Abstract
The Lewis (FUT3) and Secretor (FUT2) genes, corresponding to secretion of Lewis ABO (H) histo‐blood group antigen CA19‐9, are highly polymorphic with differences between populations. In this study, the FUT3 and FUT2 genes in 316 Chinese participants were sequenced to detect polymorphisms, and the associated CA19‐9 antigen secretion was also measured. In total, 14 genotypes of FUT3 and 10 genotypes of FUT2 were verified. Le/Le, Le/le 59,508 and Le/le 59 were the main genotypes of FUT3 with frequencies of 53.2%, 10.7% and 3.5%, respectively. Se/Se, Se/se 385 and se 385/se 385 were the main genotypes of FUT2, with frequencies of 21.4%, 18.6% and 16.2%, respectively. The alleles le 1067 and le 508 were found extensively in the Chinese population, and the frequency of allele se 385 was shown to be higher than previously reported. Phenotype analysis revealed that 9.8% of individuals were the Lewis‐negative type and 22.5% were the secretor‐negative type. Combined phenotypes showed that 3.2% of participants were of ‘double‐negative’ phenotype (le, se) and 19.3% were of single dominant non‐secretor phenotype (Le, se). Serum Lewis antigen CA19‐9 levels were significantly different between subgroups and consistent with the defined phenotype. Our study revealed the unique distribution of Lewis and Secretor polymorphisms in a large Chinese population, and decoded the combined genotypes of the two CA19‐9‐related genes.
Keywords: Chinese, FUT2, FUT3, genotype, SNP
Abbreviations
- Le
Lewis (FUT2) gene negative genotype
- Le
Lewis (FUT3) gene positive genotype
- se
Secretor (FUT2) gene negative genotype
- Se
Secretor (FUT2) gene positive genotype
- SNP
single nucleotide polymorphism
The synthesis of the Lewis ABO (H) histo‐blood group antigens requires multiple specific glycosyltransferases 1, 2, 3. The FUT3 (Lewis) and FUT2 (Secretor) genes encode an α‐(1,3/4)‐fucosyltransferase and an α‐(1,2)‐fucosyltransferase, respectively, which regulate fucose‐carbohydrate antigen synthesis by adding a fucose to precursor substrate 3, 4, 5, 6. Cooperation of the two fucosyltransferases ultimately regulates the expression of the histo‐blood group antigens, including CA19‐9 (sLea), in body fluids and on the surface of epithelial cells. A single nucleotide polymorphism (SNP) of FUT3 and FUT2 is prevalent in multiple populations and dramatically determines the fucosyltransferase activities 4, 5, 6, 7, 8. Distribution of the FUT3/FUT2 genotypes exhibits ethnic heterogeneity 9, 10 and is strongly associated with a wide range of human diseases 8, 11, 12, 13, 14.
The α‐(1,3/4)‐fucosyltransferase‐encoding gene FUT3, also known as the Lewis gene (Le), is essential for the synthesis of Lewis histo‐blood group antigens. The fucosyltransferase diverts a fucose to either the type 1 precursor or the H type 1, to form Lea or Leb, respectively. Mutations in the FUT3 gene may result in the Lewis‐null phenotype (le) 7, 15, 16, 17, 18. SNPs rs28362459 (T59G), rs812936 (T202C), rs778986 (C314T), rs3745635 (G508A) and rs3894326 (T1067G) of the FUT3 gene are the most common polymorphic loci in Asians 7, 10, 15, 18, 19. Substitution of amino acids caused by mutations T202C, C314T, G508A and T1067G leads to inactivation of the FUT3 enzyme, and mutation T59G reduces the availability of α‐(1,3/4)‐fucosyltransferase 2, 6, 11.
The FUT2 gene, also known as the Secretor gene (Se), determines the secretion status of histo‐blood group antigens 20. It encodes an α‐(1,2)‐fucosyltransferase (FUT2) that adds a fucose onto the type 1 precursor to form H type 1, the precursor of Leb. According to previous reports, non‐secretor phenotypes in Western populations are mainly caused by a homozygous loss‐of‐function mutation of FUT2 (rs601338, G428A) 5, 14, 21. However, the frequency of mutation G428A in Asians is much lower 20, 22, 23. Approximately 20% of the Asian population are non‐secretors, and homozygous missense at site 385 (rs1047781, A385T), which is the primary mutation, results in the non‐secretor phenotype 20, 22, 23, 24. Additionally, a synonymous mutation (rs281377, T357C) and a non‐synonymous mutation (rs602662, G739A) have been shown to be common in Asians 12, 20, 23, 24. The fusion gene (se fus) was found in Japanese and Korean populations, but was not detected in the Chinese population 9, 23.
Lewis‐negative individuals (the le/le genotype) have the Lewis (FUT3)‐negative phenotype, Le (a−b−), irrespective of the Se genotype. However, Lewis‐positive individuals (the Le/Le and Le/le genotypes) are divided into three Lewis‐secretor phenotypes according to distinct Secretor genotypes as follows: (a) Le (a−b+) secretors with the Se/Se or Se/se genotype; (b) Le (a+b−) non‐secretors with the se/se genotype; and (c) Le (a+b+) partial secretors having homozygosity for the weak Secretor allele 1, 2, 20, 25. Nucleotide substitutions inactivating the FUT2/3 genes have been found within various populations, and the phenotypes have been determined by the ethnic group‐specific genotypes 1, 8, 16, 19, 24, 25. Both genotypes of Lewis and Secretor are crucial for an individual's phenotype formation, and the serum CA19‐9 value will directly reflect the differences among individuals.
Knowledge of the polymorphisms of the Lewis and Secretor genes in the Chinese population will help in classifying the subgroups and defining an accurate condition for normal phenotypes. In this study, we aimed to examine the prevalence of the five major nucleotide polymorphisms of the FUT3 gene and the four main variation types of the FUT2 gene in a Chinese population. Comparison of serum CA19‐9 expression between each phenotype was performed as well. Distribution of the Lewis‐negative and secretor‐negative phenotypes in the Han ethnic population was also evaluated and compared with previous results in other populations. Since they are based on a reliable method, we expect the results will provide an important reference for disease diagnosis and therapy in the Chinese population.
Materials and methods
Participants and genomic DNA isolation
Blood samples were obtained from 316 unrelated and randomly selected healthy individuals of Han ethnicity in the eastern region of China. Either their birthplace or the paternal origin of the participants in this study was in mainland China. Oral informed consent was obtained from all participants in this study. We collected the peripheral blood in tubes containing ethylene diamine tetraacetic acid and isolated the white blood cells. Genomic DNA was extracted from white blood cells using a QIAamp DNA Blood Mini kit (Qiagen, Inc., Hilden, Germany) according to the manufacturer's instructions.
