Abstract
The mechanism for sex determination in mammalian germ cells remains unclear. Here, we reconstitute the female sex determination in mouse germ cells in vitro under a defined condition without the use of gonadal somatic cells. We show that retinoic acid (RA) and its key effector, STRA8, are not sufficient to induce the female germ‐cell fate. In contrast, bone morphogenetic protein (BMP) and RA synergistically induce primordial germ cells (PGCs)/PGC‐like cells (PGCLCs) derived from embryonic stem cells (ESCs) into fetal primary oocytes. The induction is characterized by entry into the meiotic prophase, occurs synchronously and recapitulates cytological and transcriptome progression in vivo faithfully. Importantly, the female germ‐cell induction necessitates a proper cellular competence—most typically, DNA demethylation of relevant genes—which is observed in appropriately propagated PGCs/PGCLCs, but not in PGCs/PGCLCs immediately after induction. This provides an explanation for the differential function of BMP signaling between PGC specification and female germ‐cell induction. Our findings represent a framework for a comprehensive delineation of the sex‐determination pathway in mammalian germ cells, including humans.
Keywords: bone morphogenetic protein, female germ‐cell fate, meiosis, primordial germ cell‐like cells, retinoic acid
Subject Categories: Development & Differentiation, Signal Transduction, Stem Cells
Introduction
In multicellular organisms, which are typically diploids, the germ‐cell lineage undergoes sexually dimorphic development, ensuring sexual reproduction. A key event in sexual reproduction is the meiosis, which shuffles replicated parental chromosomes through meiotic recombination and generates haploid gametes bearing novel genetic constitutions via successive reduction divisions (Handel & Schimenti, 2010; Baudat et al, 2013). The fertilization of the resultant sexually dimorphic gametes, the spermatozoa and the oocytes, creates diploid zygotes, resulting in the generation of an enormous level of genetic diversity. An improved understanding of the mechanism for sexual differentiation of germ cells would thus provide essential information on the mechanisms by which genetic diversity and organismal evolution are generated, as well as on the diseased states arising from anomalies in these processes.
The germ‐cell lineage in mice is induced in the epiblast by signaling molecules, most importantly, bone morphogenetic protein 4 (BMP4) and WNT3, and is established as primordial germ cells (PGCs) at around embryonic day (E) 7.25 (Lawson et al, 1999; Saitou et al, 2002; Ohinata et al, 2009). PGCs undergo migration and colonize embryonic gonads from around E10.5, while exhibiting rapid proliferation and epigenetic reprogramming including genomewide DNA demethylation (Seisenberger et al, 2012; Kagiwada et al, 2013). During this period, PGCs are sexually uncommitted, and their sexual fates are determined, not by their chromosomal sexes per se, but by signals/environmental effects from embryonic gonads: In XY males, the Y‐chromosome‐encoded gene, Sry, initiates transcriptional cascades specifying indifferent gonads into testes at around E10.5, while in XX females, the lack of Sry leads to the formation of ovaries (Lin & Capel, 2015). Consequently, after around E13.5, XY PGCs in the embryonic testes enter into mitotic arrest to differentiate into prospermatogonia (PSG), whereas XX PGCs in the embryonic ovaries progress into meiosis to differentiate into primary oocytes (Spiller & Bowles, 2015). It has been shown that retinoic acid (RA), apparently synthesized primarily in the mesonephric ducts, induces XX PGCs in embryonic ovaries into the female pathway by up‐regulating the expression of STRA8, a molecule essential for triggering the meiotic prophase, whereas in embryonic testes, RA is degraded by CYP26B1 strongly expressed in nascent Sertoli cells and XY PGCs ensheathed by such cells are induced into the male pathway via an as‐yet‐unknown mechanism (Baltus et al, 2006; Bowles et al, 2006; Koubova et al, 2006; Anderson et al, 2008; Dokshin et al, 2013; Soh et al, 2015).
The pursuit of a more detailed understanding of the mechanism for sexual determination of germ cells, however, has been protracted. As to the female pathway, critical questions remain unresolved, such as whether RA is sufficient for the female germ‐cell fate or other signaling pathways are necessary, and which transcription factor(s) may drive the pre‐meiotic DNA replication. This is partly due to the difficulty in analyzing complex interactions between germ cells and gonadal somatic cells as well as to the lack of an in vitro system amenable for assessing the relevant processes in a constructive fashion. On the other hand, it has been shown that mouse embryonic stem cells (ESCs)/induced pluripotent stem cells (iPSCs) are induced, by activin A (ACTA) and basic fibroblast growth factor (bFGF), into epiblast‐like cells (EpiLCs), which are in turn induced, essentially by BMP4, into PGC‐like cells (PGCLCs) with characteristics of migrating PGCs. Importantly, PGCLCs bear a robust capacity both for spermatogenesis and oogenesis, upon transplantation or aggregation with gonadal somatic cells followed by appropriate culture (Hayashi et al, 2011, 2012; Hikabe et al, 2016; Ishikura et al, 2016; Saitou & Miyauchi, 2016). More recently, a system was developed which allows PGCLCs to be propagated without gonadal somatic cells for at least 1 week, creating an opportunity to reconstitute the sex‐determination pathway of germ cells under a defined condition in vitro (Ohta et al, 2017). We here explore this possibility with a focus on the female pathway and delineate a framework for the determination of female germ‐cell fate.
Results
A system for analyzing the sex‐determination mechanism of germ cells
We have recently developed a system to propagate PGCLCs up to ~50‐fold during a 7‐day culture in the presence of stem cell factor (SCF) and the stimulators of cAMP signaling, forskolin and rolipram, on m220 feeder cells (Fig 1A) (Ohta et al, 2017). Under this condition, PGCLCs maintain the transcriptome of migrating/early gonadal PGCs, which are sexually uncommitted, while progressively erasing their DNA methylome to acquire genomewide DNA methylation levels (~5%)/patterns equivalent to those in germ cells at E13.5, which are committed either to the male or the female fate (Spiller & Bowles, 2015). Thus, DNA methylation reprogramming and sexual differentiation in germ cells are genetically separable and PGCLCs cultured under this condition may serve as a system to explore the mechanisms for sexual differentiation.
Figure 1. A screening for the factors inducing the female germ‐cell fate.
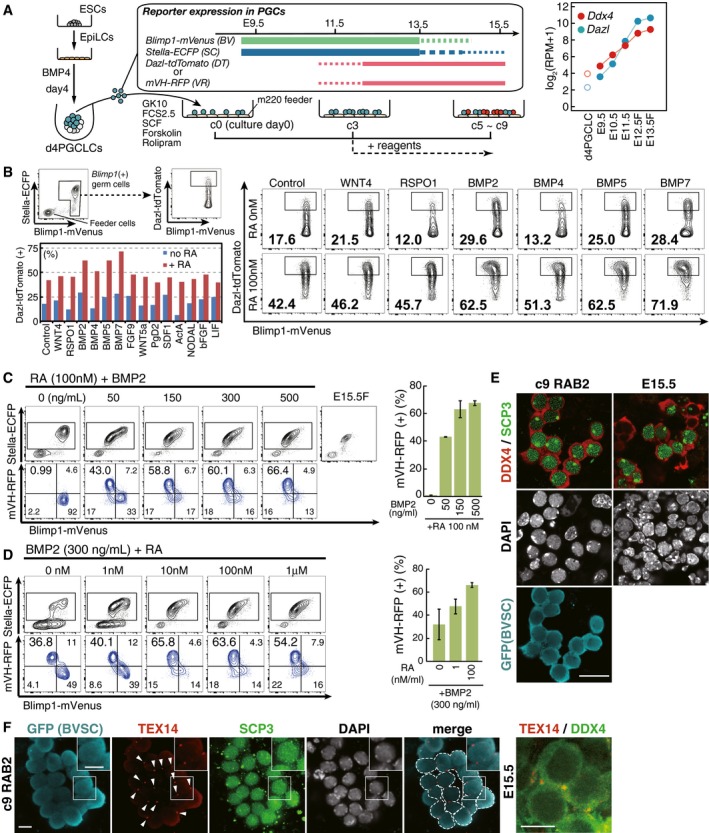
-
A(Left) Scheme for the screening of the factors inducing the female germ‐cell fate. d4/c0 PGCLCs [Blimp1‐mVenus (BV) (+) cells] induced from BV; Stella‐ECFP (SC); Dazl‐tdTomato (DT) (XY) or BVSC; mVH‐RFP (VR) (XX) ESCs were sorted by FACS onto m220 feeder cells and cultured in GMEM with 10% KSR (GK10) and 2.5% fetal calf serum (FCS) in the presence of forskolin, rolipram and SCF (Ohta et al, 2017). Cytokines/chemicals for the screening were provided from c3 (forskolin, rolipram, and SCF were provided throughout the culture). (Right) Expression of Dazl and Ddx4 in d4 PGCLCs and germ cells from E9.5 to E13.5 (female germ cells at E12.5 and E13.5) measured by RNA‐seq (Sasaki et al, 2015; Yamashiro et al, 2016; Ohta et al, 2017). The averages of two replicates are shown.
-
B(Top left) Scheme for the FACS. The DT levels among BV (+) cells were analyzed at c7. (Right) The FACS results for the cultures with the indicated conditions. The number indicates the percentage of DT (+) cells within the indicated gate. The concentration of the indicated cytokines is 500 ng/ml. See Appendix Fig S1 for larger FACS images and the data for other cytokines. (Bottom left) Summary of the screening results. Percentages of DT (+) cells among BV (+) cells under the indicated conditions are shown.
-
C, D(Left) Representative FACS plots of c9 cells (BVSCVR) under the indicated conditions. The boxed areas [SC (+) cells] in the upper plots were separated with BV and VR in the lower plots. A BVSC plot of E15.5 fetal primary oocytes is shown in (C). (Right) Percentage of the VR (+) cells under the indicated conditions. The means and the standard deviations (SDs) of two independent experiments are shown.
-
EDDX4 and SCP3 expression with DAPI in E15.5 fetal oocytes (right) and c9 RAB2 cells [BV/SC (+)] induced from BVSC ESCs (XX) (left). Scale bar, 20 μm.
-
FTEX14 expression (arrowheads) in c9 RAB2 cells [left panels: BV/SC (+)/SCP3 (+) oocyte cyst‐like structure] induced from BVSC ESCs (XX) (the boxed area is magnified in the inset) and in fetal oocytes at E15.5 (right). Scale bars, 10 μm.
We examined this possibility with a focus on differentiation into the female pathway, which is characterized by the entry into meiotic prophase. One prerequisite of the differentiation of PGCLCs into the female pathway is their acquisition of late PGC properties, characterized by the expression of genes such as Dazl and Ddx4 [also known as mouse vasa homolog (mVH)], both of which are expressed at low levels in PGCLCs/migrating PGCs and show progressive up‐regulation in germ cells up to E13.5 (Fig 1A) (Fujiwara et al, 1994; Cooke et al, 1996; Ohta et al, 2017). Moreover, Dazl has been proposed to act as a “licensing” factor for the sexual differentiation of germ cells (Lin et al, 2008; Gill et al, 2011). We therefore generated ESC lines expressing mVenus, ECFP, and tdTomato or RFP under the control of Blimp1 (also known as Prdm1), Stella (also known as Dppa3), and Dazl or Ddx4 (mVH), respectively (hereafter we designate Blimp1‐mVenus as BV, Stella‐ECFP as SC, Dazl‐tdTomato as DT, and mVH‐RFP as VR, respectively) (Fig EV1) (Materials and Methods). Blimp1 signifies PGC specification (Ohinata et al, 2005) and Stella shows expression in established PGCs (Saitou et al, 2002), while BV and SC respectively recapitulate Blimp1 and Stella expression (Ohinata et al, 2008). We also verified that DT and VR recapitulate Dazl and Ddx4 expression, respectively, from the late PGC‐stage onwards (Fig EV1C–E) (Imamura et al, 2010). We attempted to establish a condition for driving DT or VR expression in cultured PGCLCs followed by/coupled with their entry into the female fate and down‐regulation of BVSC expression. Since both XY and XX germ cells can take on the female fate (Evans et al, 1977; Taketo, 2015), we used both XY and XX ESCs as starting materials, which gave similar results (see below).
Figure EV1. BVSCDT/BVSCVR reporters and the roles of BMP ligands in the induction of female germ‐cell fate.
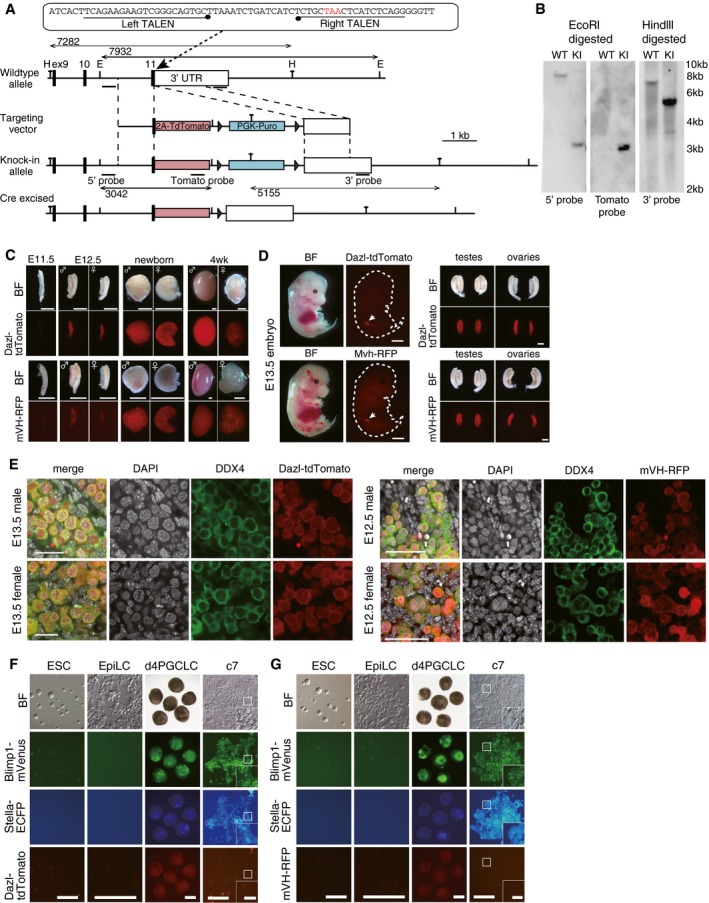
-
AThe targeting strategy to create a Dazl‐tdTomato (DT) reporter using the TALEN system (Sakuma et al, 2013). H: Hindlll site; E: EcoRl site.
-
BSouthern blot analysis of the Blimp1‐mVenus (BV); Stella‐ECFP (SC); and DT ESCs. The correct targeting was verified using the 5′‐, 3′‐, and tdTomato probes. WT: parental wild‐type cells; KI: BVSCDT knockin ESCs.
-
CDT and mVH‐RFP (VR) expression in testes and ovaries at the indicated developmental stages. BF: bright field images. Scale bars, 800 μm.
