Abstract
Deep mutational scanning is a foundational tool for addressing the functional consequences of large numbers of mutants, but a more efficient and accessible method for construction of user-defined mutagenesis libraries is needed. Here we present nicking mutagenesis, a robust, single-day, one-pot saturation mutagenesis method performed on routinely prepped plasmid dsDNA. The method can be used to produce comprehensive or single- or multi-site saturation mutagenesis libraries.
Mutational studies have been used for more than 6 decades to probe protein sequence–function relationships. Deep mutational scanning has emerged as a method to assess the effect of thousands of mutations on function through massively parallel functional screens and DNA counting via deep sequencing1. Information-rich sequence–function maps obtained from such methods allow researchers to address various aims, including the generation of biomolecular fitness landscapes2–6, therapeutic protein optimization7, and high-resolution conformational epitope mapping8. Although other technical challenges have been resolved9,10, a robust and accessible method for the construction of high-quality, user-defined mutational libraries is lacking.
Random mutagenesis methods such as error-prone PCR are hindered by limited codon sampling and imprecise control over the number of mutations introduced11. Of the comprehensive saturation mutagenesis methods published2,4,11–14, PFunkel12 offers the best combination of library coverage, mutational efficiency, control over number of mutations introduced, and scalability (Supplementary Table 1). In particular, PFunkel can be used to prepare libraries covering all possible point mutations, with most members of the library having exactly one mutation. However, PFunkel is limited by the requirement for a preparation of a uracil-containing ssDNA (dU-ssDNA) template by phage infection. dU-ssDNA yields are highly variable15, and the preparation adds at least 2 d to the mutagenesis procedure. By analogy to site-directed mutagenesis, PCR-based methods such as QuikChange have mostly supplanted Kunkel mutagenesis, which is highly efficient but also requires dU-ssDNA16.
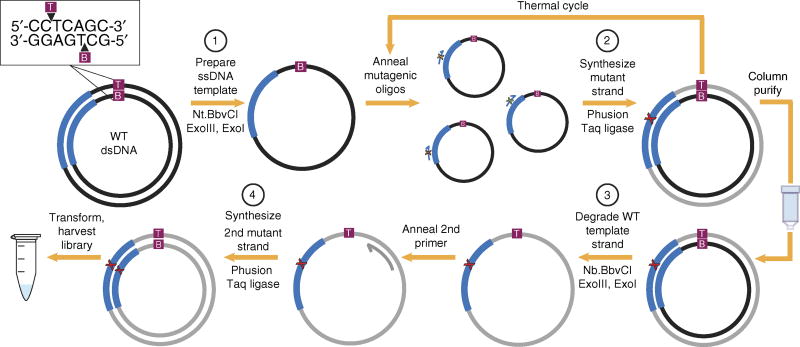
Here we present nicking mutagenesis, a method that does not rely on dU-ssDNA (Fig. 1 and Supplementary Protocols 1 and 2). Nicking mutagenesis is flexible, as any plasmid dsDNA can be used, provided it contains a 7-bp BbvCI restriction site. The key mechanism in nicking mutagenesis is the successive creation and degradation of a wild-type ssDNA template. This is accomplished through nicking, with a pair of endonucleases (Nt.BbvCI and Nb.BbvCI)17,18 that each recognize the same site but nick only one strand, followed by exonuclease digestion. First, the ssDNA template is created from a dsDNA plasmid via strand-specific nick introduced by Nt.BbvCI and selective digestion of the nicked strand with exonuclease III (step (1); Fig. 1). Mutant strands are then synthesized by thermal cycling template DNA with mutagenic oligos at a low primer-to-template ratio to promote annealing of, effectively, one primer to each template12 (step (2)). The highly processive and high-fidelity Phusion DNA polymerase extends the primer around the circular template. Taq DNA ligase closes the new strand to form a dsDNA plasmid with a mismatch at the mutational site. The heteroduplex DNA is then column purified to avoid buffer incompatibility issues and prevent potential competition between Phusion and exonuclease III.