PCR amplification of FUT3 and FUT2 genes
Both FUT3 and FUT2 genes have no intron in the open reading frame and fewer reactions are capable of amplification of the complete mutation region. Three pairs of PCR primers respectively specific for the FUT3 and FUT2 gene segments are shown in Table S1, and sequence design partly referred to those previously reported 16, 26. For each segment amplification, 20 ng of genomic DNA was combined with the primers (7.5 μm for forward and reverse) in a PCR system of final volume 25 μL. Each PCR system contained 5 mm dNTPs, 37.5 mm MgSO4, 2.5 μL 10 × PCR buffer (particular for KOD ‐Plus‐ Neo) and 0.5 U KOD ‐Plus‐ Neo (Toyobo Co., Osaka, Japan). Thirty cycles were run (2 min at 94 °C, 10 s at 98 °C, 30 s at T m and 30 s at 68 °C, where T m for 385F/385R is 62 °C, for 508F/1067R is 65.5 °C and for 21F/21R is 60 °C), and the products were isolated from agarose gels for sequencing.
Direct DNA sequencing and genotyping
The purified amplification products were sequenced directly for FUT3 and FUT2 genotyping. The anterior half‐segment of FUT3 concluding at the 59, 202 and 314 position was directly sequenced with primers 385F and 385R. The bottom half‐segment of FUT3 concluding at the 59, 202 and 314 position was directly sequenced with primers 508F and 1067R. The complete segment of FUT2 concluding at the 357, 385, 428 and 739 position was directly sequenced with primers 508F and P1R. The dideoxynucleotide termination sequencing reaction was performed by using the ABI BigDye Terminator cycle sequencing system, and the DNA sequence was analyzed by an ABI PRISM 3730 instruments (Applied Biosystems, Carlsbad, CA, USA). Sequencing data were generated by the ABI 3730 Genetic Analyzer platform and were analyzed using chromas software 27. The genotype present at each SNP site was directly determined by one or two different color peaks on the electropherogram.
CA19‐9 antigen measurement and statistical analysis
Serum CA19‐9 antigens were measured using an electrochemiluminescence immunoassay on the Roche Cobas e601 (Roche MODU D + P model, D2400‐P800) immunoassay analyzer (Roche Diagnostics, Mannheim, Germany). An obtained CA19‐9 value of less than 0.06 U·mL−1 was considered to be undetectable. The frequencies of the gene polymorphisms, as well as the genotypes and phenotypes, were determined by description analysis. Difference of CA19‐9 values among each subgroup were tested by one‐way analysis of variance and Student's t test was used to compare between each single group. Statistical analyses were performed with spss Statistics 19 (IBM Corp., Armonk, NY, USA) and P < 0.05 was defined as statistically significant.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Fudan University Shanghai Cancer Center.
Results
Distribution of FUT3 and FUT2 gene polymorphisms in a Chinese population
We amplified the coding regions containing the SNPs in the FUT3 (Lewis) and FUT2 (Secretor) genes and examined the respective polymorphisms. For the Lewis gene, nucleotide 59T>G was the most prevalent mutation, with a frequency of 39.87% (7.59% homozygous and 32.28% heterozygous). Variations 508G>A (25.00%) and 1067T>A (8.23%) manifested in a homozygous form. Variations 202T>C and 314C>T had complete linkage in 18 individuals (5.70%) and no homozygous alleles were encountered (Table 1). For the Secretor gene, four prevalent polymorphisms were also investigated. It is known that the most frequent Secretor gene mutations are detected in Asians, which as a homozygous mutation causes a non‐secretor phenotype 20, 23, 24. 385A>T was detected in 222 individuals (70.25%) with a T allele frequency of 46.04%. Variation 357A>T was detected with a frequency of 24.68%, and induced a synonymous mutation in Secretor. Mutation at nucleotides 428 and 739 were rarely detected (1.27%) and showed complete linkage (Table 1).
Table 1.
Distribution of SNPs and the allele frequencies
| Nucleotide position | SNP | Allele | ||||
|---|---|---|---|---|---|---|
| Wild‐type | Heterozygous mutant | Homozygous mutant | Primary allele | Variant allele | ||
| n (%) | n (%) | n (%) | n (%) | n (%) | ||
| Lewis | 59 | TT | TG | GG | T | G |
| 190 (60.13) | 102 (32.28) | 24 (7.59) | 482 (76.27) | 150 (23.73) | ||
| 202 | TT | TC | CC | T | C | |
| 298 (94.30) | 18 (5.70) | 0 (–) | 614 (97.15) | 18 (2.85) | ||
| 314 | CC | TC | TT | C | T | |
| 298 (94.30) | 18 (5.70) | 0 (–) | 614 (97.15) | 18 (2.85) | ||
| 508 | GG | GA | AA | G | A | |
| 237 (75.00) | 64 (20.25) | 15 (4.75) | 538 (85.13) | 94 (14.87) | ||
| 1067 | TT | TA | AA | T | A | |
| 290 (91.77) | 23 (7.28) | 3 (0.95) | 603 (95.41) | 29 (4.59) | ||
| Secretor | 375 | TT | TC | CC | T | C |
| 238 (75.32) | 70 (22.15%) | 8 (2.53) | 546 (86.39) | 86 (13.61) | ||
| 385 | AA | TA | TT | A | T | |
| 94 (29.75) | 153 (48.42%) | 69 (21.84) | 341 (53.96) | 291 (46.04) | ||
| 428 | GG | GA | AA | G | A | |
| 312 (98.73) | 4 (1.27%) | 0 (–) | 628 (99.37) | 4 (0.63) | ||
| 739 | GG | GA | AA | G | A | |
| 312 (98.73) | 4 (1.27%) | 0 (–) | 628 (99.37) | 4 (0.63) | ||
Haplotype analysis of FUT2 and FUT3 genes
Based on combinations of known alleles and their secretor phenotypes, seven haplotypes of the FUT3 gene and five haplotypes of the FUT2 gene were estimated according to each allelic polymorphism. Le and le 59,508 were present as the most common haplotypes in FUT3, with a frequency of 72.94% and 14.72%, respectively. Haplotypes le 59 and le 59,1067 appeared with an equal frequency of 4.75%. Isolated le 508 and le 1067 alleles were rare (0.32% and 0.16%, respectively) and le 59,508,1067 was not detected. le 202,314 combining the 202T>C and 314C>T variations was only found as a co‐occurrence, with a frequency of 2.85%. (Table 2).
Table 2.
