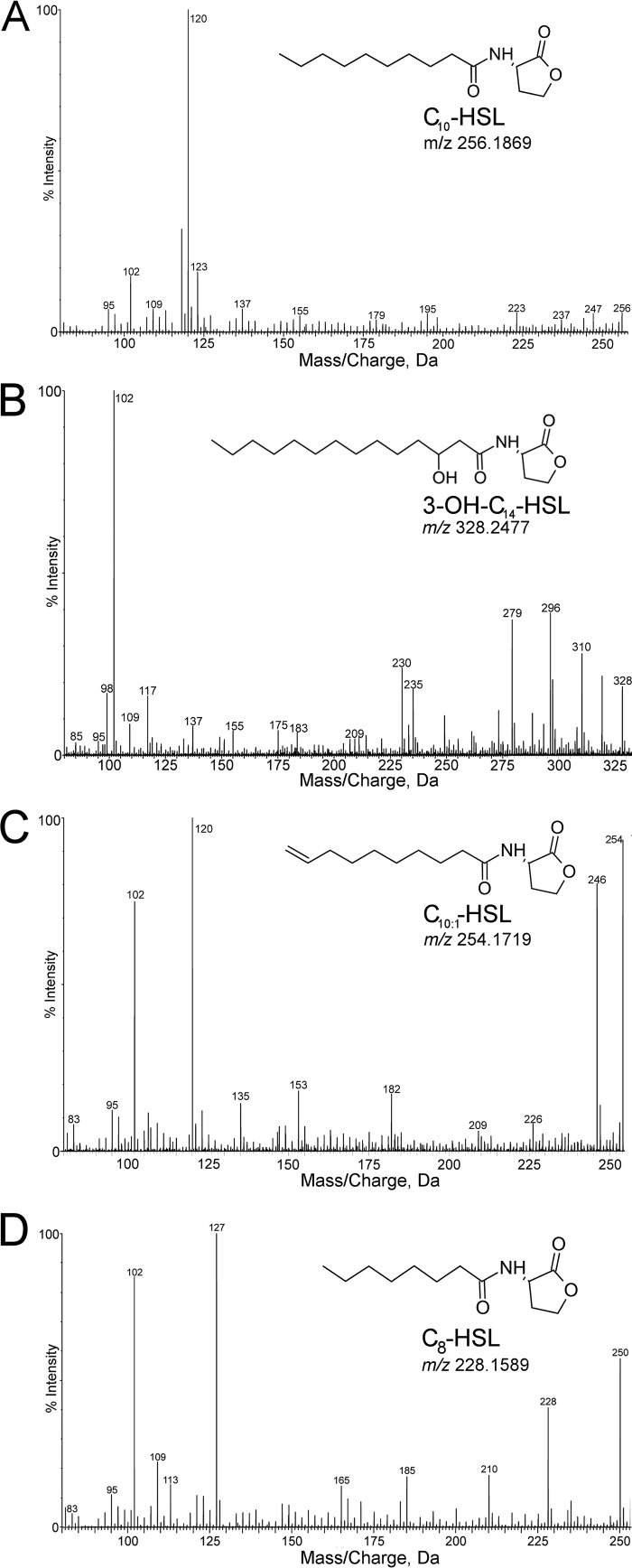
AHLs from AHL-positive nitrifiers were extracted from the batch culture supernatant and concentrated for identification by mass spectrometry. UPLC-MS analysis detected a different AHL from each species of nitrifying bacteria (Table 2 and Fig. 3). AHLs were identified based on the presence of a peak at m/z 102.055 in the extracted-ion chromatogram (XIC), which corresponds to the protonated lactone moiety. A compound with molecular ion [M + H]+ of m/z 256.1869 corresponding to C10-HSL was detected in the Nss. multiformis supernatant (Table 2 and Fig. 3A). The Nss. briensis supernatant contained a compound with [M + H]+ of m/z 328.2456, which corresponded to N-3-hydroxy-tetradecanoyl-l-homoserine lactone (3-OH-C14-HSL) (Table 2 and Fig. 3B). A compound with [M + H]+ of m/z 254.1719 corresponding to the unsaturated AHL C10:1-HSL was identified in the Nb. vulgaris supernatant (Table 2 and Fig. 3C). The Ns. moscoviensis supernatant contained a compound with [M + H]+ of m/z 228.1589, corresponding to N-octanoyl-l-homoserine lactone (C8-HSL) (Table 2 and Fig. 3D).
TABLE 2.
Chromatographic and mass spectrometric data for AHLs identified by UPLC-MS
| Bacterial species | AHL | Chemical formula | Exact mass (Da) | rT (min)a | [M + H]+ (m/z)b | Error (ppm) |
|---|---|---|---|---|---|---|
| Nss. multiformis | C10-HSL | C14H25NO3 | 255.18344 | 2.35 | 256.1869 | −3.2 |
| Nss. briensis | 3-OH-C14-HSL | C18H32NO4 | 327.24096 | 3.02 | 328.2456 | −3.2 |
| Nb. vulgaris | C10:1-HSL | C14H23NO3 | 253.16779 | 2.15 | 254.1719 | −2.1 |
| Ns. moscoviensis | C8-HSL | C12H21NO3 | 227.15214 | 2.16 | 228.1589 | −4.8 |
rT, retention time.
