Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that affect various biological processes by altering the expression of a target gene. An miRNA microarray analysis has previously revealed a significant decrease in miR-193a-3p levels in prostate cancer tissues compared with that in their benign prostate hyperplasia counterparts. However, the role of miR-193a-3p has yet to be elucidated. In the present study, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was used to evaluate the expression levels of miR-193a-3p in two human prostate cancer cell lines. Forced overexpression of miR-193a-3p was established by transfecting mimics into DU-145 and PC3 cell lines. Cell proliferation and the cell cycle were assessed using a cell viability assay, flow cytometry and a colony formation assay. In addition, the target gene of miR-193a-3p was determined by a luciferase assay, RT-qPCR and western blot analysis. The regulation of the cell cycle by miR-193a-3p was also evaluated by western blotting. The results demonstrated that miR-193a-3p expression levels were lower in prostate cancer cell lines as compared with the RWPE normal prostate epithelium cell line. Subsequent gain-of-function studies revealed that stable miR-193a-3p transfection inhibited cell viability, proliferation and colony formation, and induced G1 phase arrest in prostate cancer cells. A luciferase assay and western blot analysis identified cyclin D1 (CCND1) as a direct target gene of miR-193a-3p. In addition, the forced expression of CCND1 was able to counter the inhibitory effects of miR-193a-3p transfection in the prostate cancer cells. In summary, the results suggest that miR-193a-3p may inhibit the viability, proliferation and survival of prostate cancer cells by regulating the expression profile of CCND1, and that miR-193a-3p may be a novel therapeutic biomarker for prostate cancer.
Keywords: microRNA-193a-3p, prostate cancer, cyclin D1, cell proliferation
Introduction
Prostate cancer is one of the most common malignant tumors in males, the second most frequently diagnosed cancer, and the sixth leading cause of cancer-associated mortality in the male population in the USA (1). Typically, prostate cancer is widely considered to be a tumor of mild-to-moderate malignancy, with a 5-year survival rate of ~100% for localized tumors (1). However, the survival rate decreases to ~27.9% in the event of distant metastasis (1). Despite various advancements in therapies, the majority of patients with metastatic cancer eventually succumb to the disease. These aspects demonstrate the importance of elucidating the genetic mechanisms underlying prostate cancer, and of developing novel therapeutic targets.
MicroRNAs (miRNAs) are small (~22 nucleotides) non-coding RNA molecules that suppress gene expression by interacting with the 3′-untranslated regions (3′-UTRs) of specific target mRNAs (2). Distinct miRNA expression profiles have been identified in human prostate cancer tissues and cell lines (3–8). One of those miRNAs, miR-193a-3p, was initially identified by Lagos-Quintana et al (9) in 2003, and was revealed to be a tumor-suppressor agent (10) as well as a regulator for transdifferentiation in parietal cells (11). Lin et al (12) conducted a set of microarray assays (GEO accession: GSE36802), which identified lower expression levels of miR-193a-3p in prostate cancer tissues compared with that in benign hyperplasia prostate tissue. However, to the best of our knowledge, no previous studies have focused on the function or underlying mechanisms of miR-193a-3p in prostate cancer.
In the present study, the expression of miR-193a-3p was investigated in the DU-145 and PC3 prostate cancer cell lines. In addition, a gain-of-function study was performed to assess the effects of the forced expression of miR-193a-3p in prostate cancer cells, revealing their ability inhibit cell proliferation by regulating cyclin D1 (CCND1). These results improve the current understanding of the underlying mechanisms by which miR-193a-3p inhibits prostate cancer.
Materials and methods
Reagents, cell culture and transfection
All oligonucleotide segments were synthesized by GenePharma Co., Ltd. (Shanghai, China), including an miR-193a-3p mimic, negative control (NC) duplex and small interfering RNA (siRNA) against CCND1 (siCCND1). The sequences are listed in Table I.
Table I.
Oligonucleotide and primer sequences.
| Name | Sequence (5′-3′)a |
|---|---|
| miR-193a-3p mimic (sense) | AACUGGCCUACAAAGUCCCAGU |
| NC | ACUACUGAGUGACAGUAGA |
| siCCND1 (sense) | GGAGAACAAACAGAUCAUC |
| U6-F | TGCGGGTGCTCGCTTCGGCAGC |
| miR-193-3p-F | ACTGGCCTACAAAGTCCCAGT |
| CCND1-F | GCTGCGAAGTGGAAACCATC |
| CCND1-R | CCTCCTTCTGCACACATTTGAA |
| GAPDH-F | ACAACTTTGGTATCGTGGAAGG |
| GAPDH-R | GCCATCACGCCACAGTTTC |
| CCND1-UTR-F | ATTGCAGAGGATGTTCATAAGGCCAGTATGATTTATAAATGCAAT |
| CCND1-UTR-R | ATTGCATTTATAAATCATACTGGCCTTATGAACATCCTCTGCAAT |
| CCND1-Mut-F | ATTGCAGAGGATGTTCATAACCGGTCAATGATTTATAAATGCAAT |
| CCND1-Mut-R | ATTGCATTTATAAATCATTGACCGGTTATGAACATCCTCTGCAAT |
Target sites are in italic and bold type; mutated target sites are in bold type and underlined. miR, microRNA; NC, negative control; si, small interfering RNA; CCND1, cyclin D1; F, forward primer; R, reverse primer; UTR, untranslated region; Mut, mutant.
