Abstract
Ewing sarcoma (EWS) is a paediatric bone cancer with high metastatic potential. Cellular plasticity resulting from dynamic cytoskeletal reorganization, typically regulated via the Rho pathway, is a prerequisite for metastasis initiation. Here, we interrogated the role of the Ewing sarcoma driver oncogene EWS-FLI1 in cytoskeletal reprogramming. We report that EWS-FLI1 strongly represses the activity of the Rho-F-actin signal pathway transcriptional effector MRTFB, affecting the expression of a large number of EWS-FLI1-anticorrelated genes including structural and regulatory cytoskeletal genes. Consistent with this finding, chromatin immunoprecipitation sequencing (ChIP-seq) revealed strong overlaps in myocardin-related transcription factor B (MRTFB) and EWS-FLI1 chromatin occupation, especially for EWS-FLI1-anticorrelated genes. Binding of the transcriptional co-activator Yes-associated protein (YAP)-1, enrichment of TEAD-binding motifs in these shared genomic binding regions and overlapping transcriptional footprints of MRTFB and TEAD factors led us to propose synergy between MRTFB and the YAP/TEAD complex in the regulation of EWS-FLI1-anticorrelated genes. We propose that EWS-FLI1 suppresses the Rho-actin pathway by perturbation of a MRTFB/YAP-1/TEAD transcriptional module, which directly affects the actin-autoregulatory feedback loop. As spontaneous fluctuations in EWS-FLI1 levels of Ewing sarcoma cells in vitro and in vivo, associated with a switch between a proliferative, non-migratory EWS-FLI1-high and a non-proliferative highly migratory EWS-FLI1-low state, were recently described, our data provide a mechanistic basis for the underlying EWS-FLI1-dependent reversible cytoskeletal reprogramming of Ewing sarcoma cells.
Introduction
Ewing sarcoma (EwS) is the second most common bone malignancy affecting children and young adolescents. This highly aggressive cancer is prone to early tumour dissemination, and about a quarter of patients present with overt metastases at initial diagnosis.1 Pathways underlying metastatic processes in EwS are still poorly investigated.2 EwS is characterized by the expression of an oncogenic E-twenty-six transformation specific (ETS) fusion product, most frequently EwS-FLI1, which drives proliferation and represses differentiation via global genetic and epigenetic deregulation.3, 4, 5 EWS-FLI1 affects expression of hundreds of genes either by upregulation (EWS-FLI1-correlated genes) or repression (EWS-FLI1-anticorrelated genes).6, 7 There is emerging evidence of EWS-FLI1 regulating cellular plasticity mainly by repressing genes involved in cytoskeletal reorganization.8, 9, 10, 11 The major signalling cascade linking extracellular mechanic and chemical inputs to gene expression is the Rho-F-actin pathway upon activation of transmembrane receptors such as G-protein coupled receptors.12 Rho-downstream effectors promote polymerization of monomeric G-actin to F-actin fibres. The transcriptional co-activators myocardin-related transcription factors (MRTF) are tightly bound to G-actin in the cytoplasm. Upon depletion of G-actin, MRTFs are released and translocated to the nucleus where they interact with cofactor-dependent transcription factors, typically with serum response factor (SRF) on CCW6GG (CArG) elements. Depending on the recruited co-activator, such as the ternary complex factor family or MRTFs, SRF regulates distinct sets of target genes. MRTFs, which comprise MRTFA and MRTFB, are key effectors of the cytoskeletal autoregulatory feedback loop by mediating transcription of numerous cytoskeletal genes in a Rho-actin-dependent manner.13, 14 Furthermore, MRTFs were reported to have important roles for the metastatic propensity of aggressive cancer cell lines.15, 16
Here, we aimed at understanding the molecular mechanism by which EWS-FLI1 compromises genome-wide Rho-actin-mediated transcription in EwS. We demonstrate overall repression of MRTFA/B transcriptional activity by EWS-FLI1 and a strong overlap of MRTFB and EWS-FLI1 chromatin occupation. We show that EwS-FLI1 inhibits MRTFB-mediated co-activation of TEA domain (TEAD) and Yes-associated protein (YAP)-1-regulated EWS-FLI1-anticorrelated genes downstream of Rho leading to perturbed feedback regulation of cytoskeletal dynamics.
Results
Rho target genes are repressed by EWS-FLI1 in EwS
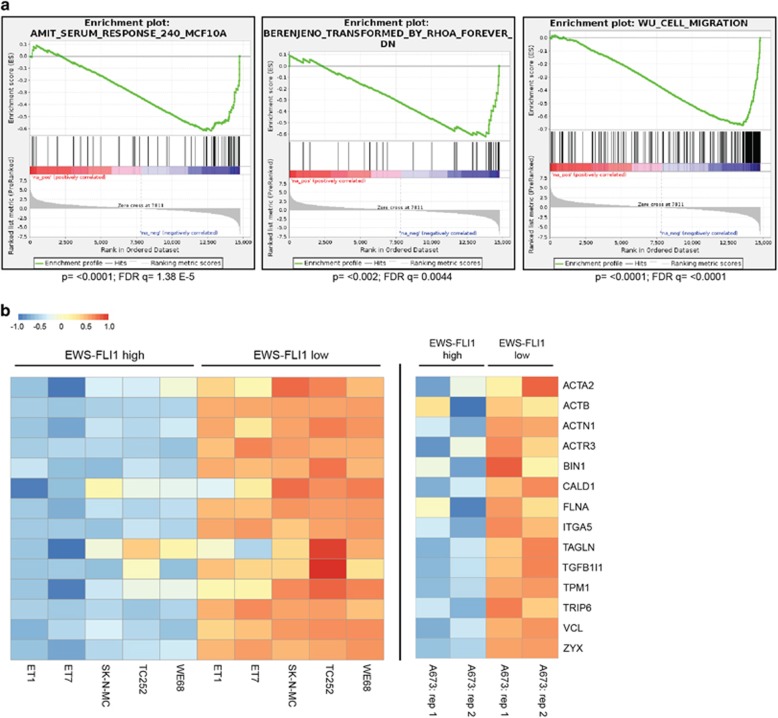
To identify pathways deregulated in EwS, we compared published gene expression data sets from EwS primary tumours17 to the putative EwS precursor mesenchymal stem cells.18, 19 Gene set enrichment analysis identified a significant over-representation of Rho pathway components among genes repressed in EwS. We found that expression of genes involved in migration and response to serum and RhoA signalling is compromised in EwS as compared to mesenchymal stem cells (Figure 1a). To validate suppression of a Rho signature in EwS, we interrogated expression of a manually curated set of 14 well-characterized Rho/SRF/MRTF target genes20, 21 in five EwS cell lines (WE68, TC252, SK-N-MC, STA-ET-7.2 and STA-ET-1)6 from previously published expression data (GSE14543) with a transient EWS-FLI1 knockdown (Figure 1b). Depletion of EWS-FLI1 resulted in upregulation of the majority of genes in this gene set in all cell lines tested. We recapitulated these results further in A673/TR/shEF EwS cells, where EWS-FLI1 levels can be modulated from endogenous (high) levels to low levels (Figure 1b and Supplementary Figure S1A). A panel of genes was additionally validated by quantitative reverse transcriptase polymerase chain reaction (qRT–PCR) upon EWS-FLI1-high and -low states in A673/TR/shEF. Expression of zyxin (ZYX), vinculin (VCL) and transgelin (TAGLN) was strongly induced under EWS-FLI1-low conditions, while no significant change in SRF mRNA was observed (Supplementary Figure S1B). These data are consistent with suppression of Rho-downstream transcriptional targets by EWS-FLI1 in EwS.
Figure 1.
