Zhou et al. discovered that the zinc finger transcription factor ZNF281 potently stimulates cardiac reprogramming by genome-wide association with GATA4 on cardiac enhancers. Concomitantly, ZNF281 suppresses expression of genes associated with inflammatory signaling, suggesting the antagonistic convergence of cardiac and inflammatory transcriptional programs.
Keywords: direct cellular reprogramming, cardiomyocytes, ZFP281, cardiac gene activation, anti-inflammation, heart regeneration
Abstract
Direct reprogramming of fibroblasts to cardiomyocytes represents a potential means of restoring cardiac function following myocardial injury. AKT1 in the presence of four cardiogenic transcription factors, GATA4, HAND2, MEF2C, and TBX5 (AGHMT), efficiently induces the cardiac gene program in mouse embryonic fibroblasts but not adult fibroblasts. To identify additional regulators of adult cardiac reprogramming, we performed an unbiased screen of transcription factors and cytokines for those that might enhance or suppress the cardiogenic activity of AGHMT in adult mouse fibroblasts. Among a collection of inducers and repressors of cardiac reprogramming, we discovered that the zinc finger transcription factor 281 (ZNF281) potently stimulates cardiac reprogramming by genome-wide association with GATA4 on cardiac enhancers. Concomitantly, ZNF281 suppresses expression of genes associated with inflammatory signaling, suggesting the antagonistic convergence of cardiac and inflammatory transcriptional programs. Consistent with an inhibitory influence of inflammatory pathways on cardiac reprogramming, blockade of these pathways with anti-inflammatory drugs or components of the nucleosome remodeling deacetylase (NuRD) complex, which associate with ZNF281, stimulates cardiac gene expression. We conclude that ZNF281 acts at a nexus of cardiac and inflammatory gene programs, which exert opposing influences on fibroblast to cardiac reprogramming.
The adult heart is among the least regenerative tissues (Xin et al. 2013; Cahill et al. 2017). In response to myocardial infarction (MI), the human heart can lose hundreds of millions of cardiomyocytes, resulting in catastrophic loss of cardiac contractility. Typically, the lost cardiomyocytes are replaced by scar tissue due to activation of endogenous fibroblasts, which account for approximately a third of the cells of the heart (Tallquist and Molkentin 2017). Reprogramming cardiac fibroblasts (CFs) to induced cardiomyocytes (iCMs) by forced expression of cardiac transcription factors represents a potential means of enhancing cardiac repair by reducing scar tissue while simultaneously generating new cardiomyocytes (Ieda et al. 2010; Qian et al. 2012; Song et al. 2012; Fu et al. 2013; Nam et al. 2013). However, low efficiency as well as the lack of understanding of the molecular basis of the reprogramming process represent challenges to its potential clinical application (Srivastava and DeWitt 2016; Kojima and Ieda 2017; Vaseghi et al. 2017).
The first cardiac reprogramming cocktail consisted of three cardiac transcription factors: GATA4, MEF2C, and TBX5 (GMT) (Ieda et al. 2010). Subsequent effort has been directed toward optimization of cardiac reprogramming by generating different cocktails that contain various combinations of proteins, microRNAs, and small molecules (Song et al. 2012; Ifkovits et al. 2014; Muraoka et al. 2014; Wang et al. 2015; Yamakawa et al. 2015; Zhao et al. 2015; Zhou et al. 2015, 2016; Abad et al. 2017; Mohamed et al. 2017). Most of these studies adopted a candidate approach to examine the effects of supplementing the GMT cocktail with other key cardiac factors or regulators that have been shown to promote other somatic cell reprogramming processes. For example, adding the cardiac transcription factor HAND2, which plays an essential role in cardiac morphogenesis (Srivastava et al. 1997; Han et al. 2006; Firulli et al. 2017), enhanced the cardiac reprogramming efficiency of GMT both in vitro and in vivo (Song et al. 2012). Addition of a cardiac microRNA, miR-133, was also shown to enhance cardiac reprogramming in mouse and human fibroblasts (Nam et al. 2013; Muraoka et al. 2014). In addition, SB431542 (a TGF-β inhibitor), and DAPT (a Notch inhibitor), which promote the reprogramming of induced pluripotent stem cells (iPSCs), also enhanced cardiac reprogramming (Ifkovits et al. 2014; Abad et al. 2017). However, these reprogramming cocktails are still relatively inefficient, and the molecular basis of the reprogramming process is poorly understood. Reprogramming of adult fibroblasts is also much less efficient compared with embryonic fibroblasts, posing challenges for potential restoration of adult cardiac function.
