ABSTRACT
Production of sulfide (H2S, HS−, and S2−) by heterotrophic bacteria during aerobic growth is a common phenomenon. Some bacteria with sulfide:quinone oxidoreductase (SQR) and persulfide dioxygenase (PDO) can oxidize self-produced sulfide to sulfite and thiosulfate, but other bacteria without these enzymes release sulfide into the medium, from which H2S can volatilize into the gas phase. Here, we report that Cupriavidus necator H16, with the fccA and fccB genes encoding flavocytochrome c sulfide dehydrogenases (FCSDs), also oxidized self-produced H2S. A mutant in which fccA and fccB were deleted accumulated and released H2S. When fccA and fccB were expressed in Pseudomonas aeruginosa strain Pa3K with deletions of its sqr and pdo genes, the recombinant rapidly oxidized sulfide to sulfane sulfur. When PDO was also cloned into the recombinant, the recombinant with both FCSD and PDO oxidized sulfide to sulfite and thiosulfate. Thus, the proposed pathway is similar to the pathway catalyzed by SQR and PDO, in which FCSD oxidizes sulfide to polysulfide, polysulfide spontaneously reacts with reduced glutathione (GSH) to produce glutathione persulfide (GSSH), and PDO oxidizes GSSH to sulfite, which chemically reacts with polysulfide to produce thiosulfate. About 20.6% of sequenced bacterial genomes contain SQR, and only 3.9% contain FCSD. This is not a surprise, since SQR is more efficient in conserving energy because it passes electrons from sulfide oxidation into the electron transport chain at the quinone level, while FCSD passes electrons to cytochrome c. The transport of electrons from the latter to O2 conserves less energy. FCSDs are grouped into three subgroups, well conserved at the taxonomic level. Thus, our data show the diversity in sulfide oxidation by heterotrophic bacteria.
IMPORTANCE Heterotrophic bacteria with SQR and PDO can oxidize self-produced sulfide and do not release H2S into the gas phase. C. necator H16 has FCSD but not SQR, and it does not release H2S. We confirmed that the bacterium used FCSD for the oxidation of self-produced sulfide. The bacterium also oxidized added sulfide. The common presence of SQRs, FCSDs, and PDOs in heterotrophic bacteria suggests the significant role of heterotrophic bacteria in sulfide oxidation, participating in sulfur biogeochemical cycling. Further, FCSDs have been identified in anaerobic photosynthetic bacteria and chemolithotrophic bacteria, but their physiological roles are unknown. We showed that heterotrophic bacteria use FCSDs to oxidize self-produced sulfide and extraneous sulfide, and they may be used for H2S bioremediation.
KEYWORDS: Cupriavidus necator H16, sulfide, flavocytochrome c, persulfide dioxygenase, sulfide dehydrogenase
INTRODUCTION
Sulfide (H2S, HS−, and S2−) is the most reduced form of sulfur in the biogeochemical cycle, and it is mainly produced by sulfur-reducing bacteria under anaerobic conditions, such as in marine sediment (1). When sulfide diffuses from the anaerobic sediment into the water interface, it is oxidized by chemolithotrophic sulfur oxidizers that use O2 or nitrate as the electron acceptor; the process conserves energy for bacterial growth (2–4). Anaerobic photosynthetic bacteria can also use the reducing power of sulfide for photosynthesis (5, 6). However, these bacteria normally grow in environments where H2S is abundant.
Animals, plants, and bacteria can also generate sulfide from cysteine metabolism under aerobic conditions (7–9). The sulfide produced has recently been reported as a new signaling molecule in mammals (10–15), and it can also protect bacteria against antibiotics by minimizing the production of hydroxyl radical, the most damaging reactive oxygen species, inside bacterial cells (16–18). Most heterotrophic bacteria produce sulfide from sulfur-containing amino acids during growth; however, some heterotrophic bacteria harbor sulfide:quinone oxidoreductase (SQR) and persulfide dioxygenase (PDO) (formerly termed sulfur dioxygenase) and oxidize self-produced sulfide (19). Bacteria without these enzymes produce sulfide and, when accumulated, sulfide is released as H2S into the gas phase (19). The SQR-PDO pathway was first discovered in human mitochondria because mutations in PDO can lead to a human disease, ethylmalonic encephalopathy (20, 21). Subsequently, the presence of PDOs as well as SQRs in heterotrophic bacteria was demonstrated (22–25); the sulfide oxidation pathway is slightly different from that reported in humans (26, 27). In bacteria, SQR oxidizes sulfide to polysulfide, which spontaneously reacts with reduced glutathione (GSH) to produce glutathione persulfide (GSSH); PDO oxidizes GSSH to sulfite, and sulfite spontaneously reacts with polysulfide to produce thiosulfate. Rhodanese, which is almost universal in bacteria, can accelerate the reaction between polysulfide and GSH (28). In Cupriavidus pinatubonensis JMP134, SQR is a fusion protein with a rhodanese domain and an SQR domain (28).
SQRs, grouped into six types, have been extensively investigated in chemolithotrophic bacteria and anaerobic photosynthetic bacteria, and their presence in heterotrophic bacteria has recently become known (29). SQRs oxidize sulfide to zero-valence sulfur, likely polysulfide (28), and pass the electrons to the electron transport system via ubiquinone (29). Similar to SQRs, flavocytochrome c sulfide dehydrogenases (FCSDs) also oxidize sulfide to zero-valence sulfur; however, the electrons enter the electron transport system at the level of cytochrome c (30). The sulfide dehydrogenase is a flavocytochrome c (FccAB) system, consisting of a large sulfide-binding flavoprotein (FccB) and a small cytochrome c (FccA) (5, 6). FCSDs are widely distributed in purple and green phototrophic bacteria and in chemolithotrophic sulfur-oxidizing bacteria (31, 32). However, the physiological role of FCSDs is still debatable. A FCSD isolated from Allochromatium (formerly Chromatium) vinosum is active in sulfide oxidation (33), but the FCSD-inactive mutant of A. vinosum does not have any apparent decrease in sulfur oxidation and sulfur-supported phototrophic growth, suggesting that SQR is the main enzyme for sulfide oxidation in the bacterium (34, 35). In chemolithotrophic sulfur-oxidizing bacteria, FccA and FccB are often referred as SoxE and SoxF, respectively, because their genes are clustered with other Sox genes involved in thiosulfate oxidation (5). In a reconstituted Sox system, however, SoxF did not stimulate sulfide oxidation (36). When soxE is not next to soxF, the gene coding for the SoxF homolog is referred to as soxJ (5), and the protein has two functions in vitro, i.e., oxidizing sulfide and enhancing the Sox system for thiosulfate oxidation (37). Sulfide oxidation by FCSDs in vivo has yet to be shown, and the physiological roles remain to be discovered.
We recently reported that most bacteria produce sulfide during heterotrophic growth under aerobic conditions (19). H2S is produced from sulfur-containing amino acids via cysteine desulfhydrase, cystathionine β-synthase (CBS), cystathionine γ-lyase (CSE), and 3-mercaptopyruvate sulfurtransferase (3MST) (7, 38). Bacteria with SQR and PDO, such as Cupriavidus pinatubonensis JMP134 and Pseudomonas aeruginosa PAO1, produce and oxidize sulfide, while bacteria without SQR and PDO produce and release H2S into the gas phase in pure cultures. Cupriavidus necator H16 is best known for its ability to produce and to store large amounts of poly[R-(−)-3-hydroxybutyrate], which can be used to make biodegradable plastics (39). We report here that C. necator H16 without SQR uses FCSD to oxidize self-produced sulfide and FCSD couples with PDO to oxidize sulfide to sulfite and thiosulfate, via a pathway similar to the SQR-PDO pathway.