Rho target genes are repressed by EwS-FLI1 in EwS. (a) Gene set enrichment analysis (GSEA) plots showing the differential analysis of 117 EwS tumours (GSE34620)17 versus mesenchymal stem cells (MSCs; GSE31215).18 Genes involved in the response to serum, RhoA or are involved in migration are repressed in the EwS tumours versus MSCs. (b) Heatmap showing a manually curated set of Rho/SRF target genes under EWS-FLI1-high or -low conditions in six different EwS cell lines: left panel: WE68, TC252, SK-N-MC, STA-ET-7.2, STA-ET-1 (data from GSE14543),6 and right panel: A673/TR/shEF, this study: two replicates. Expression values are scaled row-wise.
MRTFB knockdown antagonizes the transcriptional effects of EWS-FLI1 depletion
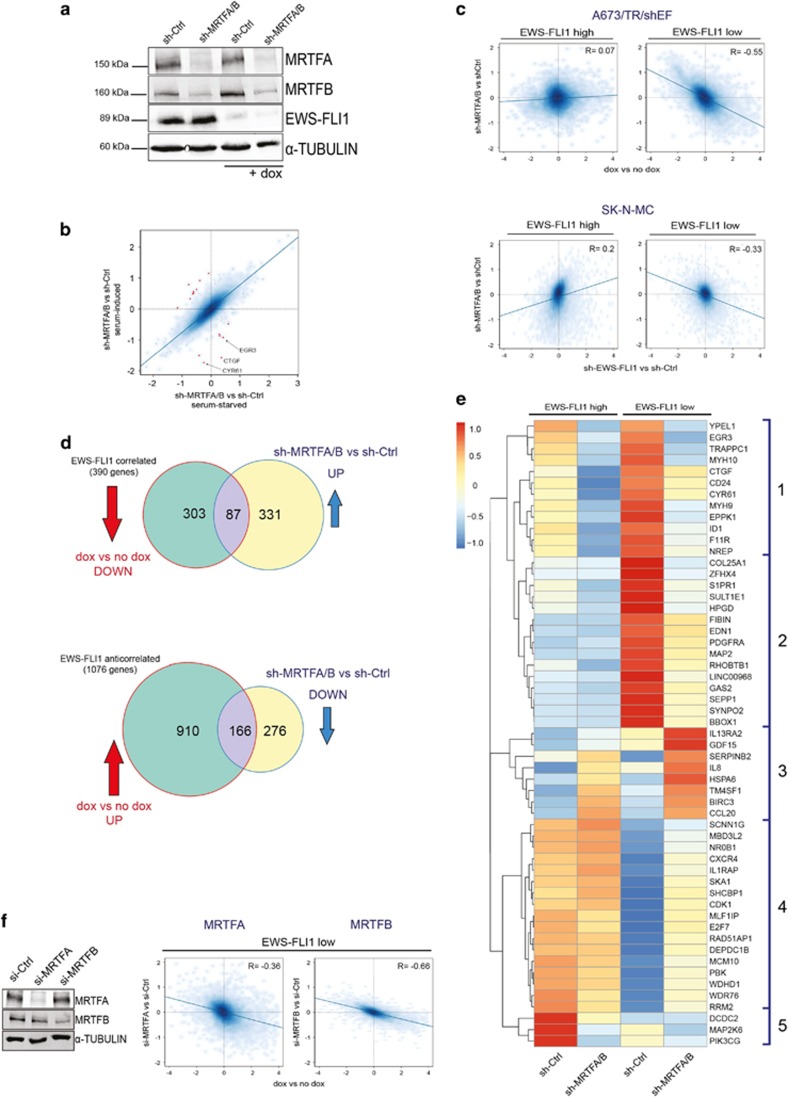
We next tested the hypothesis that EWS-FLI1 suppresses Rho target genes by interfering with the Rho transcriptional effectors MRTFA/B. To this end, gene expression analysis was performed upon modulation of MRTFA/B under EWS-FLI1-high and -low conditions. Effective reduction of MRTFA and MRTFB protein was achieved using a double-targeting short hairpin RNA vector,22 which was combined with doxycycline (dox) treatment to induce the knockdown of EWS-FLI1 (Figure 2a and Supplementary Figure S2A). In order to define the role of serum stimulation for MRTFA/B target gene expression, cells were analysed under serum-starved and serum-induced conditions (Supplementary Table 1). With the exception of only few genes, little influence of serum on overall (Supplementary Figure S2B and Supplementary Table 2) and MRTFA/B-dependent gene expression (Figure 2b and Supplementary Table 3) was observed. Differential gene expression analysis revealed that gene sets affected by EWS-FLI1 knockdown and those affected by MRTFA/B knockdown did not correlate (Figure 2c).
Figure 2.
MRTFB knockdown antagonizes the transcriptional effects of EWS-FLI1 depletion. (a) Immunoblot of MRTFA and MRTFB knockdown by transient short hairpin RNA (shRNA) transfection with and without concomitant dox-induced (48 h) EWS-FLI1 silencing. Alpha-TUBULIN was used as a loading control. (b) Scatter plot showing effects of serum on MRTFA/B-dependent gene regulation. x axis: sh-MRTFA/B versus sh-Ctrl under starved conditions. y axis: sh-MRTFA/B versus sh-Ctrl under serum-induced conditions. Red dots indicate serum-responsive MRTFA/B-regulated genes at |logFC|⩾0.8 with some examples given. (c) Comparison of effects of MRTFA/B knockdown versus EWS-FLI1 knockdown in A673/TR/shEF and SK-N-MC. Scatter plots showing log2 fold gene expression changes upon EWS-FLI1 knockdown (x axis; dox versus no dox), and upon MRTFA/B knockdown (y axis; sh-MRTFA/B versus sh-Ctrl) alone (EWS-FLI1-high) or in combination with EWS-FLI1 knockdown (EWS-FLI1-low) in A673/TR/shEF (upper panel) and SK-N-MC cell line (lower panel). (d) Inverse gene regulation by MRTFA/B and EWS-FLI1. Venn diagrams of EWS-FLI1-correlated and -anticorrelated genes (green circles) and MRTFA/B-regulated genes under EWS-FLI1-low conditions (yellow circles) in A673/TR/shEF cells. Overlapping area indicates the number of genes that are partially rescued from EWS-FLI1 knockdown effects by MRTFA/B depletion. Arrows indicate direction of gene expression change for EWS-FLI1 knockdown (dox versus no dox; red, |logFC|⩾1, P<0.05), and for combined EWS-FLI1/MRTFA/B knockdown (sh-MRTFA/B versus sh-Ctrl under dox; blue, |logFC|⩾0.7, P<0.05). (e) Heatmap and unsupervised hierarchical clustering of gene expression changes induced by MRTFA/B knockdown under EWS-FLI1-high and -low conditions (cutoff: |logFC|⩾1.5; P<0.05) from two independent experiments. (f) Gene expression effects of specific short interfering RNA (siRNA)-mediated depletion of either MRTFA or MRTFB. Left: representative immunoblot showing the individual siRNA-mediated knockdown of MRTFA and MRTFB on protein level. Right: scatter plots of gene expression (log2 fold) changes induced by EWS-FLI1 (x axis) versus combined MRTFA/B+EWS-FLI1 (y axis) silencing. The inverse correlation is higher for MRTFB (R=−0.66) than for MRTFA (R=−0.36).
