Abstract
PIWI proteins play essential roles in germ cells and stem cell lineages. In Drosophila, Piwi is required in somatic niche cells and germline stem cells (GSCs) to support GSC self‐renewal and differentiation. Whether and how other PIWI proteins are involved in GSC biology remains unknown. Here, we show that Aubergine (Aub), another PIWI protein, is intrinsically required in GSCs for their self‐renewal and differentiation. Aub needs to be loaded with piRNAs to control GSC self‐renewal and acts through direct mRNA regulation. We identify the Cbl proto‐oncogene, a regulator of mammalian hematopoietic stem cells, as a novel GSC differentiation factor. Aub stimulates GSC self‐renewal by repressing Cbl mRNA translation and does so in part through recruitment of the CCR4‐NOT complex. This study reveals the role of piRNAs and PIWI proteins in controlling stem cell homeostasis via translational repression and highlights piRNAs as major post‐transcriptional regulators in key developmental decisions.
Keywords: CCR4‐NOT deadenylation complex, germline stem cells, piRNAs, PIWI proteins, translational control
Subject Categories: Development & Differentiation, RNA Biology, Stem Cells
Introduction
The regulation of gene expression at the mRNA level is fundamental for many biological and developmental processes. In recent years, Piwi‐interacting RNAs (piRNAs) have emerged as novel key players in the regulation of gene expression at the mRNA level in several models. These 23‐ to 30‐nucleotide‐long non‐coding RNAs are loaded into specific Argonaute proteins, the PIWI proteins (Ishizu et al, 2012; Guzzardo et al, 2013). Classically, piRNAs repress transposable element expression and transposition in the germline. They are largely produced from transposable element sequences and target transposable element mRNAs by complementarity, which induces their cleavage through the endonuclease activity of PIWI proteins and represses their expression.
Recent studies have shown that piRNAs also target protein‐coding mRNAs, leading to their repression by PIWI‐dependent mRNA cleavage or via the recruitment of the CCR4‐NOT deadenylation complex. This regulation is required for embryonic patterning in Drosophila (Rouget et al, 2010; Barckmann et al, 2015), sex determination in Bombyx mori (Kiuchi et al, 2014), and degradation of spermiogenic mRNAs in mouse sperm (Gou et al, 2014; Goh et al, 2015; Watanabe et al, 2015; Zhang et al, 2015).
piRNAs involved in the regulation of protein‐coding mRNAs in Drosophila embryos are produced in the female germline and provided maternally. An open question is whether this function of the piRNA pathway in post‐transcriptional control of gene expression plays a role in the biology of germ cells and germline stem cells (GSCs). In the Drosophila ovary, two to three GSCs are localized in the anterior‐most region of each ovariole and self‐renew throughout the adult life, giving rise to all germ cells. GSCs in contact with somatic niche cells divide asymmetrically to produce a new stem cell that remains in contact with niche cells (self‐renewal) and another cell that differentiates into a cystoblast, upon losing the contact with the niche. Subsequently, the cystoblast undergoes four rounds of synchronous division with incomplete cytokinesis to produce a cyst of 16 interconnected germ cells, of which one cell is specified as the oocyte and the other 15 cells become nurse cells (Fig 1A).
Figure 1. Intrinsic role of Aub in GSC self‐renewal and differentiation.
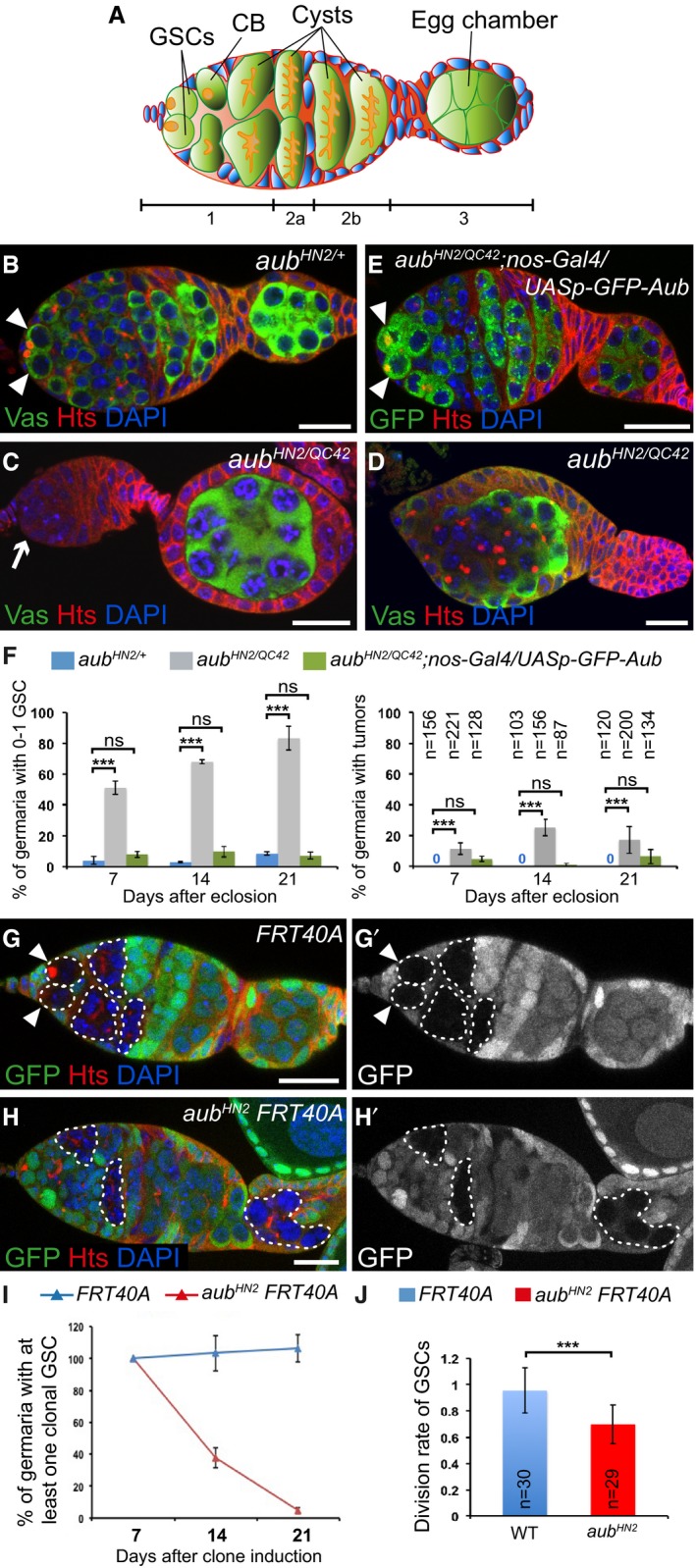
-
ASchematic diagram of a germarium showing the somatic cells (blue) and the germline cells (green). The spectrosomes and fusomes are shown in orange. The different regions of the germarium are indicated. Region 1: dividing cysts; region 2: selection of the oocyte; region 3: egg chamber with posteriorly localized oocyte. GSCs, germline stem cells; CB, cystoblast.
-
B–EImmunostaining of germaria from 7‐day‐old females with anti‐Vasa (green, B–D) or anti‐GFP (green, E), and anti‐Hts (red). DAPI (blue) was used to visualize DNA. (B) aub HN2/+ was used as a control. (C, D) Examples of aub HN2/QC42 germ cell loss and tumor, respectively. (E) Phenotypic rescue of aub HN2/QC42 with UASp‐GFP‐Aub expressed using nos‐Gal4. White arrowheads indicate GSCs; the white arrow indicates GSC loss.
-
FQuantification of mutant germaria with 0–1 GSC, or with GSC tumors shown in (B–E). The number of scored germaria (n) is indicated on the right graph. Error bars represent standard deviation. ***P‐value < 0.001, ns, non‐significant, using the χ2 test.
-
G–H’Germaria containing control (G, G’) or aub HN2 mutant (H, H’) clonal GSCs stained with anti‐GFP (green) and anti‐Hts (red), 14 days after clone induction. DAPI (blue) was used to stain DNA. Clonal cells are marked by the lack of GFP. Clonal GSCs and cysts are outlined with dashed line. White arrowheads show clonal GSCs in the control. aub mutant clonal GSCs have been lost (H, H’).
-
IQuantification of germaria containing at least one clonal GSC at 7, 14, and 21 days after clonal induction. 50 to 219 germaria were analyzed per condition. Error bars represent standard deviation.
-
JDivision rate of wild‐type and aub HN2 clonal GSCs. The number of scored germaria (n) is indicated. Error bars represent standard deviation. ***P‐value < 0.001 using the two‐tailed Student's t‐test.
Two features make Piwi unique with respect to the other two Drosophila PIWI proteins, Aubergine (Aub) and Argonaute 3 (Ago3). First, it represses transposable elements at the transcription level through a nuclear function, whereas Aub and Ago3 act by endonucleolytic cleavage of transposable element mRNAs in the cytoplasm; and second, it plays a role in the somatic and germ cells of the ovary, whereas aub and ago3 function is restricted to germ cells. piwi function in GSC biology has long been addressed. piwi is required in somatic escort cells (which surround GSCs) for GSC differentiation, as well as intrinsically in GSCs for their maintenance and differentiation (Cox et al, 1998, 2000; Jin et al, 2013; Ma et al, 2014). One molecular mechanism underlying Piwi function in GSC biology has recently been proposed to involve its direct interaction with Polycomb‐group proteins of the PRC2 complex, leading to indirect massive gene deregulation through reduced PRC2 binding to chromatin (Peng et al, 2016). Regulation of c‐Fos by Piwi at the mRNA level in somatic niche cells has also been reported to contribute to the role of Piwi in GSC maintenance and differentiation (Klein et al, 2016).
Translational control acting intrinsically in GSCs plays a major role in the switch between self‐renewal and differentiation. Two molecular pathways ensure GSC self‐renewal through translational repression of differentiation factor mRNAs: the microRNA pathway, and the translational repressors Nanos (Nos) and Pumilio (Pum; Slaidina & Lehmann, 2014). Nos and Pum bind to and repress the translation of mRNAs that encode the differentiation factors Brain tumor (Brat) and Mei‐P26, through the recruitment of the CCR4‐NOT deadenylation complex (Harris et al, 2011; Joly et al, 2013). In turn, cystoblast differentiation depends on Bag of marbles (Bam), the major differentiation factor forming a complex with Mei‐P26, Sex lethal (Sxl), and Benign gonial cell neoplasm (Bgcn) to repress nos mRNA translation; Pum interacts with Brat in these cells to repress the translation of mRNAs encoding self‐renewal factors (Li et al, 2009b, 2012, 2013; Harris et al, 2011; Chau et al, 2012).
Aub has a distinctive role in protein‐coding mRNA regulation. In the early embryo, Aub binds several hundred maternal mRNAs in a piRNA‐dependent manner and induces the decay of a large number of them during the maternal‐to‐zygotic transition (Barckmann et al, 2015). Aub‐dependent unstable mRNAs are degraded in the somatic part of the embryo and stabilized in the germ plasm. These mRNAs encode germ cell determinants, indicating an important function of Aub in embryonic patterning and germ cell development. Indeed, Aub recruits the CCR4 deadenylase to nos mRNA and contributes to its deadenylation and translational repression in the somatic part of the embryo. This Aub‐dependent repression of nos mRNA is involved in embryonic patterning (Rouget et al, 2010).
Here, we address the role of aub in GSC biology. We show that aub is autonomously required in GSCs for their self‐renewal. This aub function is independent of bam transcriptional repression in the GSCs and partly independent of activation of the Chk2‐dependent DNA damage checkpoint. Aub is also involved in GSC differentiation; aub mutant defect in GSC differentiation is less frequent and involves the Chk2‐dependent DNA damage checkpoint. Using an Aub point‐mutant form that cannot load piRNAs, we show that piRNAs are required for GSC self‐renewal. Genetic and physical interactions indicate that Aub function in GSCs involves interaction with the CCR4‐NOT deadenylation complex. Importantly, we identify Casitas B‐cell lymphoma (Cbl) mRNA as a target of Aub in GSCs. Cbl acts either as a tumor suppressor or a proto‐oncogene depending on its mutations, which lead to myeloid malignancies in humans (Sanada et al, 2009). Cbl encodes an E3 ubiquitin ligase that negatively regulates signal transduction of tyrosine kinases; it plays a role in hematopoietic stem cell homeostasis, maintaining quiescence, and preventing exhaustion of the stem cell pool (An et al, 2015). We show that Aub acts to maintain a low level of Cbl protein in GSCs and that this repression of Cbl mRNA by Aub is essential for GSC self‐renewal. Furthermore, we find that Cbl is required for GSC differentiation, thereby identifying a role for Cbl in the regulation of yet another stem cell lineage.
This study reveals the function of Aub and piRNAs in GSC self‐renewal through the translational repression of Cbl mRNA, thus highlighting the role of the piRNA pathway as a major post‐transcriptional regulator of gene expression in key developmental decisions.
