Abstract
We have developed a model system for membrane fusion that utilizes lipidated derivatives of a heterodimeric coiled-coil pair dubbed E3 (EIAALEK)3 and K3 (KIAALKE)3. In this system, peptides are conjugated to a lipid anchor via a poly(ethylene glycol) (PEG) spacer, and this contribution studies the influence of the PEG spacer length, coupled with the type of lipid anchor, on liposome–liposome fusion. The effects of these modifications on peptide secondary structure, their interactions with liposomes, and their ability to mediate fusion were studied using a variety of different content mixing experiments and CD spectroscopy. Our results demonstrate the asymmetric role of the peptides in the fusion process because alterations to the PEG spacer length affect E3 and K3 differently. We conclude that negatively charged E3 acts as a “handle” for positively charged K3 and facilitates liposome docking, the first stage of the fusion process, through coiled-coil formation. The efficacy of this E3 handle is enhanced by longer spacer lengths. K3 directs the fusion process via peptide–membrane interactions, but the length of the PEG spacer plays two competing roles: a PEG4/PEG8 spacer length is optimal for membrane destabilization; however, a PEG12 spacer increases the fusion efficiency over time by improving the peptide accessibility for successive fusion events. Both the anchor type and spacer length affect the peptide structure; a cholesterol anchor appears to enhance K3–membrane interactions and thus mediates fusion more efficiently.
Introduction
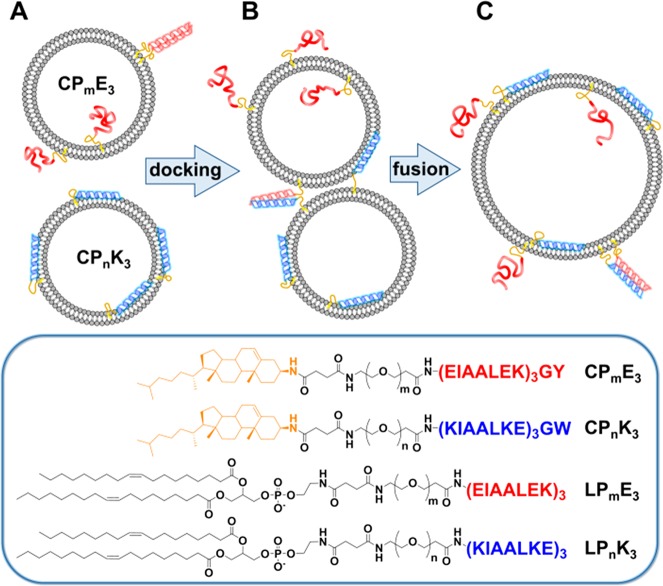
SNARE (soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein receptor) proteins are a well conserved class of fusion proteins that are primarily involved in vesicle trafficking and fusion.1−3 Complementary SNARE protein subunits are located on opposing membranes, and in the first step of the fusion process, they form a tetrameric coiled-coil, which brings the opposing membranes into close proximity. The fusion cascade, initiated by this docking process, consists of three more steps:4,5 (1) disruption of the membrane at the site of contact; (2) merging of outer membranes, i.e., lipid mixing or hemifusion; and (3) formation of a fusion pore, facilitating content mixing between the two fused compartments.6 This SNARE fusion system has been studied extensively using the natural proteins7−10 and has served as an inspiration for the design of model systems. Such systems are primarily based on phospholipid vesicles functionalized with either DNA,11−15 peptides,16−19 or small molecules20−24 that act as recognition motifs, but molecular recognition between functionalized cyclodextrin vesicles has also been reported.25−28 In our laboratory, a model system for membrane fusion has been developed with the lipidated heterodimeric coiled-coil pair E3 (EIAALEK)3 and K3 (KIAALKE)3 as fusogens.29,30 These peptides are held in lipid membranes by a lipid anchor, either cholesterol (C) or 1,2-dioleoyl-sn-glycero-3-phosphoethanol-amine (DOPE), (L), and this is conjugated to the peptide via a PEG spacer of variable length (m, n) to yield C/LPmE3 or C/LPnK3.
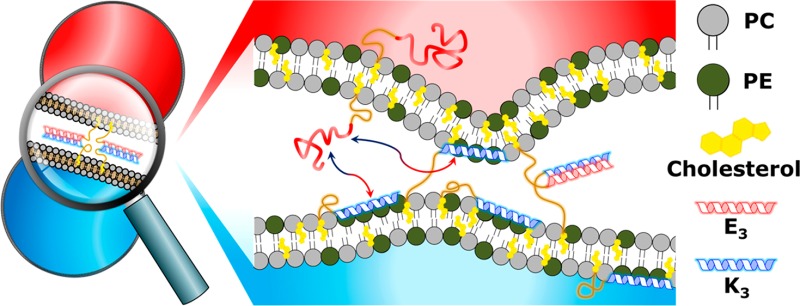
The role of these lipopeptide constructs in the fusion process is revealed by studying the relationship between rational lipopeptide modifications and their resulting influence on fusion efficiency. The consequences of the anchor type were previously investigated,31 and it was shown that only DOPE and cholesterol anchors yielded significant fusion, with the latter being the most efficient. Increasing the length of the peptide (which increases the coiled-coil binding strength and peptide–membrane interactions) also has a positive effect on the fusion efficiency.32 Conversely, the parallel or antiparallel coiled-coil orientation33 and coiled-coil oligomer state34 did not influence the rate of fusion. A recent study has also proposed an asymmetric role for the peptides in the fusion process, as illustrated in Figure 1.35 A key function of K3 is its interaction with the liposome membrane, primarily via its hydrophobic face that comprises leucine and isoleucine residues but also via the flanking lysine residues in a process known as snorkeling.35,36 This K3–membrane interaction leads to destabilization of the membrane and initiation of the fusion process, whereas E3 facilitates coiled-coil formation, bringing the membranes into close proximity, allowing fusion to occur. These different roles lead to competing peptide interactions, which provides an equilibrium between K3–E3 coiled-coil formation and K3–membrane interactions.
Figure 1.
Schematic overview of the process of peptide-mediated liposomal fusion and lipopeptide structures. The peptides are drawn in their appropriate secondary structures at distinct points in the fusion cascade: (A) liposomes functionalized with complementary peptides are initially separated; (B) liposomes dock due to coiled-coil formation, and (C) liposomes become fully fused. Peptide E3 (red) can adopt homocoils, heterocoils, and random coils, whereas peptide K3 (blue) adopts only heterocoils or membrane-immersed α-helical structures.38 The lower boxed region shows the structures of the lipopeptide constructs used in this study; succinic anhydride was used to facilitate the conjugation of the PEG spacer to the DOPE anchor; the corresponding cholesterol derivative was also employed.
In all of these liposomal fusion studies, a PEG12 spacer was used to tether the fusogenic peptides to their membranes, but it is probable that the spacer length is critical for efficient fusion. Indeed, it has been shown that the introduction of a (GGS)1–3 spacer in the natural SNARE complex reduced the overall fusogenicity of the system with increasing spacer length,37 likely because of an increase in the distance between the proteins and their membranes. The same relationship between fusion efficiency and spacer length was also reported with DNA-based fusogens;12 however, faster docking kinetics were found with increasing spacer length, which was rationalized with a diffusion-driven collision model. This demonstrates that a spacer has opposing effects at different steps in the fusion cascade, and it indicates that, in our model system, the PEG spacer length is likely to influence the fusion efficiency.
In light of these previous studies, it was decided that the influence of PEG length on fusion efficiency should be studied using our model system. Both cholesterol and DOPE anchors were employed so that separate and cumulative effects of the anchor and spacer could be studied because it is likely that the components of this model system (peptides, spacer, and anchor) do not act in isolation but have a synergistic effect on fusion. To demonstrate the asymmetry in function of the two complementary peptides, the influence of the PEG spacer length for both E3 and K3 was measured individually. It was anticipated that this approach would provide further insight as to the fusion mechanism and the roles of the different components, in addition to giving a detailed picture of the functions of the lipopeptides at different stages in the fusion process.
