Abstract
Two structurally distinct filamentous tracks, namely singlet microtubules in the cytoplasm and axonemes in the cilium, serve as railroads for long‐range transport processes in vivo. In all organisms studied so far, the kinesin‐2 family is essential for long‐range transport on axonemes. Intriguingly, in higher eukaryotes, kinesin‐2 has been adapted to work on microtubules in the cytoplasm as well. Here, we show that heterodimeric kinesin‐2 motors distinguish between axonemes and microtubules. Unlike canonical kinesin‐1, kinesin‐2 takes directional, off‐axis steps on microtubules, but it resumes a straight path when walking on the axonemes. The inherent ability of kinesin‐2 to side‐track on the microtubule lattice restricts the motor to one side of the doublet microtubule in axonemes. The mechanistic features revealed here provide a molecular explanation for the previously observed partitioning of oppositely moving intraflagellar transport trains to the A‐ and B‐tubules of the same doublet microtubule. Our results offer first mechanistic insights into why nature may have co‐evolved the heterodimeric kinesin‐2 with the ciliary machinery to work on the specialized axonemal surface for two‐way traffic.
Keywords: axonemes, heterodimeric kinesin‐2, intraflagellar transport, microtubules
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Membrane & Intracellular Transport
Introduction
One of the most visible hallmarks of eukaryotes is the employment of motor proteins that ferry cargo around the cell on tracks provided by the cytoskeleton. Many of these motors were shown to be processive, meaning that they can take many consecutive steps without dissociating from their filaments in vitro 1, 2, 3, 4, 5, 6. The homodimeric kinesin‐1 was central for the molecular understanding of how processivity is achieved. Overwhelming evidence favored the so‐called hand‐over‐hand stepping mechanism for this motor 7, 8, 9, 10, 11. Such a mechanism predicts one head domain of the double‐headed motor to move twice the distance (16 nm) of the center of mass (8 nm). Tracking the displacement of one head domain at subpixel resolution using FIONA (Fluorescence Imaging with One Nanometer Accuracy) provided direct evidence for the hand‐over‐hand stepping mechanism 12.
In contrast to homodimeric kinesin‐1, several members of the kinesin‐2 subfamily form heterodimeric motors that further associate with a non‐motor subunit to function as heterotrimeric motors in vivo 13. Previous work on heterodimeric kinesin‐2 revealed marked differences in processivity and force‐dependent behavior when compared to kinesin‐1 14, 15, 16, 17. In contrast to kinesin‐1, the kinesin‐2 processivity precipitated under load, but in load‐free fluorescence‐based assays, kinesin‐2 processivity matched that of kinesin‐1 16, 17.
Kinesin‐2 is argued to have co‐evolved with the eukaryotic cilium 18, 19, 20, 21. Indeed, kinesin‐2 dependent anterograde transport of cargo to the tip of the cilium, the so‐called intraflagellar transport (IFT), is a prerequisite to the construction of cilia in all eukaryotes 18, 19, 22, 23, 24. The retrograde IFT back to the base of the cilium is accomplished by the dynein‐2 motor 25, 26, 27, 28, 29.
Intraflagellar transport (IFT) takes place on axonemes, an elaborate microtubule structure that consists of nine peripherally arranged doublet microtubules. Each doublet microtubule is formed by the fusion of a B‐tubule to an A‐tubule 30, 31, 32. What are the functional consequences of having such an elaborate axonemal track for IFT? Previous work in Trypanosoma brucei for example showed that only a subset of doublet microtubules is available for IFT 33. Structural studies on the ciliary pore complex from Tetrahymena pyriformis revealed nine large pores that were proposed to accommodate the IFT trains on the B‐tubule 34. Indeed, kinetics and force production by dynein inner arms were shown to be affected by post‐translational modifications (PTMs) on the B‐tubule 35, 36. If, however, PTMs also control antero‐ as well as retrograde IFT is not yet known 37, 38, 39, 40.
In a remarkable recent study, doublet microtubules were found to serve as bi‐directional double‐track railways in vivo 41. Specifically, anterograde IFT trains selectively moved on the B‐tubule, whereas the retrograde transport was restricted to the neighboring A‐tubule. This partitioning in turn enabled collision‐free bi‐directional transport of large IFT trains. However, the underlying mechanisms of such directional track assignment remained elusive.
Intriguingly, some heterotrimeric kinesin‐2 motors have been adapted by evolution to also work on singlet microtubules in the cytoplasm 19, 20, 21. One of the best studied examples of kinesin‐2‐dependent transport in the cytoplasm occurs in Xenopus laevis melanophores where the KLP3A/B motor participates in the dynamic redistribution of pigment granules on the singlet microtubule network 42, 43.
Given this evolutionary specialization of heterodimeric kinesin‐2 motors to both, ciliary and cytoplasmic transport processes, here we hypothesized that kinesin‐2 may have adapted its stepping mechanism to axonemes and microtubules, respectively. By studying KLP11/20 that drives anterograde IFT trains on axonemes in Caenorhabditis elegans 44 and the KLP3A/B, we show that these kinesins differently utilize the microtubule and the axoneme lattice to processively move forward. We further show that processivity of the heterodimeric kinesin‐2 is significantly affected by one of its distal C‐terminal tail domain. The molecular features unmasked in this study provide first mechanistic clues into why nature may have specifically co‐evolved the heterodimeric kinesin‐2 to work on the axonemal lattice for two‐way traffic in vivo.
Results and Discussion
Tracking the displacement of one of the two head domains during the catalytic cycle of motor proteins such as kinesin‐1, myosin‐V, and ‐VI by FIONA technology provided strong support for the hand‐over‐hand stepping mechanism 12, 45, 46, 47. Here, we applied FIONA to two different kinesin‐2 motors, the KLP11/20 from C. elegans and the KLP3A/B kinesin‐2 from X. laevis, and compared how they stepped on microtubules and axonemes.
The two distinct subunits of the heterodimeric kinesin‐2 offer the unique opportunity to label only one subunit with a fluorophore with exquisite specificity. To obtain labeled motors, we N‐terminally fused a Halo‐tag and a SNAP‐tag to the full‐length KLP11 and the KLP3A subunits, respectively. Co‐expression of the tagged KLP11 and KLP3A subunits with their respective partner subunits KLP20 and KLP3B and subsequent fluorophore labeling resulted in heterodimeric motors that were fluorescently labeled with close to 100% specificity at only one subunit (Appendix Fig S1). Photobleaching studies of the N‐terminally labeled motors confirmed that single motors were being assayed (Appendix Fig S2).
