Abstract
The information on candidate cancer driver alterations available from public databases is often descriptive and of limited mechanistic insight, which poses difficulties for reliable distinction between true driver and passenger events. To address this challenge, we performed in-depth analysis of whole-exome sequencing data from cell lines generated by a barrier bypass-clonal expansion (BBCE) protocol. The employed strategy is based on carcinogen-driven immortalization of primary mouse embryonic fibroblasts and recapitulates early steps of cell transformation. Among the mutated genes were almost 200 COSMIC Cancer Gene Census genes, many of which were recurrently affected in the set of 25 immortalized cell lines. The alterations affected pathways regulating DNA damage response and repair, transcription and chromatin structure, cell cycle and cell death, as well as developmental pathways. The functional impact of the mutations was strongly supported by the manifestation of several known cancer hotspot mutations among the identified alterations. We identified a new set of genes encoding subunits of the BAF chromatin remodeling complex that exhibited Ras-mediated dependence on PRC2 histone methyltransferase activity, a finding that is similar to what has been observed for other BAF subunits in cancer cells. Among the affected BAF complex subunits, we determined Smarcd2 and Smarcc1 as putative driver candidates not yet fully identified by large-scale cancer genome sequencing projects. In addition, Ep400 displayed characteristics of a driver gene in that it showed a mutually exclusive mutation pattern when compared with mutations in the Trrap subunit of the TIP60 complex, both in the cell line panel and in a human tumor data set. We propose that the information generated by deep sequencing of the BBCE cell lines coupled with phenotypic analysis of the mutant cells can yield mechanistic insights into driver events relevant to human cancer development.
Introduction
During the course of their lifetime, eukaryotic cells are exposed to various mutagenic processes that cause DNA damage and mutations. Mutation analysis can help uncover specific mutational signatures associated with active or past mutational processes,1, 2, 3 as well as shed light on biological mechanisms critical for tumor development. Most alterations found in tumors are passenger mutations that accumulate during tumorigenesis but do not critically affect cell fitness. However, a small subset of alterations, so-called cancer driver mutations, can confer a selective growth advantage to a cell, which can lead to the expansion of a clonal cell population and tumor development.4 Discriminating driver from passenger events is one of the priorities in cancer research. In order to pinpoint candidate cancer driver alterations among the myriad of somatic mutations available from cancer genome sequencing studies, numerous computational approaches have been developed. These are either gene-centred methods that are based on the mutation frequency of individual genes compared with the background mutation rate5, 6, 7, 8, 9, 10 or network approaches that identify driver genes based on mutual exclusivity of genomic alterations.11, 12, 13, 14, 15, 16 Application of these approaches to mutation data generated by large sequencing consortia led to the following important observations: first, hundreds of high-confidence candidate driver genes have been extracted using these methods, many of which are novel findings.17, 18, 19 Almost 600 genes have been implicated in cancer development to date and are included in the Cancer Gene Census.20 Second, even analyses that are based on highly overlapping mutation data sets vary considerably in the candidate drivers that they identify,17, 18 raising the possibility of a sizable number of false positives among the candidate driver events.
Despite the progress made in recent years, much of the knowledge regarding candidate cancer driver alterations remains descriptive and of limited mechanistic insight, emphasizing the need for rapid experimental systems that allow efficient investigation of the functional impact of candidate driver events. The necessity of a cell to bypass senescence and become immortal in order for a tumor to develop is well established.21 Senescence bypass in rodent cells, which express telomerase and possess long telomeres, can be achieved by mutations in oncogenes and tumor suppressor genes, most importantly those belonging to the p53-p19ARF tumor suppressor pathway.22 In contrast, human cells must also reactivate telomerase in order to bypass senescence, which likely explains why immortalization following exposure of primary human cells to carcinogenic insult is difficult to achieve and has rarely been reported.23, 24, 25 Therefore, rodent cells have been extensively studied to model the events associated with cell immortalization and transformation.22, 26 However, some of the major concerns regarding their applicability include the dependence of these assays on phenotypic readouts to assess transformation and an incomplete understanding of the mechanisms. Recent advances in genome sequencing, together with the development of highly specific pharmacological inhibitors, have created exciting opportunities for mechanistic characterization of candidate cancer driver alterations using in vitro carcinogen exposure assays.
In the present study, we explored how whole-exome sequencing (WES) of carcinogen-immortalized primary mouse embryonic fibroblasts (MEFs) can be utilized to identify candidate cancer driver events. The MEF exposure system takes advantage of a biological barrier, which creates a selective pressure for clonal outgrowth of immortalized cells that have acquired a genetically-driven growth advantage. Importantly, it has already been shown that such barrier bypass-clonal expansion (BBCE) protocols select for immortalized cells with human TP53 hotspot mutations.26, 27, 28, 29 We present a comprehensive analysis of exome-wide sequencing of DNA from 26 immortalized MEF cell lines derived upon carcinogen exposure or other stimulus. Our results reveal the selection of frequent alterations in high-confidence cancer genes and genes affecting biological pathways implicated in critical steps of cell transformation, and bona fide cancer driver mutations. Based on the functional characterization of an established cancer driver mutation as well as of novel, low-frequency candidate driver alterations in a known tumor-suppressive pathway, we propose that the MEF BBCE assay coupled with massively parallel sequencing and functional validation experiments presents a valuable tool for discovery of cancer driver-like events and for characterizing their functional impact.
Results
WES of MEF clones from BBCE assays as a means to identify potential cancer driver events
While analyzing WES data from 14 immortalized cell lines to identify mutational signatures introduced by specific carcinogens, we noticed frequent mutations in known and suspected cancer driver genes.30 Therefore, we generated additional cell lines, in order to perform exome-wide, systematic sequencing analysis of cancer driver-like events followed by in-depth functional characterization of selected alterations (Figure 1). Twenty-six cell lines were derived from primary MEFs,27, 28, 29, 31, 32 19 of which following treatment with potent human carcinogens (aristolochic acid (AA), aflatoxin B1 (AFB1), benzo[a]pyrene (B[a]P), N-methyl-N’-nitro-N-nitrosoguanidine (MNNG) or ultraviolet light subclass C (UVC)). Five of the immortalized lines arose spontaneously from untreated cells and were included in the analysis. In addition, we examined two cell lines from MEFs genetically engineered to express activation-induced cytidine deaminase (Supplementary Table S1). The immortalized lines differed in their morphologies (Supplementary Data S1 and S2), suggesting that cells can take distinct immortalizing paths, each driven by different sets of acquired mutations.