We then studied the effect of combined EWS-FLI1 and MRTFA/B knockdown on gene expression. Interestingly, MRTFA/B silencing in cells expressing low levels of EWS-FLI1 significantly reversed the gene expression changes induced by EWS-FLI1 depletion. Figure 2c shows that genome-wide gene expression changes upon MRTFA/B depletion antagonized the effects of EWS-FLI1 depletion upon combined knockdown (Figure 2c). The impact of MRTFA/B knockdown on the EWS-FLI1 regulated transcriptome, and therefore the strength of the inverse correlation was similar under serum-starved (Supplementary Figure S2C) and serum-induced conditions (Figure 2c). This further suggests that serum has a small effect in these cells. The results from this RNA expression analysis in the A673/TR/shEF cell line were validated in the EwS cell line SK-N-MC by co-transfection of short-hairpin constructs targeting EWS-FLI1 and MRTFA/B (Figure 2c lower panel) followed by RNA-sequencing analysis. Even though the knockdown efficiency of EWS-FLI1 was weaker in SK-N-MC than that in A673/TR/shEF (Supplementary Figure S2D and S2A), the antagonism between MRTFA/B and EWS-FLI1 became also evident in SK-N-MC (Figure 2c). Taken together, these data suggest that a large number of EWS-FLI1 target genes are antagonistically regulated by MRTFA/B when transcription levels of the fusion oncogene are low.
Next, we categorized genes into clusters according to the differential effects of EWS-FLI1 and MRTFA/B depletion on their expression levels. At a cutoff of |logFC|>1, P<0.05 for the EWS-FLI1 knockdown and |logFC|>0.7, P<0.05 for the combined MRTFA/B-EWS-FLI1 knockdown, 390 genes were activated by EWS-FLI1 and are further referred to as EWS-FLI1-correlated target genes (Figure 2d). For 87 of these genes, MRTFA/B knockdown rescued the decline in expression upon EWS-FLI1 depletion. On the other hand, EWS-FLI1 repressed the expression of 1076 genes in A673/TR/shEF, further referred to as EWS-FLI1-anticorrelated genes, and MRTFA/B knockdown antagonized this effect for 166 of these genes. Using a more stringent cutoff for differential gene expression (|logFC|>1.5, P<0.05), prime targets of MRTFA/B regulation were identified, and subsequently unsupervised hierarchical clustering was used to identify clusters of differential EWS-FLI1/MRTFA/B response. Five distinct clusters of differential EwS-FLI1/MRTFA/B response were observed (Figure 2e). Clusters 1 and 2 represent EWS-FLI1-anticorrelated gene sets, whose activation upon EWS-FLI1 depletion was counteracted by MRTFA/B knockdown. Cluster 3 represents genes activated in response to MRTFA/B knockdown in presence of high EWS-FLI1, but even stronger under EWS-FLI1-low conditions. Cluster 4 contains EWS-FLI1-correlated target genes that show increased expression after the MRTFA/B knockdown under EWS-FLI1-low but not -high conditions. In contrast, cluster 5 comprises EWS-FLI1-correlated genes strongly affected by MRTFA/B knockdown primarily under EWS-FLI1-high conditions. The inverse effects of MRTFA/B and EWS-FLI1 were further validated by qRT–PCR for selected genes from cluster 2 (HPGD, GAS2 and MAP2) and cluster 4 (NR0B1, RAD51AP1 and MCM10;Supplementary Figure S2E).
Subsequently, we sought to separately study the effects of MRTFA and MRTFB on the EWS-FLI1 transcriptome. To this end, MRTFA and MRTFB were individually silenced using specific short interfering RNAs (Figure 2f). Under comparable knockdown efficacies, MRTFB mediated the antagonistic effect on EWS-FLI1 target gene regulation to a higher degree (R=−0.66) than MRTFA (R=−0.36) upon EWS-FLI1-low levels. Knockdown of MRTFA or MRTFB upon EWS-FLI1-high resulted in a weak positive correlation with the gene expression effects of EWS-FLI1 knockdown (Supplementary Figure S2G). The results for the individual MRTFs are consistent with the results obtained with sh-MRTFA/B; however, MRTFB can be inferred as the main driver of the transcriptional rescue from the EWS-FLI1 knockdown.
MRTFB and EWS-FLI1 overlap in chromatin binding
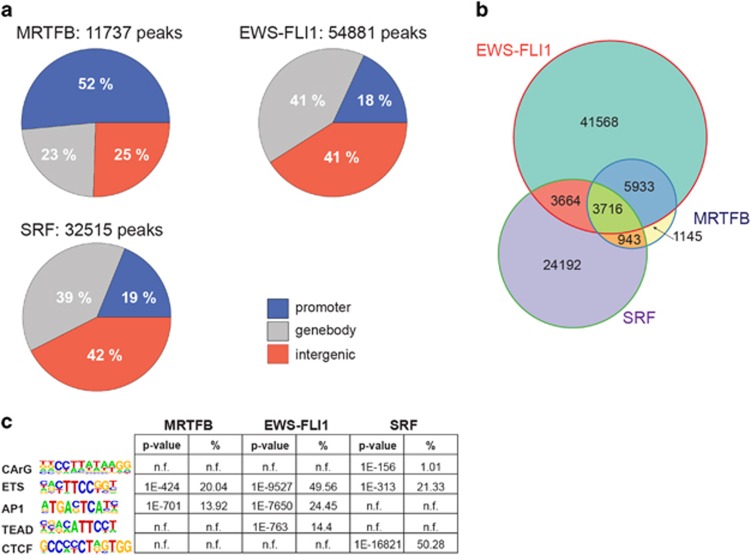
As MRTFB transcriptional effects antagonized the effects of EWS-FLI1 much stronger than MRTFA, we continued our study focusing on MRTFB. To more closely investigate the molecular mechanisms underlying repression of MRTFB transcriptional activity by EWS-FLI1, chromatin-immunoprecipitation for MRTFB, SRF and EWS-FLI1, coupled with next-generation sequencing (chromatin immunoprecipitaion sequencing (ChIP-seq)) was performed (Figure 3). As FLI1 is not expressed in EwS cells,23 a FLI1 antibody was used to specifically precipitate EWS-FLI1. The genome-wide distribution pattern observed for EWS-FLI1 chromatin binding recapitulated previously published results, with predominant binding to distal enhancers and less frequent binding in proximal gene promoter regions.7, 24, 25 The majority of MRTFB ChIP-seq peaks, in contrast, were found in proximity of transcriptional start sites (TSS; 52%). Concordant with previous studies,26 genome-wide SRF-binding distribution showed ~40% of peaks being >2 kb away from the TSS. Only ~19% of SRF peaks were found within the TSS proximity (Figure 3a). Using stringent cutoffs (q-value from model based analysis of ChIP-seq (MACS) <10E−10), 11737 MRTFB peaks, 54 881 EWS-FLI1 peaks and 32 515 SRF peaks were defined. Strikingly, the majority of MRTFB peaks (~82%) overlapped with EwS-FLI1, whereas only half as many peaks overlapped with SRF (~40%). In particular, only 8% of MRTFB-binding regions were shared exclusively with SRF, whereas 31% of MRTFB peaks overlapped with both EWS-FLI1 and SRF (Figure 3b).
Figure 3.
ChIP-seq of MRTFB and EWS-FLI1 suggests functional interaction on chromatin level. (a) Genome-wide distribution pattern of MRTFB, EWS-FLI1 and SRF ChIP-seq peaks. Peaks were defined as ‘proximal’ (−5 to +1 kb from the TSS), ‘genebody’ or ‘distal’ (all other peaks). (b) Venn diagram showing the genome-wide overlaps of EWS-FLI1, SRF and MRTFB ChIP-seq peaks. (c) Table of most enriched sequence motifs (with P-values and percentage of peaks harboring the motif) found in the MRTFB, EWS-FLI1 and SRF-binding regions (motif analysis was performed by the HOMER software, n.f. indicates that motif was not found in the HOMER analysis). For detailed HOMER motif analysis output see Supplementary Table 4.