-
D(Left) Bright field and fluorescence images of DT (top) and VR (bottom) expression in whole embryos at E13.5. Note that DT and VR show specific expression in gonads (arrows). The outlines of the embryos are delineated by dotted lines. Scale bar, 2 mm. (Right) Bright field and fluorescence images of DT (top) and VR (bottom) expression in isolated testes and ovaries at E13.5. Note that DT and VR are strongly expressed in gonads but not in mesonephroi. Scale bar, 400 μm.
-
ECo‐/specific expression of DT (left) and VR (right) in DDX4 (+) germ cells in E13.5 (left) and E12.5 (right) testes and ovaries revealed by immunofluorescence (IF) analysis. Scale bars, (left) 20 μm; (right) 50 μm.
-
F, GBright field and fluorescence images of Blimp1‐mVenus (BV); Stella‐ECFP (SC); DT (F) or BVSCVR (G) expression during PGCLC induction and culture. Note that DT and VR are expressed at low levels/not expressed in d4 and c7 PGCLCs (the boxed area is magnified in the inset; scale bar, 40 μm). Scale bars, 200 μm.
We first induced BVSCDT ESCs (XY) (Fig EV1F) into PGCLCs and isolated BV‐positive (+) day (d) 4 PGCLCs by fluorescence‐activated cell sorting (FACS) for the expansion culture. At culture day 3 (c3), when the PGCLCs were propagating exponentially, we provided the culture with a panel of cytokines that might have an impact on sex determination in the absence or presence of RA, and at c7, evaluated their effects on BV/DT expression by FACS (forskolin, rolipram and SCF were provided throughout the culture) (Fig 1A and B, Appendix Fig S1). Under the control condition (no additional cytokines and no RA), BV (+) c7 PGCLCs showed, on average, relatively low DT expression (Fig 1B, Appendix Fig S1) (Ohta et al, 2017). Interestingly, the addition of RA elevated the DT levels in the BV (+) cell population (Fig 1B, Appendix Fig S1). Notably, combined addition of RA and one of the BMPs (BMP2, 4, 5, or 7), but not the other cytokines examined, strongly activated DT, and concomitantly down‐regulated BV (Fig 1B, Appendix Fig S1), suggesting that RA and BMP induced PGCLCs into the late germ‐cell phenotype.
Bmp2 is expressed strongly in response to a key feminizing factor, Wnt4, in pre‐granulosa cells that would direct the female germ‐cell fate (Fig EV2A) (Yao et al, 2004; Jameson et al, 2012). We therefore next examined whether RA and BMP2 also elevate VR in cultured PGCLCs induced from BVSCVR ESCs (XX) (Fig EV1G). We cultured BV (+) d4 PGCLCs, provided varying concentrations of RA and BMP2 at c3, and examined their effects on BVSCVR expression at c9 (Fig 1C and D). RA alone did not significantly change the BVSC expression and did not activate VR (Fig 1C), suggesting that Dazl and Ddx4 expression are regulated differently. Remarkably, combined addition of RA and BMP2 induced BVSC down‐regulation and robust VR up‐regulation, and the percentage of VR (+) cell induction increased in a BMP dose‐dependent fashion (Fig 1C). Interestingly, BMP2 alone down‐regulated BV and activated VR, and the extent of BVSC down‐regulation and VR activation increased in an RA dose‐dependent fashion (Fig 1D) (see below).
Figure EV2. Expression of Bmps in embryonic gonads and their effects on the induction of the female germ‐cell fate.
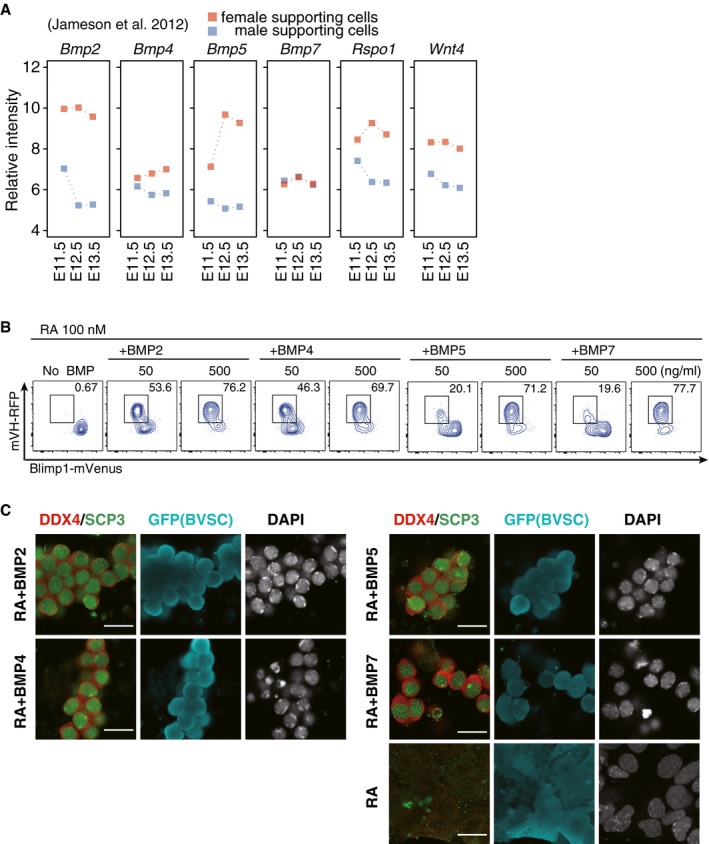
- Re‐analysis of the expressions of Bmps, Rspo1, and Wnt4 in male and female supporting cells (granulosa cells in female and Sertoli cells in male) reported in Jameson et al (2012). The average values from three replicates are shown.
- Representative FACS plots for BV; mVH‐RFP (VR) expression in c9 SC (+) cells cultured with the indicated BMPs (50 or 500 ng/ml) and RA (100 nM). Boxed areas indicate VR (+) cells and their percentages are shown.
- DDX4 and SCP3 expression in c9 Blimp1‐mVenus (BV)/Stella‐ECFP (SC) (+) cells cultured with the indicated BMPs and RA or RA alone. Scale bar, 20 μm.
By immunofluorescence (IF) analysis, we analyzed the expression of DDX4 and SCP3 (also known as SYCP3), a key component of the synaptonemal complex and a marker for meiotic prophase (Yuan et al, 2000), in cultured PGCLCs with RA and BMP2 at c9 [induced from BVSC ESCs (XX) to secure one fluorescence channel]. The BV/SC (+) cells expressed DDX4 and SCP3 in a manner highly similar to E15.5 primary oocytes: DDX4 was specifically localized in the cytoplasm and SCP3 showed a distinct pattern of localization indicative of synaptonemal complex formation. Moreover, the DDX4/SCP3 (+) cells appeared to be interconnected, reminiscent of the formation of oocyte cysts (Fig 1E) (Pepling & Spradling, 1998). Indeed, the cyst‐like structures exhibited expression and localization of TEX14, a cytoplasmic bridge marker (Greenbaum et al, 2009; Lei & Spradling, 2016), specifically at cell‐to‐cell contact sites (Fig 1F). When combined with RA, BMP4, 5, and 7 were also capable of inducing VR/DDX4 and SCP3 (+) cells (Fig EV2B and C). Thus, the combined action of RA and BMP signaling may lead cultured PGCLCs to the female fate.
BMP and RA commit PGCLCs to the female fate
We further explored the effect of RA and BMP signaling on PGCLCs induced from BVSCVR ESCs (XX), as VR exhibited a more specific response to RA and BMPs (Fig 1B and C). The culture with RA and BMP2, but not that with RA alone, from c3 onwards led to a down‐regulation of BVSC at c7 and resulted in a significant reduction of BVSC at c9 (Fig 2A). Reciprocally, under this condition, VR began to be activated by c7 and a majority (~70%) of SC (+) cells at c9 exhibited VR (Fig 2A). We confirmed that PGCLCs stimulated with RA and BMP2 elevated phosphorylated (p) SMAD1/5/8 and the expression of Id1 and Id2, immediate downstream targets of BMP signaling (Hollnagel et al, 1999; Korchynskyi & ten Dijke, 2002; Lopez‐Rovira et al, 2002), and the administration of LDN193189, a selective inhibitor of the ALK2/3 receptors (Cuny et al, 2008), blocked such effects (Fig EV3A and B), demonstrating that PGCLCs are competent in activating the BMP signaling pathway (see also below).
Figure 2. Induction of the female fate in PGCLCs by BMP and RA .
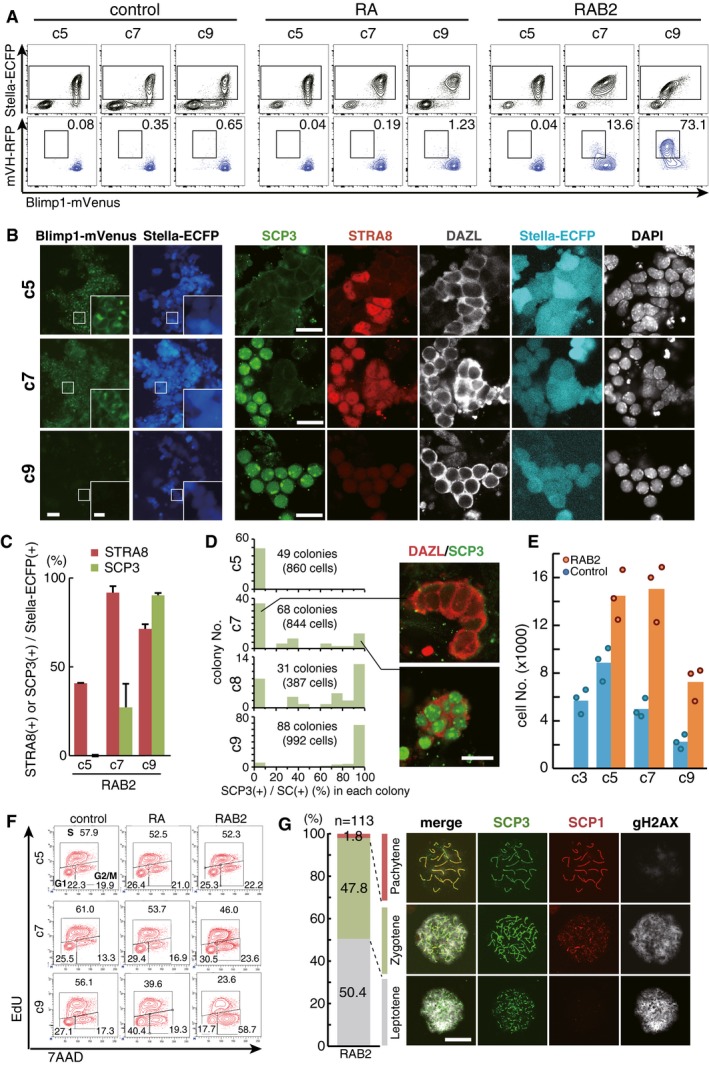
- Representative FACS plots [top: Blimp1‐mVenus (BV); Stella‐ECFP (SC) expression; bottom: BV; mVH‐RFP (VR) expression] of the PGCLC culture under the control condition (left), with RA (100 nM) (middle), and with RA (100 nM) and BMP2 (300 ng/ml) (right) at c5, c7, and c9. The SC (+) cells boxed in the top panels were analyzed in the bottom panels, with percentages of cells in the boxed areas indicated.
- (Left) BVSC fluorescence and (right) SCP3/STRA8/DAZL expression in BV/SC (+) cells during the PGCLC culture with RA (100 nM) and BMP2 (300 ng/ml) at c5, c7, and c9. The boxed areas in the left panels are magnified in the insets. Scale bars, 40 μm (left), 10 μm (left, inset), and 20 μm (right).
- The percentages of STRA8 (+) and SCP3 (+) cells among BV/SC (+) cells, based on IF analyses at c5, c7, and c9. The means and the SDs of two independent experiments are shown.
- Synchronicity of entry into the meiotic prophase. The vertical axis indicates the numbers of colonies counted. The horizontal axis indicates the percentage of SCP3 (+) cells within BV/SC (+) colonies. The colonies consisting of more than two cells were counted. Images of the representative colonies with or without SCP3 expression are shown on the right. Scale bar, 20 μm.
- The numbers of BV (+) cells during the control culture and the culture with RA (100 nM) and BMP2 (300 ng/ml). 1,500 BV (+) d4 PGCLCs were seeded at c0. One dot represents the average of five replicated culture wells, and the bar represents the mean of the dots.
- Cell cycle of PGCLCs cultured under the control condition, with RA (100 nM), and with RA (100 nM) and BMP2 (300 ng/ml) at c5, c7, and c9 as analyzed by EdU and 7AAD incorporation.
Figure EV3. Signaling requirements for female sex determination of PGCLCs.
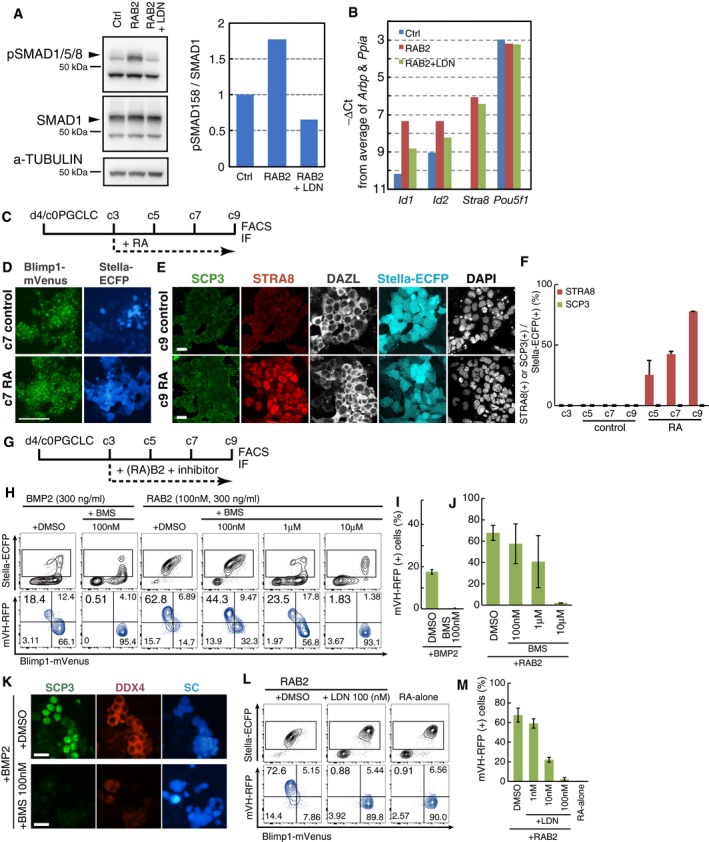
-
A(Left) Western blot analysis of the levels of phosphorylated (p) SMAD1/5/8, total SMAD1 and α‐TUBULIN (loading control) in d4 PGCLCs under the control condition, stimulated with RA (100 nM) and BMP2 (300 ng/ml) for 1 h, or with RA, BMP2, and LDN193189 (100 nM) for 1 h. (Right) Quantification of the relative levels of pSMAD1/5/8 ratio (/total SMAD) in the (left).