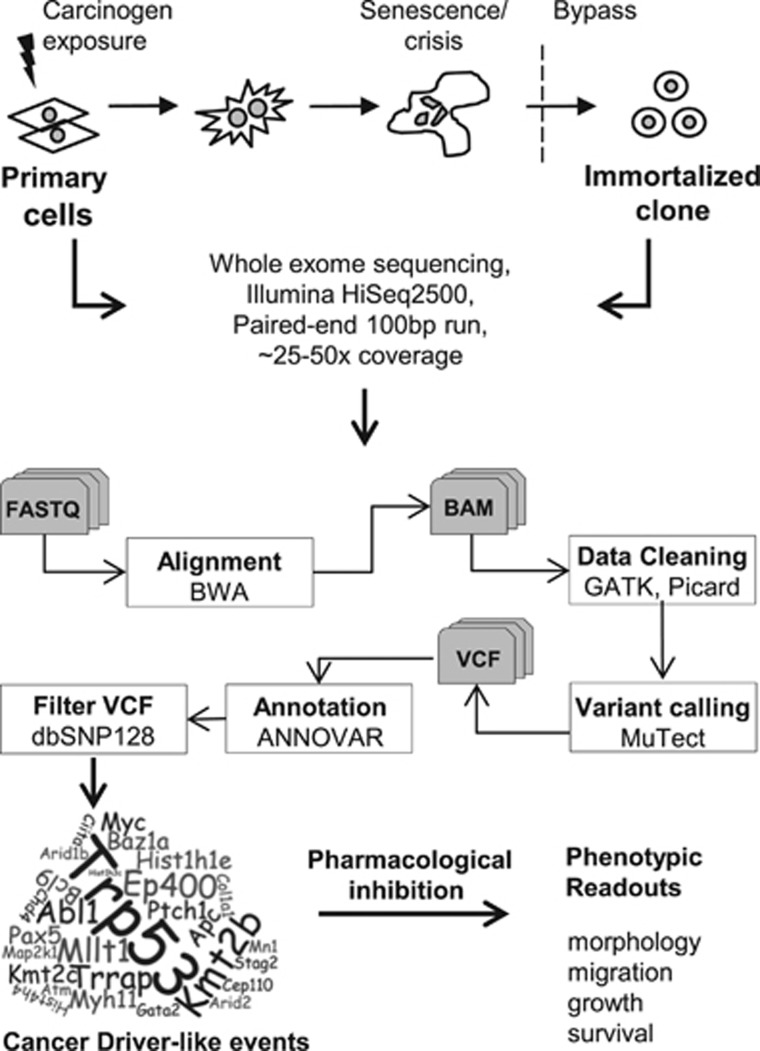
Figure 1.
Study design. MEFs were treated with carcinogens in early passage and cultivated until senescence bypass. The resulting cell lines were subjected to WES on an Illumina HiSeq2500 sequencer and data were analyzed using the indicated pipeline. Sequence variants were systematically analyzed to identify cancer driver-like events, which were further investigated in functional assays with the help of small molecule inhibitors.
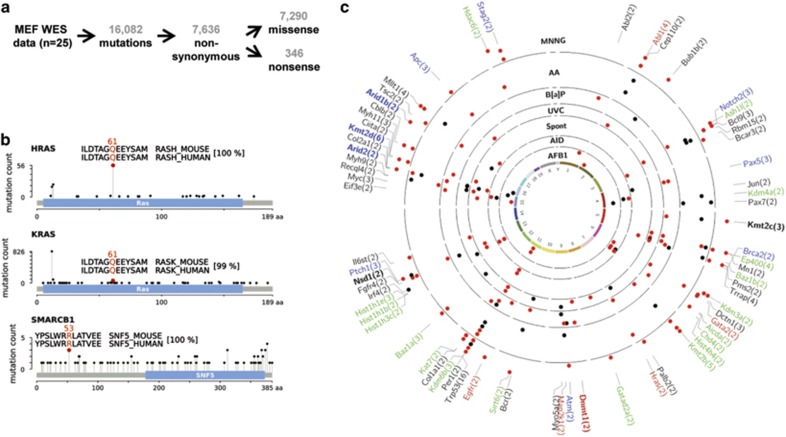
WES was carried out at a depth-of-coverage of 25-50x on primary MEFs and 26 immortalized cell lines (this study and Olivier et al.30). Twenty-five of these lines were used as a test set for analyses and one was used as a control in subsequent functional validation experiments. Owing to the clonal nature of the immortalized cell lines, sequencing at relatively low coverage was sufficient to identify high-confidence variants (Figure 1). In most cell lines, the majority of non-synonymous mutations were detected at an allelic frequency ranging from 25 to 75%, as might be expected for the accumulation of heterozygous mutations combined with clonal expansion in the MEF BBCE assay (Supplementary Figure S1). The number of mutations varied substantially depending on the compound or treatment protocol and was highest in MNNG- and benzo[a]pyrene-treated cells (Supplementary Table S2). In the analyzed set of cell lines, we identified 16 082 single base substitutions, about half of which were non-synonymous, with missense mutations accounting for the vast majority of alterations (Figure 2a, Supplementary Table S2 and Supplementary Data S3). Most carcinogen-induced mutations could be attributed to the predominant substitutions found in human tumors associated with the same exposure (Supplementary Table S2) and they amounted to about double the frequency of the two most common mutation types in spontaneously immortalized cells (69% vs 37%). Among other criteria, this exposure-specific enrichment of mutations introduced early and in a controlled manner was subsequently exploited to identify and track alterations with a potential functional impact on immortalization (see below).
Figure 2.
Global mutation analysis. (a) Overview of WES results from 25 MEF BBCE cell lines. (b) Mutations, found in MEF BBCE cell lines, which were previously identified in tumors. Plots, showing mutations in HRAS, KRAS and SMARCB1 based on TCGA data, were generated using cBioPortal.52, 53 The mutated residue in MEFs is highlighted by a red circle. Alignment of human and mouse protein sequence around the mutated residue is shown in the inset, the mutated codon is indicated above the alignment. The overall similarity of human and mouse protein sequence is indicated in square brackets. (c) Recurrently mutated cancer and epigenetic modifier genes in the MEF BBCE cell lines. Genes listed in the Cancer Gene Census (black,20), oncogenes (red) and tumor suppressor genes (blue) by Vogelstein et al.,4 and epigenetic modifiers (green,34 modified) are indicated. Epigenetic modifiers that are also listed in the Cancer Gene Census are indicated in bold black. Epigenetic modifiers that are also listed as tumor suppressor genes by Vogelstein et al.4 are in bold blue. Epigenetic modifiers that are also listed as oncogenes by Vogelstein et al.4 are in bold red. Cell lines are arranged in concentric circles and grouped by carcinogen exposure (labelled in bold black font). Red and black dots represent exposure-predominant and exposure non-predominant mutation types, respectively.