Figure 1.
Comprehensive single-site nicking mutagenesis. Wild-type (WT) plasmid dsDNA containing a 7-bp BbvCI recognition site is nicked by Nt.BbvCI. Exonuclease III (ExoIII) degrades the nicked strand to generate ssDNA template, exonuclease I (ExoI) degrades insufficiently digested DNA (step (1)). Mutagenic oligos are then added at a 1:20 ratio with template, Phusion polymerase synthesizes mutant strands, and Taq DNA ligase seals nicks (step (2)). The reaction is column purified, and then WT template strand is nicked by Nb.BbvCI and digested by ExoIII (step (3)). A second primer is added, and the complementary mutant strand is synthesized to yield mutagenized dsDNA (step (4)).
To resolve the heteroduplex, the opposite-strand nicking endonuclease, Nb.BbvCI, creates a nick in the template strand, which is subsequently degraded by exonuclease III (step (3)). A secondary primer is then added, and synthesis of the complementary mutant strand follows as above (step (4)). To reduce wild-type background, the final reaction is treated with DpnI to digest methylated and hemimethylated parental DNA. The protocol can be completed in a single day with minimal hands-on time (Supplementary Table 2).
We first optimized nicking mutagenesis using a green–white fluorescence screen based on reversion of a nonfluorescent GFP mutant (Supplementary Note 1, Supplementary Fig. 1 and Supplementary Table 3). Next, we used nicking mutagenesis to prepare comprehensive single-site saturation mutagenesis libraries for two 71-codon stretches of an aliphatic amidase encoded by the Pseudomonas aeruginosa gene amiE (residues 100–170 and 171–241 are targeted in reactions 1 and 2, respectively)19. A mixture of 71 degenerate oligo sets, each with three consecutive randomized bases (NNN) corresponding to one of the 71 codons, was used at a 1:20 (primer/template) ratio. We deep sequenced the resulting libraries to an average depth of coverage of 2,200 reads per variant and processed the data with Enrich20. We observed 100% of possible single nonsynonymous (NS) mutants (2,840 total) and 100% of possible programmed codon mutations (8,946 total) with at least 10 reads (library coverage statistics are shown in Table 1). For amiE reactions 1 and 2, respectively, 64.4% and 63.5% of library members had exactly one NS mutation. The incidence of nonprogrammed insertion or deletion (indel) mutations was 0.05% for both reactions. The frequency of individual mutations in each library followed a log-normal distribution, which is consistent with libraries prepared by PFunkel mutagenesis6,9 (Supplementary Fig. 2). In deep mutational scanning experiments, the initial library is typically sequenced at ~200-fold depth of coverage of the expected diversity. Normalization of the above sequencing results to a 200-fold depth of coverage revealed that 93.2% and 97.8% of possible NS mutations would be represented above the typical threshold of 10 sequencing reads for amiE reactions 1 and 2, respectively (Supplementary Fig. 3a). This compares favorably with PFunkel mutagenesis (91.7% using the same threshold), although the library distributions between the two methods are essentially identical (Supplementary Fig. 3b). We next assessed the libraries for off-target mutations by shotgun sequencing the input plasmid pEDA3_amiE (no intended mutations) and library dsDNA from amiE reactions 1 and 2 and found that the corresponding regions of the gene targeted by mutagenesis had significantly higher percentages of mutant alleles (P < 2.2 × 10−16) (Supplementary Fig. 4).
Table 1.
