This review by Kearse and Wilusz discusses the profound impact of non-AUG start codons in eukaryotic translation. It describes how misregulation of non-AUG initiation events contributes to multiple human diseases, including cancer and neurodegeneration, and how modulation of non-AUG usage may represent a novel therapeutic strategy.
Keywords: start codon, near-cognate, RAN translation, translation initiation, eIF2A, eIF2D
Abstract
Although it was long thought that eukaryotic translation almost always initiates at an AUG start codon, recent advancements in ribosome footprint mapping have revealed that non-AUG start codons are used at an astonishing frequency. These non-AUG initiation events are not simply errors but instead are used to generate or regulate proteins with key cellular functions; for example, during development or stress. Misregulation of non-AUG initiation events contributes to multiple human diseases, including cancer and neurodegeneration, and modulation of non-AUG usage may represent a novel therapeutic strategy. It is thus becoming increasingly clear that start codon selection is regulated by many trans-acting initiation factors as well as sequence/structural elements within messenger RNAs and that non-AUG translation has a profound impact on cellular states.
Eukaryotic genomes encode thousands of proteins with important structural and regulatory roles, and a significant amount of cellular energy is dedicated to transcription of messenger RNAs (mRNAs) and their subsequent translation into protein. Errors in translation can result in wasteful production of inactive or deleterious proteins that misfold, aggregate, lack regulation, or otherwise disrupt cellular fitness (Drummond and Wilke 2009). It is thus critical that ribosomes initiate at the appropriate codon, incorporate the appropriate amino acids into the growing polypeptide chain, and terminate only at the appropriate stop codon (Zaher and Green 2009; Rozov et al. 2016). Protein-coding sequences have traditionally been defined as uninterrupted ORFs that begin with the universal AUG start codon and end with one of three stop codons (UAA, UGA, and UAG). However, it has been known since the 1980s that translation can initiate at codons other than AUG, albeit at a much lower efficiency (Zitomer et al. 1984; Peabody 1987, 1989; Clements et al. 1988; Hann et al. 1988). In most of these cases, near-cognate codons that differ from AUG by only one nucleotide (e.g., CUG, GUG, and UUG) are used.
Given the strong evolutionary pressure that is observed across all organisms to use AUG start codons, it may appear at first glance that initiation events from non-AUG codons represent intrinsic errors of the translation machinery. This idea is directly challenged by the fact that a number of endogenous and viral proteins with important functions are derived solely from non-AUG start codons (Curran and Kolakofsky 1988; Dorn et al. 1990; Xiao et al. 1991; Chang and Wang 2004; Tang et al. 2004; Beerman and Jongens 2011; Ivanov et al. 2011). For example, DAP5 (also called eukaryotic initiation factor 4G2 [eIF4G2] or NAT1) plays a critical role in internal ribosome entry site (IRES)-mediated translation and is initiated solely from a GUG start codon in mouse and human cells (Imataka et al. 1997; Takahashi et al. 2005; Lewis et al. 2008; Marash et al. 2008; Liberman et al. 2015). Likewise, in yeast, UUG and ACG start codons are used to initiate translation of the GRS1 and ALA1 transfer RNA (tRNA) synthetases, respectively (Chang and Wang 2004; Tang et al. 2004). These non-AUG translation events represent only the tip of the iceberg, as ribosome profiling has recently revealed thousands of novel initiation events at non-AUG codons (Ingolia et al. 2009, 2011). Interestingly, not all near-cognate start codons are used with equal efficiency, with CUG generally being most efficient, followed by GUG, ACG, and AUU (Table 1). It should be noted that there is significant variation in these efficiency measurements across assays. This is likely because in vitro translation assays are strongly influenced by how the lysates are prepared (e.g., those prepared by gel filtration may lack low-molecular-weight translation factors) and the ionic concentrations used (Kozak 1989, 1990b).
Table 1.
Efficiency of non-AUG start codons in various assays
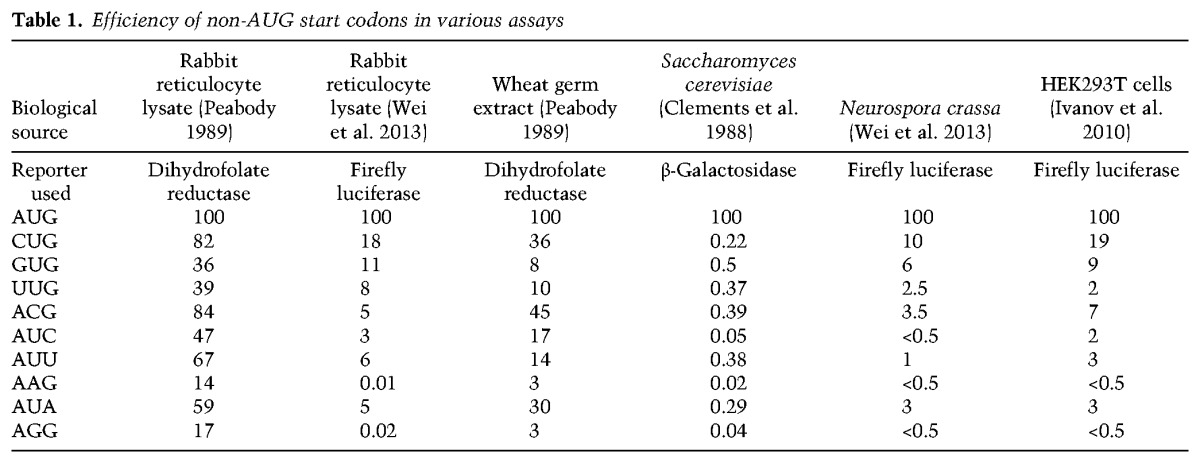
While endogenous functional non-AUG start codons may be more prevalent than previously appreciated, it should not be dismissed that such codons typically perform at a markedly reduced efficiency compared with AUG codons (Table 1). Thus, the purpose of this review is not to suggest that most proteins within cells are derived from non-AUG start codons but to highlight how alternative initiation codons can be used to increase protein isoform diversity and impact cellular processes (Touriol et al. 2003). Besides highlighting the widespread nature of non-AUG translation events in eukaryotic cells, we discuss the trans-acting proteins and the features within mRNAs that dictate start codon recognition. In some cases, a canonical scanning mechanism of translation initiation appears to be used, but there are also a number of alternative factors (which can be induced by various stresses) that alter start codon preferences. As aberrant non-AUG translation events are associated with, and likely drive, multiple human diseases, including cancer and neurodegeneration (Zu et al. 2011; Sendoel et al. 2017), there exists the intriguing possibility that modulating non-AUG translation events (e.g., using small molecule inhibitors) may have profound therapeutic effects.
Thousands of non-AUG codons can be used for translation initiation
For many years, the identification of non-AUG start codons was often fortuitous and resulted from efforts aimed at cloning genes of interest. For example, Xiao et al. (1991) observed that endogenous TEAD1 (also called TEF-1) in HeLa cells did not comigrate in SDS-PAGE with in vitro translated TEAD1 that was initiated from the predicted AUG start codon. By pursuing this observation and using mutational analysis, it was revealed that endogenous TEAD1 uses solely an upstream AUU start codon. Approaches like these are largely limited to single genes. However, a genome-wide view of translation can now be provided by ribosome profiling, in which short mRNA fragments that are protected by 80S ribosomes are purified and subjected to high-throughput sequencing (Ingolia et al. 2009, 2011; Ingolia 2010). Ribosome profiling, sometimes referred to as Ribo-seq, has revolutionized our understanding of where translating ribosomes are present across the transcriptome as well as revealed key features that influence translation patterns (for review, see Ingolia 2014; Brar and Weissman 2015; Andreev et al. 2017). For example, specific combinations of codons (often involving proline) can slow translation in both mammalian cells and yeast (Ingolia et al. 2011; Gamble et al. 2016).
By treating cells with early elongation inhibitors that block 80S ribosomes after initiation but before the first translocation cycle, ribosome profiling can be used to define translation start sites (Fig. 1). Lactimidomycin (which binds the exit site [E site] of the 60S subunit) (Schneider-Poetsch et al. 2010) or harringtonine (which binds the aminoacyl site [A site] of the 60S subunit) (Fresno et al. 1977) are used for exactly this purpose, as they have little, if any, effect on 80S ribosomes beyond the first translocation step. Thousands of previously unannotated initiation events have now been identified in mouse embryonic stem cells, ∼60% of which initiate at a non-AUG start codon (Ingolia et al. 2011). This includes >75% of upstream ORFs (uORFs) that are present in transcript leader regions (Ingolia et al. 2011). This number should not be misinterpreted to mean that most proteins in cells are derived from non-AUG translation events or that translation initiation at a non-AUG start codon is more efficient than at a canonical AUG. Instead, these data simply indicate that if one tallies up initiation sites across the transcriptome, ignoring the relative efficiency of each event, translation initiation at non-AUG start codons outweighs initiation at AUG codons.
Figure 1.
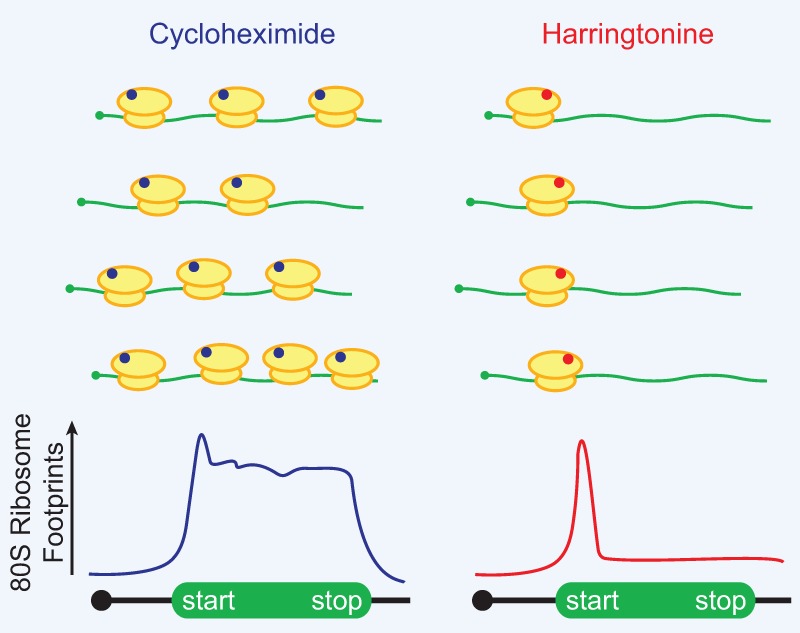
Ribosome profiling can be used to reveal translation initiation sites across the transcriptome. (Left) Treatment with cycloheximide (blue), which binds to the E site of the 60S subunit, pauses all elongating 80S ribosomes. The mapped ribosome footprints thus typically cover the entire ORF. (Right) In contrast, harringtonine (red) binds to the A site of the 60S subunit and only inhibits 80S ribosomes just after subunit joining at the start codon. Mapped ribosome footprints from harringtonine treatment are thus enriched for the start codon.