RESULTS
C. necator H16 oxidizes self-produced H2S with FCSD.
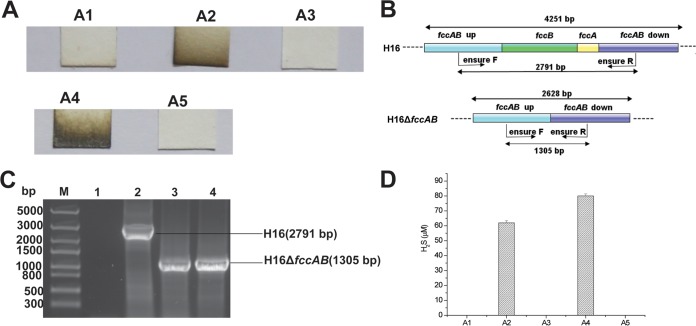
Since bacteria without SQR often release H2S during heterotrophic growth (19), C. necator H16, without SQR, was expected to produce and to release H2S during aerobic growth in LB medium, but it did not (Fig. 1A). Sequence analysis of the C. necator H16 genome revealed that the bacterium has two PDOs (NCBI accession numbers WP_011616222.1 and WP_010809980) and one FCSD, consisting of FccA (NCBI accession number WP_010812125.1) and FccB (NCBI accession number WP_011616200.1). To analyze whether the FCSD in C. necator H16 was responsible for the oxidation of self-produced H2S, the fccAB gene cluster was deleted by using homologous recombination, and the deletion was confirmed by PCR analysis (Fig. 1B and C). The C. necator H16 ΔfccAB mutant accumulated and released about 60 μM H2S (Fig. 1A and D). The estimation was performed with sulfide added to LB medium, and lead-acetate paper strips detect sulfide levels as low as 5 μM added to LB medium (19). The C. necator H16 ΔfccAB mutant with fccAB complementation consumed sulfide and did not release H2S into the gas phase (Fig. 1A and D). P. aeruginosa Pa3K (19), a mutant of P. aeruginosa PAO1 with its two sqr genes and one pdo gene deleted, produced detectable H2S. Pa3K expressing fccAB did not release H2S (Fig. 1A and D), indicating that FCSD oxidizes self-produced H2S in both C. necator H16 and the recombinant P. aeruginosa strain. The FCSD of C. necator H16 did not function in Escherichia coli BL21(DE3)(pBBR5-fccAB) (data not shown). The nonfunctional FCSD is likely due to the lack of maturation proteins for c-type cytochromes in E. coli under aerobic conditions (40, 41).
FIG 1.
Genetic analysis of fccAB in C. necator H16. (A) H2S production abilities of different bacteria. A1, C. necator H16; A2, C. necator H16 ΔfccAB; A3, C. necator H16 ΔfccAB/fccAB; A4, Pa3K; A5, Pa3K(pBBR5-fccAB). (B) Process for construction of C. necator H16 ΔfccAB. (C) Analysis of PCR fragments to confirm fccAB disruption. Lane M, molecular size standards; lane 1, product amplified with water as the template (negative control); lane 2, product amplified with C. necator H16 genomic DNA as the template; lane 3, product amplified with C. necator H16 ΔfccAB genomic DNA as the template; lane 4, product amplified with C. necator H16 ΔfccAB/fccAB genomic DNA as the template. The PCRs were performed with primers ensure F and ensure R. (D) Relative production of H2S in different strains. The data were generated by scanning and comparison to standards. The detection limit with the lead-acetate paper strips was about 5 μM.
FCSD plays a detoxification role in C. necator H16.
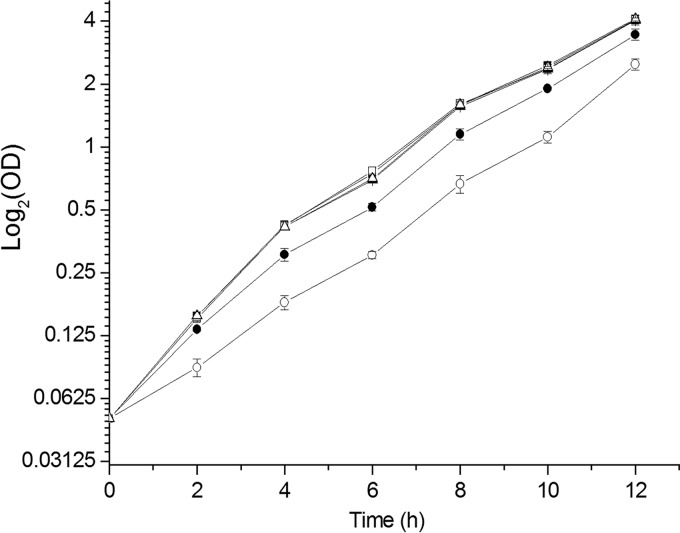
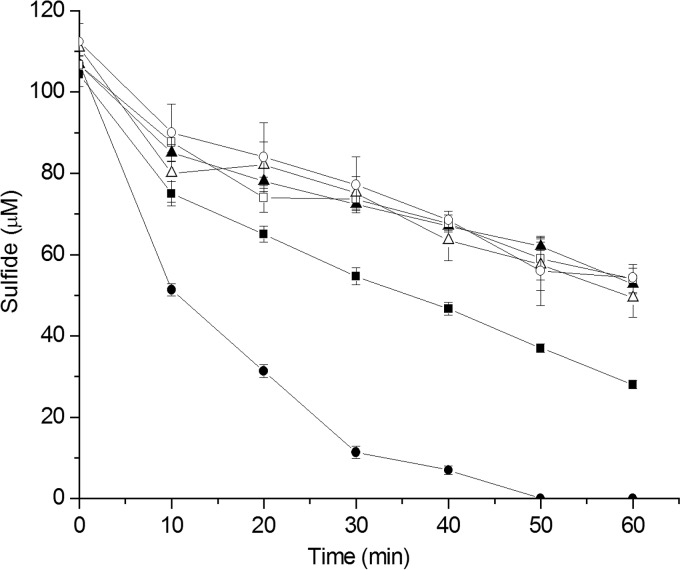
Although C. necator H16 ΔfccAB could not oxidize self-produced H2S, the mutation did not affect growth in LB medium, and the wild-type and mutant strains had similar initial growth rates of about 0.8 h−1 (Fig. 2). When 200 μM sulfide was added to the LB medium, C. necator H16 was slightly inhibited (initial growth rate of 0.71 ± 0.06 h−1) and the mutant was severely inhibited (initial growth rate of 0.41 ± 0.15 h−1). The complemented strain C. necator H16 ΔfccAB/fccAB recovered its growth rate in LB medium containing 200 μM sulfide (initial growth rate of 0.8 ± 0.13 h−1). Resting cells of C. necator H16 (3.49 ± 0.7 nmol min−1 mg [dry weight] cells−1) oxidized sulfide faster than the mutant H16 ΔfccAB (2.5 ± 0.5 nmol min−1 mg [dry weight] cells−1), and the complemented mutant H16 ΔfccAB/fccAB (6.7 ± 0.4 nmol min−1 mg [dry weight] cells−1) oxidized sulfide even faster, possibly due to the increased production of FccAB from the introduced plasmid (Fig. 3). The data confirm that FCSD oxidizes sulfide and plays a role in the detoxification of sulfide.
FIG 2.
Growth curves in LB medium and LB medium with 200 μM NaHS. Overnight cultures were inoculated into LB medium or LB medium containing 200 μM NaHS, at an initial OD600 of 0.05. Cell turbidity was assessed every 2 h. ■, C. necator H16 in LB medium; ●, C. necator H16 in LB medium with 200 μM NaHS; □, C. necator H16 ΔfccAB in LB medium; ○, C. necator H16 ΔfccAB in LB medium with 200 μM NaHS; ▲, C. necator H16 ΔfccAB/fccAB in LB medium; △, C. necator H16 ΔfccAB/fccAB in LB medium with 200 μM NaHS. All data are averages of three samples with standard deviations (error bars). All cultures reached final OD600 values of about 6.1 after 24 h of incubation.
FIG 3.
Sulfide oxidation by C. necator H16 and its mutants. Cells were suspended in 100 mM Tris buffer (pH 8.0) at an OD600 of 2, and NaHS (100 μM) was added to initiate the reaction. Controls used the same buffer without bacterial cells. ■, C. necator H16; ▲, C. necator H16 ΔfccAB; ●, C. necator H16 ΔfccAB/fccAB; □, buffer; △, heat-killed C. necator H16; ○, heat-killed C. necator H16 ΔfccAB/fccAB. All data are averages of three samples with standard deviations (error bars).