All cell lines used in this study were purchased from the Shanghai Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). The cell lines included the DU-145 and PC3 human prostate cancer cell lines, as well as the RWPE normal prostate epithelium cell line. All cell lines were cultured in RPMI-1640 medium (Takara Biotechnology Co., Ltd., Dalian, China) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 50 µg/ml streptomycin and 50 U/ml penicillin. Bovine pituitary extract (0.05 mg/ml; Thermo Fisher Scientific, Inc.) and human recombinant epidermal growth factor (5 ng/ml; Thermo Fisher Scientific, Inc.) were added to the culture medium of the RWPE cells. The cell culture environment was thermostatic at 37°C with constant humidity and 5% CO2. The cells were mainly seeded into 6-well plates at a density of 4×105 cells, lower density was used depending on certain experiments. Once the cells reached 60–70% confluency, all transfections were performed with Invitrogen Lipofectamine® 2000 Reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions, with the synthesized RNA mimics and NC. Transfected cells were cultured for 48 or 72 h at the same conditions described above.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from prostate cancer cells and normal cells with standard TRIzol® solution (Invitrogen; Thermo Fisher Scientific, Inc.). For miRNA expression, RT reactions were performed with a One Step PrimeScript miRNA cDNA Synthesis kit (Takara Biotechnology Co., Ltd.), followed by PCR with SYBR® Premix Ex Taq (Takara Biotechnology Co., Ltd). For mRNA, cDNA was synthesized from the total RNA using a PrimeScript RT Reagent Kit (Takara Biotechnology Co., Ltd.). qPCR amplification reactions for CCND1 expression were performed with SYBR® Premix Ex Taq II with ROX (Takara Biotechnology Co., Ltd.,). For miRNA and mRNA amplifications, analysis was performed with the ABI 7500 Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). According to supplier's instructions, the PCR conditions consisted of 95°C for 30 sec, followed by 40 cycles of amplification (95°C for 3 sec and 60°C for 30 sec). All fold changes were calculated using the comparative Cq (ΔΔCq) method using GAPDH for normalization (13). All relevant primer sequences are listed in Table I, and details of the reaction mixtures for RT and qPCR are listed in Tables II and III.
Table II.
Reaction mixture for reverse transcription.
| Component | Volume | Final concentration |
|---|---|---|
| 5X PrimeScript RT Master Mix | 2 µl | 1X |
| Total RNA | 500 ng | – |
| RNase Free dH2O | ≤10 µl | – |
Table III.
Reaction mixture for quantitative polymerase chain reaction (total volume, 10 µl).
| Component | Volume | Final concentration |
|---|---|---|
| SYBR Premix Ex Taq II | 5 µl | 1X |
| PCR Forward Primer | 0.4 µl | 0.4 µM |
| PCR Reverse Primer | 0.4 µl | 0.4 µM |
| ROX Reference Dye | 0.2 µl | 1X |
| cDNA solution (from reverse transcription) | 1 µl | – |
| dH2O | 3 µl | – |
Cell growth and viability assays
DU-145 and PC3 cells (6×103) were seeded into each well of a 96-well plate. Following an overnight incubation (37°C with 5% CO2), the cells were transfected with RNA duplexes (NC, miR-193a-3p or siCCND1) for 48–72 h as described in the previous section; the concentration of miR-193a-3p ranged from 0–75 nM. The medium was removed at various time points, then Cell Counting Kit-8 solution (WST-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added to each well and the cells were incubated at 37°C for 1 h. The absorbance of the solution was determined spectrophotometrically at 450 nm with an MRX II absorbance reader (Dynex Technologies, Inc., Chantilly, VA, USA).
Colony formation assay
The cells were harvested 24 h following RNA treatment (50 nM NC or 50 nM miR-193a-3p), resuspended in RPMI-1640 supplemented with 10% FBS, and seeded into 6-well plates at a density of 500 cells/well. The cultures were maintained under standard conditions (37°C and 5% CO2) for 14 days. The colonies were then fixed with 50% methanol for 15 min and stained with 5% crystal violet solution for 20 min. All colonies were scanned into a computer, then colonies visible to unaided eyes were counted and plotted for both cell lines.
Cell cycle analysis
Cells were harvested at 48 h following transfection and fixed in 75% ethanol at −20°C. Following fixing overnight, the cells were washed twice with phosphate-buffered saline (PBS) and stained with DNA Prep Stain (Beckman Coulter, Inc., Brea, CA, USA) for 30 min. Cell cycle analysis was performed using the BD LSRII Flow Cytometry system with FACSDiva software version 6.0 (BD Biosciences, Franklin Lakes, NJ, USA). The data were analyzed with the ModFit LT 5.0 software package (Verity Software House, Topsham, ME, USA).