KLP3A/B and KLP11/20 motors are highly processive
Properties of single KLP11/20 from C. elegans have previously been studied in optical tweezers assays 14. In these experiments, full‐length KLP11/20 that was attached to micron‐sized polystyrene beads displayed significantly shorter run lengths than kinesin‐1 under comparable load 14, 48.
Here, we employed total internal reflection fluorescence (TIRF) microscopy to investigate the transport parameters of the full‐length KLP3A/B kinesin‐2 from X. laevis (Fig 1A–C) and KLP11/20 from C. elegans (Fig 1D–F). An advantage of this setup is that the motors do not have to be coupled to macroscopic beads. At saturating ATP concentrations, we found that KLP11/20 and KLP3A/B were highly processive, traveling several micrometers before dissociating from the microtubule (Fig 1C and F, and Appendix Figs S3 and S4). The processivity of the full‐length KLP11/20 and KLP3A/B motors also exceeded that of a C‐terminally truncated KIF3A/B motor from mouse in previous TIRF assays 17. The discrepancies between the fluorescence‐ and optical tweezers‐based assays prompted us to search for possible explanations next.
Figure 1. Single‐molecule transport parameters of the full‐length KLP3A/B and KLP11/20 motors on microtubules and axonemes.
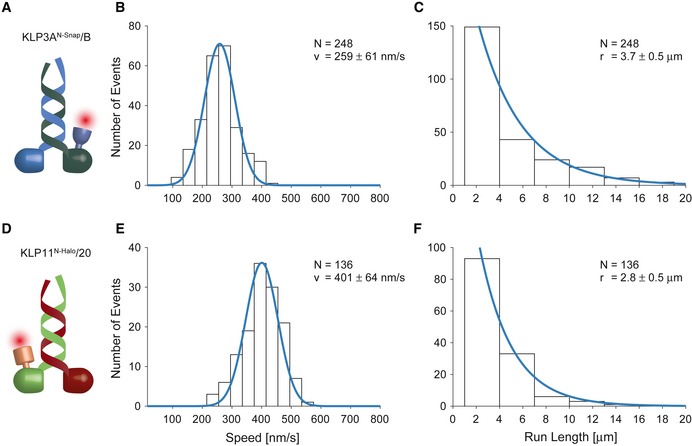
-
A–CThe movement of single KLP3A/B motors fluorescently labeled at the KLP3A subunit was tracked on surface‐attached microtubules at saturating ATP conditions. The velocity (B) and run length data (C) were fit to a Gaussian and single‐exponential distribution, respectively.
-
D–FThe corresponding analysis of the KLP11/20 motor fluorescently labeled at the KLP11 subunit moving on surface‐attached axonemes.
The distal C‐terminal random‐coil tail is necessary for full processivity of kinesin‐2
In contrast to the optical tweezers‐based assays where the motors are attached to micron‐sized beads with their C‐terminus, both C‐termini of the heterodimeric kinesin‐2 motors remained free in our assays. We therefore asked whether a free C‐terminus was necessary for full processivity at the example of the KLP3A/B motor. To this end, we cut off the predicted C‐terminal random‐coil domains from KLP3A and, separately, from KLP3B. It is important to note that heterodimerization of the kinesin‐2 motors is initiated at the C‐terminal end of the predicted coiled‐coil domains 49, 50; the C‐terminal truncations of the kinesin‐2 motor were thus limited to the predicted random‐coil domains (or tail domains; Appendix Fig S5A). We therefore truncated the KLP3A and KLP3B subunits at the highly conserved proline residues that demarcate the end of the predicted coiled‐coil and the start of the random‐coil domains (Appendix Fig S5B). Co‐expression of the truncated constructs with their full‐length partner subunits created two heterodimeric kinesin‐2 motors lacking the C‐terminal random‐coil regions on either subunit (Appendix Fig S6A). As done for the full‐length KLP3A/B motor, the truncated constructs were labeled specifically at the KLP3A subunit via an N‐terminal SNAP‐tag (Appendix Fig S6A). Photobleaching studies confirmed that we detected single motor molecules (Appendix Fig S6B).
Deletion of the C‐terminal tail domain of the KLP3A subunit marginally affected speed and processivity of KLP3A/B (Figs 1B and C, and 2A). Intriguingly, however, deletion of the corresponding region in the KLP3B subunit reduced the run length by ~50% without substantially affecting the velocity of the motor (Fig 2B). To probe if the absence of the C‐terminal B‐tail curtails the processivity of the motor because the A‐tail can more efficiently auto‐inhibit the two head domains 51, we removed both C‐termini of the heterodimeric KLP3A/B motor. Simultaneous removal of both random‐coil domains did not impact the processivity of the construct when compared to the removal of the B‐random coil alone (Fig 2B and C). Indeed, the run length of a truncated heterodimeric KIF3A/B from mouse that lacked its respective C‐terminal random‐coil domains was consistent with that of KLP3AN‐Snap 1‐597/B1‐592 motor also lacking its C‐termini (Fig 2C) 17. These results indicate that the tail domain of KLP3B, but not KLP3A, has the ability to modulate the processivity of the motor. It will be interesting to see whether the C‐terminal random‐coil domain of the KLP3A also has an assigned role to modulate the motor's kinetics other than its processivity.
Figure 2. Impact of the C‐terminal random‐coil domains on the run length of the KLP3A/B motor.
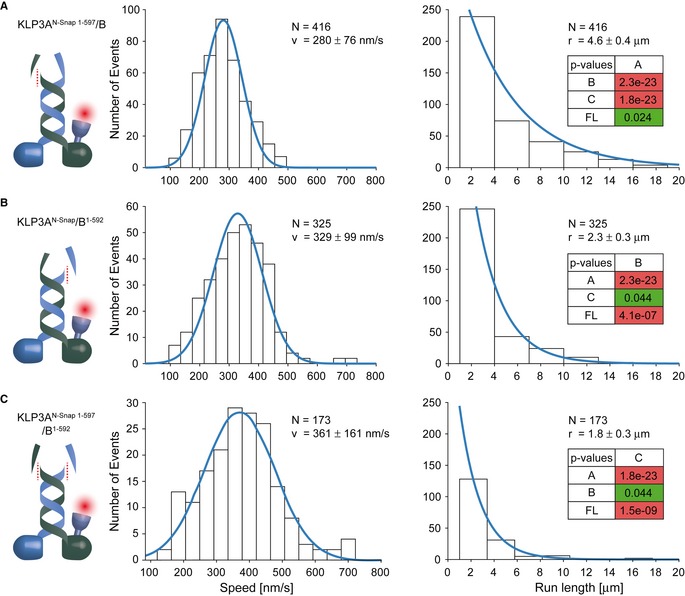
- Removal of the random‐coil domain of the KLP3A subunit failed to interfere with the transport parameters of the KLP3A/B motor.