P. aeruginosa Pa3K with FCSD oxidizes sulfide to sulfane sulfur.
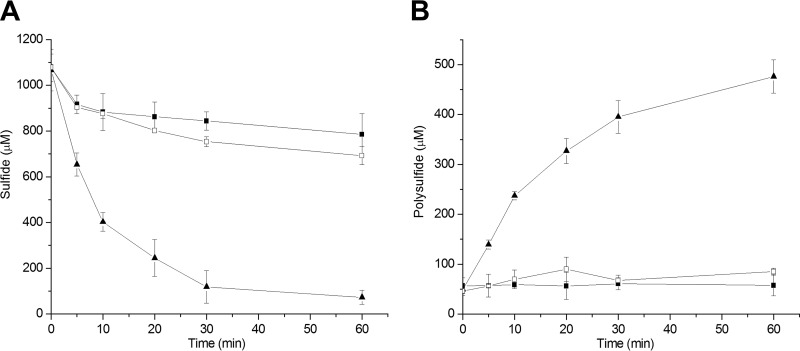
P. aeruginosa strains Pa3K and Pa3K(pBBR5-fccAB) were cultured, harvested, and resuspended at an optical density at 600 nm (OD600) of 2 in 100 mM Tris buffer (pH 8.0). A cell suspension of Pa3K(pBBR5-fccAB) oxidized 1 mM sulfide in 60 min, with an initial rate of 81.9 ± 2 nmol min−1 mg (dry weight) cells−1, and the control PA3K strain had a minimal initial rate of sulfide oxidation of 2.3 ± 1.6 nmol min−1 mg (dry weight) cells−1 (Fig. 4). Sulfane sulfur, including polysulfide, was produced. If the sulfane sulfur produced is in the form of disulfide (HSSH), then 1 mM sulfide is expected to produce 0.5 mM sulfane sulfur. Disulfide and trisulfide were detectable during sulfide oxidation by Pa3K(pBBR5-fccAB) (data not shown), similar to levels produced by recombinant E. coli with cloned C. pinatubonensis sqr (28). Sulfide levels were also decreased in the control experiment with P. aeruginosa Pa3K, but the small loss could be due to volatilization, autoxidation (42), and fortuitous activities of certain enzymes, such as superoxide dismutase (43). Polysulfide production was not detectable for the control P. aeruginosa Pa3K strain during incubation with 1 mM sulfide (Fig. 4). Sulfite and thiosulfate levels remained low, at less than 50 μM and 25 μM, respectively, during the course of analysis for P. aeruginosa Pa3K and the recombinant strains with fccAB or sqr.
FIG 4.
Effect of FccAB expression in P. aeruginosa Pa3K on H2S oxidization. The cell suspension in 100 mM Tris buffer (pH 8.0), at an OD600 of 2, oxidized 1 mM sulfide. Sulfide (A) and polysulfide (B) were consumed or produced by Pa3K (■), Pa3K(pBBR5-fccAB) (▲), and heat-killed Pa3K(pBBR5-fccAB) (□). All data are averages of at least three samples with standard deviations (error bars).
FCSD and PDO collectively oxidize H2S to sulfite and thiosulfate in P. aeruginosa Pa3K.
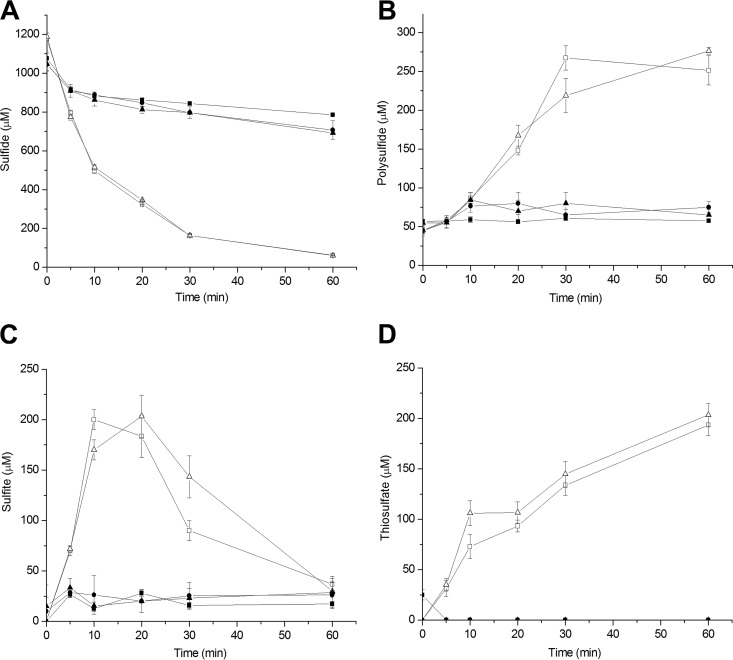
C. necator H16 has two pdo genes, Cnpdo1 and Cnpdo2. The genes were separately cloned with FccAB in P. aeruginosa Pa3K. Resting Pa3K(pBBR5-fccAB-Cnpdo1) and Pa3K(pBBR5-fccAB-Cnpdo2) cells, containing FCSD and PDO, oxidized sulfide to sulfane sulfur, sulfite, and thiosulfate in 60 min, with initial rates of sulfide oxidation of 81.9 ± 2 nmol min−1 mg (dry weight) cells−1 and 81.0 ± 3.9 nmol min−1 mg (dry weight) cells−1, respectively (Fig. 5). The bacteria with different PDOs showed similar catalytic activities during sulfide oxidation. The bacteria with PDOs accumulated polysulfide more slowly than did Pa3K(pBBR5-fccAB), without PDO, and they produced 250 to 275 μM polysulfide after 60 min (Fig. 5). Interestingly, sulfite was rapidly produced to about 200 μM at 10 min and then levels gradually decreased to about 36 ± 10 μM at 60 min (Fig. 5C). During the same period, thiosulfate levels increased to 203 ± 11 μM (Fig. 5D), due the spontaneous reaction of sulfite with polysulfide (28). When the loss of sulfide from the control was subtracted, 824 μM sulfide was consumed by the bacteria with FccAB and PDO. Given that thiosulfate contains two sulfur atoms and polysulfide has two or more sulfur atoms, the sulfur balance is close.
FIG 5.
Effect of FccAB and PDO coexpression in P. aeruginosa Pa3K on H2S oxidization. Sulfide was added at 1 mM to initiate the reaction. Sulfide (A), polysulfide (B), sulfite (C), and thiosulfate (D) were consumed or produced by Pa3K (■), Pa3K(pBBR5-fccAB-Cnpdo1) (□), Pa3K(pBBR5-fccAB-Cnpdo2) (△), heat-killed Pa3K(pBBR5-fccAB-Cnpdo1) (▲), and heat-killed Pa3K(pBBR5-fccAB-Cnpdo2) (●). All data are averages of at least three samples with standard deviations (error bars).
Distribution of FccAB in bacteria.
Of the 46 reported FccBs (5, 29), 34 were reported with accession numbers and were retrieved from GenBank. After duplicate sequences were removed, 32 sequences were used for a BLAST search of a microbial genomic protein sequence set of 4,929 bacterial genomes (NCBI database, updated to 15 April 2016). A total of 351 candidates were identified, and phylogenetic tree analysis narrowed the list to 257 proteins. Since all of the seed FccBs contained the FCSD flavin-binding domain (pfam09242) and FadH2 domain (COG0446), these domains were also used to check the candidates. Finally, 240 candidates were identified as FccBs, and all were from Gram-negative bacteria. In contrast, SQRs are also present in Gram-positive bacteria or archaea (19). The 240 proteins were from 190 Gram-negative bacterial genomes, representing 3.85% of the 4,929 sequenced bacterial genomes, including 160 Proteobacteria, 10 Chlorobia, and 10 Deinococcus genomes and genomes from a few Aquificales, Cytophagia, and Deferribacterales species (Table 1). Of the 190 bacteria with FccBs, 34 contained more than one FccB and 121 (63.7%) also carried SQR. The frequent presence of both FccB and SQR within single bacteria is a surprise, as both are involved in sulfide oxidation.