Nicking mutagenesis library coverage statistics
| Theoretical | amiE reaction 1 | amiE reaction 2 | bla | |
|---|---|---|---|---|
| Sequencing reads after quality filtering20 (fold coverage) | 4,273,346 (941) | 5,378,051 (1,184) | 414,417 (74) | |
| Number of transformants | 1.3 × 107 | 1.4 × 107 | 1.5 × 105 | |
| Number of mutated codons | 71 | 71 | 88 | |
| Total plasmid length (nt) | 4,612 | 4,612 | 6,907 | |
| Percentage of reads with: | ||||
|
| ||||
| 0 nonsynonymous mutations | 1.6 | 27.2 | 26.3 | 30.1 |
| 1 nonsynonymous mutation | 98.4 | 64.4 | 63.5 | 59.9 |
| Multiple nonsynonymous mutations | 0 | 8.4 | 10.2 | 9.7 |
| Frameshift mutation | 0 | 0.05 | 0.05 | 0.34 |
| Percentage of mutant codons with: | ||||
|
| ||||
| 1-bp substitution | 14.3 | 32.2 | 31.4 | 25.4 |
| 2-bp substitution | 42.9 | 32.8 | 31.5 | 41.7 |
| 3-bp substitution | 42.9 | 35.0 | 37.1 | 33.0 |
| Percentage of possible codon substitutions observed: | ||||
|
| ||||
| 1-bp substitution | 100.0 | 100.0 | 99.7 | |
| 2-bp substitution | 100.0 | 100.0 | 83.5 | |
| 3-bp substitution | 100.0 | 100.0 | 77.8 | |
| All substitutions | 100.0 | 100.0 | 83.4 | |
| Coverage of possible single amino acid substitutions with ≥5 reads | 100.0 | 100.0 | 91.5 | |
| Coverage of possible programmed mutant codons with ≥5 reads | 100.0 | 100.0 | 75.4 | |
To demonstrate performance on larger plasmids, we used nicking mutagenesis to prepare a comprehensive single-site saturation mutagenesis library for an 88-codon stretch of the Escherichia coli gene bla, encoding TEM-1 β-lactamase, from a 6.9-kb plasmid and sequenced to 74-fold coverage of codon space. We observed nearly identical library composition with 91.5% coverage of possible amino acid substitutions (Table 1), which is consistent with expected coverage at this depth of sequencing (Supplementary Fig. 5). Of note, we observed an order of magnitude fewer transformants when preparing this library compared to the amiE library, consistent with larger plasmids having lower transformation efficiency. One potential strategy to improve transformation efficiency is to use ultracompetent cells. Alternatively, the library can be constructed on a smaller plasmid and then transferred to a desired plasmid via subcloning.
To further expand the utility of nicking mutagenesis, we developed a single- and multisite protocol (Supplementary Fig. 6 and Supplementary Protocol 2). For this protocol, we added primer at a 5:1 (primer/template) molar ratio and altered the thermal cycling steps for mutant-strand synthesis (Online Methods). We tested the protocol by performing three single- and one triple-mutation nicking mutagenesis reactions on bla from a 4.8-kb plasmid. Sanger sequencing of two clones from each of the three single-site reactions revealed that five of the six clones contained a single mutation. For the multi-site reaction, five out of ten sequenced clones contained the desired three programmed mutations.
Robust and effective molecular biology methods are characterized by their ease of adoption in laboratories outside of where they were developed. To evaluate the accessibility of nicking mutagenesis, an external lab (A.A. and K.E.J.T.) tested the method by performing single-site nicking mutagenesis on the positive-control plasmid pEDA5_GFPmut3_Y66H with the restore-to-function oligo GFP_H66Y. The resulting mutational efficiency, calculated by counting fluorescent (mutant) and nonfluorescent (wild-type) colonies, was 86.8% ± 6.1 s.d. (n = 3 independent experiments).
We have demonstrated a one-pot, single-day method for the preparation of comprehensive single- and multisite saturation mutagenesis libraries from plasmid dsDNA (see Supplementary Table 4 for details about cost). The utility of nicking mutagenesis is not limited to saturation mutagenesis. Codon substitutions are user defined, making it possible to restrict diversity to specific residues such as hydrophobic or charged substitutions. An inherent limitation is that if a plasmid contains multiple BbvCI nicking sites, all of them must be in the same orientation. In the human genome, BbvCI has a mean distance between sites of 2,058 bp; thus, a considerable fraction of human genes will have nicking sites. Solutions include cloning the gene of interest into a plasmid with a compatible nicking-site orientation or using custom gene synthesis to remove extra BbvCI sites.