The association between MRTFB and EWS-FLI1 binding was also substantiated by results obtained from HOMER de novo motif analysis. ETS and AP-1 motifs, preferentially associated with EWS-FLI1 binding,3, 27 were also found among the most significant motifs enriched in the MRTFB ChIP-seq (Figure 3c). Motif analysis also revealed an over-representation of binding motifs for TEAD transcription factors in EWS-FLI1 ChIP-seq peaks. The most prominent sequence motifs associated with SRF binding in EwS cells were CTCF and ETS, and only to a much lesser extend CArG, the canonical SRF-binding motif. Taken together, the results obtained from ChIP-seq analysis suggest an interaction on the chromatin level between EWS-FLI1 and MRTFB that is independent of SRF.
MRTFB binding and TEAD motifs are significantly enriched in distal regions of EWS-FLI1-anticorrelated target genes
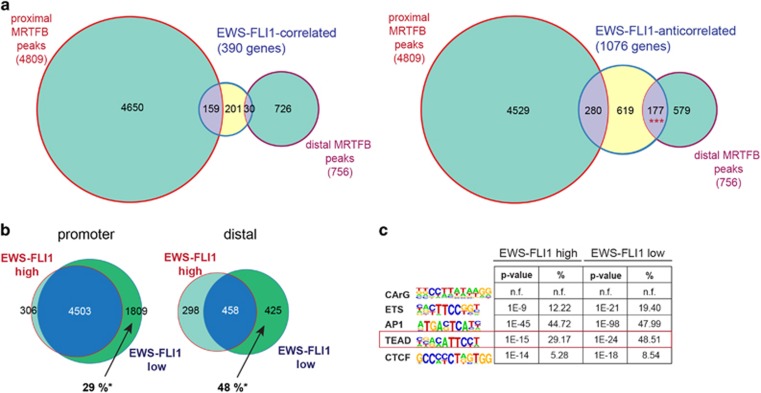
Given the opposing transcriptional effects upon MRTFB and EWS-FLI1 depletion, we tested for MRTFB direct binding to EWS-FLI1 target genes. RNA expression data from EWS-FLI1 knockdown experiments were integrated with MRTFB ChIP-seq data. We detected binding of MRTFB to EWS-FLI1-correlated as well as -anticorrelated gene sets (Figure 4a). A high number of MRTFB ChIP-seq peaks (4809) were found around gene promoters (proximal peaks), but only ~3% (159 peaks) and 6% (280 peaks) of binding regions were found in EWS-FLI1-correlated and EWS-FLI1-anticorrelated genes, respectively. For distal peaks however, although less numerous (756 peaks), a more substantial fraction (23%) was found to be associated with EWS-FLI1-anticorrelated targets. This association of distal MRTFB peaks with EWS-FLI1-anticorrelated genes was significant (P<10−20, hypergeometric test), whereas all other overlaps were not (Figure 4a).
Figure 4.
MRTFB binding and TEAD motifs are significantly enriched in distal regions of EWS-FLI1-anticorrelated target genes. (a) Venn diagram representing the overlap of MRTFB ChIP-seq peaks (green circles) in promoter (proximal) or distal gene regions of EWS-FLI1-correlated or EWS-FLI1-anticorrelated genes (yellow circles). Significant enrichment of the MRTFB binding was solely found in distal genomic regions of EWS-FLI1-anticorrelated genes (lower right, ***P=1.039E−25). (b) MRTFB ChIP-seq peaks in proximal (−5 to +1 kb from the TSS) and distal gene regions under EWS-FLI1-high and -low conditions. * percentage of newly appearing MRTFB peaks as compared to all MRTFB peaks upon EWS-FLI1-low. (c) HOMER motif analysis of MRTFB ChIP-seq peaks in EWS-FLI1-anticorrelated genes under EWS-FLI1-high and -low conditions. The peak set was filtered allowing for only one peak per gene (the nearest to the TSS). Motif analysis was performed by the HOMER software, n.f. indicates that motif was not found in the HOMER analysis. For detailed HOMER motif analysis output see Supplementary Table 4.
To study the effect of EWS-FLI1 on MRTFB chromatin occupancy, MRTFB ChIP-seq was additionally performed upon EWS-FLI1-low state. We found that MRTFB peak numbers generally increased under low compared to high EWS-FLI1 levels. Of note, under EWS-FLI1-low conditions, MRTFB chromatin occupancy increased to a higher extent in distal enhancer than in proximal promoter regions (48% versus 29% Figure 4b), indicating EWS-FLI1 interference with MRTFB chromatin binding.
Motif analysis was repeated for the specific subset of MRTFB peaks, which were associated with distal regions of EWS-FLI1-anticorrelated genes. We identified AP-1 and TEAD motifs to be most prominently enriched. Notably, the significance of this association increased upon EWS-FLI1-low conditions (Figure 4c). Representative examples for increased MRTFB ChIP-seq signals upon EWS-FLI1-low as compared to -high levels are illustrated in Supplementary Figure S3B. In distal MRTFB-binding regions associated with EWS-FLI1-correlated gene sets, no significant enrichment in TEAD motifs upon EWS-FLI1-high or -low conditions was observed (Supplementary Table 4).
TEAD depletion recapitulates the effects of MRTFB knockdown on the EWS-FLI1 transcriptome
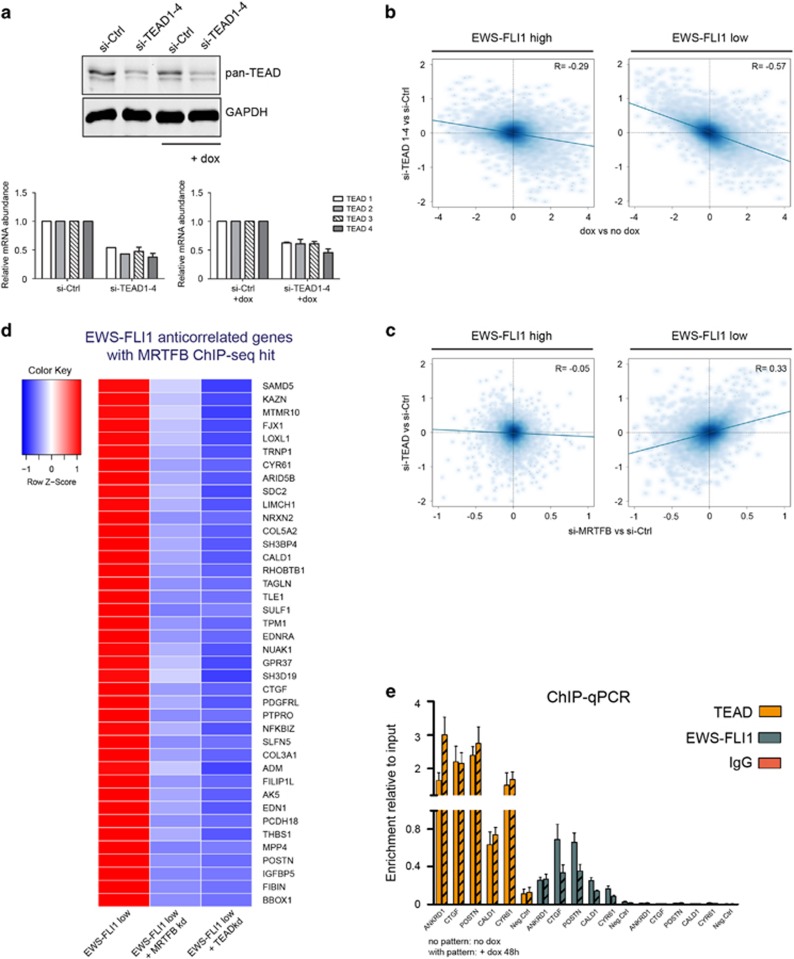
Co-occurrence of MRTFB peaks with TEAD motifs in regions assigned to EWS-FLI-anticorrelated genes, and the fact that several genes inversely regulated by EWS-FLI1 and MRTFB are known target genes of the Hippo/YAP-TAZ/TEAD signalling pathway (CYR61, CTGF, SERPINE1)28 led us to investigate a potential association of MRTFB with TEAD transcription factors. The TEAD transcription factor family comprises four members, TEAD1–4, which require co-activation for their transcriptional activity commonly provided by YAP-1 and its paralogue the transcriptional co-activator with PDZ-binding motif TAZ (WWTR1). Direct interactions between YAP, TAZ and TEADs with MRTFs have already been described.29, 30, 31 TEADs are, furthermore, known to associate with AP-1 transcription factors at distal enhancers, regulating motility and proliferation.28, 32 To elucidate the role of TEADs in MRTFB-mediated transcriptional regulation in EwS cells, we analysed the expression signature of combinatorial knockdown of all four TEAD transcription factors (TEAD1–4) upon EWS-FLI1-high and -low conditions. Depletion of TEAD1–4 by ~50% was achieved by pooling short interfering RNAs directed against TEAD1, 2, 3 and 4 transcripts (Figure 5a). Strikingly, similar to MRTFA/B knockdown, silencing of TEAD1–4 antagonized EWS-FLI1 modulation-mediated transcriptional effects. In addition, unlike MRTFA/B knockdown, depletion of TEAD1–4 in presence of EWS-FLI1 also resulted in gene expression changes in the opposite direction as caused by EWS-FLI1 knockdown, although to a lesser extent than observed under EWS-FLI1-low conditions (R=−0.29 versusR=−0.57; Figure 5b). Comparison of differential gene regulation upon TEAD and MRTFB knockdown showed that there was a positive correlation between gene regulatory signatures of MRTFB and TEAD depletion at low EWS-FLI1 levels (R=0.33) but not at EWS-FLI1-high levels (R=−0.05; Figure 5c). Furthermore, analysis of the respective TEAD target gene spectra by Gene set enrichment analysis revealed significant enrichment of genes involved in migration, response to serum or RhoA, which we previously observed in the comparison of EwS primary tumours to mesenchymal stem cells (Supplementary Figure S4B).