Western blot analysis
PC3 and DU-145 cells were lysed with radioimmunoprecipitation assay buffer solution (Beyotime Institute of Biotechnology, Nantong, China) following 48 h of transfection on ice. The supernatants were collected following centrifugation at 13,000 × g at 4°C for 20 min and the protein concentration was determined using a bicinchoninic acid protein assay kit (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany). Supernatant samples (2 µl) were heated at 99°C for 5 min prior to loading and were separated by 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The proteins were transferred to a polyvinylidene difluoride membrane in transfer buffer containing 48 mmol/l Tris-HCl, 39 mmol/l glycine, 0.037% SDS and 20% methanol at 4°C for 45 min. The membranes were blocked with 5% non-fat milk for 1 h prior to incubation overnight with primary antibodies against CCND1 and GAPDH (catalog nos. EPR2241 and EPR16891; Epitomics; Abcam, Cambridge, MA, USA) at a dilution of 1:1,000. Following three washes in TBS, the membranes were then incubated with an horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (catalog no. 7074; Cell Signaling Technology, Inc., Boston, MA, USA) at room temperature for 1 h. Following three further washes in TBS, the proteins were detected and visualized using an electrochemiluminescence system (Pierce; Thermo Fisher Scientific, Inc.).
Plasmid construction and luciferase reporter assay
Bioinformatic screening was performed in order to initially identify the binding target of miR-193a-3p. TargetScan (http://www.targetscan.org) analysis revealed CCND1 as a potential binding site of miR-193a-3p. Two pairs of oligonucleotide fragments containing the miRNA target region and mutant miRNA target region of interest were designed and purchased from Sangon Biotech Co., Ltd. (Shanghai, China; Table I). The oligonucleotide pair was annealed at 90°C for 3 min and cooled to 37°C for 15 min prior to being inserted into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega Corporation, Madison, WI, USA) between the SacI and SalI sites. All the insertions were confirmed using sequencing to identify any mutations.
PC3 and DU-145 cells (7×104 cells/well) were cultured in a 24-well plate for 24 h prior to transfection. The pmirGLO vectors containing the wild type (Wt) 3′-UTR regions were co-transfected with miR-193a-3p-expressing or NC vectors. The co-transfection process was duplicated with vectors including a mutant (Mut) 3′-UTR region. The cells were harvested following a 24-h incubation at 37°C and luciferase activity was assayed using a Dual-Luciferase Reporter Assay System (Promega Corporation). Renilla luciferase (with the cloned 3′-UTR) activity was normalized to Firefly luciferase activity. Multiple (n≥3) independent experiments were conducted and the means and standard deviations were calculated from duplicate wells. The expression of luciferase was analyzed as described below.
CCND1 rescue experiments
The pIRES-enhanced green fluorescent protein (EGFP)-CCND1 plasmid (GeneChem Co., Ltd., Shanghai, China) was constructed following the insertion of the human CCND1 coding sequence into a pIRES-EGFP vector and confirmed by sequencing. The cells were co-transfected with either miR-193-3p mimics or NC with pIRES-EGFP-CCND1 or an empty pIRES-EGPF vector. The cells were harvested at 48 h post-transfection and flow cytometry was used to analyze the cell cycle. In addition, CCND1 expression levels were determined by western blotting.
Statistical analysis
The data are presented as the mean ± standard deviation of three independent experiments. The variations between the cell samples were analyzed using a Student's t-test. All data analysis was performed using Prism version 6 (GraphPad Software, Inc., La Jolla, CA, USA) for Windows. P<0.05 was considered to indicate a statistically significant result.
Results
miR-193-3p is downregulated in prostate cancer cell lines
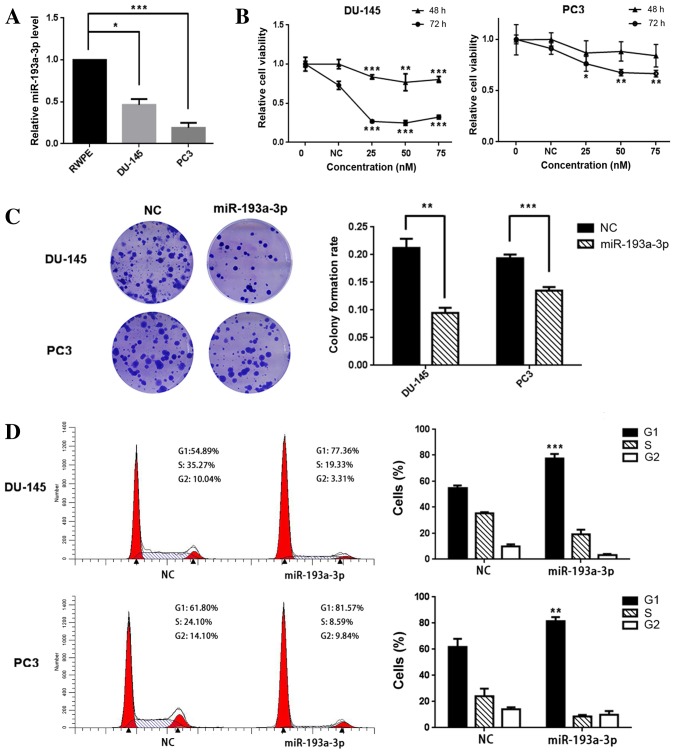
To investigate the role of miR-193a-3p in human prostate cancer, RT-qPCR was used to evaluate the gene expression levels in PC3 and DU-145 cell lines and the RWPE non-malignant cell line. The results demonstrated that the expression levels of miR-193a-3p in PC3 and DU-145 cells were significantly reduced, compared with those in RWPE cells; the relative expression levels of DU-145 and PC3 were 46.3% (P=0.018) and 19.0% (P<0.001), respectively, relative to that in RWPE cells (Fig. 1A).