Distribution of Lewis and Secretor genotypes
| Lewis (FUT3) | Secretor (FUT2) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | Phenotype | Distribution n (%) | Allele | Frequency (%) | Genotype | Phenotype | Distribution n (%) | Allele | Frequency (%) |
| Le/Le | Le | 176 (55.70) | Le | 72.94 | Se/se 385 | Se | 114 (36.08) | Se | 46.20 |
| Le/le 59,508 | Le | 56 (17.72) | le 59,508 | 14.72 | se 385/se 385 | se | 69 (21.84) | se 385 | 40.19 |
| Le/le 59 | Le | 24 (7.59) | le 59 | 4.75 | Se/Se | Se | 55 (17.41) | se 357,385 | 5.85 |
| le 59,508/le 59,508 | le | 15 (4.75) | le 59,1067 | 4.27 | Se/se 357,385 | Se | 36 (11.39) | Se 357 | 7.12 |
| Le/le 59,1067 | Le | 15 (4.75) | le 202,314 | 2.85 | Se/Se 357 | Se | 31 (9.81) | se 357,428,739 | 0.63 |
| Le/le 202,314 | Le | 11 (3.48) | le 1067 | 0.32 | Se 357/Se 357 | Se | 6 (1.90) | ||
| le 59,1067/le 59,508 | le | 5 (1.58) | le 508 | 0.16 | se 385/se 357,428,739 | se | 2 (0.63) | ||
| le 59/le 202,314 | le | 4 (1.27) | Se 357/se 357,385 | Se | 1 (0.32) | ||||
| le 59,1067/le 59,1067 | le | 3 (0.95) | Se 357/se 357,428,739 | Se | 1 (0.32) | ||||
| le 59,508/le 202,314* | le | 2 (0.63) | Se/se 357,428,739 | Se | 1 (0.32) | ||||
| Le/le 1067 | Le | 2 (0.63) | |||||||
| le 59/le 59 | le | 1 (0.32) | |||||||
| le 59,1067/le 202,314* | le | 1 (0.32) | |||||||
| Le/le 508 | Le | 1 (0.32) | |||||||
Presumed genotypes.
For the FUT2 gene, Se and se 385 were the most prevalent haplotypes at a frequency of 46.20% and 40.19%, respectively, and thus accounted for 86% of all allele counts. The rarely detected Se 357 and se 357,385 were present at a frequency of 7.12% and 5.85%, respectively. We also noticed that sole variations of se 428 and se 739 were hardly encountered in the Chinese population and only appeared as allele se 357,428,739 at a low frequency of 0.63% (Table 2).
Distribution of FUT2 and FUT3 genotypes
For FUT3 (Lewis) genotypes, Le/Le, Le/le 59,508 and Le/le 59 were demonstrated to be the most common positive genotypes (giving a Lewis‐functional phenotype), and were detected with a frequency of 55.70%, 17.72% and 7.59%, respectively. Thirty‐one individuals (9.82%) exhibited Lewis‐negative genotypes (giving the Lewis‐null phenotype), in which le 59,508/le 59,508 (2.17%) was shown to be the most prevalent (Fig. 1A and Table 2).
Figure 1.
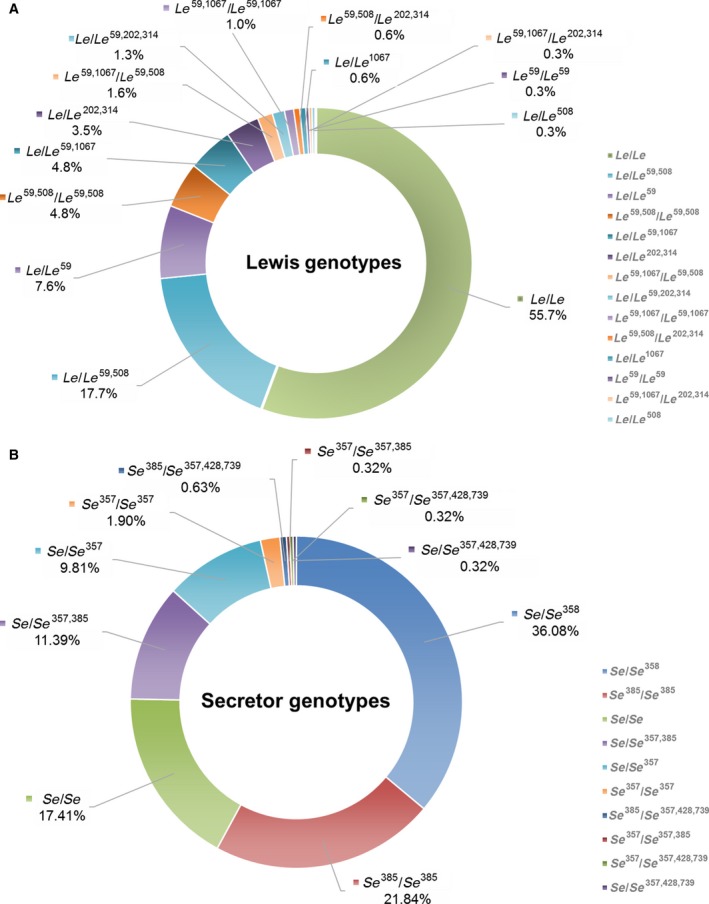
Distribution and frequencies of the FUT3 (Lewis) and FUT2 (Secretor) genotypes in a Chinese population. Doughnut charts representing genotypes for (A) FUT3 (Lewis) and (B) FUT2 (Secretor).
For FUT2 (Secretor) genotypes, allele se 385 was the major factor giving rise to 97% of non‐secretor phenotypes. In the secretor‐positive cohort, Se/se 385 was confirmed to be the most prevalent genotype, with a frequency of 36.08%, and individuals with the Secretor wild‐type (Se/Se) were at 17.41%. The non‐secretor phenotype was found at a frequency of 22.46% and mostly (21.84%) was identified as se 385/se 385. Another non‐secretor genotype, se 385 /se 357,428,739, was much rarer (0.63%). se 428 was previously reported as the major mutation to cause the non‐secretor phenotype and to be widely distributed in other races 28, but was not detected in Chinese. In addition, variation at position 428 was also present in conjugation with the 357 and 739 variation, and completely co‐present with 739 (Fig. 1B and Table 2).
A cluster analysis based on each SNP was performed by combining genotypes of the studied objects (Fig. 1). In total, 55 combined genotypes were encountered of which 18 types (84.9%) were present with a frequency greater than 1% (Table 3). As shown in the genotypes distributions in Fig. 1, subjects homozygous for the functional allele are marked as Le/Le or Se/Se, homozygous mutated loss‐of‐function alleles are denoted as le/le or se/se, and heterozygotes mutated genotypes are represented by Le/le or Se/se. The particular nucleic acid site variations of the Lewis, Secretor and combined genotypes is showed in Table S2.
Table 3.