Hotspot mutations and recurrently mutated cancer genes
The clustering of mutations in certain regions of a gene (hotspots) is potentially indicative of cancer driver events. Interestingly, we identified well-known human cancer hotspot and cancer-related mutations in several cell lines: missense mutations in Hras (c.A182T/p.Q61L) and Kras (c.A182G/p.Q61R), and a mutation in the gene encoding the chromatin remodeling factor Smarcb1 (c.G158A/p.R53Q) (Figure 2b). The applicability of the human mutation data to the mouse proteins is supported by the sequence homology of the proteins between the two species (Figure 2b) and is well-established for the Ras Q61L and Q61R mutations.33
Natural selection favors cells with functional mutations in driver genes, which explains why these genes are recurrently mutated in cancer. Within our set of 25 cell lines, we identified 1 231 recurrently mutated genes, potentially suggesting their selective enrichment during the immortalization process. In a proof-of-principle analysis, we cross-referenced the recurrently mutated genes in the MEFs with human cancer genes4, 20 and high-confidence epigenetic modifier genes (due to their emergence as critical cancer drivers).34, 35 In total, 67 cancer and epigenetic modifier genes were found recurrently mutated in the MEF cell lines (Figure 2c). Besides TP53, the mutated status of which was used in most cell lines as an indicator of clonality and thus preferentially chosen for WES, several other well-established tumor suppressors (for example, Apc, Atm, Brca2 and Ptch1) and oncogenes (for example, Hras, Abl1, Egfr and Myc) were among the recurrently affected genes. Recurrent mutations were also found in a number of genes encoding epigenetic modifiers, most frequently affecting histone H3K4-methylation, histone acetylation and ATP-dependent chromatin remodeling (for example, Kmt2b, Kmt2d, Ep400 and Baz1a) (Figure 2c).
Cancer driver events contribute to the deregulation of critical biological processes such as cell proliferation, apoptosis or DNA repair.4, 21 To assess whether similar processes are also affected in the MEF BBCE assay system, we analyzed the non-synonymous mutations from each one of the 25 cell lines for commonly targeted biological processes and pathways, integrating DAVID and Ingenuity Pathway Analysis (see Materials and Methods for details). In concordance with the importance of deregulating cell proliferation, apoptosis and DNA repair during cellular transformation, we identified these processes among the most frequently and recurrently altered in the immortalized cell lines (Supplementary Data S4). In addition, cell matrix organization, transcription/chromatin structure, pluripotency, cancer-related signaling pathways and drug metabolism (probably due to selective pressure inflicted by carcinogen treatment) were also affected in the majority of cell lines. These findings suggest that MEF BBCE assays are applicable to the identification of cancer driver-like events.
A systematic prioritization scheme for high-confidence candidate driver events
To prioritize potential driver alterations among the hundreds of mutations present in each cell line, we devised a ranking system to select alterations of high interest and explore their functional impact on cell transformation and immortal cell growth. For this proof-of-concept analysis we focused solely on mutations affecting Cancer Gene Census20 and epigenetic modifier genes. Alterations were scored based on multiple criteria, including allelic frequency and potential functional impact (hotspot, truncation, functional domain and predicted deleterious in silico) (see Materials and Methods for details). The inclusion of exposure-specific mutation types into the prioritization criteria allowed us to select for mutations introduced early in the experiment, increasing the likelihood that they contributed to the immortalization process. Using this strategy, we identified candidate driver events in several exposed cell lines, and we focused our subsequent analyses on two cell lines, derived from AA and MNNG treatment (AA_2; MNNG_4). As shown in Table 1, the highest-scoring mutations were distributed among biological processes closely linked to cell transformation and cancer development. They affected well-known cancer genes, such as TP53, Hras, Jak2, Apc, Atm or Brca1, or genes that have previously been implicated in the regulation of cellular senescence. We derived multiple cultures by single-cell subcloning to confirm the overall clonal nature of the AA_2 and MNNG_4 cell lines by Sanger sequencing (Supplementary Figure S2 and S3). With the exception of three mutations, all tested candidate driver events were found consistently altered across the subclones. These results confirm the clonality of AA_2 and MNNG_4 regarding the highest-scoring mutations, permitting follow-up studies on the interplay of co-occurring alterations.
Table 1. Candidate driver mutations in two immortalized MEF cell lines.
| Cell line | Gene symbol | Function | Transcript ID | cDNA change | AA change | Mutated in human tumors (COSMIC, %) | Known to be involved in senescence |
|---|---|---|---|---|---|---|---|
| AA_2 | Cbx7 | PRC1 complex | NM_144811 | c.T32A | p.F11Y | 0.2 | |
| Cdkn1aa | Cell cycle | NM_007669.5 | c.94-2A>T | splice site | 0.3 | Yes | |
| Ep400a | TIP60 complex | NM_029337 | c.A970T | p.R324X | 2.2 | Yes | |
| Ext1 | Glycosaminoglycan metabolism | NM_010162 | c.A103T | p.S35C | 0.5 | ||
| Ext1a | c.A1036T | p.R346X | |||||
| Hrasa | MAPK signaling | NM_001130443 | c.A182T | p.Q61L | 2.7 | ||
| Jak2 | JAK-STAT signaling | NM_001048177 | c.A2479T | p.I827L | 30.0 | ||
| Smarcc1a | BAF complex | NM_009211 | c.A356T | p.H119L | 0.7 | Yes | |
| Smyd1 | H3K4 methylation | NM_009762 | c.T1072A | p.S358T | 0.7 | ||
| Tp53a | Transcription, DNA repair | NM_000546.4 | c.A391T | p.N131Y | 26.8 | Yes | |
| Tp53a | c.A871T | p.K291X | |||||
| MNNG_4 | Apca | Wnt signaling | NM_007462 | c.C8278T | p.P2760S | 10.9 | Yes |
| Atma | DNA repair | NM_007499 | c.C3092T | p.T1031I | 4.2 | Yes | |
| Baz1aa | ACF complex | NM_013815 | c.G392A | p.R131K | 0.8 | ||
| Brca1 | DNA repair | NM_009764 | c.C4322T | p.P1441L | 1.3 | Yes | |
| Gatad2a | Histone deacetylation | NM_001113345 | c.G243A | p.M81I | 0.3 | ||
| Jak1a | JAK-STAT signaling | NM_146145 | c.C1327T | p.P443S | 1.0 | ||
| Jak1a | c.C1286T | p.P429L | |||||
| Kmt2aa | H3K4 methylation | NM_001081049 | c.C9755T | p.P3252L | 1.6 | ||
| Prdm1 | Transcription | NM_007548 | c.C2420T | p.P807L | 1.1 | ||
| Setd1aa | H3K4 methylation | NM_178029 | c.G1499A | p.S500N | 1.2 | ||
| Setd1aa | c.G5095A | p.D1699N | |||||
| Sin3ba | HDAC | NM_009188 | c.C2441T | p.T814I | 0.7 | Yes | |
| Smarcd2a | BAF complex | NM_031878 | c.G497A | p.G166E | 0.3 | ||
| Trrapa | TIP60 complex | NM_001081362 | c.G6952A | p.V2318M | 2.6 | ||
| Tp53a | Transcription, DNA repair | NM_000546.4 | c.C454T | p.P152S | 26.8 | Yes | |
| Tp53a | c.C476T | p.A159V |
Validated by Sanger sequencing (Supplementary Figures S2 and S3).