Figure 1.
(A) miR-193a-3p is downregulated in prostate cancer cell lines. Reverse transcription-quantitative polymerase chain reaction, using U6 for normalization, was utilized to assess the expression levels of miR-193a-3p in DU-145 and PC3 prostate cancer cells and RWPE non-cancerous prostate cells, revealing decreased expression levels in the cancerous cell lines compared with the non-cancerous cells (*P<0.05 and ***P<0.001). (B) Cell viability/cell growth assays revealed that the relative cell viabilities (with cell viability at 0 nM regarded as 1.0) of the miR-193a-3p-transfected groups of DU-145 and PC3 cells were lower, compared with the NC-transfected groups (*P<0.05, **P<0.01 and ***P<0.001 vs. NC). An optimal concentration for miR-193a-3p was identified between 25 and 50 nM for DU-145 and PC3 cells. (C) Colony formation assays indicated that the colony formation rate was lower for miR-193a-3p-transfected groups compared with NC-transfected groups (**P<0.01 and ***P<0.001); representative wells are shown. (D) Cell cycle profiles in DU-145 and PC3 cells are shown. Overexpression of miR-193a-3p induced a significant G1-phase arrest in DU-145 and PC3 cells (**P<0.01 and ***P<0.001 vs. NC). Representative histograms are depicted and data are presented as the average of repeated experiments. miR, microRNA; NC, negative control.
miR-193-3p suppresses prostate cancer cell proliferation and colony formation, and induces G1-phase arrest
In order to investigate whether miR-193a-3p functions as a tumor-suppressor or enhancer, PC3 and DU-145 cells were transfected with miR-193a-3p mimics. miR-193a-3p transfection was found to suppress the growth and viability of the prostate cancer cells (Fig. 1B); compared with the NC, 25 nM miR-193a-3p was able to reduce PC3 cell viability by ~13% (P=0.083) and ~24% (P=0.021) at 48 and 72 h post-transfection, respectively, and the corresponding results for DU-145 were ~17% (P<0.001) and ~63% (P<0.001), respectively. Similarly, the colony formation ability was inhibited in the two prostate cancer lines; the colony formation rates of miR-193a-3p-transfected cells were lower, compared with those in the NC-transfected cells, in the two cancer cell lines [59.1% (P=0.007) and 31.6% (P<0.001), respectively; Fig. 1C].
In order to investigate the mechanisms underlying miR-193a-3p-mediated cell growth suppression, flow cytometry was used to analyze the distribution of the cell cycle in PC3 and DU-145 cells following their transfection with miR-193a-3p. The cell cycle distribution demonstrated a significant G1-phase arrest in the DU-145 and PC3 cells [54.89–77.36% (P<0.001) and 61.80–81.57% (P=0.004), respectively; Fig. 1D]. The results indicate that miR-193a-3p is able to suppress the growth of prostate cancer cells by arresting the cells in the G1 phase.
miR-193-3p inhibits the expression of CCND1
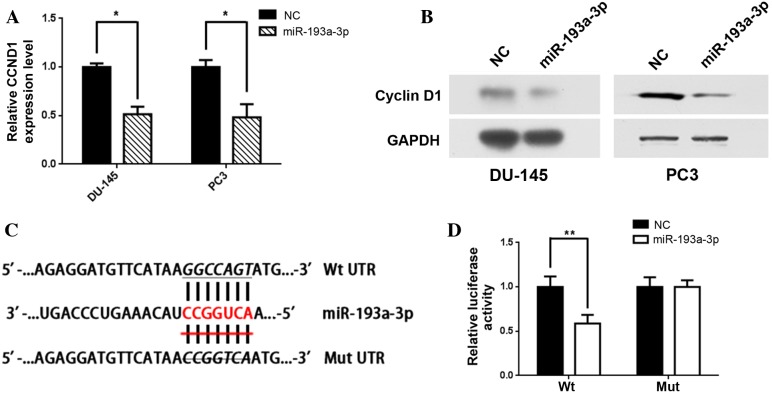
TargetScan provided 221 potential downstream targets with conserved sites for miR-193a-3p. Among the genes predicted, CCND1 exhibited a significant decline in mRNA expression levels in the PC3 and DU-145 cells. Furthermore, RT-qPCR revealed that the relative mRNA expression levels of CCND1 were significantly inhibited following transfection with miR-193a-3p as compared with the NC [relative expression, 0.52 (P=0.03) and 0.49 (P=0.04), respectively; Fig. 2A], and western blot analysis also indicated that CCND1 protein levels were markedly suppressed in the cell lines transfected with miR-193a-3p (Fig. 2B).
Figure 2.