Results
Aub is intrinsically required in GSCs for their self‐renewal and differentiation
Aub and Ago3 are expressed in GSCs, and we addressed their function in GSC biology (Brennecke et al, 2007; Gunawardane et al, 2007). GSCs can be recognized by their anterior localization in the germarium as well as the presence of the spectrosome, an anteriorly localized spherical organelle in contact with the niche, which is enriched in cytoskeletal proteins (Fig 1A). Cystoblasts also contain a spectrosome that is randomly located in the cell, whereas cells in differentiating cysts are connected by the fusome, a branched structure derived from the spectrosome (Fig 1A). GSCs and differentiating cells were analyzed by immunostaining with an anti‐Hts antibody that labels the spectrosome and fusome, and anti‐Vasa, a marker of germ cells.
We used aub HN2 and aub QC42 strong or null alleles (Schupbach & Wieschaus, 1991) to address the role of Aub in GSC biology. Immunostaining of ovaries with anti‐Hts and anti‐Vasa revealed strong defects in both GSC self‐renewal and differentiation, in aub HN2/QC42 mutant ovaries at 7, 14, and 21 days. A large proportion of aub HN2/QC42 germaria had 0–1 GSC indicating GSC loss, and this defect increased over time (Fig 1B, C and F). A lower proportion of germaria showed differentiation defects, observed as tumors containing undifferentiated cells with spectrosomes (Fig 1D). This phenotype did not markedly increase with time (Fig 1F). Both aub HN2/QC42 phenotypes were almost completely rescued following expression of GFP‐Aub with the germline driver nos‐Gal4, indicating that both defects were due to aub loss of function in germ cells (Fig 1E and F).
Because GSC loss was the most prominent defect in aub mutant ovaries, we focused on this phenotype. We used clonal analysis as an independent evidence to confirm the intrinsic role of aub in GSCs for their self‐renewal. Wild‐type and aub mutant clonal GSCs were generated using the FLP‐mediated FRT recombination system (Golic & Lindquist, 1989) and quantified at three time points after clone induction. We first verified that aub clonal GSCs did not express Aub (Fig EV1A and A’). The percentage of germaria with wild‐type clonal GSCs was stable over time (Fig 1G and I). In contrast, the percentage of germaria with aub mutant clonal GSCs strongly decreased with increasing time after clone induction, showing that aub mutant GSCs cannot self‐renew (Fig 1H and I). The presence of aub mutant clonal differentiated cysts marked with fusomes indicated that aub mutant GSCs were lost by differentiation (Fig 1H). To confirm this conclusion, we used anti‐cleaved Caspase 3 staining to record cell death and address whether the loss of aub mutant GSCs could be due to apoptosis. The number of GSCs expressing cleaved Caspase 3 was low and similar in control (aub HN2/+) and aub mutant GSCs (Fig EV1B–D), indicating that aub mutant GSCs did not undergo cell death.
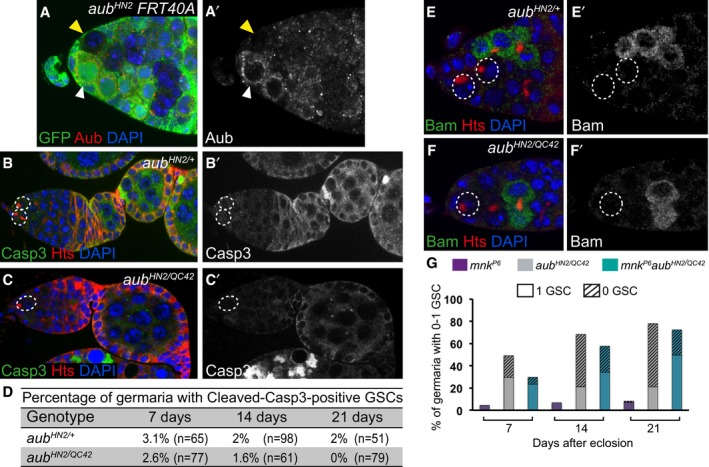
Figure EV1. GSC loss in aub mutant is independent of apoptosis and of Bam expression.
-
A, A’aub HN2 clonal GSCs do not express Aub protein. Immunostaining of mosaic germaria with anti‐GFP (green) and anti‐Aub (red) 7 days after clonal induction. DAPI (blue) was used to visualize DNA. The white and yellow arrowheads indicate the control aub HN2/+ and the aub HN2 clonal GSCs, respectively.
-
B–C’aub mutant GSCs do not die by apoptosis. Immunostaining of control aub HN2/+ (B, B’) and mutant aub HN2/QC42 (C, C’) germaria with anti‐cleaved Caspase3 (green) and anti‐Hts (red). DAPI (blue) was used to visualize DNA. GSCs are outlined.
-
DQuantification of germaria shown in (B–C’) with cleaved Caspase3‐positive GSCs in 7‐, 14‐ and 21‐day‐old females. The percentages are not statistically different using the χ2 test.
-
E–F’aub mutant GSCs do not express Bam. Immunostaining of control aub HN2/+ (E, E’) and mutant aub HN2/QC42 (F, F’) germaria with anti‐Bam (green) and anti‐Hts (red). DAPI (blue) was used to visualize DNA. GSCs are outlined.
- G
Next, we asked whether the GSC self‐renewal defect in aub mutant ovaries could result from Bam expression in GSCs. Anti‐Bam immunostaining of aub mutant ovaries demonstrated that aub mutant GSCs did not express Bam (100% n = 90, Fig EV1E–F’).
Finally, we determined the division rate of aub HN2 GSCs by counting the number of cysts produced by a clonal marked mutant GSC and dividing it by the number of cysts produced by a control unmarked GSC in the same germarium (Jin & Xie, 2007). As expected, the division rate of wild‐type GSCs (FRT40A chromosome) was close to 1 (0.95), whereas that of aub HN2 mutant GSCs was 0.67, indicating a slower division rate in aub mutant GSCs (Fig 1J).
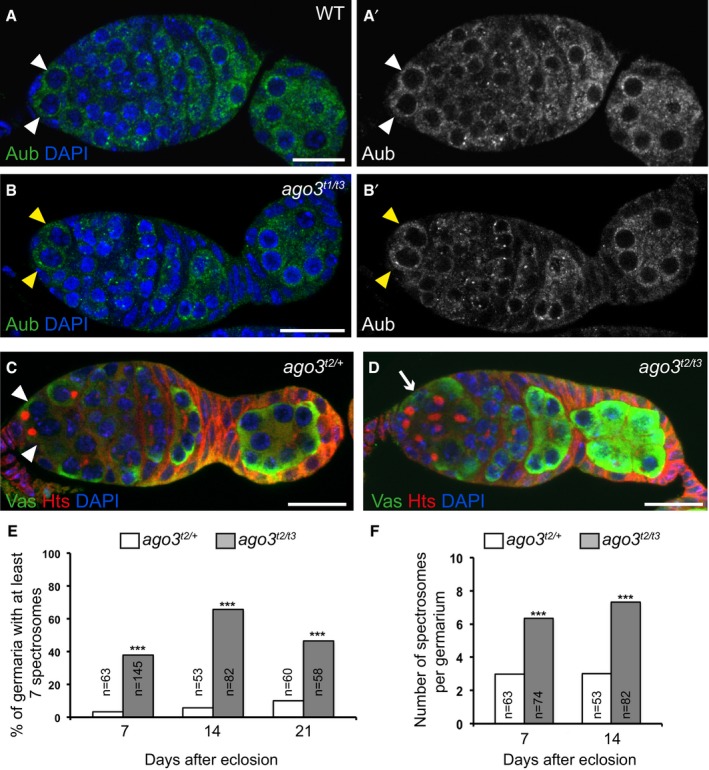
To address the role of Ago3 in GSC biology, we used ago3 mutant alleles that contain premature stop codons, ago3 t1, ago3 t2 , and ago3 t3 (Li et al, 2009a). No GSC loss was recorded in the mutant combination ago3 t2 / t3 in 7‐, 14‐, or 21‐day‐old females, showing that GSC self‐renewal was not affected in the ago3 mutant. We checked that Aub expression was similar in wild‐type and ago3 mutant GSCs (Fig EV2A–B’). In contrast, ago3 t2/t3 females showed a prominent defect in GSC differentiation, with a large proportion of germaria having a higher number of undifferentiated germ cells with a spectrosome (two to four in control germaria, versus six to nine in ago3 t2/t3 germaria; Fig EV2C–F).
Figure EV2. GSC tumor phenotype in ago3 mutant germaria.
-
A–B’Immunostaining of wild‐type (A, A’) and ago3 t1/t3 mutant (B, B’) germaria with anti‐Aub (green) showing that Aub expression is not affected in ago3 mutant GSCs. DAPI (blue) was used to visualize DNA. White and yellow arrowheads indicate wild‐type and mutant GSCs, respectively.
-
C, DImmunostaining of control ago3 t2/+ (C) and ago3 t2/t3 mutant (D) germaria with anti‐Vasa (green) and anti‐Hts (red). DAPI (blue) was used to visualize DNA. White arrowheads indicate GSCs, and the white arrow indicates the increased number of cells with spectrosomes.
-
E, FQuantification of germaria shown in (C, D) with increased number of spectrosomes (E) and quantification of spectrosomes per germarium (F). The number of scored germaria (n) is indicated. ***P‐value < 0.001 using the χ2 test in (E) and the two‐tailed Student's t‐test in (F).
Together, these results demonstrate that Aub is required intrinsically in GSCs for their self‐renewal and differentiation. Aub maintains GSCs by preventing their differentiation independently of Bam expression. In contrast, Ago3 is specifically involved in GSC differentiation.
Aub function in GSC self‐renewal is partly independent of Chk2
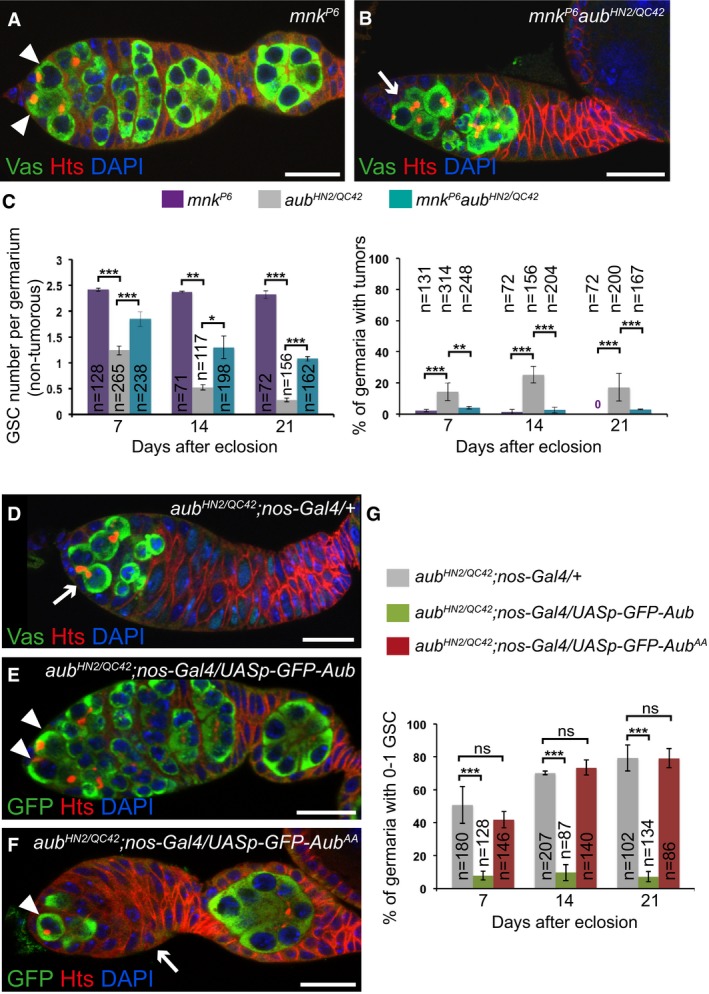
The Chk2‐dependent DNA damage checkpoint is activated in several piRNA pathway mutants, leading to developmental defects during mid‐oogenesis and, in turn, defective dorsoventral and anteroposterior embryonic patterning (Klattenhoff et al, 2007; Pane et al, 2007). These developmental defects are partially rescued in double mutants of Chk2 kinase (mnk mutant) and different piRNA pathway components. DNA damage in piRNA pathway mutants is thought to result from transposable element transposition. Mobilization of P‐elements in crosses that induce hybrid dysgenesis (i.e., the crossing of females devoid of P‐elements with males that contain P‐elements) leads to a block in GSC differentiation, which is partially rescued by mutation in Chk2 (Rangan et al, 2011). To address whether the defects in GSC self‐renewal and differentiation in aub mutant ovaries might be due to activation of the Chk2‐dependent checkpoint, we analyzed mnk p6 aub HN2/QC42 double‐mutant ovaries using anti‐Hts and anti‐Vasa immunostaining. The aub mutant tumor phenotype of undifferentiated cell accumulation was almost completely rescued by mnk p6 , demonstrating that this phenotype depended on Chk2 (Fig 2A–C). In contrast, the GSC loss phenotype was only partially rescued by mnk p6 and remained visible in a large proportion of double mutant germaria (Figs 2A–C and EV1G), showing that this defect was in part independent of Chk2 checkpoint activation.