Oncogenic Ras Q61 hotspot mutations mediate increased proliferation in cells derived from BBCE assays
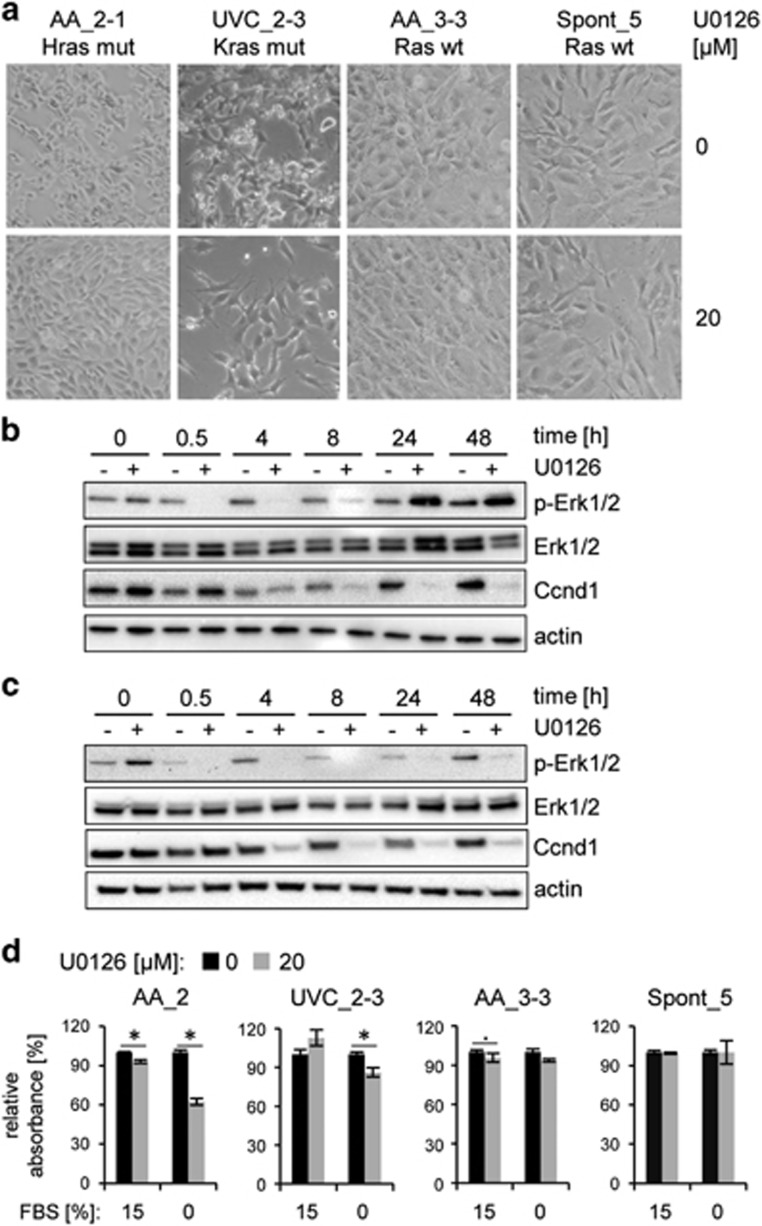
Hras (c.A182T/p.Q61L) and Kras (c.A182G/p.Q61R) are well-characterized driver mutations, which result in constitutive activation of the Ras signaling pathway. Comparison of single-cell subclones harboring Hras and Kras mutations (AA_2-1 and UVC_2-3, respectively), with two immortal clones lacking activating Ras mutations (AA_3-3, Spont_5) revealed clear differences in cell morphology (Figure 3a, top panel). AA_2-1 and, to a slightly lesser extent, UVC_2-3 grew in multilayers, and the cells appeared less tightly attached to the surface than AA_3-3 and Spont_5, reminiscent of a partial transition towards anchorage-independent growth. Moreover, the doubling times of AA_2-1 and UVC_2-3 were approximately 12 h, whereas the other two cell lines had doubling times of around 24 h, which reflects the standard generation time for most immortalized MEFs in this study. These differences could, at least in part, be due to constitutive activation of the Ras pathway in the mutant cell lines. Therefore, we treated the cells with the Mek inhibitor U0126 to inhibit the Ras/Raf/Mek/Erk signaling pathway and observed decreased phospho-Erk levels immediately after start of treatment, followed by a slightly delayed downregulation of the Ras pathway target Ccnd1 (Figures 3b and c). Treatment of cells with 20 μM U0126 for 24 h almost completely reverted the AA_2-1 and UVC_2-3 phenotypes, but it had no effect on the morphology of the control cells (Figure 3a). Next, we determined the proliferation rate of AA_2, UVC_2-3, AA_3-3 and Spont_5 in response to Mek inhibitor treatment (Figure 3d). In contrast to the other cell lines, AA_2 showed a small (7%) but statistically significant decrease in proliferation upon treatment. Given the role of Ras in translating exogenous mitogenic signals, we hypothesized that the effect of the inhibitor on mutant Ras proteins would be potentiated in conditions that limit such signals. Indeed, upon serum starvation, Mek inhibitor treatment resulted in a significant decrease in cell proliferation in the two Ras mutant compared with the wild-type cell lines (Figure 3d). In fact, the IC50 of the Ras mutant cells was about twofold lower than for the wild-type lines (30.9 and 35.3 μM for Ras mutant versus 55.2 & 57.9 μM for Ras wt). These results suggest that only the mutant cell lines have developed a dependency on the Ras signaling pathway and this might be partly responsible for their overall increased proliferation rate.
Figure 3.
Morphology and survival of Ras-mutant and Ras-wild-type cell lines upon Mek inhibitor treatment. (a) Morphology of Ras-mutant (AA_2-1, UVC_2-3) and Ras-wild-type (AA_3-3, Spont_5) cells after 24 h treatment with 20 μM of Mek inhibitor U0126. Immunoblot showing levels of Erk1/2 phosphorylation, total Erk1/2 protein and a target of MAP kinase pathway (Ccnd1) in AA_2-1 (b) and UVC_2-3 (c) cells upon Mek inhibitor U0126 treatment. The immunoblot was carried out using whole-cell protein extracts. Actin was used as loading control. The blot was cropped for display. (d) Proliferation of Ras-mutant and Ras-wild-type cells after 24 h treatment with 20 μM of Mek inhibitor U0126 in complete (15% fetal bovine serum, FBS) and serum-free (0% FBS) growth medium. Relative absorbance (related to treatment with dimethyl sulfoxide (DMSO) carrier), indicative of cell viability, was measured. Columns represent the mean value and s.e.m. derived from three independent experiments. Significance of two sample two-tailed Wilcoxon test is displayed;. = P<0.05 and * = P<0.01.