To validate the performance of nicking mutagenesis we used ‘testers’ from an external lab; we propose this practice to enhance reproducibility and accessibility of new molecular biology methods. To aid in method adoption, practice, and troubleshooting, plasmid pEDA5_GFPmut3_Y66H has been deposited to the Addgene repository (http://www.addgene.org; catalog number 80085).
ONLINE METHODS
Reagents
All chemicals were purchased from Sigma-Aldrich unless otherwise noted. All enzymes were purchased from New England BioLabs. All mutagenic oligos were designed using the QuikChange Primer Design Program (Agilent). Mutagenic oligos and sequencing primers were ordered from Integrated DNA Technologies.
Plasmid construction
All primer sequences used in this work are listed in Supplementary Table 3. Plasmid pEDA5_GFPmut3_Y66H was prepared by modification of pJK_proB_GFPmut3 as described in Bienick et al.19 by a single Kunkel16 reaction with two mutagenic primers: one encoding a BbvCI site (primer pED_BbvCI) and the second to introduce a Tyr66His point mutation (primer GFP_Y66H). pEDA3_amiE was constructed by altering pJK_proK17_amiE as described in Bienick et al.19 with a single Kunkel16 reaction with two primers: one encoding a BbvCI site (pED_BbvCI) and the second encoding a mutated ribosome binding sequence (pED_kRBS3). pEDA5_GFPmut3_Y66H has been deposited in the Addgene repository (http://www.addgene.org; catalog number 80085).
Plasmid pSALECT-wtTEM1/csTEM1 was created as follows. Overhang PCR was used to add in XhoI and BbvCI sites after the existing NdeI site and before the original stop codon of plasmid pSALECT-EcoBam (Addgene, 59705). A truncation mutant of TEM-1 lacking residues 2–23 (Δ2–23) was cloned in frame between the NdeI and XhoI sites. A codon-swapped Δ2–23 truncation of wild-type TEM-1 with a C-terminal His6 tag and double stop codon was ordered as a gBlock (IDT) and was cloned in frame between the XhoI and BbvCI sites. This second TEM-1 is a C-terminal fusion to the wild-type TEM-1.
Plasmid pETconNK-TEM1(S70A, D179G) was created as follows. Gibson assembly was used to remove the ampicillin-resistance gene from pETcon(−) (Addgene 41522) and insert a kanamycin-resistance gene with a 3′ BbvCI site on the coding strand. A Δ2–23 truncation of TEM-1 with point mutations encoding S70A and D179G was cloned in frame between the NdeI and XhoI sites.
Comprehensive nicking mutagenesis optimization
The final optimized comprehensive nicking mutagenesis protocol is supplied in Supplementary Protocol 1 and at Protocol Exchange21. 1× CutSmart buffer (NEB) was used as an enzyme diluent when necessary. Two reactions were set up as follows: 0.76 pmol pEDA5_GFPmut3_Y66H was incubated with 10 U each Nt.BbvCI and exonuclease III in 1× CutSmart buffer (20 µL final volume) for 60 min at 37 °C then at 80 °C for 20 min (heat kill). 40 U DpnI was added, and the reaction was incubated at 37 °C for 60 min then at 80 °C for 20 min (heat kill). One reaction was then column purified by Zymo Clean and Concentrator (5:1 v/v ratio of binding buffer to sample), eluted in 6 µL nuclease-free H2O (NFH2O, Integrated DNA Technologies), transformed into XL1-Blue electrocompetent cells, and dilution plated. The following was added to the second reaction: 200 U Taq DNA ligase, 2 U Phusion high-fidelity DNA polymerase, 20 µL 5× Phusion HF buffer, 20 µL 50 mM DTT, 1 µL 50 mM NAD+, 2 µL 10 mM dNTPs, 29 µL NFH2O (final reaction volume, 100 µL). The tube was placed into a preheated (98 °C) thermal cycler set with the following program: 98 °C for 2 min, 15 cycles of 98 °C for 30 s (denature), 55 °C for 45 s (anneal oligos), 72 °C for 7 min (extension), and final incubation at 45 °C for 20 min to complete ligation. The reaction was column purified, transformed, and dilution plated as described above.