-
BqPCR analysis of the levels of the indicated genes in d4c5 PGCLCs cultured under the control condition, stimulated with RA (100 nM) and BMP2 (300 ng/ml) from c3, or with RA, BMP2, and LDN193189 (100 nM) from c3. For each gene examined, the ∆CT from the average CT values of the two independent housekeeping genes Arbp and Ppia (set as 0) were calculated and plotted.
-
CScheme for the experiments in (D–F).
-
D, EBlimp1‐mVenus (BV); Stella‐ECFP (SC) fluorescence (D) and IF images for SCP3, STRA8, DAZL, and BV/SC (E) of c7 (D) or c9 (E) cells cultured without (control) or with RA. Scale bars, 100 μm in (D) and 20 μm in (E).
-
FThe percentages of STRA8 (+) and SYCP3 (+) cells among BV/SC (+) cells. The percentages of STRA8 (+) and SCP3 (+) cells among BV/SC (+) cells, based on IF analyses at c5, c7, and c9. The means and the SDs of two independent experiments are shown.
-
GScheme for the experiments in (H–M).
-
HRepresentative BVSC (top) or BV; mVH‐RFP (VR) (bottom) FACS plots of c9 cells cultured under the indicated conditions. SC (+) cells (boxed cells in the top panels) were analyzed in the bottom panels. The percentages of cells plotted in each quadrant are indicated in the bottom panels.
-
I, JThe percentages of the VR (+) cells under the indicated conditions. The means and SDs of two independent experiments are shown.
-
KSCP3 and DDX4 expression in c9 BV/SC (+) cells cultured under the indicated conditions. Scale bars, 20 μm.
-
LRepresentative BVSC (top) or BVVR (bottom) FACS plots of c9 cells cultured under the indicated conditions. SC (+) cells (boxed cells in the top panels) were analyzed in the bottom panels. The percentages of cells plotted in each quadrant are indicated in the bottom panels.
-
MThe percentages of the VR (+) cells under the indicated conditions. The means and SDs of two independent experiments are shown.
IF analyses revealed that PGCLCs cultured with RA and BMP2 began to express STRA8, a key meiosis inducer (Baltus et al, 2006; Anderson et al, 2008; Dokshin et al, 2013; Soh et al, 2015), as early as c5 [~40.7%/SC (+) cells], and at c7 the vast majority (~91.7%) of SC (+) cells exhibited STRA8 expression, and some of them (~27.1%) were SCP3 (+) (Fig 2B and C). Remarkably, at c9, more than 90% of SC (+) cells expressed SCP3 with synaptonemal complex‐like structure formation, and STRA8 began to wane (Fig 2B and C). Interestingly, SCP3 was up‐regulated in a synchronous fashion in all the SC (+) cells that comprised distinct colonies at a given time during the culture (Fig 2B and D). From c5 to c9, SC (+) cells exhibited clear positivity for DAZL (Fig 2B).
Meiotic entry is characterized by pre‐meiotic DNA replication and arrest at the four‐chromosome (4C) state (Baltus et al, 2006). In the presence of RA and BMP2, BV/SC (+) cells exhibited an enhanced proliferation by c5, appeared to cease proliferation by c7, and declined to approximately half their peak number by c9 (Fig 2E). The cell‐cycle analysis revealed that at c7, a substantial fraction of BV/SC (+) cells were either replicating their DNAs (~46.0%) or in the 4C state (~23.6%) (Fig 2F). At c9, remarkably, the majority (~58.7%) of BV/SC (+) cells were in the 4C state (Fig 2F). This was not the case when PGCLCs were cultured under the control condition or with RA alone (Fig 2F). Collectively, these findings indicate that PGCLCs cultured with RA and BMP2 take on the female fate and enter into meiotic prophase. Indeed, the spread analyses revealed that the SC (+) cells progressed up to the pachytene stage (leptotene: ~50.4%; zygotene: ~47.8%; pachytene: ~1.8%) of the meiotic prophase (Fig 2G).
Under our condition, RA alone was insufficient to induce the female germ‐cell fate: Although PGCLCs cultured with RA activated DT/DAZL to a certain extent and up‐regulated STRA8 [~77.7% of BV/SC (+) cells expressed STRA8], RA did not activate VR/DDX4 or SCP3 (Figs 1B and C, 2A, and EV3C–F), and the BV/SC (+) cells with RA addition appeared to be cycling even at c9 (Fig 2F). Unexpectedly, BMP2 alone induced VR and SCP3 (+) meiocytes at c9, albeit less effectively than RA and BMP2 (Figs 1D, and EV3G–I and K). Since our culture included 10% knockout serum replacement (KSR) and 2.5% fetal calf serum (FCS) (Fig 1A) (Ohta et al, 2017), we reasoned that while the presence of BMP activity in such components might be negligible, the presence of RA activity (Hore et al, 2016) would confer onto the PGCLCs the ability to take on the female fate in response to exogenous BMP signaling. Accordingly, the culture of PGCLCs with BMP2 and an inhibitor for the RA receptor (RAR) (BMS493) (Koubova et al, 2006) abrogated the up‐regulation of VR/DDX4 as well as the induction of SCP3 (Fig EV3G–K). Similarly, the inhibition of BMP signaling by LDN193189 blocked the VR activation and meiotic entry elicited by RA and BMP2 in a dose‐dependent fashion (Fig EV3G, L and M). We conclude that RA and BMP signaling are necessary and, most likely, sufficient to induce PGCLCs into the female pathway and that RA or STRA8 alone is incapable of such induction.
BMP and RA commit PGCs to the female fate
We next examined the roles of RA and BMP signaling in female sex determination in PGCs. We isolated PGCs at E11.5 from Stella‐EGFP (SG) transgenic embryos (Payer et al, 2006; Seki et al, 2007) by FACS and cultured them with RA, BMP2, or both for 4 days (Fig 3A). While neither the control culture nor the culture with RA alone induced SCP3 or significantly affected the SG and DDX4 expression, the culture with RA and BMP2 substantially increased the induction of SCP3 (+)/DDX4 strongly (+)/SG‐negative (−) cells (~65.6%) (Fig 3B and C). The culture with BMP2 alone also induced SCP3 (+)/DDX4 strongly (+)/SG‐negative (−) cells (~22.9%) (Fig 3B and C).
Figure 3. Induction of the female fate in PGCs by BMP and RA .
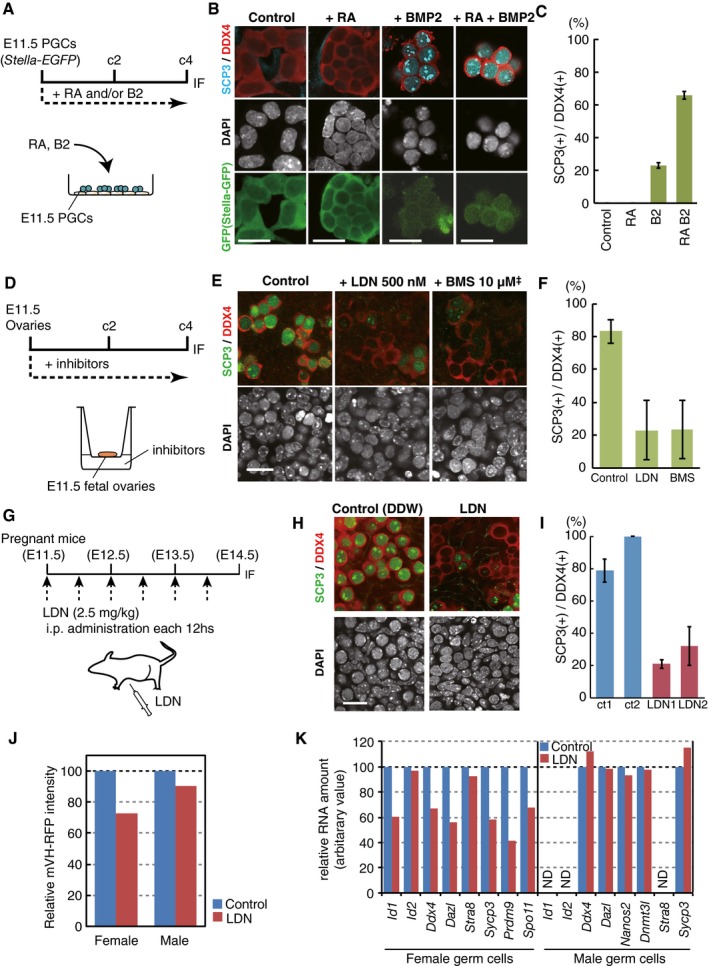
- Scheme of the PGC culture. Stella‐EGFP (SG) (+) cells were cultured with forskolin, rolipram, and SCF on m220 feeder cells, and RA (100 nM) and/or BMP2 (300 ng/ml) were provided at c0.
- DDX4/SCP3/SG expression under the indicated conditions at c4. Scale bar, 20 μm.
- The percentages of SCP3 (+) cells among DDX4 (+) cells. The means and SDs of duplicated wells are shown.
- Scheme of the E11.5 fetal ovary culture. BMS493 (10 μM1) or LDN1931189 (500 nM) was provided at c0.
- DDX4/SCP3 expression under the indicated conditions at c4. Scale bar, 20 μm.2
- The percentages of SCP3 (+) cells among DDX4 (+) cells. The means and SDs of two independent experiments are shown.
- Scheme of the LDN1931189 administration (2.5 mg/kg, every 12 h) in pregnant mice.
- DDX4/SCP3 expression at E14.5 in embryonic ovaries administered water (control) or LDN1931189 as in (G). Scale bar, 20 μm.
- The percentages of SCP3 (+) cells among DDX4 (+) cells. The means and SDs in two anterior and two posterior regions of the gonads are shown.
- Relative mVH‐RFP (VR) intensities at E14.5 analyzed by FACS of female or male VR (+) cells administered water (control) or LDN1931189 as in (G). The VR intensity of the control cells is set as 100.
- Relative expression levels of the indicated genes at E14.5 analyzed by qPCR in female or male VR (+) cells administered water (control) or LDN1931189 as in (G). The levels in the control cells are set as 100. ND, not detected.
We performed a culture of whole embryonic ovaries at E11.5 and examined the effects of inhibitors of RA or BMP signaling on the induction of female germ‐cell fate. The results showed that either type of inhibitor abrogated progression to the female germ‐cell fate, as represented by the expression of SCP3 (Fig 3D–F). Similarly, inhibition of BMP signaling under the in vivo condition via the administration of LDN193189 to pregnant females every 12 h from E11.5 to E14.0 led to a severe impairment in the progression to the female germ‐cell fate at E14.5 (Fig 3G–I). We isolated female or male VR (+) cells from LDN‐administered embryos by FACS at E14.5 and examined the expression of key genes for female or male germ‐cell development, respectively, by quantitative (q) PCR. This analysis revealed that the LDN‐treated female cells exhibited an impaired up‐regulation of key genes for late PGC/female germ‐cell development, such as Ddx4, Dazl, Sycp3, Prdm9, and Spo11, whereas, interestingly, male germ‐cell development appeared unaffected (Fig 3J and K). We conclude that both PGCs and PGCLCs require RA and BMP signaling to acquire the female germ‐cell fate.
Transcription dynamics during female sex determination of PGCLCs/PGCs
Next, we analyzed the transcription dynamics during the female sex determination of PGCs/PGCLCs. For this purpose, we isolated total RNAs from PGCs [Stella‐EGFP (+) cells at E9.5, E10.5, E11.5, E12.5 female, E12.5 male] (Kagiwada et al, 2013), fetal primary oocytes [Stella‐EGFP (+) at E13.5 (Kagiwada et al, 2013), DDX4‐RFP (+) at E14.5, E15.5], PSG [Stella‐EGFP (+) at E13.5 (Kagiwada et al, 2013), DDX4‐RFP (+) at E14.5, E15.5], PGCLCs cultured under the control condition (d4/c0, c3, c5, c7, c9) (Ohta et al, 2017), PGCLCs cultured with RA from c3 onwards (c5 RA, c7 RA, c9 RA), and PGCLCs cultured with RA and BMP2 from c3 onwards (c5 RAB2, c7 RAB2, c9 RAB2), and analyzed their transcriptome by RNA‐sequencing (RNA‐seq) (Nakamura et al, 2015).
Unsupervised hierarchical clustering (UHC) revealed that sexually undifferentiated PGCs until E11.5 were clustered closely with d4 PGCLCs, and then with c3‐c9 PGCLCs and with RA‐ or RAB2‐stimulated PGCLCs at c5 (c5 RA, c5 RAB2) (Fig 4A). Fetal primary oocytes (E14.5, E15.5) and PSG (E14.5, E15.5) formed distinct clusters, respectively, and remarkably, RAB2‐stimulated PGCLCs at c9 (c9 RAB2) were clustered tightly with fetal primary oocytes (Fig 4A). Germ cells that initiated sexual differentiation (male and female germ cells at E12.5, E13.5) and RAB2‐stimulated PGCLCs at c7 (c7 RAB2) and RA‐simulated PGCLCs at c7/c9 (c7/c9RA) formed distinct clusters showing a property intermediate between sexually undifferentiated PGCs/PGCLCs and fetal primary oocytes/c9 RAB2 cells (Fig 4A). Consistently, principal component analyses (PCA) clustered sexually undifferentiated PGCs and d4/c3‐c9 PGCLCs closely together, highlighted a progressive transition of female and male germ‐cell properties along their development, and defined a parallel progression of PGCLCs cultured with RAB2 along the female pathway, with c9 RAB2 cells clustered with fetal primary oocytes at E14.5/E15.5 (Fig 4B). In contrast, PGCLCs cultured with RA acquired the properties of fetal oocytes only partially (Fig 4B). Thus, cultured PGCLCs stimulated with RA and BMP2 recapitulate the transcriptome progression of the female pathway to form fetal primary oocytes.
Figure 4. Transcriptome during female sex determination of PGCLCs/PGCs.