As these data challenge the prevailing model of eukaryotic initiation, there has naturally been debate about whether all the footprints obtained from ribosome profiling experiments represent true translation signals (Guttman et al. 2013). For example, it is now clear that the inclusion of translation inhibitors (such as cycloheximide, which helps trap 80S ribosomes on mRNAs) during the preparation and lysis of the cells can sometimes adversely influence the data (Gerashchenko and Gladyshev 2014; Lareau et al. 2014; Hussmann et al. 2015; Andreev et al. 2017). Furthermore, the addition of early elongation inhibitors such as lactimidomycin or harringtonine causes large increases in free ribosomal subunits, which likely influence initiation to some degree. Indeed, some putative initiation events identified using ribosome profiling have not been able to be experimentally confirmed and thus likely represent false positives (Zhang and Hinnebusch 2011). Nevertheless, a growing number of functional peptides generated from non-AUG start codons have been documented (Starck et al. 2008, 2012; Slavoff et al. 2013), and continuing improvements to the methods provide increasingly high-confidence data sets. For example, computational approaches now take into account both fragment size and 3-nucleotide (nt) periodicity (indicative of decoding the genetic code) (Ingolia 2014; Brar and Weissman 2015; Fields et al. 2015), and orthogonal approaches, such as mass spectrometry and the insertion of small epitope tags into endogenous genes (Menschaert et al. 2013; Slavoff et al. 2013; Ingolia et al. 2014), are increasingly being used for validation.
Start codon recognition is tightly controlled by multiple initiation factors
The mechanism of canonical eukaryotic translation initiation and the role of individual eIFs have been reviewed extensively (Sonenberg and Hinnebusch 2009; Jackson et al. 2010; Lorsch and Dever 2010; Hinnebusch 2017). Briefly, for the ribosome to identify a suitable start codon, most eukaryotic translation is thought to follow the scanning model of initiation (Fig. 2). The ternary complex (TC) is first formed when GTP-bound eIF2 (a heterotrimer of α, β, and γ subunits) binds the initiator methionyl-tRNA (Met-tRNAiMet). The TC then interacts with the 40S ribosomal subunit and a number of eIFs, including eIF1, eIF1A, eIF5, and the eIF3 complex (13 subunits [a–m] in humans), to form the 43S preinitiation complex (PIC). Once assembled, the 43S PIC is recruited to the 7-methylguanosine (m7G) cap at the 5′ ends of mRNAs by the eIF4F complex, which consists of the cap-binding protein eIF4E, the DEAD-box helicase eIF4A, and the scaffolding protein eIF4G that binds PABP at the mRNA 3′ end, thereby generating a 48S-activated mRNA. In an ATP-dependent manner, the PIC then scans 5′ to 3′ in search of a start codon, which is most often the first AUG. Once the anti-codon of Met-tRNAiMet (bound by eIF2) base-pairs with the start codon in the peptidyl site (P site) of the scanning PIC, GTP is hydrolyzed by eIF2, and Pi is released. This step is promoted by eIF5, the GTPase-activating protein for eIF2. Subsequent displacement of eIF1, eIF1A, eIF2•GDP, eIF3, and eIF5 along with an additional GTP hydrolysis event by eIF5B allows for joining of the large 60S ribosomal subunit, thereby forming the complete 80S ribosome and allowing the second codon to be decoded in the A site.
Figure 2.
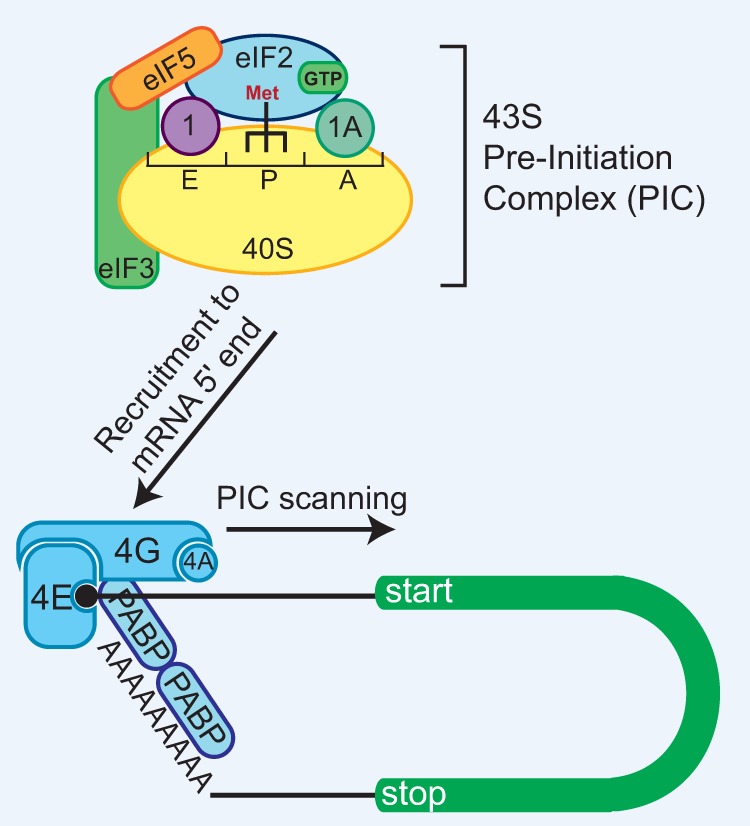
Canonical eukaryotic translation uses a scanning model of initiation. The 43S PIC consists of the 40S small ribosomal subunit, the TC (eIF2•GTP•Met-tRNAiMet), eIF1, eIF1A, eIF3, and eIF5. The PIC is recruited to the 5′ ends of mRNAs by the eIF4F complex, which consists of the cap-binding protein eIF4E, the DEAD-box helicase eIF4A, and the scaffolding protein eIF4G that binds PABP at the mRNA 3′ end. The PIC then scans in an ATP-dependent manner in a 5′-to-3′ direction in search of a start codon. Base pairing between the anti-codon of the Met-tRNAiMet and the start codon causes GTP hydrolysis by eIF2 and translation initiation. (1) eIF1; (1A) eIF1A; (4A) eIF4A; (4E) eIF4E; (4G) eIF4G.
The eIFs that are responsible for start codon recognition were first identified in Saccharomyces cerevisiae using a strain that harbored a start codon mutation (AUG to ACG) in the HIS4 gene (Donahue and Cigan 1988; Castilho-Valavicius et al. 1990). When cultured in medium lacking the essential amino acid histidine, these cells survived only if they used an in-frame near-cognate UUG codon that is present at the third codon of the mutant ORF. This is because initiation from the UUG codon generates a slightly truncated but functional His4 protein that supports histidine biosynthesis. By characterizing spontaneous revertants that grew despite the absence of histidine in the medium, five suppressor of initiation (SUI) codon mutation genes were identified: eIF1 (SUI1), all three (α, β, and γ) subunits of eIF2 (SUI2, SUI3, and SUI4, respectively), and eIF5 (SUI5) (Donahue and Cigan 1988; Castilho-Valavicius et al. 1990; Huang et al. 1997). Subsequent experiments identified further mutations (termed suppressor of Sui phenotype [Ssu]) in these same eIFs that caused decreased initiation from a UUG start codon, showcasing how tightly start codon recognition is regulated (Martin-Marcos et al. 2011, 2014; Singh et al. 2012; Saini et al. 2014). In fact, all of the eIFs found in the PIC (Fig. 2), including eIF1A and eIF3, have important roles in start codon recognition (Valasek et al. 2004; Fekete et al. 2005; Saini et al. 2010; Hinnebusch 2017).
Consistent with GTP hydrolysis by eIF2 and subsequent Pi release serving as key steps in start codon recognition, mutations in eIF2 and eIF5 that stimulate premature GTPase activity and Pi release result in increased initiation at non-AUG codons, such as UUG (Huang et al. 1997). It was thus proposed early on that mutations in eIF1, eIF1A, and eIF3 likely influence start codon recognition by forcing conformational changes in eIF2 and/or eIF5 that drive premature GTP hydrolysis and/or Pi release. This is certainly still plausible; however, recent high-resolution structural data have now made it clear that many of these factors also contact the context nucleotides (e.g., “Kozak sequence”) (discussed below) immediately upstream of and downstream from the start codon. These initiation factors thus influence the overall conformation of the PIC, causing the stringency of start codon recognition to be strengthened or weakened depending on the sequence/structure of the mRNA (Hussain et al. 2014; Llacer et al. 2015; Hinnebusch 2017). It should also be highlighted that eIFs, including eIF1 and eIF5, can be post-translationally modified, and, at least for eIF1, these modifications affect start codon fidelity (Majumdar et al. 2002; Homma et al. 2005). Additional trans-acting factors, such as ABC50 (also known as ABCF1), can also bind eIF2 to regulate start codon recognition (Stewart et al. 2015).
Alterations in eIF stoichiometry are further known to influence start codon recognition patterns. For example, addition of extra eIFs caused shifts in the relative amounts of AUG versus non-AUG initiation in rabbit reticulocyte lysates (Barth-Baus et al. 2013). Using a reporter mRNA that could initiate at either an AUG or an upstream CUG start codon, these in vitro experiments showed that the addition of extra eIF1 and eIF1A proteins strengthened start codon fidelity, thereby resulting in less initiation at the CUG codon and more at the AUG. In contrast, when additional eIF5 and eIF5B was added, start codon fidelity was loosened, resulting in more initiation at the CUG codon and less at the AUG (Barth-Baus et al. 2013). Overexpression of eIF5 similarly increases non-AUG translation in human HEK293T cells (Nanda et al. 2009; Loughran et al. 2012). In yeast, haploinsufficiency of eIF1, but not eIF1A or eIF5, is known to increase non-AUG initiation (Takacs et al. 2011). Consistent with these data, overexpression of eIF1 in mammalian cells inhibited initiation from non-AUG codons (Ivanov et al. 2010), highlighting the importance of proper eIF1 expression for start codon recognition.
RNA sequences and structures affect start codon utilization
Landmark experiments by Kozak's laboratory (Kozak 1984, 1986a,b) in the 1980s provided some of the earliest clues about how sequences and secondary structures surrounding the start codon can influence initiation. Focusing first on the preproinsulin gene, Kozak identified a functional role for the nucleotide at position −3 (relative to the A of the AUG start codon) (Kozak 1984) and then further defined additional nucleotides upstream of and downstream from the start codon that influence initiation across a variety of vertebrate mRNAs (Kozak 1986b, 1987, 1989). These critical nucleotides are now commonly referred to as the “Kozak sequence.” In mammals, the consensus Kozak sequence is C(A/G)CCAUGG, where the AUG start codon is underlined (Kozak 1989), and the flanking nucleotides have a larger influence on recognition of non-AUG start codons than on AUG start codons. In particular, the purine at position −3 and the guanosine at position +4 provide stabilizing interactions with the PIC (Pisarev et al. 2006; Hussain et al. 2014; Hinnebusch 2017). The yeast consensus Kozak sequence (AAAAAUG) (Hamilton et al. 1987; Shabalina et al. 2004) likewise primarily only affects initiation at non-AUG start codons, not at AUG start codons (Zitomer et al. 1984; Donahue and Cigan 1988; Chen et al. 2008).