TABLE 1.
Distribution of FccA and FccB in different bacterial phyla
| Taxon | No. of genomes |
||
|---|---|---|---|
| fccB | Linked fccA and fccB | More than one fccB | |
| Alphaproteobacteria | 56 | 49 | 11 |
| Betaproteobacteria | 72 | 64 | 5 |
| Gammaproteobacteria | 27 | 23 | 7 |
| Epsilonproteobacteria | 5 | 1 | 0 |
| Aquificales | 7 | 1 | 4 |
| Chlorobia | 10 | 10 | 6 |
| Cytophagia | 1 | 1 | 0 |
| Deferribacterales | 2 | 2 | 0 |
| Deinococcus | 10 | 0 | 1 |
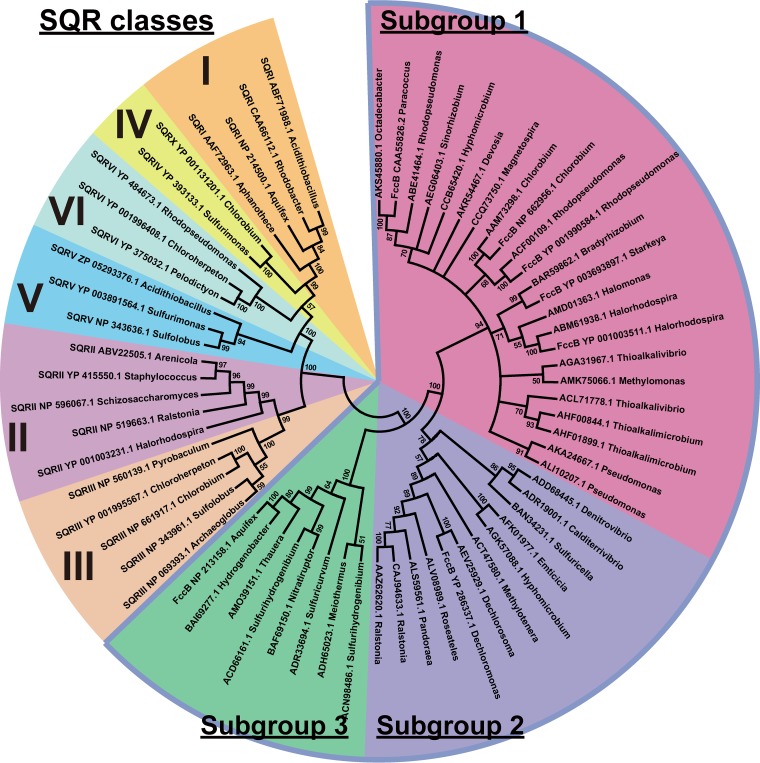
FccB and SQR are evolutionarily related, and both contain flavin adenine dinucleotide (29). However, SQR transfers the electrons from sulfide oxidation to quinone, whereas FccB transfers the electrons to cytochrome c. Since FccBs are often used as outgroups for SQR phylogenetic analysis (5, 29), SQRs were used as outgroups for FccB phylogenetic analysis in this study. The 240 sequences were further assembled into 34 unique groups by using the CD-HIT program, with 50% identity as the cutoff value (44). With the same strategy, the 32 FccB seed sequences and the 102 SQR sequences were clustered into 7 and 22 unique groups, respectively. One representative sequence from each unique group was selected. Thus, 41 FccBs together with FccBs from C. necator H16 and C. pinatubonensis JMP134 were used to build a phylogenetic tree with 22 SQRs as outgroups (MEGA 7.0) (Fig. 6). FccBs were divided into 3 subgroups on the phylogenetic tree.
FIG 6.
Phylogenetic analysis of FccBs and SQRs. The tree was generated with a neighbor-joining method by using MEGA 7 software. Proteins are listed with their accession numbers and organism origins.
The distribution of FccA near FccB was checked; 114 of the 138 FccBs in subgroup 1 and 68 of the 72 FccBs in subgroup 2 had fccA located within 5 loci of the corresponding fccB gene within the chromosome. However, only 6 of the 30 FccBs in subgroup 3 had their genes next to fccA within the chromosome.
The association of fccB with the sox genes coding for the SOX enzyme system was also investigated. Thirty strains contained the full set of sox genes, including soxA, soxB, soxC, soxD, soxX, soxY, and soxZ. Twenty-one strains had the sox genes without soxC and soxD, and the Sox system without SoxC and SoxD also oxidizes thiosulfate (45). An additional 67 strains had fccB near soxYZ. Thus, a total of 118 of 190 strains had soxYZ near fccB.
The association of fccB with the pdo genes was analyzed. Of the 190 strains, 137 contained PDOs. All of the green sulfur bacteria and most of the purple sulfur bacteria did not have PDOs. Most of the heterotrophic bacteria possessed PDOs.
DISCUSSION
We reported previously that significant proportions of heterotrophic bacteria with SQR and PDO are able to oxidize self-produced H2S. These bacteria are rather common in nature, such as Bacillus spp. and Pseudomonas spp. in soil and Roseobacter spp. in marine environments (19). Here we demonstrated that bacteria with FCSD and PDO also have the ability to oxidize self-produced H2S. These heterotrophic bacteria are likely to oxidize sulfide in the environment, especially where organic compounds are seasonally abundant, such as in garden soil or during algal blooms. The oxidation is significant, facilitating the geochemical cycling of sulfur, as chemical oxidation of sulfide is relatively slow, with a half-life of 26 h in seawater at 25°C (46).
Although the sulfide oxidation activity of FCSD in vitro has been shown, its in vivo activity and physiological functions in sulfide oxidation have not been demonstrated (33). Our data show that FCSD also oxidizes sulfide in vivo. In C. necator H16, FCSD oxidizes self-produced and extraneous sulfide, preventing volatilization of H2S (Fig. 1A). It also plays a detoxification role, as the fccAB mutant grows slowly in the presence of sulfide levels as low as 200 μM (Fig. 2).
FCSD and SQR both oxidize sulfide to polysulfide in bacteria (Fig. 4) (19), but SQR is more common. A total of 1,014 of 4,929 sequenced bacterial genomes contain SQR, while only 190 possess FccB. One possible reason for its abundance is that SQR is more efficient in conserving energy, as it passes the electrons from sulfide oxidation to the electron transport chain at the level of quinone, while FCSD transfers electrons to the electron transport chain at the level of cytochrome c. The transport of electrons from ubiquinones to O2 pumps more protons across the membrane than does that from cytochrome c to O2. Thus, it has been reported that SQR is the primary enzyme for sulfide oxidation in A. vinosum, which contains both SQR and FCSD (5, 34, 35).
All reported FCSDs are in the periplasm (5, 34). FccBs containing flavin adenine diphosphate are thought to be folded and assembled with the flavin in the cytoplasm and then translocated to the periplasm by the TAT transport system (47). Indeed, FccB of C. necator H16 has a typical TAT signal peptide with a twin arginine motif at the N terminus, suggesting that it is transported by the TAT system. SignalP analysis identified a typical Sec-dependent signal peptide at the N terminus of FccA (48), suggesting that apo-FccA is transported by the Sec system into the periplasm, where FccA is assembled with heme to generate the cytochrome c (49). A cytoplasmic location of PDOs has been proposed because PDOs usually do not have the signal peptide for protein trafficking across the cytoplasmic membrane into the periplasm and their activity requires Fe2+, which is readily oxidized to Fe3+ outside the reducing cytoplasm (23). Thus, the scarce Fe2+ in the periplasm will likely not satisfy the Fe2+ requirement of PDOs unless Fe2+ is tightly bound to the enzyme, and it must be regenerated when oxidized to Fe3+. On the basis of our data, we propose that the pathway of sulfide oxidation by the concerted actions of FCSD and PDO in P. aeruginosa Pa3K is similar to the SQR-PDO pathway (28). FCSD oxidizes sulfide to polysulfide in the periplasmic space (Fig. 7), polysulfide is transported into the cytoplasm and spontaneously reacts with GSH to produce GSSH, which is oxidized by PDO to sulfite, and sulfite chemically reacts with polysulfide to produce thiosulfate. Due to the lack of a rhodanese domain in FCSD, more thiosulfate was produced, similar to the C. pinatubonensis SQR-PDO system when the rhodanese activity is inactivated (28). It is currently unclear whether polysulfide simply diffuses into the cells or is transported by membrane proteins.