Distribution of combined genotype of the FUT3 and FUT2 genes and the corresponding phenotype. Le, Se represents the double‐positive genotype; Le, se represents Lewis positive and synchronously secretor negative; le, Se represents Lewis negative and synchronously secretor positive; le, se represents the double‐negative genotype. *Genotypes with frequency less than 1% were omitted
| Combined Lewis and Secretor | Phenotype | Distribution* n (%) | Combination | |
|---|---|---|---|---|
| Phenotype | Frequency (%) | |||
| Le/Le and Se/se 385 | Le, Se | 60 (19) | Le, Se | 70.9 |
| Le/Le and se 385/se 385 | Le, se | 38 (12) | Le, se | 19.3 |
| Le/Le and Se/Se | Le, Se | 27 (8.5) | le, Se | 6.6 |
| Le/le 59,508 and Se/se 385 | Le, Se | 23 (7.3) | le, se | 3.2 |
| Le/Le and Se/se 357,385 | Le, Se | 23 (7.3) | Sum | 100.0 |
| Le/Le and Se/Se 357 | Le, Se | 20 (6.3) | Particular | |
| Le/le 59 and Se/se 385 | Le, Se | 13 (4.1) | Phenotype | Frequency (%) |
| Le/le 59,508 and Se/Se | Le, Se | 11 (3.5) | Lewis‐null (le) | 9.8 |
| Le/le 59,508 and se 385/se 385 | Le, se | 9 (2.9) | Non‐secretor (se) | 22.5 |
| le 59,508/le 59,508 and se 385/se 385 | le, se | 7 (2.2) | ||
| le 59,508/le 59,508 and Se/se 385 | le, Se | 5 (1.6) | ||
| Le/le 59,508 and Se/Se 357 | Le, Se | 5 (1.6) | ||
| Le/le 59,508 and Se/se 357,385 | Le, Se | 5 (1.6) | ||
| Le/le 59,508 and se 385/se 385 | Le, se | 5 (1.6) | ||
| Le/le 59 and Se/Se | Le, Se | 5 (1.6) | ||
| Le/le 59,1067 and Se/se 385 | Le, Se | 4 (1.3) | ||
| Le/le 202,314 and Se/Se | Le, Se | 4 (1.3) | ||
| Le/Le and Se 357/Se 357 | Le, Se | 4 (1.3) | ||
Combination of FUT2 and FUT3 phenotype and corresponding CA19‐9 value
Corresponding to each genotype, the phenotype of single individuals was investigated. Four combined phenotypes were identified according to Lewis (FUT3) and Secretor (FUT2) genotypes (Fig. 2), namely Le, Se; Le, se; le, Se; and le, se. Among the population, 226 participants (71.52%) were detected as Le, Se (the double‐positive phenotype), which manifested as the highest frequency. Le, se (Lewis positive and synchronously secretor negative) and le, Se (Lewis negative and synchronously secretor positive) were present in 62 individuals at a frequency of 19.62% and in 18 individuals at a frequency of 7.28%, respectively. The double‐negative phenotype le, se was shown in 3.17% (10 of 316) of individuals. Table 3 summarized the 18 high‐frequency genotypes and corresponding phenotypes.
Figure 2.
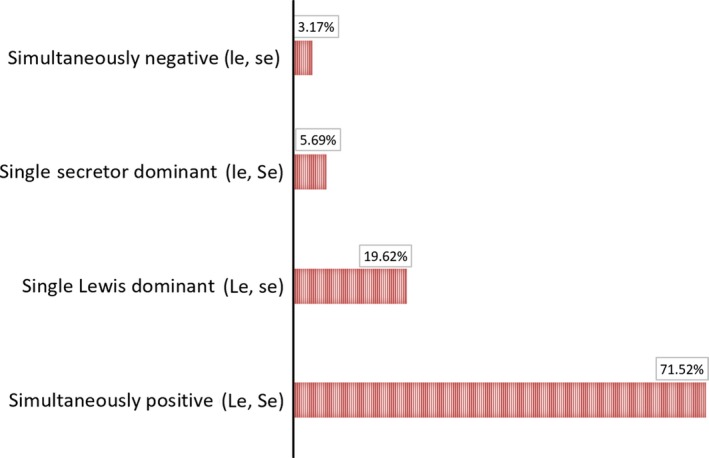
Distribution of combined phenotypes in Chinese population.
The serum CA19‐9 value of each participant was measured and compared between genotypes. Comparison between the Lewis‐functional phenotypes showed that CA19‐9 values in participants with the Le/Le genotype were significantly higher than ones with the Le/le genotype (mean 13.07 vs 9.37 U·mL−1, P < 0.05). Otherwise, CA19‐9 values in the Lewis‐null phenotype group were completely undetectable (CA19‐9 value < 0.06 U·mL−1) (Fig. 3A). Grouping by secretor phenotype, participants with the se/se phenotype showed a much higher CA19‐9 value (mean 17.27 U·mL−1) than the Se/se type (mean 9.43 U·mL−1) and Se/Se type (mean 7.38 U·mL−1) (P < 0.0001) (Fig. 3B). For the combined Lewis and secretor phenotypes, the Le/se group showed the highest CA19‐9 value compared with other groups (mean 20 U·mL−1, 95% CI: 17.95–22.05 U·mL−1) (P < 0.0001) (Table 4). Both le/se and le/Se types showed an undetectable CA19‐9 value (Fig. 3C and Table 4).
Figure 3.