Experimental m/z values of protonated molecules.
FIG 3.
UPLC-MS chromatograms and structures of AHLs produced by NH3-oxidizing and NO2−-oxidizing nitrifying bacteria. Shown are extracted-ion chromatogram (XIC) fragmentation pattern spectra for protonated molecules ([M + H]+), chemical structures, and m/z of C10-HSL produced by the AOB Nss. multiformis (A), 3-OH-C14-HSL produced by the AOB Nss. briensis (B), C10:1-HSL produced by the NOB Nb. vulgaris (C), and C8-HSL produced by the NOB Ns. moscoviensis (D).
Further analysis of the XICs of each AHL confirmed the identity of the chemical structures. The m/z values listed below are rounded to the nearest whole number. The structures of C10-HSL and C10:1-HSL were compared to previously reported XICs with similar fragmentation patterns (31). The XIC for C10-HSL showed characteristic [M + H − H2O]+, [M + H − 101]+, and [M + H − 101 − H2O]+ peaks at m/z 238, 155, and 137, respectively (Fig. 3A). The XIC of C10:1-HSL showed peaks at m/z 236, 153, and 135, corresponding to [M + H − H2O]+, [M + H − 101]+, and [M + H − 101 − H2O]+, respectively, representing the loss of two H molecules due to the carbon-carbon double bond (C=C bond) (Fig. 3C). The position of the C=C bond in the acyl chain was determined to be between carbon 9 and carbon 10 based on the ion fragmentation model presented in a previous study (38). However, we were unable to clarify the isomeric form of the C=C bond of C10:1-HSL. While the XIC for 3-OH-C14-HSL differed from previously reported results, both XICs showed m/z 209, corresponding to [M + H − 101 − H2O], and different MS methods might account for other differences (Fig. 3B) (39). The XIC of the Ns. moscoviensis AHL C8-HSL showed [M + H − H2O]+, [M + H − 101]+, and [M + H − 101 − H2O]+ at m/z 210, 127, and 109, respectively, matching previously reported results (Fig. 3D) (39). In addition, a previous study showed that the A. tumefaciens bioassay used to quantify AHL concentrations is particularly sensitive to C8-HSL, which explains the enhanced detection of AHLs in Ns. moscoviensis batch cultures (36).
DISCUSSION
Overview.
In this study, genomic databases were searched for AHL synthase and receptor genes to comprehensively screen for the distribution of putative AHL QS genes in nitrifying bacteria. Based on our search and subsequent phylogenetic analyses, we selected 10 nitrifying bacteria, both with and without putative AHL synthase genes, to screen for AHL production during chemolithoautotrophic growth. Our results suggested that the presence of an AHL synthase gene homolog is required for, but does not guarantee, the production of AHLs by nitrifying bacteria under the conditions tested. In particular, we were unable to detect AHLs from pure cultures of Nm. europaea, an AOB without an AHL synthase gene, despite previous reports of AHL production, and we were also unable to detect AHLs from pure cultures of Nc. mobilis and Nb. hamburgensis, NOB that have AHL synthase genes (18, 28). Finally, we identified AHLs produced by Nss. multiformis, Nss. briensis, Nb. vulgaris, and Ns. moscoviensis and measured cell density-dependent increases in AHL production during batch culturing. Our results suggest that these nitrifiers may have functional AHL QS systems. Our work confirmed and expanded on previous reports of AHL QS genes and phenotypes for the genera Nitrosospira and Nitrobacter, identified AHL production in the genus Nitrospira, and suggested that AHL QS genes may be widespread in nitrifying bacteria.
Evolution of AHL QS in nitrifying bacteria.
Our analysis of the evolutionary history of putative autoinducer genes in nitrifying bacteria has expanded on previous work focused on Nb. winogradskyi and QS-proficient Rhizobiales (31). Similar to previous work, our analysis identified both a Nitrobacter-specific clade (clade 4) and a Ti plasmid-associated clade (clade 3) of autoinducer synthase homologs (31). By expanding our phylogenetic analysis to other genera and species of AOB and NOB, we identified several more clades specific to each group of nitrifiers and their environments (clades 1, 2, and 5). Our analysis suggested an ancient origin of QS genes in nitrifying bacteria, without recent horizontal gene transfer, since the autoinducer synthase gene phylogeny closely resembles the 16S rRNA phylogeny (34).