Consistent with the scanning model (Fig. 2), mRNA structures either upstream of or immediately downstream from start codons can influence initiation efficiency by affecting the movement of the PIC. For example, initiation from a non-AUG start codon or an AUG in a poor context can become more efficient when a strong thermostable hairpin is placed downstream from the start codon (Kozak 1990a). Importantly, this stimulatory effect is sensitive to the placement of the hairpin: When a hairpin was placed 14 nt downstream from a reporter mRNA start codon, thereby stalling the start codon in the P site of the PIC, translation initiation from this codon increased in vitro (Kozak 1990a). This structure-dependent influence on initiation has now been confirmed to affect non-AUG start codons on endogenous mRNAs (Kearse et al. 2016; Liang et al. 2017) as well as AUG start codons on viral mRNAs (Ventoso et al. 2006; Toribio et al. 2016). For example, the PTEN gene, a tumor suppressor gene that is frequently mutated in many cancers, generates an alternative protein isoform, PTENβ, by using an upstream AUU start codon (Liang et al. 2017). In addition to requiring a favorable Kozak sequence, efficient PTENβ translation requires an evolutionarily conserved hairpin that begins 18 nt downstream from the AUU. Removal of this hairpin from reporter mRNAs eliminated initiation from the PTENβ start codon but had no effect on the downstream canonical AUG start codon (Liang et al. 2017). Non-AUG translation at ACG, GUG, and GCG start codons in the FMR1 5′ leader likewise strongly increases when the downstream CGG repeat forms highly stable hairpins and G-quadruplexes (Kearse et al. 2016). A recent genome-wide analysis shows that mRNA secondary structures are commonly present downstream from non-AUG start codons and AUG codons in poor contexts, suggesting that structure may commonly be used to bolster translation initiation from weak initiation sites (Lee et al. 2012).
Met-tRNAiMet is generally, but not always, used for initiation
If a non-AUG start codon is used for initiation, what aminoacyl-tRNA (aa-tRNA) is used? This appears to depend on both the start codon and the type of initiation mechanism used. The canonical initiation pathway used by most genes uses eIF2 to deliver Met-tRNAiMet to the P site of the ribosome (Fig. 2). Given that eIF2 has high specificity for Met-tRNAiMet and does not significantly bind other tRNAs (Kolitz and Lorsch 2010), initiation using eIF2 should use Met-tRNAiMet exclusively. Indeed, early in vitro work using [35S]-labeled Met-tRNAiMet demonstrated that reporter mRNAs harboring a CUG, GUG, ACG, UUG, AGG, AAG, AUA, AUC, or AUU start codon all gave rise to full-length proteins containing [35S]-labeled methionine, strongly suggesting that non-AUG translation can use Met-tRNAiMet for initiation (Peabody 1989). Recent mass spectrometry data further support Met-tRNAiMet usage for initiation at ACG and AUU codons (Liang et al. 2017; Sellier et al. 2017).
Although translation initiation generally requires eIF2 to deliver Met-tRNAiMet, it is now recognized that at least two other eIFs, eIF2A and eIF2D, can also be used for initiation at non-AUG codons (Fig. 3). Interestingly, eIF2A (not to be confused with eIF2α) can initiate translation in a GTP-independent manner and has a relaxed stringency for binding tRNAs, as it can directly bind the charged and uncharged forms of tRNAiMet with equal affinity in vitro (Merrick and Anderson 1975; Zoll et al. 2002; Komar et al. 2005; Kim et al. 2011; Starck et al. 2012). eIF2A also may be able to bind and deliver Leu-tRNA for initiation at CUG and UUG codons, although the exact affinity of eIF2A for charged tRNALeu has not yet been measured (Starck et al. 2012, 2016; Liang et al. 2014; Sendoel et al. 2017). It thus remains possible that eIF2A may simply promote the recruitment of Leu-tRNA to the 40S ribosomal subunit rather than directly bind it for delivery. Using an ELISA (enzyme-linked immunosorbent assay)-based assay to detect small reporter peptides encoded by either an AUG or CUG start codon, Starck et al. (2012) surprisingly showed that a CUG start codon can be initiated, most likely by eIF2A, as much as ∼40% more efficiently with Leu-tRNACUG than with Met-tRNAiMet. Indeed, depletion of eIF2A reduced initiation at the CUG start codon but had no effect on initiation at AUG codons (Starck et al. 2012). Nevertheless, eIF2A appears to have only a highly specialized role in translation, as the eIF2A mouse knockout has no apparent phenotype (Golovko et al. 2016). Considering that eIF2A is up-regulated upon stress and, in some cases, can function in IRES-mediated initiation (Komar et al. 2005; Kim et al. 2011; Starck et al. 2016; Kwon et al. 2017), it is possible that eIF2A may help enable non-AUG translation to become favored when canonical translation (which normally outcompetes non-AUG translation) is down-regulated.
Figure 3.
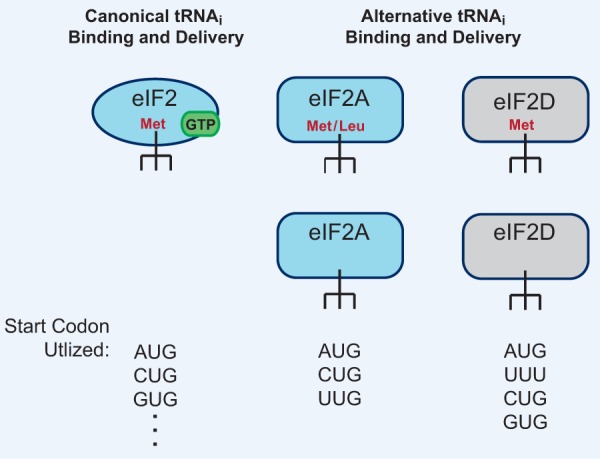
Canonical and alternative initiator tRNAi-binding eIFs are differentially regulated and have different tRNAi-binding stringency. (Left) Canonical translation uses eIF2 in a GTP-dependent manner to deliver the canonical Met-tRNAiMet to the P site of the 40S ribosomal subunit. This allows initiation at AUG start codons as well as at non-AUG start codons, albeit at a reduced efficiency. (Right) eIF2A and eIF2D are somewhat similar to eIF2 but are GTP-independent and have the capability to bind both charged and uncharged forms of tRNAiMet. eIF2A can also use Leu-tRNACUG and initiate at CUG and UUG start codons in some mRNAs in vitro and in vivo. eIF2D, which regulates reinitiation, can initiate at AUG codons in vitro as well as use other aa-tRNAs for initiation in a selective manner.
Like eIF2A, eIF2D (sometimes referred to as ligatin [LGTN]) can also deliver Met-tRNAiMet (as well as the uncharged form) in a GTP-independent manner to the P site of the 40S ribosomal subunit (Fig. 3; Dmitriev et al. 2010; Skabkin et al. 2013; Zinoviev et al. 2015; Weisser et al. 2017). eIF2D has a large role in reinitiation and ribosome recycling in vivo but can also function in initiation at non-AUG start codons in vitro in a somewhat selective manner. For example, it was shown that eIF2D can initiate at a GUG codon using Val-tRNAGUG on the Sindbis virus 26S mRNA but not on the hepatitis C virus (HCV) IRES (Dmitriev et al. 2010; Skabkin et al. 2010). Interestingly, eIF2D was identified as an autoantigen in human hepatocellular carcinoma patients (Wang et al. 2002), suggesting a possible functional role for this initiation factor in tumor development. It should also be noted that the MCT-1/DENR heterodimer (which is homologous to the N-terminal and C-terminal domains of eIF2D, respectively) has also been shown to regulate reinitiation and ribosome recycling (Reinert et al. 2006; Schleich et al. 2014, 2017; Janich et al. 2015; Haas et al. 2016). While in vitro experiments have shown that MCT-1/DENR can promote eIF2-independent recruitment of Met-tRNAiMet to the P site of the 40S ribosomal subunit on viral mRNAs (Skabkin et al. 2010, 2013), it is not yet clear whether it can bind/recruit other aa-tRNAs and/or initiate at non-AUG start codons.
In some cases, such as the cricket paralysis virus (CrPV) IRES (Jan and Sarnow 2002; Jan et al. 2003; Pestova and Hellen 2003), Taura syndrome virus (TSV) IRES (Cevallos and Sarnow 2005; Koh et al. 2014), and the Plautia stali intestine virus (PSIV) IRES (Sasaki and Nakashima 2000), an aa-tRNA in the P site is not required for initiation. These viral RNA sequences instead fold into structures that directly bind the 40S ribosomal subunit independent of any eIFs or an aa-tRNA in the P site. Upon formation of the 80S•IRES complex, a pseudoknot of the IRES is localized in the P site on the small subunit in a manner mimicking Met-tRNAiMet and the AUG codon. This conformation allows the cognate aa-tRNA to be delivered into the A site, thus forming the first amino acid of the nascent polypeptide and initiating translation (Fernandez et al. 2014; Koh et al. 2014). So far, this mode of initiation has been reported only for this class of viral IRES elements, but these results clearly demonstrate that translation initiation can occur without the canonical Met-tRNAiMet.
Usage of non-AUG initiation codons changes during development and upon stress
Ribosome profiling enables snapshots of translation to be taken, thereby revealing insights into how endogenous genes, including those containing non-AUG start codons, are regulated in response to developmental or external cues. Highlighting this approach, Brar et al. (2012) generated >25 ribosome profiling libraries to reveal temporal changes in translation as S. cerevisiae sporulate and transition from exponential vegetative growth to meiotic growth. As expected, there were broad changes in gene expression at both the mRNA and translation levels, with some genes displaying stage-specific meiotic translation. Furthermore, a number of previously unannotated translation events, including from antisense RNAs, intergenic regions, and uORFs, were found to be specific to meiotic cells. Only 5% of ribosome footprints mapped outside annotated ORFs in vegetative cells, but this number increased to a staggering ∼30% in meiotic cells (Brar et al. 2012). This suggests that there may be a profound shift in meiotic cells away from canonical translation, although functional roles (if any) for the majority of these novel ORFs remain unknown.