Figure 5.
Effect of TEAD transcription factors on the EWS-FLI1-regulated transcriptome. (a) Western blot analysis showing combined knockdown of all four TEAD transcription factors (TEAD1–4) using siRNA targeting each individual TEAD mRNA. TEAD1–4 factors are recognized by a pan-TEAD antibody. Relative mRNA levels demonstrate that the knockdown efficiency of the four TEAD factors is comparable. Mean±s.e.m. of two biological replicates is shown. (b) Scatter plots showing the effects of the TEAD1–4 knockdown on EWS-FLI1-regulated genes upon EWS-FLI1-high (left plot; R=−0.29) and EWS-FLI1-low (right plot; R=−0.57). TEAD1–4 knockdown antagonizes the effect of EWS-FLI1 depletion, however, more potently upon EWS-FLI1 low conditions. (c) Comparison of gene expression changes induced by MRTFB knockdown (y axis) versus TEAD1–4 depletion (x axis) upon high (left scatter plot; R=−0.05) or low (right scatter plot; R=0.33) EWS-FLI1 levels. Effects of the MRTFB and TEAD1–4 knockdown correlate upon EWS-FLI1-low state only. (d) Heatmap of ‘inversely regulated’ EWS-FLI1/MRTFB target genes (genes for which expression is rescued after the combined EWS-FLI1/MRTFB or EWS-FLI1/TEAD knockdown in comparison to the single EWS-FLI1 knockdown), which were further filtered according to the presence of significant MRTFB ChIP-seq hits, and the presence of TEAD-binding motifs. Gene expression cutoff: EWS-FLI1 knockdown (dox versus no dox) |logFC|⩾1, P<0.05; MRTFB or TEAD knockdown upon EWS-FLI1-low conditions (si-MRTFB+dox/si-TEAD+dox versus si-Ctrl+dox): |logFC|⩽0.4, P<0.1. (e) ChIP–qPCR of for TEAD, FLI1 and IgG ChIP for selected ‘inversely regulated’ genes. ANKRD1 was used as a positive control for the TEAD ChIP–qPCR. Mean±s.e.m. of three biological replicates is shown.
These data strongly suggest a synergistic regulation of a set of genes by MRTFB and TEAD, associated with the Rho pathway, which is impeded in the presence of EWS-FLI1. Taking into account the inverse correlation in gene expression observed for EWS-FLI1 versus MRTFB or TEAD modulation, and ChIP-seq data for MRTFB, as well as presence of TEAD motifs in ChIP-seq peaks, a set of target genes for this regulatory module was defined (Figure 5d). Expression of this panel of genes increased when EWS-FLI1 levels were low, but decreased again upon additional MRTFB or TEAD knockdown. This gene set included the ‘bona fide’ YAP/TAZ-TEAD target genes CYR61 and CTGF. To corroborate a direct functional interaction between MRTFB and TEAD, several candidates from this TEAD/MRTFB target gene list were analysed by ChIP coupled with qRT–PCR using primers flanking MRTFB ChIP-seq hits. TEAD ChIP was enriched for ANKRD1 and all tested MRTFB/EWS-FLI1 targets, enrichments were slightly amplified upon low EWS-FLI1 levels (Figure 5e). EWS-FLI1 ChIP showed enrichments in all tested regions with decreased occupancy upon knockdown of EWS-FLI1 for most of them (Figure 5e). Furthermore, YAP-1 was found to be enriched in the respective MRTFB/TEAD/EWS-FLI1 regions as demonstrated by ChIP–qPCR (Supplementary Figure S4C).
Taken together, MRTFB and TEAD bind to EWS-FLI1-anticorrelated targets, especially when the expression of the fusion oncogene is low. Expression of this set of MRTFB-TEAD bound EWS-FLI1-anticorrelated genes is repressed by EWS-FLI1 but re-activated by MRTFB/TEAD upon EWS-FLI1 depletion. DAVID functional annotation analysis of the inversely EwS-FLI1/TEAD-regulated genes demonstrated that cell migration (P=3.5E−3), adhesion (P=3.9E−5), and regulation of the extracellular matrix (P=1E−10) were significantly enriched in this gene set. This indicates that the interaction of MRTFB and TEAD on a transcriptional level, perturbed by EWS-FLI1, is a network hub for the regulation of cytoskeletal processes such as migration and adhesion in EwS.
Discussion
Early metastasis onset is still the major clinical challenge in the treatment of EwS patients. Several studies proposed that high EWS-FLI1 suppresses migration and adhesion by disrupting EwS cell morphology.8, 9, 10 It was suggested that EWS-FLI1 mediates resistance to cell detachment-induced anoikis through suppression of actin-fibre formation. Knockdown of EWS-FLI1 resulted in a more mesenchymal phenotype of EwS cells including increased expression of genes involved in adhesion and cell architecture. Most recently, oscillation of EwS cells between a proliferative, non-migratory, EWS-FLI1-high small cell phenotype and a non-proliferative, highly migratory EWS-FLI1-low state with re-programmed actin cytoskeleton was described.11 This finding is consistent with EWS-FLI1-repressing cytoskeletal genes (for example, zyxin, α5-integrin) and the formation of actin-rich cytoskeletal structures such as focal adhesions and stress fibres.9, 10 Being mainly regulated by Rho signalling, the actin-cytoskeleton has an important role in microenvironmental signal integration with gene expression via activation of the transcriptional co-activators MRTF. MRTFA and MRTFB typically engage with SRF and activate transcription of cytoskeletal genes in response to serum stimulation. Interestingly, we found little influence of serum in EwS cells presumably due to the induction of growth arrest.33 A recent study in mouse fibroblasts (NIH3T3) demonstrated serum inducibility for 960 SRF target genes, most of them regulated by MRTF.26 Given the discrepancy in serum responsiveness between EwS cells and other cell types, it is conceivable that EwS cells have, to some degree, become independent from extracellular growth signals due to their EWS-FLI1 oncogene addiction.