Novel BAF complex mutations confer sensitivity to Ezh2 inhibition in a Ras-dependent manner in carcinogen-immortalized MEFs
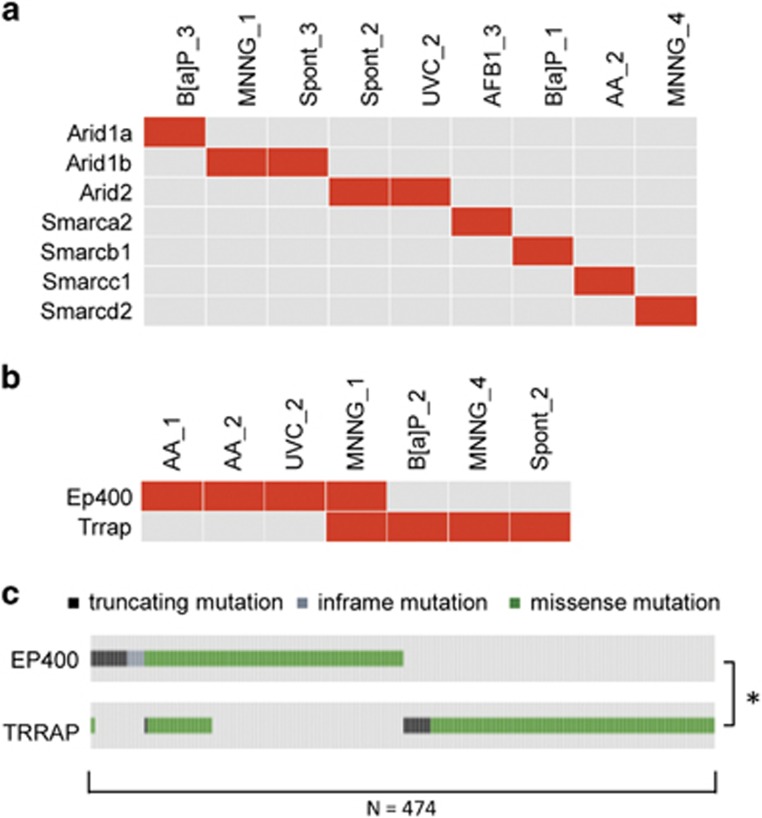
Interestingly, we observed alterations in different subunits of the SWI/SNF (BAF) chromatin remodeling and TIP60 histone acetyltransferase complexes among the mutations identified in the MEF cell lines. Furthermore, all BAF, and the majority of TIP60 complex mutations, were mutually exclusive across the BBCE cell line panel (Figures 4a and b). A similar mutual exclusivity was observed for genes encoding subunits of these multi-protein complexes upon analysis of data sets available through The Cancer Genome Atlas (TCGA) (Figure 4c and Supplementary Figure S4), suggesting that destabilization of a single complex subunit is sufficient to modify its activity.
Figure 4.
Mutual exclusivity analysis of mutations in BAF and TIP60 complex genes. (a) BAF complex subunits mutated in MEF BBCE cell lines. (b) TIP60 complex subunits Ep400 and Trrap mutated in MEF BBCE cell lines. (c) TIP60 complex subunits Ep400 and Trrap mutated in human sequencing studies included in cBioPortal. Result of χ2-test indicated; *P<0.01.
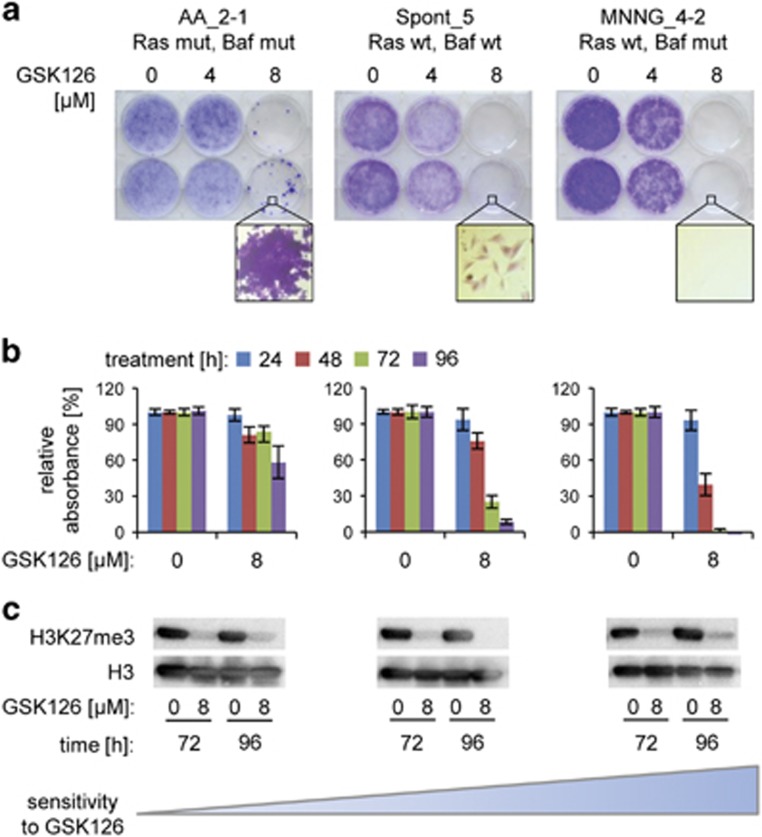
Loss-of-function mutations in subunits of the BAF complex are found in a large number of human cancers.36 Intriguingly, we observed an equally frequent rate of non-synonymous mutations in genes encoding BAF complex subunits in our set of MEF lines (9 out of 26 cell lines). Moreover, previous work revealed an increased dependence of BAF mutant animal tumors and human cancer cell lines on the PRC2 histone methyltransferase complex and this dependency was alleviated in human cancer cells with oncogenic RAS mutations.37, 38 We set out to test whether we could recapitulate this functional relationship using immortalized MEF cell lines harboring previously untested BAF complex mutations, either alone (Smarcd2–MNNG_4) or in combination with oncogenic Ras (Smarcc1, Hras–AA_2). Spontaneously immortalized Spont_5 cells (BAF and Ras wild type), MNNG_4-2 (BAF mutant and Ras wild type) and AA_2-1 (BAF and Ras mutant) were treated with Ezh2 inhibitor (GSK126), and cell viability was assessed using MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) and colony formation assays (Figures 5a and b). Spont_5 cells exhibited a significant decrease in cell viability following treatment, but both assays indicated that a fraction of cells survived. In contrast, the MNNG_4-2 cell line was highly sensitive to GSK126 treatment and showed no remaining viability under the same treatment conditions. The observed GSK126 sensitivity of both BAF wild-type and mutant cells, with a more pronounced effect in the mutants, recapitulates previous findings in MEFs isolated from wild-type and Arid1a mutant animals.38 Finally, AA_2-1, which harbors BAF and Ras alterations, was much more resistant to Ezh2 inhibitor treatment. The same order of sensitivity was observed in both the MTT and colony formation assays. An analogous trend of decreased sensitivity of a BAF/Ras mutant compared with multiple BAF mutant cell lines upon treatment with GSK126 was observed in a replicate experiment with an independent set of cell lines (Arid2, Kras–UVC_2 vs Arid1b–MNNG_1; Smarca2–AFB1_3; Smarcb1–BaP_1) (Supplementary Figure S5). Immunoblotting analysis showed a decrease in H3K27me3 levels upon inhibitor treatment in all tested cell lines (Figure 5c and Supplementary Figure S5c). Taken together, we show that mutations in several previously untested BAF complex subunits consistently confer (oncogenic) Ras-dependent sensitivity to Ezh2 inhibition, consistent with findings for other BAF complex subunits in human cancer cell lines.