- In contrast, removing the corresponding domain of the KLP3B subunit reduced the run length by ˜50% without substantially affecting the velocity of the KLP3A/B motor.
- Removal of both random‐coil domains had the same effect as the removal of the KLP3B C‐terminal domain (B).
Taken together, our results argue that the C‐terminal tail of KLP3B augments processivity, possibly by tethering the motor to the microtubule 52, which prevents premature dissociation. Attaching the motor at its C‐terminus to beads in previous optical tweezers studies therefore likely curtailed the processivity of the kinesin‐2 motor.
Kinesin‐2 adapted its steps specifically to axonemes and microtubules
It is well established that kinesin‐1 moves on microtubules according to the hand‐over‐hand stepping mechanism by swinging the trailing head domain forward, ahead of the other, by ~16 nm 12, 53. These steps occur exclusively along one protofilament, restricting the kinesin‐1 motor to a straight path on the microtubule lattice in vitro 53, 54. In stark contrast to kinesin‐1, beads coated with multiple kinesin‐2 motors spiraled around microtubules with a characteristic, left‐handed pitch, suggesting that single kinesin‐2 motors may possess an intrinsic propensity to switch protofilaments 55. Indeed, a single, truncated kinesin‐2 motor from mouse was shown to take sidesteps on the microtubule lattice when it encountered road blocks in vitro 56.
These observations provoked the question how kinesin‐1 and kinesin‐2 motors accomplish efficient intracellular transport on microtubules, which are known to be decorated with many factors such as microtubule‐associated proteins (MAPs) in vivo 57, 58, 59. For example, the direct comparison between kinesin‐1 and kinesin‐2 motors showed that Tau, a MAP known to heavily decorate axonal microtubules in vivo, affected kinesin‐1's but not kinesin‐2's processivity in vitro 60. Even though kinesin‐1 can also overcome permanent obstacles in vitro 61, kinesin‐2 appears to do so more efficiently via its higher propensity to switch protofilaments when compared to kinesin‐1 56, 60.
The proposed capability of kinesin‐2 to take off‐axis steps predicts the step size of the motor to significantly differ from the 16 nm on‐axis steps, which kinesin‐1 for example displays. Here, we directly tested this hypothesis by specifically tracking one head domain of two heterodimeric kinesin‐2 motors. To this end, the N‐terminally fluorophore‐labeled KLP11 and KLP3A subunits of the respective heterodimeric motors were tracked with nanometer resolution. Limiting ATP concentrations were used to sufficiently slow down the motors. Under these conditions, displacements of individual head domains could be resolved.
The respective step sizes of the KLP11/20 and KLP3A/B motors centered around ~13 nm on microtubules and thus significantly deviated from kinesin‐1's ~16 nm on‐axis steps (Figs 3A and B, and EV1, and Appendix Fig S8) 12, 53. A ~13 nm displacement is inconsistent with protofilament tracking because it does not reflect the ~8 nm periodicity of the microtubule track. Instead, these results implicate that the two kinesin‐2 motors take sidesteps to the neighboring protofilaments (Appendix Fig S8). The respective step sizes are consistent with a hand‐over‐hand mechanism, as is the double‐exponential decay of the observed dwell times. The dwell time distribution indicates the existence of two substeps, one from the fluorescently labeled and the other from the unlabeled and thus “hidden” head (Fig 3E and F) 12, 46. We conclude that individual KLP11/20 and KLP3A/B kinesin‐2 motors possess an inherent propensity to take sidesteps to the neighboring protofilament as previously suggested from multiple‐motor assays 55.
Figure 3. Kinesin‐2 motors differentiate between the microtubule and axoneme surface to take processive steps.
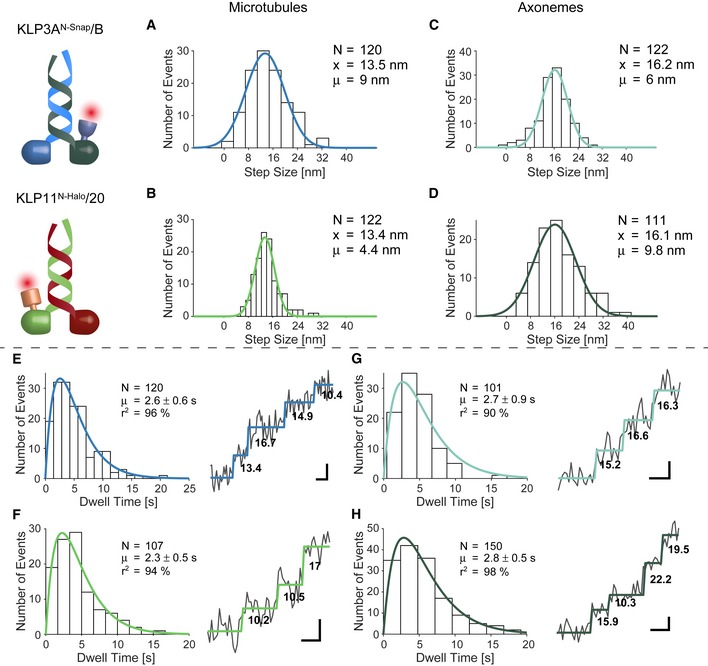
-
A, BThe step size distribution centered around ˜13 nm when walking on microtubules. Together with the double‐exponential decay of the dwell times (E, F), these results support a hand‐over‐hand type stepping of the respective motors.
-
C, DTracking on axonemes increased step size distribution to ˜16 nm which is consistent with protofilament tracking.
-
E, FDouble‐exponential decay of the dwell times and raw stepping data with detected steps.
-
G, HDouble‐exponential decay of the dwell times again argues for a hand‐over‐hand stepping mechanism on axonemes.
Figure EV1. Single fluorophore tracking of KLP11/20 and KLP3A/B motors at limiting ATP concentrations.
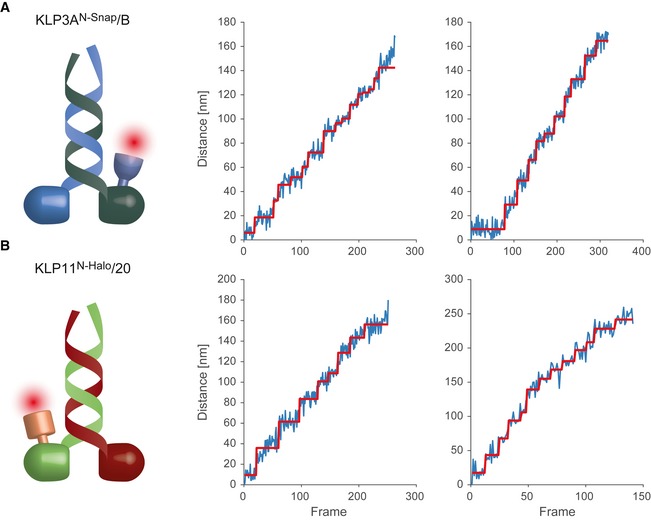
-
A, BPhotons from a single fluorophore detected on the CCD are localized and tracked over time. Localization is achieved by fitting a two‐dimensional Gaussian profile to the spot. The Euclidean distance of each location to the position in the first frame is calculated and plotted in blue. Steps in the data are detected by a gliding t‐value algorithm and plotted in red. Frames have a cycle time of 505 ms.