Experimental Section
Materials
Fmoc-protected amino acids, rink amide resin, and O-(1H-6-chlorobenzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HCTU) were purchased from NovaBioChem. Diisopropylethylamine (DIPEA), piperidine, acetic anhydride, N-methylpyrrolidine (NMP), dimethylformamide (DMF), acetonitrile, and trifluoroacetic acid (TFA) were obtained from Biosolve. Dichloromethane (DCM), diethyl ether, triisopropylsilane (TIS), trimethylamine (TEA), cholesterol, trimethylphosphine (1 M in toluene), (1H-benzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate (PyBOP), succinic anhydride, and sulforhodamine B were obtained from Sigma-Aldrich. 1,2-Dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC) and 1,2-dioleoyl-sn-glycero-3-phosphatidylethanolamine (DOPE) were purchased from Avanti Polar Lipids. The Fmoc-NH-(PEG)2/8/12/16-COOH spacers were purchased from Iris Biotech. The N3-(PEG)4-COOH spacer was synthesized according to a literature procedure, as reported previously.39,40 Cholesteryl-4-amino-4-oxobutanoic acid was synthesized according to a literature procedure described in the SI.41 PBS buffer contains 5 mM KH2PO4, 15 mM K2HPO4, and 150 mM NaCl at pH 7.4. Data analysis and visualization were performed using OriginPro 9.1.
PEG-Peptide Synthesis
Peptide synthesis was performed on a CEM Liberty 1 on a 100 μmol scale using a Rink amide resin of 0.55–0.73 mmol/g substitution. Amino acid activation was achieved using HCTU/DIPEA in DMF. Fmoc deprotection was carried out using two cycles of 20% piperidine in DMF. All automated reactions were carried out at 70–80 °C with the use of a microwave. The N-terminal free amine was used to couple the PEG spacer through standard solid-phase chemistry using PyBOP/DIPEA activation. With the exception of N3-PEG4-COOH-containing lipopeptides, Fmoc deprotection of the PEG component was achieved using 20% piperidine for 20 min to obtain a free amine at the N-terminus. N3-PEG4-COOH-containing lipopeptides were reduced using two cycles of PMe3 in dioxane/water (4:1).
DOPE Anchor Attachment
Succinic anhydride was coupled to the N-terminal free amine using a succinic anhydride/TEA (10 equiv/10 equiv) mixture in NMP. The reaction was left overnight on a shaker at room temperature. The resin was washed thoroughly with NMP, DCM, and NMP to remove excess reactants. Next, the resin (1 equiv) was activated using PyBOP/DIPEA (3 equiv/5 equiv) in DCM/DMF (1:1) for 1 h. DOPE (2 equiv) was dissolved in a minimal volume (2 mL) of DCM/DMF (1:1), and a small amount of TEA (2 equiv) was added to improve solubility. After 1 h, this DOPE solution was added to the resin and warmed to 50 °C, and the reaction was left overnight on a shaker. The resin was washed thoroughly with DMF and DCM to remove excess reactants.
Cholesterol Anchor Attachment
Cholesteryl-4-amino-4-oxobutanoic acid (1.2 equiv) was activated with PyBOP/DIPEA (3 equiv/5 equiv) in DCM/DMF 1:1 and added to the peptide-bound resin, and the reaction was left for 2 days on a shaker. The resin was washed thoroughly with DMF and DCM to remove excess reactants.
Cleavage
The lipopeptides were cleaved from the resin and side-chain deprotected using a mixture of TFA/H2O/TIS (95:2.5:2.5 v/v) for 2 h. The cholesterol-anchored peptides were precipitated in cold diethyl ether, followed by centrifugation and drying under vacuum. DOPE-anchored peptides were concentrated in vacuo, dissolved in a minimal amount of acetic acid/H2O, and subsequently purified.
Purification
RP-HPLC was performed with a Shimadzu HPLC system with two LC-8A pumps and an SPD-10AVP UV–vis detector. Sample elution was monitored by UV detection at 214 and 256 nm. Samples were eluted with a linear gradient from 10 to 90% (v/v) A to B, with A being H2O, 0.1% (v/v) TFA, and B being MeCN, 0.1% (v/v) TFA. Purification of DOPE-lipopeptides was performed on a Vydac C4 reversed-phase column (214TP1022, 22 mm diameter, 250 mm length, 10.00 μM particle size) at a flow rate of 20 mL min–1. Purification of cholesterol lipopeptides was performed on a Phenomenex C18 reversed-phase column (21.2 mm diameter, 150 mm length, 5.00 μM particle size) with a flow rate of 15 mL min–1. Collected fractions were tested for >95% purity using LC-MS with a Gemini C18 column and freeze dried.
Liposome Preparation
A 1 mM stock solution containing DOPC/DOPE/cholesterol (50:25:25 mol %) lipids in 1:1 (v/v) methanol/chloroform was prepared for all fusion experiments. Lipopeptides were dissolved in 1:1 (v/v) chloroform/methanol at 50 μM concentration. In general, 1 mol % of the lipopeptide solution was mixed with the liposome solution, and the solvent was removed under a stream of air. The dry lipid/peptide layer containing lipopeptides K3 was rehydrated with PBS, and PBS buffer containing 20 mM sulforhodamine B was added to the dried lipid/peptide layers containing lipopeptide E3. These solutions were subsequently sonicated for 5–10 min at 55 °C to yield ∼100-nm-diameter liposomes, verified by dynamic light scattering (DLS).29 The solutions with K3-functionalized liposomes were used without further purification, whereas sulforhodamine B-containing liposomes were purified using a Sephadex G25 column to remove any nonencapsulated sulforhodamine B. The purification dilution factor was 1:3.
Fluorescence Spectroscopy
Content-mixing experiments were performed on a TECAN Infinite M1000 PRO fluorimeter using a 96-well plate at 25 °C. The percentage of fluorescence increase, %F, was calculated as
Content Mixing Experiments
The sulforhodamine B fluorescence intensity, F(t), at 580 nm was monitored in a continuous fashion for 30 min after mixing nonfluorescent K3 liposomes with sulforhodamine B-loaded E3 liposomes. F0 was obtained by measuring the emission of sulforhodamine B-loaded E3 liposomes to which an equal amount of PBS was added, and Fmax was obtained by measuring the emission of plain liposomes loaded with 10 mM sulforhodamine B. [Total lipid] = 0.1 mM, in PBS pH 7.4.
Multiple Round Content Mixing Experiments
Experimental settings of regular content mixing experiments were used as described above. E3-decorated, dye-loaded liposomes were prepared with a total lipid concentration of 0.05 mM in PBS. K3-decorated liposomes were prepared with a total lipid concentration of 0.5 mM, in PBS. Total experimental mixtures for equimolar fusion experiments contained 100 μL of E3 liposomes, 10 μL of K3 liposomes, and 90 μL of PBS. Total experimental mixtures for fusion experiments in a 10:1 ratio contained 100 μL of E3 liposomes and 100 μL of K3 liposomes.
Circular Dichroism Spectroscopy
CD spectra were obtained using a Jasco J-815 spectropolarimeter equipped with a Peltier temperature controller. The ellipticity, given as the mean residue molar ellipticity, [θ] (deg cm2 dmol–1), is calculated using the following equation
where is the observed ellipticity (mdeg), n is the number of peptide residues, l is the path length of the cuvette (cm), and c is the peptide concentration (mM). Spectra were recorded from 260 to 200 nm at 20 °C. Data points were collected with a 1 nm bandwidth at 1 nm intervals using a scan speed of 1 nm s–1. Each spectrum was an average of five scans. For analysis, each spectrum had the background spectrum (plain liposomes in PBS) subtracted. The percentage of α-helicity was calculated using the predicted value [θ]222 = −40 000(1 – 4.6/n) as 100% of the value for an α-helical peptide of n residues.42 [Total lipid] = 0.5 mM, with 1 mol % lipopeptide, in PBS at pH 7.4.
Results and Discussion
Design of the Study
Both DOPE and cholesterol have been demonstrated to be effective anchor moieties, so using both anchors, a PEG series of 0, 4, 8, and 12 repeating units was synthesized initially. During the course of this study, the spacer series was extended to cover a significant range close to the maximum fusion efficiency, and LP2E3/K3 and CP16E3/K3 derivatives were subsequently synthesized and evaluated (Table 1).