The discovery of putative AHL synthase and receptor genes within the genus Nitrospira is particularly interesting, as AHL QS genes had not been previously characterized for these NOB in the literature. Surprisingly, we found putative QS genes only within Nitrospira species belonging to the previously described lineage II that includes both NOB and comammox bacteria (14, 15, 40, 41). Our findings suggest that a fundamentally different selective pressure led to AHL QS genes being only either acquired or maintained in one of the six lineages of Nitrospira. Future experiments are needed to determine the purpose of putative AHL QS genes in Nitrospira and confirm if other less-studied Nitrospira lineages also contain putative AHL QS genes.
Determination of whether chemical language exists during nitrification.
Our survey of AHL QS in nitrifying bacteria forms a foundation for future studies on chemical communication in environmental bacteria. While many studies have focused on model heterotroph bacteria that are amenable to genetic manipulation, many important environmental bacteria, such as nitrifiers, are understudied (24). The goal of this study was to identify which nitrifying bacteria produce AHLs and which AHLs are produced. By screening nitrifiers closely related to species with previously characterized AHL QS genes (Nitrosospira and Nitrobacter species), our study identified potential themes in the chemical signaling of clades 1 and 4. Nss. multiformis and Nss. briensis produced drastically different AHLs, suggesting that although their AHL synthases show 88% identity at the amino acid level, their AHL synthases function differently, possibly due to different fatty acid pools within the cell (42). In addition, based on data from previous studies, the Nss. multiformis AHL synthase NmuI produced different AHLs in pure culture (C10-HSL) than when the gene was expressed in Escherichia coli (C14-HSL and 3-oxo-C14-HSL) (30). On the other hand, Nb. vulgaris produced the same unsaturated AHL, C10:1-HSL, as previously characterized for Nb. winogradskyi, and the Nb. vulgaris AHL synthase gene is 94% identical (31). These results suggest the likelihood that there is an amino acid identity threshold for AHL synthases to produce the same AHLs.
A coupled AHL synthase gene and an AHL receptor gene generally suggest a within-species function for QS (43). However, if different species of bacteria produce similar AHLs, we hypothesize that they could also perceive each other's AHLs when growing in close proximity. Previous work suggested that AHLs with similar tail lengths can be identified by noncognate receptor proteins (43). Of the AHL-producing nitrifiers known, AHL cross talk between the AOB Nss. multiformis and the NOB Nb. winogradskyi and Nb. vulgaris may be possible, as they all utilize a similar AHL with a 10-carbon acyl tail and can be found in soil environments (12, 13, 17, 31). In addition, AHL receptor homologs vary in signal specificity, so possible cross talk with a Nitrospira species that produces C8-HSL, similar to Ns. moscoviensis, may also be possible (43).
Implications for future studies of AHL QS in environmental and engineered systems.
In environmental systems such as soils, AHL QS offers an attractive model for potential chemical communication between AOB and NOB. We are particularly interested in how AOB and ammonia-oxidizing archaea remain coupled with NOB to avoid the accumulation of nitrite in the environment, especially considering the role of nitrite in the production of nitrogen oxide gases (7–9, 44). AHL QS may serve to facilitate nitrification coupling or, alternatively, may contribute or reduce nitrogen oxide gas emissions indirectly. Research has shown that nitrite accumulates transiently in soils under some circumstances, and whether or not there might be a role for AHL QS in the recovery of NO2− consumption is unknown (7–9). Indeed, in a previous study involving quorum-quenching mRNA-Seq techniques, the results suggested that AHL QS might play a role in nitrogen oxide emissions from NO2− metabolism by the NOB Nb. winogradskyi (22).
There is also an opportunity for future studies of the contribution of AHL QS to engineered systems such as wastewater treatment systems. Most wastewater treatment systems depend on biofilm consortiums of heterotrophs and nitrifiers to effectively treat wastewater, and QS can play an important role in biofilm development (45). In addition, previous studies suggested a role for AHL QS in mixed populations carrying out anaerobic ammonium oxidation (anammox) (46, 47). Although there has been recent interest in cycles of AHL production and destruction, via quorum quenching, in wastewater treatment systems, most of those studies focused on heterotrophs (45). Future studies that focus on the potential for AHL QS in nitrifying bacteria are needed.
MATERIALS AND METHODS
Chemicals.
N-Decanoyl-dl-homoserine lactone (C10-HSL), analytical-grade acetonitrile, and formic acid were purchased from Sigma-Aldrich (St. Louis, MO). High-performance liquid chromatography (HPLC)-grade ethyl acetate and acetic acid were purchased from EMD Chemicals (Darmstadt, Germany) and VWR International (Radnor, PA), respectively.
Bacterial strains and growth medium.