Figure 2. The role of Aub in GSC self‐renewal is partially independent of Chk2 and requires its loading with piRNAs.
-
A, BImmunostaining of germaria with anti‐Vasa (green) and anti‐Hts (red). DAPI (blue) was used to visualize DNA. Examples of mnk P6 and mnk P6 aub HN2/QC42 germaria are shown. White arrowheads indicate GSCs; the white arrow indicates GSC loss.
-
CQuantification of the number of GSCs per non‐tumorous germarium, and of germaria with GSC tumors, in the indicated genotypes. The number of scored germaria (n) is indicated. Error bars represent standard deviation. The two‐tailed Student's t‐test and the χ2 test were used in the left and right panels, respectively. ***P‐value < 0.001, **P‐value < 0.01, *P‐value < 0.05.
-
D–FImmunostaining of germaria from 7‐day‐old females with anti‐Vasa or anti‐GFP (green) and anti‐Hts (red). DAPI (blue) was used to visualize DNA. aub HN2/QC42; nos‐Gal4/+ was used as a negative control. Examples of rescue in aub HN2/QC42; nos‐Gal4/UASp‐GFP‐Aub germarium (E), and of lack of rescue in aub HN2/QC42; nos‐Gal4/UASp‐GFP‐Aub AA germarium (F). White arrowheads indicate GSCs; white arrows indicate GSC loss in (D) and germ cell loss in (F).
-
GQuantification of mutant germaria with 0–1 GSC shown in (D–F). The number of scored germaria (n) is indicated. Error bars represent standard deviation. ***P‐value < 0.001, ns, non‐significant, using the χ2 test.
These results reveal that the mild defect in GSC differentiation in the aub mutant is due to activation of the DNA damage checkpoint. In contrast, the prominent GSC self‐renewal defect is partly independent of the DNA damage checkpoint activation, consistent with an additional more direct role of Aub in GSC self‐renewal.
Aub loading with piRNAs is required for GSC self‐renewal
To determine whether the role of Aub in GSC self‐renewal depends on its loading with piRNAs, we used an Aub double point mutant in the PAZ domain that is unable to bind piRNAs (AubAA; Barckmann et al, 2015). In contrast to UASp‐GFP‐Aub, which was able to rescue the GSC loss phenotype in aub mutant flies when expressed with nos‐Gal4 (Fig 1E and F), expression of UASp‐GFP‐Aub AA at similar levels (Fig EV3A–C) failed to rescue this phenotype (Fig 2D–G). These data indicate that Aub loading with piRNAs is required for GSC self‐renewal. To confirm this result, we used a piRNA pathway mutant in which piRNA biogenesis is strongly compromised. In the absence of Ago3, piRNAs are produced through Aub/Aub homotypic ping‐pong (Li et al, 2009a) and these piRNAs could be used in Aub‐dependent regulation in GSCs, consistent with the lack of GSC self‐renewal defect in ago3 mutant. In contrast, armitage (armi) encodes an RNA helicase that is essential for piRNA biogenesis (Cook et al, 2004; Malone et al, 2009). Immunostaining of armi 1/72 ovaries with anti‐Hts and anti‐Vasa revealed a GSC loss that increased with time (Fig EV3D–F), showing a function for armi in GSC self‐renewal.
Figure EV3. Expression of GFP‐Aub transgenes in GSCs, and GSC loss phenotype in armi mutant.
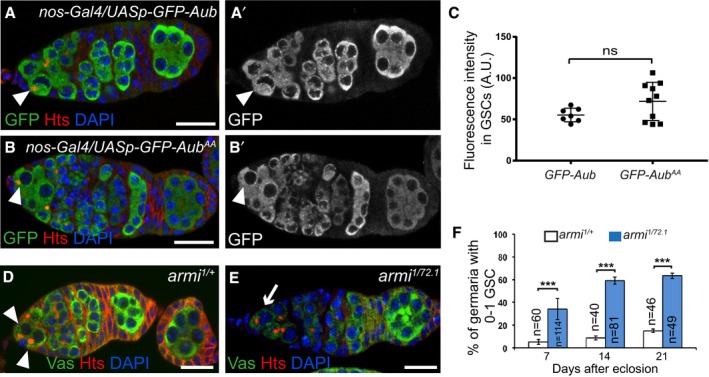
-
A–B’GFP‐Aub and GFP‐AubAA are expressed at similar levels in GSCs using the nos‐Gal4 driver. Immunostaining of nos‐Gal4/UASp‐GFP‐Aub (A, A’) and nos‐Gal4/UASp‐GFP‐Aub AA (B, B’) ovaries with anti‐GFP (green) and anti‐Hts (red). DAPI (blue) was used to visualize DNA. White arrowheads indicate GSCs.
-
CQuantification of GFP‐Aub and GFP‐AubAA protein levels in GSCs using fluorescence intensity of immunostaining with anti‐GFP shown in (A’, B’). Fluorescence intensity was measured in arbitrary units using the ImageJ software. Horizontal bars correspond to the mean and standard deviation. ns, non‐significant using the two‐tailed Student's t‐test. The number of cells analyzed is shown in the figure as dots or squares.
-
D, EGSC self‐renewal defect in armi mutant. Immunostaining of control armi 1/+ (D) and armi 1/72.1 mutant (E) germaria with anti‐Vasa (green) and anti‐Hts (red). DAPI (blue) was used to visualize DNA. White arrowheads indicate GSCs; the white arrow indicates reduced number of GSCs.
-
FQuantification of germaria with 0–1 GSC shown in (D, E), in 7‐, 14‐ and 21‐day‐old females. The number of scored germaria (n) is indicated. Error bars represent standard deviation. ***P‐value < 0.001 using the χ2 test.
We conclude that Aub function in GSC self‐renewal depends on its loading with piRNAs.
Aub interacts with the CCR4‐NOT complex for GSC self‐renewal
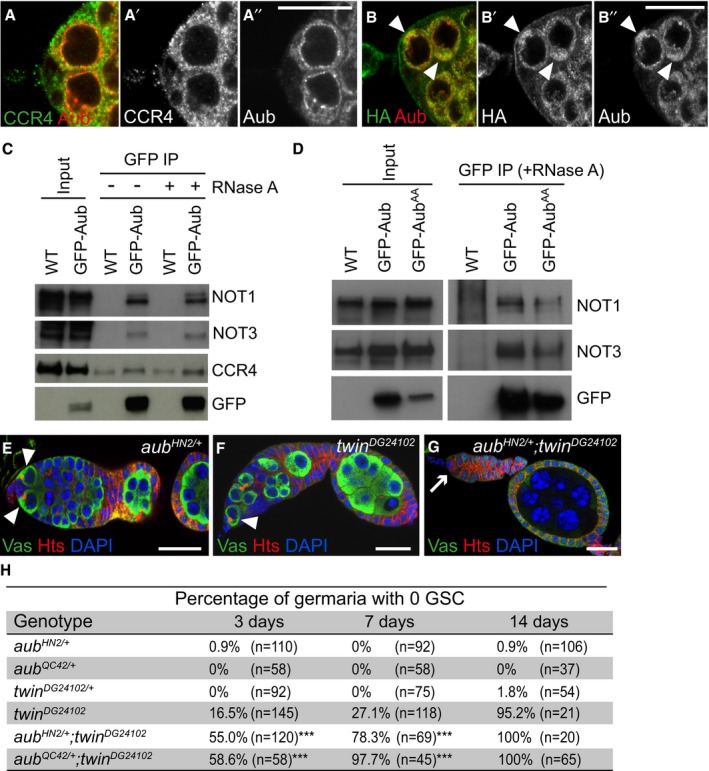
Previous reports have shown that PIWI proteins can recruit the CCR4‐NOT deadenylation complex to repress mRNAs at the post‐transcriptional level (Rouget et al, 2010; Gou et al, 2014). In the Drosophila embryo, Aub is in complex with CCR4, independently of RNA (Rouget et al, 2010). To address whether Aub might act through a similar mode of action in GSCs, we analyzed Aub and CCR4 colocalization. CCR4 is present diffusely in the cytoplasm and accumulates in cytoplasmic foci, in GSCs (Joly et al, 2013). Aub also has a diffuse distribution in the cytoplasm and is present in foci that surround the nucleus collectively referred to as “nuage” (Harris & Macdonald, 2001). Colocalization occurred in diffusely distributed pools of proteins and occasionally in foci (Fig 3A–A’’), consistent with deadenylation not taking place in foci (Joly et al, 2013). We then overexpressed CCR4‐HA in GSCs using nos‐Gal4 and found that CCR4‐HA was able to recruit Aub in discrete cytoplasmic regions where CCR4‐HA had accumulated, consistent with the presence of CCR4 and Aub in the same complex in GSCs (Fig 3B–B’’). Coimmunoprecipitation experiments in ovaries revealed that GFP‐Aub was able to coprecipitate the NOT1, NOT3, and CCR4 subunits of the CCR4‐NOT deadenylation complex, either in the presence or absence of RNA (Fig 3C). We checked that the coprecipitation of CCR4‐NOT subunits was maintained with the mutant form of Aub that does not load piRNAs, GFP‐AubAA (Fig 3D).
Figure 3. Physical and genetic interaction between Aub and the CCR4‐NOT complex.
-
A–B’’Immunostaining of wild‐type (A–A’’) or nos‐Gal/UASp‐CCR4‐HA (B–B’’) germaria with anti‐Aub (red), and anti‐CCR4 or anti‐HA (green). GSCs are shown in (A–A’’). White arrowheads in (B–B’’) indicate cytoplasmic accumulation of CCR4‐HA colocalized with Aub in GSCs.
-
CCo‐immunoprecipitation (IP) of NOT1, NOT3, and CCR4 with GFP‐Aub in ovaries. Wild‐type (WT, mock IP) or nos‐Gal4/UASp‐GFP‐Aub (GFP‐Aub) ovarian extracts were immunoprecipitated with anti‐GFP, either in the absence or the presence of RNase A. Western blots were revealed with anti‐GFP, anti‐NOT1, anti‐NOT3, and anti‐CCR4. Inputs are extracts prior to IP.
-
DCo‐IP of NOT1 and NOT3 with GFP‐Aub or GFP‐AubAA in ovaries. Wild‐type (WT, mock IP), nos‐Gal4/UASp‐GFP‐Aub (GFP‐Aub), or nos‐Gal4/UASp‐GFP‐Aub AA (GFP‐AubAA) ovarian extracts were immunoprecipitated with anti‐GFP in the presence of RNase A. Western blots were revealed with anti‐GFP, anti‐NOT1, and anti‐NOT3. Inputs are extracts prior to IP.
-
E–GGenetic interaction between aub and twin in GSC self‐renewal. Immunostaining of germaria with anti‐Vasa (green) and anti‐Hts (red). DAPI (blue) was used to visualize DNA. Examples of aub HN2/+, twin DG24102 and aub HN2/+; twin DG24102 germaria are shown. White arrowheads indicate GSCs; the white arrow indicates GSC loss.
-
HQuantification of mutant germaria with no GSC in 3‐, 7‐, and 14‐day‐old females of the genotypes shown in (D–F). The number of scored germaria (n) is indicated. ***P‐value < 0.001 using the χ2 test.
The twin gene that encodes the CCR4 deadenylase is essential for GSC self‐renewal (Joly et al, 2013; Fu et al, 2015). To genetically determine whether aub acts together with twin in GSC self‐renewal, we tested whether GSC loss in the hypomorphic allele twin DG24102 (Joly et al, 2013) might be enhanced by reducing the gene dosage of aub. GSC loss in twin DG24102 was accelerated in the presence of heterozygous aub HN2 or aub QC42 mutations, consistent with a role for Aub and CCR4 in the same molecular pathway for GSC self‐renewal (Fig 3E–H).
Together, these results show that Aub and the CCR4‐NOT complex physically interact in GSCs, and cooperate for GSC self‐renewal.
Cbl is an mRNA target of Aub in GSCs
To identify mRNA targets of Aub in GSCs, we looked for candidate genes with a reported role in GSC biology or other stem cell lineages (Appendix Fig S1A). Eight genes were selected, five of which produce mRNAs that directly interact with Aub in embryos (Barckmann et al, 2015). Antibody staining in ovaries containing clonal aub mutant GSCs was used to record potential increased levels of the corresponding proteins in mutant GSCs as compared to control (Appendix Fig S1A–C’’). We found a mild increase in Mei‐P26 and Fused protein levels in aub mutant GSCs, and a more prominent increase in Nos levels. Increased Nos protein levels in aub mutant GSCs suggest that the direct regulation of nos mRNA by Aub occurring in the early embryo is maintained in other biological contexts (Rouget et al, 2010).