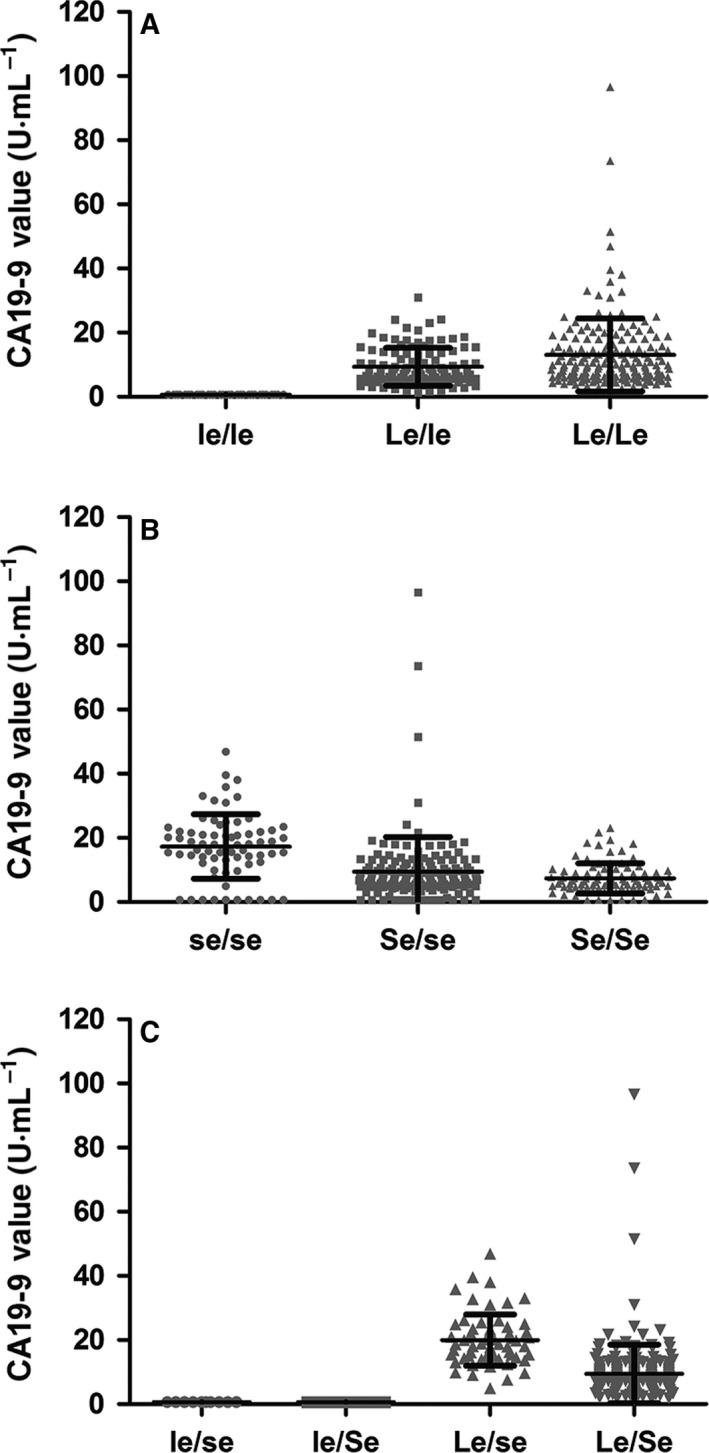
CA19‐9 values for different phenotypes. The comparison of serum antigen expression was conducted between Lewis phenotypes (A), secretor phenotypes (B) and combined phenotypes (C).
Table 4.
Distribution of phenotypes and corresponding CA19‐9 values
| Phenotype | No. of values | CA19‐9 value (U·mL−1) | 95% CI | One‐way analysis of variance (P value) | ||||
|---|---|---|---|---|---|---|---|---|
| Min | Median | Max | Mean | |||||
| Lewis | le/le | 31 | < 0.6 | < 0.6 | < 0.6 | < 0.6 | < 0.6 | < 0.0001 |
| Le/le | 107 | 1.39 | 7.61 | 30.96 | 9.373 | 8.25–10.5 | ||
| Le/Le | 178 | 1.72 | 9.8 | 96.53 | 13.07 | 11.38–14.76 | ||
| Secretor | se/se | 71 | 0.6 | 17.8 | 46.82 | 17.27 | 14.89–19.64 | < 0.0001 |
| Se/se | 153 | 0.6 | 7.38 | 96.53 | 9.43 | 7.71–11.15 | ||
| Se/Se | 92 | 0.6 | 6.495 | 23.03 | 7.383 | 6.41–8.359 | ||
| Combined | le/se | 10 | < 0.6 | < 0.6 | < 0.6 | < 0.6 | < .6 | < 0.0001 |
| le/Se | 22 | < 0.6 | < 0.6 | < 0.6 | < 0.6 | < 0.6 | ||
| Le/se | 61 | 4.94 | 18.72 | 46.82 | 20 | 17.95–22.05 | ||
| Le/Se | 224 | 1.39 | 7.375 | 96.53 | 9.417 | 8.22–10.62 | ||
Population differentiation of the genotype in FUT2 and FUT3 alleles
In the present study, we encountered seven kinds of negative Lewis haplotypes and four kinds of negative Secretor haplotypes. Figure 4 presents the ethnic specificity of putative allelic frequencies among various populations including Korean 9, 29, Chinese 10, 22, 23, 30, Japanese 6, 25, Thai 26, Caucasian 15, 31 and others. Distribution of the FUT3 genotype is approximatively consistent with reported data in Chinese and other Asian populations. Nevertheless, no Asian population study has confirmed the existence of le 508 previously, which is at a frequency of 0.16% in the present study and 0.70% in an Amazonian population 16. le 59,508 is abundantly distributed in most populations with a frequency from 14% to 31%, but rarely found in Caucasians (1.0% and 1.5%). In contrast, le 202,314 is present in Asians at a much low frequency compared with Caucasians 15, 31 (Fig. 4A).
Figure 4.
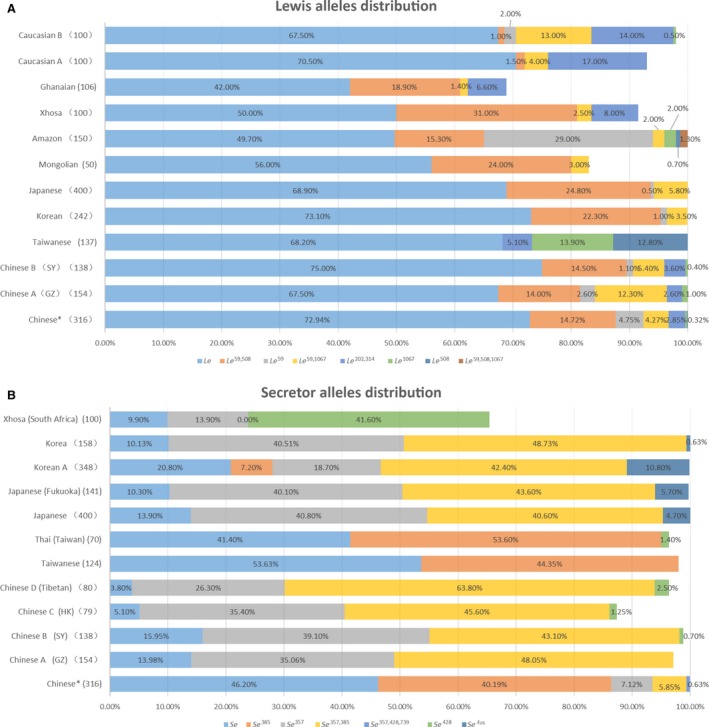
Distribution of genotypes and phenotypes in different ethnic and geographic groups. Comparison of Lewis (A) and Secretor (B) allele frequencies among different populations. (*Population in present study; a: Liu et al. 10; b: Liu et al. 19; c: Park et al. 29; d: Narimatsu et al. 25; e: Soejima et al. 31; f: Corvelo et al. 16; g: Pang et al. 15; h: Liu et al. 23; i: Yip et al. 22; j: Pang et al. 30 ; k: Chang et al. 26; l: Park et al. 9; m: Koda et al. 6; n: Liu et al. 28.)