Next, we assessed whether KLP11/20 and KLP3A/B motors walk differently on axonemes than on single microtubules. Remarkably, the mean displacements of individual motors increased from ~13 to ~16 nm when allowed to walk on axonemes (Fig 3C and D). This step size is consistent with efficient on‐axis protofilament tracking and a hand‐over‐hand stepping mechanism. The dwell time distribution again displayed a double‐exponential decay for both motors on axonemes further supporting an hand‐over‐hand stepping mechanism (Fig 3G and H).
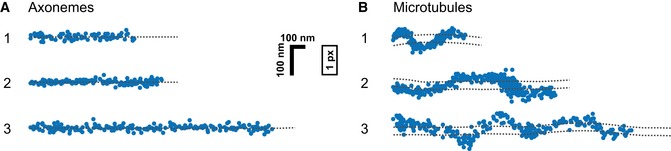
The observed stepping behavior implicates that single kinesin‐2 molecules have an intrinsic propensity to take sidesteps by switching protofilaments on singlet microtubules; on axonemes, however, they faithfully track one protofilament following a straight path. If true, these respective stepping modes should be reflected in the trajectories of the motors moving on microtubules versus axonemes. Following KLP11/20 over time on axonemes indeed resulted in straight trajectories consistent with the 16 nm on‐axis displacements (Figs 4A and EV2A, and Appendix Fig S9). In contrast, on singlet microtubules with rotational symmetry, the motors followed sinusoid‐like paths consistent with sidestepping (Figs 4B and EV2B, and Appendix Figs S8 and S10). These correspond to the projection of a helical motion of an individual motor around the tube‐shaped microtubule lattice.
Figure 4. Example trajectories of single KLP11/20 molecules tracked on axonemes (A) and microtubules (B).
-
A, BOn the rotationally symmetric microtubule filaments, the motors show a sinusoidal trajectory, corresponding to a periodic helical path around the symmetry axis of the filament. The axoneme is lacking this symmetry due to the combination of the A‐ and B‐tubules into a doublet microtubule along with the interconnection between the respective doublet tubules. Without a symmetry axis, the periodic path is lost and the motor is restricted to a straight path. The dashed lines underlining the respective trajectories have been obtained from a subpixel detection of filament positions (see Materials and Methods for details). Images of the filaments are shown in Appendix Fig S9, and respective kymographs are shown in Fig EV2.
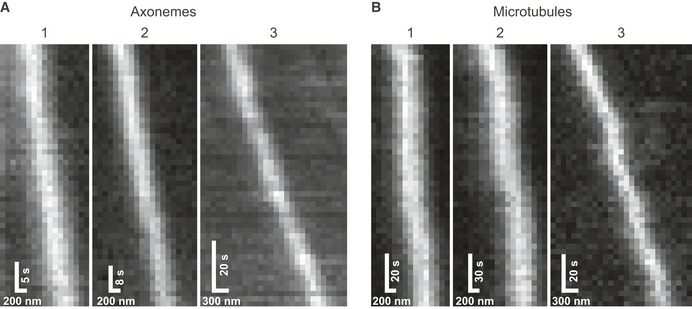
Figure EV2. Kymographs of KLP11/20 walking on axonemes (A) and microtubules (B).
-
A, BThe numbers correspond to the runs shown in Fig 4A and B.
In fact, the sole assumption of a 50% probability of sidestepping was sufficient to recapitulate the experimentally determined step size of the motor (Figs EV3, and 3A and B). Remarkably, this assumption alone sufficed to also recapitulate the motors characteristic trajectories observed above (Figs EV3 and 4B). Taken together, these results strongly argue for an adaption of the motor's trajectory dependent on the respective track employed.
Figure EV3. Dependence between the trajectory and the step size distribution of a motor taking sidesteps.
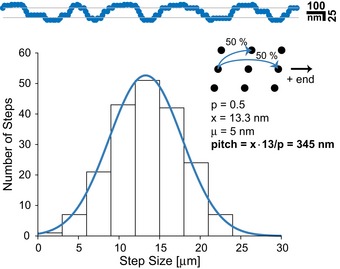
The stepping of a motor was modeled on a cylinder‐shaped geometry with binding sites distributed as expected for the microtubule lattice 72. A sidestepping probability of 50% was assumed, and 200 steps were simulated. The data were projected onto a plane to simulate a microscope image. For the histogram, step sizes were blurred by a Gaussian distribution of width 3 nm corresponding to an estimated detection accuracy. The pitch for a 13 protofilament microtubule is calculated from the mean step size (x) and the probability for sidesteps (P).
Why do the motor's stepping patterns differ on the two respective tracks? The nine peripheral microtubule doublets of the axoneme are connected among others via densely packed nexin and dynein arms 30, 31, 32, 62, 63. We suggest that kinesin‐2's inherent ability to side‐track on the microtubule lattice (Fig EV3) in a left‐handed manner 55 will eventually restrict the motor next to these impenetrable structural demarcations on the axoneme surface forcing the motor to take on‐axis ~16‐nm steps (Fig 5). This process ensures that all kinesin‐2‐powered intraflagellar trains segregate to one side of the doublet microtubule, the B‐tubule, during anterograde transport toward the tip of the cilium 41. Kinesin‐1, in contrast, would fail to segregate to one of the two tubules because it cannot directionally switch protofilaments 12, 54, 55, 61. Instead, it would walk straight ahead and eventually collide with dynein‐powered trains coming from the tip of the cilium. Our results therefore offer a simple mechanistic explanation for how step adaptation of kinesin‐2 motors enables collision‐free bi‐directional transport on axonemes in the cilium 41. We speculate that dynein‐2, like kinesin‐2, may also exhibit an intrinsic left‐handed bias during retrograde transport toward the base of the cilium (Fig 5). In doing so, it would efficiently avoid collisions with kinesin‐2 powered trains by spatially partitioning them to the opposite side of the doublet microtubule. Taken together, this work provides first mechanistic insights into why heterodimeric kinesin‐2 has specifically co‐evolved with the IFT machinery to work on axonemes 19, 20, 21.