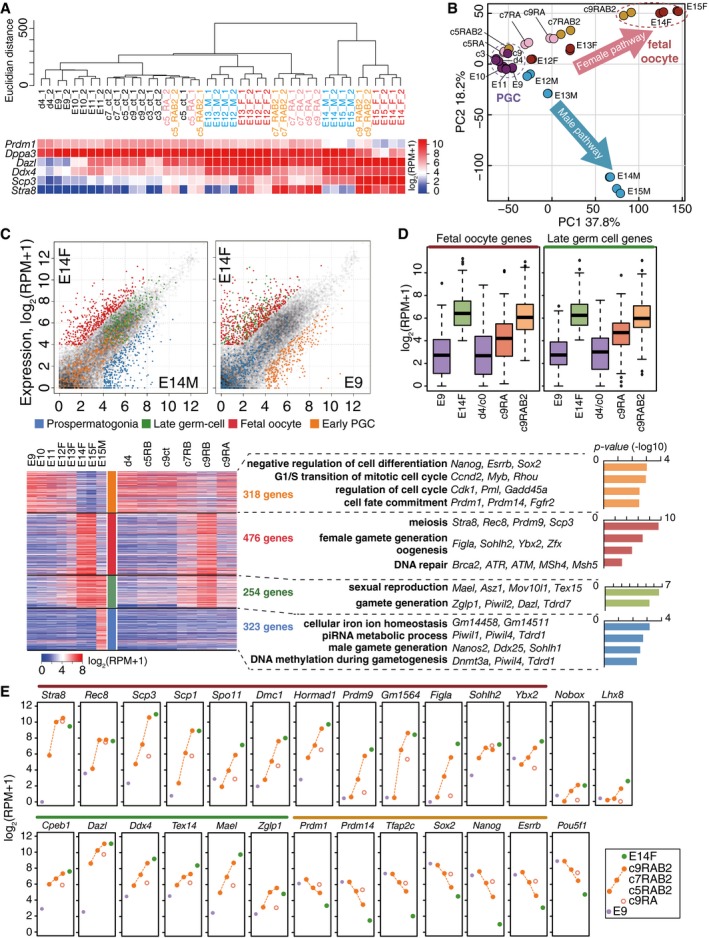
- UHC (Ward method) of the indicated cells using genes with log2(RPM+1) > 2 in at least one sample (15,849 genes) and a heat map of the expression levels of key genes. ct: cultured PGCLCs without RA or BMP2. The color coding is as indicated.
- PCA of germ cells in vivo (E9.5–E11.5 PGCs, E12.5–E15.5 male and female germ cells) and in vitro (cultured PGCLCs). A purple dotted circle clusters PGCs (E9.5, E10.5, E11.5) and PGCLCs (c0, c3, c9). A red dotted circle clusters fetal oocytes (E14.5, E15.5 female germ cells) and c9 RAB2 cells. Blue, red, pink, and yellow represent male germ cells, female germ cells, PGCLCs cultured with RA, and PGCLCs cultured with RAB2, respectively.
- (Top) Scatter‐plot comparison of gene expressions between E14.5 male and female germ cells (left) and between E14.5 female and E9.5 germ cells (right). Orange, green, red, blue, and gray dots indicate early PGC genes [318 genes: log2 fold‐change: E9.5 – male/female E14.5 > 2, log2(RPM+1) at E9.5 > 4], late germ‐cell genes [254 genes: log2 fold‐change: male/female E14.5 – E9.5 > 2, log2(RPM+1) at male/female E14.5 > 4], fetal oocyte genes [476 genes: log2 fold‐change: female E14.5 – male E14.5 > 2, female E14.5 – E9.5 > 2, log2(RPM+1) at female E14.5 > 4], PSG genes [323 genes: log2 fold‐change: male E14.5 – female E14.5 > 2, male E14.5 – E9.5 > 2, log2(RPM+1) at male E14.5 > 4], and non‐classified genes, respectively. (Bottom left) A heat map of the expression of early PGC genes (orange), late germ‐cell genes (green), fetal oocyte genes (red), and PSG genes (blue) (in total: 1,371 genes) in germ cells in vivo and in vitro as indicated. (Bottom right) GO enrichments (P‐values indicated) and key genes for each gene class.
- Box plots of the levels of fetal oocyte genes (left) and late germ‐cell genes (right) in the indicated cells [the averages (horizontal lines), 25th and 75th percentiles (boxes), and 5th and 95th percentiles (error bars) are shown].
- Expression [log2(RPM+1)] of key genes during female sex determination of PGCLCs/PGCs. The averages of two replicates are shown. Purple, green, and orange filled circles and red open circles represent E9.5 PGC, E14.5 fetal oocytes, c9 RAB2 cells, and c9 RA cells, respectively. The lines above the gene name are color‐coded as in (C).
To facilitate understanding of the gene‐expression dynamics during the sexual differentiation of germ cells/PGCLCs, we defined four classes of gene sets, early PGC genes (318 genes), late germ‐cell genes (254 genes), fetal oocyte genes (476 genes), and PSG genes (323 genes), that characterize the developmental stages of these cell types (Fig 4C) (Dataset EV1). Early PGC genes were enriched with genes bearing gene ontology (GO) functional terms such as “negative regulation of cell differentiation/regulation of cell cycle” (Prdm1, Prdm14, Tfap2c, Nanog, Sox2, etc.); late germ‐cell genes were enriched with genes for “sexual reproduction/gamete generation” (Dazl, Ddx4, Piwil2, Mael, Mov10l1, etc.); fetal oocyte genes were enriched with genes for “meiosis/female gamete generation” (Stra8, Rec8, Sycp3, Dmc1, Sycp1, etc.); and PSG genes were enriched with genes for “piRNA metabolic process/male gamete generation” (Nanos2, Dnmt3l, Tdrd9, Tdrd5, Piwil1, etc.) (Fig 4C).
As shown in Fig 4C and D, PGCLCs cultured with RAB2 progressively acquired late germ‐cell and fetal oocyte genes, while down‐regulating early PGC genes. In contrast, PGCLCs cultured with RA manifested such progressions only partially (Fig 4C and D): For example, c9 RAB2 cells up‐regulated key genes for meiosis (Stra8, Rec8, Sycp3, Sycp1, Spo11, Dmc1, Hormad1, Prdm9) (all included in fetal oocyte genes) and oocyte development (Figla, Ybx2, Nobox, Cpeb1) to levels similar to those in E14.5/E15.5 fetal oocytes (Fig 4E). In contrast, c9 RA cells failed to exhibit sufficient acquisition of such genes, although they up‐regulated Stra8 and Rec8, genes responding to RA even in a heterologous cellular context (Oulad‐Abdelghani et al, 1996; Mahony et al, 2011) (Fig 4E). Consistent with the role of BMP signaling in female germ‐cell specification, PGCLCs cultured with RAB2 and developing female germ cells expressed receptors and key targets for the BMP signaling in a similar manner (Fig EV4A).
Figure EV4. RA induces ectopic developmental genes.
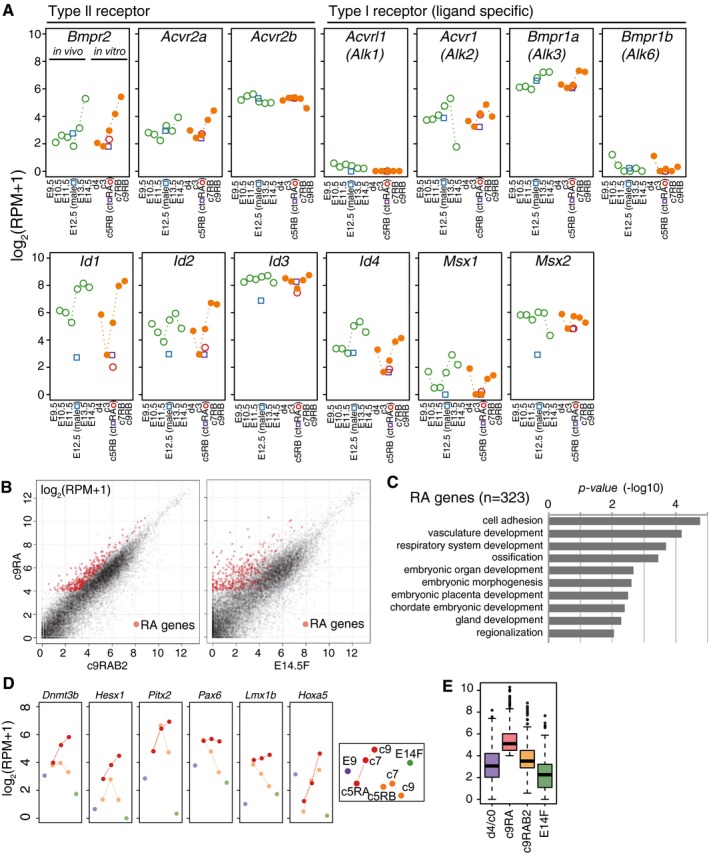
- Expression [log2(RPM+1)] of receptors and representative targets of the BMP signaling during female sex determination of PGCLCs (orange filled circles)/PGCs (green open circles). The averages of two replicates are shown. Expression levels in E12.5 male germ cells (blue open squares), d4c5 control cells (purple open squares), and d4c5 RA cells (red open circles) are also shown.
- Scatter‐plot comparisons of gene expression (left) between c9 RAB2 cells and c9 RA cells, or (right) between E14.5 primary oocytes and c9 RA cells. The ectopic RA genes (323 genes: log2(RPM+1) > 4; log2 fold‐change: c9 RA – c9 RAB2 > 1 and c9 RA – d4 PGCLCs > 1) are shown in red.
- GO enrichments in the RA genes. P‐values are indicated.
- Expression [log2(RPM+1)] of selected RA genes in the indicated cells (the color coding is as indicated).
- Box plots of the levels of the RA genes in the indicated cells [the averages (horizontal lines), 25th and 75th percentiles (boxes), and 5th and 95th percentiles (error bars) are shown].
We identified genes up‐regulated in c9 RA cells compared to c9 RAB2 cells (323 genes: RA genes) (Fig EV4B) (Dataset EV1). Such genes were also up‐regulated compared to fetal primary oocytes at E14.5/E15.5 and were enriched with those for “cell adhesion/vasculature development/embryonic organ development” (Hoxa5, Hesx1, Pax6, Lmx1b, Pitx2, Dnmt3b, etc.) (Fig EV4B–E). Thus, BMP signaling is critical not only for robustly driving the female pathway, but also for repressing the inappropriate developmental program elicited by RA.
Role of STRA8 in fetal primary oocyte development
We next evaluated the effect of the loss of Stra8 during the fetal primary oocyte differentiation from PGCLCs. We generated several lines of Stra8‐knockout BVSC ESCs (XY) using the CRISPR/Cas9 system (Ran et al, 2013a,b) and confirmed the frame shift deletions within the targeted exon and loss of STRA8 expression in these lines (Fig EV5A–C). Three independent lines [Stra8‐knockout (SK) 1, 2, 3] exhibited essentially identical phenotypes (Fig EV5D), and we therefore present the results using the representative line, SK1. Unlike wild‐type PGCLCs, SK1 cells cultured with RAB2 continued to retain robust BVSC expression until c7 and exhibited a modest down‐regulation of BVSC only at c9 (Fig 5A). Compared to wild‐type cells, SK1 cells proliferated less effectively in response to RAB2 and yet continued to exhibit a cycling profile even at c9 (Fig 5B and C), indicating their failure to progress into the meiotic prophase. This finding is in good agreement with the fact that Stra8‐knockout germ cells fail to undergo pre‐meiotic DNA replication and are eliminated thereafter (Baltus et al, 2006; Dokshin et al, 2013).
Figure EV5. Generation of the Stra8‐knockout (SK) ESCs.
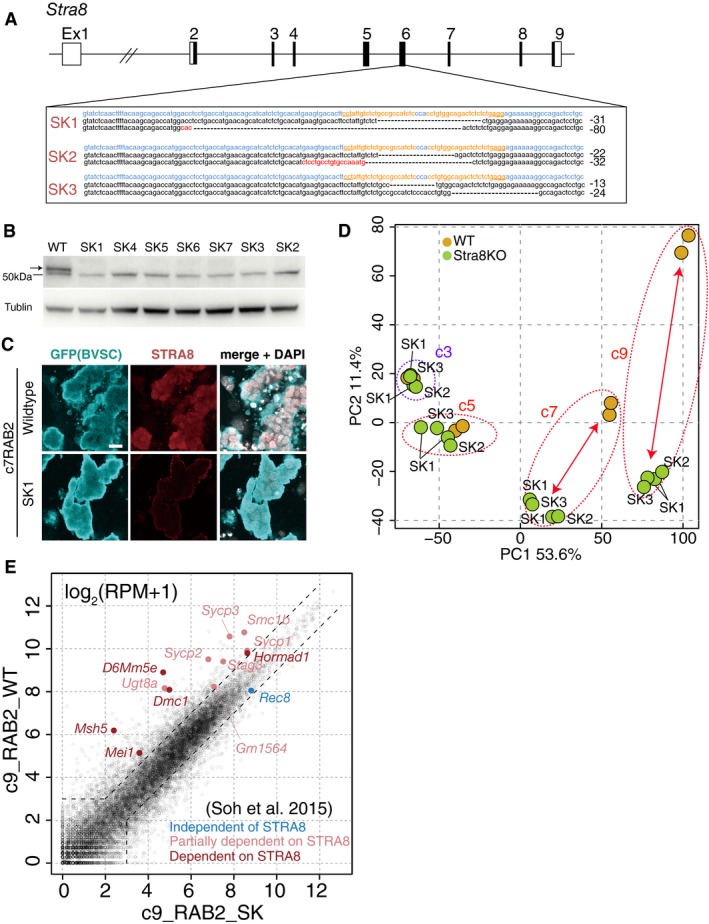
- Generation of three independent Stra8‐knockout ESC lines (SK1, 2, 3) by CRISPR‐disruption of exon 6. The parental line was BDF1‐2‐1 Blimp1‐mVenus (BV); Stella‐ECFP (SC) ESCs (XY). Nucleotides colored blue, orange, and red indicate the wild‐type sequences, guide RNA regions for CRISPR and insertion mutations, respectively. The deleted regions are indicated by hyphens. The numbers of deleted bases is indicated on the right.
- Western blot analysis of STRA8 expression in wild‐type (WT) and Stra8‐knockout (SK) lines. The upper band indicated by an arrow corresponds to STRA8, and the lower band is a non‐specific signal. α‐TUBULIN was used as a loading control.
- Validation of the loss of STRA8 expression in c7 RAB2 BV/SC (+) SK1 cells. Scale bar, 20 μm.
- PCA of the WT and SK1, 2, and 3 RAB2 cells at c3, c5, c7, and c9. Dotted circles represent samples under the same culture periods. Arrows highlight the differences between the WT and SK cells.
- Scatter‐plot comparison of gene expression between the WT and SK1 c9 RAB2 cells and the expression of genes reported to be independent (blue), partially dependent (pink), and dependent (red) on Stra8 (Soh et al, 2015).
Figure 5. STRA8 function in female sex determination.
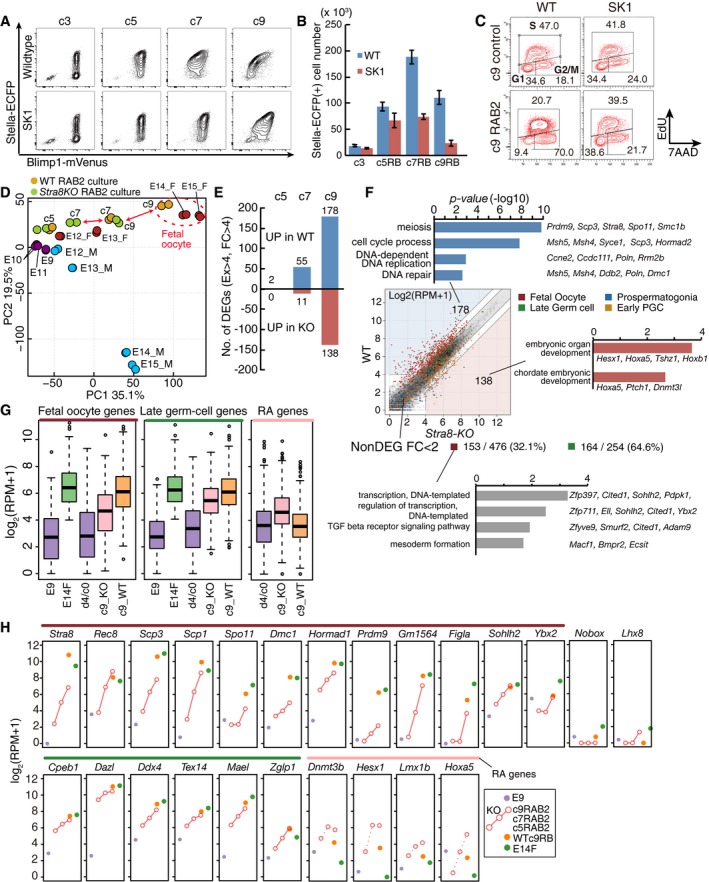
- Representative FACS plots (BVSC) of wild‐type (WT) and Stra8‐knockout (SK1) PGCLCs at c3, c5, c7, and c9 with RAB2.