In this study we provide evidence for a new network hub involved in the extracellular signalling inhibition by EWS-FLI1 in EwS cells. Our data suggest that downstream Rho/F-actin-signalling MRTFs function as co-activators for TEAD and that EWS-FLI1 binds to MRTFB/TEAD target genes and attenuates their transcriptional activation. As YAP-1 and TEAD chromatin occupancy did not significantly increase upon EWS-FLI1 knockdown, the fusion oncogene is unlikely to compete with YAP-1 or TEAD DNA binding. However, ChIP-seq enrichments for MRTFB to the respective regions were increased upon EWS-FLI1-low levels. It is therefore conceivable that EWS-FLI1 interferes with the recruitment of MRTFB to the YAP-1–TEAD complex. A recent study in breast cancer cells further corroborates this hypothesis by demonstrating that MRTFA and especially MRTFB are substantial for YAP-1/TEAD target gene activation via direct interaction and recruitment of other transcriptional co-activators.29
MRTFB and SRF chromatin-binding regions shared relatively little overlap and were not enriched for the same DNA motifs in our study. In contrast to other data sets where the SRF consensus CArG motif was found in ~50% of SRF-binding peaks (HOMER motif re-analysis of SRF ChIP-seq data from Encode (https://www.encodeproject.org/targets/SRF-human/), data not shown), in EwS cells the CArG motif was rarely present in the SRF-binding regions. Instead, we found an overwhelming enrichment of CTCF motifs, which are frequently associated with gene insulators.34 Earlier studies reported CTCF motif enrichments for SRF in neuron-specific lineages,35 and a direct interaction of SRF and CTCF was demonstrated via the chromodomain helicase-DNA-binding protein 8 (CHD8), important for protecting smooth muscle alpha cells from apoptosis.36 Depending on the cellular context it is possible that SRF might have distinct roles and hence might act differently in EwS cells as compared to other cell types.
MRTFs do not carry a DNA-binding domain but are recruited to chromatin through interaction with DNA-bound transcription factors. Recent studies discovered a direct association of the transcriptional co-activators MRTFA/B and the TEAD co-activators YAP-129, 31 and TAZ,30 downstream of Rho-actin regulation. In fact, MRTFs and YAP-1/TAZ exhibit crosstalk on multiple levels, mutually affecting their nucleocytoplasmic distribution, expression and transcriptional activity.31, 37 Several studies indicated that, in order to fully activate gene expression of TEAD target genes, MRTF and YAP-1/TAZ are required.29, 31 It was furthermore shown that MRTFA/B interaction with YAP-TEAD via NcoA3 recruitment is essential for target gene activation having a role in breast cancer metastasis.29 Our data suggest that recruitment of MRTFB to sites of TEAD and YAP-1 binding is hindered by EWS-FLI1. The enrichment of TEAD motifs was predominantly found in distal regions of EWS-FLI1-anticorrelated genes. TEADs have been reported to associate with the transcription factor AP-1 (Fos-Jun) at distal enhancers of genes involved in oncogenic growth in several tumour cell lines.28, 32 Notably, we found co-enrichment of AP-1 motifs with TEAD motifs in MRTFB-enriched sites of EWS-FLI1-anticorrelated genes and AP-1 has previously been demonstrated to bind to EWS-FLI1 at genomic regions equipped with AP-1-binding motifs.27 Hence, AP-1 might provide a link to TEAD occurrence at EWS-FLI1-bound regions associated with EWS-FLI1-anticorrelated genes.
A prerequisite for MRTFB/YAP-1 activity is their nuclear translocation upon signal-induced F-actin polymerization,13, 38, 39 which can be inhibited by pre-treatment with Latrunculin B. Previous studies reported EWS-FLI1-dependent perturbation of the actin cytoskeleton. To exclude that EWS-FLI1 represses TEAD target genes solely by prohibiting the release of transcriptional co-activators YAP-1 and/or MRTFB to the nucleus, we used confocal immunofluorescence microscopy and immunoblotting to visualize the subcellular localization of MRTFB and YAP-1 in A673/TR/shEF cells in absence and presence of serum. Under serum-starved conditions, both proteins were retained in the cytoplasm. However, MRTFB and YAP-1 readily translocated to the nucleus upon 60 min of serum stimulation in the absence, but not in presence of Latrunculin B despite the continuous presence of EWS-FLI1 (Supplementary Figure S5). Consistent with cytoplasmic retention of co-activators in response to Latrunculin B treatment, a significant reduction in CTGF, ANKRD1 and CYR61 gene expression was observed (Supplementary Figure S5D). Our data therefore suggest that EWS-FLI1-induced cytoskeletal perturbation is not sufficient to explain aberrant downstream gene regulation, as MRTFA/B and YAP-1 signal-related nuclear localization was functional. We therefore propose interference of EWS-FLI1 with MRTFB recruitment to a transcriptional module including YAP-1/TEAD as the main mechanism of cytoskeletal target gene dysregulation in EwS.
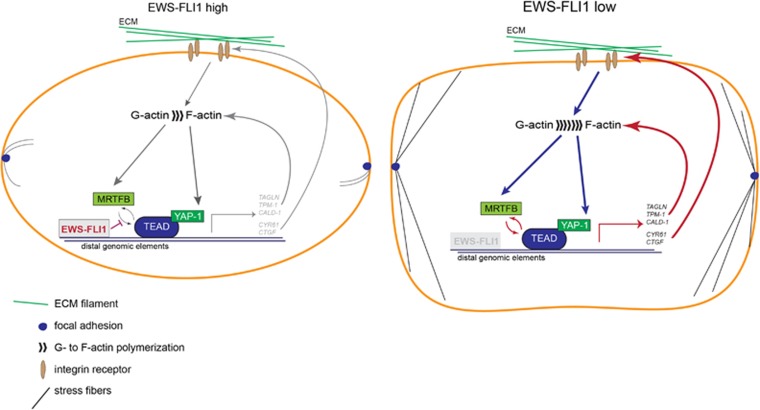
Taken together, this study supports a model for EWS-FLI1 perturbing feedback regulation of genes involved in Rho signalling via the actin cytoskeleton by a transcription modulatory mechanism (Figure 6). The EWS-FLI1-low state drastically alters EwS cell morphology (Supplementary Figure S5E). We hypothesize that this change in morphology is due to the perturbation of transcriptional complex formation of MRTFB with YAP-1/TEAD by EWS-FLI1. Among MRTFB- and TEAD-regulated genes we identified several key factors of the Rho pathway involved in upstream regulation (for example, CYR61, CTGF) and actin-fibre formation and stability (for example, TAGLN, CALD-1 and TPM-1) to be repressed by EWS-FLI1 (Figure 5d). Given the established role of Rho signalling in cellular plasticity, this study provides novel molecular insights into the nature of initial events of metastasis in EwS.
Figure 6.