Figure 5. Proposed model for two‐way traffic on a single microtubule doublet.
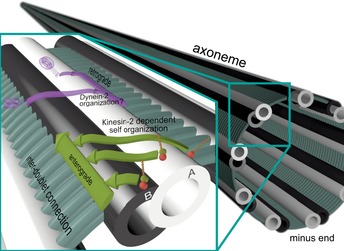
The organization of large intraflagellar trains for a collision‐free bi‐directional transport on a single microtubule doublet within such a spatially restricted environment as seen in vivo poses a remarkable challenge. With the inter‐doublet connections acting as a structural barrier, the motors’ intrinsic ability to switch protofilaments to the left 55, 71 would allow a collision‐free two‐way traffic by separating the large intraflagellar trains to either side of the doublet microtubule. This model may explain why nature has not recycled any other efficient transporter as kinesin‐1 but opted to co‐evolve heterodimeric kinesin‐2 motors with an intrinsic left‐handedness. Whether dynein‐2 displays an exclusive left‐handedness is, however, not yet known.
Materials and Methods
DNA constructs and design
The full‐length heterodimeric kinesin‐2 constructs from X. laevis (KLP3A/B) and C. elegans (KLP11/20) were obtained as described previously 55.
DNA manipulations were done according to standard procedures and as instructed by manufacturers. The truncated constructs were PCR‐amplified using suitable PCR oligos. Full‐length and truncated constructs are listed below:
KLP3AN‐SNAP C‐FLAG
KLP3BC‐His
KLP3A1‐597 N‐SNAP C‐FLAG
KLP3B1‐592 C‐His
KLP20C‐FLAG
KLP11N‐Halo C‐His
All constructs contained a C‐terminal FLAG‐ or His‐tag, respectively, for affinity‐tag purification and were subjected to DNA sequencing to guarantee that no secondary mutations were introduced. Recombinant Bacmids and first virus generations were generated according to standard protocols provided by the manufacturer (Thermofisher). Subsequent viral amplification for protein expression was done as described previously 64.
Protein expression, purification, and fluorescent labeling
All proteins were expressed using the Baculovirus Expression System (Thermofisher) in insect cells [Spodoptera frugiperda (Sf9)] according to the manufacturer's instructions.
Heterodimeric motor proteins were Flag‐tagged (DYKDDDDK) at one of the C‐terminal ends to facilitate purification. The following protocol refers to 50 ml suspension culture at 2 × 106 cells/ml. For protein purification, virus‐infected insect cells were pelleted after 48‐h incubation at 28°C by centrifuging for 10 min at 2,600 g. Cells were carefully lysed in lysis buffer [50 mM PIPES, pH 6.9, 300 mM potassium acetate, 1 mM MgCl2, 1 mM DTT, 0.1 mM ATP, 0.5% Triton X‐100, Complete Protease Inhibitor Cocktail (Roche)]. Lysed cells were pelleted by centrifugation for 10 min at 40,000 g. The supernatant was incubated with 50 μl ANTI‐Flag M2 Affinity Agarose gel (Sigma) for 90 min. The beads were washed three times with 1 ml wash buffer 1 (80 mM PIPES, pH 6.9, 500 mM potassium acetate, 1 mM MgCl2, 1 mM DTT, 5 μM ATP, 0.1% Tween‐20, 1 mM EGTA) and three times with 1 ml wash buffer 2 (80 mM PIPES, pH 6.9, 200 mM potassium acetate, 1 mM MgCl2, 1 mM DTT, 0.1 mM ATP, 0.1% Tween‐20, 1 mM EGTA). Subsequently, the beads were incubated on a rotator for 40 min at 4°C with 100 μl wash buffer 2 containing either 1 mM HaloTag® AlexaFluor®660 ligand (for the labeling of the Halo‐tagged KLP11/20 protein) or 1 mM SNAP surface® Alexa Fluor®647 ligand (for labeling of the SNAP‐tagged KLP3A/B protein). The protein was eluted in 100 μl elution buffer [80 mM PIPES, pH 6.9, 200 mM potassium acetate, 1 mM MgCl2, 1 mM DTT, 0,1 mM ATP, 0,1% Tween‐20, 1 mM EGTA, 0.5 mg/ml 1× Flag Peptide (Sigma)] for 1 h. This protocol leads to a typical range of protein yields of 0.1–0.2 mg/ml.
Photobleaching experiments
In order to verify that single molecules were measured in the microscopic assays, bleaching step analysis was performed on all constructs. Kinesin was adsorbed to coverslips via non‐specific interactions, and fluorescence was observed in TIRF illumination. Spots for analysis were chosen for all constructs by intensity, and the intensity values for a 3 × 3 pixel window were summarized. Performing a gliding t‐value test with a corresponding threshold identified the bleaching steps 65.
Single‐molecule assays
Transport parameters at saturating ATP concentrations
Tubulin isolated from porcine brain 66 was polymerized and incubated at 35°C in BRB80 (80 mM PIPES, pH 6.9, 2 mM MgCl2, 1 mM EGTA, 1 mM DTT) containing 1 mM GTP. Taxol (5 μM) was added after incubation for 30 min, and microtubules were kept at 35°C 67. Axonemes were isolated from sea urchin sperm as described previously 68.
A flow chamber (V = 5 μl) was created by fusing a Parafilm cut‐out between a slide and a cover slip. For experiments on microtubules, the chamber was coated with 1 mg/ml biotinylated BSA, 1 mg/ml streptavidin (Sigma), and biotinylated, fluorescently (Alexa Fluor®555 dye, Thermofisher) labeled microtubules, respectively, with intervening 2‐min‐long incubation and subsequent washing steps using BRB80 buffer (80 mM PIPES, pH 6.9, 2 mM MgCl2, 1 mM EGTA, 1 mM DTT, 5 μM paclitaxel) containing 7 mg/ml BSA. For experiments on axonemes, the chamber was coated via unspecific binding of the filaments and subsequently washed using BRB80/BSA to block the surface. Finally, a motility buffer (see below) containing a desired concentration of motor proteins was perfused into the flow chamber.
The fluorescently labeled KLP3A/B proteins were recorded over time (cycle time 206 ms) using an objective‐type Leica DMI6000 B TIRF microscope (Leica, Germany), equipped with a plan objective lens (100×, N.A. 1.47 Oil), and a back‐illuminated Andor U897 EMCCD camera (Andor, UK). Excitation was achieved with the help of solid‐state laser at 561‐ and 635‐nm wavelength, and frames were recorded and analyzed with AF 6000 software (Leica, Germany). The motility buffer used was as follows: BRB80 buffer, pH 6.9, 10 mM ATP, 0.4% glucose, 20% sucrose, 10% glycerin, 0.2% Tween‐20, 0.145 mg/ml glucose oxidase (Sigma), 0.0485 mg/ml catalase (Sigma), 5 μM paclitaxel, 100 mM potassium acetate.