Bacterial strains used in this study are outlined in Table 1. All nitrifying bacteria were cultivated in different formulations of mineral salts medium as previously described, with minor modifications (48–53). Nitrosospira species were cultivated at 28°C in mineral salts medium containing 2.5 to 10 mM (NH4)2SO4, 15 mM HEPES, 3.2 mM Na2CO3, 0.75 mM MgSO4, 1 mM KCl, 10 μM FeCl3, 16.7 μM EDTA, 0.2 mM CaCl2, 100 μM KH2PO4, 10 μM NaH2PO4, 1 μM CuSO4, 0.6 μM Na2MoO4, 1.59 μM MnCl2, 0.6 μM CoCl2, and 0.96 μM ZnSO4. The medium pH was adjusted to 8.0 with KOH. Nitrosococcus oceani was cultivated at 30°C in the same mineral salts medium with the addition of 0.5 mM NaCl, 50 μM KH2PO4, and 5 μM NaH2PO4, and the pH was adjusted to 7.5. The pH of actively growing cultures was adjusted with 1 M Na2CO3, and 0.4 μg ml−1 phenol red was added to monitor the pH. Nitrosomonas species were cultivated at 30°C in mineral salts medium containing 2.5 to 12.5 mM (NH4)2SO4, 4.72 mM Na2CO3, 0.75 mM MgSO4, 1 mM KCl, 10 μM FeCl3, 16.7 μM EDTA, 0.2 mM CaCl2, 25 mM KH2PO4, 2.5 mM NaH2PO4, 1 μM CuSO4, 0.6 μM Na2MoO4, 1.59 μM MnCl2, 0.6 μM CoCl2, and 0.96 μM ZnSO4, and the pH was adjusted to 8. All NOB were cultivated at 28°C, except for Nitrospira moscoviensis, which was cultivated at 37°C. Nitrobacter hamburgensis was cultivated in mineral salts medium containing 60 mM NaNO2, 3.5 mM KH2PO4, 0.52 mM NaH2PO4, 0.75 mM MgSO4, 10 μM FeCl3, 16.7 μM EDTA, 0.28 Na2CO3, 0.2 mM CaCl2, 1 μM CuSO4, 0.6 μM Na2MoO4, 1.59 μM MnCl2, 0.6 μM CoCl2, and 0.96 μM ZnSO4, and the pH was adjusted to 8.0. Nitrobacter vulgaris, Nitrospira defluvii, and Ns. moscoviensis were cultivated in mineral salts medium containing 100 μM to 30 mM NaNO2, 70 μM CaCO3, 8.6 mM NaCl, 0.2 mM MgSO4, 1.1 mM KH2PO4, 0.2 μM MnSO4, 0.8 μM H3BO3, 0.15 μM ZnSO4, 32 nM (NH4)6Mo7O24, 3.5 μM FeSO4, and 0.1 μM CuSO4, and the pH was adjusted to 7.5. Nitrococcus mobilis and Nitrospina gracilis were cultivated in undefined seawater medium consisting of 70% 0.2-μm-filtered, autoclaved seawater with the addition of 34 μM CaCl2, 406 μM MgSO4, 3.6 μM FeCl3, 6 μM EDTA, 12.5 μM KH2PO4, 103 nM Na2MoO4, 253 nM MnCl2, 2.1 nM CoCl2, 87 nM ZnSO4, and 24 nM CuSO4. Seawater medium contained 100 to 30 mM NaNO2 depending on the culture activity, and the pH was adjusted to 7.5. Agrobacterium tumefaciens KYC55(pJZ372)(pJZ384)(pJZ410) was cultivated, preinduced, and used for the detection of AHLs as described previously (36, 54).
Growth conditions.
Batch cultures of nitrifying bacteria were inoculated to an OD600 of 0.001 and grown in Erlenmeyer flasks in a rotatory shaker at 100 rpm in mineral salts medium as outlined above. Experimental cultures were monitored at regular intervals to check the OD600, and the growth medium was analyzed for NO2− concentrations by the Griess assay (55) and for the presence of AHL (bioassay).
Bioinformatic analyses.
The National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/), the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/), and the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were used to identify LuxI autoinducer synthase homologs and LuxR receptor/transcription factor homologs in nitrifiers and other bacteria (see Data Set S1 in the supplemental material) (56, 57). MUSCLE was used for multiple alignments of amino acid sequences (58, 59), and phylogenetic analyses were conducted by using MEGA7 (60). The evolutionary history of LuxI and LuxR homologs in nitrifying bacteria was inferred by using the maximum likelihood method, based on the JTT matrix model (61). Phylogenetic trees were visualized by using FigTree 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
AHL bioassay.
AHLs were extracted by liquid-liquid extraction from 10 ml of the culture supernatant with acidified ethyl acetate and concentrated as previously described (31). Extracts were screened for AHLs by a broad-range bioassay (36, 54) and quantified as relative nanomolar concentrations of C10-HSL by comparison to known concentrations of C10-HSL standards, as described previously (22).
UPLC-MS.
AHL structures were identified as previously described, with modifications (31). The supernatants of replicate 1-liter batch cultures were collected by centrifugation during peak AHL accumulation and extracted with acidified ethyl acetate. Extracts were evaporated under filtered air and reconstituted in water containing 20% (vol/vol) acetonitrile and 0.1% (vol/vol) formic acid.