The three most common FUT2 alleles in the present study were Se, se 385 and Se 357, with a frequency of 46.20%, 40.19% and 7.12%, respectively, and the distribution is highly distinctive in multiple Asian populations including four Chinese groups 22, 23, 30, except for one group in Taiwan 26 (Fig. 4B). We have noticed that allele se 385 was found at an especially common rate in the current study compared with other populations. On the other hand, the frequency of se 357,428 in the present population is much lower, while the frequency of Se is much higher. No se fus was detected in Asians except for Japanese (4.7–5.7%). In contrast to Xhosa and Caucasians, se 428 in Asians is rare and previous studies revealed a very low frequency. However, our data showed that 428 absolutely conjugated with other mutations in Chinese rather than being an isolated allele. se 357,428,739 was not reported before in other studies, mainly accounted for by a deficiency in nucleotide 739 detection. The variations of Lewis and Secretor allele distributions among different populations are shown Table S3.
Discussion
The distribution of genotype has been reported to have ethnic specificity, and genetic heterogeneity in Lewis‐null and non‐secretor individuals is obviously present in different populations 2, 15, 17, 18, 32. Even in the Chinese population, the frequency of phenotypes varies with subject selection, study range, detected position and investigation method 10, 19, 22, 26, 30. Integral and systemic comprehension of the FUT3 (Lewis) and FUT2 (Secretor) genotypes in the Chinese population has been insufficient.
In a large cohort of 316 Chinese participants, we performed accurate genotyping and a combined analysis of nine SNPs reported previously in the fucosyltransferase genes 8, 10, 16, 19, 23, 24. False‐negative reactions are often produced in Lewis blood typing due to weak hemagglutination reactions, which in the main is accounted for by adsorption of glycolipids and by erythrocyte levels 2, 16, 25. For further study, FUT2/FUT3 genotyping is the most accurate method for distinguishing the phenotypes 6, 7, 11, 20. Accordingly, by direct sequencing, five major SNPs of the FUT3 gene and four major SNPs of the FUT2 gene were detected. Additionally, we also discovered 18 primary genotypes and four phenotypes in the Chinese cohort by cluster analysis. It was known that Lewis genotypes in Asians are inconsistent with the polymorphism in Europe and America, and our study has shown the unique distribution of the FUT3/FUT2 gene polymorphism and allied genotype frequencies in a randomly recruited Chinese population. We analyzed the coding DNA sequence range and in FUT3 did not detect any SNPs other than those previously reported, but three individuals with heterozygous 571C>T (rs1800028) in FUT2 were detected.
In the present population, the ‘wild‐type’ (Le/Le) FUT3 gene was detected at a frequency of 55.70%, which is consistent with previous reports in Chinese and Asians. Compared with Caucasians, allele le 59,508 is more abundant and le 202,314 is much rarer in Asians 15, 31. The Amazonian population shows a particular enrichment of the le 59 allele compared with other populations 16. Allele le 1067 and allele le 508 were reported to be absent in Asians; nevertheless, they were detected at the frequency of 0.32% and 0.16%, respectively, in the present Chinese population. This discrepancy could be attributed to the sensitivity of the detection method. The two alleles were reported with much higher frequency in a study that included three Asian populations, which was mostly caused by omitted the detection of le 59 19.
The mutated allele le 59 was shown to have the highest prevalence (40.19%) in the present participants, which homozygotes generating an amino acid variation in the transmembrane domain. Variation 59T>G leading to an L20R amino acid substitution was reported to be responsible for a Lewis‐negative phenotype probably accounted for by a reduction in Golgi retention 3, 7, 33. We classified the isolated 59 mutated allele in the Lewis‐negative group (marked as le 59), even though multiple studies have counted the allele in the Lewis‐positive genotype 10, 15, 16. Variations of nucleotides 202 and 314 have usually been reported as present together 32, 34, and we found that both T202C and C314T were present at frequencies of 5.70% in the Chinese population and no isolated alleles were found. Similar correlation of two single mutations in FUT2 was also demonstrated for 428 and 739, at a lower frequency of 1.27%.
Earlier reports have revealed that differential histo‐blood group antigen expression (such as CA19‐9) has been insufficiently attributed to FUT3 variation. Enzyme FUT2 competitively binds the substrate from FUT3 and the genotype also influences histo‐blood group antigens status. Multiple polymorphic nucleotides in the FUT2 gene reflecting the secretor (Se) and non‐secretor (se) type have been reported with ethnic specificity 5, 22. Homozygous 428G>A (non‐secretor allele se 428) was the first identified missense mutation and was reported in approximately 20% of Caucasians 5, 20. In Asians, variations 357C>T and 385A>T were common, but variation 428G>A was rare 9, 23, 24, 30. se 385 is reported as the most prevalent allele in Asian populations and showed the highest frequency (40.19%) in the current population. Although the allele se 385 was definitively associated with the non‐secretor phenotype, it was rarely present in European populations (at a frequency of 0.4%) 24. In comparison with other populations including Chinese, allele Se (wild‐type) and allele se 385 (non‐secretor type) appeared at obviously higher frequencies in the current study. Simultaneously, functional allele Se 357 and non‐secretor se 357,385 were much rarer than in other populations (Fig. 4B).
As the synthesized product, the Lewis ABO (H) histo‐blood group antigen is routinely utilized as a clinical diagnosis biomarker, especially for malignant gastrointestinal tumor indication 35. It was previously known that individuals with a Lewis‐negative blood group are not able to synthesize Lewis antigens, and that the FUT2/3 gene status determines Lewis antigen synthesis and secretion 25, 36. In the present study, 9.8% of individuals detected as the Lewis‐negative genotype were confirmed as Lewis‐null and had undetectable serum sLea (CA19‐9) antigen, unrelated to the Secretor genotype. Moreover, we perceived no Lea, Leb antigen synthase, and 22.5% of individuals detected as the Secretor‐negative genotype were perceived as non‐secretor and had no Leb antigen synthase 25. Additionally, 19.3% of participants with the ‘single Lewis dominant genotype’ (Le, se) were detected with significantly more elevated sLea than those of the ‘simultaneously positive type’ (Le, Se) (mean 20 vs 9.42 U·mL−1, P < 0.0001). A classic study showed that nine groups divided from 400 normal individuals by Le/Se genotype were detected with discrepant serum CA19‐9 and DU‐PAN‐2 values, and DU‐PAN‐2 measurement was more useable for colorectal cancer diagnosis in Le‐negative patients 25. Recently, Wannhoff et al. 1 have found that differentiation between three FUT2/3 phenotypes improves the clinical practicability of gene‐based cut‐off values of CA19‐9 for cholangiocarcinoma diagnosis. The application of FUT2/3 genotype‐based cut‐offs improved sensitivity to 82.4% and 100.0% in the intermediate and high biosynthesis groups, respectively 1. The indicator sensitivity of Lewis antigens would be significantly increased based on the determination of the FUT2/3 genotype, which may have wide application in tumor diagnosis.