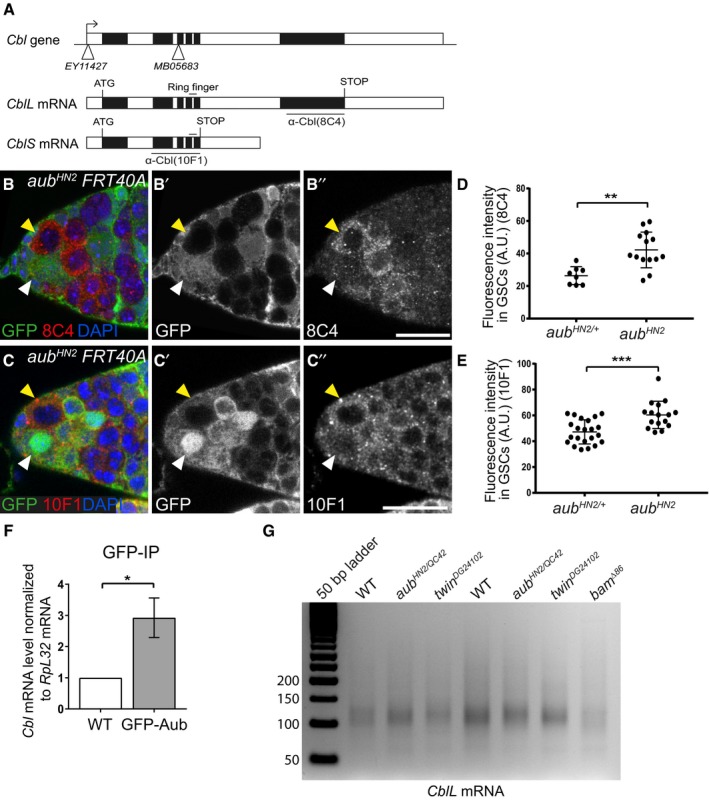
Cbl protein displayed the highest increased levels in aub mutant GSCs compared to control. We thus focused on the possible regulation of the Cbl proto‐oncogene by Aub. Cbl encodes two isoforms through alternative splicing: a long isoform (CblL) and a short isoform (CblS), both of which contain the N‐terminal phosphotyrosine binding domain that binds phosphotyrosine kinases, in addition to a ring finger domain that acts as an E3 ubiquitin ligase (Fig 4A; Robertson et al, 2000). We used two available monoclonal antibodies directed against either the long Cbl isoform (8C4) or both isoforms (10F1; Pai et al, 2006), to analyze the deregulation of Cbl in aub mutant GSCs. Cbl protein levels were significantly increased in aub mutant GSCs as observed with either antibody, although a stronger effect was revealed with the 8C4 antibody (specific to CblL; Fig 4B–E). These results suggest the regulation of CblL mRNA by Aub in the GSCs and are consistent with the reported mRNA expression of CblL in germaria and CblS in later stages of oogenesis (Pai et al, 2006). Quantification of CblL mRNA in germaria, using RT–qPCR, showed that the increased Cbl protein levels in aub mutant did not result from increased mRNA levels (Fig EV4A).
Figure 4. Aub represses Cbl expression in the GSCs.
-
AGenomic organization of the Cbl locus and Cbl mRNAs. Open boxes represent UTRs and introns, and black boxes are exons. The insertion points in the Cbl EY11427 and Cbl MB05683 mutants are represented by white triangles. The region encoding the E3 ubiquitin ligase domain (Ring finger) is indicated. The regions used to raise the 8C4 and 10F1 monoclonal antibodies are underlined.
-
B–C’’Immunostaining of mosaic germaria with anti‐GFP (green), to identify clonal cells by the lack of GFP, and either 8C4 (B–B’’) or 10F1 (C–C’’) monoclonal anti‐Cbl (red). DAPI was used to visualize DNA. White arrowheads indicate aub HN2/+ control GSCs; yellow arrowheads indicate clonal mutant aub HN2 GSCs. Scale bars: 10 μm.
-
D, EQuantification of Cbl protein levels in aub HN2/+ and aub HN2 mutant GSCs using fluorescence intensity of immunostaining with 8C4 or 10F1. Fluorescence intensity was measured in arbitrary units using the ImageJ software. Horizontal bars correspond to the mean and standard deviation. **P‐value < 0.01, ***P‐value < 0.001 using the two‐tailed Student's t‐test. The number of cells analyzed is indicated as the dots in the figure itself.
-
FRNA immunoprecipitation (IP) with anti‐GFP antibody in wild‐type (mock IP) and nos‐Gal4/UASp‐GFP‐Aub ovarian extracts. Cbl mRNA was quantified using RT–qPCR. Normalization was with RpL32 mRNA. Mean of three biological replicates. The error bar represents standard error to the mean. *P‐value < 0.05 using the two‐tailed Student's t‐test.
-
GePAT assay of CblL mRNA. Ovaries from 1‐day‐old (germarium to stage 8) wild‐type, aub and twin mutant females, and from 4‐ to 7‐day‐old bam mutant females were used.
Source data are available online for this figure.
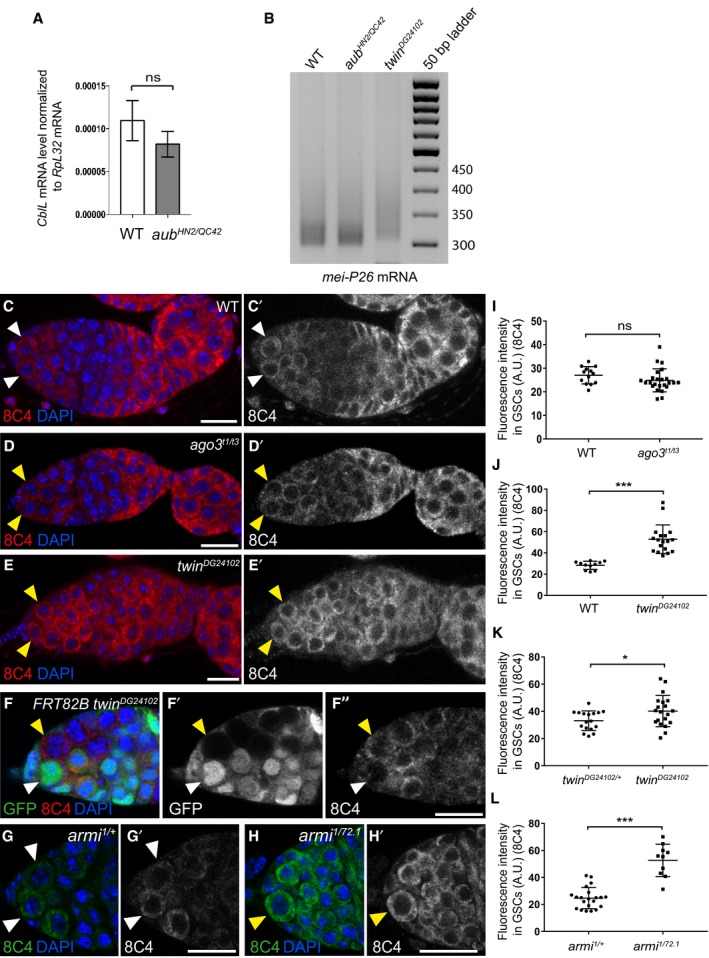
Figure EV4. Cbl protein levels in ago3, twin, and armi mutant GSCs.
-
AQuantification by RT–qPCR of CblL mRNA in dissected germaria/early egg chambers from wild‐type and aub HN2/QC42 females. Normalization was with RpL32 mRNA. Mean of three biological replicates. Error bars represent standard error to the mean. ns, non‐significant using the two‐tailed Student's t‐test.
-
BePAT assay of mei‐P26 mRNA showing elongated poly(A) tail in twin mutant. Ovaries from 1‐day‐old (germarium to stage 8) wild‐type, aub and twin mutant females were used.
-
C–E’Immunostaining of wild‐type (C, C’), ago3 mutant (D, D’), and twin mutant (E, E’) germaria with anti‐Cbl 8C4 antibody (red). DAPI (blue) was used to visualize DNA. White and yellow arrowheads indicate wild‐type and mutant GSCs, respectively.
-
F–F’’Immunostaining of twin DG24102 mosaic germaria 7 days after clone induction, with anti‐GFP (green) and 8C4 anti‐Cbl antibody (red). DAPI (blue) was used to visualize DNA. White arrowheads indicate control (twin DG24102/+) GSCs; yellow arrowheads indicate twin DG24102 mutant GSCs.
-
G–H’Immunostaining of control armi 1/+ (G, G’) and armi 1/72.1 mutant (H, H’) germaria with anti‐Cbl 8C4 antibody (green). DAPI (blue) was used to visualize DNA. White and yellow arrowheads indicate wild‐type and mutant GSCs, respectively.
-
I–LQuantification of Cbl protein levels in GSCs using fluorescence intensity of immunostaining with 8C4 antibody. Fluorescence intensity was measured in arbitrary units using ImageJ. Horizontal bars correspond to the mean and standard deviation. ***P‐value < 0.001, *P‐value < 0.05, ns, non‐significant, using the two‐tailed Student's t‐test. The number of cells analyzed is shown in the figure as dots or squares (I, J).
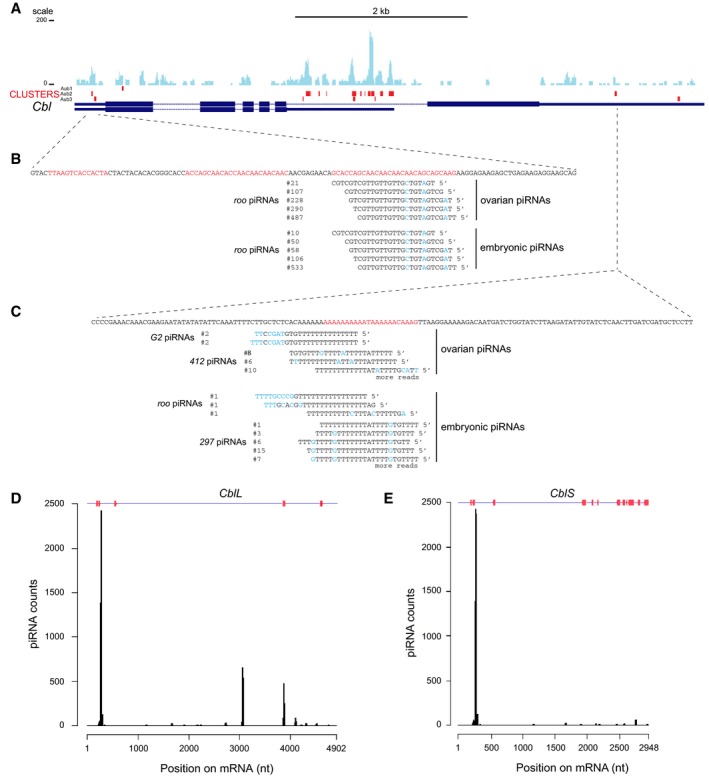
We used RNA immunoprecipitation with Aub to confirm the potential regulation of Cbl by Aub at the mRNA level. GFP‐Aub protein was immunoprecipitated from UASp‐GFP‐Aub/nos‐Gal4 or wild‐type (mock immunoprecipitation) ovaries. Quantification of Cbl mRNA by RT–qPCR revealed that it was enriched in GFP‐Aub immunoprecipitates as compared to the mock immunoprecipitates (Fig 4F). Consistent with Aub interaction with Cbl mRNA, Aub iCLIP experiments in 0‐ to 2‐h embryos have revealed the direct binding of Aub to Cbl mRNA (Barckmann et al, 2015). In addition, a recent study independently reported the role of Aub in GSC self‐renewal and differentiation (Ma et al, 2017). GFP‐Aub iCLIP experiments in cultured GSCs were performed in this study, and we found statistically significant GFP‐Aub crosslinks in Cbl mRNA 5′‐ and 3′UTRs, demonstrating Aub direct binding to Cbl mRNA in GSCs (Fig 5A). The other mRNAs identified as potential targets of Aub, nos, mei‐P26, and fused were also significantly crosslinked by GFP‐Aub in GSCs (Appendix Fig S2).
Figure 5. GFP‐Aub iCLIP in GSCs and targeting by piRNAs, in Cbl mRNAs.
-
AGFP‐Aub crosslinks in cultured GSCs (Ma et al, 2017), in Cbl mRNAs. Thin boxes are 5′‐ and 3′UTRs, lines are introns, and thick boxes are exons. The coverage of Cbl mRNAs with iCLIP reads is shown in blue. Significant crosslink clusters are shown in red.
-
B, CSequences of crosslinked regions in 5′UTR (B) and 3′UTR (C) of CblL mRNA. The nucleotides (nt) in significant crosslink clusters are in red. The sequence, occurrence, and origin of anti‐sense piRNAs from ovarian and embryonic libraries are indicated. Mismatched nt are in blue.
-
D, ECoverage of CblL (D) and CblS (E) mRNAs with ovarian and embryonic piRNAs. Significant crosslink clusters are indicated in red at the top of the graphs.