CCND1 is a direct target of miR-193a-3p. (A) Reverse transcription-quantitative polymerase chain reaction revealed that overexpression of miR-193a-3p significantly decreased CCND1 expression levels in DU-145 and PC3 cells (*P<0.05). (B) Western blot analysis was performed to detect the expression of CCND1 or GAPDH following the transfection of DU-145 and PC3 cells with miR-193a-3p mimics. (C) A seed region predicted in TargetScan in the Wt 3′-UTR of CCND1 is depicted (top) above the corresponding miR-193a-3p sequence. The Mut sequence used is shown at the bottom (crossed). (D) PC3 cells were co-transfected with 50 nM of miR-193a-3p mimics or NC oligonucleotides, as well as 100 ng plasmid with either Wt or Mut 3′-UTR of CCND1. The relative firefly luciferase activity, normalized with renilla luciferase, was evaluated 48 h post-transfection (**P<0.01). All data are presented as the mean + standard deviation. CCND1, cyclin D1; miR, microRNA; UTR, untranslated region; Wt, wild type; Mut, mutant type; NC, negative control.
In order to determine whether CCND1 is a direct target of miR-193a-3p, the 3′-UTR of CCND1 was inserted into a pmirGLO Dual-Luciferase miRNA Target Expression Vector downstream of the firefly luciferase. A second vector in which the putative binding site of miR-193a-3p in the 3′-UTR was mutated was also constructed (Fig. 2C). The results indicated that co-transfection of the Wt 3′-UTR and miR-193a-3p in PC3 cells significantly suppressed luciferase activity compared with cells co-transfected with the Wt 3′-UTR and NC (P=0.004). However, co-transfection with Mut 3′-UTR and miR-193a-3p was not observed to alter luciferase activity (P=0.22) in PC3 cells (Fig. 2D).
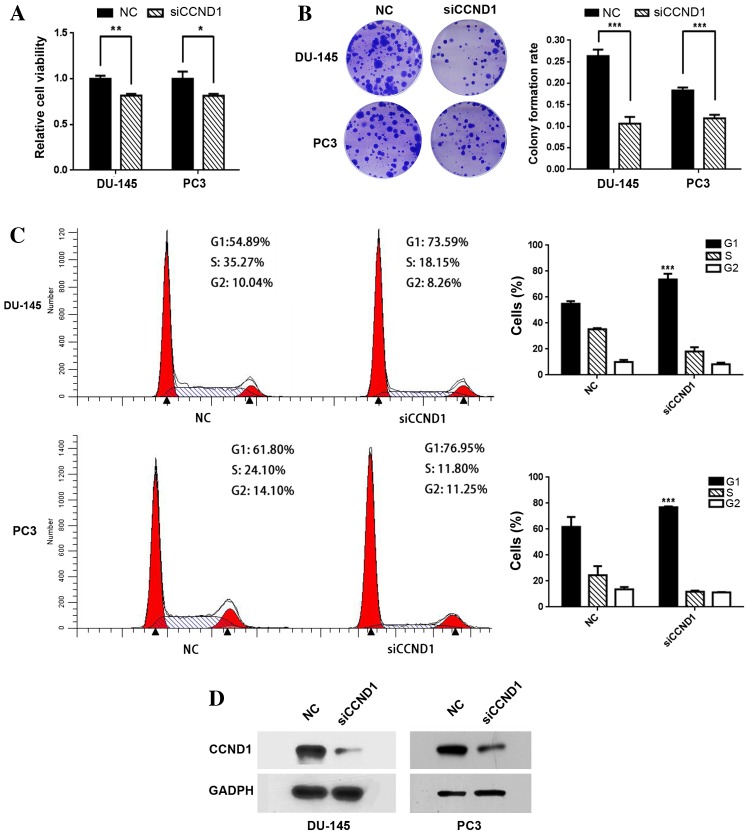
Furthermore, the knockdown of CCND1 by siCCND1 suppressed cell viability in the two cancer cell lines [reduced by 18.6% in DU-145 (P=0.005) and 18.5% (P=0.013) in PC3; shown in Fig. 3A]. In addition, cell colony counts were significantly decreased by siCCND1 transfection [reduced by 60.6% (P<0.001) in DU-145 and 36.3% (P<0.001) in PC3, respectively; Fig. 3B] The results also indicate that siCCND1 may induce G1/S phase arrest, leading to a significant increase in the proportion of G1 phase cells (P<0.001 in both cell lines; Fig. 3C). Western blot analysis demonstrated that siCCND1 had a similar effect to transfection with miR-193a-3p, inhibiting CCND1 expression (Fig. 3D). The results suggest that the knockdown of CCND1 may mimic the biological effect of miR-193a-3p.
Figure 3.
Knockdown of CCND1 has a similar effect to miR-193a-3p transfection. (A) Knockdown of miR-193a-3p by siCCND1 suppressed prostate cancer cell growth; siCCND1 induced a reduction in the viability of DU-145 and PC3 cells at 48 h post-transfection by ~18.6 and ~18.5%, respectively (*P<0.05 and **P<0.01). (B) siCCND1 reduced the colony-formation rate in DU-145 and PC3 cells (representative wells are presented) (***P<0.001). (C) siCCND1 reduced the protein expression levels of CCND1 in the cells. All data are presented as the mean + standard deviation of at least three repeated experiments (***P<0.001 vs. NC). CCND1, cyclin D1; miR, microRNA; siCCND1, small interfering RNA targeting CCND1; UTR, untranslated region; NC, negative control. (D) Knockdown of CCND1 induced a significant G1 phase arrest (representative histograms are depicted).