- The numbers of Stella‐ECFP (SC) (+) cells estimated by FACS during the culture as in (A). The initial BV (+) cell numbers (c0) were 5,000. The means and SDs of two independent experiments are shown.
- Cell cycle of WT and SK1 PGCLCs cultured under the control condition (top) or with RAB2 (bottom) at c9 as analyzed by EdU and 7AAD incorporation.
- PCA of the indicated cells. Arrows highlight differences between the WT and SK1 cells. A red dotted circle clusters fetal oocytes (E14.5, E15.5 female germ cells) and c9 RAB2 cells.
- The number of DEGs between WT and SK1 cells [log2(RPM+1) > 4, log2(fold‐change) > 2] cultured with RAB2 at c5, c7, and c9.
- (Top) Scatter‐plot comparison of gene expression between WT and SK1 c9 RAB2 cells and selected GO terms for DEGs. Four gene classes are color‐coded as indicated. (Bottom) Non‐DEGs [log2(fold‐change) < 1] among fetal oocyte genes or late germ‐cell genes between WT and SK1 c9 RAB2 cells and their selected GO terms. In total, 153 of 476 (˜32.1%) and 164 of 254 (˜64.6%) genes are non‐DEGs for fetal oocyte genes and late germ‐cell genes, respectively.
- Box plots of the levels of fetal oocyte genes, late germ‐cell genes, and RA genes in the indicated cells [the averages (horizontal lines), 25th and 75th percentiles (boxes), and 5th and 95th percentiles (error bars) are shown]. c9_KO, c9 SK1 cells with RAB2; c9_WT, c9 WT cells with RAB2.
- Expression [log2(RPM+1)] of key genes in WT and SK1 cells during the RAB2 culture. The averages of two replicates are shown. The color coding is as indicated.
We determined the transcriptome of SK1 cells. PCA revealed that SK1 RAB2 cells progressed along the female pathway in a protracted manner and, at c9, acquired a property similar to wild‐type c7 cells, which was closer to fetal primary oocytes at E13.5 (Fig 5D). Compared to wild‐type cells, SK1 cells exhibited a growing number of differentially expressed genes (DEGs) from c7 onwards (Fig 5E) (Dataset EV2), and the genes that were not fully up‐regulated in c9 SK1 cells (178 genes) were highly enriched with those for “meiosis/cell‐cycle process” (Prdm9, Sycp3, Spo11, Smc1b, Msh4, Msh5, Dmc1, Ccdc111, Poln, etc.) (Fig 5F), and all twelve genes that have been reported to be dependent on Stra8 (Soh et al, 2015) were down‐regulated in c9 SK1 cells (Fig EV5E).
However, it is of note that c9 SK1 cells up‐regulated 32.1% of fetal oocyte genes (153/476 genes) in a relatively normal fashion, including those associated with oocyte development, such as Ybx2 and Sohlh2 (Fig 5F–H). Interestingly, genes aberrantly up‐regulated in c9 SK1 cells were enriched with those for “embryonic organ development/chordate embryonic development” and were overlapped with the RA genes aberrantly up‐regulated in c9 RA cells (Figs 5F and G, and EV4B–E). On the other hand, c9 SK1 cells gained late germ‐cell genes in a relatively normal manner [164/254 genes (64.6%)] (Fig 5F–H) (Dataset EV2). Thus, in cooperation with an effector(s) of BMP signaling, STRA8 ensures sufficient expression levels of some of the genes involved in meiosis, in addition to repressing unnecessary developmental pathways elicited by RA.
Cellular competence for inducing the female fate in response to RA and BMP
Bone morphogenetic protein signaling specifies epiblasts/EpiLCs into PGCs/PGCLCs, but it does not directly induce them into the female fate (Hayashi et al, 2011). We therefore attempted to define a cellular context that leads to the female fate in response to RA and BMP signaling. We cultured d4/c0 or c7 PGCLCs with RA and BMP2 for 2 days and evaluated their responses by examining their transcriptomes (Fig 6A and B). In comparison with the control, c7 PGCLCs with RAB2 exhibited a substantial number of DEGs (up: 218 genes; down: 56 genes), with up‐regulated genes enriched for “meiosis”, and progressed along the female pathway (Fig 6C and D) (Dataset EV3). In contrast, d4/c0 PGCLCs with RAB2 showed only a slight change in gene expression (up: seven genes; down: two genes) and did not progress toward the female fate (Fig 6B and C) (Dataset EV3).
Figure 6. Competence for the female germ‐cell fate.
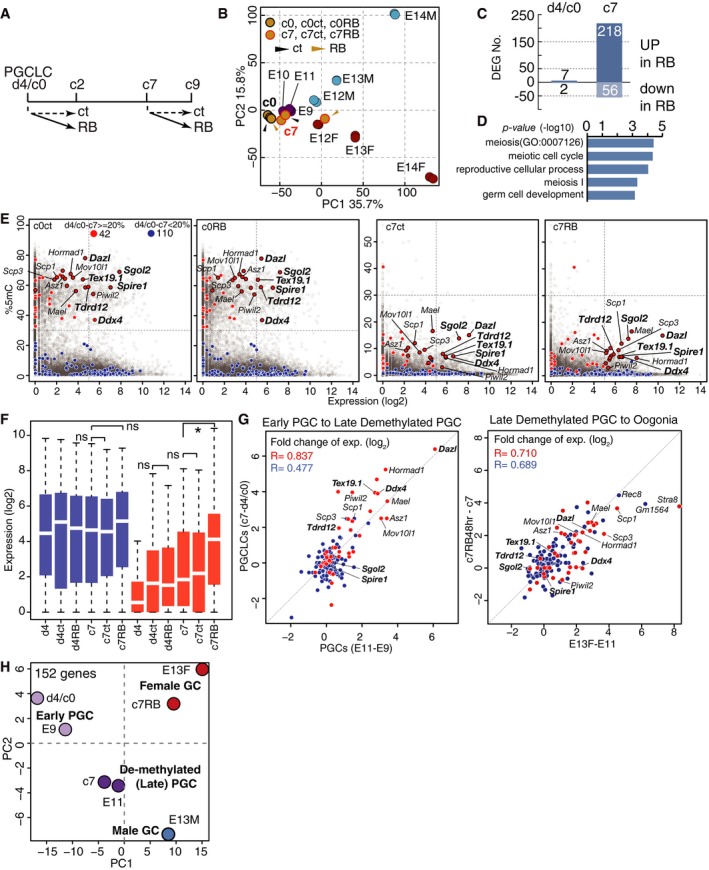
- Scheme of the experiments. Ct: control; RB: RAB2 culture for 48 h from d4/c0 or c7.
- PCA of the indicated cells. Yellow circles outlined in black or red represent d4/c0, c0 Ct and c0 RB or c7, c7 Ct, and c7 RB cells, respectively. Black or yellow arrowheads indicate cells with Ct or RB culture.
- The numbers of DEGs between c0 RB and Ct cultures (left) and between c7 RB and Ct cultures (right).
- GO terms for the 218 genes up‐regulated in c7 RB cells compared to c7 Ct cells.
- Scatter‐plot representation of the relationship between expression in c0 Ct/RB (left) and c7 Ct/RB (right) cells and promoter‐5mC levels in c0 (left) and c7 (right) cells (Shirane et al, 2016; Ohta et al, 2017). Red and blue circles indicate “meiosis” genes (GO: 0007126) (D), whose % 5mC differences between d4/c0 and c7 PGCLCs are > 20% (42 genes) or < 20% (110 genes), respectively. Red circles outlined in black indicate genes with a log2(RPM+1) > 5 in C7 RB cells. Boldface types indicate genes with log2(RPM+1) > 5 in c0 RB cells.
- Box plots of the levels of “meiosis” genes in the indicated cells [the averages (horizontal lines), 25th and 75th percentiles (boxes), and 5th and 95th percentiles (error bars) are shown]. *Statistically significant difference [Student's t‐test, P < 0.05]. The color coding is as in (E).
- Scatter‐plot representations of the relationship of the expression differences of “meiosis” genes (left) during early‐to‐late (E9.5 to E11.5) PGC transition with those during d4/c0‐to‐c7 PGCLC culture, and (right) during late PGC‐to‐E13.5 female‐germ‐cell transition with those during c7 to c7 RAB2 PGCLC culture. The color coding and boldface types are as in (E). Correlation coefficients are indicated.
- PCA of E9.5 PGCs, E11.5 PGCs, E13.5 female and male germ cells, d4/c0PGCLCs, c7 PGCLCs, and c7 RAB2 PGCLCs based on the “meiosis” gene expression.
We reasoned that the DNA demethylation of key genes for meiosis during the expansion culture might underlie the acquisition of the competence by PGCLCs to respond to RA and BMP2. We analyzed the relationship between the 5‐methylcytosine (5mC) levels of promoters and the expression levels of genes categorized as those involved in “meiosis” [GO terms: meiotic nuclear division GO: 0007126]. Among the 152 such genes (Dataset EV3), 42 genes, including Stra8, Spo11, Sycp3, Dpep3, Dazl, Ddx4, and Piwil2, exhibited promoter demethylation of > 20% between d4/c0 and c7. On the other hand, 110 genes did not show significant promoter 5mC‐level change (< 20%) during the culture and consisted of those involved in more general processes such as “DNA repair” and “response to DNA damage stimulus” (Mlh1, Brca2, Fanca, Cdc20, Plk1, etc.) (Fig 6E) (Dataset EV3). In d4/c0 PGCLCs, the 42 genes showed high promoter 5mC levels and no/low expression, while the 110 genes were barely methylated and exhibited varying expression levels, the distribution of which was similar to that of all genes with low promoter 5mC levels (Fig 6E and F). In c7 PGCLCs, the 42 genes as well as nearly all other genes were demethylated, and some of the 42 genes such as Dazl and Hormad1 were partially up‐regulated, whereas the 110 genes remained un‐methylated and retained an expression‐level distribution similar to that in d4/c0 PGCLCs (Fig 6E and F) (Ohta et al, 2017). Remarkably, in response to RA and BMP2 at c7, the 42 genes exhibited specific and robust activation, whereas the 110 genes showed only modest up‐regulation (Fig 6E and F). Collectively, these findings indicate that progressive promoter DNA demethylation of relevant genes during PGCLC expansion induces a basal activation or a permissive state for activation of such genes, which in turn acquire fully activated states in response to RA and BMP signaling to create the female germ‐cell fate.
To validate the relevance of this notion to germ‐cell development in vivo, we compared the differences in expression of the 152 genes between E9.5 and E11.5 PGCs with the differences in the expression of these genes between d4/c0 and c7 PGCLCs; we then compared the differential expression of the 152 genes between E11.5 PGCs and E13.5 fetal primary oocytes and that between c7 PGCLCs and c7 PGCLCs stimulated by BMP2 and RA for 2 days. As shown in Fig 6G, the expression differences (up‐regulation) of the 42 genes between successive stages of germ‐cell and PGCLC development were highly correlated. Consistently, PCA using the 152 genes clustered d4/c0 PGCLCs closely with E9.5 PGCs, c7 PGCLCs closely with E11.5 PGCs, and c7 PGCLCs cultured with RAB2 for 48 h closely with E13.5 female germ cells (Fig 6H). We conclude that the PGCLC‐based in vitro system precisely recapitulates the mechanism for the acquisition of the female germ‐cell fate in vivo.
Discussion
The mechanism for sex determination of germ cells that occurs in the middle of development has been difficult to analyze, due to the complexity of germ‐soma interactions, and the redundancy/earlier requirements of relevant factors during development. For example, RA is present throughout the body and acts through three receptors with similar function, RARα, β, and γ (Rhinn & Dolle, 2012); BMP signaling is essential for PGC specification (Lawson et al, 1999; Ying & Zhao, 2001); Bmp2 is required for amnion/chorion and cardiac development and Bmp2‐knockout embryos die by E11.5 prior to sex determination (Zhang & Bradley, 1996); and both Bmp2 and Bmp5 are expressed in pre‐granulosa cells (Fig EV2A) (Jameson et al, 2012). Our in vitro system overcomes such difficulties and allows analyses of the sex determination of germ cells both in a reductive and a constructive fashion, demonstrating that female germ‐cell specification requires multiple signaling inputs and a proper epigenetic background in an integrated fashion (Fig 7). Our data also demonstrate unambiguously that both XY and XX germ cells enter into the female pathway in a similar manner.
Figure 7. Models for the mechanism of female sex determination in mouse germ cells.
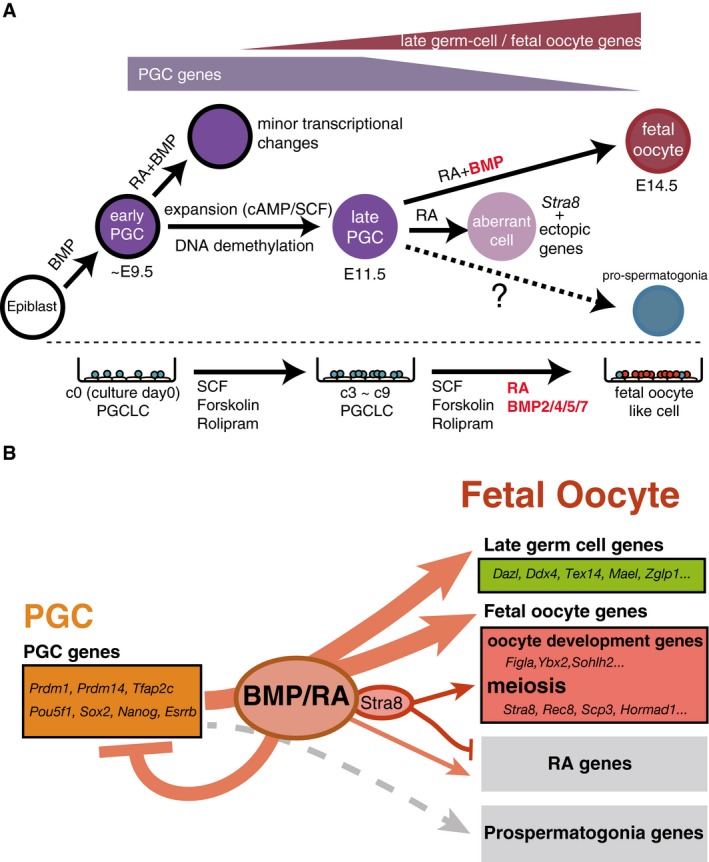
- A model for the cell fate transitions and signaling requirements for the differentiation of fetal oocytes from the epiblast. PGC specification from the epiblast depends on the BMP signaling (Lawson et al, 1999; Saitou et al, 2002; Ohinata et al, 2009). The maturation from early‐to‐late PGCs depends on the DNA demethylation of key promoters, likely through passive as well as active mechanisms (Yamaguchi et al, 2012; Kagiwada et al, 2013), coupled with PGC propagation through SCF and cAMP signaling. The differentiation from late PGCs to fetal primary oocytes depends on the BMP and RA signaling.