Table 1. Overview of the Synthesized Derivatives of Peptides E3 and K3 Used in This Studya.
| spacer | DOPE anchor | LPEGd | cholesterol anchor | LPEGd |
|---|---|---|---|---|
| noneb | LP0E3/LP0K3 | 0.5 | CP0E3/CP0K3 | –0.5 |
| PEG2c | LP2E3/LP2K3 | 1.6 | n.a. | n.a. |
| PEG4 | LP4E3/LP4K3 | 2.3 | CP4E3/CP4K3 | 1.3 |
| PEG8 | LP8E3/LP8K3 | 3.9 | CP8E3/CP8K3 | 2.9 |
| PEG12 | LP12E3/LP12K3 | 5.4 | CP12E3/CP12K3 | 4.4 |
| PEG16c | n.a. | n.a. | CP16E3/CP16K3 | 5.8 |
Peptide purities of >95% were confirmed using LC-MS for all constructs.
The peptide is connected to the anchor via a succinic diamide, with a length of 0.5 nm.
An LP2 spacer and a CP16 spacer were added to the PEG series to cover a significant range close to the maximum fusion efficiency.
LPEG is the distance between the N-terminus of the peptide and the surface of the membrane in nanometers. For DOPE anchors, this is the maximum theoretical chain length of the PEG spacer (nm), as estimated by AVOGADRO,43 and for cholesterol anchors, this is the maximum theoretical chain length, 1.0 nm,44 due to the intrinsic membrane insertion property of cholesterol in DOPC/DOPE membranes.
To visualize the influence of the PEG spacer length, its stretched end-to-end distance was calculated with AVOGADRO43 software, and from this value, the distance between the peptide and the surface of the membrane was derived and termed LPEG. It should be noted that for longer linkers this effective distance may be reduced because it is unlikely that PEG will retain a stretched conformation. Because the membrane surface is defined by the polar headgroups, it was assumed that in the case of a 75% DOPC/DOPE membrane and a lipopeptide DOPE anchor the PEG length and the peptide–membrane distance are equal. For cholesterol-containing DOPC/DOPE membranes, it is known that cholesterol is buried in the membrane,44 so to calculate LPEG for cholesterol-anchored peptides, the average thickness of the membrane hydration layer (1.0 nm)45 was subtracted because cholesterol resides at the glycerol level in the phospholipid membrane.46
Designing experiments to obtain insight as to the details of the fusion process is challenging because of complex short-lived intermediate stages and fast dynamics of fusion events. In addition, it is possible that the PEG spacer will have opposing effects during the fusion process.30 Therefore, it is necessary to identify the impact of these effects on the fusion efficiency.
The cumulative effect of the combination of PEG spacer length and anchor type on lipopeptide-mediated membrane fusion was studied using a content mixing assay. Further experiments were then conducted to determine the effects of the PEG spacer length at different stages in the fusion process. Peptide-ratio-dependent fusion provided insights as to the influence of the equilibria between different peptide conformations on the fusion mechanism. Furthermore, an experiment was designed to measure the ability of the lipopeptides to mediate multiple rounds of liposomal fusion. The influence of membrane tethering on the peptide structure was measured using circular dichroism (CD) spectroscopy. The results here, supported by previous studies,35,38,47 suggest that the process of membrane fusion is rate-limited by equilibria between different peptide conformations in the initial fusion stages and that the PEG length influences these equilibria and therefore the extent of fusion.
Content Mixing Assay
The final stage of a full fusion process between two membranes is content mixing. Although efficient and leakage-free content mixing remains a challenge for many fusion model systems, it has been successfully demonstrated using this system in previous studies. To facilitate this process, a quantitative content mixing model was developed by encapsulating water-soluble dye sulforhodamine B.48−53 This dye self-quenches at high concentrations, but dilution results in the relief of self-quenching and an increase in fluorescence. Liposomes incorporating E3 lipopeptides were loaded with 20 mM sulforhodamine B in PBS and mixed with K3-functionalized buffer-containing liposomes. The effect of the anchor type (with a PEG12 spacer length) on the fusion efficiency was demonstrated previously, resulting in a variable degree of membrane fusion between LP12E3/K3 and CP12E3/K3.31 Here, the influence of both the spacer length and anchor type on content mixing (as a measure of full fusion) was determined (Figures 2 and S1).
Figure 2.
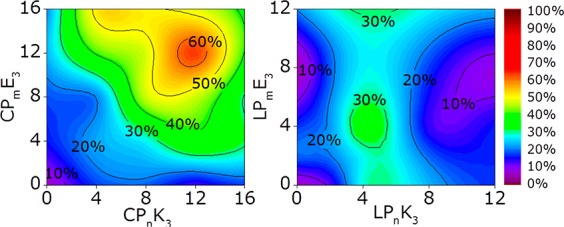
Content mixing between E3- and K3-decorated liposomes as a function of PEG spacer length (m/n) as indicated by an increase in sulforhodamine B emission after 30 min. (Left) Cholesterol-anchored Pm/n series and (right) DOPE-anchored Pm/n peptides. [Total lipid] = 0.1 mM, with 1 mol % lipopeptide, PBS pH 7.4.
It is evident that liposomal fusion with CPmE3/CPnK3 is most efficient if both lipopeptide constructs use a PEG12 spacer, with fusion being less efficient if shorter, or longer, spacers are employed. For DOPE-anchored peptides, a PEG4 spacer is necessary for the K3 construct, but the length of the spacer for E3 appears to be irrelevant. Furthermore, fusion mediated by cholesterol-anchored peptides is twice as efficient as DOPE-peptide-mediated fusion.
This spacer-dependent trend for DOPE-peptide-mediated fusion shows that the LPEG of peptide K3 highly influences the fusion efficiency, indicating that the K3 peptide is crucial for efficient fusion and that the linker length strongly influences the efficiency of this process. The observed differences in the PEG spacer length preference between cholesterol- and DOPE-anchored peptides cannot be explained by differences in LPEG because their various spacers all cover the same distance range (0–6 nm), although cholesterol is bound more tightly to the membrane than is DOPE.31 Because liposomal membranes already contain 25 mol % cholesterol, significant influences in general membrane characteristics attributed to cholesterol were not expected.53,54 However, recent studies on the influence of the lipid anchor on membrane-tethered PEG spacers showed that a cholesterol anchor increased the flexibility of the spacers55 and influenced their function.56 This reveals that the anchor type does affect the behavior of the other components (spacer and peptide) in this construct, and it is hypothesized that the cholesterol anchor enhances the membrane interacting and disrupting role of K3 in the fusion process, as shown in Figure 1, which may explain why increased levels of content mixing are observed with cholesterol-anchored peptides.
Circular Dichroism Spectroscopy
Because LP12E3 and LP12K3 are known to adopt distinct structures when tethered to lipid membranes,35,38 the influence of linker length on the peptide secondary structure was examined using circular dichroism (CD) spectroscopy. In addition to exploring these linker effects, both anchors were tested to determine whether these also influence the peptide structure. The membrane-tethered peptide constructs were tested in isolation and when mixed to determine whether heteromeric coiled-coil species were formed. The results are illustrated in Table 2 and Figures 3 and S4. A θ222/θ208 ratio ≥1 is indicative of coiled-coil formation; however, this ratio can be skewed by the inherent scattering properties of liposomes. Although θ222 is a measure of helicity, both peptide–membrane interactions and coiled-coil formation contribute to the observed helicity. Therefore, identifying whether peptides are folded as α-helices or coiled coils is nontrivial.
Table 2. Degree of Peptide Helicity As Determined by Circular Dichroism.
| E3 |
K3 |
E3 + K3 |
||||
|---|---|---|---|---|---|---|
| anchor + spacer | θ222/θ208 | %Ha | θ222/θ208 | %Ha | θ222/θ208 | %Ha |
| acylb | 0.4 | 22 | 0.4 | 21 | 1.0 | 66 |
| CP0 | 0.9 | 16 | 0.8 | 79 | 0.8 | 52 |
| CP4 | 1.5 | 46 | 1.2 | 82 | 0.9 | 65 |
| CP8 | 0.9 | 52 | 1.0 | 60 | 1.2 | 71 |
| CP12 | 0.6 | 51 | 1.1 | 52 | 1.3 | 71 |
| CP16 | n.a.c | 7 | n.a.c | 28 | 1.3 | 48 |
| LP0 | 1.0 | 41 | 1.0 | 41 | 1.0 | 42 |
| LP2 | 1.1 | 33 | 1.0 | 43 | 1.1 | 40 |
| LP4 | 1.0 | 43 | 1.0 | 54 | 1.0 | 62 |
| LP8 | 0.9 | 37 | 1.0 | 63 | 1.1 | 62 |
| LP12 | 1.0 | 48 | 1.0 | 48 | 1.1 | 60 |
The percentage of α-helicity was calculated using the formula [θ]222 = −40 000(1 – 4.6/n) to obtain a 100% helicity value for an α-helical peptide of n residues.42
Measurements with acylated peptides, in PBS pH 7.4, without vesicles.