Samples were separated on an Acquity UPLC BEH C18 column (2.1 by 50 mm, with a 1.7-μm particle size; Waters, Milford, MA) using an Acquity I-class UPLC instrument (Waters, Milford, MA) at a total flow rate of 0.4 ml min−1. The elution program consisted of an aqueous solution of 5% (vol/vol) acetonitrile for 0.2 min, followed by a linear increase to 90% (vol/vol) acetonitrile over 3 min and a hold of 1 min at 90% acetonitrile, which was followed by reequilibration for 1 min with 5% acetonitrile. Mass spectrometry was performed by using a Synapt G2 instrument (Waters, Milford, MA) in positive-ion ionization (ES+) mode. The instrument was operated at a source temperature of 120°C, a desolution temperature of 550°C, and a spray voltage of 2.8 kV. MS and product ion data were acquired by MSe continuum. MSe function 1 (low energy) was set to a trap collision energy of 4 V and a transfer collision energy of 6 V. MSe function 2 (high energy) was set to a ramp collision energy of 20 to 30 V, and the ramp transfer collision energy was off. Product ion spectra were searched for a protonated ([M + H+]) lactone moiety (C4H8NO2 [M + H]+, m/z 102.055) since AHLs consistently fragment to this product ion. Data were analyzed by using MassLynx V4.1 (Waters, Milford, MA). Chemical structures were drawn with ChemSketch Freeware 2012 (ACD Labs, Ontario, Canada).
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeanette Norton, Lisa Stein, and Martin Klotz for providing pure cultures of NH3-oxidizing bacteria; Jun Zhu for providing Agrobacterium tumefaciens bioassay strain KYC55(pJZ372)(pJZ384)(pJZ410) for this and previous projects; Jeffrey Morré for mass spectrometry expertise; and Andrew Giguere, Anne Taylor, Neeraja Vajrala, and Ashley Waggoner for helpful advice. We acknowledge the Biomolecular Mass Spectrometry Core of the Environmental Health Sciences Core Center at OSU.
This work was supported by USDA-NIFA postdoctoral fellowship award no. 2016-67012-24691 (B.L.M.) and the Oregon Agricultural Experiment Station (L.A.S.-S.). Mass spectrometry was supported in part by award no. P30ES000210 from the NIEHS and NIH.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01540-17.
REFERENCES
- 1.Ward BB, Arp DJ, Klotz MG (ed). 2011. Nitrification. ASM Press, Washington, DC. [Google Scholar]
- 2.Galloway JN, Townsend AR, Erisman JW, Bekunda M, Cai Z, Freney JR, Martinelli LA, Seitzinger SP, Sutton MA. 2008. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320:889–892. doi: 10.1126/science.1136674. [DOI] [PubMed] [Google Scholar]
- 3.Schreiber F, Wunderlin P, Udert KM, Wells GF. 2012. Nitric oxide and nitrous oxide turnover in natural and engineered microbial communities: biological pathways, chemical reactions, and novel technologies. Front Microbiol 3:372. doi: 10.3389/fmicb.2012.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Law Y, Ni BJ, Lant P, Yuan Z. 2012. N2O production rate of an enriched ammonia-oxidising bacteria culture exponentially correlates to its ammonia oxidation rate. Water Res 46:3409–3419. doi: 10.1016/j.watres.2012.03.043. [DOI] [PubMed] [Google Scholar]
- 5.Mellbye BL, Giguere A, Chaplen F, Bottomley PJ, Sayavedra-Soto LA. 2016. Steady-state growth under inorganic carbon limitation conditions increases energy consumption for maintenance and enhances nitrous oxide production in Nitrosomonas europaea. Appl Environ Microbiol 82:3310–3318. doi: 10.1128/AEM.00294-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Court MN, Stephen RC, Waid JS. 1962. Toxic effect of urea on plants: nitrite toxicity arising from use of urea as a fertilizer. Nature 194:1263–1265. doi: 10.1038/1941263a0. [DOI] [Google Scholar]
- 7.Venterea RT. 2007. Nitrite-driven nitrous oxide production under aerobic soil conditions: kinetics and biochemical controls. Glob Change Biol 13:1798–1809. doi: 10.1111/j.1365-2486.2007.01389.x. [DOI] [Google Scholar]
- 8.Gelfand I, Yakir D. 2008. Influence of nitrite accumulation in association with seasonal patterns and mineralization of soil nitrogen in a semi-arid pine forest. Soil Biol Biochem 40:415–424. doi: 10.1016/j.soilbio.2007.09.005. [DOI] [Google Scholar]
- 9.Giguere AT, Taylor AE, Suwa Y, Myrold DD, Bottomley PJ. 2017. Uncoupling of ammonia oxidation from nitrite oxidation: impact upon nitrous oxide production in non-cropped Oregon soils. Soil Biol Biochem 104:30–38. doi: 10.1016/j.soilbio.2016.10.011. [DOI] [Google Scholar]