- A model for the roles of BMP and RA signaling. BMP and RA signaling contribute to the repression of early PGC genes (e.g., Prdm1, Prdm14, Tfap2c, Pou5f1, Sox2, Nanog, and Esrrb) and to the up‐regulation of late germ‐cell genes (e.g., Ddx4, Dazl, Piwil2, Mov10l1, and Mael) and fetal oocyte genes (e.g., Stra8, Rec8, Sycp3, Hormad1 as meiosis genes, and Figla, Ybx2, Sohlh2 as oocyte‐development genes). With BMP signaling, STRA8 promotes the expression of meiosis genes and inhibits the ectopic expression of developmental genes induced by RA signaling (RA genes). STRA8 does not have a significant impact on late germ‐cell genes and oocyte‐development genes. Without BMP signaling, STRA8 cannot fully up‐regulate meiosis genes or induce meiosis entry. BMP and RA signaling do not up‐regulate prospermatogonia genes.
Classical observations that ectopic germ cells take on the female fate have led to a proposition that the female fate is a default pathway in germ‐cell development (Zamboni & Upadhyay, 1983; McLaren & Southee, 1997). Identification of RA as a critical inducer of the female germ‐cell fate has revised this concept, and at the same time, has provided a rationale for the classical observations, since RA is both present in embryonic ovaries and also broadly distributed throughout the body, yet, importantly, is actively degraded by CYP26B1 in nascent Sertoli cells in embryonic testes (Bowles et al, 2006; Koubova et al, 2006). However, we found that RA and its key downstream effector, STRA8, are not sufficient to induce the female fate in competent PGC(LC)s (Fig EV3C–F), and that the combined action of RA and BMP signaling robustly induces competent PGC(LC)s into primary oocytes (Figs 2, 3, 4). Accordingly, pre‐granulosa cells that are likely instructive in specifying the female fate express high levels of Bmp2 and Bmp5 (Fig EV2A) (Jameson et al, 2012)—particularly Bmp2 in response to Wnt4, a key feminizing signal (Yao et al, 2004)—and a recent report demonstrated a critical function of Smad4, a transducer of BMP/TGFβ signaling, in the female sex determination of germ cells (Wu et al, 2016). Moreover, the fact that all the BMP ligands (BMP2, 4, 5, and 7) that activate BMP receptor type II (BMPRII)‐ALK3 signaling are capable of inducing the female fate (Fig EV2B and C) would also explain the tendency for the ectopic germ cells to adopt the female fate (Zamboni & Upadhyay, 1983; McLaren & Southee, 1997).
The mechanism by which RA and BMP signaling elicit the female germ‐cell program, by activating late germ‐cell genes, including Dazl and Ddx4, and fetal oocyte genes, remains unknown and warrants precise investigation. During fetal oocyte development, both gene sets exhibit progressive and monotonous up‐regulation (Figs 1A and 4). The provision of RA signaling alone in PGCLCs resulted in full activation of STRA8, yet only partially up‐regulated both gene sets, and ectopically up‐regulated somatic genes (RA genes) (Fig EV4), and consequently, the PGCLCs failed to initiate pre‐meiotic DNA replication. On the other hand, in the presence of RA and BMP signaling, Stra8‐knockout PGCLCs acquired late germ‐cell genes in a relatively normal manner, but failed to gain fetal oocyte genes (in particular, genes for meiosis) and to repress the RA‐induced ectopic somatic program, which also resulted in a failure of PGCLCs to undergo pre‐meiotic DNA replication (Fig 5). Thus, a key downstream effector(s) of BMP signaling may cooperate with STRA8 to fully activate late germ‐cell and fetal oocyte genes and to repress the RA‐induced somatic program. Such a mechanism is also critical for repressing early PGC genes (Figs 4 and 5). Identification of a key downstream effector(s) of BMP signaling and investigation of its mechanisms of action, including the relationship with STRA8, will be a critical challenge in order to clarify the mechanism for female germ‐cell specification. Importantly, the late germ‐cell genes are robustly and monotonously up‐regulated during PSG development as well (Fig 4) (Ohta et al, 2017), suggesting that they can be activated in a distinctive fashion by the mechanism specifying male germ‐cell fate.
A previous study has shown a critical role of the Polycomb repressive complex 1 (PRC1) in coordinating the timing of sexual differentiation of female germ cells (Yokobayashi et al, 2013). In good agreement, while a majority of the 524 genes aberrantly up‐regulated in female germ cells deficient in Rnf2, a key component of PRC1, at E12.5, were those involved in basic cellular functions (“cell cycle”, “protein dephosphorylation”, etc.), as many as 97 genes (18.5%) were late germ‐cell (Mael, Mov10l1, Tdrd7, Tex11, Tex14, etc.) or fetal oocyte genes (Stra8, Rec8, Sycp1, Sycp3, Smc1b, Hormad2, Sohlh2, etc.) (Appendix Fig S2A) (Yokobayashi et al, 2013). Furthermore, we have shown that c7, but not d4/c0, PGCLCs were capable of immediately responding to RA and BMP signaling to up‐regulate genes involved in “meiosis”, and this difference can be explained by the promoter demethylation of relevant genes, likely through passive as well as active mechanisms (Seisenberger et al, 2012; Yamaguchi et al, 2012; Kagiwada et al, 2013), during the expansion culture (Fig 6) (Ohta et al, 2017). This would also explain why early PGCs, which are specified by BMP signaling, are not directly induced into the meiotic program. In further support of this notion, we found that in the male pathway, KIT (+) spermatogonia at postnatal day 7 (P7), a precursor for meiosis in the first wave of spermatogenesis, exhibited low/no 5mC levels in the promoters of the relevant 42 genes, despite the fact that such cells re‐acquired high global 5mC levels during their development (Appendix Fig S2B) (Kubo et al, 2015; Ishikura et al, 2016). Interestingly, these genes exhibited significant demethylation during the transition from KIT (−) spermatogonia at P7 (Appendix Fig S2B) (Kubo et al, 2015). Thus, multiple layers of epigenetic regulations underlie the timely activation of the female germ‐cell fate, and the epigenetic requirements for meiotic entry appear to be in common, at least in part, between male and female germ cells.
During the 9‐day culture period with RA and BMP2 from c3 onward, d4/c0 PGCLCs progressed up to the pachytene stage (~1.8%). We found that the addition of RA as early as c0 followed by the combined provision of BMP2 from c3 onward resulted in an enhanced maturation of PGCLCs as primary oocytes both at the cytological (leptotene: ~34.9%; zygotene: ~53.4%; pachytene: ~11.6%) and the transcriptome levels (Appendix Fig S2C and D), suggesting that under an appropriate condition, it might be possible to extend the maturation of primary oocytes even further without gonadal somatic cells, a possibility that warrants future investigation. It is also interesting to note that the pachytene meiocyte‐like cells induced from male PGCLCs exhibited a sex body‐like structure positive for γH2AX (Appendix Fig S2E). The in vitro system that we have established recapitulates key processes of germ‐cell development, for example, PGC specification, PGC propagation, epigenetic reprogramming and female sex determination, under a defined condition, and should serve as a critical foundation to explore the mechanisms underpinning such salient processes. Moreover, our strategy could be applied to exploration of the mechanism for male sex determination as well as to the development of an equivalent culture of PGCLCs of other key species, including humans.
Materials and Methods
Animals
All animal experiments were performed under the ethical guidelines of Kyoto University. The BVSC (Acc. No. BV, CDB0460T; SC CDB0465T: http://www.cdb.riken.jp/arg/TG%20mutant%20mice%20list.html), Stella‐EGFP (SG), and mVH‐RFP (VR) transgenic mice were established as reported previously (Payer et al, 2006; Seki et al, 2007; Ohinata et al, 2008; Imamura et al, 2010) and were maintained on a largely C57BL/6 background. The Dazl‐tdTomato (DT) mice were generated by injecting the BVSCDT ESCs (XY) into the blastocysts (ICR) followed by their transfer to foster mothers. The ICR mice were purchased from SLC (Shizuoka, Japan). Noon of the day when a copulation plug was identified was designated as embryonic day (E) 0.5.
For administering LDN‐193189 into pregnant females (ICR), LDN‐193189 (sml0559; Sigma‐Aldrich) was dissolved in sterile water and 2.5 mg of the LDN‐193189 per kg body weight was injected intra‐peritoneally every 12 h from E11.5 to E14.
ESC culture/derivation and PGCLC induction/culture
H18 BVSC ESCs (XX) (Hayashi et al, 2012), R8 BVSC ESCs (XY) (Hayashi et al, 2011), L9 BVSCVR ESCs (XX), L5 BVSCVR ESCs (XY), BVSCDT ESCs (XY; a subline of R8), BDF1‐2‐1 BVSC ESCs (XY) (Ohta et al, 2017), and Stra8‐knockout BVSC ESCs (SK1, 2, 3; XY; sublines of BDF1‐2‐1) were used for this study. L5 and L9 BVSCVR ESCs were established from blastocysts obtained through matings of the VR females (Imamura et al, 2010) with the BVSC males (Ohinata et al, 2008), according to a procedure described previously, and were adapted to a feeder‐free condition (Hayashi et al, 2011). The procedures for the establishment of BVSCDT and Stra8‐knockout ESCs are described below in the “Establishment of the BVSCDT ESCs” and “Establishment of Stra8‐knockout ESCs” sections, respectively.
The ESC culture and PGCLC induction were performed as described previously (Hayashi et al, 2011; Hayashi & Saitou, 2013) with a few modifications. ESCs were maintained under a 2i+LIF condition on a dish coated with poly‐l‐ornithine (0.01%; Sigma) and laminin (300 ng/ml; BD Biosciences) or on mouse embryonic fibroblasts (MEFs). EpiLCs were induced by plating 1.0 × 105 ESCs on wells of a 12‐well plate coated with human plasma fibronectin (16.7 μg/ml; Millipore) in N2B27 medium containing activin A (20 ng/ml; PeproTech), bFGF (12 ng/ml; Life Technology) and KSR (1%; Thermo Fisher). At 42–46 h after the EpiLC induction, PGCLCs were induced under a floating condition by plating 2.0 × 103 EpiLCs in wells of a low‐cell‐binding 96‐well Lipidure‐coat plate (Thermo Fisher) in 200 μl of GK15 medium with BMP4 (500 ng/ml; R&D Systems), LIF (1,000 U/ml; Merck Millipore), SCF (100 ng/ml; R&D Systems), and EGF (50 ng/ml; R&D Systems). The GK15 medium was composed of GMEM (Thermo Fisher) with 15% KSR, 0.1 mM NEAA, 1 mM sodium pyruvate, 0.1 mM 2‐mercaptoethanol, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 2 mM l‐glutamine.
The PGC/PGCLC culture was performed as described previously (Ohta et al, 2017). Briefly, d4 PGCLC aggregates were collected, washed with PBS, and dissociated with TrypLE Express (Thermo Fisher). They were then washed with DMEM/F12 + 0.1% BSA (Gibco) and filtered by a cell strainer (BD Bioscience) to remove large clumps of cells. Next, the samples were centrifuged, resuspended in 0.1% BSA‐PBS, and sorted by FACS (Aria III; BD Bioscience). The BV (+) cells were plated onto mitomycin C (MMC)‐treated m220 feeder cells in the PGC/PGCLC‐expansion medium. The PGC/PGCLC‐expansion medium was composed of GMEM with 10% KSR, 2.5% FBS, 0.1 mM NEAA, 1 mM sodium pyruvate, 2 mM l‐glutamine, 0.1 mM 2‐mercaptoethanol, 100 U/ml penicillin, 0.1 mg/ml streptomycin, 10 μM forskolin, and 10 μM rolipram. The entire medium was changed every 2 days from c3. The cytokines/chemicals for the induction of the female fate were provided from c3 to the end of the culture. The concentrations of RA and BMP used were 100 nM and 300 ng/ml, respectively, unless otherwise specified. Bright field and fluorescence images were captured using an IX73 inverted microscope (Olympus).
Establishment of the BVSCDT ESCs
To construct the donor vector for the generation of Dazl‐tdTomato (DT) knockin ESCs, the homology arms of Dazl (the fragments from 1,048 bp upstream and to 1,247 bp downstream of the stop codon, respectively) were amplified from the genomic DNA of R8 BVSC ESCs by PCR (Primers), and were subcloned into the pCR2.1 vector (TOPO TA Cloning; Life Technologies). The P2A‐tdTomato fragment with the Pgk‐Puro cassette flanked by the LoxP sites was amplified by PCR from the vector reported previously (Sasaki et al, 2015) and inserted in‐frame at the 3′‐end of the Dazl coding sequence of the subcloned vector containing the homology arms, using a GeneArt Seamless Cloning & Assembly Kit (Life Technologies). The stop codon was removed for the expression of the in‐frame fusion protein.
The TALEN constructs targeting the sequences adjacent to the stop codon of Dazl were generated using a GoldenGate TALEN and TAL Effector kit (Addgene #1000000016) as described previously (Sakuma et al, 2013; Sasaki et al, 2015). The activities of the TALENs were evaluated by a single‐strand annealing (SSA) assay.
The donor vector (5 μg) and the TALEN plasmids (10 μg each) were introduced into the R8 BVSC ESCs by electroporation using a NEPA21 type II electroporator (Nepagene). Single colonies were picked up after puromycin selection, and random or targeted integrations were evaluated by PCR (Primers), followed by verification by Southern blot analyses. The line with the correct targeting was transfected with a plasmid expressing Cre recombinase to remove the Pgk‐Puro cassette.
Establishment of Stra8‐knockout ESCs
The vector expressing the Cas9 nickase (Addgene #42335) fused in frame with a reporter gene GSG‐p2A‐mCherry was created (the mCherry used contained a silent mutation, 432G>A). Two pairs of oligonucleotides (Primers) for targeting exon 6 of Stra8 were annealed, phosphorylated, and ligated separately to the above‐mentioned vector digested by Bbsl (NEB), according to the reported protocols (Ran et al, 2013a,b). The activities of the nickases were evaluated by the SSA assay. The pair of nickase plasmids (200 ng each) was introduced into the BDF1‐2‐1 BVSC ESCs by electroporation using the NEPA21 type II electroporator. The ESCs were dissociated 2 days after the transfection, and single cells expressing high levels of mCherry, which are expected to also express high levels of the Cas9 nickase, were sorted by FACS and seeded onto MEFs in single wells of 96‐well plates so that each well contained a single clone. The clones were cultured and expanded, and the disruption of the Stra8 loci in the clones was assessed by Sanger sequencing of the PCR products of the relevant region (Primers). The Stra8 knockout was confirmed by Western blot and IF analyses.