The optimization experiment including addition of exonuclease I was performed as described below with the following modifications. Single mutagenic primer GFP_H66Y, which restores the wild-type chromophore sequence, was used at a 1:20 primer/template ratio. The reaction was column purified and transformed into XL1 Blue electrocompetent cells as above. Green fluorescent (mutant) and white (parental) colonies were counted to calculate transformational and mutational efficiencies.
Comprehensive nicking mutagenesis of amiE and bla
Three separate reactions, targeting residues 100–170 and 171–241 of amiE and 201–289 of TEM-1, were performed. Mutagenic oligos programming degenerate codons (NNN) for each reaction were mixed in equimolar amounts to a final concentration of 10 µM. 20 µL each primer mix was added to a phosphorylation reaction containing 2.4 µL T4 polynucleotide kinase buffer, 1 µL 10 mM ATP, 10 U T4 polynucleotide kinase and incubated for 1 h at 37 °C. Secondary primer pED_2ND was phosphorylated in a reaction containing 18 µL NFH2O, 2 µL T4 polynucleotide kinase buffer, 7 µL 100 µM secondary primer, 1 µL 10 mM ATP, and 10 U T4 polynucleotide kinase. The reaction was incubated for 1 h at 37 °C. Phosphorylated NNN and secondary primers were diluted 1:1,000 and 1:20, respectively, in NFH2O.
ssDNA template was prepared in a reaction containing 0.76 pmol plasmid dsDNA, 2 µL NEB CutSmart buffer, 10 U Nt.BbvCI, 10 U exonuclease III, 20 U exonuclease I, and NFH2O to a final reaction volume of 20 µL in a PCR tube. The following thermal cycle program was used: 37 °C for 60 min, 80 °C for 20 min (heat kill), hold at 4–10 °C. Next, for mutant-strand synthesis, the following was added to each PCR tube on ice: 20 µL 5× Phusion HF buffer, 20 µL 50 mM DTT, 1 µL 50 mM NAD+, 2 µL 10 mM dNTPs, 4.3 µL 1:1,000 diluted phosphorylated NNN mutagenic oligos, and 26.7 µL NFH2O (final reaction volume, 100 µL). The tube contents were mixed, spun down for 10 s at 1,000 × g, and placed on ice. 200 U Taq DNA ligase and 2 U Phusion high-fidelity DNA polymerase were added to each reaction, mixed, spun down for 10 s at 1,000 × g, and placed into a preheated (98 °C) thermal cycler set with the following program: 98 °C for 2 min, 15 cycles of 98 °C for 30 s (denature), 55 °C for 45 s (anneal oligos), 72 °C for 7 min (extension). An additional 4.3 µL oligos was added at the beginnings of the sixth and eleventh cycles. A final incubation at 45 °C for 20 min was then done to complete ligation. Each reaction was then column purified using a Zymo Clean and Concentrator kit (5:1 ratio of DNA binding buffer to sample). Each reaction was eluted in 15 µL NFH2O, and 14 µL was transferred to a fresh PCR tube.
Next, for the template degradation reaction, the following was added to each tube: 2 µL 10× NEB CutSmart buffer, 1 U Nb.BbvCI, 2 U exonuclease III, and 20 U exonuclease I (20 µL final volume). The following thermocycler program was used: 37 °C for 60 min, 80 °C for 20 min (heat kill), hold at 4–10 °C. To synthesize the second (complementary) mutant strand, the following was added to each reaction: 20 µL 5× Phusion HF buffer, 20 µL 50 mM DTT, 1 µL 50 mM NAD+, 2 µL 10 mM dNTPs, 3.3 µL 1:20 diluted phosphorylated secondary primer (0.38 pmol), and 27.7 µL NFH2O (final reaction volume of 100 µL). The tube contents were mixed, spun down for 10 s at 1,000 × g, and placed on ice. 200 U Taq DNA ligase and 2 U Phusion high-fidelity DNA polymerase were added to each reaction, mixed, spun down for 10 s at 1,000 × g, and placed into a preheated (98 °C) thermal cycler set with the following program: 98 °C for 30 s, 55 °C for 45 s, 72 °C for 10 min (can be extended for longer constructs), and 45 °C for 20 min.