We then analyzed the potential deregulation of Cbl mRNA in the absence of CCR4 deadenylase. We performed Cbl immunostaining using the 8C4 antibody, in twin mutant ovaries, as well as in ovaries containing clonal twin mutant GSCs. Similar to the results observed in aub mutant GSCs, the levels of CblL protein were increased in twin mutant GSCs in both experimental conditions (Fig EV4C, C’, E–F”, J and K). To address whether the regulation of Cbl mRNA by Aub and CCR4 occurred at the level of poly(A) tail length, we measured the poly(A) tail of CblL mRNA in early ovaries using ePAT assays. ePAT assays from bam Δ86 ovaries that only contained undifferentiated GSC‐like cells confirmed the presence of the long CblL mRNA in these cells (Fig 4G). CblL poly(A) tails were not notably affected in twin and aub mutant early ovaries as compared to wild‐type (Fig 4G), whereas mei‐P26 used as a control mRNA undergoing deadenylation by CCR4 (Joly et al, 2013) had longer poly(A) tails in twin mutant (Fig EV4B). This suggested that CblL mRNA regulation by Aub/CCR4‐NOT did not involve deadenylation. Indeed, the CCR4‐NOT complex has the capacity to repress mRNA translation independently of its role in deadenylation, through the recruitment of translational repressors (Chekulaeva et al, 2011; Chen et al, 2014; Mathys et al, 2014).
Taken together, these results reveal Cbl mRNA as a direct target of Aub/CCR4‐NOT‐dependent translational repression in GSCs.
piRNAs are involved in Cbl mRNA regulation by Aub
We used embryonic (germline) and ovarian (somatic and germline) piRNA libraries to identify piRNAs complementary to Cbl mRNA. Strikingly, using strong complementarities (0–3 mismatches, or 20‐nt seed/16‐nt seed without mismatch in the seed, Barckmann et al, 2015), transposable element piRNA target sites were found overlapping significant Aub crosslinks identified in GSCs, in CblL mRNA 5′‐ and 3′UTRs (Fig 5B and C). Mapping of complementary piRNAs to the entire length of Cbl mRNAs identified discrete peaks overlapping significant crosslinks in CblL (Fig 5D). These peaks were reduced, however, in CblS 3′UTR that was heavily bound by Aub (Fig 5E), possibly due to a different mode of Aub binding in this region (e.g., involving reduced piRNA base‐pairing and/or additional RNA binding proteins).
To functionally address the role of piRNAs in the regulation of Cbl mRNA by Aub, we analyzed Cbl protein levels in GSCs from armi mutant females, in which piRNA biogenesis is strongly affected (Malone et al, 2009). Immunostaining of armi mutant germaria with anti‐Cbl antibody 8C4 revealed a significant increase in CblL levels in armi mutant GSCs (Fig EV4G–H’ and L). In contrast, CblL levels were not increased in GSCs from ago3 mutant, in which piRNAs were produced, and which did not display GSC loss (Fig EV4C–D’ and I).
We conclude that piRNAs that target CblL mRNA by complementarity guide Aub interaction with Cbl.
Regulation of Cbl mRNA by Aub is required for GSC self‐renewal
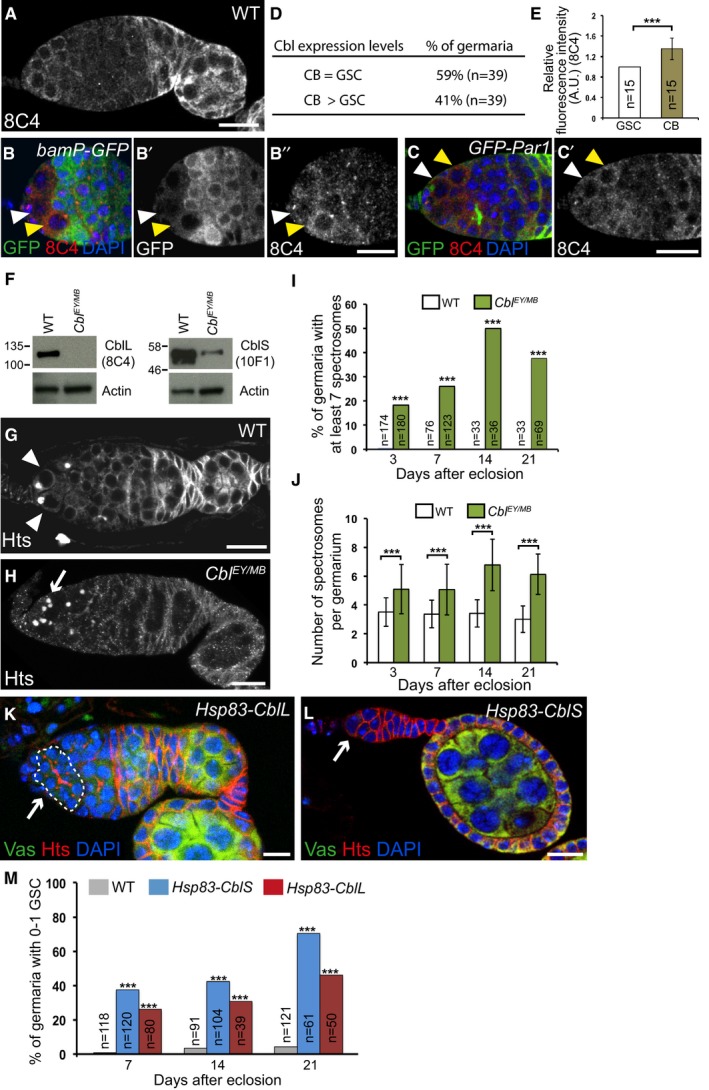
To determine whether translational repression of Cbl mRNA by Aub is relevant to GSC self‐renewal, we analyzed the function of Cbl in GSCs. We first determined the expression pattern of Cbl in the germarium using immunostaining. The bamP‐GFP reporter was used to mark the cystoblast to 8‐cell cysts (Chen & McKearin, 2003). Co‐staining of both Cbl antibodies with GFP revealed the presence of Cbl at low levels in GSCs, cystoblasts, and early (2‐cell) cysts. This was followed by reduced levels in the remaining dividing and differentiating cysts up to region 2a of the germarium, and stronger expression starting in region 2b in both somatic and germ cells (Fig 6A–B’’). Strikingly, co‐staining with anti‐Cbl and anti‐GFP antibodies of germaria expressing GFP‐Par1 to visualize spectrosomes revealed a transient increase (visible in ≈40% of germaria) of Cbl protein levels in cystoblasts as compared to GSCs (Fig 6C–E, Appendix Fig S3A and B).
Figure 6. Cbl is required for GSC differentiation.
-
A–ECbl expression in germaria. (A) Immunostaining of wild‐type germaria with anti‐Cbl 8C4. (B–B’’) Immunostaining of bamP‐GFP germaria that express GFP under the bam promoter, with anti‐GFP (green) and anti‐Cbl 8C4 (red). (C–C’) Immunostaining of germaria expressing GFP‐Par1 to label spectrosomes with anti‐GFP (green) and anti‐Cbl 8C4 (red). DAPI (blue) was used to visualize DNA. White arrowheads indicate GSCs; yellow arrowheads indicate cystoblasts. (D) Quantification of germaria showing similar or increased Cbl protein levels in cystoblasts as compared to GSCs. (E) Quantification of increased Cbl protein levels in cystoblasts using fluorescence intensity of immunostaining with 8C4 antibody. Fluorescence intensity was measured in arbitrary units using ImageJ, in germaria showing increased Cbl levels in cystoblasts. The number of scored cells (n) is indicated. Intensity in GSCs was set to 1. The error bar represents standard deviation. ***P‐value < 0.001 using the two‐tailed Student's t‐test.
-
FWestern blots of protein extracts from wild‐type and Cbl mutant ovaries showing CblL (left panel) and CblS (right panel) revealed with 8C4 and 10F1 antibodies, respectively. A low level of CblS remained, while no CblL was present in the Cbl EY11427/MB05683 combination. The 10F1 antibody recognizes very poorly CblL in western blot.
-
G–JCbl is required for germline differentiation. Immunostaining of wild‐type and Cbl EY11427/MB05683 germaria with anti‐Hts to visualize spectrosomes and fusomes. White arrowheads indicate GSCs; the white arrow indicates increased number of spectrosomes. (I) Quantification of germaria with increased number of spectrosomes, and (J) quantification of spectrosomes per germarium. The number of scored germaria (n) is indicated in (I). Error bars represent standard deviation. ***P‐value < 0.001 using the χ2 test in (I), and the two‐tailed Student's t‐test in (J).
-
K–MCbl induces GSC differentiation. Immunostaining of germaria overexpressing Cbl with Hsp83‐CblL (K) or Hsp83‐CblS (L) with anti‐Vasa (green) and anti‐Hts (red). DNA (blue) was revealed with DAPI. White arrows indicate a cyst in the GSC niche (K, outlined) or the loss of GSCs and germ cells (L). (M) Quantification of germaria with 0–1 GSC in the indicated genotypes. The number of scored germaria (n) is indicated. ***P‐value < 0.001 using the χ2 test.
Null alleles of Cbl are larval/pupal lethal (Pai et al, 2000, 2006). To address a potential role for Cbl in GSC biology in adults, we used two Cbl insertion alleles: Cbl EY11427, which contains a P‐UAS insertion in Cbl 5′UTR (Bellen et al, 2004), and Cbl MB05683, which contains a Minos‐based insertion in the third exon (Metaxakis et al, 2005; Fig 4A). Cbl EY11427/MB05683 transheterozygotes mostly died at the pupal stage, with a small number of adult escapers surviving for 3–4 days at 25°C. The number of escapers and their survival time increased at 22°C. Western blots of ovaries from these escapers revealed that the CblL isoform was absent in this mutant combination and that CblS levels were strongly reduced (Fig 6F). Anti‐Hts immunostaining of ovaries from 3‐, 7‐, 14‐, and 21‐day‐old Cbl EY11427/MB05683 mutant females revealed a defect in GSC differentiation. The number of undifferentiated germ cells containing a spectrosome in Cbl mutant germaria was higher than in the wild‐type and increased with time (two to four spectrosomes in the wild‐type versus six to nine in Cbl mutant; Fig 6G–J).
In a reverse experiment, we overexpressed the long or short isoforms of Cbl in germ cells using the Hsp83‐CblL and Hsp83‐CblS transgenes (Pai et al, 2006) and analyzed germaria using immunostaining with anti‐Hts and anti‐Vasa. Consistent with a role for Cbl in GSC differentiation, overexpressing Cbl in the germline led to a GSC loss that increased with time (Fig 6K–M). Germaria with a lower number of germ cells were also visible (Appendix Fig S3C and D). These results identify a new Cbl function in GSC homeostasis.
Finally, we addressed whether the translational repression of Cbl mRNA by Aub in the GSCs has a functional role in their self‐renewal. If increased Cbl protein levels in aub mutant ovaries contribute to the aub GSC loss phenotype, we would expect to reduce this phenotype by reducing Cbl gene dosage. We used both Cbl EY11427 and Cbl MB05683 heterozygous mutants in combination with aub HN2/QC42 and examined the germaria of these females at three time points by immunostaining with anti‐Hts and anti‐Vasa. Strikingly, the GSC loss phenotype in aub mutants was significantly rescued in the presence of both Cbl heterozygous mutants (Fig 7A–C). These results show that the regulation of Cbl by Aub is essential for GSC self‐renewal.
Figure 7. Regulation of Cbl by Aub in the GSCs is essential for their self‐renewal.
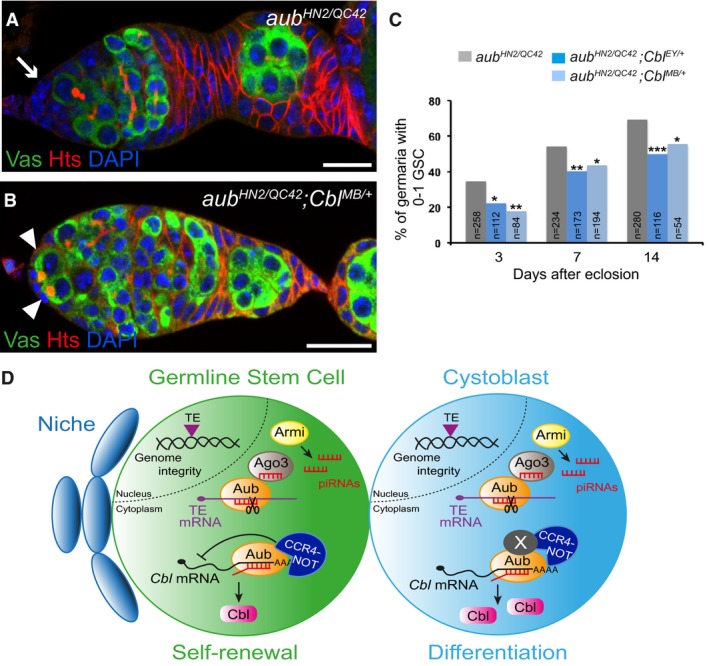
-
A, BImmunostaining of germaria from aub HN2/QC42 (A) and aub HN2/QC42; Cbl MB/+ (B) females with anti‐Vasa (green) and anti‐Hts (red). DAPI (blue) was used to visualize DNA. The white arrow indicates the lack of GSCs; white arrowheads indicate GSCs. Scale bars: 10 μm.