Ex vivo culture of PGCs or embryonic gonads
For the PGC culture, embryonic gonads (not sex‐typed) of the SG mice at E11.5 were dissected and dissociated. SG (+) PGCs were sorted by FACS, plated onto m220 feeder cells, and cultured in the PGC/PGCLC‐expansion medium. The reagents for the induction of the female fate were provided from c0.
For the culture of the embryonic gonads, embryonic ovaries with mesonephroi at E11.5 [sex‐typed by PCR (Primers)] were dissected out and cultured under a gas–liquid interface condition on the culture inserts (353095; BD Falcon). The medium used was DMEM with 10% FBS, 100 U/ml penicillin, 0.1 mg/ml streptomycin and 2 mM l‐glutamine. Chemical inhibitors were provided with the medium from c0.
Fluorescence‐activated cell sorting, cell‐cycle analysis, and cell counting
The preparation of d4/c0 PGCLCs for FACS was described in “ESC culture/derivation and PGCLC induction/culture”. To isolate the germ cells in vivo, embryonic gonads of BVSC, VR, or SG mice were dissected and processed for FACS according to the procedure described for d4/c0 PGCLCs. Cultured PGCLCs were also prepared in a similar manner, except that after dissociation, they were washed with 0.1% BSA‐DMEM containing 100 μg/ml DNasel (Sigma‐Aldrich) to digest DNA lysed out from dead cells to prevent the formation of cell/debris clumps. The fluorescence activities of BV/SG, SC, or DT/VR were detected in the FITC, Horizon V500 or PE‐Texas Red channel, respectively. The FACS data were analyzed using the FlowJo or FACS Diva software packages.
The cell‐cycle analysis was performed using a Click‐iT EdU Flow Cytometry Assay Kit (C10424; Thermo Fischer Scientific) according to the manufacturer's instructions. Cultured PGCLCs were treated with 10 μg/ml of EdU for 30 min to 2 h and were analyzed by FACS.
Cultured PGCLCs were stained with a chicken anti‐GFP antibody followed by an Alexa Fluor 633 goat anti‐chicken antibody and were analyzed using a Cellavista instrument (SynenTec) (Ohta et al, 2017).
Cytokines/chemicals
The cytokines/chemicals used to screen for activities involved in the induction of the female fate were as follows (Fig 1): 100 nM all‐trans retinoic acid, 500 ng/ml WNT4 (R&D Systems), 500 ng/ml RSPO1 (R&D Systems), 100 ng/ml FGF9 (R&D Systems), 500 ng/ml PgD2 (Cayman), 25 ng/ml activin A, 100 ng/ml NODAL (R&D Systems), 500 ng/ml SDF1 (R&D Systems), 50 ng/ml bFGF, 500 ng/ml BMP2 (R&D Systems), 500 ng/ml BMP4 (R&D Systems), 500 ng/ml BMP5 (R&D Systems), 500 ng/ml BMP7 (R&D Systems), 250 ng/ml WNT5a (R&D Systems), and 1,000 U/ml LIF. In the signal inhibition experiments, LDN193189 (#04‐0074; Stemgent) and BMS493 (B6688; Sigma‐Aldrich), which were dissolved in DMSO, were used as an ALK2/3 inhibitor and an RAR inhibitor, respectively.
Immunofluorescence (IF) analysis
Immunofluorescence (IF) analysis was performed as described previously (Hayashi et al, 2012). The following primary antibodies were used: chicken anti‐GFP (ab13970; Abcam), rabbit anti‐DDX4 (ab13840; Abcam), mouse anti‐DDX4 (ab27591; Abcam), rabbit anti‐DAZL (ab34129; Abcam), goat anti‐DAZL (sc‐27333; Santa Cruz), rabbit anti‐STRA8 (ab49602; Abcam), mouse anti‐SYCP3 (ab97672; Abcam), and rabbit anti‐TEX14 (ab41733; Abcam) IgG. The following secondary antibodies were used: Alexa Fluor 488 goat anti‐mouse or ‐chicken IgG; Alexa Fluor 568 goat anti‐rabbit IgG and Alexa Fluor 633 goat anti‐mouse, ‐chicken IgG; Alexa Fluor 488 donkey anti‐mouse IgG; Alexa Fluor 568 donkey anti‐rabbit IgG and Alexa Fluor 633 donkey anti‐goat IgG. IF images were captured using a confocal microscope [FV1000 (Olympus) or LSM 780 (Zeiss)].
Spread analysis of the meiotic chromosomes in fetal oocyte‐like cells
Spread preparations and IF analyses were performed as described previously with minor modifications (Yamashiro et al, 2016). c9 RAB2 cells were sorted by FACS, and the sorted cells were washed with PBS and treated in a hypotonic‐extraction solution at 25°C for 1 h. The primary antibodies used were as follows: goat anti‐SCP3 (1:250; sc‐20845; Santa Cruz), mouse anti‐γH2AX (1:1,000; 05‐636; Millipore), and rabbit anti‐SCP1 antibody (1:250; NB300‐229; Novus). The secondary antibodies used were as follows: Alexa Fluor 488 donkey anti‐goat IgG (A11055; Thermo Fisher), Alexa Fluor 568 donkey anti‐rabbit IgG (A10042; Thermo Fisher), and Alexa 647 donkey anti‐mouse IgG (A31571; Thermo Fisher). We counted SYCP3 (+) cells and the definition of the meiosis stages was as follows: leptotene: γH2AX (+) and SYCP1 (−); zygotene: γH2AX(+) and SYCP1 (+); pachytene: SYCP1(++) in at least 80% of the chromosomes.
Southern blot analysis
Southern blot analysis was performed as described previously (Nakaki et al, 2013). Briefly, 10 μg of genomic DNA was digested with the restriction enzymes and the resulting DNA fragments were electrophoresed in 0.9% agarose gel, transferred to the Hybond N+ membrane (RPN303B; GE Healthcare), and baked for the cross‐link. The DIG‐labeled probes for tdTomato, and the five and three prime sides of the relevant region of Dazl were generated by PCR (PCR DIG Labeling Mix; Sigma‐Aldrich) (Primers). Images were captured using an LAS4000 (Fujifilm).
Western blot analysis
For the Western blot analysis, 5 × 104 cells were lysed with the SDS sample buffer [62.5 mM Tris–HCl (pH 6.8), 2% SDS, 10% glycerol, 0.025% bromophenol blue and 0.14 M –mercaptoethanol] at 90°C for 5 min. For the detection of phosphorylated (p) SMAD1/5/8, cells were treated with the PhosSTOP (Roche) and the complete protease inhibitor cocktail (Roche) prior to the lysis. The extracted proteins were separated on SuperSep Ace 10–20% gels (Wako), blotted onto the iBlot2 PVDF transfer membrane (Thermo Fisher) by an iBlot2 dry blotting system (Thermo Fisher), and incubated with primary antibodies: rabbit anti‐STRA8 IgG (ab49405; Abcam), mouse anti‐α‐TUBULIN (T9026; Sigma‐Aldrich), rabbit anti‐pSMAD1/5/8 IgG (#9511; CST), or rabbit anti‐SMAD1 IgG (#9743S CST). The primary antibodies were detected with the goat anti‐rabbit or sheep anti‐mouse IgG conjugated with HRP (A0545, M8642; Sigma‐Aldrich) followed by detection using Chemi‐Lumi One Super (Nacalai). The capture and analysis of chemiluminescence images were performed with a Fusion Solo system and Fusion‐Capt software (M&S Instruments).
Transcriptome analysis
E14.5 and E15.5 male and female germ cells were collected as VR (+) cells and cultured PGCLCs as SC (+) cells by FACS, and total RNAs of the samples were extracted and purified using an RNeasy Micro Kit (74004; QIAGEN) according to the manufacturer's instructions. The cDNAs synthesis, library construction, and analysis by Nextseq500 (Illumina) from 1 ng of total RNAs from each sample were performed according to the methods described previously (Nakamura et al, 2015; Ishikura et al, 2016). The cDNAs of germ cells from E9.5 to E13.5 prepared in previous studies (Kagiwada et al, 2013; Ohta et al, 2017) were also analyzed by the Nextseq500 sequencer system.
All the read data were converted into expression levels as described previously (Nakamura et al, 2015). Briefly, all reads were treated with cutadapt‐1.3 (Martin, 2011) to remove the V1 and V3 adaptor sequences and the poly‐A sequences. The resulting reads of 30‐bp or longer were mapped onto the mm10 genome using TopHat1.4.1/Bowtie1.0.1 with the “‐no‐coverage‐search” option (Kim et al, 2013). Mapped reads were then converted to the expression level (RPM) using cufflinks‐2.2.0 with the “‐compatible‐hits‐norm”, “‐no‐length‐correction”, “‐max‐mle‐iterations 50000” and “library‐type fr‐secondstrand” options, and mm1‐reference gene annotation with up to 10‐kb extension at the 3′‐ends. The entire gene set analyzed was the same as that in our previous study (Ishikura et al, 2016).
Transcriptome analysis was performed using the R software package, version 3.2.1, with the gplots package and Microsoft Excel. First, the raw expression data were converted into log2(RPM+1) values, and genes with expression values > 2 in at least one sample were defined as expressed, unless otherwise specified. Data processing (e.g., identification of DEGs) was performed by using average values of biological replicates, except for the construction of the heat maps, clustering, and PCA. To create heat maps, the gplots package (heatmap.2) was used. Gene ontology (GO) analysis was performed by using the DAVID 6.7 website (https://david.ncifcrf.gov) (Huang da et al, 2009).
Quantitative (q) PCR
qPCR was performed using CFX384 (Bio‐Rad) and Power SYBR Green (ABI, Foster City, CA) according to the manufacturer's instruction. Template cDNAs were prepared as described in the “Transcriptome analysis” section, and the primers used are listed in the Primers.
Analysis of the DNA methylation of promoters
Whole‐genome bisulfite sequencing (WGBS) data of ESCs, EpiLCs, and PGCLCs were obtained from our previous studies (Shirane et al, 2016; Ohta et al, 2017), and those of KIT (−) and KIT (+) spermatogonia were obtained from a previous study (Kubo et al, 2015). We here defined promoters as regions between 900 bp upstream and 400 bp downstream of the transcription start sites (TSSs), and all the percent 5‐methylcytosine values (% 5mC) were calculated from the average of CpGs, in which the read depth was between 4 and 200, as previously described (Ishikura et al, 2016). We used the genes with promoters bearing at least one CpG site for the analysis in this study.
Primers
Primer sequences used in this study were as follows: for the sex typing: Uba1 forward: TGGATGGTGTGGCCAATGCYCT; reverse: CCACCTGCACGTTGCCCTTKGTGCCCAG. For the Dazl‐tdTomato vector construction: Dazl forward: AGAATCTGGTCCACAGGAAAAGGGCCTGAT; reverse: AGGCTTAGCCCCTGAGCTGCTTGTTAACTG. Genotype forward: AATAGACCCTAACCAGTTGGTGCAT; reverse: AAGAGTAAGTGGAATCCATGTTTGAAGGA. Random‐integration check forward: CCGGATGAATGTCAGCTACTGGGCTATCTG; reverse: TTCGGGGCGAAAACTCTCAAGGATCTTACC. For the generation of Stra8‐knockout cells: Nickase1 forward: CACCgGAGATGGCGGCAGAGACAAT; reverse: AAACATTGTCTCTGCCGCCATCTCc. Nickase2 forward: CACCgCCTGTGGCAGACTCTCTCTG; reverse: AAACCAGAGAGAGTCTGCCACAGGc. Sequence check forward: TAAGGCCAGGGGAAGGCAGAC; reverse: CGAAGGGCCATCTCACAGGGTC. For the Southern blot analysis of the Dazl‐tdTomato locus: 5′ probe forward: GGAGGCAAAGACAGGATCCTAAGCAAACTA; reverse: TCCCTAGTGCTCTATAAAATTGAGGTAATT. tdTomato probe forward: CCGCCGACATCCCCGATTAC; reverse: GCAGTTGCACGGGCTTCTTG. 3′ probe forward: ACCAGAAAACTAAGAAGTGTCCTGAA; reverse: AGACAGGACAGTAAGTCAGTGATGCT. For the qPCR analyses: Id1 forward: CAACAGAGCCTCACCCTCTC; reverse: AGAAATCCGAGAAGCACGAA. Id2 forward: CACAAAGGTGGAGCGTGAATAC; reverse: GCATTCAGTAGGCTCGTGTCAA. Ddx4 forward: CAGCTTCAGTAGCAGCACAAG; reverse: CATGACTCGTCATCAACTGGA. Dazl forward: GATGGACATGAGATCATTGGAC; reverse: ATACCAGGGAGCAATCCTGAC. Stra8 forward: GCCGGAGAAGGAGGAGATTAAA; reverse: AGCAGCCTTTCTCAATGAGTCT. Sycp3 forward: GTGTTGCAGCAGTGGGAAC; reverse: GCTTTCATTCTCTGGCTCTGA. Prdm9 forward: CCTGGCTACAAATTCTCATTTTC; reverse: TGTTTTTGTTTGTTTTGTTTTTGAGGA. Spo11 forward: AGCATGAAGTGTCTCACTAGCA; reverse: CATTAACAGGGCAAGGCACCTA. Nanos2 forward: AGAGAAGAATGCCAGTTGGGTT; reverse: ACAACGCTTTATTCAGCAGCAG. Dnmt3l forward: TCGGGTTTCTCTCCTGTTTG; reverse: GTTATCCCACCGGGAACTTG. Ppia forward: TTACCCATCAAACCATTCCTTCT; reverse: AACCCAAAGAACTTCAGTGAGAGC. Rplp0 forward: CAAAGCTGAAGCAAAGGAAGAG; reverse: AATTAAGCAGGCTGACTTGGTTG. Pou5f1 forward: GATGCTGTGAGCCAAGGCAAG; reverse: GGCTCCTGATCAACAGCATCAC.
Accession numbers
The accession numbers of the data used in this study are as follows: the RNA‐seq data of E10.5 and E11.5 PGCs and E13.5 female germ cells in Fig 4 (GEO: GSE74094) (Yamashiro et al, 2016), the RNA‐seq data of E9.5 PGCs and E12.5 female germ cells in Fig 4 (GEO: GSE87644) (Ohta et al, 2017), the RNA‐seq data of d4 PGCLCs in Fig 4 (GEO: GSE67259) (Sasaki et al, 2015), the microarray data of E11.5, E12.5, and E13.5 male and female supporting cells (GEO: GSE27715) (Jameson et al, 2012), the WGBS data of ESCs, EpiLCs, and d4 PGCLCs (DDBJ: DRA003471) (Shirane et al, 2016), the WGBS data of c7 PGCLCs (DDBJ: DRA005166) (Ohta et al, 2017), and the WGBS data of P7 KIT− SG and KIT+ SG (DDBJ: DRA002477) (Kubo et al, 2015).