Many (but certainly not all) of the translated uORFs in meiotic cells use non-AUG start codons. uORFs have generally been thought to play an inhibitory role and act competitively to prevent translation of the downstream AUG-encoded ORF (Sachs and Geballe 2006; Chew et al. 2016; Johnstone et al. 2016). Indeed, about half of the AUG-encoded uORFs were associated with translational repression of their downstream ORFs in meiosis (Brar et al. 2012). In stark contrast, translation of non-AUG-encoded uORFs was correlated primarily with increased translation of their downstream ORFs. The underlying mechanism responsible for this positive correlation remains unknown, but it is possible that these non-AUG uORFs may directly prime translation of their downstream ORF, or, alternatively, the uORF-encoded polypeptides may have functional roles (Brar et al. 2012). Given that nearly 50% of RNA polymerase II transcription is devoted to production of ribosomal proteins when yeast are grown under vegetative conditions (Warner 1999), it is tempting to speculate that changes in ribosomal protein (or eIF) stoichiometry and/or post-translational modifications may be responsible for the drastic changes in translation patterns observed in meiotic cells. Consistent with this idea, it has been shown recently that ribosome composition and ribosome protein stoichiometry can influence translation of particular classes of mRNAs (Xue et al. 2015; Shi et al. 2017; Simsek et al. 2017).
Upon experiencing stress, eukaryotic cells generally respond by down-regulating translation initiation, thereby saving their metabolic energy and limiting the production of what could be deleterious or toxic proteins (Fulda et al. 2010; Pakos-Zebrucka et al. 2016). Multiple stress conditions (e.g., ER or oxidative stress) converge on the inhibitory phosphorylation of eIF2α. Under these conditions, eIF2 cannot exchange GDP for GTP and thus cannot bind Met-tRNAiMet for a subsequent round of initiation. This causes nearly all translation initiation events to become strongly inhibited, but there are some mRNAs whose translation is increased—most notably genes with regulatory uORFs such as GCN4 (which has multiple AUG uORFs) (Hinnebusch 2005), ATF4 (which also has multiple AUG uORFs) (Wek et al. 2006), and BiP (Kaufman 1999; Starck et al. 2016). BiP is an ER chaperone protein that senses misfolded proteins, and thus induction of its expression is critical for helping restore ER homeostasis. Starck et al. (2016) showed recently that BiP translation is regulated by two non-AUG uORFs (UUG and CUG) that are up-regulated during stress and dependent on eIF2A. Knocking down eIF2A prior to ER stress led to decreased levels of BiP, which were further reduced when the UUG-encoded uORF was deleted. These results suggest that both eIF2A and the non-AUG uORFs are required for the increased expression of BiP during ER stress (Starck et al. 2016). As various stresses beyond ER stress lead to increases in eIF2A protein levels (Kim et al. 2011; Starck et al. 2016; Sendoel et al. 2017), induction of non-AUG translation appears to be a common stress response mechanism.
In mammalian cells, heat shock causes translation of MRPL18 (which encodes a mitochondrial large ribosomal subunit protein) to no longer initiate at the annotated AUG start codon but instead initiate at a downstream CUG codon (Zhang et al. 2015) (Fig. 4). The truncated MRPL18 protein that is generated lacks the N-terminal mitochondrial targeting signal and thus is not incorporated into mitochondrial ribosomes. Instead, the truncated MRPL18 protein is incorporated into cytoplasmic ribosomes. These newly defined “hybrid” ribosomes are functional and are required for increased synthesis of the Hsp70 chaperone protein during heat shock (Zhang et al. 2015). This is perhaps because the presence of MRPL18 permits directed ribosome recruitment to Hsp70 mRNA, similar to what has been reported previously for RpL38 and the Hox genes (Xue et al. 2015). Regardless of the exact underlying mechanism, the MRPL18 locus beautifully demonstrates how changes in the efficiency of non-AUG start codon usage can have a profound impact on cell survival and homeostasis.
Figure 4.
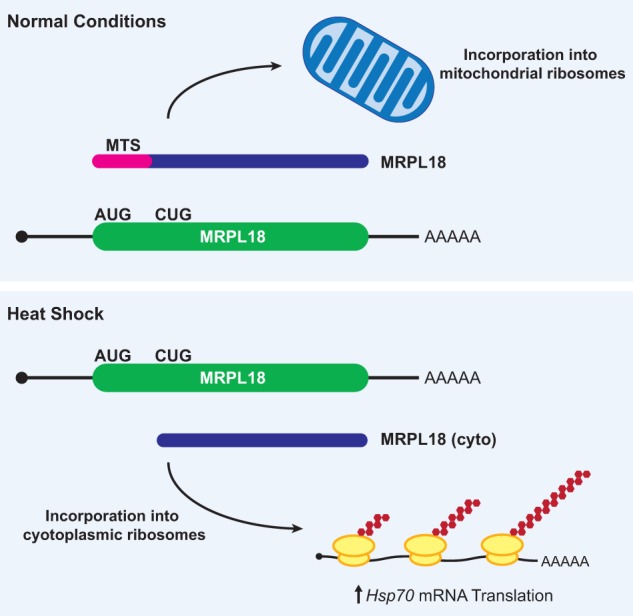
Heat shock causes the production of a CUG-initiated MRPL18 protein that becomes incorporated into cytoplasmic ribosomes. (Top) Under normal growth conditions, the MRPL18 protein is synthesized from a canonical AUG start codon and includes the mitochondrial targeting signal (MTS; pink), which enables transport into mitochondria and subsequent incorporation into mitochondrial ribosomes. (Bottom) Heat shock causes a switch in the preferred start codon, resulting in initiation at a downstream CUG in the MRPL18 mRNA. The truncated MRPL18 (cyto) protein isoform lacks the MTS and is instead incorporated into cytoplasmic ribosomes, creating “hybrid ribosomes” that are required for increased Hsp70 mRNA translation during heat shock.
Non-AUG translation events can drive cancer progression
Alternative protein isoforms derived from non-AUG start codons have also been found to be generated from numerous cancer-relevant genes and act to either promote (e.g., fibroblast growth factor 2 [FGF2]) or inhibit (e.g., c-myc) cancer growth (Hann et al. 1988; Hann 1994; Touriol et al. 2003; Ivanov et al. 2011). For example, FGF2 controls cell proliferation, differentiation, and angiogenesis (Turner and Grose 2010), and multiple protein isoforms can be generated from this gene via the use of alternative start codons. Whereas the canonical AUG start codon generates an 18-kDa protein isoform that is mostly cytoplasmic or secreted, at least four upstream in-frame CUG codons can also be used to generate longer isoforms that localize to the nucleus (Renko et al. 1990; Bugler et al. 1991; Arnaud et al. 1999). Intriguingly, the CUG-encoded protein isoforms are unique to transformed cells (Vagner et al. 1996). Supporting their functional relevance, ectopic expression of these CUG-encoded isoforms caused cell immortalization in culture and increased the tumorigenic properties of these cells when they were injected into mice (Couderc et al. 1991; Quarto et al. 1991).
The c-myc proto-oncogene likewise regulates cell proliferation and transformation. Two protein isoforms can be generated from c-myc through the usage of different start codons: c-myc 2 (p64) uses a canonical AUG start codon, whereas c-myc 1 (p67) uses an upstream in-frame CUG codon (Hann et al. 1988). At low cell density, the CUG-encoded c-myc 1 protein is only 10%–15% as abundant as the AUG-encoded c-myc 2. However, as cell density increases, the availability of amino acids, specifically methionine, becomes limiting, and the CUG-encoded c-myc 1 isoform becomes preferentially translated (Hann et al. 1992). The underlying mechanism is not fully clear, but this reduction in methionine availability may cause activation of the GCN2 kinase, reduction in TC levels, and/or a reduction in charged Met-tRNA levels, leading to a change in start codon usage. Once generated, the unique N terminus of c-myc 1 causes this isoform to have additional DNA-binding capabilities that allow it to regulate genes that inhibit cell growth (Hann et al. 1994). Indeed, overexpression of this CUG-encoded isoform (but not the AUG-encoded isoform) inhibits growth of cultured cells. Inactivation of c-myc 1 is observed in some tumor cells (e.g., in human Burkitt's lymphomas) (Hann and Eisenman 1984; Hann et al. 1988), suggesting that the inability to generate this CUG-encoded protein may provide a selective growth advantage.
The mechanisms by which individual non-AUG translation events become activated are still poorly understood, but there are likely global changes in start codon fidelity in cancer cells. Many eIFs, including the cap-binding protein eIF4E (Mamane et al. 2004), are often misregulated in cancers (for review, see Spilka et al. 2013; Chu et al. 2016), and imbalances of eIFs are known to influence overall initiation fidelity and start codon choice in various systems (as discussed above). Perhaps the strongest correlation between non-AUG translation and cancer comes from recent work in squamous cell carcinomas (SCCs) (Sendoel et al. 2017). Sendoel et al. (2017) identified eIF2A as an essential factor in tumor initiation that promotes the translation of many non-AUG uORFs that act to positively regulate expression of their downstream cancer-related ORFs (Fig. 5). Remarkably, deleting eIF2A protected mice from tumors. In stark contrast, depletion of the canonical eIF2 initiation factor largely had no effect on oncogenic growth but did affect the growth of normal cells (Sendoel et al. 2017). As discussed earlier (Fig. 3), eIF2A can initiate at CUG and UUG start codons (both encode Leu) and deliver/promote the recruitment of Leu-tRNACUG for initiation (Starck et al. 2012, 2016; Liang et al. 2014). Considering that the eIF2A locus is amplified in 29% of patients with lung SCC, 15% of patients with head and neck SCC, and 15% of patients with oesophageal carcinoma (Sendoel et al. 2017), it appears likely that misregulation of eIF2A and non-AUG uORFs may play a more general role in cancer development.
Figure 5.
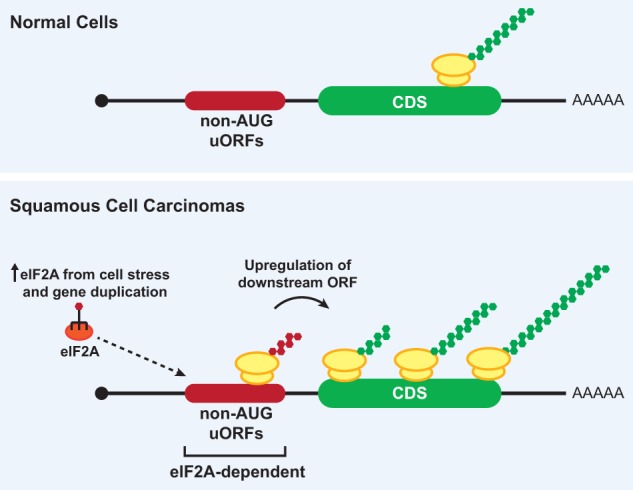
eIF2A regulates translation of non-AUG uORFs in SCCs. (Top) In normal cells, translation of non-AUG uORFs is limited, and downstream ORFs are translated at basal levels. (Bottom) In some SCCs, cell stress and/or gene duplication events cause elevated eIF2A levels. This results in increased expression of non-AUG uORFs in oncogenic mRNAs (e.g., Ctnnb1) and subsequent up-regulation of the downstream oncoprotein ORFs. Upon depleting eIF2A in SCC models, premalignant and malignant progression is greatly reduced.