- 10.Sayavedra-Soto LA, Arp DJ. 2011. Ammonia-oxidizing bacteria: their biochemistry and molecular biology, p 11–37. In Ward BB, Arp DJ, Klotz MG (ed), Nitrification. ASM Press, Washington, DC. [Google Scholar]
- 11.Hidetoshi U, Martens-Habbena W, Stahl DA. 2011. Physiology and genomics of ammonia-oxidizing archaea, p 117–155. In Ward BB, Arp DJ, Klotz MG (ed), Nitrification. ASM Press, Washington, DC. [Google Scholar]
- 12.Starkenburg SR, Spieck E, Bottomley PJ. 2011. Metabolism and genomics of nitrite-oxidizing bacteria: emphasis on studies of pure cultures and of Nitrobacter species, p 267–293. In Ward BB, Arp DJ, Klotz MG (ed), Nitrification. ASM Press, Washington, DC. [Google Scholar]
- 13.Daims H, Lucker S, Paslier DL, Wagner M. 2011. Diversity, environmental genomics, and ecophysiology of nitrite-oxidizing bacteria, p 295–322. In Ward BB, Arp DJ, Klotz MG (ed), Nitrification. ASM Press, Washington, DC. [Google Scholar]
- 14.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. 2015. Complete nitrification by Nitrospira bacteria. Nature 528:504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Kessel MA, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJ, Kartal B, Jetten MS, Lucker S. 2015. Complete nitrification by a single microorganism. Nature 528:555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starkenburg SR, Chain PS, Sayavedra-Soto LA, Hauser L, Land ML, Larimer FW, Malfatti SA, Klotz MG, Bottomley PJ, Arp DJ, Hickey WJ. 2006. Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl Environ Microbiol 72:2050–2063. doi: 10.1128/AEM.72.3.2050-2063.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norton JM, Klotz MG, Stein LY, Arp DJ, Bottomley PJ, Chain PS, Hauser LJ, Land ML, Larimer FW, Shin MW, Starkenburg SR. 2008. Complete genome sequence of Nitrosospira multiformis, an ammonia-oxidizing bacterium from the soil environment. Appl Environ Microbiol 74:3559–3572. doi: 10.1128/AEM.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starkenburg SR, Larimer FW, Stein LY, Klotz MG, Chain PS, Sayavedra-Soto LA, Poret-Peterson AT, Gentry ME, Arp DJ, Ward B, Bottomley PJ. 2008. Complete genome sequence of Nitrobacter hamburgensis X14 and comparative genomic analysis of species within the genus Nitrobacter. Appl Environ Microbiol 74:2852–2863. doi: 10.1128/AEM.02311-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koch H, Lucker S, Albertsen M, Kitzinger K, Herbold C, Spieck E, Nielsen PH, Wagner M, Daims H. 2015. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc Natl Acad Sci U S A 112:11371–11376. doi: 10.1073/pnas.1506533112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice MC, Norton JM, Valois F, Bollmann A, Bottomley PJ, Klotz MG, Laanbroek HJ, Suwa Y, Stein LY, Sayavedra-Soto L, Woyke T, Shapiro N, Goodwin LA, Huntemann M, Clum A, Pillay M, Kyrpides N, Varghese N, Mikhailova N, Markowitz V, Palaniappan K, Ivanova N, Stamatis D, Reddy TB, Ngan CY, Daum C. 2016. Complete genome of Nitrosospira briensis C-128, an ammonia-oxidizing bacterium from agricultural soil. Stand Genomic Sci 11:46. doi: 10.1186/s40793-016-0168-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellbye BL, Davis EW II, Spieck E, Chang JH, Bottomley PJ, Sayavedra-Soto LA. 2017. Draft genome sequence of Nitrobacter vulgaris strain Ab1, a nitrite-oxidizing bacterium. Genome Announc 5:e00290-17. doi: 10.1128/genomeA.00290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellbye BL, Giguere AT, Bottomley PJ, Sayavedra-Soto LA. 2016. Quorum quenching of Nitrobacter winogradskyi suggests that quorum sensing regulates fluxes of nitrogen oxide(s) during nitrification. mBio 7:e01753-16. doi: 10.1128/mBio.01753-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hense BA, Kuttler C, Muller J, Rothballer M, Hartmann A, Kreft JU. 2007. Does efficiency sensing unify diffusion and quorum sensing? Nat Rev Microbiol 5:230–239. doi: 10.1038/nrmicro1600. [DOI] [PubMed] [Google Scholar]