Negative ratio due to the positive θ208 value caused by a scattering artifact. [Total lipid] = 0.5 mM, with 1 mol % lipopeptides, PBS pH 7.4.
Figure 3.
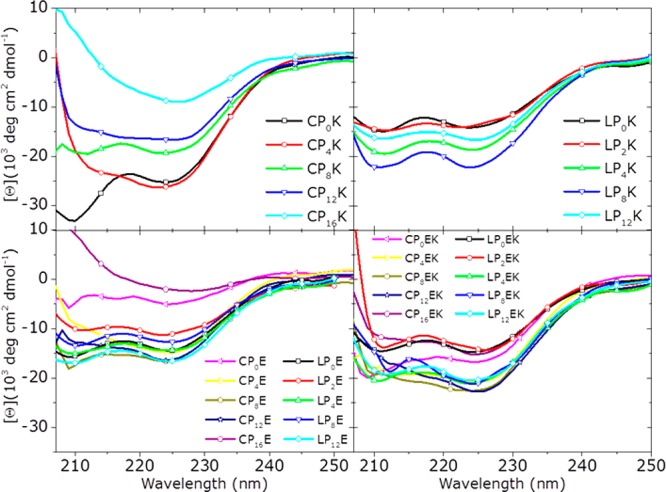
CD spectra of liposome-tethered lipopeptides. [Total lipid] = 0.5 mM, with 1 mol % lipopeptides, PBS pH 7.4.
It was demonstrated previously that the α-helical conformation of membrane-tethered K3 is stabilized via spontaneous incorporation into the membrane,38,47 so a correlation between the observed helicity of the peptides and the relative strength of this membrane interaction may be inferred. For CPnK3 derivatives, increasing helicity upon decreasing spacer length shows that peptides that are in closer proximity to the membrane experience more stabilization by, and therefore are likely to interact more strongly with, the membrane. The helicity of LPn-anchored K3 peptides is lower compared to that of the CPnK3 peptides and does not change significantly with changing PEG lengths. This indicates that the peptide membrane interaction is less pronounced for LPn lipopeptides and that the peptide conformation is not significantly affected by varying the spacer length. The decreased fusion efficiency measured when liposomes are decorated with LPm/n anchored peptides is likely a result of this weaker membrane interaction of LPnK3, which provides further evidence for a correlation between the strength of the peptide–membrane interaction and the fusion efficiency of peptide-decorated liposomes.
To explain this influence of anchor type on peptide–membrane interactions, it is necessary to examine the characteristics of the membrane and the tethered lipopeptides. When peptide K3 interacts with the membrane, it tends to be surrounded by lipids with smaller headgroups (i.e., DOPE and cholesterol).35,57,58 The accumulation of DOPE in the proximity of peptide K3 destabilizes the membrane, promoting the formation of a protrusion centered on the peptide’s position,35 and supports the highly curved membranes of fusion intermediates.53,59 The cholesterol anchor could enhance the membrane insertion of peptide CPnK3 as a result of its relatively small headgroup, and because cholesterol promotes the chain flexibility of conjugated PEG spacers,55 these factors could increase the tendency of the conjugated peptides to incorporate into the supporting membrane.
Peptide E3 adopts structures with relatively small amounts of helicity, supporting previous findings that peptide E3 does not significantly interact with DOPC/DOPE/cholesterol membranes and adopts a mixture of weakly helical homodimers and random coils in solution.38 Although α-helical peptide stabilization by PEG has been reported,60 here these PEG–peptide interactions are not likely to be significant because of the short length of the used PEG spacers.
The structural characteristics of the mixture of E3- and K3-decorated liposomes show the ability of the peptides to form a coiled coil. The helicity measured in a mixture of LPmE3- and LPnK3-decorated liposomes is, for all m/n, higher than the helicity of the separated LPmE3- and LPnK3-functionalized liposomes and demonstrates the coiled-coil formation between peptides tethered to different liposomes. The absence of coiled-coil formation for CP0E3 + CP0K3 as measured by the θ222/θ208 ratio supports the previous observation that this lipopeptide pair does not mediate fusion efficiently, most likely because of the peptides being held too close to the membrane to be able to form a coiled coil efficiently, thus preventing liposome docking. For cholesterol-anchored peptides with elongated spacers, coiled coils are formed, which correlates to the efficient fusion observed from the content-mixing experiments.
Time-Resolved Rate of Fusion
Experiments using model systems incorporating DNA-based fusogens showed that whereas the increasing spacer length decreased the fusion efficiency, the docking of liposomes (i.e., the initial stage of the fusion process) was accelerated with longer spacers.12 To separate these influences of time and LPEG on the fusion efficiency of this model system, it is necessary to identify and isolate distinct time periods in the fusion experiment. During the initial minutes of the content-mixing assays, the rate of fusion is linear and provides a reliable correlation between LPEG and the docking/fusion efficiency. This linearity is largely due to the fact that fusion events occur with unfused liposomes randomly dispersed in a well-mixed experimental solution. After a few minutes, the experimental mixture becomes increasingly more complex as larger fusion products appear and the available number of unfused liposomes decreases.30
Because CPmE3/CPnK3-mediated liposomal fusion is significantly more efficient than LPmE3/LPnK3-mediated liposomal fusion, the fusion efficiency with CPmE3/CPnK3 was continuously measured for all possible combinations of m/n for 30 min. To compare the different stages in the fusion experiment, the fusion rate was determined after 4 and 30 min (Figure 4). Complete data sets, including control experiments that demonstrate a fluorescence increase, are obtained only when both peptide-functionalized liposome populations are present and are available in Supporting Information Figures S1 and S2.
Figure 4.
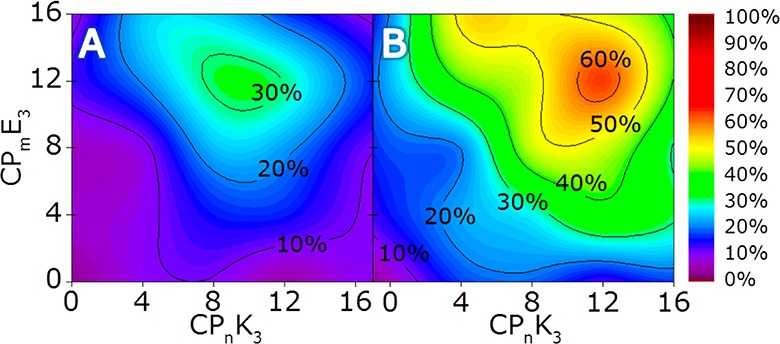
Time-resolved rate of fusion (measured using content-mixing assays) with CPmE3- and CPnK3-decorated liposomes. Data is presented as a function of m/n, indicated by an increase in sulforhodamine B emission after (A) 4 and (B) 30 min. [Total lipid] = 0.1 mM, with 1 mol % lipopeptide, PBS pH 7.4.
After 4 min, all fusion events involving the CP12E3 lipopeptide showed the highest fusion efficiency, regardless of the length of the PEG spacer in the CPnK3 construct, although a CP8K3/CP12E3 combination gives the highest initial fusion efficiency. Liposomal docking is dependent on the length of CPmE3 because peptide K3 is fully incorporated into the membrane prior to fusion, as evidenced by infrared reflection absorption spectroscopy (IRRAS) measurements preformed previously.35,38 This indicates that the LPEG of E3 is critical for liposome docking, with longer linkers being more efficient. This supports the theory that E3 acts as a handle for K3, searching for a K3 peptide to form a coiled coil with, a process resulting in liposome docking.
It has been shown that, during liposomal docking, peptide K3 is in dynamic equilibrium between a heterodimeric coiled-coil structure and a membrane-immersed α-helix; the length of the CPnK3 spacer could strongly influence this equilibrium. Membrane immersion of multiple K3 peptides in both liposomes creates the necessary protrusions to initiate fusion and simultaneously dictates the maximum distance between connected liposomes.35 The optimal initial fusion efficiency, with an equimolar ratio of both peptides, obtained with a CP8K3 a spacer length of 2.9 nm is optimal for bringing both membranes into close proximity and overcoming the energy barriers in the fusion process.