The accession number of the RNA‐seq data of E9.5, E10.5, and E11.5 PGCs, E12.5, E13.5, E14.5, and E15.5 male and female germ cells, and cultured PGCLCs under the described conditions is GSE94136 (the GEO database).
Author contributions
HM conducted all the experiments and analyzed the data. HO contributed to the PGCLC propagation, SN contributed to the analyses of meiotic cells, FN and KS contributed to the genome editing, and KH contributed to the PGCLC induction. YY contributed to the analysis of the WGBS data, and YY, TN, and TY contributed to the RNA‐seq. MS conceived the project, and HM and MS designed the experiments and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Review Process File
Acknowledgements
We thank K. Abe, T. Noce, and Mitsubishi Chemical Corporation for providing us with the mVH‐RFP mice. We are grateful to the members of the Saitou Laboratory for their discussion of this study. We thank Y. Nagai, R. Kabata, N. Konishi, Y. Sakaguchi, and M. Kawasaki of the Saitou Laboratory, and T. Sato and M. Kabata of the Yamamoto Laboratory for their technical assistance. This work was supported in part by JST‐ERATO to M.S. (JPMJER1104) and by a Grant‐in‐Aid for Specially Promoted Research from JSPS to M.S. (17H06098) and by a Grant‐in‐Aid for JSPS to H.O. (JP15H05636). H.M. is a fellow of the Takeda Science Foundation.
See also: ME Gill & AHFM Peters (November 2017)
Notes
Correction added on 11 October 2017 after first online publication: Concentration corrected from 10 mM to 10 μM.
Correction added on 11 October 2017 after first online publication: Concentration corrected from 10 mM to 10 μM.
References
- Anderson EL, Baltus AE, Roepers‐Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AM, Page DC (2008) Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci USA 105: 14976–14980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltus AE, Menke DB, Hu YC, Goodheart ML, Carpenter AE, de Rooij DG, Page DC (2006) In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet 38: 1430–1434 [DOI] [PubMed] [Google Scholar]
- Baudat F, Imai Y, de Massy B (2013) Meiotic recombination in mammals: localization and regulation. Nat Rev Genet 14: 794–806 [DOI] [PubMed] [Google Scholar]
- Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, Yashiro K, Chawengsaksophak K, Wilson MJ, Rossant J, Hamada H, Koopman P (2006) Retinoid signaling determines germ cell fate in mice. Science 312: 596–600 [DOI] [PubMed] [Google Scholar]
- Cooke HJ, Lee M, Kerr S, Ruggiu M (1996) A murine homologue of the human DAZ gene is autosomal and expressed only in male and female gonads. Hum Mol Genet 5: 513–516 [DOI] [PubMed] [Google Scholar]
- Cuny GD, Yu PB, Laha JK, Xing X, Liu JF, Lai CS, Deng DY, Sachidanandan C, Bloch KD, Peterson RT (2008) Structure‐activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg Med Chem Lett 18: 4388–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokshin GA, Baltus AE, Eppig JJ, Page DC (2013) Oocyte differentiation is genetically dissociable from meiosis in mice. Nat Genet 45: 877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans EP, Ford CE, Lyon MF (1977) Direct evidence of the capacity of the XY germ cell in the mouse to become an oocyte. Nature 267: 430–431 [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Komiya T, Kawabata H, Sato M, Fujimoto H, Furusawa M, Noce T (1994) Isolation of a DEAD‐family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Proc Natl Acad Sci USA 91: 12258–12262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill ME, Hu YC, Lin Y, Page DC (2011) Licensing of gametogenesis, dependent on RNA binding protein DAZL, as a gateway to sexual differentiation of fetal germ cells. Proc Natl Acad Sci USA 108: 7443–7448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum MP, Iwamori N, Agno JE, Matzuk MM (2009) Mouse TEX14 is required for embryonic germ cell intercellular bridges but not female fertility. Biol Reprod 80: 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel MA, Schimenti JC (2010) Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 11: 124–136 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M (2011) Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146: 519–532 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Ogushi S, Kurimoto K, Shimamoto S, Ohta H, Saitou M (2012) Offspring from oocytes derived from in vitro primordial germ cell‐like cells in mice. Science 338: 971–975 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Saitou M (2013) Generation of eggs from mouse embryonic stem cells and induced pluripotent stem cells. Nat Protoc 8: 1513–1524 [DOI] [PubMed] [Google Scholar]
- Hikabe O, Hamazaki N, Nagamatsu G, Obata Y, Hirao Y, Hamada N, Shimamoto S, Imamura T, Nakashima K, Saitou M, Hayashi K (2016) Reconstitution in vitro of the entire cycle of the mouse female germ line. Nature 539: 299–303 [DOI] [PubMed] [Google Scholar]
- Hollnagel A, Oehlmann V, Heymer J, Ruther U, Nordheim A (1999) Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem 274: 19838–19845 [DOI] [PubMed] [Google Scholar]
- Hore TA, von Meyenn F, Ravichandran M, Bachman M, Ficz G, Oxley D, Santos F, Balasubramanian S, Jurkowski TP, Reik W (2016) Retinol and ascorbate drive erasure of epigenetic memory and enhance reprogramming to naive pluripotency by complementary mechanisms. Proc Natl Acad Sci USA 113: 12202–12207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Imamura M, Aoi T, Tokumasu A, Mise N, Abe K, Yamanaka S, Noce T (2010) Induction of primordial germ cells from mouse induced pluripotent stem cells derived from adult hepatocytes. Mol Reprod Dev 77: 802–811 [DOI] [PubMed] [Google Scholar]
- Ishikura Y, Yabuta Y, Ohta H, Hayashi K, Nakamura T, Okamoto I, Yamamoto T, Kurimoto K, Shirane K, Sasaki H, Saitou M (2016) In vitro derivation and propagation of spermatogonial stem cell activity from mouse pluripotent stem cells. Cell Rep 17: 2789–2804 [DOI] [PubMed] [Google Scholar]
- Jameson SA, Natarajan A, Cool J, DeFalco T, Maatouk DM, Mork L, Munger SC, Capel B (2012) Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet 8: e1002575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M (2013) Replication‐coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J 32: 340–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korchynskyi O, ten Dijke P (2002) Identification and functional characterization of distinct critically important bone morphogenetic protein‐specific response elements in the Id1 promoter. J Biol Chem 277: 4883–4891 [DOI] [PubMed] [Google Scholar]
- Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC (2006) Retinoic acid regulates sex‐specific timing of meiotic initiation in mice. Proc Natl Acad Sci USA 103: 2474–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo N, Toh H, Shirane K, Shirakawa T, Kobayashi H, Sato T, Sone H, Sato Y, Tomizawa S, Tsurusaki Y, Shibata H, Saitsu H, Suzuki Y, Matsumoto N, Suyama M, Kono T, Ohbo K, Sasaki H (2015) DNA methylation and gene expression dynamics during spermatogonial stem cell differentiation in the early postnatal mouse testis. BMC Genom 16: 624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, Korving JP, Hogan BL (1999) Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev 13: 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Spradling AC (2016) Mouse oocytes differentiate through organelle enrichment from sister cyst germ cells. Science 352: 95–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Gill ME, Koubova J, Page DC (2008) Germ cell‐intrinsic and ‐extrinsic factors govern meiotic initiation in mouse embryos. Science 322: 1685–1687 [DOI] [PubMed] [Google Scholar]
- Lin YT, Capel B (2015) Cell fate commitment during mammalian sex determination. Curr Opin Genet Dev 32: 144–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Rovira T, Chalaux E, Massague J, Rosa JL, Ventura F (2002) Direct binding of Smad1 and Smad4 to two distinct motifs mediates bone morphogenetic protein‐specific transcriptional activation of Id1 gene. J Biol Chem 277: 3176–3185 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco‐Rodriguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS (2001) Recombinational DNA double‐strand breaks in mice precede synapsis. Nat Genet 27: 271–276 [DOI] [PubMed] [Google Scholar]
- Mahony S, Mazzoni EO, McCuine S, Young RA, Wichterle H, Gifford DK (2011) Ligand‐dependent dynamics of retinoic acid receptor binding during early neurogenesis. Genome Biol 12: R2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011) Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet J 17: 10 [Google Scholar]
- McLaren A, Southee D (1997) Entry of mouse embryonic germ cells into meiosis. Dev Biol 187: 107–113 [DOI] [PubMed] [Google Scholar]
- Meuwissen RL, Offenberg HH, Dietrich AJ, Riesewijk A, van Iersel M, Heyting C (1992) A coiled‐coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J 11: 5091–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaki F, Hayashi K, Ohta H, Kurimoto K, Yabuta Y, Saitou M (2013) Induction of mouse germ‐cell fate by transcription factors in vitro . Nature 501: 222–226 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Yabuta Y, Okamoto I, Aramaki S, Yokobayashi S, Kurimoto K, Sekiguchi K, Nakagawa M, Yamamoto T, Saitou M (2015) SC3‐seq: a method for highly parallel and quantitative measurement of single‐cell gene expression. Nucleic Acids Res 43: e60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y, Payer B, O'Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, Saitou M, Surani MA (2005) Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436: 207–213 [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Sano M, Shigeta M, Yamanaka K, Saitou M (2008) A comprehensive, non‐invasive visualization of primordial germ cell development in mice by the Prdm1‐mVenus and Dppa3‐ECFP double transgenic reporter. Reproduction 136: 503–514 [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M (2009) A signaling principle for the specification of the germ cell lineage in mice. Cell 137: 571–584 [DOI] [PubMed] [Google Scholar]
- Ohta H, Kurimoto K, Okamoto I, Nakamura T, Yabuta Y, Miyauchi H, Yamamoto T, Okuno Y, Hagiwara M, Shirane K, Sasaki H, Saitou M (2017) In vitro expansion of mouse primordial germ cell‐like cells recapitulates an epigenetic blank slate. EMBO J 36: 1888–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulad‐Abdelghani M, Bouillet P, Decimo D, Gansmuller A, Heyberger S, Dolle P, Bronner S, Lutz Y, Chambon P (1996) Characterization of a premeiotic germ cell‐specific cytoplasmic protein encoded by Stra8, a novel retinoic acid‐responsive gene. J Cell Biol 135: 469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Chuva de Sousa Lopes SM, Barton SC, Lee C, Saitou M, Surani MA (2006) Generation of stella‐GFP transgenic mice: a novel tool to study germ cell development. Genesis 44: 75–83 [DOI] [PubMed] [Google Scholar]
- Pepling ME, Spradling AC (1998) Female mouse germ cells form synchronously dividing cysts. Development 125: 3323–3328 [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F (2013a) Double nicking by RNA‐guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013b) Genome engineering using the CRISPR‐Cas9 system. Nat Protoc 8: 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn M, Dolle P (2012) Retinoic acid signalling during development. Development 139: 843–858 [DOI] [PubMed] [Google Scholar]
- Saitou M, Barton SC, Surani MA (2002) A molecular programme for the specification of germ cell fate in mice. Nature 418: 293–300 [DOI] [PubMed] [Google Scholar]
- Saitou M, Miyauchi H (2016) Gametogenesis from pluripotent stem cells. Cell Stem Cell 18: 721–735 [DOI] [PubMed] [Google Scholar]
- Sakuma T, Hosoi S, Woltjen K, Suzuki K, Kashiwagi K, Wada H, Ochiai H, Miyamoto T, Kawai N, Sasakura Y, Matsuura S, Okada Y, Kawahara A, Hayashi S, Yamamoto T (2013) Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells 18: 315–326 [DOI] [PubMed] [Google Scholar]
- Sasaki K, Yokobayashi S, Nakamura T, Okamoto I, Yabuta Y, Kurimoto K, Ohta H, Moritoki Y, Iwatani C, Tsuchiya H, Nakamura S, Sekiguchi K, Sakuma T, Yamamoto T, Mori T, Woltjen K, Nakagawa M, Yamamoto T, Takahashi K, Yamanaka S et al (2015) Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell 17: 178–194 [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W (2012) The dynamics of genome‐wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 48: 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M (2007) Cellular dynamics associated with the genome‐wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 134: 2627–2638 [DOI] [PubMed] [Google Scholar]
- Shirane K, Kurimoto K, Yabuta Y, Yamaji M, Satoh J, Ito S, Watanabe A, Hayashi K, Saitou M, Sasaki H (2016) Global landscape and regulatory principles of DNA methylation reprogramming for germ cell specification by mouse pluripotent stem cells. Dev Cell 39: 87–103 [DOI] [PubMed] [Google Scholar]
- Soh YQ, Junker JP, Gill ME, Mueller JL, van Oudenaarden A, Page DC (2015) A gene regulatory program for meiotic prophase in the fetal ovary. PLoS Genet 11: e1005531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller CM, Bowles J (2015) Sex determination in mammalian germ cells. Asian J Androl 17: 427–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketo T (2015) The role of sex chromosomes in mammalian germ cell differentiation: can the germ cells carrying X and Y chromosomes differentiate into fertile oocytes? Asian J Androl 17: 360–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Fukuda K, Kato Y, Zhou Z, Deng CX, Saga Y (2016) Sexual fate change of XX germ cells caused by the deletion of SMAD4 and STRA8 independent of somatic sex reprogramming. PLoS Biol 14: e1002553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, Shen L, Inoue A, Diep D, Zhang K, Zhang Y (2012) Tet1 controls meiosis by regulating meiotic gene expression. Nature 492: 443–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro C, Hirota T, Kurimoto K, Nakamura T, Yabuta Y, Nagaoka SI, Ohta H, Yamamoto T, Saitou M (2016) Persistent requirement and alteration of the key targets of PRDM1 during primordial germ cell development in mice. Biol Reprod 94: 7 [DOI] [PubMed] [Google Scholar]
- Yao HH, Matzuk MM, Jorgez CJ, Menke DB, Page DC, Swain A, Capel B (2004) Follistatin operates downstream of Wnt4 in mammalian ovary organogenesis. Dev Dyn 230: 210–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y, Zhao GQ (2001) Cooperation of endoderm‐derived BMP2 and extraembryonic ectoderm‐derived BMP4 in primordial germ cell generation in the mouse. Dev Biol 232: 484–492 [DOI] [PubMed] [Google Scholar]
- Yokobayashi S, Liang CY, Kohler H, Nestorov P, Liu Z, Vidal M, van Lohuizen M, Roloff TC, Peters AH (2013) PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature 495: 236–240 [DOI] [PubMed] [Google Scholar]
- Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, Hoog C (2000) The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol Cell 5: 73–83 [DOI] [PubMed] [Google Scholar]
- Zamboni L, Upadhyay S (1983) Germ cell differentiation in mouse adrenal glands. J Exp Zool 228: 173–193 [DOI] [PubMed] [Google Scholar]
- Zhang H, Bradley A (1996) Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development 122: 2977–2986 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Review Process File