In conclusion, this genotyping study of FUT2 and FUT3 indicated the particular distribution of polymorphisms in the Chinese population. Clarifying individuals’ FUT2/3 genotype by sequencing might facilitate clinical classification and accurate diagnosis. Frequencies of each allele and the association with phenotype should be investigated in an extended population and at additional polymorphism positions, and the characteristics of separate subgroups warrants further study.
Author contributions
MG and CL designed the study; HC, CY and YL carried out experiments; RL and WS provided the materials; KJ, ZW and JL analyzed the sequencing data; QN and GL analyzed experimental results and conducted statistical analysis; MG and GL wrote the manuscript.
Supporting information
Table S1. Primers sequence for PCR amplification of the FUT3 (Lewis) and FUT2 (Secretor) genes.
Table S2. Particular nucleic acid sites variation of Lewis, Secretor and combined genotypes.
Table S3. Comparison of Lewis and Secretor allele frequencies among different populations.
Acknowledgements
We thank Dr Mengyun Wang (Shanghai cancer center, Fudan University, China) for kind assistance with the technology. We are also grateful to Dr Chenyue Zhang (Shanghai Cancer Center, Fudan University, China) for her contribution to the preparation of this manuscript. This study was supported in part by the Sino‐German Center (Grant no. GZ857), by the Science Foundation of Shanghai (Grant no. 14QA1400900) and by the National Science Foundation of China (Grant no. 81101807). The Sino‐German Center for Research Promotion is jointly established by National natural science foundation of China (NSFC) and German science foundation (DFG), (China & German).
Contributor Information
Chen Liu, Email: liuchen@fudanpci.org.
Xianjun Yu, Email: yuxianjun@fudanpci.org.
References
- 1. Wannhoff A, Hov JR, Folseraas T, Rupp C, Friedrich K, Anmarkrud JA, Weiss KH, Sauer P, Schirmacher P, Boberg KM et al (2013) FUT2 and FUT3 genotype determines CA19‐9 cut‐off values for detection of cholangiocarcinoma in patients with primary sclerosing cholangitis. J Hepatol 59, 1278–1284. [DOI] [PubMed] [Google Scholar]
- 2. Nishihara S, Narimatsu H, Iwasaki H, Yazawa S, Akamatsu S, Ando T, Seno T and Narimatsu I (1994) Molecular genetic analysis of the human Lewis histo‐blood group system. J Biol Chem 269, 29271–29278. [PubMed] [Google Scholar]
- 3. Nishihara S, Hiraga T, Ikehara Y, Iwasaki H, Kudo T, Yazawa S, Morozumi K, Suda Y and Narimatsu H (1999) Molecular behavior of mutant Lewis enzymes in vivo. Glycobiology 9, 373–382. [DOI] [PubMed] [Google Scholar]
- 4. Henry S, Oriol R and Samuelsson B (1995) Lewis histo‐blood group system and associated secretory phenotypes. Vox Sang 69, 166–182. [DOI] [PubMed] [Google Scholar]
- 5. Kelly RJ, Rouquier S, Giorgi D, Lennon GG and Lowe JB (1995) Sequence and expression of a candidate for the human Secretor blood group alpha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme‐inactivating nonsense mutation commonly correlates with the non‐secretor phenotype. J Biol Chem 270, 4640–4649. [DOI] [PubMed] [Google Scholar]
- 6. Koda Y, Soejima M, Liu Y and Kimura H (1996) Molecular basis for secretor type alpha(1,2)‐fucosyltransferase gene deficiency in a Japanese population: a fusion gene generated by unequal crossover responsible for the enzyme deficiency. Am J Hum Genet 59, 343–350. [PMC free article] [PubMed] [Google Scholar]
- 7. Mollicone R, Reguigne I, Kelly RJ, Fletcher A, Watt J, Chatfield S, Aziz A, Cameron HS, Weston BW and Lowe JB (1994) Molecular basis for Lewis alpha(1,3/1,4)‐fucosyltransferase gene deficiency (FUT3) found in Lewis‐negative Indonesian pedigrees. J Biol Chem 269, 20987–20994. [PubMed] [Google Scholar]
- 8. Hu DY, Shao XX, Xu CL, Xia SL, Yu LQ, Jiang LJ, Jin J, Lin XQ and Jiang Y (2014) Associations of FUT2 and FUT3 gene polymorphisms with Crohn's disease in Chinese patients. J Gastroenterol Hepatol 29, 1778–1785. [DOI] [PubMed] [Google Scholar]
- 9. Park KU, Song J, Han KS and Kim JQ (2005) The fusion allele of the FUT2 (secretor type alpha(1,2)‐fucosyltransferase) gene at a high frequency and a new se385 allele in a Korean population. Ann Hematol 84, 656–660. [DOI] [PubMed] [Google Scholar]
- 10. Liu YH, Koda Y, Soejima M, Pang H, Wang B and Kimura H (1999) Lewis (FUT3) genotypes in two different Chinese populations. J Forensic Sci 44, 82–86. [PubMed] [Google Scholar]
- 11. Koda Y, Kimura H and Mekada E (1993) Analysis of Lewis fucosyltransferase genes from the human gastric mucosa of Lewis‐positive and ‐negative individuals. Blood 82, 2915–2919. [PubMed] [Google Scholar]
- 12. Folseraas T, Melum E, Rausch P, Juran BD, Ellinghaus E, Shiryaev A, Laerdahl JK, Ellinghaus D, Schramm C, Weismuller TJ et al (2012) Extended analysis of a genome‐wide association study in primary sclerosing cholangitis detects multiple novel risk loci. J Hepatol 57, 366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Franke A, McGovern DP, Barrett JC, Wang K, Radford‐Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R et al (2010) Genome‐wide meta‐analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet 42, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McGovern DP, Jones MR, Taylor KD, Marciante K, Yan X, Dubinsky M, Ippoliti A, Vasiliauskas E, Berel D, Derkowski C et al (2010) Fucosyltransferase 2 (FUT2) non‐secretor status is associated with Crohn's disease. Hum Mol Genet 19, 3468–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pang H, Liu Y, Koda Y, Soejima M, Jia J, Schlaphoff T, Du Toit ED and Kimura H (1998) Five novel missense mutations of the Lewis gene (FUT3) in African (Xhosa) and Caucasian populations in South Africa. Hum Genet 102, 675–680. [DOI] [PubMed] [Google Scholar]