Figure 5.
Effect of Ezh2 inhibitor treatment in BAF-mutant, BAF-wild-type and BAF/Ras double-mutant MEF BBCE cell lines. (a) Cells mutant in Ras and BAF (AA_2-1), wild type in Ras and BAF (Spont_5), and cells with BAF mutation in a wild-type Ras background (MNNG_4-2) were seeded at low density in standard six-well plates and treated with Ezh2 inhibitor GSK126 for 7 days. Cells were then visualized using crystal violet. The result is representative of three independent experiments. (b) Cells mutant in Ras and BAF (AA_2-1), wild type in Ras and BAF (Spont_5), and cells with BAF mutation in a wild-type Ras background (MNNG_4-2) were plated in 96-well plates and treated with GSK126 for up to 96 h. Relative absorbance (related to treatment with dimethyl sulfoxide (DMSO) carrier), indicative of cell viability was measured at 24, 48, 72 and 96 h. Columns represent mean value and s.e.m.derived from three independent experiments. (c) Immunoblots for the H3K27me3 chromatin mark in all tested cell lines upon GSK126 treatment. Immunoblot was carried out using acid-extracted histones. Histone H3 immunoblot was performed to control for the baseline level of the protein. The blots were cropped for display.
Discussion
Building on knowledge gained from single-gene sequencing studies, large-scale tumor sequencing efforts have transformed the field of cancer genetics over the last years. This has led to an explosion in the number of genes implicated in cancer development. In this study we report that a simple, cell-based in vitro carcinogen exposure assay, combined with massively parallel sequencing, can contribute to the identification of candidate cancer driver events from human tumor sequencing data. As a proof-of-concept, our results demonstrate the functional impact of known activating mutations in Ras genes on the immortalized cell phenotype and, potentially, on subsequent steps leading to transformation. Furthermore, we show that mutations in BAF complex genes that were not implicated as putative drivers by cancer genome sequencing studies (Smarcc1 and Smarcd2), recapitulate cancer-specific sensitivity to Ezh2 inhibition, suggesting that BBCE assays can be utilized to identify and study the functional impact of candidate cancer driver events.
Tumor-sequencing databases put the mutation frequency of all three human RAS genes combined between 9 and 30% (all cancer types), with 98% of mutations affecting amino acid residues G12, G13 or Q61.39 The Q61L and Q61R alterations, found in AA_2 and UVC_2, respectively, are among the most highly transforming mutations in this residue in NIH-3T3 cells.40 They lock Ras in a constitutively active state and Q61L is the most frequent HRAS mutation in human prostate adenocarcinoma.41 We note that the Q61L and Q61R Ras protein alterations found in our in vitro exposure system are the most predominant cancer-associated changes identified at this position.41 The observed Ras hotspot mutations provide support to the notion that the MEF BBCE protocol can specifically select for cancer-associated genetic alterations.
Genome-sequencing studies established the BAF ATP-dependent chromatin remodeling complex as one of the most commonly mutated human tumor suppressors.42, 43 Mutations in several subunits of the complex (SMARCB1, SMARCA4, SMARCA2, ARID1A and PBRM1) sensitize cancer cells to either inactivation of the EZH2 Polycomb protein, or treatment with an EZH2 inhibitor, and this effect is alleviated upon co-occurring RAS mutation.37, 38 Similarly, independently derived MEF cell lines harboring BAF complex mutations (Arid1b, Smarcd2, Smarca2 and Smarcb1) displayed high sensitivity to Ezh2 inhibitor treatment, whereas the two cell lines with co-occurring BAF and Ras mutations present in our cell panel (Arid2-Kras and Smarcc1-Hras) were relatively unresponsive (Figure 5 and Supplementary Figure S5). Our studies using in vitro immortalization assays therefore considerably expand the set of known BAF complex mutants exhibiting a cancer-related interplay with PRC2 histone methyltransferase activity. Interestingly, among the tested BAF complex mutants, neither SMARCD2 nor SMARCC1 has previously been described as a candidate driver gene in human tumor-sequencing studies and, therefore, these genes have not been captured in the Sanger COSMIC Cancer Gene Census. Compared with other genes encoding BAF subunits, both are mutated at relatively low frequencies. However, recent saturation analysis suggests the existence of many more infrequently mutated cancer drivers18 and the significant positive selection of Smarcd2 and Smarcc1 in the in vitro assay, combined with the results from the Ezh2 inhibitor experiment, certainly warrants additional studies regarding their role in the development of particular cancer types.
Our finding that more than 30% of cell lines derived from MEF BBCE assays harbor non-synonymous mutations in BAF subunits approximates the prevalence of non-synonymous mutations in human cancers (>20%). Similarities between the mutation profiles of BBCE cell lines and human cancers are further highlighted by the presence of a known cancer hotspot mutation in Smarcb1 among the in vitro-induced alterations and by the non-overlapping nature of mutations in BAF complex components in exposed MEFs (Figure 4a). The latter is in agreement with the recently described non-overlapping nature of mutations in BAF subunits in sequencing data derived from more than 3000 TCGA samples.19 Intriguingly, we also observed mutual exclusivity of mutations in genes encoding the Ep400 and Trrap subunits of the TIP60 histone acetyltransferase complex in our cell line panel as well as in a set of 474 TCGA samples (Figure 4). Although the Ep400 subunit of the TIP60 complex is not an established cancer driver, its mutual exclusivity with Trrap mutations is consistent with the notion from cancer genome sequencing that the selection of a mutation in a single component is sufficient to alter the activity of a pathway or protein complex, while obviating the need for additional changes,19 and with network approaches that identify driver genes based on mutual exclusivity of genomic alterations.11, 12, 13, 14, 15, 16 Identification and functional characterization of candidate driver events beyond what has been highlighted by cancer genome sequencing projects (Smarcc1, Smarcd2 and Ep400) justifies the utilization of BBCE assays to both model and examine cancer driver events. Extended analysis of existing BBCE lines and sequencing of additional cell lines is warranted to further investigate broader commonalities between MEFs and human tumors.
It is important to keep in mind that the presence of heterozygous or homo/hemizygous TP53 mutations was used as an indicator of clonality for choosing cell lines for exome sequencing. Therefore, most candidate driver events we identified act as such in the context of TP53 alterations. This situation resembles what is commonly found in human tumors. Some immortalized cell lines, however, retain wild-type p53. It will be interesting to investigate whether in this context other key regulators of the p53 pathway are affected,44 and whether CRISPR/Cas9-mediated correction of potential driver events in the same gene, protein complex or pathway results in diverse functional outcomes depending on the p53 status.