Repeat-associated non-AUG (RAN) translation is associated with many neurodegenerative diseases
A number of neurological diseases, so-called nucleotide repeat disorders, are caused by the expansion of nucleotide repeats (Orr and Zoghbi 2007; Almeida et al. 2013). This includes Huntington's disease, which is caused by expansion of a CAG repeat sequence in the coding sequence of the Huntingtin (HTT) gene, and the fragile X disorders, which are due to CGG repeat expansions in the 5′ leader (untranslated region) of the FMR1 gene. Expansion of these repeats can cause RAN translation (Green et al. 2016; Cleary and Ranum 2017) as well as other effects, such as protein loss of function, protein gain of function, and/or RNA gain of function (Orr and Zoghbi 2007; Todd and Paulson 2010; Almeida et al. 2013). In all known cases of RAN translation, non-AUG start codons are used to initiate translation of repeat proteins in multiple reading frames from the expanded nucleotide repeat itself. Once generated, these translation products appear to be toxic to cells in multiple ways, as they impair the ubiquitin-proteasome system, alter ribosomal RNA synthesis, and block nuclear import (Kwon et al. 2014; Oh et al. 2015; Yamakawa et al. 2015; Zhang et al. 2016; Cleary and Ranum 2017).
Toxic polypeptides generated by RAN translation were first identified from expanded CAG repeats in ATXN8 and DMPK as well as CUG repeats in ATXN8OS (Zu et al. 2011). Subsequent work from many laboratories has now shown that RAN translation produces neurotoxic proteins from expanded GGGGCC and CCCCGG repeats from the C9orf72 locus (associated with amyotrophic lateral sclerosis and frontotemporal dementia [ALS/FTD]), CGG and CCG repeats from the FMR1 locus (associated with fragile X-associated tremor/ataxia syndrome [FXTAS]), CAG repeats from the HTT locus (associated with Huntington's disease), and UGGAA repeats from the SCA31 locus (associated with spinocerebellar ataxia type 31) (Ash et al. 2013; Mori et al. 2013; Todd et al. 2013; Zu et al. 2013; Banez-Coronel et al. 2015; Krans et al. 2016; Ishiguro et al. 2017).
A better understanding of how RAN translation occurs has come from recent work focused on the CGG repeats in the 5′ leader of FMR1 (Fig. 6). A series of reporters was generated with the luciferase ORF placed downstream from a FMR1 5′ leader that contained either normal or disease-associated lengths of CGG repeats (Kearse et al. 2016). Upon mutating the start codon of luciferase to eliminate canonical translation of the luciferase ORF, only the reporters with expanded repeats produced polypeptides through RAN translation. These RAN-translated proteins were generated from multiple reading frames in vitro, in HeLa cells, and in cultured primary neurons, although at orders of magnitude less efficiently compared with a canonical luciferase reporter. In the case of FMR1 and CGG repeats, RAN translation is 5′ cap-, eIF4E-, and eIF4A-dependent and therefore resembles the scanning mechanism used in canonical translation except that either an ACG, GUG, or GCG codon is used for initiation (Kearse et al. 2016). Subsequent mass spectrometry confirmed that this ACG codon is used for initiation (incorporating methionine) when the endogenous FMR1 mRNA contains an expanded repeat (Sellier et al. 2017). RAN translation at expanded CAG repeats from the SCA3, SCA8, DM1, HD, and HDL2 disease loci is likewise initiated at upstream AUU or AUC codons in vitro, and, at least for AUU, [35S]-labeled Met-tRNAiMet was incorporated (Zu et al. 2011).
Figure 6.
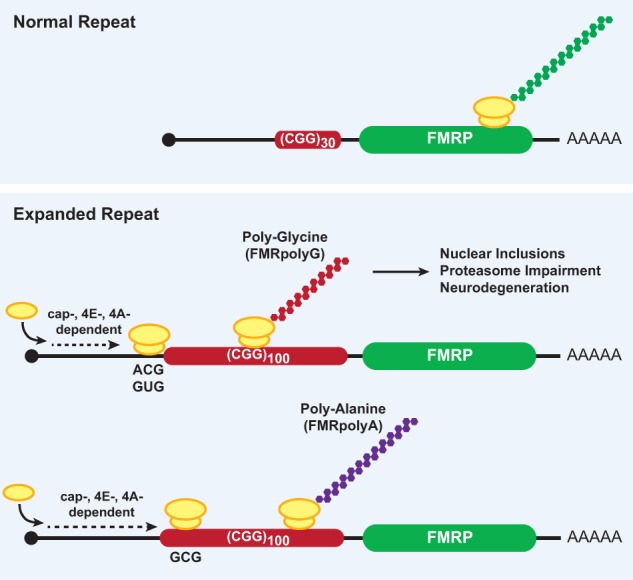
RAN translation at CGG repeats in FMR1 produces neurotoxic proteins from multiple reading frames. The fragile X disorders are caused by a CGG nucleotide repeat expansion in the 5′ leader of the FMR1 gene, which encodes FMRP. Unaffected patients have a mean repeat length of ∼30 (top), whereas fragile X disorder patients have expanded repeats of >55 (bottom), and there is a correlation between repeat size and disease severity. CGG repeat expansion stimulates a noncanonical form of protein synthesis (called RAN translation) in multiple reading frames that uses a 5′ cap-, eIF4E-, and eIF4A-dependent scanning mode of initiation. RAN translation of the CGG repeat produces two toxic homopolymeric proteins from separate reading frames: (1) a polyglycine protein (FMRpolyG) that initiates at ACG and GUG codons upstream of the repeat in the +1 reading frame and (2) a polyalanine protein (FMRpolyA) that most likely initiates within the repeat at a GCG codon from the +2 reading frame.
It is still largely unclear why expansion of nucleotide repeats beyond a critical threshold is required for RAN translation. It may be that the expanded repeat RNA forms a specific structure that binds trans-acting factors. Indeed, expanded repeats are known to be able to sequester RNA-binding proteins (Todd and Paulson 2010). It is also possible that the complex secondary structure of the expanded repeat (Cammas and Millevoi 2017) is directing initiation by altering ribosome scanning or recruitment. It is noteworthy that IRES-mediated mechanisms do not appear to account for RAN translation.
Targeting non-AUG translation may represent a promising therapeutic strategy
Small molecule compounds that inhibit protein synthesis not only have provided important insights into how the translation machinery functions but have great potential in the treatment of human diseases that are marked by dysregulated translation, including cancer (Lindqvist and Pelletier 2009; Blanchard et al. 2010; Garreau de Loubresse et al. 2014; Bhat et al. 2015). A key issue, however, is the need for sufficient specificity so that only deleterious translation events are targeted. Most common protein synthesis inhibitors target either the ribosome itself (e.g., cycloheximide, emetine, or anisomycin) or the eIFs (e.g., hippuristanol and Rocaglamide A [RocA]) (Bordeleau et al. 2006; Lindqvist et al. 2008; Iwasaki et al. 2016). Drugs that bind the ribosome itself should inhibit all forms of cytoplasmic translation, whereas hippuristanol and RocA should affect only eIF4A- and scanning-dependent initiation events (note that RocA further requires GA motifs for activity). Nevertheless, most cellular translation is scanning-dependent, and thus these drugs have rather limited specificity.
Considering that non-AUG translation events can be regulated quite differently from canonical AUG start codons and that decreasing non-AUG translation by knocking out eIF2A reduced cancer progression (Sendoel et al. 2017), it has been proposed that pharmacologically modulating noncanonical initiation codons may represent a novel therapeutic strategy. In an effort to identify such inhibitors, Takacs et al. (2011) screened >55,000 natural compounds in yeast for effects on a dual-luciferase reporter that expressed UUG-encoded firefly luciferase and AUG-encoded renilla luciferase. This effort resulted in the identification of two structurally similar quinolone-containing compounds (NSC218351 and NSC92218), which increased translation of the UUG-encoded reporter and concomitantly decreased translation of the AUG-encoded reporter (Takacs et al. 2011). Using the same dual-luciferase reporter but with other non-AUG start codons, it was determined that both compounds also result in increased initiation from CUG, GUG, AUA, AUC, and ACG (but not AUU) start codons (Takacs et al. 2011).
The specific mechanisms of action of NSC218351 or NSC92218 are not yet fully understood, but genetic and biochemical experiments have provided important clues (Takacs et al. 2011). Overexpression of eIF1 suppressed the effects of NSC218351 and NSC92218, whereas haploinsufficiency of eIF1, eIF1A, or eIF5 in diploid yeast caused increased sensitivity. These results strongly suggest that the compounds interact with the PIC. However, there was no change in eIF1 affinity for the 40S subunit or in the rate of release of eIF1 upon start codon recognition when either compound was present. Further work is required to clarify the detailed mechanism as well as determine whether these compounds are effective in mammalian cells.
As translation initiation at CUG codons can use either Met-tRNAiMet (bound by eIF2) or Leu-tRNACUG (bound by eIF2A), it is perhaps not surprising that CUG initiation is selectively resistant against a few inhibitors (Starck et al. 2008, 2012). For example, NSC119893, which inhibits Met-tRNAiMet association with eIF2, inhibited only AUG, but not CUG, initiation in vivo (Starck et al. 2012). Likewise, bruceantin, which binds the P site of the 60S subunit where Met-tRNAiMet binds, was shown in vitro to inhibit AUG, but not CUG, initiation from a short reporter mRNA (Starck et al. 2008). Edeine, which binds the E site of the eukaryotic 40S subunit to interfere with start codon recognition and subunit joining, inhibited AUG initiation by ∼90% but CUG initiation by only ∼25% in vitro (Starck et al. 2008). This suggests that CUG initiation, at least in vitro, may be partially independent of the canonical scanning mechanism and that it may be possible to specifically modulate these events while not affecting AUG translation. The underlying mechanism of edeine is not clear but could be due to Leu-tRNACUG binding in a unique conformation within the ribosome.
The nucleic acid intercalating agent acriflavine appears to inhibit CUG initiation more efficiently than AUG initiation in vitro and in vivo (Starck et al. 2008, 2012). Unfortunately, the exact mechanism of action is not known. Acriflavine has been reported to inhibit the aminoacylation of tRNAs in vitro (Birkmayer and Balda 1970; Novac et al. 2004), but it is not immediately obvious how such an activity could make the drug more specific toward CUG initiation. Whether acriflavine inhibits initiation at other non-AUG start codons has not been tested, but this molecule could be a valuable starting point for developing a more potent CUG/non-AUG-specific inhibitor.