-
CQuantification of mutant germaria with 0–1 GSC in the indicated genotypes. The number of scored germaria (n) is indicated. ***P‐value < 0.001, **P‐value < 0.01, *P‐value < 0.05, using the χ2 test.
-
DModel of Aub function in GSCs. Aub is required intrinsically in GSCs for their self‐renewal and differentiation. Aub function in self‐renewal depends on translational repression of Cbl mRNA in GSCs through the recruitment of the CCR4‐NOT complex. In cystoblasts, this translational repression is decreased, likely through the implication of at least another factor (X). As is the case for other translational controls in the GSC lineage, Aub/CCR4‐NOT acts in fine‐tuning Cbl levels. Aub function in GSC differentiation depends on activation of the Chk2 DNA damage checkpoint, consistent with a role in transposable element (TE) repression to maintain genome integrity; Ago3 has the same role in GSC differentiation.
Together, these data identify a new role for Cbl in GSC differentiation and reveal an essential role of Cbl mRNA translational repression by Aub for GSC self‐renewal.
Discussion
Here, we have demonstrated that aub is required intrinsically in GSCs for their self‐renewal. The main phenotype of aub mutant GSCs is a reduced capacity to self‐renew, leading to their progressive loss by differentiation. Our results show that this aub function depends only in part on activation of the DNA damage response mediated by Chk2 kinase and involves the regulation of protein‐coding genes at the mRNA level.
We provide evidence that the Aub mechanism of action involves the recruitment of the CCR4‐NOT deadenylation complex (Fig 7D). Aub and subunits of CCR4‐NOT, NOT1, NOT3, and CCR4 form a complex in ovaries. Moreover, aub heterozygous mutants significantly increase the GSC loss phenotype of a twin hypomorphic mutant. Translational repression plays an essential role in GSC biology, for both GSC self‐renewal and differentiation. Through their interaction with CCR4‐NOT, the translational repressors Nos and Pum repress the translation of differentiation factors in GSCs for their self‐renewal (Joly et al, 2013); in turn, Pum and Brat recruit CCR4‐NOT in the cystoblasts for their differentiation by repressing the translation of self‐renewal factors (Harris et al, 2011; Newton et al, 2015). Therefore, the CCR4‐NOT complex is central to mRNA regulation for GSC homeostasis. We have identified Aub as a novel interactor of the CCR4‐NOT deadenylation complex involved in translational repression of Cbl mRNA for GSC self‐renewal. Interestingly, repression of Cbl mRNA by Aub/CCR4‐NOT does not involve poly(A) tail shortening. This is consistent with the reported role of the CCR4‐NOT complex in translational repression, independent of deadenylation. In this mode of regulation, the CCR4‐NOT complex serves as a platform to recruit translational repressors such as DDX6/Me31B and Cup (Igreja & Izaurralde, 2011; Chen et al, 2014; Mathys et al, 2014). Whether CCR4‐NOT mediates deadenylation or translational repression might depend on the set of RNA binding proteins involved in its recruitment to specific mRNAs. mei‐P26 mRNA to which CCR4‐NOT is recruited by Nos and Pum, in addition to Aub, undergoes deadenylation, whereas Cbl mRNA does not. In addition, a role in translational regulation has been proposed for two mouse PIWI proteins, MILI and MIWI. MILI associates with the translation factor eIF3A, while both MILI and MIWI associate with the cap‐binding complex (Grivna et al, 2006; Unhavaithaya et al, 2009). Similarly, Aub might also regulate mRNA translation through direct interaction with translation factors. Aub and translation initiation factors were recently reported to coprecipitate when overexpressed in S2 cells (Ma et al, 2017).
A major point addressed here is the characterization of the role of the piRNA pathway in GSC biology. Until now, the function of Piwi had been thoroughly analyzed in GSCs. Piwi is involved in somatic niche cells for GSC differentiation, as well as in GSCs for their maintenance and differentiation (Cox et al, 1998, 2000; Jin et al, 2013; Ma et al, 2014). Strikingly, the molecular mechanisms underlying the somatic role of Piwi for GSC differentiation are not related to transposable element repression, but gene regulation. Piwi interacts with components of the PRC2 complex, thus limiting PRC2 binding to chromatin and transcriptional repression (Peng et al, 2016). Piwi also represses c‐Fos mRNA via its processing into piRNAs (Klein et al, 2016). Cutoff, a piRNA pathway component that is required for piRNA production, is reported to play a role in GSC self‐renewal and differentiation, with a partial rescue of the differentiation defects by mutation in the Chk2 kinase (Chen et al, 2007; Pane et al, 2011). Here, we describe the GSC phenotypes in three additional piRNA pathway mutants: aub, ago3, and armi (Fig 7D). Notably, the three mutants display different defects in GSC biology. Specifically, ago3 mutants only have differentiation defects, whereas the most prominent phenotype of aub and armi mutants is GSC loss. This suggests that different molecular pathways affect GSC biology in these mutants. Importantly, the effect of transposition per se on GSC homeostasis has been analyzed using P‐element mobilization in PM hybrid dysgenesis crosses and corresponds to defects in GSC differentiation (Rangan et al, 2011). These defects are partially rescued by Chk2 mutation, indicating that they arise following DNA damage. Accordingly, differentiation defects in aub mutant GSCs are almost completely rescued by Chk2 mutation and might result from transposition. In contrast, GSC loss, the main phenotype in aub mutants, is less strongly rescued by the Chk2 mutant, suggesting that it does not only depend on transposable element mobilization or DNA damage. These results are consistent with our identification of Cbl mRNA regulation by Aub for GSC self‐renewal and might be explained by both roles of Aub in transposition repression and cellular mRNA regulation for GSC self‐renewal.
Aub function in transposable element regulation occurs through direct cleavage of transposable element mRNAs, guided by complementary piRNAs (Brennecke et al, 2007). Thus, the contribution of Aub/CCR4‐NOT interaction to transposable element regulation is expected to be minor. Nonetheless, CCR4 was reported to specifically regulate Het‐A transposable element (Morgunova et al, 2015). Whether Het‐A repression does contribute to CCR4 function in GSC self‐renewal, in addition to its major role in mRNA regulation, remains unknown. The twin GSC loss phenotype was reported to be partially rescued upon Chk2 downregulation using RNAi; however, the rescue was not robust and would require validation using mutants (Fu et al, 2015).
Post‐transcriptional regulation of cellular mRNAs by Aub and other PIWI proteins involves piRNAs that target mRNAs by complementarity (Rouget et al, 2010; Gou et al, 2014; Barckmann et al, 2015; Goh et al, 2015). Here, we show that: (i) the GSC loss in aub mutant is not rescued with AubAA that is unable to bind piRNAs, (ii) germline piRNAs have the potential to target Cbl mRNAs at Aub binding sites, and (iii) Armi, which has an essential function in piRNA biogenesis, is required for Cbl regulation in GSCs. These results are consistent with a role of piRNAs in the regulation of mRNAs by Aub in GSCs. These findings broaden the developmental functions of Aub and piRNAs as regulators of gene expression in various biological processes, and highlight their key role in developmental transitions.
A recent study reporting the role of Aub in mRNA regulation in GSCs suggested that this Aub function was independent of piRNAs (Ma et al, 2017). This was based on the lack of piRNA target site enrichment in the regions bound by Aub within 3′UTRs of Aub target mRNAs. This conclusion might differ upon utilization of different piRNA complementarities. Strikingly, this study also reported the GSC loss phenotype of a mutant for dunce (dnc), another Aub target mRNA (Ma et al, 2017). This dnc mutation, dnc ΔpiR1 (called dnc 3′utrΔ1 in Ma et al, 2017) is a CRISPR‐based deletion of a piRNA target site (Barckmann et al, 2015). Thus, the GSC loss phenotype of this mutant supports the role of piRNAs in mRNA regulation by Aub in GSCs.
Importantly, our study reveals a new function for Cbl in GSC biology. Overexpression and mutant analysis show the implication of Cbl in GSC differentiation. Cbl is a tumor suppressor gene encoding an E3 ubiquitin ligase that binds and represses receptor tyrosine kinases. In particular, Cbl regulates epidermal growth factor receptor (Egfr) signaling through ubiquitination and degradation of activated Egfr (Mohapatra et al, 2013). In Drosophila, the regulation of Gurken/Egfr signaling by Cbl is involved in dorsoventral patterning during oogenesis (Pai et al, 2000, 2006; Chang et al, 2008). In humans, mutations in Cbl that disrupt E3 ubiquitin ligase activity lead to myeloid neoplasms (Sanada et al, 2009). Cbl plays a key role in hematopoietic stem cell homeostasis, in the maintenance of their quiescence and their long‐term self‐renewal capacity (An et al, 2015). Our data thus add a biological function for Cbl to yet another stem cell lineage.
Other E3 ubiquitin ligases are known to play important roles in GSC biology. Mei‐P26 and Brat, two members of the conserved Trim‐NHL family of proteins, contain E3 ubiquitin ligase domains and have roles in stem cell lineages. Mei‐P26 in particular is involved in GSC self‐renewal and differentiation, and this dual function partly depends on a very tight regulation of its levels in these cells (Neumuller et al, 2008; Li et al, 2012; Joly et al, 2013). Smurf is another E3 ubiquitin ligase that plays a major role in GSC differentiation. Specifically, Smurf associates with the Fused serine/threonine kinase in cystoblasts to degrade the Thickveins receptor and thus repress BMP signaling, the main signaling pathway in the GSC lineage. This mechanism generates a steep gradient of BMP activity between GSCs and cystoblasts (Xia et al, 2010). Cbl might participate in the regulation of Egfr signaling or other pathways in the GSC lineage. Although the role of Egfr in adult GSC biology in the ovary has not yet been addressed, this signaling pathway is involved in regulating the primordial germ cell number in the larval gonad (Gilboa & Lehmann, 2006) and in the GSC mitotic activity in adult males (Parrott et al, 2012).
Our data highlight an important role of Aub in fine‐tuning Cbl levels for GSC self‐renewal. Intriguingly, a recent study reported a functional link between Aub and dFMR1 (Fragile X mental retardation protein; Bozzetti et al, 2015). dFMR1 was described previously to regulate Cbl mRNA during oogenesis (Epstein et al, 2009) and to play a role in GSC biology (Yang et al, 2007), thus pointing to the possibility that Aub and dFMR1 might cooperate for Cbl regulation. Further studies will be required to address this question.
Translational regulation is known to be central for cell fate choices in adult stem cell lineages. RNA binding proteins and microRNAs have a recognized regulatory function in female GSCs to trigger cell fate changes through cell‐specific regulation of mRNA targets. Here, we reveal piRNAs as an additional layer of translational regulators for GSC biology. PIWI proteins and piRNAs are stem cell markers in somatic stem cells of higher organisms, as well as in pluripotent stem cells involved in regeneration in lower organisms (Juliano et al, 2011). This function of piRNAs in translational control is likely to be conserved in these stem cell lineages and might play a key role in stem cell homeostasis.
PIWI proteins and piRNAs are upregulated in a number of cancers, and functional studies in Drosophila have shown that this upregulation participates in cancer progression (Janic et al, 2010; Fagegaltier et al, 2016). This suggests that the role of piRNAs in the translational control of cellular mRNA targets might also be crucial to cancer.
Materials and Methods
Drosophila stocks and genetics
The w 1118 stock was used as a control. The following mutant alleles and transgenic lines were used: aub HN2 and aub QC42 (Schupbach & Wieschaus, 1991), mnk P6 (Abdu et al, 2002 ), twin DG24102 (Joly et al, 2013), armi 1 (Tomari et al, 2004), armi 72.1 (Cook et al, 2004), ago3 t1, ago3 t2 and ago3 t3 (Li et al, 2009a), nos‐Gal4:VP16 (Rorth, 1998), UASp‐CCR4‐HA (Semotok et al, 2005), UASp‐GFP‐Aub (Harris & Macdonald, 2001), UASp‐GFP‐Aub AA (Barckmann et al, 2015), Cbl MB05683 and Cbl EY11427 (Bloomington Drosophila Stock Center), Hsp83‐CblL and Hsp83‐CblS (Pai et al, 2006), bam Δ86 (McKearin & Ohlstein, 1995), bamP‐GFP (Chen & McKearin, 2003), and GFP‐Par1 (Pubq‐GFP‐Par1, a gift from A. González‐Reyes). The recombinant chromosomes mnk P6 aub HN2 and mnk P6 aub QC42 (Klattenhoff et al, 2007) were used. Adult females were dissected 3, 7, 14, or 21 days after eclosion. To generate mitotic germline clones, the following stocks were used: hs‐flp 1112, aub HN2 FRT40A (this study), ubi‐nls‐GFP FRT40A, FRT40A, FRT82B twin DG24102 and FRT82B ubi‐nls‐GFP. Clones were induced in 3‐day‐old females with two 1‐h heat shocks at 37°C per day, separated by an 8‐h recovery period at 25°C, during three consecutive days. Ovaries were dissected 7, 14, or 21 days after the final heat shock.