FIG 7.
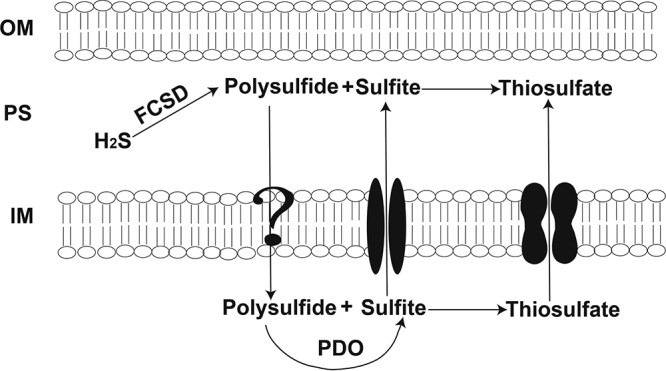
Proposed model for sulfide oxidation by FCSD and PDO in C. necator H16. Sulfide is oxidized by FCSD to polysulfide in the periplasmic space (PS). Polysulfide moves into the cytoplasm in an unknown way, and it is oxidized by PDO to produce sulfite in the cytoplasm. Sulfite is transported into the periplasmic space by a transport protein. Polysulfide and sulfite react to produce thiosulfate in both the periplasmic space and the cytoplasm. OM, outer membrane; IM, inner membrane.
The slow movement of polysulfide from the periplasm into the cytoplasm is implied by the transitory accumulation of sulfite during the initial sulfide oxidation (Fig. 5C). Sulfite is produced by PDO in the cytoplasm and exported into the periplasm, which has a pH value lower than that of the cytoplasm. The chemical reaction between sulfite and polysulfide is slower at low pH, because the protonated forms of the species are less reactive. Thus, the slow reaction and rapid diffusion out of the periplasm through porins on the outer membrane allow the escape of sulfite into the medium. However, sulfite can diffuse back into the periplasm or even be transported into the cytoplasm; in both spaces, sulfite reacts with polysulfide to produce thiosulfate, with subsequent decreases in sulfite levels and increases in thiosulfate levels (Fig. 5C and D). Thus, a model of sulfide oxidation in Gram-negative bacteria with FCSD in the periplasm and PDO in the cytoplasm is proposed (Fig. 7).
Phylogenetic analyses divided FccBs into three subgroups and the SQR outgroups into six subgroups (Fig. 6). The six classes of SQRs were the same as reported previously (29). The three subgroups of FccBs were also well conserved within taxonomic classes (Table 2). The subgroup 1 FccBs were mainly distributed in Alphaproteobacteria, Chlorobia, and Gammaproteobacteria. Most Chlorobia and Gammaproteobacteria members are green or purple sulfur bacteria that use H2S for anaerobic photosynthesis (5). Some Alphaproteobacteria members are common heterotrophs, such as Rhodopseudomonas spp., Bradyrhizobium spp., and Sinorhizobium spp. Rhodopseudomonas spp. are purple nonsulfur bacteria that are also able to grow phototrophically under anaerobic conditions (50). The subgroup 2 FccBs were primarily present in Betaproteobacteria, most of which are heterotrophs, including C. necator H16. The subgroup 3 FccBs were well conserved in autotrophic bacteria of Aquificales, Epsilonproteobacteria, and Deferribacterales, which can oxidize H2 or H2S, and they were also present in Thermus spp. of the Deinococcus-Thermus phylum, which are often isolated from hot springs or hot composts where H2S is abundant (51). FccBs were previously named SoxJ, SoxF, and FccB, according to whether their genes are associated with the sox genes or FccA is present (5). Here, we established a phylogenetic tree by using the FccBs identified from the whole GenBank database, and the previously reported SoxJ, SoxF, and FccB were randomly distributed in the subgroups (Fig. 6). Thus, the previously reported SoxJ and SoxF proteins are all FccBs. FccBs in each of the phylogenetic subgroups are well conserved at the taxonomic level (Table 2).
TABLE 2.
Taxonomic distribution of FccB subgroups
| Taxon | No. of FccB proteins | FccB subgroup |
|---|---|---|
| Alphaproteobacteria | 68 | 1 |
| Aquificales | 1 | 1 |
| Betaproteobacteria | 9 | 1 |
| Chlorobia | 16 | 1 |
| Epsilonproteobacteria | 1 | 1 |
| Gammaproteobacteria | 43 | 1 |
| Alphaproteobacteria | 1 | 2 |
| Betaproteobacteria | 68 | 2 |
| Cytophagia | 1 | 2 |
| Gammaproteobacteria | 2 | 2 |
| Aquificales | 12 | 3 |
| Betaproteobacteria | 1 | 3 |
| Deinococci | 11 | 3 |
| Epsilonproteobacteria | 4 | 3 |
As reported, the fccA and sox genes frequently appeared near the fccB genes (5). We found that the fccA gene was often associated with fccB genes coding for subgroup 1 and 2 FccBs but not subgroup 3 FccBs. Since FccB is known to reduce other cytochrome c forms in vivo (31, 32), it may use alternative cytochrome c for electron transfer, reflecting the divergent evolution of FccBs. In contrast, the sox genes were less commonly associated with fccB, especially the full set of the Sox system; interestingly, fccB is often near soxYZ. The common presence of soxYZ with fccB (in 118 of 190 strains) suggests that SoxYZ may serve as a carrier for the sulfane sulfur produced by FccB. This possibility needs further investigation.
In summary, C. necator H16 without SQR actively oxidizes self-produced sulfide, and it uses FCSD to oxidize sulfide to polysulfide in the periplasm. The polysulfide produced is either transported or diffused into the cytoplasm, where it reacts with GSH to produce GSSH and PDO oxidizes GSSH to sulfite (28). Sulfite is exported to the periplasm, where it spontaneously reacts with polysulfide to generate thiosulfate (Fig. 7). Most of the heterotrophic bacteria containing FCSD also had PDOs, and the two enzymes may function for sulfide oxidation in other heterotrophic bacteria. Heterotrophic bacteria with FCSD may have the potential to be used for bioremediation of H2S.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 3. C. pinatubonensis JMP134 and C. necator H16 were cultivated in LB medium at 30°C. E. coli and P. aeruginosa were cultured in LB medium at 37°C or as stated. C. necator H16 was also grown in minimal salt medium (MM) supplemented with 1% sodium gluconate during the process of gene inactivation (52). Gentamicin, tetracycline, and kanamycin were added as needed. The culture growth was monitored with OD600 measurements, and the growth rate was calculated via the change in log2 OD600 per hour.
TABLE 3.
Bacteria and plasmids used in this study
| Strain or plasmid | Genotype and/or descriptiona | Source or reference |
|---|---|---|
| Strain | ||
| E. coli DH5α | Cloning strain | Novagen |
| E. coli S17-1 | RP4 derivative integrated in chromosome | 56 |
| E. coli BL21(DE3) | Cloning strain | Novagen |
| C. pinatubonensis JMP134 | Wild type | Ron L. Crawford |
| C. necator H16 | Wild type | ATCC 17699 |
| P. aeruginosa PAO1 | Wild type | ATCC 15692 |
| Pa3K | Mutant of P. aeruginosa PAO1 in which sqr1, sqr2, and pdo were deleted | This study |
| Plasmid | ||
| pBBR1MCS-3 | Tetr, Plac; expression vector | 57 |
| pBBR1MCS-5 | Gmr, Plac; expression vector | 57 |
| pK18mobsacBtet | Kmr and Tetr, sacB, RP4 oriT, ColE1 ori; suicide vector | This study |
| pBBR5-fccAB | pBBR1MCS-5 containing fccAB from C. necator H16 | This study |
| pBBR5-fccAB-pdo1 | pBBR1MCS-5 containing fccAB and pdo1 from C. necator H16 | This study |
| pBBR5-fccAB-pdo2 | pBBR1MCS-5 containing fccAB and pdo2 from C. necator H16 | This study |
| pBBR3-fccAB | pBBR1MCS-3 containing fccAB from C. necator H16 | This study |
| pBBR3-fccAB-pdo1 | pBBR1MCS-3 containing fccAB and pdo1 from C. necator H16 | This study |
| pBBR3-fccAB-pdo2 | pBBR1MCS-3 containing fccAB and pdo2 from C. necator H16 | This study |
| pK18mobsacBtet-ΔfccAB | Tetr; suicide vector for deletion of fccAB of C. necator H16 | This study |
Tetr, tetracycline resistance; Gmr, gentamycin resistance; Kmr, kanamycin resistance.