After 30 min, CP12K3 + CP12E3-decorated liposomes show the highest overall fusion efficiency. These results oppose a diffusion-driven collision model with the docking of liposomes as the rate-limiting step37 because that implies a linear correlation between the observed fusion efficiency and the PEG spacer length. Additionally, fusion with CP16E3 is less efficient compared to that with CP12E3, which also demonstrates that the rate-limiting step in this fusion system is not liposomal docking. The shift in fusion efficiency to K3 peptides functionalized with elongated spacers can be caused by different phenomena: liposomal diffusion, better peptide accessibility, and the occurrence of multiple rounds of fusion, all of which can be tested by changing the conditions of the content-mixing experiments (vide infra).
Although the fusion process is not driven by diffusion, liposomal diffusion could still have a minor effect on the fusion efficiency, although only over longer time scales when the number of liposomes decreases as a result of fusion. With decreased liposomal concentration, liposomes functionalized with elongated PEG spacers have a higher probability of finding unfused liposomes with available coiled-coil binding partners. This is illustrated by data showing that initially there is no difference between the fusion of liposomes functionalized with CP4K3 and liposomes bearing CP8E3 or CP16E3, but over time, fusion with CP16E3 -decorated liposomes is more efficient.
Influence of the CPE/CPK Ratio on Fusion
The liposomal fusion efficiency mediated by LP12E3/LP12K3 is highly dependent on peptide concentration;30 at concentrations of ≥0.5 mol %, efficient lipid mixing has been reported. To date, both liposome populations used in the fusion studies were functionalized with LP12E3 or LP12K3 in an equimolar ratio. A ratio ≠1 should support the asymmetric function of the lipopeptides in the fusion process; however, the roles of both peptides will be affected differently by altering their concentrations.
To test this hypothesis, the concentration of CPmE3 was lowered to 0.5 mol % whereas [CPnK3] was maintained at 1 mol %. Here, changes in the fusion efficiency will provide more detailed insights as to the peptide’s roles in the fusion process. Furthermore, lowering the concentration of CPmE3 will affect the handle function of E3, and it is expected that the change in the peptide ratio enhances the impact of LPEG of CPnK3 on the fusion process. Because the most striking differences in efficiency are found with elongated CPmE3 spacers, only liposomal fusion with CP12E3- and CP16E3-decorated liposomes was studied (Figures 5 and S3)
Figure 5.
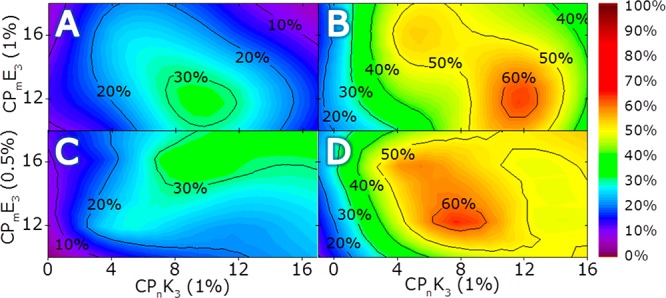
Peptide-ratio-dependent and time-resolved rate of fusion (monitored using a content mixing assay) with CPmE3- and CPnK3-decorated liposomes as a function of m/n and the peptide ratio. The fusion of liposomes decorated with 1 mol % CPmE3 and 1 mol % CPnK3 is measured after (A) 4 and (B) 30 min and is taken as a reference plot from Figure 2. The fusion of liposomes decorated with 0.5 mol % CPmE3 and 1 mol % CPnK3 measured after (C) 4 and (D) 30 min. The change in E3 concentration affects the trends in fusion efficiency as is evident if plots A and C, and B and D are compared. [Total lipid] = 0.1 mM, PBS pH 7.4.
As is evident from Figure 5, the influence of CPnK3 on the fusion process is enhanced by lowering the concentration of CPmE3, and the resulting fusion efficiencies reveal that CP8K3, with a calculated LPEG of 2.9 nm, is optimal for fusion. Remarkably, initial fusion efficiencies were shifted to elongated spacers with decreasing CPmE3 concentration, which supports the handle function of E3 for liposomal docking and demonstrates the effect of the diffusion-driven collision of liposomes. Over time, however, no significant difference between CP12E3 and CP16E3 was observed, indicating a minor role for LPEG of E3 under these conditions. The impact of LPEG of K3 is demonstrated by the shift in fusion efficiency to CP8K3-decorated liposomes.
In the presence of membrane-tethered E3, K3 will either form a coiled-coil with E3 or insert in the lipid membrane. It has been demonstrated previously that the insertion of unbound peptide K3 in a membrane is favored over coiled-coil formation, and the presence of membrane-tethered E3 greatly enhances the speed of this process.35 This means that, when the concentration of CPmE3 is lowered, the initial docking of both liposomes via a transient coiled-coil interaction is still efficient, but the equilibrium between coiled-coil formation and membrane insertion of CPnK3 (in the opposing liposomal membrane; if not, membrane fusion would be less efficient) shifts to membrane insertion, which in turn results in higher fusion efficiencies for shorter spacers. These results demonstrate that peptide equilibria between different conformations after initial docking strongly influences the fusion efficiency and that the interaction between lipopeptide K3 and the phospholipid membrane is crucial for membrane fusion.
Multiple Rounds of Fusion Assay
Finally, a different content mixing experiment was conducted using CPmE3/CPnK3 (m/n = 8, 12, 16) in order to visualize the influence of the PEG spacer length on the efficiency of multiple rounds of fusion. To measure this parameter, we used a population of (self-quenched) fluorescent, CPmE3-decorated liposomes with excess CPnK3-decorated plain liposomes that should result in a higher loss of self-quenching, with respect to the standard content mixing assay using an equimolar mixture of fluorescent and nonfluorescent liposomes, upon every round of fusion. Therefore, CPnK3-decorated plain liposomes were mixed with sulforhodamine B-loaded CPmE3-decorated liposomes in two ratios (1:1 and 10:1), and the fusion efficiencies were determined by measuring the fluorescence increase after 30 min (Table 3).
Table 3. Multiple Rounds of Fusion Assay.
| functionalized liposomes | CP8K3 + |
CP12K3 + |
CP16K3 + |
||||||
|---|---|---|---|---|---|---|---|---|---|
| [CPnK3]/[CPmE3]a | CP8E3 | CP12E3 | CP16E3 | CP8E3 | CP12E3 | CP16E3 | CP8E3 | CP12E3 | CP16E3 |
| 1b | 37 | 38 | 33 | 37 | 47 | 29 | 25 | 47 | 29 |
| 10 | 41 | 41 | 36 | 76 | 111 | 59 | 40 | 72 | 42 |
| Δ%c | 10% | 7% | 8% | 104% | 135% | 104% | 61% | 53% | 46% |
Fusion efficiency is indicated by an increase in sulforhodamine B emission after 30 min, with K3-decorated liposomes in 1- or 10-fold excess with respect to dye-loaded, E3-decorated liposomes.
Fusion efficiency percentages are normalized to efficiency percentages reported for the equimolar mixture at a total lipid concentration of 0.1 mmol, as shown in Figure 4.
Differences in the fluorescence increase between both experiments are calculated as a measure of multiple rounds of fusion, and the value of the normalized equimolar mixing entry used is 100%. [Total lipid E3-decorated liposomes] = 0.05 mM, with 1 mol % lipopeptides, PBS pH 7.4.
Remarkably, the efficiency of multiple rounds of fusion appears to be dictated by the PEG spacer length used for peptide K3 only and is not affected significantly by LPEG of CPmE3, confirming that K3 controls the fusion process while E3 serves only to mediate the coiled-coil formation, which leads to liposome docking. The fluorescence increase measured with the fusion of CPmE3-decorated liposomes and a 10-fold excess of CP8K3-decorated liposomes was as efficient as the fusion experiment with an equimolar mixture of the peptide-functionalized liposomes. This shows that the abundant availability of CP8K3-decorated liposomes does not yield a higher fusion efficiency. For CP12K3-decorated liposomes, fusion experiments with a 10-fold excess of K3-decorated liposomes yielded a fluorescence increase of 115 ± 18%, demonstrating the fusion of one CPmE3-decorated liposome with multiple CP12K3-decorated liposomes. For CP16K3, the multiple-round fusion efficiency was 53 ± 7%, also indicating the presence of multiple rounds of fusion with this lipopeptide.