Immunostaining and image analysis
Ovaries were dissected at room temperature in PBS supplemented with 0.1% Tween‐20 (PBT), fixed with 4% paraformaldehyde, blocked with PBS containing 10% BSA for 1 h and incubated in primary antibodies with PBT 1% BSA overnight at 4°C. Primary antibodies were then washed three times with PBT 1% BSA for 10 min at room temperature. Secondary antibodies were diluted in PBT 0.1% BSA and were incubated for 4 h at room temperature. Secondary antibodies were then washed three times in PBT for 10 min. Primary antibodies were used at the following concentrations: mouse anti‐Hts [1B1; Developmental Studies Hybridoma Bank (DSHB), University of Iowa] 1/100; rabbit anti‐Vasa (Santa Cruz Biotechnology) 1/1,000; rat anti‐Vasa (DSHB) 1/50; rabbit anti‐cleaved Caspase 3 (Biolabs) 1/300; rabbit anti‐Bam (a gift from D. Chen) 1/2,000; rabbit anti‐Aub (ab17724; Abcam) 1/500; mouse anti‐Aub (4D10, Gunawardane et al, 2007) 1/1500; mouse anti‐HA (ascite produced from 12CA5, Joly et al, 2013) 1/2,000; rabbit anti‐GFP (A6455; Invitrogen) 1/500; mouse anti‐Cbl (8C4; DSHB) 1/50; mouse anti‐Cbl (10F1; a gift from LM. Pai) 1/300; rabbit anti‐Nanos (a gift from A. Nakamura), 1/1,000; rabbit anti‐Brat (Betschinger et al, 2006) 1/300; rabbit anti‐Mei‐P26 (Liu et al, 2009) 1/100; mouse anti‐Fused (22F10; DSHB) 1/100; rabbit anti‐Pgc (Hanyu‐Nakamura et al, 2008) 1/1,000; rabbit anti‐Bruno (Sugimura & Lilly, 2006) 1/3,000; rabbit anti‐Lola (a gift from E. Giniger, Giniger et al, 1994) 1/100. Secondary antibodies (Alexa 488‐ and Cy3‐conjugated; Jackson ImmunoResearch) were used at 1/300. DNA staining was performed using DAPI at 0.5 μg/ml. Images were captured with a Leica SP8 confocal microscope and analyzed using the ImageJ software. Fluorescence intensity was measured with ImageJ software in wild‐type, heterozygous, or mutant GSCs. For each GSC, the mean fluorescence intensity was determined using three independent quantifications in three different cytoplasmic regions in the same confocal section; the number of cells analyzed (n) is indicated in the bar graphs, or each cell analyzed is shown as a dot or a square on the graphs.
Coimmunoprecipitations and Western blots
Protein coimmunoprecipitations were performed using 60 ovaries per experiment from w 1118, nos‐Gal4/UASp‐GFP‐Aub, or nos‐Gal4/UASp‐GFP‐Aub AA 3‐day‐old females. Ovaries were homogenized in 600 μl of DXB‐150 (25 mM Hepes‐KOH pH 6.8, 250 mM sucrose, 1 mM MgCl2, 1 mM DTT, 150 mM NaCl, 0.1% Triton X‐100) containing cOmplete™ EDTA‐free Protease Inhibitor Cocktail (Roche) and either RNase Inhibitor (0.25 U/μl; Promega) or RNase A (0.1 U/μl; Sigma). 50 μl of Dynabeads Protein G (Invitrogen) was incubated with 15 μl of mouse anti‐GFP antibody (3E6; Invitrogen) for 1 h on a wheel at room temperature. Protein extracts were cleared on 30 μl of Dynabeads Protein G previously equilibrated with DXB‐150 for 30 min at 4°C. The pre‐cleared protein extracts were incubated with Dynabeads Protein G bound to mouse anti‐GFP antibody for 3 h at 4°C. The beads were then washed seven times with DXB‐150 for 10 min at room temperature. Proteins were eluted in NUPAGE buffer supplemented with 100 mM DTT at 70°C. Western blots were performed as previously reported (Benoit et al, 1999) with antibodies used at the following concentrations: rabbit anti‐GFP (Invitrogen) 1/1,000; rabbit anti‐CCR4 (Temme et al, 2004) 1/1,000; rabbit anti‐NOT3 (Jeske et al, 2006) 1/2,000; mouse anti‐NOT1 (Temme et al, 2010) 1/250; mouse anti‐Cbl (8C4; DSHB) 1/1,000; mouse anti‐Cbl (10F1) 1/1,000; and rabbit anti‐actin (Sigma) 1/2,500.
RNA‐immunoprecipitations and RT–qPCR
For RNA‐immunoprecipitations (RNA‐IP), protein extracts were performed using 300 ovaries from w 1118 or nos‐Gal4/UASp‐GFP‐Aub females. Ovaries were homogenized in 600 μl of DXB‐150 containing cOmplete™ EDTA‐free Protease Inhibitor Cocktail (Roche) and RNase Inhibitor (0.25 U/μl; Promega). 50 μl of Protein G Mag Sepharose (GE Healthcare) was incubated with 2 μg mouse anti‐GFP antibody (3E6; Invitrogen) for 3 h on a wheel at 4°C. Protein extracts were cleared on 50 μl of Protein G Mag Sepharose previously equilibrated with DXB‐150 for 30 min at 4°C. The pre‐cleared protein extracts were incubated with Protein G Mag Sepharose bound to mouse anti‐GFP antibody for 3 h at 4°C. The beads were then washed seven times with DXB‐150 for 10 min at room temperature. RNA was prepared using TRIzol (Invitrogen), followed by DNA removal with TURBO DNA‐free (Ambion). The total RNA amount was used for reverse transcription; RT–qPCR was performed with the LightCycler System (Roche Molecular Biochemical) using three independent RNA extractions. RT–qPCR to quantify CblL mRNA levels was performed as for the RNA‐IP, except that 1 μg RNA was used for reverse transcription and RNA was prepared from germaria/early egg chambers dissected from 20 ovaries. Primers used for RT–qPCR were as follows.
| Cbl | Forward CGAACTGAAGGCCATATTCC |
| Cbl | Reverse TGTGCTGTTACCGAAGTTGC |
| CblL | Forward CGTTGTGGACGCTTTCGATC |
| CblL | Reverse CGTTGTGGACGCTTTCGATC |
| RpL32 | Forward CTTCATCCGCCACCAGTC |
| RpL32 | Reverse CGACGCACTCTGTTGTCG |
Poly(A) tail assays
ePAT assays were performed as previously described (Chartier et al, 2015) using the following primers.
| CblL | CACGTCATGTAACCGAACAAATC |
| mei‐P26 | CCTCTCTCTTTGTTGAAATCACAAAATGG |
Bioinformatics and statistics
To map iCLIP reads to the genome, PCR amplification was eliminated by merging into one, duplicate sequences that shared the same random barcodes. Mapping of reads from the three GFP‐Aub iCLIP replicates (Ma et al, 2017) was performed using Bowtie. Reads were mapped allowing unique hits and no mismatches (Bowtie parameters ‐v 0 ‐m 1). The sequencing coverage at each nucleotide position was computed using BEDTools. For prediction of piRNA target sites on mRNAs, we used a pool of piRNAs from ovaries sequenced in published libraries [GSM327620, GSM327621, GSM327622, GSM327623, GSM327624 (Brennecke et al, 2008); GSM872307 (Zhang et al, 2012); GSM548585 (Rozhkov et al, 2010); and GSM948741 (Olivieri et al, 2012)]. This led to a total of 3,042,979 non‐redundant ovarian piRNA sequences. The pool of embryonic piRNA sequences was described previously (Barckmann et al, 2015). Bowtie was used with different complementarities to identify piRNA target sites on transcripts, as follows. Bowtie with option “‐v 0”, “‐v 1”, “‐v 2”, or “‐v 3” was used to identify piRNAs complementary to mRNAs with up to 0, 1, 2, or 3 mismatch(es), respectively. For complementarities with a seed (16‐nt seed, or 20‐nt seed without mismatch), we did not use quality values; therefore, the sum of the quality values at all mismatched read positions (−e/−maqerr) was set to an arbitrary value of 2,000, which disabled the quality values. Furthermore, −l (length of the seed) and −n (number of mismatches within the seed) were set to different values. The option “–nofw” was used to search only for reverse complementarity between piRNAs and mRNAs. Statistical tests were performed using the Excel, GraphPad Prism, or online χ2 test (http://www.aly-abbara.com/utilitaires/statistiques/khi_carre.html) softwares.
Author contributions
PR‐R and AC performed the experiments and analyzed the data; SP performed bioinformatic analyses; MS analyzed the data; PR‐R and MS designed the study and wrote the manuscript; all authors discussed the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Source Data for Figure 3C and D
Source Data for Figure 4G
Source Data for Figure 6F
Acknowledgements
We are very grateful to D Chen, E Giniger, A Gonzalez‐Reyes, J Knoblich, A Nakamura, LM Pai, M Siomi, and P Zamore for their gifts of fly stocks or antibodies. We thank AL Finoux and W Joly for initial work in this study. This work was supported by the CNRS UMR9002, ANR (ANR‐2010‐BLAN‐1201 01 and ANR‐15‐CE12‐0019‐01), and FRM (“Equipe FRM 2013 DEQ20130326534”). PRR held a salary from the Labex EpiGenMed/University of Montpellier and from the Fondation ARC. SP held a salary from FRM “Projets Innovants” and “Equipe FRM 2013”.