To degrade methylated and hemimethylated wild-type DNA, 40 U DpnI was added to each reaction and incubated at 37 °C for 1 h. The final reaction was column purified using the Zymo Clean and Concentrator-5 kit as described above and eluted in 6 µL NFH2O. The entire 6 µL was transformed into 40 µL of XL1-Blue electroporation-competent cells (Agilent) and plated on Corning square bioassay dishes (Sigma-Aldrich, 245 mm × 245 mm × 25 mm). The following day, colonies were scraped with 15 mL of TB and vortexed, and 1 mL was removed and mini-prepped using a Qiagen Mini-Prep Kit.
Single- and multi-site nicking mutagenesis
The final optimized single- and multi-site nicking mutagenesis protocol is supplied in Supplementary Protocol 2. Mutagenic primers were phosphorylated separately following the protocol described above for the secondary primer, then diluted 1:20 with NFH2O. For multi-site nicking mutagenesis, 2 µL each primer was mixed in a single tube and diluted to a final volume of 40 µL. ssDNA template preparation was performed as described above. For mutant-strand synthesis, oligos were annealed in the absence of polymerase as suggested by Firnberg et al.11. 3.3 µL 1:20 phosphorylated oligos (single or mixed), 10 µL 5× Phusion HF buffer, and 16.7 µL NFH2O were added to the appropriate tube. Oligos were annealed with the following thermocycler program: 98 °C for 2 min, decrease to 55 °C over 15 min, 55 °C for 5 min, and hold at 55 °C. While the reactions were held on the block, the following was added to each tube from a master mix: 20 µL 5× Phusion HF buffer, 20 µL 50 mM DTT, 1 µL 50 mM NAD+, 2 µL 10 mM dNTPs, and 11 µL NFH2O (final reaction volume of 100 µL). The tube contents were mixed by pipetting, then 200 U Taq DNA ligase and 2 U Phusion high-fidelity DNA polymerase were added to each reaction, mixed, spun down for 10 s at 1,000 × g, and returned to the thermocycler for the following program: 72 °C for 10 min, 45 °C for 20 min. The remainder of the protocol proceeded as described in the comprehensive protocol (Supplementary Protocol 1).
DNA deep sequencing and analysis
Plasmids obtained after transformation of the reaction mix and miniprep were used for deep sequencing analysis of library coverage. Samples were prepared for deep sequencing using ‘method B’ from Kowalsky et al.9. Sequences of PCR primers are listed in Supplementary Table 3. Samples for shotgun sequencing were prepared at the Michigan State University sequencing core (approximate median insert size of 360 bp). amiE libraries were sequenced on an Illumina MiSeq with 250-bp paired-end (PE) reads at the University of Illinois Chicago sequencing core. All other samples were sequenced on an Illumina MiSeq with 300-bp PE reads at Michigan State University. Read statistics are given in Table 1. Raw FASTQ files were analyzed with Enrich software20 with modifications as described in Kowalsky et al.9. Analysis of libraries for frameshift and off-target mutations was done using the Burrows–Wheeler Aligner22 followed by processing with SAMtools23. Library statistics (Table 1) and read coverage plots (Supplementary Figs. 2 and 5a) were obtained using custom scripts freely available at GitHub (https://github.com/JKlesmith/Deep_Sequencing_Analysis).
Statistics
For analysis of shotgun sequencing data, the mean of the background subtracted per-position percentage mutant allele values for amiE reactions 1 and 2 at positions inside and outside the targeted region for mutagenesis were computed. Welch two-sample t-tests were performed using R statistical software (https://www.r-project.org/) to calculate significance between averages from the inside regions and the outside regions for reaction 1 (P value < 2.2 × 10−16, t = −14.846, df = 697.06) and reaction 2 (P value < 2.2 × 10−16, t = −19.259, df = 214).