Inhibitors to RAN translation at GGGGCC (causes ALS/FTD) and CGG (causes FXTAS) nucleotide repeats are perhaps the most medically promising inhibitors of non-AUG translation because of their noted selectivity. Both of these expanded repeats form very stable hairpins and G quadruplexes (Cammas and Millevoi 2017), and small molecule compounds that bind the repeats caused marked improvements in a number of molecular and cellular phenotypes (Disney et al. 2012; Su et al. 2014). For example, these compounds inhibited production of RAN translation proteins, although the effects were not equal across all reading frames of the GGGGCC repeat or antisense CCCCGG repeat (Su et al. 2014). Most importantly, canonical translation of reporter mRNAs in cells appeared unaffected by these compounds (Su et al. 2014). As these compounds increase the melting temperature of the expanded repeats in vitro, RAN translation may be inhibited because the ribosome cannot elongate through the stabilized G-rich repeat secondary structures. However, it remains equally plausible that the effects may occur at the initiation stage, as the stabilized secondary structure could sterically prevent recognition of the non-AUG start codons (e.g., by placing the P site of the PIC over a start codon in a poor context).
Perspectives and concluding remarks
It is now clear that multiple alternative initiation mechanisms can be used in eukaryotic cells. These include IRES-mediated initiation, eIF3-directed cap binding, reinitiation, the use of alternative eIFs to bind tRNAi (Fig. 3), the recruitment of the ribosome via N6-methyladenosine (m6A) mRNA modifications, and the usage of non-AUG start codons (Hellen and Sarnow 2001; Touriol et al. 2003; Ivanov et al. 2011; Starck et al. 2012; Skabkin et al. 2013; Wang et al. 2015; Zhou et al. 2015; Zinoviev et al. 2015; Lee et al. 2016; Johnson et al. 2017). Especially in certain cell states (e.g., during meiosis or cell stress), ribosomes often initiate from near-cognate codons to give rise to functional proteins that regulate key cellular processes, thereby contributing to protein diversity (Fig. 4; Ivanov et al. 2011; Crosby et al. 2015). Thus, it is not surprising that misregulation of non-AUG translation can stimulate cancer malignancy and neurodegeneration. Nevertheless, one should keep in mind that only a handful of non-AUG translation events have yet to be orthogonally validated or characterized in detail. It still remains very possible that some initiation events that have been identified in genome-wide approaches, such as ribosome profiling, may be very rare or inconsequential. Western blot analysis and/or mass spectrometry experiments can provide strong confirmatory support for novel ORFs, but proteins that are expressed at very low levels may not be detected with current technologies. This is not to say that such proteins are not potentially biologically relevant, as some cellular factors (e.g., some microRNA and transcription factors) can be found at only a few molecules per cell and still have important functional roles (Zenklusen et al. 2008; Vaquerizas et al. 2009; Parasramka et al. 2012; Marinov et al. 2014; Denzler et al. 2016).
As non-AUG initiation events are influenced by both the translation machinery and the mRNA sequence/structure, an intriguing question is whether we can accurately predict non-AUG translation events in any given transcript. In silico tools using algorithms that are based on ribosome profiling data sets are now available to help predict true non-AUG start codons (Reuter et al. 2016), and the inclusion of additional data, including secondary structure predictions (Kochetov et al. 2007) and evolutionary conservation (Ivanov et al. 2011, 2017), likely can improve accuracy. However, as noted in the multiple examples that were highlighted throughout this review, cellular conditions (e.g., stress) and disease mutations significantly influence start codon usage. Unfortunately, it is difficult to account for these features in prediction software, but this may become more feasible as additional new insights into the underlying molecular mechanisms of non-AUG translation are revealed.
In summary, it is now clear that translation can initiate at non-AUG codons across eukaryotes to impact both normal and disease biology. Recent work has revealed many surprises, including the ability of Leu-tRNACUG to be used for initiation and the critical role that RAN translation plays in neurodegenerative diseases. Going forward, it will be of great interest to further define exactly how the various alternative initiation pathways become specifically induced or repressed, depending on cell state. This will likely reveal novel insights into how non-AUG translation is used to expand the functional coding potential of our genome. Furthermore, such studies may also suggest novel therapeutic strategies that can be used to treat a variety of diseases that are marked by dysregulated non-AUG translation events.
Acknowledgments
We thank Aaron Goldstrohm, Daniel Goldman, Deirdre Tatomer, and all members of the Wilusz laboratory for their suggestions and comments. J.E.W. is supported by National Institutes of Health grants R00-GM104166 and R35-GM119735 and National Cancer Institute Cancer Center Support grant NCI-P30-CA016520. J.E.W. is a Rita Allen Foundation Scholar. M.G.K. is supported by National Institutes of Health grant K99-GM126064.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.305250.117.
References
- Almeida B, Fernandes S, Abreu IA, Macedo-Ribeiro S. 2013. Trinucleotide repeats: a structural perspective. Front Neurol 4: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, O'Connor PB, Loughran G, Dmitriev SE, Baranov PV, Shatsky IN. 2017. Insights into the mechanisms of eukaryotic translation gained with ribosome profiling. Nucleic Acids Res 45: 513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud E, Touriol C, Boutonnet C, Gensac MC, Vagner S, Prats H, Prats AC. 1999. A new 34-kilodalton isoform of human fibroblast growth factor 2 is cap dependently synthesized by using a non-AUG start codon and behaves as a survival factor. Mol Cell Biol 19: 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW III, Rademakers R, et al. 2013. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77: 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banez-Coronel M, Ayhan F, Tarabochia AD, Zu T, Perez BA, Tusi SK, Pletnikova O, Borchelt DR, Ross CA, Margolis RL, et al. 2015. RAN translation in Huntington disease. Neuron 88: 667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth-Baus D, Bhasker CR, Zoll W, Merrick WC. 2013. Influence of translation factor activities on start site selection in six different mRNAs. Translation 1: e24419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman RW, Jongens TA. 2011. A non-canonical start codon in the Drosophila fragile X gene yields two functional isoforms. Neuroscience 181: 48–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. 2015. Targeting the translation machinery in cancer. Nat Rev Drug Discov 14: 261–278. [DOI] [PubMed] [Google Scholar]
- Birkmayer GD, Balda BR. 1970. Proflavine inhibition of protein synthesis in malignant hamster melanoma. FEBS Lett 11: 221–223. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Cooperman BS, Wilson DN. 2010. Probing translation with small-molecule inhibitors. Chem Biol 17: 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J. 2006. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol 2: 213–220. [DOI] [PubMed] [Google Scholar]
- Brar GA, Weissman JS. 2015. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol 16: 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335: 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugler B, Amalric F, Prats H. 1991. Alternative initiation of translation determines cytoplasmic or nuclear localization of basic fibroblast growth factor. Mol Cell Biol 11: 573–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammas A, Millevoi S. 2017. RNA G-quadruplexes: emerging mechanisms in disease. Nucleic Acids Res 45: 1584–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho-Valavicius B, Yoon H, Donahue TF. 1990. Genetic characterization of the Saccharomyces cerevisiae translational initiation suppressors sui1, sui2 and SUI3 and their effects on HIS4 expression. Genetics 124: 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevallos RC, Sarnow P. 2005. Factor-independent assembly of elongation-competent ribosomes by an internal ribosome entry site located in an RNA virus that infects penaeid shrimp. J Virol 79: 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KJ, Wang CC. 2004. Translation initiation from a naturally occurring non-AUG codon in Saccharomyces cerevisiae. J Biol Chem 279: 13778–13785. [DOI] [PubMed] [Google Scholar]
- Chen SJ, Lin G, Chang KJ, Yeh LS, Wang CC. 2008. Translational efficiency of a non-AUG initiation codon is significantly affected by its sequence context in yeast. J Biol Chem 283: 3173–3180. [DOI] [PubMed] [Google Scholar]
- Chew GL, Pauli A, Schier AF. 2016. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat Commun 7: 11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Cargnello M, Topisirovic I, Pelletier J. 2016. Translation initiation factors: reprogramming protein synthesis in cancer. Trends Cell Biol 26: 918–933. [DOI] [PubMed] [Google Scholar]
- Cleary JD, Ranum LP. 2017. New developments in RAN translation: insights from multiple diseases. Curr Opin Genet Dev 44: 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements JM, Laz TM, Sherman F. 1988. Efficiency of translation initiation by non-AUG codons in Saccharomyces cerevisiae. Mol Cell Biol 8: 4533–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couderc B, Prats H, Bayard F, Amalric F. 1991. Potential oncogenic effects of basic fibroblast growth factor requires cooperation between CUG and AUG-initiated forms. Cell Regul 2: 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby MA, Gramates LS, Dos Santos G, Matthews BB, St Pierre SE, Zhou P, Schroeder AJ, Falls K, Emmert DB, Russo SM, et al. 2015. Gene model annotations for Drosophila melanogaster: the rule-benders. G3 (Bethesda) 5: 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J, Kolakofsky D. 1988. Ribosomal initiation from an ACG codon in the Sendai virus P/C mRNA. EMBO J 7: 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler R, McGeary SE, Title AC, Agarwal V, Bartel DP, Stoffel M. 2016. Impact of microRNA levels, target-site complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Mol Cell 64: 565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disney MD, Liu B, Yang WY, Sellier C, Tran T, Charlet-Berguerand N, Childs-Disney JL. 2012. A small molecule that targets r(CGG)(exp) and improves defects in fragile X-associated tremor ataxia syndrome. ACS Chem Biol 7: 1711–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN. 2010. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem 285: 26779–26787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue TF, Cigan AM. 1988. Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol Cell Biol 8: 2955–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn P, DaSilva L, Martarano L, Derse D. 1990. Equine infectious anemia virus tat: insights into the structure, function, and evolution of lentivirus trans-activator proteins. J Virol 64: 1616–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. 2009. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet 10: 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete CA, Applefield DJ, Blakely SA, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. 2005. The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. EMBO J 24: 3588–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V. 2014. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell 157: 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields AP, Rodriguez EH, Jovanovic M, Stern-Ginossar N, Haas BJ, Mertins P, Raychowdhury R, Hacohen N, Carr SA, Ingolia NT, et al. 2015. A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol Cell 60: 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresno M, Jimenez A, Vazquez D. 1977. Inhibition of translation in eukaryotic systems by harringtonine. Eur J Biochem 72: 323–330. [DOI] [PubMed] [Google Scholar]
- Fulda S, Gorman AM, Hori O, Samali A. 2010. Cellular stress responses: cell survival and cell death. Int J Cell Biol 2010: 214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble CE, Brule CE, Dean KM, Fields S, Grayhack EJ. 2016. Adjacent codons act in concert to modulate translation efficiency in yeast. Cell 166: 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreau de Loubresse N, Prokhorova I, Holtkamp W, Rodnina MV, Yusupova G, Yusupov M. 2014. Structural basis for the inhibition of the eukaryotic ribosome. Nature 513: 517–522. [DOI] [PubMed] [Google Scholar]