- 24.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. 2013. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu Rev Microbiol 67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 25.Hense BA, Schuster M. 2015. Core principles of bacterial autoinducer systems. Microbiol Mol Biol Rev 79:153–169. doi: 10.1128/MMBR.00024-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellbye B, Schuster M. 2011. More than just a quorum: integration of stress and other environmental cues in acyl-homoserine lactone signaling networks. In Storz G, Hengge R (ed), Bacterial stress responses, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 27.Mellbye B, Schuster M. 2014. Physiological framework for the regulation of quorum sensing-dependent public goods in Pseudomonas aeruginosa. J Bacteriol 196:1155–1164. doi: 10.1128/JB.01223-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burton EO, Read HW, Pellitteri MC, Hickey WJ. 2005. Identification of acyl-homoserine lactone signal molecules produced by Nitrosomonas europaea strain Schmidt. Appl Environ Microbiol 71:4906–4909. doi: 10.1128/AEM.71.8.4906-4909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, Hauser L, Hooper A, Klotz M, Norton J, Sayavedra-Soto L, Arciero D, Hommes N, Whittaker M, Arp D. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J Bacteriol 185:6496. doi: 10.1128/JB.185.21.6496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao J, Ma A, Zhuang X, Zhuang G. 2014. An N-acyl homoserine lactone synthase in the ammonia-oxidizing bacterium Nitrosospira multiformis. Appl Environ Microbiol 80:951–958. doi: 10.1128/AEM.03361-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellbye BL, Bottomley PJ, Sayavedra-Soto LA. 2015. Nitrite-oxidizing bacterium Nitrobacter winogradskyi produces N-acyl-homoserine lactone autoinducers. Appl Environ Microbiol 81:5917–5926. doi: 10.1128/AEM.01103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen Q, Gao J, Liu J, Liu S, Liu Z, Wang Y, Guo B, Zhuang X, Zhuang G. 2016. A new acyl-homoserine lactone molecule generated by Nitrobacter winogradskyi. Sci Rep 6:22903. doi: 10.1038/srep22903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nasuno E, Kimura N, Fujita MJ, Nakatsu CH, Kamagata Y, Hanada S. 2012. Phylogenetically novel LuxI/LuxR-type quorum sensing systems isolated using a metagenomic approach. Appl Environ Microbiol 78:8067–8074. doi: 10.1128/AEM.01442-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray KM, Garey JR. 2001. The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology 147:2379–2387. doi: 10.1099/00221287-147-8-2379. [DOI] [PubMed] [Google Scholar]
- 35.White CE, Winans SC. 2007. Cell-cell communication in the plant pathogen Agrobacterium tumefaciens. Philos Trans R Soc Lond B Biol Sci 362:1135–1148. doi: 10.1098/rstb.2007.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu J, Chai Y, Zhong Z, Li S, Winans SC. 2003. Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl Environ Microbiol 69:6949–6953. doi: 10.1128/AEM.69.11.6949-6953.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu J, Beaber JW, More MI, Fuqua C, Eberhard A, Winans SC. 1998. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J Bacteriol 180:5398–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cataldi TR, Bianco G, Abate S, Losito I. 2011. Identification of unsaturated N-acylhomoserine lactones in bacterial isolates of Rhodobacter sphaeroides by liquid chromatography coupled to electrospray ionization-hybrid linear ion trap-Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom 25:1817–1826. doi: 10.1002/rcm.5054. [DOI] [PubMed] [Google Scholar]
- 39.Girard L, Blanchet E, Intertaglia L, Baudart J, Stien D, Suzuki M, Lebaron P, Lami R. 2017. Characterization of N-acyl homoserine lactones in Vibrio tasmaniensis LGP32 by a biosensor-based UHPLC-HRMS/MS method. Sensors (Basel) 17:906. doi: 10.3390/s17040906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nowka B, Off S, Daims H, Spieck E. 2015. Improved isolation strategies allowed the phenotypic differentiation of two Nitrospira strains from widespread phylogenetic lineages. FEMS Microbiol Ecol 91:fiu031. doi: 10.1093/femsec/fiu031. [DOI] [PubMed] [Google Scholar]
- 41.Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. 1995. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp nov and its phylogenetic relationship. Arch Microbiol 164:16–23. doi: 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- 42.Schaefer AL, Taylor TA, Beatty JT, Greenberg EP. 2002. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J Bacteriol 184:6515–6521. doi: 10.1128/JB.184.23.6515-6521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hawver LA, Jung SA, Ng WL. 2016. Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol Rev 40:738–752. doi: 10.1093/femsre/fuw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giguere AT, Taylor AE, Myrold DD, Bottomley PJ. 2015. Nitrification responses of soil ammonia-oxidizing archaea and bacteria to ammonium concentrations. Soil Sci Soc Am J 79:1366–1374. doi: 10.2136/sssaj2015.03.0107. [DOI] [Google Scholar]