Restoration of CCND1 expression partially rescues miR-193-3p-induced cell cycle arrest
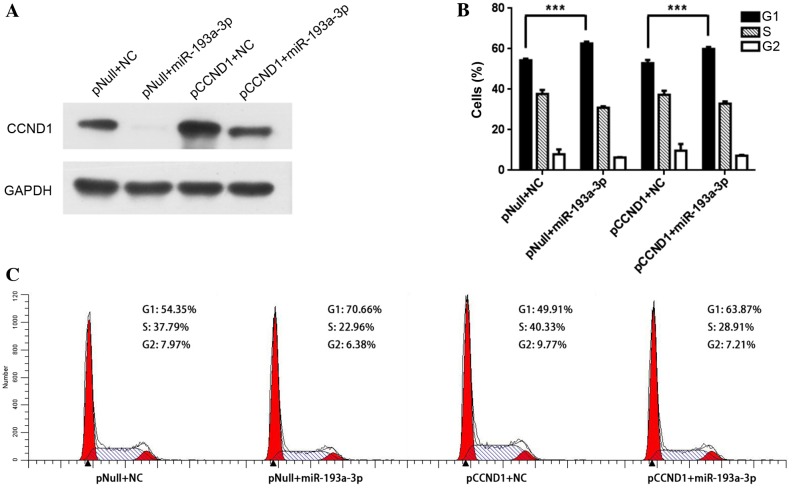
To investigate whether forced CCND1 expression is able to counter the cell cycle arrest induced by miR-193a-3p, the human CCND1 coding sequence was inserted into a pIRES-EGFP vector. Western blot analysis revealed that CCND1 expression was restored following treatment with the pIRES-EGFP-CCND1 vector in miR-193a-3p-transfected cells, compared to treatment with the empty vector (Fig. 4A). Furthermore, the cell cycle was evaluated by flow cytometry following the co-transfection of PC3 cells with miR-193a-3p mimics or NC, and pIRES-EGFP-CCND1 or empty pIRES-EGPF vectors. The results demonstrated that restoration of CCND1 expression is able to partially reverse the miR-193a-3p-induced cell cycle arrest (Fig. 4B and C).
Figure 4.
Rescued CCND1 expression reverses miR-193a-3p-induced G1 phase arrest. (A) PC3 cells were co-transfected with miR-193a-3p mimics or NC oligonucleotides, and pCCND1 or pNull. The expression of CCND1 and GAPDH was detected by western blot analysis. (B) Independent of CCND1 transfection, miR-193a-3p causes significant G1 phase arrest (***P<0.001). (C) Flow cytometry indicated that the induced expression of CCND1 partially attenuated the cell cycle arrest in miR-193a-3p-transfected PC3 cells. All data are presented as the mean + standard deviation. CCND1, cyclin D1; miR, microRNA; NC, negative control; pNull, empty vector; pCCND1, pIRES-EGFP-CCND1.
Discussion
miRNAs have previously been indicated to be promising diagnostic and prognostic factors in tumor studies (14), and dysregulated miRNA expression profiles have been observed in several previous studies of prostate cancer (15–17). miRNAs have been reported to have vital roles in prostate cancer cell proliferation, progression and metastasis (18). Microarray analysis has indicated that miR-193a-3p is a specific miRNA that has decreased expression in prostate cancer tissues (GEO accession: GSE36802). Similar results were obtained in the current study from quantification experiments in the PC3 and DU-145 prostate cancer cell lines. However, the specific function of miR-193a-3p in prostate cancer has yet to be elucidated. Therefore, a series of experiments were conducted to study the function of miR-193a-3p in prostate cancer.
In order to identify how prostate cancer cell function is altered by miR-193a-3p, gain-of-function studies were conducted in PC3 and DU-145 cells. The results of the cell viability and colony formation assays demonstrated that transfection with miR-193a-3p markedly suppressed cell proliferation in PC3 and DU-145 cells. Furthermore, flow cytometry revealed both PC3 and DU-145 exhibited significant G1/S phase arrest following the transfection of miR-193a-3p, which may be one of the mechanisms underlying miR-193a-3p-mediated growth inhibition. The results suggested that miR-193a-3p had a profound effect on the cell cycle, proliferation and survival in prostate cancer.
Subsequently, the aim of the current study was to identify the molecular mechanisms underlying the alterations in cell function induced by miR-193a-3p. RT-qPCR revealed reduced expression levels of CCND1 and the luciferase assay further demonstrated that CCND1 is a target of miR-193a-3p. Furthermore, the knockdown of CCND1 exhibited a similar effect to the transfection of miR-193a-3p, inducing the inhibition of cell growth and initiating G1 phase arrest. Restored CCND1 expression was able to partially reverse the miR-193a-3p-induced cell cycle arrest. In addition, co-transfection with pIRES-EGFP-CCND1 partially rescued the miR-193a-3p-induced G1 phase arrest. Therefore, the results of the present study indicated that miR-193a-3p is capable of suppressing the proliferation of prostate cancer cells by targeting CCND1.