Testing of H2S production.
The method used for the detection of H2S production was the same as described previously (19). Briefly, bacteria were transferred into 2 ml of LB medium in a 15-ml glass tube, and a paper strip with lead acetate was affixed at the top of the tube with a rubber stopper. The culture was incubated with shaking for 48 h, and then the paper strip was photographed to detect any lead(II)-sulfide black precipitates, as a measure of H2S production. NaHS added to LB medium was used to generate black precipitates on the paper strips to estimate the amount of H2S produced.
Deletion of fccAB from C. necator H16.
The method used for the knockout of fccAB was the same as described previously (19, 53). Briefly, 1-kb regions immediately upstream and downstream of fccAB were amplified from genomic DNA of C. necator H16 via PCR with the primer sets fccuf/fccur and fccdf/fccdr (Table 4). Then the two fragments were joined and cloned into pK18mobsacBtet at the EcoRI site by using the In-fusion kit (TaKaRa, Beijing, China), to produce pK18mobsacBtet-ΔH16fccAB in E. coli S17-1. The plasmid pK18mobsacBtet-ΔH16fccAB was transferred into C. necator H16 by conjugation. The plasmid pK18mobsacBtet-ΔH16fccAB was integrated into the chromosome of C. necator H16 by the first crossover and selected on MM plates with sodium gluconate as the sole carbon source. Cells with the second crossover to generate the deletion were selected by culture on low-salt LB medium plates supplemented with 12% sucrose. The deletions were validated by PCR and DNA sequencing.
TABLE 4.
Primers used in this study
| Primer name | Sequence | Source |
|---|---|---|
| Fccf | CACACAGGAAACAGCTATGACAATCTTCCCCCGGCC | This study |
| fccr | TTCCATTCGCCATTCACTACCCCAGCATCTCCGCCC | This study |
| Pdo1f | GGGCGGAGATGCTGGGGTAGACGAAAAACAAGAGGGCAGCC | This study |
| Pdo1r | TTCCATTCGCCATTCATCAGGCGCCGTGCGGCACG | This study |
| Pdo2f | GGGCGGAGATGCTGGGGTAGGCATCCAAGGAGGCGAGCATG | This study |
| Pdo2r | TTCCATTCGCCATTCATCAGATGACGTCCAGAGGGATT | This study |
| Fccr2 | CTACCCCAGCATCTCCGCCC | This study |
| pBBRf | TGAATGGCGAATGGAAATTGTAAG | This study |
| pBBRr | AGCTGTTTCCTGTGTGAAATTGTTATC | This study |
| fccuf | CAGGAAACAGCTATGACATGATTACGAATTGCAATTCTGCCAACGTGATCGCCGC | This study |
| fccur | CCCAGCATCTCCGCTGTCATGGTCTCGCTCCGTGTAGGG | This study |
| fccdf | GCGAGACCATGACAGCGGAGATGCTGGGGTAGGTGCAGG | This study |
| fccdr | TTCAGGATCCCCGGGTACCGAGCTCGAATTACAGCTCGATCACGCTTTCGTCCGC | This study |
| Ensure F | GGTCGACCGGGCCGATCTTC | This study |
| Ensure R | GACCCTGCTGATGACCGCCG | This study |
Complementation of fccAB.
All primers are listed in Table 4. The fccA and fccB genes were amplified by using PCR with the C. necator H16 genome as the template and fccf and fccr as the primers. Linearized pBBR1MCS-3 was obtained via PCR with pBBR1MCS-3 as the template and pBBRf and pBBRr as the primers; the PCR product was treated with DpnΙ to degrade the template plasmid and was gel purified. Finally, the fccAB fragment and pBBR1MCS-3 were linked by using the In-fusion kit (TaKaRa). The resulting plasmid was cloned into E. coli and then transferred into C. necator H16 ΔfccAB via electroporation. For overexpression, pBBR1MCS-5-fccAB was constructed using the same method as described above.
Expression of fccAB and pdo of C. necator H16 in P. aeruginosa.
The fragments Cnpdo1 and Cnpdo2 were PCR amplified with the primer pairs pdo1f/pdo1r and pdo2f/pdo2r, containing 15- to 20-bp extensions overlapping the adjacent fragment or the cloning site. For coexpression of fccAB and Cnpdo1, the primers pBBRf and fccr2 were used to amplify pBBR1MCS-5-fccAB(pBBR5-fccAB). The fragment Cnpdo1 was cloned into pBBR5-fccAB by using the In-fusion kit, to generate pBBR5-fccAB-Cnpdo1. Using the same approach, pBBR5-fccAB-Cnpdo2 was constructed. The resulting plasmids pBBR5-fccAB, pBBR5-fccAB-Cnpdo1, and pBBR5-fccAB-Cnpdo2 were transferred into Pa3K via electroporation.
Whole-cell analysis of sulfide oxidation.
Cells of Pa3K and recombinant Pa3K(pBBR5-fccAB), Pa3K(pBBR5-fccAB-Cnpdo1), and Pa3K(pBBR5-fccAB-Cnpdo2) strains were grown in LB medium without induction to an OD600 of 3, harvested by centrifugation (6,000 × g for 10 min), and suspended in 100 mM Tris buffer (pH 8) at an OD600 of 2. For the heat-killed control, the cell suspension was heated in boiling water for 10 min and cooled to room temperature. Two milliliters of the cell suspension was transferred to a 15-ml capped centrifuge tube, and freshly prepared NaHS was added to initiate the reaction. The tube was incubated at 30°C, with shaking at 200 rpm. The pH was selected to minimize H2S from rapid valorization, as the dominant species of sulfide is HS− at pH 8 (54). The sulfide, polysulfide, sulfite, and thiosulfate levels were analyzed at various times.
Analytical procedures.
The sulfide, polysulfide, sulfite, and thiosulfate levels were analyzed as described previously (28). Briefly, sulfide levels were analyzed by a colorimetric method; sulfite and thiosulfate levels were determined by ion chromatography as described previously, with minor modifications. Specific polysulfide species from sulfide oxidation by whole cells were also detected, after monobromobimane derivatization, by using high-performance liquid chromatography (HPLC) (28).
Analysis of FccB in sequenced bacterial genomes.
A microbial genomic protein sequence set from NCBI (updated to 15 April 2016) was downloaded for the FccB search. The query sequences for FccBs were reported FccBs (5, 29) and were used to search the database with the standalone BLASTP algorithm, using conventional criteria (E value of ≤1e−10, coverage of ≥50%, and identity of ≥30%), to obtain FccB candidates from a total of 4,929 bacterial genomes. The candidates were combined with the seed FccBs for phylogenetic tree analysis using ClustalW for alignment and MEGA 7.0 for neighbor-joining tree building, with pairwise deletion, p-distance distribution, and bootstrap analysis of 1,000 repeats as parameters (55). The candidates that were in the same clade as the seed FccBs were picked for further analysis. The seed FccBs were searched for in the Conserved Domain Database (CDD) at the NCBI website. All seed FccBs contained the FCSD flavin-binding domain (pfam09242) and FadH2 domain (COG0446); therefore, these two motifs were used as standard features for further filtration of FccB candidates. The identified FccB sequences and seed sequences were separately grouped into unique groups by using the CD-HIT program, with identity of >50% as the threshold (44). Published SQR sequences were also collected (5, 29) and grouped into unique groups to be used as outgroups. One representative sequence from each unique group was selected and used for phylogenetic analysis (MEGA 7.0).