Experiments on KLP11/20 were conducted with a cycle time of 206 ms on a custom‐built setup. A 642‐nm laser is coupled into an Apo N 100× HOTIRF objective (Olympus K.K) off‐axis for total internal reflection illumination. Gathered light is split from the laser light with a Laser Quad Band Filter (89901 Chroma Technology GmbH), and residual laser light is removed by a notch filter (Semrock Inc). The light is then focused on an iXon888 Ultra (Andor Technology Ltd) emCCD camera with a system of lenses of 150‐ and 250‐mm focal length (Qioptiq Photonics GmbH), yielding a pixel size of 93 nm. Fused silica coverslips are used with this setup (V‐A Optical Labs Inc.). The motility buffer used was as follows: BRB80 buffer, pH 6.9, 2 mM ATP, 0.145 mg/ml glucose oxidase, 0.0485 mg/ml catalase, 0.4% glucose, 100 mM potassium acetate.
The velocities and run lengths were analyzed with custom‐written programs using the MATLAB software (Mathworks Inc.). Runs were considered processive with a minimal run length of 1 μm. The run length data were fit to a truncated (x 0 = 1 μm) single‐exponential distribution. Parts of the distance over time data were considered for speed calculation if a linear fit of at least six frames resulted in a r 2‐value > 95%.
Statistical significance between datasets was determined by a two‐sample t‐test performed in R. Random resampling of the data by bootstrapping (R = 1,000) resulted in P‐values for comparison of the means. P‐values of < 0.01 were used to reject the null hypothesis 69.
Step detection at limiting ATP concentrations
For step detection experiments, the ATP concentration was reduced to 0.4 μM, in order to reduce the speed of motors and enable detection of single steps. The lactate dehydrogenase/pyruvate/phosphoenolpyruvate (PEP: 3.2 mg NADH, 6.2 mg PEP & 6.8 μl LDH in 133 μl BRB80) system guaranteed stable ATP concentrations 14, 70. Movies were recorded with an exposure time of 500 ms resulting in a cycle time of 505 ms. The electron‐magnifying gain was set to 118 from the first register, and data were digitalized at 30 MHz with the 16 bit analogue to digital converter. 500 frames were recorded before changing the position in the sample.
Data analysis of the step size distributions
The experiments for step size detection were performed on the custom‐built setup as described above. Movies were analyzed by a custom routine, implemented using MATLAB. A least‐squares fit procedure was used to fit a Gaussian profile to the data, with a starting point deduced from the initial detection of the brightest pixel. This fit provided a subpixel accuracy position of the spot which was stored for every frame. The distance of the spots in the subsequent frames was then calculated with respect to the position in the first frame. The gliding t‐value approach was used in order to detect steps 65. The individual sizes of steps were calculated from the mean distances before and after each step. The threshold for the step detection algorithm was 1.7 for the “MinPeakProminence” parameter of the “findpeaks” function in MATLAB, that is performed directly on the t‐values. The window size was adjusted to the speed of the motors and was 13 frames for 11/20 on AX, 11 for 11/20 on MT, 11 for 3A/B on AX and 25 for 3A/B on MT. Dwell times were calculated by calculating the times between the occurrences of steps.
Path detection at intermediate ATP concentrations
For the detection of motor path, an ATP concentration of 1 μM was used as a tradeoff between length of the runs and detection accuracy. Buffer conditions, ATP regeneration system, and camera settings were used as described for the step detection experiments. Microtubules and axonemes were labeled with an Atto 488 dye, and imaged after movies of the HaloTag® AlexaFluor®660 labeled motors were recorded. The position of the filaments was detected with an accuracy below the size of a pixel performing a Gaussian fit on the lateral profiles of the filaments. A moving average filter with a window size of 5 pixels (465 nm) was performed on the position data to account for the persistence length of the filaments. Walking paths and filaments were positioned along the filament axis according to the position on the camera; the lateral position was set to the x‐axis of the plot. Kymographs were prepared using a standard kymograph algorithm with a line width of 3 pixels implemented in MATLAB.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on request.
Author contributions
WLS, GM, and ZÖ planned the experiments. WLS built the custom TIRF setup for the FIONA experiments and wrote all the customized MATLAB routines. WLS and GM collected and analyzed data. ZÖ and FM‐P wrote the manuscript with contributions of WLS and GM.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
The research leading to these results has received funding from the European Research Council (GA no. 335623) to Willi Stepp & Zeynep Ökten and Deutsche Forschungsgemeinschaft SFB863 to Georg Merck & Zeynep Ökten. We thank Julia Maxi Kanold and the Department of Zoology at the University of Stuttgart for the preparation and supply of sea urchin sperm of Arbacia lixula for axoneme preparation. We would further like to acknowledge Angela Oberhofer for discussions throughout the progress of this work and for critically reading the manuscript.