- 45.Huang J, Shi Y, Zeng G, Gu Y, Chen G, Shi L, Hu Y, Tang B, Zhou J. 2016. Acyl-homoserine lactone-based quorum sensing and quorum quenching hold promise to determine the performance of biological wastewater treatments: an overview. Chemosphere 157:137–151. doi: 10.1016/j.chemosphere.2016.05.032. [DOI] [PubMed] [Google Scholar]
- 46.De Clippeleir H, Defoirdt T, Vanhaecke L, Vlaeminck SE, Carballa M, Verstraete W, Boon N. 2011. Long-chain acylhomoserine lactones increase the anoxic ammonium oxidation rate in an OLAND biofilm. Appl Microbiol Biotechnol 90:1511–1519. doi: 10.1007/s00253-011-3177-7. [DOI] [PubMed] [Google Scholar]
- 47.Tang X, Liu S, Zhang Z, Zhuang G. 2015. Identification of the release and effects of AHLs in anammox culture for bacteria communication. Chem Eng J 273:184–191. doi: 10.1016/j.cej.2015.03.045. [DOI] [Google Scholar]
- 48.Watson SW, Waterbury JB. 1971. Characteristics of two marine nitrite oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen nov sp. Arch Microbiol 77:203–230. [Google Scholar]
- 49.Krummel A, Harms H. 1982. Effect of organic-matter on growth and cell yield of ammonia-oxidizing bacteria. Arch Microbiol 133:50–54. doi: 10.1007/BF00943769. [DOI] [Google Scholar]
- 50.Norton JM, Alzerreca JJ, Suwa Y, Klotz MG. 2002. Diversity of ammonia monooxygenase operon in autotrophic ammonia-oxidizing bacteria. Arch Microbiol 177:139–149. doi: 10.1007/s00203-001-0369-z. [DOI] [PubMed] [Google Scholar]
- 51.Spieck E, Lipski A. 2011. Cultivation, growth physiology, and chemotaxonomy of nitrite-oxidizing bacteria. Methods Enzymol 486:109–130. doi: 10.1016/B978-0-12-381294-0.00005-5. [DOI] [PubMed] [Google Scholar]
- 52.Nowka B, Daims H, Spieck E. 2015. Comparison of oxidation kinetics of nitrite-oxidizing bacteria: nitrite availability as a key factor in niche differentiation. Appl Environ Microbiol 81:745–753. doi: 10.1128/AEM.02734-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pérez J, Buchanan A, Mellbye B, Ferrell R, Chang JH, Chaplen F, Bottomley PJ, Arp DJ, Sayavedra-Soto LA. 2015. Interactions of Nitrosomonas europaea and Nitrobacter winogradskyi grown in co-culture. Arch Microbiol 197:79–89. doi: 10.1007/s00203-014-1056-1. [DOI] [PubMed] [Google Scholar]
- 54.Joelsson AC, Zhu J. 2006. LacZ-based detection of acyl-homoserine lactone quorum-sensing signals. Curr Protoc Microbiol Chapter 1:Unit 1C.2. doi: 10.1002/9780471729259.mc01c02s3. [DOI] [PubMed] [Google Scholar]
- 55.Hood-Nowotny R, Hinko-Najera Umana N, Inselbacher E, Oswald-Lachouani P, Wanek W. 2010. Alternative methods for measuring inorganic, organic, and total dissolved nitrogen in soil. Soil Sci Soc Am J 74:1018–1027. doi: 10.2136/sssaj2009.0389. [DOI] [Google Scholar]
- 56.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, Raytselis Y, Sayers EW, Tao T, Ye J, Zaretskaya I. 2013. BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41:W29–W33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. [DOI] [PubMed] [Google Scholar]
- 62.Stein LY, Arp DJ, Berube PM, Chain PS, Hauser L, Jetten MS, Klotz MG, Larimer FW, Norton JM, Op den Camp HJ, Shin M, Wei X. 2007. Whole-genome analysis of the ammonia-oxidizing bacterium, Nitrosomonas eutropha C91: implications for niche adaptation. Environ Microbiol 9:2993–3007. doi: 10.1111/j.1462-2920.2007.01409.x. [DOI] [PubMed] [Google Scholar]
- 63.Klotz MG, Arp DJ, Chain PS, El-Sheikh AF, Hauser LJ, Hommes NG, Larimer FW, Malfatti SA, Norton JM, Poret-Peterson AT, Vergez LM, Ward BB. 2006. Complete genome sequence of the marine, chemolithoautotrophic, ammonia-oxidizing bacterium Nitrosococcus oceani ATCC 19707. Appl Environ Microbiol 72:6299–6315. doi: 10.1128/AEM.00463-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bock E, Sundermeyerklinger H, Stackebrandt E. 1983. New facultative lithoautotrophic nitrite-oxidizing bacteria. Arch Microbiol 136:281–284. doi: 10.1007/BF00425217. [DOI] [Google Scholar]
- 65.Bock E, Koops HP, Moller UC, Rudert M. 1990. A new facultatively nitrite oxidizing bacterium, Nitrobacter vulgaris sp. nov. Arch Microbiol 153:105–110. doi: 10.1007/BF00247805. [DOI] [Google Scholar]
- 66.Lucker S, Nowka B, Rattei T, Spieck E, Daims H. 2013. The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol 4:27. doi: 10.3389/fmicb.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lucker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, Rattei T, Damste JS, Spieck E, Le Paslier D, Daims H. 2010. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci U S A 107:13479–13484. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.