Although it has been shown that CP8K3 is most efficient in mediating the early rounds of fusion, it is not able to mediate multiple rounds of fusion in the setting described above. To address this, the behavior of the peptides just before the second fusion event, when both E3 and K3 are tethered to the same membrane and are approached by an unfused K3-decorated liposome, has to be considered. E3 tethered to the membrane of the fluorescent liposome should be accessible to unfused K3-bearing liposomes, but efficient fusion occurs only when these liposomes are functionalized with CP12K3 lipopeptides. It is possible that the LPEG distance is again crucial by shifting the peptide equilibria between membrane-incorporated and coiled-coil conformations and/or by bridging the gap between opposite membranes, thus enabling or disabling the next round of liposomal fusion.
Implications for the Initial Stages of Liposomal Fusion
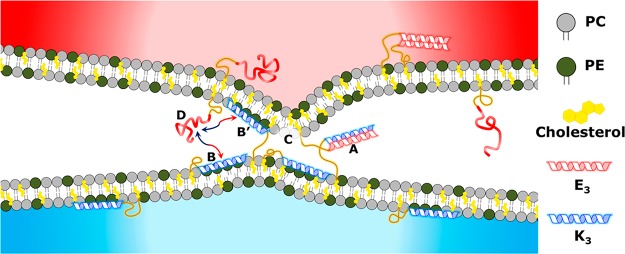
On the basis of these results, the following mechanism for the initial stages of E3-K3-mediated liposomal fusion is proposed: Initially, lipopeptide K3 is fully immersed in the liposomal membrane, and lipopeptide E3 adopts a largely unfolded state. Interliposomal coiled-coil formation occurs when liposomes collide and lipopeptide E3 binds transiently with lipopeptide K3. K3–membrane interactions are favored over coiled-coil formation; therefore, peptides are in an equilibrium, switching between the different states. Once folded as a coiled-coil, K3 is in close proximity to both membranes and hence can interact with either. Immersing itself in the opposite membrane will force both membranes into even closer proximity, destabilizing both membranes by forming protrusions, and will lead to the dehydration of both membrane solvation layers, thus providing the necessary initial conditions for the merging of the membrane bilayers35,58 (Figure 6).
Figure 6.
Peptide equilibria prior to fusion. A transient coiled-coil complex (A) forms upon contact of separate liposome populations. Peptide K3 can then immerse in either membrane (B–B′). Membrane insertion into the opposing membrane will force both membranes into closer proximity, and this interaction subsequently destabilizes the membrane by the formation of protrusions (C), providing the necessary conditions for membrane merging and fusion. Equilibria of peptide interactions among coiled-coil formation (A), unfolded E3 peptides (D), and membrane-inserted K3 peptides (B–B′) are strongly affected by the length of the PEG spacer used for both E3 and K3. As a consequence, the mechanism of liposomal fusion is dictated by peptide equilibria affecting the function of peptide K3, and a difference of 1 or 2 nm in the length of the spacers employed strongly influences the efficiency of the peptides to mediate multiple rounds of fusion.
Conclusions
This study has demonstrated that the membrane interaction propensity of K3, a factor crucial to efficient membrane fusion, is dependent on both the anchor and PEG spacer length employed. When conjugated to a cholesterol anchor, K3 shows increasing helicity with decreasing PEG length, indicating a more profound peptide–membrane interaction for shorter spacers. In contrast, the LPnK3 derivatives do not affect the peptide structure significantly, and the helical secondary structure of all LPnK3 derivatives appears to be less membrane-stabilized when compared to that of CP12K3 lipopeptides. The influence of the anchor on the peptide–membrane interaction is likely caused by altering the local membrane characteristics and is therefore also a factor in the fusion efficiency. Cholesterol-anchored lipopeptides are more efficient in mediating fusion than DOPE-anchored lipopeptides, as evidenced by the content mixing assays.
The intrinsic stability of liposomes prevents coiled-coil formation when peptides are tethered directly to the membrane, (i.e., when a spacer is not used) and lessens the fusion efficiency when very long spacer lengths are applied. Changing the length of the spacer region between E3 and K3 independently demonstrated that the peptides clearly play different roles in the fusion process, as shown with the various content-mixing experiments that reflect the initial stages of fusion. It can be concluded that peptide E3 acts as a handle for peptide K3 to enable initial liposomal docking via coiled-coil formation, and a CP12E3 or CP16E3 construct is most efficient.
After coiled-coil formation, peptide K3 migrates to the lipid membrane,35 and lowering the concentration of CPmE3 at the liposomal surface from 1 to 0.5% revealed the effect of K3 equilibria between coiled-coil formation and K3–membrane interaction on the rate and efficiency of liposomal fusion.35 This change in the CPmE3/CPnK3 ratio increased the probability of peptide K3 immersing itself in the opposing membrane directly after coiled-coil formation, which in turn significantly increased the fusion efficiency for liposomes with shorter spacers, showing the strong influence of LPEG on the rate of membrane fusion.
Multiple rounds of liposomal fusion were observed only when liposomes were functionalized with CP12K3 and CP16K3, whereas no apparent influence of CPmE3 on the fusion efficiency was measured. Here, the fusion efficiency depends only on the spacer length of lipopeptide K3. This pronounced influence of the PEG spacer illustrates the extreme sensitivity of the fusion system to LPEG and to the dynamics of the involved peptides in the initial stages of fusion.
The differences in the (initial) rate of fusion revealed that lipopeptides LP4K3 and CP8K3 are most efficient in mediating fusion compared to other LPnK3/CPnK3 derivatives, which means that a distance of ±2 nm between peptide K3 and the membrane is most favorable, and this lipopeptide is an ideal candidate for further investigations in similar fusion systems. The length of the spacer strongly influences the speed and efficiency of the fusion process and highlights the differing roles of the peptides in the fusion mechanism. These factors should be considered during the development of existing and novel molecular-recognition-based fusion systems.
Acknowledgments
We thank Dr. J. Raap for critical reading.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.langmuir.7b02931.
Analysis of the lipopeptides, complete data sets of content mixing fusion experiments, and CD spectra (PDF)
Author Present Address
† Center for Nanomedicine and Department of Anesthesiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02115, United States.
Author Present Address
‡ Institute of Organic Chemistry, University of Duisburg-Essen, D-45117 Essen, Germany.
G.A.D. was funded by The Netherlands Organization for Scientific Research (NWO) via a ChemThem grant to A.K., H.R.Z. and J.V. were funded by the European Research Council via an ERC starting grant to A.K., A.L.B. is funded by a VENI grant from the NWO. A.K. acknowledges the support of a NWO VICI grant.
The authors declare no competing financial interest.