Model of Rho/F-actin pathway repression by EWS-FLI1 via MRTFB/YAP-1/TEAD transcriptional perturbation. A673/TR/shEF: left: EWS-FLI1-low state: EWS-FLI1 binds to distal genomic elements of EWS-FLI1-anticorrelated target genes and thereby likely hinders MRTFB/YAP-1/TEAD interaction and transcriptional activation of genes involved in actin–cytoskeletal structure (TAGLN, TPM-1 and CALD-1) or are involved in binding to the extracellular matrix (ECM) and insulin growth factors (IGF; CYR61, CTGF). Low expression of these genes might affect cytoskeletal integrity due to compromised actin polymerization and upstream signalling via ECM–receptor interactions. As a consequence, cells exhibit few focal adhesions and stress fibres (F-actin filaments) and F-actin polymerization is reduced.9, 10 Right: upon knockdown of EWS-FLI1 (EWS-FLI1-low), expression of MRTFB/YAP-1/TEAD target genes, which regulate cytoskeletal key factors, is increased and thereby likely promoting signalling via Rho both upstream and downstream of actin polymerization. Consequently, cell morphology is drastically altered with increased numbers of focal adhesions and stress fibres as compared to the high EWS-FLI1 cell state.
Materials and methods
Cell culture
The dox-inducible A673/TR/shEF cell line was previously described.40 Knockdown of EWS-FLI1 was achieved by addition of 1 μg/ml dox to the medium (24–72 h). The SK-N-MC EwS cell line was kindly provided by J Biedler (Memorial Sloan-Kettering Cancer Centre, New York, NY, USA). Cell lines were authenticated by STR profiling and regularly tested for mycoplasma (Mykoalert detection kit; Lonza, Basel, Switzerland). For serum induction cells were starved overnight in 0.2% fetal bovine serum medium with subsequent serum stimulation for 60 min with 20% fetal bovine serum medium. Latrunculin B (Cayman Chemicals, Ann Arbor, MI, USA) was added in serum-free DMEM at 1 μM concentration to the overnight starved cells 30 min before serum induction. Vehicle controls were treated with dimethyl sulfoxide (<0.05%).
Transfection experiments
To simultaneously knockdown MRTFA and MRTFB, the pLKO sh-MKL-1/2 plasmid (Addgene, Cambridge, MA, USA) was transiently transfected into cells. A control plasmid (sh-NS) containing an inert short hairpin RNA (CAACAAGATGAAGAGCACCAA) was used. For combined knockdown of EwS-FLI1, dox was added 48 h prior to harvest. To transiently silence EWS-FLI1 in SK-N-MC, the sh-EF30 pSUPER plasmid and, as a control, a scrambled-short hairpin RNA control plasmid41 were used. Transfections were carried out using Lipofectamine Plus reagent (Invitrogen, Groningen, the Netherlands). Transfections with short interfering RNA were carried out using Oligofectamine reagent (Invitrogen).
Protein extraction and immunoblot
Protein extraction and immunoblotting were performed according to standard procedures. Antibodies used are listed in Supplementary Table 5. Protein quantification from three biological replicates was achieved by measurement of protein band intensities using the LI-COR Odyssey Infrared Imaging System (LI-COR Biosciences, Bad Homburg, Germany).
Oligonucleotides
Information on all oligonucleotides used in this study (qPCR, ChIP–qPCR and short interfering RNA) is enclosed in Supplementary Table 6.
Gene expression data
Total RNA was isolated using the RNeasy Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, and biological duplicates were analysed either by Affymetrix expression array or RNA-sequencing. Affymetrix HGU-133-PLUS2 CEL files were normalized using ‘frma',42 genes with z-scores <2 (‘barcode’ of the ‘frma’ package) in all samples were excluded, and per gene the probe set with the highest variance across samples was chosen for further analysis. Differential gene expression analysis was performed using ‘limma’.43 Sequencing was done on a HiSeq2000, yielding >17Mio 50 bp single end reads for all samples. After quality control (QC) (FASTQC, RNA-SeqQC, http://www.bioinformatics.babraham.ac.uk/projects), reads were aligned to hs37d5 (ftp://ftp.1000genomes.ebi.ac.uk/) using STAR.44 Counts for Ensembl (GRCh37.75) genes were obtained using the R-package ‘Rsubread’. For genes with counts per million aligned reads>5 in more than two samples, differential expression analysis was performed by ‘edgeR’ and ‘voom’.45 A summary of all gene expression data obtained in this study can be found in Supplementary Table 7.
ChIP–qPCR and ChIP-Seq
Approximately 10 million cells were collected and double crosslinked using disuccinimidyl glutarate (Thermo Fisher Scientific, Waltham, MA, USA) and formaldehyde (Thermo Fisher Scientific). ChIP was performed essentially as described previously.46 Antibodies used are described in Supplementary Table 5.
For ChIP-PCR three biological replicates were performed per experiment and analysed by qRT–PCR using SYBR-Green (Thermo Fisher Scientific). Data were normalized to the input control according to the following equation: normalized to Input= 2 × (average Ct input−average Ct IP). IgG controls were performed for every ChIP experiment.
All reagents for ChIP-seq were from Illumina (New England Biolabs, Frankfurt, Germany). ChIP-DNA (5 ng) was used for library prep with the NEBNext Ultra DNA Library Prep Kit, the NEBNext Multiplex Oligos were used for adapter labelling in a multiplex sample preparation. The NEBNext Q5 Hot Start HIFI PCR Master mix was used and no size selection of adapter-ligated DNA was performed before sequencing, which was performed in biological duplicates for each experimental condition on an Illumina Hi-Seq 2000. For FLI1 and SRF ChIP, 50 bp single reads, and for MRTFB ChIP, 100 bp paired-end reads were obtained, yielding a minimum of 28 million reads for each sample. Reads were aligned to hs37d5 (ftp://ftp.1000genomes.ebi.ac.uk/) using the bwa aligner,47 and peaks were called by MACS2 with default parameters48 using a large ChIP-input sample from Tomazou et al.7 R-package ‘DiffBind’49 was used to construct a consensus peak matrix from all ChIP experiments yielding a list of 50 798 peak regions. Only peaks were retained that yielded a MACS2 log10 q-value>10 in both replicas. The combined peak matrix is reported as Supplementary Table 8.
Peak annotation and motif analysis
Peak annotation and motif analysis were performed with HOMER (http://homer.salk.edu/homer/).50 Peaks were defined as ‘proximal’ (−5 to+1 kb around the TSS) and ‘distal’ (all other peaks). For the calculations in Figure 4C the peak set was filtered so that for each gene only the nearest to the TSS peak was retained.
Gene set enrichment analysis and gene ontology analyses
Gene set enrichment analysis was performed using the command-line tool and MSigDb gene sets from the Broad Institute (http://software.broadinstitute.org/gsea).51 For the analysis in Figure 1a, 117 Ewing tumours from Postel-Vinayet al.17 (GEO: GSE34620) were compared to paediatric mesenchymal stem cells18 (GEO: GSE31215).
Gene ontology analysis of gene expression data was performed using the DAVID bioinformatics database (https://david.ncifcrf.gov/).52
Statistics
P-values were calculated from three independent experiments (mean±s.e.m.) using the Graph Pad-software (http://www.graphpad.com/;Graph Pad Prism Software Inc., La Jolla, CA, USA). If not indicated otherwise, data were analysed using the two-tailed one-sample t-test including Welch’s correction, setting the hypothetical mean value to 1. P-values of ⩽0.05 were considered significant. ***P⩽0.001; **P⩽0.01; *P⩽0.05.
Data availability
All genomic data obtained from Affymetrix arrays, RNA-sequencing or ChIP-seq were submitted to the Gene Expression Omnibus from NCBI (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE92741.
Acknowledgments
This study was supported by the Austrian Science Fund grant I1125-B19, the Liddy Shriver Initiative and by the European Commission Framework Program 7 grant 259348 ('ASSET').