EMBO Reports (2017) 18: 1947–1956
References
- 1. Block SM, Goldstein LS, Schnapp BJ (1990) Bead movement by single kinesin molecules studied with optical tweezers. Nature 348: 348–352 [DOI] [PubMed] [Google Scholar]
- 2. Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE (1999) Myosin‐V is a processive actin‐based motor. Nature 400: 590–593 [DOI] [PubMed] [Google Scholar]
- 3. Howard J, Hudspeth AJ, Vale RD (1989) Movement of microtubules by single kinesin molecules. Nature 342: 154–158 [DOI] [PubMed] [Google Scholar]
- 4. Svoboda K, Schmidt CF, Schnapp BJ, Block SM (1993) Direct observation of kinesin stepping by optical trapping interferometry. Nature 365: 721–727 [DOI] [PubMed] [Google Scholar]
- 5. Woehlke G, Schliwa M (2000) Walking on two heads: the many talents of kinesin. Nat Rev Mol Cell Biol 1: 50–58 [DOI] [PubMed] [Google Scholar]
- 6. Thorn KS, Ubersax JA, Vale RD (2000) Engineering the processive run length of the kinesin motor. J Cell Biol 151: 1093–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cross RA (1995) On the hand‐over‐hand footsteps of kinesin heads. J Muscle Res Cell Motil 16: 91–94 [DOI] [PubMed] [Google Scholar]
- 8. Guydosh NR, Block SM (2009) Direct observation of the binding state of the kinesin head to the microtubule. Nature 461: 125–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Asbury CL, Fehr AN, Block SM (2003) Kinesin moves by an asymmetric hand‐over‐hand mechanism. Science 302: 2130–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaseda K, Higuchi H, Hirose K (2003) Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat Cell Biol 5: 1079–1082 [DOI] [PubMed] [Google Scholar]
- 11. Schief WR, Clark RH, Crevenna AH, Howard J (2004) Inhibition of kinesin motility by ADP and phosphate supports a hand‐over‐hand mechanism. Proc Natl Acad Sci USA 101: 1183–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yildiz A, Tomishige M, Vale RD, Selvin PR (2004) Kinesin walks hand‐over‐hand. Science 303: 676–678 [DOI] [PubMed] [Google Scholar]
- 13. Cole DG, Chinn SW, Wedaman KP, Hall K, Vuong T, Scholey JM (1993) Novel heterotrimeric kinesin‐related protein purified from sea urchin eggs. Nature 366: 268–270 [DOI] [PubMed] [Google Scholar]
- 14. Brunnbauer M, Mueller‐Planitz F, Kosem S, Ho TH, Dombi R, Gebhardt JC, Rief M, Okten Z (2010) Regulation of a heterodimeric kinesin‐2 through an unprocessive motor domain that is turned processive by its partner. Proc Natl Acad Sci USA 107: 10460–10465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Milic B, Andreasson JOL, Hogan DW, Block SM (2017) Intraflagellar transport velocity is governed by the number of active KIF17 and KIF3AB motors and their motility properties under load. Proc Natl Acad Sci USA 114: E6830–E6838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Andreasson JO, Shastry S, Hancock WO, Block SM (2015) The mechanochemical cycle of mammalian kinesin‐2 KIF3A/B under load. Curr Biol 25: 1166–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guzik‐Lendrum S, Rank KC, Bensel BM, Taylor KC, Rayment I, Gilbert SP (2015) Kinesin‐2 KIF3AC and KIF3AB can drive long‐range transport along microtubules. Biophys J 109: 1472–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao C, Omori Y, Brodowska K, Kovach P, Malicki J (2012) Kinesin‐2 family in vertebrate ciliogenesis. Proc Natl Acad Sci USA 109: 2388–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scholey JM (2013) Kinesin‐2: a family of heterotrimeric and homodimeric motors with diverse intracellular transport functions. Annu Rev Cell Dev Biol 29: 443–469 [DOI] [PubMed] [Google Scholar]
- 20. Scholey JM (2003) Intraflagellar transport. Annu Rev Cell Dev Biol 19: 423–443 [DOI] [PubMed] [Google Scholar]
- 21. Mitchell DR (2007) The evolution of eukaryotic cilia and flagella as motile and sensory organelles. Adv Exp Med Biol 607: 130–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pedersen LB, Rosenbaum JL (2008) Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol 85: 23–61 [DOI] [PubMed] [Google Scholar]
- 23. Scholey JM (2008) Intraflagellar transport motors in cilia: moving along the cell's antenna. J Cell Biol 180: 23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Verhey KJ, Dishinger J, Kee HL (2011) Kinesin motors and primary cilia. Biochem Soc Trans 39: 1120–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gibbons BH, Asai DJ, Tang WJ, Hays TS, Gibbons IR (1994) Phylogeny and expression of axonemal and cytoplasmic dynein genes in sea urchins. Mol Biol Cell 5: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tanaka Y, Zhang Z, Hirokawa N (1995) Identification and molecular evolution of new dynein‐like protein sequences in rat brain. J Cell Sci 108(Pt 5): 1883–1893 [DOI] [PubMed] [Google Scholar]
- 27. Pazour GJ, Dickert BL, Witman GB (1999) The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol 144: 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Porter ME, Bower R, Knott JA, Byrd P, Dentler W (1999) Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol Biol Cell 10: 693–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Signor D, Wedaman KP, Orozco JT, Dwyer ND, Bargmann CI, Rose LS, Scholey JM (1999) Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans . J Cell Biol 147: 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maheshwari A, Obbineni JM, Bui KH, Shibata K, Toyoshima YY, Ishikawa T (2015) alpha‐ and beta‐tubulin lattice of the axonemal microtubule doublet and binding proteins revealed by single particle cryo‐electron microscopy and tomography. Structure 23: 1584–1595 [DOI] [PubMed] [Google Scholar]
- 31. Song YH, Mandelkow E (1995) The anatomy of flagellar microtubules: polarity, seam, junctions, and lattice. J Cell Biol 128: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sui H, Downing KH (2006) Molecular architecture of axonemal microtubule doublets revealed by cryo‐electron tomography. Nature 442: 475–478 [DOI] [PubMed] [Google Scholar]
- 33. Absalon S, Blisnick T, Kohl L, Toutirais G, Dore G, Julkowska D, Tavenet A, Bastin P (2008) Intraflagellar transport and functional analysis of genes required for flagellum formation in trypanosomes. Mol Biol Cell 19: 929–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ounjai P, Kim KD, Liu H, Dong M, Tauscher AN, Witkowska HE, Downing KH (2013) Architectural insights into a ciliary partition. Curr Biol 23: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kubo T, Yanagisawa HA, Yagi T, Hirono M, Kamiya R (2010) Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner‐arm dyneins. Curr Biol 20: 441–445 [DOI] [PubMed] [Google Scholar]
- 36. Suryavanshi S, Edde B, Fox LA, Guerrero S, Hard R, Hennessey T, Kabi A, Malison D, Pennock D, Sale WS et al (2010) Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr Biol 20: 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Multigner L, Pignot‐Paintrand I, Saoudi Y, Job D, Plessmann U, Rudiger M, Weber K (1996) The A and B tubules of the outer doublets of sea urchin sperm axonemes are composed of different tubulin variants. Biochemistry 35: 10862–10871 [DOI] [PubMed] [Google Scholar]