Cyclins are a set of regulatory subunits of holoenzymes that regulate the progression of cells through the cell cycle. When it was first discovered in 1991 (19), CCND1 was considered to be a regulator of the cell cycle. In more recent studies, CCND1 has also been identified as an oncogene that is frequently dysregulated in breast cancer (20) and non-Hodgkin's lymphoma (21), as well as other types of cancer. During normal physiological functioning, CCND1 serves as a key sensor and integrator of extracellular signals, mediating cell function through the binding of cyclin-dependent kinases. CCND1 also induces the sequential inactivation of the cell cycle-inhibitory function of retinoblastoma protein (Rb) (22), which serves as a gatekeeper of the G1 phase in the cell cycle; passage through this restriction point leads to DNA synthesis and G1/S phase transition (23). Therefore, inhibiting CCND1 expression by miR-193a-3p transfection may increase the proportion of prostate cancer cells in the G1 phase, which is concordant with the results of the present study. However, CCND1 may have additional oncogenic effects beyond its role in the cell cycle. Studies in solid tumor models have demonstrated that CCND1 is able to function as a transcription regulator by interacting with certain transcription factors, and chromatin-remodeling and histone-modifying enzymes (22,24,25). CCND1 may also promote chromosome instability by binding to specific genes that regulate chromosome segregation and chromatin reorganization (26). Notably, CCND1 has previously been implicated in promoting DNA repair by binding to recombination-activating gene 1 and by homologous DNA recombination (27), which is also considered an anti-apoptotic effect. A previous study reported that cytoplasmic CCND1 was involved in the estradiol-induced DNA damage response; CCND1 binds estrogen receptor α at the cytoplasmic membrane and augments protein kinase B (Akt) phosphorylation (Ser473) and histone γ H2AX foci formation (28). In the present study, decreased expression levels of CCND1 were observed following transfection with miR-193a-3p and the luciferase assay also identified that miR-193a-3p inhibits CCND1 expression by directly binding to the 3′-UTR at a post-transcriptional level. Tian et al (29) suggested that miR-19b, miR-23b, miR-26a and miR-92a may promote prostate cancer cell proliferation by co-regulating the expression of phosphatase and tensin homolog, phosphoinositide 3-kinase/Akt and CCND1 in vitro. Therefore, the suppression of cell proliferation by inhibiting CCND1 expression in prostate cancer is a promising therapeutic target.
In conclusion, the current study identified the downregulation of miR-193a-3p in certain prostate cancer cell lines and revealed that miR-193a-3p may function as a potential tumor-suppressor in prostate cancer, which is able to induce G1-phase arrest by targeting CCND1. Although further studies are required to identify other targets of miR-193a-3p, the results demonstrated that the restoration of miR-193-3p expression may be a novel therapeutic strategy for preventing the progression of prostate cancer.
Acknowledgements
This study was supported by grants from the following institutions: The Qianjiang Talent Project of Technology Office in Zhejiang Province (grant no. 2013R10045); the Scientific Research Foundation of the Ministry of Public Health of China (grant no. WKJ2012-2-009); the Zhejiang Province Key Project of Science and Technology (grant no. 2014C4008-2); the National Natural Science Foundation of China (grant nos. 81372773 and 81101717); and the Scientific Research Fund of Zhejiang Provincial Education Department (grant no. Y201120149).
Glossary
Abbreviations
- miRNA
microRNA
- miR-193a-3p
microRNA-193a-3p
- CCND1
cyclin D1
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- NC
negative control
- Wt
wild type
- Mut
mutant type
References
- 1.NCI: SEER stat fact sheets: Prostate cancer. In: Surveillance, epidemiology and end results program. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE36802. National Institute of Health. 2013 2013 Sep 4; Accessed. [Google Scholar]
- 2.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Fendler A, Jung M, Stephan C, Erbersdobler A, Jung K, Yousef GM. The antiapoptotic function of miR-96 in prostate cancer by inhibition of FOXO1. PLoS One. 2013;8:e80807. doi: 10.1371/journal.pone.0080807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kojima S, Enokida H, Yoshino H, Itesako T, Chiyomaru T, Kinoshita T, Fuse M, Nishikawa R, Goto Y, Naya Y, et al. The tumor-suppressive microRNA-143/145 cluster inhibits cell migration and invasion by targeting GOLM1 in prostate cancer. J Hum Genet. 2014;59:78–87. doi: 10.1038/jhg.2013.121. [DOI] [PubMed] [Google Scholar]