Supplementary Material
Acknowledgments
We thank M. Ostermeier (John Hopkins University) for the oligo sets used in creation of the bla libraries. This research was partially supported by a fellowship from Michigan State University under the Training Program in Plant Biotechnology for Health and Sustainability (T32-GM110523 to E.E.W.), a Howard Hughes Medical Institute Gilliam fellowship (to A.A.), US Department of Agriculture National Institute of Food and Agriculture award 2016-67011-24701 (to J.R.K.), and the US National Science Foundation Career Award 1254238 CBET (to T.A.W.).
Footnotes
Accession codes. Sequence Read Archive: Sequencing data have been deposited under accession numbers SRR4105481, SRR4105482, SRR4105483, SRR4105484, SRR4105485, and SRR4105486.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
E.E.W. and T.A.W. wrote the manuscript with input from all coauthors; E.E.W., T.A.W., J.R.K., and J.A.S. conceived the method; E.E.W., J.R.K., J.A.S., and T.A.W. devised experiments; E.E.W., J.R.K., and A.A. performed experiments; K.E.J.T. and A.A. tested the method.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Fowler DM, Fields S. Nat. Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hietpas RT, Jensen JD, Bolon DNA. Proc. Natl. Acad. Sci. USA. 2011;108:7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Firnberg E, Labonte JW, Gray JJ, Ostermeier M. Mol. Biol. Evol. 2014;31:1581–1592. doi: 10.1093/molbev/msu081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melnikov A, Rogov P, Wang L, Gnirke A, Mikkelsen TS. Nucleic Acids Res. 2014;42:e112. doi: 10.1093/nar/gku511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stiffler MA, Hekstra DR, Ranganathan R. Cell. 2015;160:882–892. doi: 10.1016/j.cell.2015.01.035. [DOI] [PubMed] [Google Scholar]
- 6.Klesmith JR, Bacik JP, Michalczyk R, Whitehead TA. ACS Synth. Biol. 2015;4:1235–1243. doi: 10.1021/acssynbio.5b00131. [DOI] [PubMed] [Google Scholar]
- 7.Whitehead TA, et al. Nat. Biotechnol. 2012;30:543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kowalsky CA, et al. J. Biol. Chem. 2015;290:26457–26470. doi: 10.1074/jbc.M115.676635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowalsky CA, et al. PLoS One. 2015;10:e0118193. doi: 10.1371/journal.pone.0118193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fowler DM, Stephany JJ, Fields S. Nat. Protoc. 2014;9:2267–2284. doi: 10.1038/nprot.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitzman JO, Starita LM, Lo RS, Fields S, Shendure J. Nat. Methods. 2015;12:203–206. doi: 10.1038/nmeth.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firnberg E, Ostermeier M. PLoS One. 2012;7:e52031. doi: 10.1371/journal.pone.0052031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain PC, Varadarajan R. Anal. Biochem. 2014;449:90–98. doi: 10.1016/j.ab.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Fowler DM, et al. Nat. Methods. 2010;7:741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 16.Kunkel TA. Proc. Natl. Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan S-H, Stoddard BL, Xu SY. Nucleic Acids Res. 2011;39:1–18. doi: 10.1093/nar/gkq742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heiter DF, Lunnen KD, Wilson GG. J. Mol. Biol. 2005;348:631–640. doi: 10.1016/j.jmb.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 19.Bienick MS, et al. PLoS One. 2014;9:e109105. doi: 10.1371/journal.pone.0109105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fowler DM, Araya CL, Gerard W, Fields S. Bioinformatics. 2011;27:3430–3431. doi: 10.1093/bioinformatics/btr577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wrenbeck E, Klesmith J, Stapleton J, Whitehead T. Protocol Exchange. 2016 http://dx.doi.org/10.1038/protex.2016.061.
- 22.Li H, Durbin R. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, et al. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.