- Gerashchenko MV, Gladyshev VN. 2014. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res 42: e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovko A, Kojukhov A, Guan BJ, Morpurgo B, Merrick WC, Mazumder B, Hatzoglou M, Komar AA. 2016. The eIF2A knockout mouse. Cell Cycle 15: 3115–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KM, Linsalata AE, Todd PK. 2016. RAN translation-What makes it run? Brain Res 1647: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. 2013. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154: 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas MA, Ngo L, Li SS, Schleich S, Qu Z, Vanyai HK, Cullen HD, Cardona-Alberich A, Gladwyn-Ng IE, Pagnamenta AT, et al. 2016. De novo mutations in DENR disrupt neuronal development and link congenital neurological disorders to faulty mRNA translation re-initiation. Cell Rep 15: 2251–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton R, Watanabe CK, de Boer HA. 1987. Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res 15: 3581–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann SR. 1994. Regulation and function of non-AUG-initiated proto-oncogenes. Biochimie 76: 880–886. [DOI] [PubMed] [Google Scholar]
- Hann SR, Eisenman RN. 1984. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol 4: 2486–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann SR, King MW, Bentley DL, Anderson CW, Eisenman RN. 1988. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt's lymphomas. Cell 52: 185–195. [DOI] [PubMed] [Google Scholar]
- Hann SR, Sloan-Brown K, Spotts GD. 1992. Translational activation of the non-AUG-initiated c-myc 1 protein at high cell densities due to methionine deprivation. Genes Dev 6: 1229–1240. [DOI] [PubMed] [Google Scholar]
- Hann SR, Dixit M, Sears RC, Sealy L. 1994. The alternatively initiated c-Myc proteins differentially regulate transcription through a noncanonical DNA-binding site. Genes Dev 8: 2441–2452. [DOI] [PubMed] [Google Scholar]
- Hellen CU, Sarnow P. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev 15: 1593–1612. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. 2017. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem Sci 42: 589–611. [DOI] [PubMed] [Google Scholar]
- Homma MK, Wada I, Suzuki T, Yamaki J, Krebs EG, Homma Y. 2005. CK2 phosphorylation of eukaryotic translation initiation factor 5 potentiates cell cycle progression. Proc Natl Acad Sci 102: 15688–15693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HK, Yoon H, Hannig EM, Donahue TF. 1997. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev 11: 2396–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T, Llacer JL, Fernandez IS, Munoz A, Martin-Marcos P, Savva CG, Lorsch JR, Hinnebusch AG, Ramakrishnan V. 2014. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell 159: 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussmann JA, Patchett S, Johnson A, Sawyer S, Press WH. 2015. Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet 11: e1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H, Olsen HS, Sonenberg N. 1997. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J 16: 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT. 2010. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol 470: 119–142. [DOI] [PubMed] [Google Scholar]
- Ingolia NT. 2014. Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet 15: 205–213. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. 2014. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep 8: 1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro T, Sato N, Ueyama M, Fujikake N, Sellier C, Kanegami A, Tokuda E, Zamiri B, Gall-Duncan T, Mirceta M, et al. 2017. Regulatory role of RNA chaperone TDP-43 for RNA misfolding and repeat-associated translation in SCA31. Neuron 94: 108–124.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, Sachs MS, Atkins JF. 2010. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc Natl Acad Sci 107: 18056–18060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Firth AE, Michel AM, Atkins JF, Baranov PV. 2011. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res 39: 4220–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Wei J, Caster SZ, Smith KM, Michel AM, Zhang Y, Firth AE, Freitag M, Dunlap JC, Bell-Pedersen D, et al. 2017. Translation initiation from conserved non-AUG codons provides additional layers of regulation and coding capacity. MBio 8: e00844-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Floor SN, Ingolia NT. 2016. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature 534: 558–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Sarnow P. 2002. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol 324: 889–902. [DOI] [PubMed] [Google Scholar]
- Jan E, Kinzy TG, Sarnow P. 2003. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc Natl Acad Sci 100: 15410–15415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janich P, Arpat AB, Castelo-Szekely V, Lopes M, Gatfield D. 2015. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res 25: 1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AG, Grosely R, Petrov AN, Puglisi JD. 2017. Dynamics of IRES-mediated translation. Philos Trans R Soc Lond B Biol Sci 372: 20160177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone TG, Bazzini AA, Giraldez AJ. 2016. Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J 35: 706–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman RJ. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev 13: 1211–1233. [DOI] [PubMed] [Google Scholar]
- Kearse MG, Green KM, Krans A, Rodriguez CM, Linsalata AE, Goldstrohm AC, Todd PK. 2016. CGG repeat-associated non-AUG translation utilizes a cap-dependent scanning mechanism of initiation to produce toxic proteins. Mol Cell 62: 314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Park SM, Park JH, Keum SJ, Jang SK. 2011. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J 30: 2454–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochetov AV, Palyanov A, Titov II, Grigorovich D, Sarai A, Kolchanov NA. 2007. AUG_hairpin: prediction of a downstream secondary structure influencing the recognition of a translation start site. BMC Bioinformatics 8: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CS, Brilot AF, Grigorieff N, Korostelev AA. 2014. Taura syndrome virus IRES initiates translation by binding its tRNA–mRNA-like structural element in the ribosomal decoding center. Proc Natl Acad Sci 111: 9139–9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolitz SE, Lorsch JR. 2010. Eukaryotic initiator tRNA: finely tuned and ready for action. FEBS Lett 584: 396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA, Gross SR, Barth-Baus D, Strachan R, Hensold JO, Goss Kinzy T, Merrick WC. 2005. Novel characteristics of the biological properties of the yeast Saccharomyces cerevisiae eukaryotic initiation factor 2A. J Biol Chem 280: 15601–15611. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1984. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature 308: 241–246. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1986a. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci 83: 2850–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1986b. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283–292. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15: 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1989. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol Cell Biol 9: 5073–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1990a. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci 87: 8301–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1990b. Evaluation of the fidelity of initiation of translation in reticulocyte lysates from commercial sources. Nucleic Acids Res 18: 2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krans A, Kearse MG, Todd PK. 2016. Repeat-associated non-AUG translation from antisense CCG repeats in fragile X tremor/ataxia syndrome. Ann Neurol 80: 871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. 2014. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345: 1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon OS, An S, Kim E, Yu J, Hong KY, Lee JS, Jang SK. 2017. An mRNA-specific tRNAi carrier eIF2A plays a pivotal role in cell proliferation under stress conditions: stress-resistant translation of c-Src mRNA is mediated by eIF2A. Nucleic Acids Res 45: 296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Hite DH, Hogan GJ, Brown PO. 2014. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife 3: e01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. 2012. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci 109: E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Kranzusch PJ, Doudna JA, Cate JH. 2016. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536: 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SM, Cerquozzi S, Graber TE, Ungureanu NH, Andrews M, Holcik M. 2008. The eIF4G homolog DAP5/p97 supports the translation of select mRNAs during endoplasmic reticulum stress. Nucleic Acids Res 36: 168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, He S, Yang J, Jia X, Wang P, Chen X, Zhang Z, Zou X, McNutt MA, Shen WH, et al. 2014. PTENα, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab 19: 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Chen X, Yin Q, Ruan D, Zhao X, Zhang C, McNutt MA, Yin Y. 2017. PTENβ is an alternatively translated isoform of PTEN that regulates rDNA transcription. Nat Commun 8: 14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman N, Gandin V, Svitkin YV, David M, Virgili G, Jaramillo M, Holcik M, Nagar B, Kimchi A, Sonenberg N. 2015. DAP5 associates with eIF2β and eIF4AI to promote Internal Ribosome Entry Site driven translation. Nucleic Acids Res 43: 3764–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist L, Pelletier J. 2009. Inhibitors of translation initiation as cancer therapeutics. Future Med Chem 1: 1709–1722. [DOI] [PubMed] [Google Scholar]
- Lindqvist L, Oberer M, Reibarkh M, Cencic R, Bordeleau ME, Vogt E, Marintchev A, Tanaka J, Fagotto F, Altmann M, et al. 2008. Selective pharmacological targeting of a DEAD box RNA helicase. PLoS One 3: e1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llacer JL, Hussain T, Marler L, Aitken CE, Thakur A, Lorsch JR, Hinnebusch AG, Ramakrishnan V. 2015. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell 59: 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, Dever TE. 2010. Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. J Biol Chem 285: 21203–21207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran G, Sachs MS, Atkins JF, Ivanov IP. 2012. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res 40: 2898–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar R, Bandyopadhyay A, Deng H, Maitra U. 2002. Phosphorylation of mammalian translation initiation factor 5 (eIF5) in vitro and in vivo. Nucleic Acids Res 30: 1154–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. 2004. eIF4E—from translation to transformation. Oncogene 23: 3172–3179. [DOI] [PubMed] [Google Scholar]
- Marash L, Liberman N, Henis-Korenblit S, Sivan G, Reem E, Elroy-Stein O, Kimchi A. 2008. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol Cell 30: 447–459. [DOI] [PubMed] [Google Scholar]
- Marinov GK, Williams BA, McCue K, Schroth GP, Gertz J, Myers RM, Wold BJ. 2014. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res 24: 496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Marcos P, Cheung YN, Hinnebusch AG. 2011. Functional elements in initiation factors 1, 1A, and 2β discriminate against poor AUG context and non-AUG start codons. Mol Cell Biol 31: 4814–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Marcos P, Nanda JS, Luna RE, Zhang F, Saini AK, Cherkasova VA, Wagner G, Lorsch JR, Hinnebusch AG. 2014. Enhanced eIF1 binding to the 40S ribosome impedes conformational rearrangements of the preinitiation complex and elevates initiation accuracy. RNA 20: 150–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menschaert G, Van Criekinge W, Notelaers T, Koch A, Crappe J, Gevaert K, Van Damme P. 2013. Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol Cell Proteomics 12: 1780–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC, Anderson WF. 1975. Purification and characterization of homogeneous protein synthesis initiation factor M1 from rabbit reticulocytes. J Biol Chem 250: 1197–1206. [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. 2013. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339: 1335–1338. [DOI] [PubMed] [Google Scholar]