Several key characteristics of the BBCE assay highlight its applicability as a promising in vitro screening strategy for characterizing candidate driver events. These include the barrier bypass step (biological selection), defined exposure conditions (identification of early driver mutations), applicability of genome editing technologies (CRISPR/Cas9) and a favorable experimental timeline (6–8 weeks). The use of exogenous human liver S9 fraction to activate pro-carcinogens and MEF culture under physiological oxygen conditions to reduce the background mutation rate should further improve the efficiency and stringency of MEF BBCE assays. In the future, the assessment of tumorigenicity in nude mice of immortalized cell lines that have undergone massively parallel sequencing analysis and the development of clonal expansion assays using human cells23, 45 or organoids46 will be promising new avenues for detailed characterization of candidate driver events.
Materials and methods
Cell lines
Sixteen of the 26 Hupki (humanized p53 knock-in) MEF cell lines (Supplementary Table S1) were generated from carcinogen-exposed and -unexposed primary MEFs.27, 28, 29, 30, 31, 32 The additional 10 cell lines were established for this study using the same procedure.47, 48 Briefly, carcinogen-exposed or untreated primary cells were cultivated until senescence bypass and immortalized cell lines with non-synonymous TP53 mutations were preferentially chosen for WES (Supplementary Table S1). Clonal populations were generated from cell lines AA_2, AA_3, MNNG_4, MNNG_1, AFB1_3, BaP_1 and UVC_2 by dilution cloning.
Whole-exome sequencing (WES) and data processing
Sequencing data used in this study were generated previously for 14 of the 26 cell lines;30 for this report, 12 additional cell lines were sequenced and the data processed as described in Olivier et al.30 An average of 51.44 million reads (100 bp) were sequenced per sample, of which 98% were mapped, 75% on target (mm9 reference genome), with a mean depth-of-coverage of 54 (software used: BWA-MEM v0.7.15, GATK v3.6-0, Picard tools v2.4.1 (Broad Institute, Cambridge, MA, USA)). Bam files were uploaded to NCBI BioProjects web site, accession number PRJNA238303. Variants were called with MuTect software (version 1.1.4, Broad Institute) using default parameters. Each immortalized cell line was compared to multiple primary cultures and only overlapping calls were considered, to ensure robust variant calling and exclude germline variants.
Pathway analysis
RefSeq-annotated mouse genes containing non-synonymous single-base substitutions were analyzed using DAVID49 and Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA, USA). If RefSeq gene names were not recognized, aliases were used. Gene Ontology and KEGG pathways were interrogated by DAVID using relaxed criteria, as deregulation of biological processes in transformed cells can occur in the absence of multiple hits. Ingenuity Pathway Analysis was run with default settings and canonical pathways were extracted using either standard (P<0.05) or relaxed criteria (P<0.175). The identified biological processes and pathways were prioritized based on recurrence among cell lines and cancer relevance.
Identification of candidate cancer driver mutations
Variants were filtered for exonic non-synonymous and splicing mutations, and cross-referenced with cancer-related and chromatin associated genes.4, 20, 34, 50 Mutations were prioritized using a simple scoring system. A score of 1 was added if the mutation was of the exposure-predominant type (likely introduced early in the assay). A score of 1 was added if the mutation was in a known human hotspot, if it was truncating or affected a splice site. A score of 0.5 was added if the mutation was located in a functional domain and if the mutation was predicted deleterious in the protein by SIFT via Variant Effect Predictor.51 A score of 0.5 was also added if the allelic frequency of the mutation was higher than 25%.
Sanger sequencing
Sequencing primers are listed in Supplementary Table S3.
Inhibitor treatment
Cells were treated with 20 μM Mek inhibitor U0126 (Sigma, St Louis, MO, USA), 4, 8 and 16 μM Ezh2 inhibitor GSK126 (Xcess Biosciences, San Diego, CA, USA), and dimethyl sulfoxide carrier (Sigma).
MTT proliferation assay
Cells were plated in 96-well plates and treated as indicated. Cell viability was measured using CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Fitchburg, WI, USA). Plates were incubated for 2 h at 37 °C and absorbance was measured at 492 nm.
Colony formation assay
Cells were seeded in six-well plates to ensure ~5000 cells at treatment onset for each cell line and condition. Colonies were visualized after 7 days using crystal violet staining.
Immunoblotting and antibodies
Electrophoresis was performed using 4–20% Mini-Protean TGX Precast Protein Gels (BioRad, Hercules, CA, USA). The following antibodies were used for immunoblotting: phospho-Erk1/2 (9101, 1:1000, Cell Signaling, Danvers, MA, USA), total Erk1/2 (9102, 1:1000, Cell Signaling), histone H3K27me3 (ab6002, 1:4000, Abcam, Cambridge, UK), histone H3 (ab1791, 1:20 000, Abcam), Ccnd1 (NCL-L-CYCLIN D1-GM, 1:100, Leica Biosystems-Novocastra, Wetzlar, Germany), Actin (08691001, 1:25 000, MP Biomedicals, Santa Ana, CA, USA).
Mutation analysis in human tumor data
Published studies included in cBioPortal for Cancer Genomics52, 53 were mined for samples with non-synonymous mutations and small indels in the Ep400 and Trrap subunits of the TIP60 complex and in BAF complex subunits. Duplicates and samples missing mutation annotation were excluded. Data were visualized using OncoPrinter version 1.0.1. Mutual exclusivity was tested using χ2-test.
Acknowledgments
We thank the group of Hiroyuki Marusawa, Kyoto University, Japan, for providing cells from Hupki-activation-induced cytidine deaminase transgenic mice, Adriana Heguy (NYU Genome Technology Center) for next-generation sequencing support, Christine Carreira (IARC) for antibodies, the Centre Léon Bérard in Lyon, France, for providing computational capacity, and Leigh Ellis, Roswell Park Cancer Institute for technical advice. This work was supported by INCa–INSERM Plan Cancer 2015 ENV201507 Grant to JZ; Ministry of Education, Youth and sports of the Czech Republic (LQ1604), European Regional Development Fund (CZ.1.05/1.1.00/02.0109), Czech Science Foundation GAČR (GAČR 16-27790A) and Ministry of Health/Grant Agency for Health Research of the Czech Republic (AZV 16-27790A) to TS; and Charles University in Prague institutional financing ([LH15170, UNCE 204021) and the Mobility fund, First Faculty of Medicine, Charles University in Prague, Czech Republic to KV. NYU Technology Center is funded in part by grant NIH/NCI P30 CA016087-33.