- 38. Kann ML, Prigent Y, Levilliers N, Bre MH, Fouquet JP (1998) Expression of glycylated tubulin during the differentiation of spermatozoa in mammals. Cell Motil Cytoskeleton 41: 341–352 [DOI] [PubMed] [Google Scholar]
- 39. Huitorel P, White D, Fouquet JP, Kann ML, Cosson J, Gagnon C (2002) Differential distribution of glutamylated tubulin isoforms along the sea urchin sperm axoneme. Mol Reprod Dev 62: 139–148 [DOI] [PubMed] [Google Scholar]
- 40. Johnson KA (1998) The axonemal microtubules of the Chlamydomonas flagellum differ in tubulin isoform content. J Cell Sci 111(Pt 3): 313–320 [DOI] [PubMed] [Google Scholar]
- 41. Stepanek L, Pigino G (2016) Microtubule doublets are double‐track railways for intraflagellar transport trains. Science 352: 721–724 [DOI] [PubMed] [Google Scholar]
- 42. Nascimento AA, Roland JT, Gelfand VI (2003) Pigment cells: a model for the study of organelle transport. Annu Rev Cell Dev Biol 19: 469–491 [DOI] [PubMed] [Google Scholar]
- 43. Tuma MC, Zill A, Le Bot N, Vernos I, Gelfand V (1998) Heterotrimeric kinesin II is the microtubule motor protein responsible for pigment dispersion in Xenopus melanophores. J Cell Biol 143: 1547–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Signor D, Wedaman KP, Rose LS, Scholey JM (1999) Two heteromeric kinesin complexes in chemosensory neurons and sensory cilia of Caenorhabditis elegans . Mol Biol Cell 10: 345–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yildiz A, Park H, Safer D, Yang Z, Chen LQ, Selvin PR, Sweeney HL (2004) Myosin VI steps via a hand‐over‐hand mechanism with its lever arm undergoing fluctuations when attached to actin. J Biol Chem 279: 37223–37226 [DOI] [PubMed] [Google Scholar]
- 46. Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR (2003) Myosin V walks hand‐over‐hand: single fluorophore imaging with 1.5‐nm localization. Science 300: 2061–2065 [DOI] [PubMed] [Google Scholar]
- 47. Okten Z, Churchman LS, Rock RS, Spudich JA (2004) Myosin VI walks hand‐over‐hand along actin. Nat Struct Mol Biol 11: 884–887 [DOI] [PubMed] [Google Scholar]
- 48. Schnitzer MJ, Visscher K, Block SM (2000) Force production by single kinesin motors. Nat Cell Biol 2: 718–723 [DOI] [PubMed] [Google Scholar]
- 49. Vukajlovic M, Dietz H, Schliwa M, Okten Z (2011) How kinesin‐2 forms a stalk. Mol Biol Cell 22: 4279–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. De Marco V, Burkhard P, Le Bot N, Vernos I, Hoenger A (2001) Analysis of heterodimer formation by Xklp3A/B, a newly cloned kinesin‐II from Xenopus laevis . EMBO J 20: 3370–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hackney DD, Baek N, Snyder AC (2009) Half‐site inhibition of dimeric kinesin head domains by monomeric tail domains. Biochemistry 48: 3448–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Girotra M, Srivastava S, Kulkarni A, Barbora A, Bobra K, Ghosal D, Devan P, Aher A, Jain A, Panda D et al (2017) The C‐terminal tails of heterotrimeric kinesin‐2 motor subunits directly bind to alpha‐tubulin1: possible implications for cilia‐specific tubulin entry. Traffic 18: 123–133 [DOI] [PubMed] [Google Scholar]
- 53. Yildiz A, Tomishige M, Gennerich A, Vale RD (2008) Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell 134: 1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ray S, Meyhofer E, Milligan RA, Howard J (1993) Kinesin follows the microtubule's protofilament axis. J Cell Biol 121: 1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brunnbauer M, Dombi R, Ho TH, Schliwa M, Rief M, Okten Z (2012) Torque generation of kinesin motors is governed by the stability of the neck domain. Mol Cell 46: 147–158 [DOI] [PubMed] [Google Scholar]
- 56. Hoeprich GJ, Mickolajczyk KJ, Nelson SR, Hancock WO, Berger CL (2017) The axonal transport motor kinesin‐2 navigates microtubule obstacles via protofilament switching. Traffic 18: 304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dehmelt L, Halpain S (2005) The MAP2/Tau family of microtubule‐associated proteins. Genome Biol 6: 204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mandelkow E, Mandelkow EM (1995) Microtubules and microtubule‐associated proteins. Curr Opin Cell Biol 7: 72–81 [DOI] [PubMed] [Google Scholar]
- 59. Maiato H, Sampaio P, Sunkel CE (2004) Microtubule‐associated proteins and their essential roles during mitosis. Int Rev Cytol 241: 53–153 [DOI] [PubMed] [Google Scholar]
- 60. Hoeprich GJ, Thompson AR, McVicker DP, Hancock WO, Berger CL (2014) Kinesin's neck‐linker determines its ability to navigate obstacles on the microtubule surface. Biophys J 106: 1691–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schneider R, Korten T, Walter WJ, Diez S (2015) Kinesin‐1 motors can circumvent permanent roadblocks by side‐shifting to neighboring protofilaments. Biophys J 108: 2249–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Oda T, Yanagisawa H, Kikkawa M (2015) Detailed structural and biochemical characterization of the nexin‐dynein regulatory complex. Mol Biol Cell 26: 294–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ishikawa T (2015) Cryo‐electron tomography of motile cilia and flagella. Cilia 4: 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fitzgerald DJ, Berger P, Schaffitzel C, Yamada K, Richmond TJ, Berger I (2006) Protein complex expression by using multigene baculoviral vectors. Nat Methods 3: 1021–1032 [DOI] [PubMed] [Google Scholar]
- 65. Carter NJ, Cross RA (2005) Mechanics of the kinesin step. Nature 435: 308–312 [DOI] [PubMed] [Google Scholar]
- 66. Mandelkow EM, Herrmann M, Ruhl U (1985) Tubulin domains probed by limited proteolysis and subunit‐specific antibodies. J Mol Biol 185: 311–327 [DOI] [PubMed] [Google Scholar]
- 67. Oberhofer A, Spieler P, Rosenfeld Y, Stepp WL, Cleetus A, Hume AN, Mueller‐Planitz F, Okten Z (2017) Myosin Va's adaptor protein melanophilin enforces track selection on the microtubule and actin networks in vitro . Proc Natl Acad Sci USA 114: E4714–E4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gibbons IR, Fronk E (1979) A latent adenosine triphosphatase form of dynein 1 from sea urchin sperm flagella. J Biol Chem 254: 187–196 [PubMed] [Google Scholar]
- 69. Thompson AR, Hoeprich GJ, Berger CL (2013) Single‐molecule motility: statistical analysis and the effects of track length on quantification of processive motion. Biophys J 104: 2651–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kosem S, Okten Z, Ho TH, Trommler G, Koonce MP, Samereier M, Muller‐Taubenberger A (2013) A non‐mitotic CENP‐E homolog in Dictyostelium discoideum with slow motor activity. Biochem Biophys Res Commun 431: 490–495 [DOI] [PubMed] [Google Scholar]
- 71. Can S, Dewitt MA, Yildiz A (2014) Bidirectional helical motility of cytoplasmic dynein around microtubules. Elife 3: e03205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chretien D, Wade RH (1991) New data on the microtubule surface lattice. Biol Cell 71: 161–174 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on request.