Supplementary Material
References
- Hu C.; Ahmed M.; Melia T. J.; Sollner T. H.; Mayer T.; Rothman J. E. Fusion of cells by flipped SNAREs. Science 2003, 300 (5626), 1745–1749. 10.1126/science.1084909. [DOI] [PubMed] [Google Scholar]
- Maximov A.; Tang J.; Yang X. F.; Pang Z. P. P.; Sudhof T. C. Complexin Controls the Force Transfer from SNARE Complexes to Membranes in Fusion. Science 2009, 323 (5913), 516–521. 10.1126/science.1166505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof T. C.; Rothman J. E. Membrane Fusion: Grappling with SNARE and SM Proteins. Science 2009, 323 (5913), 474–477. 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R.; Lang T.; Sudhof T. C. Membrane fusion. Cell 2003, 112 (4), 519–533. 10.1016/S0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- Chernomordik L. V.; Kozlov M. M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008, 15 (7), 675–683. 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T.; Zemelman B. V.; McNew J. A.; Westermann B.; Gmachl M.; Parlati F.; Sollner T. H.; Rothman J. E. SNAREpins: Minimal machinery for membrane fusion. Cell 1998, 92 (6), 759–772. 10.1016/S0092-8674(00)81404-X. [DOI] [PubMed] [Google Scholar]
- Chen Y. A.; Scales S. J.; Scheller R. H. Sequential SNARE assembly underlies priming and triggering of exocytosis. Neuron 2001, 30 (1), 161–170. 10.1016/S0896-6273(01)00270-7. [DOI] [PubMed] [Google Scholar]
- Rizo J.; Sudhof T. C. Snares and Munc18 in synaptic vesicle fusion. Nat. Rev. Neurosci. 2002, 3 (8), 641–653. 10.1038/nrn898. [DOI] [PubMed] [Google Scholar]
- Jena B. P. Role of SNAREs in membrane fusion. Adv. Exp. Med. Biol. 2011, 713, 13–32. 10.1007/978-94-007-0763-4_3. [DOI] [PubMed] [Google Scholar]
- Han J.; Pluhackova K.; Bockmann R. A. The Multifaceted Role of SNARE Proteins in Membrane Fusion. Front. Membr. Physiol. Membr. Biophys. 2017, 8, 5. 10.3389/fphys.2017.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel G.; Simonsson L.; Campbell R. A.; Hook F. Determinants for membrane fusion induced by cholesterol-modified DNA zippers. J. Phys. Chem. B 2008, 112 (28), 8264–8274. 10.1021/jp802005b. [DOI] [PubMed] [Google Scholar]
- Chan Y. H.; van Lengerich B.; Boxer S. G. Effects of linker sequences on vesicle fusion mediated by lipid-anchored DNA oligonucleotides. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (4), 979–984. 10.1073/pnas.0812356106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen U.; Vogel S. DNA-Controlled Assembly of Liposomes in Diagnostics. Methods Enzymol. 2009, 464, 233–248. 10.1016/S0076-6879(09)64012-X. [DOI] [PubMed] [Google Scholar]
- Ries O.; Loffler P. M. G.; Vogel S. Convenient synthesis and application of versatile nucleic acid lipid membrane anchors in the assembly and fusion of liposomes. Org. Biomol. Chem. 2015, 13 (37), 9673–9680. 10.1039/C5OB01207D. [DOI] [PubMed] [Google Scholar]
- Meng Z.; Yang J.; Liu Q.; de Vries J. W.; Gruszka A.; Rodríguez-Pulido A.; Crielaard B.; Kros A.; Hermann A. Efficient Fusion of Liposomes by Nucleobase Quadruple-Anchored DNA. Chem. - Eur. J. 2017, 23, 9391. 10.1002/chem.201701379. [DOI] [PubMed] [Google Scholar]
- Lygina A. S.; Meyenberg K.; Jahn R.; Diederichsen U. Transmembrane Domain Peptide/Peptide Nucleic Acid Hybrid as a Model of a SNARE Protein in Vesicle Fusion. Angew. Chem., Int. Ed. 2011, 50 (37), 8597–8601. 10.1002/anie.201101951. [DOI] [PubMed] [Google Scholar]
- Kashiwada A.; Matsuda K.; Mizuno T.; Tanaka T. Construction of a pH-responsive artificial membrane fusion system by using designed coiled-coil polypeptides. Chem. - Eur. J. 2008, 14 (24), 7343–7350. 10.1002/chem.200701726. [DOI] [PubMed] [Google Scholar]
- Meyenberg K.; Lygina A. S.; van den Bogaart G.; Jahn R.; Diederichsen U. SNARE derived peptide mimic inducing membrane fusion. Chem. Commun. (Cambridge, U. K.) 2011, 47 (33), 9405–9407. 10.1039/c1cc12879e. [DOI] [PubMed] [Google Scholar]
- Rauschenberg M.; Bomke S.; Karst U.; Ravoo B. J. Dynamic Peptides as Biomimetic Carbohydrate Receptors. Angew. Chem., Int. Ed. 2010, 49 (40), 7340–7345. 10.1002/anie.201002847. [DOI] [PubMed] [Google Scholar]
- Richard A.; Marchi-Artzner V.; Lalloz M. N.; Brienne M. J.; Artzner F.; Gulik-Krzywicki T.; Guedeau-Boudeville M. A.; Lehn J. M. Fusogenic supramolecular vesicle systems induced by metal ion binding to amphiphilic ligands. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (43), 15279–15284. 10.1073/pnas.0406625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y.; Luo Y.; Bong D. Membrane activation: selective vesicle fusion via small molecule recognition. J. Am. Chem. Soc. 2006, 128 (45), 14430–14431. 10.1021/ja0644576. [DOI] [PubMed] [Google Scholar]
- Kashiwada A.; Tsuboi M.; Takamura N.; Brandenburg E.; Matsuda K.; Koksch B. Design and Characterization of Endosomal-pH-Responsive Coiled Coils for Constructing an Artificial Membrane Fusion System. Chem. - Eur. J. 2011, 17 (22), 6179–6186. 10.1002/chem.201003392. [DOI] [PubMed] [Google Scholar]
- Marchi-Artzner V.; Gulik-Krzywicki T.; Guedeau-Boudeville M. A.; Gosse C.; Sanderson J. M.; Dedieu J. C.; Lehn J. M. Selective adhesion, lipid exchange and membrane-fusion processes between vesicles of various sizes bearing complementary molecular recognition groups. ChemPhysChem 2001, 2 (6), 367–376. . [DOI] [PubMed] [Google Scholar]
- Ma M. M.; Bong D. Controlled Fusion of Synthetic Lipid Membrane Vesicles. Acc. Chem. Res. 2013, 46 (12), 2988–2997. 10.1021/ar400065m. [DOI] [PubMed] [Google Scholar]
- Voskuhl J.; Ravoo B. J. Molecular recognition of bilayer vesicles. Chem. Soc. Rev. 2009, 38 (2), 495–505. 10.1039/B803782P. [DOI] [PubMed] [Google Scholar]
- Voskuhl J.; Fenske T.; Stuart M. C.; Wibbeling B.; Schmuck C.; Ravoo B. J. Molecular recognition of vesicles: host-guest interactions combined with specific dimerization of zwitterions. Chem. - Eur. J. 2010, 16 (28), 8300–8306. 10.1002/chem.201000623. [DOI] [PubMed] [Google Scholar]
- Nalluri S. K. M.; Ravoo B. J. Light-Responsive Molecular Recognition and Adhesion of Vesicles. Angew. Chem., Int. Ed. 2010, 49 (31), 5371–5374. 10.1002/anie.201001442. [DOI] [PubMed] [Google Scholar]
- Nalluri S. K. M.; Bultema J. B.; Boekema E. J.; Ravoo B. J. Photoresponsive Molecular Recognition and Adhesion of Vesicles in a Competitive Ternary Supramolecular System. Chem. - Eur. J. 2011, 17 (37), 10297–10303. 10.1002/chem.201100789. [DOI] [PubMed] [Google Scholar]
- Marsden H. R.; Elbers N. A.; Bomans P. H. H.; Sommerdijk N.; Kros A. A Reduced SNARE Model for Membrane Fusion. Angew. Chem., Int. Ed. 2009, 48 (13), 2330–2333. 10.1002/anie.200804493. [DOI] [PubMed] [Google Scholar]
- Marsden H. R.; Korobko A. V.; Zheng T. T.; Voskuhl J.; Kros A. Controlled liposome fusion mediated by SNARE protein mimics. Biomater. Sci. 2013, 1 (10), 1046–1054. 10.1039/c3bm60040h. [DOI] [PubMed] [Google Scholar]
- Versluis F.; Voskuhl J.; van Kolck B.; Zope H.; Bremmer M.; Albregtse T.; Kros A. In situ modification of plain liposomes with lipidated coiled coil forming peptides induces membrane fusion. J. Am. Chem. Soc. 2013, 135 (21), 8057–8062. 10.1021/ja4031227. [DOI] [PubMed] [Google Scholar]
- Zheng T.; Voskuhl J.; Versluis F.; Zope H. R.; Tomatsu I.; Marsden H. R.; Kros A. Controlling the rate of coiled coil driven membrane fusion. Chem. Commun. (Cambridge, U. K.) 2013, 49 (35), 3649–3651. 10.1039/c3cc38926j. [DOI] [PubMed] [Google Scholar]