- 5.Arora S, Saini S, Fukuhara S, Majid S, Shahryari V, Yamamura S, Chiyomaru T, Deng G, Tanaka Y, Dahiya R. MicroRNA-4723 inhibits prostate cancer growth through inactivation of the abelson family of nonreceptor protein tyrosine kinases. PLoS One. 2013;8:e78023. doi: 10.1371/journal.pone.0078023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Li B, Li L, Wang T. MicroRNA-497 suppresses proliferation and induces apoptosis in prostate cancer cells. Asian Pac J Cancer Prev. 2013;14:3499–3502. doi: 10.7314/APJCP.2013.14.6.3499. [DOI] [PubMed] [Google Scholar]
- 7.He L, Yao H, Fan LH, Liu L, Qiu S, Li X, Gao JP, Hao CQ. MicroRNA-181b expression in prostate cancer tissues and its influence on the biological behavior of the prostate cancer cell line PC-3. Genet Mol Res. 2013;12:1012–1021. doi: 10.4238/2013.April.2.17. [DOI] [PubMed] [Google Scholar]
- 8.Mao Y, Chen H, Lin Y, Xu X, Hu Z, Zhu Y, Wu J, Xu X, Zheng X, Xie L. MicroRNA-330 inhibits cell motility by downregulating Sp1 in prostate cancer cells. Oncol Rep. 2013;30:327–333. doi: 10.3892/or.2013.2452. [DOI] [PubMed] [Google Scholar]
- 9.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 11.Kietzmann L, Guhr SS, Meyer TN, Ni L, Sachs M, Panzer U, Stahl RA, Saleem MA, Kerjaschki D, Gebeshuber CA, Meyer-Schwesinger C. MicroRNA-193a regulates the transdifferentiation of human parietal epithelial cells toward a podocyte phenotype. J Am Soc Nephrol. 2015;26:1389–1401. doi: 10.1681/ASN.2014020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin PC, Chiu YL, Banerjee S, Park K, Mosquera JM, Giannopoulou E, Alves P, Tewari AK, Gerstein MB, Beltran H, et al. Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression. Cancer Res. 2013;73:1232–1244. doi: 10.1158/0008-5472.CAN-12-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Allegra A, Alonci A, Campo S, Penna G, Petrungaro A, Gerace D, Musolino C. Circulating microRNAs: New biomarkers in diagnosis, prognosis and treatment of cancer (review) Int J Oncol. 2012;41:1897–1912. doi: 10.3892/ijo.2012.1647. [DOI] [PubMed] [Google Scholar]
- 15.Tay FC, Lim JK, Zhu H, Hin LC, Wang S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv Drug Deliv Rev. 2015;81:117–127. doi: 10.1016/j.addr.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Wojcicka A, de la Chapelle A, Jazdzewski K. MicroRNA-related sequence variations in human cancers. Hum Genet. 2014;133:463–469. doi: 10.1007/s00439-013-1397-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim WT, Kim WJ. MicroRNAs in prostate cancer. Prostate Int. 2013;1:3–9. doi: 10.12954/PI.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiang XF, Zhang ZW, Liu Q, Sun N, Pan LL, Shen J, Li T, Yun C, Li H, Shi LH. MiR-20a promotes prostate cancer invasion and migration through targeting ABL2. J Cell Biochem. 2014;115:1269–1276. doi: 10.1002/jcb.24778. [DOI] [PubMed] [Google Scholar]
- 19.Peters G. The D-type cyclins and their role in tumorigenesis. J Cell Sci Suppl. 1994;18:89–96. doi: 10.1242/jcs.1994.Supplement_18.13. [DOI] [PubMed] [Google Scholar]
- 20.Lee EY, Muller WJ. Oncogenes and tumor suppressor genes. Cold Spring Harb Perspect Biol. 2010;2:a003236. doi: 10.1101/cshperspect.a003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jares P, Colomer D, Campo E. Molecular pathogenesis of mantle cell lymphoma. J Clin Invest. 2012;122:3416–3423. doi: 10.1172/JCI61272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: Normal and abnormal functions. Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 23.Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 24.Bienvenu F, Jirawatnotai S, Elias JE, Meyer CA, Mizeracka K, Marson A, Frampton GM, Cole MF, Odom DT, Odajima J, et al. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature. 2010;463:374–378. doi: 10.1038/nature08684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aggarwal P, Vaites LP, Kim JK, Mellert H, Gurung B, Nakagawa H, Herlyn M, Hua X, Rustgi AK, McMahon SB, Diehl JA. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell. 2010;18:329–340. doi: 10.1016/j.ccr.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casimiro MC, Crosariol M, Loro E, Ertel A, Yu Z, Dampier W, Saria EA, Papanikolaou A, Stanek TJ, Li Z, et al. ChIP sequencing of cyclin D1 reveals a transcriptional role in chromosomal instability in mice. J Clin Invest. 2012;122:833–843. doi: 10.1172/JCI60256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jirawatnotai S, Hu Y, Michowski W, Elias JE, Becks L, Bienvenu F, Zagozdzon A, Goswami T, Wang YE, Clark AB, et al. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature. 2011;474:230–234. doi: 10.1038/nature10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, Chen K, Jiao X, Wang C, Willmarth NE, Casimiro MC, Li W, Ju X, Kim SH, Lisanti MP, et al. Cyclin D1 integrates estrogen-mediated DNA damage repair signaling. Cancer Res. 2014;74:3959–3970. doi: 10.1158/0008-5472.CAN-13-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian L, Fang YX, Xue JL, Chen JZ. Four microRNAs promote prostate cell proliferation with regulation of PTEN and its downstream signals in vitro. PLoS One. 2013;8:e75885. doi: 10.1371/journal.pone.0075885. [DOI] [PMC free article] [PubMed] [Google Scholar]