The 190 strains containing these 240 FccB proteins were manually checked for their heterotrophy with respect to the representative strains at the genus level, as described previously (19). Whether fccA appeared within 5 loci of the 240 fccB genes was also checked; to do so, the accession numbers of five genes around fccB were collected and used for the CDD search to check for cytochrome c553 (COG5863), using default parameters. If the transcription directions were the same and the intergenic region was smaller than 500 bp, then the cytochrome c gene was considered to be fccA.
Whether genes encoding the Sox enzyme system appeared within 10 loci of the 240 fccB genes in the genome was also checked. To find the conserved domains of these proteins, we collected all of the sequences of the Sox proteins from the KEGG gene database and used them to search the CDD with default parameters. All of the proteins in the Sox system had the conserved domain feature; SoxA (TIGR04484), SoxB (TIGR04486), SoxC (TIGR04555), SoxX (TIGR04485), SoxY (pfam13501), SoxZ (pfam08770), and SoxD (COG3474, COG4654, or COG3258) were used to check the corresponding proteins. The accession numbers of 10 genes around fccB were collected and used to search the CDD with default parameters. The nearby genes coding for proteins with the conserved Sox domains were identified. Genes coding for SQRs and PDOs within the same genome were detected as described previously (19).
ACKNOWLEDGMENTS
This work was financially supported by grants from the National Natural Science Foundation of China (grant 21477062), the National Key Research and Development Program of China (grant 2016YFA0601103), the Natural Science Foundation of Shandong Province (grant ZR2014CM003), and the State Key Laboratory of Microbial Technology at Shandong University.
We declare we have no conflicts of interest regarding the contents of this article.
REFERENCES
- 1.Wasmund K, Mussmann M, Loy A. 2017. The life sulfuric: microbial ecology of sulfur cycling in marine sediments. Environ Microbiol Rep 9:323–344. doi: 10.1111/1758-2229.12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kodama Y, Watanabe K. 2003. Isolation and characterization of a sulfur-oxidizing chemolithotroph growing on crude oil under anaerobic conditions. Appl Environ Microbiol 69:107–112. doi: 10.1128/AEM.69.1.107-112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangold S, Valdes J, Holmes DS, Dopson M. 2011. Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Front Microbiol 2:17. doi: 10.3389/fmicb.2011.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Liu S, Liu X, Li X, Wen Q, Lin J. 2014. Identification and characterization of an ETHE1-like sulfur dioxygenase in extremely acidophilic Acidithiobacillus spp. Appl Microbiol Biotechnol 98:7511–7522. doi: 10.1007/s00253-014-5830-4. [DOI] [PubMed] [Google Scholar]
- 5.Gregersen LH, Bryant DA, Frigaard NU. 2011. Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Front Microbiol 2:116. doi: 10.3389/fmicb.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kostanjevecki V, Brige A, Meyer TE, Cusanovich MA, Guisez Y, van Beeumen J. 2000. A membrane-bound flavocytochrome c-sulfide dehydrogenase from the purple phototrophic sulfur bacterium Ectothiorhodospira vacuolata. J Bacteriol 182:3097–3103. doi: 10.1128/JB.182.11.3097-3103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke PH. 1953. Hydrogen sulphide production by bacteria. J Gen Microbiol 8:397–407. doi: 10.1099/00221287-8-3-397. [DOI] [PubMed] [Google Scholar]
- 8.Papenbrock J, Riemenschneider A, Kamp A, Schulz-Vogt HN, Schmidt A. 2007. Characterization of cysteine-degrading and H2S-releasing enzymes of higher plants: from the field to the test tube and back. Plant Biol 9:582–588. doi: 10.1055/s-2007-965424. [DOI] [PubMed] [Google Scholar]
- 9.Stipanuk MH, Beck PW. 1982. Characterization of the enzymic capacity for cysteine desulphhydration in liver and kidney of the rat. Biochem J 206:267–277. doi: 10.1042/bj2060267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao XH, Krokowski D, Guan BJ, Bederman I, Majumder M, Parisien M, Diatchenko L, Kabil O, Willard B, Banerjee R, Wang B, Bebek G, Evans CR, Fox PL, Gerson SL, Hoppel CL, Liu M, Arvan P, Hatzoglou M. 2015. Quantitative H2S-mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. eLife 4:e10067. doi: 10.7554/eLife.10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura H. 2010. Hydrogen sulfide: from brain to gut. Antioxid Redox Signal 12:1111–1123. doi: 10.1089/ars.2009.2919. [DOI] [PubMed] [Google Scholar]
- 12.Mishanina TV, Libiad M, Banerjee R. 2015. Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat Chem Biol 11:457–464. doi: 10.1038/nchembio.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH. 2009. H2S signals through protein S-sulfhydration. Sci Signal 2:ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishida M, Sawa T, Kitajima N, Ono K, Inoue H, Ihara H, Motohashi H, Yamamoto M, Suematsu M, Kurose H, van der Vliet A, Freeman BA, Shibata T, Uchida K, Kumagai Y, Akaike T. 2012. Hydrogen sulfide anion regulates redox signaling via electrophile sulfhydration. Nat Chem Biol 8:714–724. doi: 10.1038/nchembio.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang R. 2012. Physiological implications of hydrogen sulfide: a whiff exploration that blossomed. Physiol Rev 92:791–896. doi: 10.1152/physrev.00017.2011. [DOI] [PubMed] [Google Scholar]
- 16.Mironov A, Seregina T, Nagornykh M, Luhachack LG, Korolkova N, Lopes LE, Kotova V, Zavilgelsky G, Shakulov R, Shatalin K, Nudler E. 2017. Mechanism of H2S-mediated protection against oxidative stress in Escherichia coli. Proc Natl Acad Sci U S A 114:6022–6027. doi: 10.1073/pnas.1703576114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shatalin K, Shatalina E, Mironov A, Nudler E. 2011. H2S: a universal defense against antibiotics in bacteria. Science 334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 18.Wu LP, Xun LR, Xu L, Hussain A, Shi BY. 2015. A study on neonatal tolerance against Graves' disease in BALB/c mice. Chin Med J (Engl) 128:3243–3246. doi: 10.4103/0366-6999.170274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia Y, Lü C, Hou N, Xin Y, Liu J, Liu H, Xun L. 2017. Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J doi: 10.1038/ismej.2017.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hildebrandt TM, Grieshaber MK. 2008. Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J 275:3352–3361. doi: 10.1111/j.1742-4658.2008.06482.x. [DOI] [PubMed] [Google Scholar]
- 21.Barth M, Ottolenghi C, Hubert L, Chretien D, Serre V, Gobin S, Romano S, Vassault A, Sefiani A, Ricquier D, Boddaert N, Brivet M, de Keyzer Y, Munnich A, Duran M, Rabier D, Valayannopoulos V, de Lonlay P. 2010. Multiple sources of metabolic disturbance in ETHE1-related ethylmalonic encephalopathy. J Inherit Metab Dis 33:443–453. doi: 10.1007/s10545-010-9227-y. [DOI] [PubMed] [Google Scholar]