- Versluis F.; Dominguez J.; Voskuhl J.; Kros A. Coiled-coil driven membrane fusion: zipper-like vs. non-zipper-like peptide orientation. Faraday Discuss. 2013, 166, 349–359. 10.1039/c3fd00061c. [DOI] [PubMed] [Google Scholar]
- Zheng T. T.; Bulacu M.; Daudey G.; Versluis F.; Voskuhl J.; Martelli G.; Raap J.; Sevink G. J. A.; Kros A.; Boyle A. L. A non-zipper-like tetrameric coiled coil promotes membrane fusion. RSC Adv. 2016, 6 (10), 7990–7998. 10.1039/C5RA26175A. [DOI] [Google Scholar]
- Rabe M.; Aisenbrey C.; Pluhackova K.; de Wert V.; Boyle A. L.; Bruggeman D. F.; Kirsch S. A.; Bockmann R. A.; Kros A.; Raap J.; Bechinger B. A Coiled-Coil Peptide Shaping Lipid Bilayers upon Fusion. Biophys. J. 2016, 111 (10), 2162–2175. 10.1016/j.bpj.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandberg E.; Killian J. A. Snorkeling of lysine side chains in transmembrane helices: how easy can it get?. FEBS Lett. 2003, 544 (1–3), 69–73. 10.1016/S0014-5793(03)00475-7. [DOI] [PubMed] [Google Scholar]
- McNew J. A.; Weber T.; Engelman D. M.; Sollner T. H.; Rothman J. E. The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion. Mol. Cell 1999, 4 (3), 415–421. 10.1016/S1097-2765(00)80343-3. [DOI] [PubMed] [Google Scholar]
- Rabe M.; Schwieger C.; Zope H. R.; Versluis F.; Kros A. Membrane Interactions of Fusogenic Coiled-Coil Peptides: Implications for Lipopeptide Mediated Vesicle Fusion. Langmuir 2014, 30 (26), 7724–7735. 10.1021/la500987c. [DOI] [PubMed] [Google Scholar]
- Cubberley M. S.; Iverson B. L. (1)H NMR investigation of solvent effects in aromatic stacking interactions. J. Am. Chem. Soc. 2001, 123 (31), 7560–7563. 10.1021/ja015817m. [DOI] [PubMed] [Google Scholar]
- Shirude P. S.; Kumar V. A.; Ganesh K. N. BisPNA targeting to DNA: Effect of neutral loop on DNA duplex strand invasion by aepPNA-N7G/aepPNA-C substituted peptide nucleic acids. Eur. J. Org. Chem. 2005, 24, 5207–5215. 10.1002/ejoc.200500544. [DOI] [Google Scholar]
- Sun Q.; Cai S.; Peterson B. R. Practical Synthesis of 3 beta-Amino-5-cholestene and Related 3 beta-Halides Involving i-Steroid and Retro-i-Steroid Rearrangements. Org. Lett. 2009, 11 (3), 567–570. 10.1021/ol802343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. H.; Yang J. T.; Chau K. H. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry 1974, 13 (16), 3350–3359. 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- Hanwell M. D.; Curtis D. E.; Lonie D. C.; Vandermeersch T.; Zurek E.; Hutchison G. R.. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminf. 2012, 4, 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel A.; Ben-Tal N.; May S. Interactions of cholesterol with lipid bilayers: the preferred configuration and fluctuations. Biophys. J. 2001, 81 (2), 643–658. 10.1016/S0006-3495(01)75729-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwang T. J.; Fletcher W. R.; Lane T. J.; Johal M. S. Quantification of the layer of hydration of a supported lipid bilayer. Langmuir 2010, 26 (7), 4598–4601. 10.1021/la100275v. [DOI] [PubMed] [Google Scholar]
- Tu K.; Klein M. L.; Tobias D. J. Constant-pressure molecular dynamics investigation of cholesterol effects in a dipalmitoylphosphatidylcholine bilayer. Biophys. J. 1998, 75 (5), 2147–2156. 10.1016/S0006-3495(98)77657-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabe M.; Zope H. R.; Kros A. Interplay between Lipid Interaction and Homo-coiling of Membrane-Tethered Coiled-Coil Peptides. Langmuir 2015, 31 (36), 9953–9964. 10.1021/acs.langmuir.5b02094. [DOI] [PubMed] [Google Scholar]
- Kyoung M.; Srivastava A.; Zhang Y. X.; Diao J. J.; Vrljic M.; Grob P.; Nogales E.; Chu S.; Brunger A. T. In vitro system capable of differentiating fast Ca2+-triggered content mixing from lipid exchange for mechanistic studies of neurotransmitter release. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (29), E304–E313. 10.1073/pnas.1107900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J.; Grob P.; Cipriano D. J.; Kyoung M.; Zhang Y.; Shah S.; Nguyen A.; Padolina M.; Srivastava A.; Vrljic M.; Shah A.; Nogales E.; Chu S.; Brunger A. T. Synaptic proteins promote calcium-triggered fast transition from point contact to full fusion. eLife 2012, 1, e00109. 10.7554/eLife.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyoung M.; Zhang Y.; Diao J.; Chu S.; Brunger A. T. Studying calcium-triggered vesicle fusion in a single vesicle-vesicle content and lipid-mixing system. Nat. Protoc. 2013, 8 (1), 1–16. 10.1038/nprot.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y.; Zhao L.; Bu B.; Lou X.; Li D.; Ji B.; Liu J.; Diao J.; Shin Y. K. Lipid molecules influence early stages of yeast SNARE-mediated membrane fusion. Phys. Biol. 2015, 12 (2), 025003. 10.1088/1478-3975/12/2/025003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.; Bahreman A.; Daudey G.; Bussmann J.; Olsthoorn R. C.; Kros A. Drug Delivery via Cell Membrane Fusion Using Lipopeptide Modified Liposomes. ACS Cent. Sci. 2016, 2 (9), 621–630. 10.1021/acscentsci.6b00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton B. S.; Warner J. M.; Wu Z. Y.; Nikolaus J.; Wei G.; Wagnon E.; Baddeley D.; Karatekin E.; O’Shaughnessy B. Cholesterol Increases the Openness of SNARE-Mediated Flickering Fusion Pores. Biophys. J. 2016, 110 (7), 1538–1550. 10.1016/j.bpj.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. T.; Kreutzberger A. J. B.; Lee J.; Kiessling V.; Tamm L. K. The role of cholesterol in membrane fusion. Chem. Phys. Lipids 2016, 199, 136–143. 10.1016/j.chemphyslip.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe K.; Higashi K.; Watabe K.; Kobayashi A.; Limwikrant W.; Yamamoto K.; Moribe K. Effects of the PEG molecular weight of a PEG-lipid and cholesterol on PEG chain flexibility on liposome surfaces. Colloids Surf., A 2015, 474, 63–70. 10.1016/j.colsurfa.2015.03.006. [DOI] [Google Scholar]
- Kusumoto K.; Akita H.; El-Sayed A.; Harashima H. Effect of the Anchor in Polyethylene Glycol-Lipids on the Transfection Activity of PEGylated Cationic Liposomes Encapsulating DNA. Biol. Pharm. Bull. 2012, 35 (4), 445–448. 10.1248/bpb.35.445. [DOI] [PubMed] [Google Scholar]
- Pluhackova K.; Wassenaar T. A.; Kirsch S.; Bockmann R. A. Spontaneous Adsorption of Coiled-Coil Model Peptides K and E to a Mixed Lipid Bilayer. J. Phys. Chem. B 2015, 119 (12), 4396–4408. 10.1021/acs.jpcb.5b00434. [DOI] [PubMed] [Google Scholar]
- Woo S. Y.; Lee H. All-atom simulations and free-energy calculations of coiled-coil peptides with lipid bilayers: binding strength, structural transition, and effect on lipid dynamics. Sci. Rep. 2016, 6, 22299. 10.1038/srep22299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia R. A.; Pantazatos S. P.; Pantazatos D. P.; MacDonald R. C. Cholesterol stabilizes hemifused phospholipid bilayer vesicles. Biochim. Biophys. Acta, Biomembr. 2001, 1511 (2), 264–270. 10.1016/S0005-2736(01)00283-8. [DOI] [PubMed] [Google Scholar]
- Klok H. A.; Vandermeulen G. W.; Nuhn H.; Rosler A.; Hamley I. W.; Castelletto V.; Xu H.; Sheiko S. S. Peptide mediated formation of hierarchically organized solution and solid state polymer nanostructures. Faraday Discuss. 2005, 128, 29–41. 10.1039/B402273D. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.