- Nanda JS, Cheung YN, Takacs JE, Martin-Marcos P, Saini AK, Hinnebusch AG, Lorsch JR. 2009. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J Mol Biol 394: 268–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novac O, Guenier AS, Pelletier J. 2004. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res 32: 902–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SY, He F, Krans A, Frazer M, Taylor JP, Paulson HL, Todd PK. 2015. RAN translation at CGG repeats induces ubiquitin proteasome system impairment in models of fragile X-associated tremor ataxia syndrome. Hum Mol Genet 24: 4317–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. 2007. Trinucleotide repeat disorders. Annu Rev Neurosci 30: 575–621. [DOI] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. 2016. The integrated stress response. EMBO Rep 17: 1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasramka MA, Dashwood WM, Wang R, Saeed HH, Williams DE, Ho E, Dashwood RH. 2012. A role for low-abundance miRNAs in colon cancer: the miR-206/Kruppel-like factor 4 (KLF4) axis. Clin Epigenetics 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody DS. 1987. Translation initiation at an ACG triplet in mammalian cells. J Biol Chem 262: 11847–11851. [PubMed] [Google Scholar]
- Peabody DS. 1989. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem 264: 5031–5035. [PubMed] [Google Scholar]
- Pestova TV, Hellen CU. 2003. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev 17: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV. 2006. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev 20: 624–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarto N, Talarico D, Florkiewicz R, Rifkin DB. 1991. Selective expression of high molecular weight basic fibroblast growth factor confers a unique phenotype to NIH 3T3 cells. Cell Regul 2: 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert LS, Shi B, Nandi S, Mazan-Mamczarz K, Vitolo M, Bachman KE, He H, Gartenhaus RB. 2006. MCT-1 protein interacts with the cap complex and modulates messenger RNA translational profiles. Cancer Res 66: 8994–9001. [DOI] [PubMed] [Google Scholar]
- Renko M, Quarto N, Morimoto T, Rifkin DB. 1990. Nuclear and cytoplasmic localization of different basic fibroblast growth factor species. J Cell Physiol 144: 108–114. [DOI] [PubMed] [Google Scholar]
- Reuter K, Biehl A, Koch L, Helms V. 2016. PreTIS: a tool to predict non-canonical 5′ UTR translational initiation sites in human and mouse. PLoS Comput Biol 12: e1005170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. 2016. New structural insights into translational miscoding. Trends Biochem Sci 41: 798–814. [DOI] [PubMed] [Google Scholar]
- Sachs MS, Geballe AP. 2006. Downstream control of upstream open reading frames. Genes Dev 20: 915–921. [DOI] [PubMed] [Google Scholar]
- Saini AK, Nanda JS, Lorsch JR, Hinnebusch AG. 2010. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNA(i)(Met) binding to the ribosome. Genes Dev 24: 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini AK, Nanda JS, Martin-Marcos P, Dong J, Zhang F, Bhardwaj M, Lorsch JR, Hinnebusch AG. 2014. Eukaryotic translation initiation factor eIF5 promotes the accuracy of start codon recognition by regulating Pi release and conformational transitions of the preinitiation complex. Nucleic Acids Res 42: 9623–9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki J, Nakashima N. 2000. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc Natl Acad Sci 97: 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleich S, Strassburger K, Janiesch PC, Koledachkina T, Miller KK, Haneke K, Cheng YS, Kuechler K, Stoecklin G, Duncan KE, et al. 2014. DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature 512: 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleich S, Acevedo JM, Clemm von Hohenberg K, Teleman AA. 2017. Identification of transcripts with short stuORFs as targets for DENR*MCTS1-dependent translation in human cells. Sci Rep 7: 3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol 6: 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Buijsen RA, He F, Natla S, Jung L, Tropel P, Gaucherot A, Jacobs H, Meziane H, Vincent A, et al. 2017. Translation of expanded CGG repeats into FMRpolyG is pathogenic and may contribute to fragile X tremor ataxia syndrome. Neuron 93: 331–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, Levorse J, Dill BD, Schramek D, Molina H, et al. 2017. Translation from unconventional 5′ start sites drives tumour initiation. Nature 541: 494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Ogurtsov AY, Rogozin IB, Koonin EV, Lipman DJ. 2004. Comparative analysis of orthologous eukaryotic mRNAs: potential hidden functional signals. Nucleic Acids Res 32: 1774–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Fujii K, Kovary KM, Genuth NR, Rost HL, Teruel MN, Barna M. 2017. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol Cell 67: 71–83.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Tiu GC, Flynn RA, Byeon GW, Leppek K, Xu AF, Chang HY, Barna M. 2017. The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell 169: 1051–1065.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, Watanabe R, Chowdhury W, Hiraishi H, Murai MJ, Yamamoto Y, Miles D, Ikeda Y, Asano M, Asano K. 2012. Sequential eukaryotic translation initiation factor 5 (eIF5) binding to the charged disordered segments of eIF4G and eIF2β stabilizes the 48S preinitiation complex and promotes its shift to the initiation mode. Mol Cell Biol 32: 3978–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. 2010. Activities of ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev 24: 1787–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Hellen CU, Pestova TV. 2013. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol Cell 51: 249–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, Saghatelian A. 2013. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol 9: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilka R, Ernst C, Mehta AK, Haybaeck J. 2013. Eukaryotic translation initiation factors in cancer development and progression. Cancer Lett 340: 9–21. [DOI] [PubMed] [Google Scholar]
- Starck SR, Ow Y, Jiang V, Tokuyama M, Rivera M, Qi X, Roberts RW, Shastri N. 2008. A distinct translation initiation mechanism generates cryptic peptides for immune surveillance. PLoS One 3: e3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Jiang V, Pavon-Eternod M, Prasad S, McCarthy B, Pan T, Shastri N. 2012. Leucine–tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science 336: 1719–1723. [DOI] [PubMed] [Google Scholar]
- Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, Martins-Green M, Shastri N, Walter P. 2016. Translation from the 5′ untranslated region shapes the integrated stress response. Science 351: aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JD, Cowan JL, Perry LS, Coldwell MJ, Proud CG. 2015. ABC50 mutants modify translation start codon selection. Biochem J 467: 217–229. [DOI] [PubMed] [Google Scholar]
- Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P, et al. 2014. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron 83: 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs JE, Neary TB, Ingolia NT, Saini AK, Martin-Marcos P, Pelletier J, Hinnebusch AG, Lorsch JR. 2011. Identification of compounds that decrease the fidelity of start codon recognition by the eukaryotic translational machinery. RNA 17: 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Maruyama M, Tokuzawa Y, Murakami M, Oda Y, Yoshikane N, Makabe KW, Ichisaka T, Yamanaka S. 2005. Evolutionarily conserved non-AUG translation initiation in NAT1/p97/DAP5 (EIF4G2). Genomics 85: 360–371. [DOI] [PubMed] [Google Scholar]
- Tang HL, Yeh LS, Chen NK, Ripmaster T, Schimmel P, Wang CC. 2004. Translation of a yeast mitochondrial tRNA synthetase initiated at redundant non-AUG codons. J Biol Chem 279: 49656–49663. [DOI] [PubMed] [Google Scholar]
- Todd PK, Paulson HL. 2010. RNA-mediated neurodegeneration in repeat expansion disorders. Ann Neurol 67: 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Oh SY, Krans A, He F, Sellier C, Frazer M, Renoux AJ, Chen KC, Scaglione KM, Basrur V, et al. 2013. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78: 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toribio R, Diaz-Lopez I, Boskovic J, Ventoso I. 2016. An RNA trapping mechanism in Alphavirus mRNA promotes ribosome stalling and translation initiation. Nucleic Acids Res 44: 4368–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touriol C, Bornes S, Bonnal S, Audigier S, Prats H, Prats AC, Vagner S. 2003. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol Cell 95: 169–178. [DOI] [PubMed] [Google Scholar]
- Turner N, Grose R. 2010. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 10: 116–129. [DOI] [PubMed] [Google Scholar]
- Vagner S, Touriol C, Galy B, Audigier S, Gensac MC, Amalric F, Bayard F, Prats H, Prats AC. 1996. Translation of CUG- but not AUG-initiated forms of human fibroblast growth factor 2 is activated in transformed and stressed cells. J Cell Biol 135: 1391–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG. 2004. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol Cell Biol 24: 9437–9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. 2009. A census of human transcription factors: function, expression and evolution. Nat Rev Genet 10: 252–263. [DOI] [PubMed] [Google Scholar]
- Ventoso I, Sanz MA, Molina S, Berlanga JJ, Carrasco L, Esteban M. 2006. Translational resistance of late alphavirus mRNA to eIF2α phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev 20: 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Han KJ, Pang XW, Vaughan HA, Qu W, Dong XY, Peng JR, Zhao HT, Rui JA, Leng XS, et al. 2002. Large scale identification of human hepatocellular carcinoma-associated antigens by autoantibodies. J Immunol 169: 1102–1109. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. 2015. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440. [DOI] [PubMed] [Google Scholar]
- Wei J, Zhang Y, Ivanov IP, Sachs MS. 2013. The stringency of start codon selection in the filamentous fungus Neurospora crassa. J Biol Chem 288: 9549–9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisser M, Schafer T, Leibundgut M, Bohringer D, Aylett CHS, Ban N. 2017. Structural and functional insights into human re-initiation complexes. Mol Cell 67: 447–456.e7. [DOI] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. 2006. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 34: 7–11. [DOI] [PubMed] [Google Scholar]
- Xiao JH, Davidson I, Matthes H, Garnier JM, Chambon P. 1991. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell 65: 551–568. [DOI] [PubMed] [Google Scholar]
- Xue S, Tian S, Fujii K, Kladwang W, Das R, Barna M. 2015. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 517: 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa M, Ito D, Honda T, Kubo K, Noda M, Nakajima K, Suzuki N. 2015. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum Mol Genet 24: 1630–1645. [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R. 2009. Fidelity at the molecular level: lessons from protein synthesis. Cell 136: 746–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH. 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15: 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Hinnebusch AG. 2011. An upstream ORF with non-AUG start codon is translated in vivo but dispensable for translational control of GCN4 mRNA. Nucleic Acids Res 39: 3128–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gao X, Coots RA, Conn CS, Liu B, Qian SB. 2015. Translational control of the cytosolic stress response by mitochondrial ribosomal protein L18. Nat Struct Mol Biol 22: 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, Katzman RB, Gass J, Murray ME, Shinohara M, et al. 2016. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci 19: 668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. 2015. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526: 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoviev A, Hellen CU, Pestova TV. 2015. Multiple mechanisms of reinitiation on bicistronic calicivirus mRNAs. Mol Cell 57: 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitomer RS, Walthall DA, Rymond BC, Hollenberg CP. 1984. Saccharomyces cerevisiae ribosomes recognize non-AUG initiation codons. Mol Cell Biol 4: 1191–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoll WL, Horton LE, Komar AA, Hensold JO, Merrick WC. 2002. Characterization of mammalian eIF2A and identification of the yeast homolog. J Biol Chem 277: 37079–37087. [DOI] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. 2011. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci 108: 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. 2013. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci 110: E4968–E4977. [DOI] [PMC free article] [PubMed] [Google Scholar]