- 22.Guimaraes BG, Barbosa RL, Soprano AS, Campos BM, de Souza TA, Tonoli CC, Leme AF, Murakami MT, Benedetti CE. 2011. Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J Biol Chem 286:26148–26157. doi: 10.1074/jbc.M111.234039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, Xin Y, Xun L. 2014. Distribution, diversity, and activities of sulfur dioxygenases in heterotrophic bacteria. Appl Environ Microbiol 80:1799–1806. doi: 10.1128/AEM.03281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen J, Keithly ME, Armstrong RN, Higgins KA, Edmonds KA, Giedroc DP. 2015. Staphylococcus aureus CstB is a novel multidomain persulfide dioxygenase-sulfurtransferase involved in hydrogen sulfide detoxification. Biochemistry 54:4542–4554. doi: 10.1021/acs.biochem.5b00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen J, Peng H, Zhang Y, Trinidad JC, Giedroc DP. 2016. Staphylococcus aureus sqr encodes a type II sulfide:quinone oxidoreductase and impacts reactive sulfur speciation in cells. Biochemistry 55:6524–6534. doi: 10.1021/acs.biochem.6b00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson MR, Melideo SL, Jorns MS. 2012. Human sulfide:quinone oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite. Biochemistry 51:6804–6815. doi: 10.1021/bi300778t. [DOI] [PubMed] [Google Scholar]
- 27.Kabil O, Banerjee R. 2012. Characterization of patient mutations in human persulfide dioxygenase (ETHE1) involved in H2S catabolism. J Biol Chem 287:44561–44567. doi: 10.1074/jbc.M112.407411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xin Y, Liu H, Cui F, Liu H, Xun L. 2016. Recombinant Escherichia coli with sulfide:quinone oxidoreductase and persulfide dioxygenase rapidly oxidises sulfide to sulfite and thiosulfate via a new pathway. Environ Microbiol 18:5123–5136. doi: 10.1111/1462-2920.13511. [DOI] [PubMed] [Google Scholar]
- 29.Marcia M, Ermler U, Peng G, Michel H. 2010. A new structure-based classification of sulfide:quinone oxidoreductases. Proteins 78:1073–1083. doi: 10.1002/prot.22665. [DOI] [PubMed] [Google Scholar]
- 30.Sorokin DY, de Jong GAH, Robertson LA, Kuenen GJ. 1998. Purification and characterization of sulfide dehydrogenase from alkaliphilic chemolithoautotrophic sulfur-oxidizing bacteria. FEBS Lett 427:11–14. doi: 10.1016/S0014-5793(98)00379-2. [DOI] [PubMed] [Google Scholar]
- 31.Kusai A, Yamanaka T. 1973. Cytochrome c (553, Chlorobium thiosulfatophilum) is a sulphide-cytochrome c reductase. FEBS Lett 34:235–237. doi: 10.1016/0014-5793(73)80801-4. [DOI] [PubMed] [Google Scholar]
- 32.Visser JM, de Jong GAH, Robertson LA, Kuenen JG. 1997. A novel membrane-bound flavocytochrome c sulfide dehydrogenase from the colourless sulfur bacterium Thiobacillus sp. W5. Arch Microbiol 167:295–301. doi: 10.1007/s002030050447. [DOI] [PubMed] [Google Scholar]
- 33.Yamanaka T, Fukumori Y, Okunuki K. 1979. Preparation of subunits of flavocytochromes c derived from Chlorobium limicola f. thiosulfatophilum and Chromatium vinosum. Anal Biochem 95:209–213. doi: 10.1016/0003-2697(79)90207-0. [DOI] [PubMed] [Google Scholar]
- 34.Frigaard NU, Dahl C. 2009. Sulfur metabolism in phototrophic sulfur bacteria. Adv Microb Physiol 54:103–200. doi: 10.1016/S0065-2911(08)00002-7. [DOI] [PubMed] [Google Scholar]
- 35.Reinartz M, Tschape J, Bruser T, Truper HG, Dahl C. 1998. Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch Microbiol 170:59–68. doi: 10.1007/s002030050615. [DOI] [PubMed] [Google Scholar]
- 36.Rother D, Henrich HJ, Quentmeier A, Bardischewsky F, Friedrich CG. 2001. Novel genes of the sox gene cluster, mutagenesis of the flavoprotein SoxF, and evidence for a general sulfur-oxidizing system in Paracoccus pantotrophus GB17. J Bacteriol 183:4499–4508. doi: 10.1128/JB.183.15.4499-4508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogawa T, Furusawa T, Shiga M, Seo D, Sakurai H, Inoue K. 2010. Biochemical studies of a soxF-encoded monomeric flavoprotein purified from the green sulfur bacterium Chlorobaculum tepidum that stimulates in vitro thiosulfate oxidation. Biosci Biotechnol Biochem 74:771–780. doi: 10.1271/bbb.90815. [DOI] [PubMed] [Google Scholar]
- 38.Morra MJ, Dick WA. 1991. Mechanisms of H2S production from cysteine and cystine by microorganisms isolated from soil by selective enrichment. Appl Environ Microbiol 57:1413–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pohlmann A, Fricke WF, Reinecke F, Kusian B, Liesegang H, Cramm R, Eitinger T, Ewering C, Potter M, Schwartz E, Strittmatter A, Voss I, Gottschalk G, Steinbuchel A, Friedrich B, Bowien B. 2006. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat Biotechnol 24:1257–1262. doi: 10.1038/nbt1244. [DOI] [PubMed] [Google Scholar]
- 40.Grove J, Tanapongpipat S, Thomas G, Griffiths L, Crooke H, Cole J. 1996. Escherichia coli K-12 genes essential for the synthesis of c-type cytochromes and a third nitrate reductase located in the periplasm. Mol Microbiol 19:467–481. doi: 10.1046/j.1365-2958.1996.383914.x. [DOI] [PubMed] [Google Scholar]
- 41.Thony-Meyer L, Fischer F, Kunzler P, Ritz D, Hennecke H. 1995. Escherichia coli genes required for cytochrome c maturation. J Bacteriol 177:4321–4326. doi: 10.1128/jb.177.15.4321-4326.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen KY, Morris JC. 1972. Kinetics of oxidation of aqueous sulfide by O2. Environ Sci Technol 6:529–537. doi: 10.1021/es60065a008. [DOI] [Google Scholar]
- 43.Searcy DG, Whitehead JP, Maroney MJ. 1995. Interaction of Cu,Zn superoxide dismutase with hydrogen sulfide. Arch Biochem Biophys 318:251–263. doi: 10.1006/abbi.1995.1228. [DOI] [PubMed] [Google Scholar]
- 44.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 45.Welte C, Hafner S, Krätzer C, Quentmeier A, Friedrich CG, Dahl C. 2009. Interaction between Sox proteins of two physiologically distinct bacteria and a new protein involved in thiosulfate oxidation. FEBS Lett 583:1281–1286. doi: 10.1016/j.febslet.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 46.Millero FJ, Hubinger S, Fernandez M, Garnett S. 1987. Oxidation of H2S in seawater as a function of temperature, pH, and ionic strength. Environ Sci Technol 21:439–443. doi: 10.1021/es00159a003. [DOI] [PubMed] [Google Scholar]
- 47.Stanley NR, Palmer T, Berks BC. 2000. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J Biol Chem 275:11591–11596. doi: 10.1074/jbc.275.16.11591. [DOI] [PubMed] [Google Scholar]
- 48.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 49.Sanders C, Turkarslan S, Lee DW, Daldal F. 2010. Cytochrome c biogenesis: the Ccm system. Trends Microbiol 18:266–274. doi: 10.1016/j.tim.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oda Y, Star B, Huisman LA, Gottschal JC, Forney LJ. 2003. Biogeography of the purple nonsulfur bacterium Rhodopseudomonas palustris. Appl Environ Microbiol 69:5186–5191. doi: 10.1128/AEM.69.9.5186-5191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beffa T, Blanc M, Lyon PF, Vogt G, Marchiani M, Fischer JL, Aragno M. 1996. Isolation of Thermus strains from hot composts (60 to 80°C). Appl Environ Microbiol 62:1723–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Louie TM, Webster CM, Xun L. 2002. Genetic and biochemical characterization of a 2,4,6-trichlorophenol degradation pathway in Ralstonia eutropha JMP134. J Bacteriol 184:3492–3500. doi: 10.1128/JB.184.13.3492-3500.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harighi B. 2009. Genetic evidence for CheB- and CheR-dependent chemotaxis system in A. tumefaciens toward acetosyringone. Microbiol Res 164:634–641. doi: 10.1016/j.micres.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 54.Guenther EA, Johnson KS, Coale KH. 2001. Direct ultraviolet spectrophotometric determination of total sulfide and iodide in natural waters. Anal Chem 73:3481–3487. doi: 10.1021/ac0013812. [DOI] [PubMed] [Google Scholar]
- 55.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teng F, Murray BE, Weinstock GM. 1998. Conjugal transfer of plasmid DNA from Escherichia coli to enterococci: a method to make insertion mutations. Plasmid 39:182–186. doi: 10.1006/plas.1998.1336. [DOI] [PubMed] [Google Scholar]
- 57.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]