Author contributions
MK and HH: Designed the study, acquired data and led the analysis and interpretation of results, wrote the manuscript and approved the final version. JZ and MH: Conceived and designed the study and interpreted the results, revised the manuscript and approved the final version. MO: Designed the study and analysed data, revised the manuscript and approved the final version. MA, AW, KV, SB and SV: Acquired and analysed data, revised the manuscript and approved the final version. TS and ZH: Interpreted the results, revised the manuscript and approved the final version.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Hollstein M, Alexandrov LB, Wild CP, Ardin M, Zavadil J. Base changes in tumour DNA have the power to reveal the causes and evolution of cancer. Oncogene 2017; 36: 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet 2014; 15: 585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev 2014; 24: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW. Cancer genome landscapes. Science 2013; 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn A, Simon R. Identifying cancer driver genes in tumor genome sequencing studies. Bioinformatics 2011; 27: 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees ND, Zhang Q, Kandoth C, Wendl MC, Schierding W, Koboldt DC et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res 2012; 22: 1589–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez A, Lopez-Bigas N. Functional impact bias reveals cancer drivers. Nucleic Acids Res 2012; 40: e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics 2013; 29: 2238–2244. [DOI] [PubMed] [Google Scholar]
- Reimand J, Bader GD. Systematic analysis of somatic mutations in phosphorylation signaling predicts novel cancer drivers. Mol Syst Biol 2013; 9: 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeang CH, McCormick F, Levine A. Combinatorial patterns of somatic gene mutations in cancer. FASEB J 2008; 22: 2605–2622. [DOI] [PubMed] [Google Scholar]
- Miller CA, Settle SH, Sulman EP, Aldape KD, Milosavljevic A. Discovering functional modules by identifying recurrent and mutually exclusive mutational patterns in tumors. BMC Med Genomics 2011; 4: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Cerami E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res 2012; 22: 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandin F, Upfal E, Raphael BJ. De novo discovery of mutated driver pathways in cancer. Genome Res 2012; 22: 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang S, Wu LY, Zhang XS. Efficient methods for identifying mutated driver pathways in cancer. Bioinformatics 2012; 28: 2940–2947. [DOI] [PubMed] [Google Scholar]
- Leiserson MD, Blokh D, Sharan R, Raphael BJ. Simultaneous identification of multiple driver pathways in cancer. PLoS Comput Biol 2013; 9: e1003054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborero D, Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Kandoth C, Reimand J et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep 2013; 3: 2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014; 505: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Vandin F, Wu HT, Dobson JR, Eldridge JV, Thomas JL et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet 2015; 47: 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R et al. A census of human cancer genes. Nat Rev Cancer 2004; 4: 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Odell A, Askham J, Whibley C, Hollstein M. How to become immortal: let MEFs count the ways. Aging (Albany NY) 2010; 2: 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stampfer MR, Bartley JC. Induction of transformation and continuous cell lines from normal human mammary epithelial cells after exposure to benzo[a]pyrene. Proc Natl Acad Sci USA 1985; 82: 2394–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson PL, Vrba L, Stampfer MR, Futscher BW. Exome-wide mutation profile in benzo[a]pyrene-derived post-stasis and immortal human mammary epithelial cells. Mutat Res Genet Toxicol Environ Mutagen 2014; 775-776: 48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med 2002; 347: 1593–1603. [DOI] [PubMed] [Google Scholar]
- vom Brocke J, Schmeiser HH, Reinbold M, Hollstein M. MEF immortalization to investigate the ins and outs of mutagenesis. Carcinogenesis 2006; 27: 2141–2147. [DOI] [PubMed] [Google Scholar]
- Liu Z, Hergenhahn M, Schmeiser HH, Wogan GN, Hong A, Hollstein M. Human tumor p53 mutations are selected for in mouse embryonic fibroblasts harboring a humanized p53 gene. Proc Natl Acad Sci USA 2004; 101: 2963–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Muehlbauer KR, Schmeiser HH, Hergenhahn M, Belharazem D, Hollstein MC. p53 mutations in benzo(a)pyrene-exposed human p53 knock-in murine fibroblasts correlate with p53 mutations in human lung tumors. Cancer Res 2005; 65: 2583–2587. [DOI] [PubMed] [Google Scholar]
- Nedelko T, Arlt VM, Phillips DH, Hollstein M. TP53 mutation signature supports involvement of aristolochic acid in the aetiology of endemic nephropathy-associated tumours. Int J Cancer 2009; 124: 987–990. [DOI] [PubMed] [Google Scholar]
- Olivier M, Weninger A, Ardin M, Huskova H, Castells X, Vallee MP et al. Modelling mutational landscapes of human cancers in vitro. Sci Rep 2014; 4: 4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmeyer N, Schmeiser HH, Muehlbauer KR, Belharazem D, Knyazev Y, Nedelko T et al. Further studies with a cell immortalization assay to investigate the mutation signature of aristolochic acid in human p53 sequences. Mutat Res 2006; 608: 163–168. [DOI] [PubMed] [Google Scholar]
- Reinbold M, Luo JL, Nedelko T, Jerchow B, Murphy ME, Whibley C et al. Common tumour p53 mutations in immortalized cells from Hupki mice heterozygous at codon 72. Oncogene 2008; 27: 2788–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westcott PM, Halliwill KD, To MD, Rashid M, Rust AG, Keane TM et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature 2015; 517: 489–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez A, Jene-Sanz A, Lopez-Bigas N. The mutational landscape of chromatin regulatory factors across 4,623 tumor samples. Genome Biol 2013; 14: r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet 2013; 14: 765–780. [DOI] [PubMed] [Google Scholar]
- Helming KC, Wang X, Roberts CW. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell 2014; 26: 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 2010; 18: 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Kim W, Howard TP, Vazquez F, Tsherniak A, Wu JN et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med 2015; 21: 1491–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 2014; 13: 828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der CJ, Finkel T, Cooper GM. Biological and biochemical properties of human rasH genes mutated at codon 61. Cell 1986; 44: 167–176. [DOI] [PubMed] [Google Scholar]
- Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res 2012; 72: 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 2013; 45: 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS ONE 2013; 8: e55119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Shin JS, Moon JH, Hong SW, Jung DJ, Kim JH et al. Foxp3 is a key downstream regulator of p53-mediated cellular senescence. Oncogene 2017; 36: 219–230. [DOI] [PubMed] [Google Scholar]
- Poon SL, Pang ST, McPherson JR, Yu W, Huang KK, Guan P et al. Genome-wide mutational signatures of aristolochic acid and its application as a screening tool. Sci Transl Med 2013; 5: 197ra01. [DOI] [PubMed] [Google Scholar]
- Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, Sasaki N et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 2016; 538: 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis JE. Cell Biology: A Laboratory Handbook3rd edElsevier Academic Press: Amsterdam, 2006. 4. [Google Scholar]
- Liu Z, Belharazem D, Muehlbauer KR, Nedelko T, Knyazev Y, Hollstein M. Mutagenesis of human p53 tumor suppressor gene sequences in embryonic fibroblasts of genetically-engineered mice. Genet Eng (NY) 2007; 28: 45–54. [DOI] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 2003; 4: P3. [PubMed] [Google Scholar]
- Cancer Gene Census online. http://cancer.sanger.ac.uk/census.
- Ensembl Variant Effect Predictor web interface. http://www.ensembl.org/vep.
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.