- 16. Corvelo TC, de Loiola Rdo S, Aguiar DC, de Matos Gde C and de Brito DC (2013) The Lewis histo‐blood group system: molecular analysis of the 59T>G, 508G>A, and 1067T>A polymorphisms in an Amazonian population. PLoS One 8, e69908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elmgren A, Rydberg L and Larson G (1993) Genotypic heterogeneity among Lewis negative individuals. Biochem Biophys Res Commun 196, 515–520. [DOI] [PubMed] [Google Scholar]
- 18. Orntoft TF, Vestergaard EM, Holmes E, Jakobsen JS, Grunnet N, Mortensen M, Johnson P, Bross P, Gregersen N, Skorstengaard K et al (1996) Influence of Lewis alpha1‐3/4‐L‐fucosyltransferase (FUT3) gene mutations on enzyme activity, erythrocyte phenotyping, and circulating tumor marker sialyl‐Lewis a levels. J Biol Chem 271, 32260–32268. [DOI] [PubMed] [Google Scholar]
- 19. Liu TC, Chang JG, Lin SF, Chang WC, Yang TY, Lin CL, Wang NM and Tsai CH (2000) Lewis (FUT3) genotypes in Taiwanese, Thai, and Filipino populations. Ann Hematol 79, 599–603. [DOI] [PubMed] [Google Scholar]
- 20. Kudo T, Iwasaki H, Nishihara S, Shinya N, Ando T, Narimatsu I and Narimatsu H (1996) Molecular genetic analysis of the human Lewis histo‐blood group system. II. Secretor gene inactivation by a novel single missense mutation A385T in Japanese nonsecretor individuals. J Biol Chem 271, 9830–9837. [DOI] [PubMed] [Google Scholar]
- 21. Weiss FU, Schurmann C, Guenther A, Ernst F, Teumer A, Mayerle J, Simon P, Volzke H, Radke D, Greinacher A et al (2015) Fucosyltransferase 2 (FUT2) non‐secretor status and blood group B are associated with elevated serum lipase activity in asymptomatic subjects, and an increased risk for chronic pancreatitis: a genetic association study. Gut 64, 646–656. [DOI] [PubMed] [Google Scholar]
- 22. Yip SP, Lai SK and Wong ML (2007) Systematic sequence analysis of the human fucosyltransferase 2 (FUT2) gene identifies novel sequence variations and alleles. Transfusion 47, 1369–1380. [DOI] [PubMed] [Google Scholar]
- 23. Liu YH, Koda Y, Soejima M, Pang H, Wang BJ, Kim DS, Oh HB and Kimura H (1999) The fusion gene at the ABO‐secretor locus (FUT2): absence in Chinese populations. J Hum Genet 44, 181–184. [DOI] [PubMed] [Google Scholar]
- 24. Ferrer‐Admetlla A, Sikora M, Laayouni H, Esteve A, Roubinet F, Blancher A, Calafell F, Bertranpetit J and Casals F (2009) A natural history of FUT2 polymorphism in humans. Mol Biol Evol 26, 1993–2003. [DOI] [PubMed] [Google Scholar]
- 25. Narimatsu H, Iwasaki H, Nakayama F, Ikehara Y, Kudo T, Nishihara S, Sugano K, Okura H, Fujita S and Hirohashi S (1998) Lewis and secretor gene dosages affect CA19‐9 and DU‐PAN‐2 serum levels in normal individuals and colorectal cancer patients. Cancer Res 58, 512–518. [PubMed] [Google Scholar]
- 26. Chang JG, Yang TY, Liu TC, Lin TP, Hu CJ, Kao MC, Wang NM, Tsai FJ, Peng CT and Tsai CH (1999) Molecular analysis of secretor type alpha(1,2)‐fucosyltransferase gene mutations in the Chinese and Thai populations. Transfusion 39, 1013–1017. [DOI] [PubMed] [Google Scholar]
- 27. Goodstadt L and Ponting CP (2001) CHROMA: consensus‐based colouring of multiple alignments for publication. Bioinformatics 17, 845–846. [DOI] [PubMed] [Google Scholar]
- 28. Liu Y, Koda Y, Soejima M, Pang H, Schlaphoff T, du Toit ED and Kimura H (1998) Extensive polymorphism of the FUT2 gene in an African (Xhosa) population of South Africa. Hum Genet 103, 204–210. [DOI] [PubMed] [Google Scholar]
- 29. Park HD, Park KU, Song J, Ki CS, Han KS and Kim JQ (2010) The relationship between Lewis/Secretor genotypes and serum carbohydrate antigen 19‐9 levels in a Korean population. Korean J Lab Med 30, 51–57. [DOI] [PubMed] [Google Scholar]
- 30. Pang H, Koda Y, Soejima M, Fujitani N, Ogaki T, Saito A, Kawasaki T and Kimura H (2001) Polymorphism of the human ABO‐Secretor locus (FUT2) in four populations in Asia: indication of distinct Asian subpopulations. Ann Hum Genet 65, 429–437. [DOI] [PubMed] [Google Scholar]
- 31. Soejima M, Munkhtulga L, Iwamoto S and Koda Y (2009) Genetic variation of FUT3 in Ghanaians, Caucasians, and Mongolians. Transfusion 49, 959–966. [DOI] [PubMed] [Google Scholar]
- 32. Elmgren A, Borjeson C, Svensson L, Rydberg L and Larson G (1996) DNA sequencing and screening for point mutations in the human Lewis (FUT3) gene enables molecular genotyping of the human Lewis blood group system. Vox Sang 70, 97–103. [DOI] [PubMed] [Google Scholar]
- 33. Mollicone R, Cailleau A and Oriol R (1995) Molecular genetics of H, Se, Lewis and other fucosyltransferase genes. Transfus Clin Biol 2, 235–242. [DOI] [PubMed] [Google Scholar]
- 34. Elmgren A, Mollicone R, Costache M, Borjeson C, Oriol R, Harrington J and Larson G (1997) Significance of individual point mutations, T202C and C314T, in the human Lewis (FUT3) gene for expression of Lewis antigens by the human alpha(1,3/1,4)‐fucosyltransferase, Fuc‐TIII. J Biol Chem 272, 21994–21998. [DOI] [PubMed] [Google Scholar]
- 35. Luo G, Liu C, Guo M, Cheng H, Lu Y, Jin K, Liu L, Long J, Xu J, Lu R et al (2017) Potential Biomarkers in Lewis Negative Patients With Pancreatic Cancer. Ann Surg 265, 800–805. [DOI] [PubMed] [Google Scholar]
- 36. Vestergaard EM, Hein HO, Meyer H, Grunnet N, Jorgensen J, Wolf H and Orntoft TF (1999) Reference values and biological variation for tumor marker CA 19‐9 in serum for different Lewis and secretor genotypes and evaluation of secretor and Lewis genotyping in a Caucasian population. Clin Chem 45, 54–61. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers sequence for PCR amplification of the FUT3 (Lewis) and FUT2 (Secretor) genes.
Table S2. Particular nucleic acid sites variation of Lewis, Secretor and combined genotypes.
Table S3. Comparison of Lewis and Secretor allele frequencies among different populations.