The EMBO Journal (2017) 36: 3194–3211
References
- Abdu U, Brodsky M, Schupbach T (2002) Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of Gurken through Chk2/Mnk. Curr Biol 12: 1645–1651 [DOI] [PubMed] [Google Scholar]
- An W, Nadeau SA, Mohapatra BC, Feng D, Zutshi N, Storck MD, Arya P, Talmadge JE, Meza JL, Band V, Band H (2015) Loss of Cbl and Cbl‐b ubiquitin ligases abrogates hematopoietic stem cell quiescence and sensitizes leukemic disease to chemotherapy. Oncotarget 6: 10498–10509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barckmann B, Pierson S, Dufourt J, Papin C, Armenise C, Port F, Grentzinger T, Chambeyron S, Baronian G, Desvignes JP, Curk T, Simonelig M (2015) Aubergine iCLIP reveals piRNA‐dependent decay of mRNAs involved in germ cell development in the early embryo. Cell Rep 12: 1205–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans‐Holm M, Hiesinger PR, Schulze KL, Rubin GM, Hoskins RA, Spradling AC (2004) The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B, Nemeth A, Aulner N, Kuhn U, Simonelig M, Wahle E, Bourbon HM (1999) The Drosophila poly(A)‐binding protein II is ubiquitous throughout Drosophila development and has the same function in mRNA polyadenylation as its bovine homolog in vitro . Nucleic Acids Res 27: 3771–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA (2006) Asymmetric segregation of the tumor suppressor brat regulates self‐renewal in Drosophila neural stem cells. Cell 124: 1241–1253 [DOI] [PubMed] [Google Scholar]
- Bozzetti MP, Specchia V, Cattenoz PB, Laneve P, Geusa A, Sahin HB, Di Tommaso S, Friscini A, Massari S, Diebold C, Giangrande A (2015) The Drosophila fragile X mental retardation protein participates in the piRNA pathway. J Cell Sci 128: 2070–2084 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ (2007) Discrete small RNA‐generating loci as master regulators of transposon activity in Drosophila . Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ (2008) An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WL, Liou W, Pen HC, Chou HY, Chang YW, Li WH, Chiang W, Pai LM (2008) The gradient of Gurken, a long‐range morphogen, is directly regulated by Cbl‐mediated endocytosis. Development 135: 1923–1933 [DOI] [PubMed] [Google Scholar]
- Chartier A, Klein P, Pierson S, Barbezier N, Gidaro T, Casas F, Carberry S, Dowling P, Maynadier L, Bellec M, Oloko M, Jardel C, Moritz B, Dickson G, Mouly V, Ohlendieck K, Butler‐Browne G, Trollet C, Simonelig M (2015) Mitochondrial dysfunction reveals the role of mRNA poly(A) tail regulation in oculopharyngeal muscular dystrophy pathogenesis. PLoS Genet 11: e1005092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau J, Kulnane LS, Salz HK (2012) Sex‐lethal enables germline stem cell differentiation by down‐regulating Nanos protein levels during Drosophila oogenesis. Proc Natl Acad Sci USA 109: 9465–9470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W (2011) miRNA repression involves GW182‐mediated recruitment of CCR4‐NOT through conserved W‐containing motifs. Nat Struct Mol Biol 18: 1218–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, McKearin DM (2003) A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130: 1159–1170 [DOI] [PubMed] [Google Scholar]
- Chen Y, Pane A, Schupbach T (2007) Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila . Curr Biol 17: 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Boland A, Kuzuoglu‐Ozturk D, Bawankar P, Loh B, Chang CT, Weichenrieder O, Izaurralde E (2014) A DDX6‐CNOT1 complex and W‐binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell 54: 737–750 [DOI] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE (2004) The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 116: 817–829 [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H (1998) A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self‐renewal. Genes Dev 12: 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H (2000) piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127: 503–514 [DOI] [PubMed] [Google Scholar]
- Epstein AM, Bauer CR, Ho A, Bosco G, Zarnescu DC (2009) Drosophila Fragile X protein controls cellular proliferation by regulating cbl levels in the ovary. Dev Biol 330: 83–92 [DOI] [PubMed] [Google Scholar]
- Fagegaltier D, Falciatori I, Czech B, Castel S, Perrimon N, Simcox A, Hannon GJ (2016) Oncogenic transformation of Drosophila somatic cells induces a functional piRNA pathway. Genes Dev 30: 1623–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Geng C, Wang H, Yang Z, Weng C, Li H, Deng L, Liu L, Liu N, Ni J, Xie T (2015) Twin promotes the maintenance and differentiation of germline stem cell lineage through modulation of multiple pathways. Cell Rep 13: 1366–1379 [DOI] [PubMed] [Google Scholar]
- Gilboa L, Lehmann R (2006) Soma‐germline interactions coordinate homeostasis and growth in the Drosophila gonad. Nature 443: 97–100 [DOI] [PubMed] [Google Scholar]
- Giniger E, Tietje K, Jan LY, Jan YN (1994) lola encodes a putative transcription factor required for axon growth and guidance in Drosophila . Development 120: 1385–1398 [DOI] [PubMed] [Google Scholar]
- Goh WS, Falciatori I, Tam OH, Burgess R, Meikar O, Kotaja N, Hammell M, Hannon GJ (2015) piRNA‐directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev 29: 1032–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic KG, Lindquist S (1989) The FLP recombinase of yeast catalyzes site‐specific recombination in the Drosophila genome. Cell 59: 499–509 [DOI] [PubMed] [Google Scholar]
- Gou LT, Dai P, Yang JH, Xue Y, Hu YP, Zhou Y, Kang JY, Wang X, Li H, Hua MM, Zhao S, Hu SD, Wu LG, Shi HJ, Li Y, Fu XD, Qu LH, Wang ED, Liu MF (2014) Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res 24: 680–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H (2006) MIWI associates with translational machinery and PIWI‐interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci USA 103: 13415–13420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC (2007) A slicer‐mediated mechanism for repeat‐associated siRNA 5’ end formation in Drosophila . Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Guzzardo PM, Muerdter F, Hannon GJ (2013) The piRNA pathway in flies: highlights and future directions. Curr Opin Genet Dev 23: 44–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanyu‐Nakamura K, Sonobe‐Nojima H, Tanigawa A, Lasko P, Nakamura A (2008) Drosophila Pgc protein inhibits P‐TEFb recruitment to chromatin in primordial germ cells. Nature 451: 730–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM (2001) Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128: 2823–2832 [DOI] [PubMed] [Google Scholar]
- Harris RE, Pargett M, Sutcliffe C, Umulis D, Ashe HL (2011) Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev Cell 20: 72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igreja C, Izaurralde E (2011) CUP promotes deadenylation and inhibits decapping of mRNA targets. Genes Dev 25: 1955–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizu H, Siomi H, Siomi MC (2012) Biology of PIWI‐interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev 26: 2361–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C (2010) Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila . Science 330: 1824–1827 [DOI] [PubMed] [Google Scholar]
- Jeske M, Meyer S, Temme C, Freudenreich D, Wahle E (2006) Rapid ATP‐dependent deadenylation of nanos mRNA in a cell‐free system from Drosophila embryos. J Biol Chem 281: 25124–25133 [DOI] [PubMed] [Google Scholar]
- Jin Z, Xie T (2007) Dcr‐1 maintains Drosophila ovarian stem cells. Curr Biol 17: 539–544 [DOI] [PubMed] [Google Scholar]
- Jin Z, Flynt AS, Lai EC (2013) Drosophila piwi mutants exhibit germline stem cell tumors that are sustained by elevated Dpp signaling. Curr Biol 23: 1442–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly W, Chartier A, Rojas‐Rios P, Busseau I, Simonelig M (2013) The CCR4 deadenylase acts with Nanos and Pumilio in the fine‐tuning of Mei‐P26 expression to promote germline stem cell self‐renewal. Stem Cell Reports 1: 411–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano C, Wang J, Lin H (2011) Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet 45: 447–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuchi T, Koga H, Kawamoto M, Shoji K, Sakai H, Arai Y, Ishihara G, Kawaoka S, Sugano S, Shimada T, Suzuki Y, Suzuki MG, Katsuma S (2014) A single female‐specific piRNA is the primary determiner of sex in the silkworm. Nature 509: 633–636 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis‐Schultz N, Koppetsch BS, Cook HA, Theurkauf WE (2007) Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell 12: 45–55 [DOI] [PubMed] [Google Scholar]
- Klein JD, Qu C, Yang X, Fan Y, Tang C, Peng JC (2016) c‐Fos repression by piwi regulates Drosophila ovarian germline formation and tissue morphogenesis. PLoS Genet 12: e1006281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, Kittler EL, Zapp ML, Klattenhoff C, Schulz N, Theurkauf WE, Weng Z, Zamore PD (2009a) Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Minor NT, Park JK, McKearin DM, Maines JZ (2009b) Bam and Bgcn antagonize Nanos‐dependent germ‐line stem cell maintenance. Proc Natl Acad Sci USA 106: 9304–9309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Maines JZ, Tastan OY, McKearin DM, Buszczak M (2012) Mei‐P26 regulates the maintenance of ovarian germline stem cells by promoting BMP signaling. Development 139: 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Carreira‐Rosario A, Maines JZ, McKearin DM, Buszczak M (2013) Mei‐p26 cooperates with Bam, Bgcn and Sxl to promote early germline development in the Drosophila ovary. PLoS One 8: e58301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Han H, Lasko P (2009) Vasa promotes Drosophila germline stem cell differentiation by activating mei‐P26 translation by directly interacting with a (U)‐rich motif in its 3’ UTR. Genes Dev 23: 2742–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Wang S, Do T, Song X, Inaba M, Nishimoto Y, Liu LP, Gao Y, Mao Y, Li H, McDowell W, Park J, Malanowski K, Peak A, Perera A, Li H, Gaudenz K, Haug J, Yamashita Y, Lin H et al (2014) Piwi is required in multiple cell types to control germline stem cell lineage development in the Drosophila ovary. PLoS One 9: e90267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhu X, Han Y, Story B, Do T, Song X, Wang S, Zhang Y, Blanchette M, Gogol M, Hall K, Peak A, Anoja P, Xie T (2017) Aubergine controls germline stem cell self‐renewal and progeny differentiation via distinct mechanisms. Dev Cell 41: 157–169 e155 [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ (2009) Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H, Basquin J, Ozgur S, Czarnocki‐Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W (2014) Structural and biochemical insights to the role of the CCR4‐NOT complex and DDX6 ATPase in microRNA repression. Mol Cell 54: 751–765 [DOI] [PubMed] [Google Scholar]
- McKearin D, Ohlstein B (1995) A role for the Drosophila bag‐of‐marbles protein in the differentiation of cystoblasts from germline stem cells. Development 121: 2937–2947 [DOI] [PubMed] [Google Scholar]
- Metaxakis A, Oehler S, Klinakis A, Savakis C (2005) Minos as a genetic and genomic tool in Drosophila melanogaster . Genetics 171: 571–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra B, Ahmad G, Nadeau S, Zutshi N, An W, Scheffe S, Dong L, Feng D, Goetz B, Arya P, Bailey TA, Palermo N, Borgstahl GE, Natarajan A, Raja SM, Naramura M, Band V, Band H (2013) Protein tyrosine kinase regulation by ubiquitination: critical roles of Cbl‐family ubiquitin ligases. Biochem Biophys Acta 1833: 122–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgunova V, Akulenko N, Radion E, Olovnikov I, Abramov Y, Olenina LV, Shpiz S, Kopytova DV, Georgieva SG, Kalmykova A (2015) Telomeric repeat silencing in germ cells is essential for early development in Drosophila . Nucleic Acids Res 43: 8762–8773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA (2008) Mei‐P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454: 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton FG, Harris RE, Sutcliffe C, Ashe HL (2015) Coordinate post‐transcriptional repression of Dpp‐dependent transcription factors attenuates signal range during development. Development 142: 3362–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Senti KA, Subramanian S, Sachidanandam R, Brennecke J (2012) The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila . Mol Cell 47: 954–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai LM, Barcelo G, Schupbach T (2000) D‐cbl, a negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell 103: 51–61 [DOI] [PubMed] [Google Scholar]
- Pai LM, Wang PY, Chen SR, Barcelo G, Chang WL, Nilson L, Schupbach T (2006) Differential effects of Cbl isoforms on Egfr signaling in Drosophila . Mech Dev 123: 450–462 [DOI] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T (2007) zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell 12: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Jiang P, Zhao DY, Singh M, Schupbach T (2011) The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J 30: 4601–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott BB, Hudson A, Brady R, Schulz C (2012) Control of germline stem cell division frequency–a novel, developmentally regulated role for epidermal growth factor signaling. PLoS One 7: e36460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Liu N, Lin H (2016) Piwi maintains germline stem cells and oogenesis in Drosophila through negative regulation of Polycomb group proteins. Nat Genet 48: 283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, Lehmann R (2011) piRNA production requires heterochromatin formation in Drosophila . Curr Biol 21: 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H, Hime GR, Lada H, Bowtell DD (2000) A Drosophila analogue of v‐Cbl is a dominant‐negative oncoprotein in vivo . Oncogene 19: 3299–3308 [DOI] [PubMed] [Google Scholar]
- Rorth P (1998) Gal4 in the Drosophila female germline. Mech Dev 78: 113–118 [DOI] [PubMed] [Google Scholar]
- Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M (2010) Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467: 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Aravin AA, Zelentsova ES, Schostak NG, Sachidanandam R, McCombie WR, Hannon GJ, Evgen'ev MB (2010) Small RNA‐based silencing strategies for transposons in the process of invading Drosophila species. RNA 16: 1634–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S, Tamura A, Honda H, Sakata‐Yanagimoto M, Kumano K, Oda H, Yamagata T, Takita J, Gotoh N, Nakazaki K, Kawamata N, Onodera M, Nobuyoshi M, Hayashi Y, Harada H et al (2009) Gain‐of‐function of mutated C‐CBL tumour suppressor in myeloid neoplasms. Nature 460: 904–908 [DOI] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E (1991) Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129: 1119–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA (2005) Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol 15: 284–294 [DOI] [PubMed] [Google Scholar]
- Slaidina M, Lehmann R (2014) Translational control in germline stem cell development. J Cell Biol 207: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura I, Lilly MA (2006) Bruno inhibits the expression of mitotic cyclins during the prophase I meiotic arrest of Drosophila oocytes. Dev Cell 10: 127–135 [DOI] [PubMed] [Google Scholar]
- Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E (2004) A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila . EMBO J 23: 2862–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, Simonelig M, Wahle E (2010) Subunits of the Drosophila CCR4‐NOT complex and their roles in mRNA deadenylation. RNA 16: 1356–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD (2004) RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 116: 831–841 [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi‐Miyagawa S, Nakano T, Lin H (2009) MILI, a PIWI‐interacting RNA‐binding protein, is required for germ line stem cell self‐renewal and appears to positively regulate translation. J Biol Chem 284: 6507–6519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Cheng EC, Zhong M, Lin H (2015) Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res 25: 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Jia S, Huang S, Wang H, Zhu Y, Mu Y, Kan L, Zheng W, Wu D, Li X, Sun Q, Meng A, Chen D (2010) The Fused/Smurf complex controls the fate of Drosophila germline stem cells by generating a gradient BMP response. Cell 143: 978–990 [DOI] [PubMed] [Google Scholar]
- Yang L, Duan R, Chen D, Wang J, Chen D, Jin P (2007) Fragile X mental retardation protein modulates the fate of germline stem cells in Drosophila . Hum Mol Genet 16: 1814–1820 [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, Weng Z, Theurkauf WE (2012) UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 151: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Kang JY, Gou LT, Wang J, Xue Y, Skogerboe G, Dai P, Huang DW, Chen R, Fu XD, Liu MF, He S (2015) MIWI and piRNA‐mediated cleavage of messenger RNAs in mouse testes. Cell Res 25: 193–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File
Source Data for Figure 3C and D
Source Data for Figure 4G
Source Data for Figure 6F