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Bernstein M, Kovar H, Paulussen M, Randall RL, Schuck A, Teot LA et al. Ewing's sarcoma family of tumors: current management. Oncologist 2006; 11: 503–519. [DOI] [PubMed] [Google Scholar]
- Lawlor ER, Sorensen PH. Twenty years on: what do we really know about ewing sarcoma and what is the path forward? Crit Rev Oncog 2015; 20: 155–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar H. Downstream EwS/FLI1 - upstream Ewing's sarcoma. Genome Med 2010; 2: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spraker H, Price S, Chaturvedi A, Schiffman J, Jones K, Lessnick S et al. The clone wars – revenge of the metastatic rogue state: the sarcoma paradigm. Front Oncol 2012; 2: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomey EC, Schiffman JD, Lessnick SL. Recent advances in the molecular pathogenesis of Ewing's sarcoma. Oncogene 2010; 29: 4504–4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer M, Ban J, Kofler R, Walker B, Davis S, Meltzer P et al. A molecular function map of Ewing's sarcoma. PLoS ONE 2009; 4: e5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomazou EM, Sheffield NC, Schmidl C, Schuster M, Schonegger A, Datlinger P et al. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep 2015; 10: 1082–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsellem V, Kryszke MH, Hervy M, Subra F, Athman R, Leh H et al. The actin cytoskeleton-associated protein zyxin acts as a tumor suppressor in Ewing tumor cells. Exp Cell Res 2005; 304: 443–456. [DOI] [PubMed] [Google Scholar]
- Chaturvedi A, Hoffman LM, Jensen CC, Lin YC, Grossmann AH, Randall RL et al. Molecular dissection of the mechanism by which EwS/FLI expression compromises actin cytoskeletal integrity and cell adhesion in Ewing sarcoma. Mol Biol Cell 2014; 25: 2695–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi A, Hoffman LM, Welm AL, Lessnick SL, Beckerle MC. The EwS/FLI oncogene drives changes in cellular morphology, adhesion, and migration in Ewing sarcoma. Genes Cancer 2012; 3: 102–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzetti GA, Laud-Duval K, van der Ent W, Brisac A, Irondelle M, Aubert S et al. Cell-to-cell heterogeneity of EWSR1-FLI1 activity determines proliferation/migration choices in Ewing sarcoma cells. Oncogene 2017; e-pub ahead of print 30 January 2017; doi:10.1038/onc.2016.498. [DOI] [PMC free article] [PubMed]
- Asparuhova MB, Gelman L, Chiquet M. Role of the actin cytoskeleton in tuning cellular responses to external mechanical stress. Scand J Med Sci Sports 2009; 19: 490–499. [DOI] [PubMed] [Google Scholar]
- Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol 2010; 11: 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posern G, Treisman R. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol 2006; 16: 588–596. [DOI] [PubMed] [Google Scholar]
- Hu Q, Guo C, Li Y, Aronow BJ, Zhang J. LMO7 mediates cell-specific activation of the Rho-myocardin-related transcription factor-serum response factor pathway and plays an important role in breast cancer cell migration. Mol Cell Biol 2011; 31: 3223–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol 2009; 11: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postel-Vinay S, Veron AS, Tirode F, Pierron G, Reynaud S, Kovar H et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet 2012; 44: 323–327. [DOI] [PubMed] [Google Scholar]
- Riggi N, Suva ML, De Vito C, Provero P, Stehle JC, Baumer K et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev 2010; 24: 916–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell 2007; 11: 421–429. [DOI] [PubMed] [Google Scholar]
- Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol 2007; 292: C70–C81. [DOI] [PubMed] [Google Scholar]
- Selvaraj A, Prywes R. Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol Biol 2004; 5: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Vasishtha M, Prywes R. Activation and repression of cellular immediate early genes by serum response factor cofactors. J Biol Chem 2010; 285: 22036–22049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Owen LA, Trem DJ, Wong JS, Whangbo JS, Golub TR et al. Expression profiling of EwS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell 2006; 9: 405–416. [DOI] [PubMed] [Google Scholar]
- Bilke S, Schwentner R, Yang F, Kauer M, Jug G, Walker RL et al. Oncogenic ETS fusions deregulate E2F3 target genes in Ewing sarcoma and prostate cancer. Genome Res 2013; 23: 1797–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suva ML et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 2014; 26: 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N et al. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev 2014; 28: 943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Denny CT, Wisdom R. Cooperative DNA binding with AP-1 proteins is required for transformation by EWS-Ets fusion proteins. Mol Cell Biol 2006; 26: 2467–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol 2015; 17: 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Hwang D, Lee D, Kim JH, Kim SY, Lim DS. MRTF potentiates TEAD-YAP transcriptional activity causing metastasis. EMBO J 2017; 36: 520-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speight P, Kofler M, Szaszi K, Kapus A. Context-dependent switch in chemo/mechanotransduction via multilevel crosstalk among cytoskeleton-regulated MRTF and TAZ and TGFbeta-regulated Smad3. Nat Commun 2016; 7: 11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu OM, Miyamoto S, Brown JH. Myocardin-related transcription factor a and yes-associated protein exert dual control in G protein-coupled receptor- and RhoA-mediated transcriptional regulation and cell proliferation. Mol Cell Biol 2015; 36: 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li H, Rajurkar M, Li Q, Cotton JL, Ou J et al. Tead and AP1 coordinate transcription and motility. Cell Rep 2016; 14: 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwentner R, Papamarkou T, Kauer MO, Stathopoulos V, Yang F, Bilke S et al. EWS-FLI1 employs an E2F switch to drive target gene expression. Nucleic Acids Res 2015; 43: 2780–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 2009; 19: 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan AL, Benner C, Heinz S, Huang W, Xie L, Miano JM et al. Serum response factor utilizes distinct promoter- and enhancer-based mechanisms to regulate cytoskeletal gene expression in macrophages. Mol Cell Biol 2011; 31: 861–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenberg JM, Hoggatt AM, Chen M, Touw K, Jones R, Herring BP. Regulation of serum response factor activity and smooth muscle cell apoptosis by chromodomain helicase DNA-binding protein 8. Am J Physiol Cell Physiol 2010; 299: C1058–C1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Chan SW, Guo F, Toloczko A, Cui L, Hong W. MRTF/SRF dependent transcriptional regulation of TAZ in breast cancer cells. Oncotarget 2016; 7: 13706–13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M et al. Role of YAP/TAZ in mechanotransduction. Nature 2011; 474: 179–183. [DOI] [PubMed] [Google Scholar]
- Yu OM, Brown JH. G protein–coupled receptor and rhoa-stimulated transcriptional responses: links to inflammation, differentiation, and cell proliferation. Mol Pharmacol 2015; 88: 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo J, Garcia-Aragoncillo E, Azorin D, Agra N, Sastre A, Gonzalez-Mediero I et al. Cholecystokinin down-regulation by RNA interference impairs Ewing tumor growth. Clin Cancer Res 2007; 13: 2429–2440. [DOI] [PubMed] [Google Scholar]
- Ban J, Bennani-Baiti IM, Kauer M, Schaefer KL, Poremba C, Jug G et al. EWS-FLI1 Suppresses NOTCH-Activated p53 in Ewing’s Sarcoma. Cancer Res 2008; 68: 7100–7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA). Biostatistics 2010; 11: 242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014; 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. BioTechniques 2005; 39: 715–725. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 2012; 481: 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 2003; 4: R60. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomic data obtained from Affymetrix arrays, RNA-sequencing or ChIP-seq were submitted to the Gene Expression Omnibus from NCBI (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE92741.