Previously, we screened a library of protein kinases and discovered that AKT1 dramatically accelerates and amplifies the cardiac reprogramming process (Zhou et al. 2015). Our optimal cocktail, which contains AKT1, GATA4, HAND2, MEF2C, and TBX5 (which we refer to as AGHMT), converts ∼50% of mouse embryonic fibroblasts (MEFs) to iCMs (Zhou et al. 2015). However, the efficiency of conversion of adult fibroblasts, such as adult tail-tip fibroblasts (TTFs) and CFs, is still low.
Given that most known regulators of cardiac development or iPSC reprogramming have been tested extensively in previous studies and that none was able to efficiently reprogram adult fibroblasts to iCMs, we sought to identify previously unidentified regulators of cardiac reprogramming. Here we describe our findings from an unbiased screen of 1052 ORF cDNAs encoding 786 transcription factors, epigenetic regulators, cytokines, and nuclear receptors to augment AGHMT-dependent cardiac reprogramming of adult TTFs. This screen led to the discovery of 49 activators and 129 inhibitors of cardiac reprogramming, many of which participate in various signaling pathways and biological processes, including the TGF-β and Notch signaling pathways, which have been shown to be important for cardiac reprogramming (Ifkovits et al. 2014; Abad et al. 2017). Interestingly, several factors involved in the inflammatory response were identified as inhibitors of cardiac reprogramming, indicating that the inflammatory response might act as a barrier for adult cardiac reprogramming. The strongest activator of cardiac reprogramming, Krüppel-type zinc finger transcription factor 281 (ZNF281), does not have a characterized function in cardiac development and thus would not have been anticipated to impinge on the mechanisms of fibroblast-to-cardiomyocyte reprogramming. We show that ZNF281 enhances cardiac reprogramming by associating with GATA4 on cardiac enhancers and inhibiting inflammatory signaling, which antagonizes cardiac reprogramming. Consistent with this notion, treatment of adult fibroblasts with anti-inflammatory drugs dramatically increased cardiac reprogramming efficiency. Our findings identify ZNF281 as a robust and efficient activator of adult cardiac reprogramming and highlight the importance of inflammatory signaling in the governance of cardiac gene expression.
Results
Identification of regulators of cardiac reprogramming in adult fibroblasts
Our previously optimized cardiac reprogramming protocol with five factors (AGHMT, also called 5F) was able to reprogram ∼3% of adult TTFs to iCMs as measured by activation of a cardiac-specific αMHC-GFP transgene and cardiac troponin T (cTnT) immunostaining (Zhou et al. 2015). To identify additional regulators of cardiac reprogramming, we created a retroviral expression library consisting of 1052 ORF cDNAs encoding 786 human transcription factors, cytokines, epigenetic regulators, and nuclear receptors (Supplemental Table S1). We initially screened this expression library for activators and inhibitors of cardiac reprogramming by expressing individual cDNAs together with 5F in isolated TTFs from αMHC-GFP mice, as schematized in Figure 1A. After 9 d, a high-throughput cell analyzer system was used to image and quantify cardiac reprogramming based on αMHC-GFP and cTnT expression. Activators were defined as genes that increased αMHC-GFP or cTnT expression with a Z-score ≥2, whereas genes with Z-scores −2 or lower for these cardiac markers were defined as inhibitors. This screen revealed 49 potential activators and 129 potential inhibitors of cardiac reprogramming (Fig. 1B,C; Supplemental Table S2).
Figure 1.
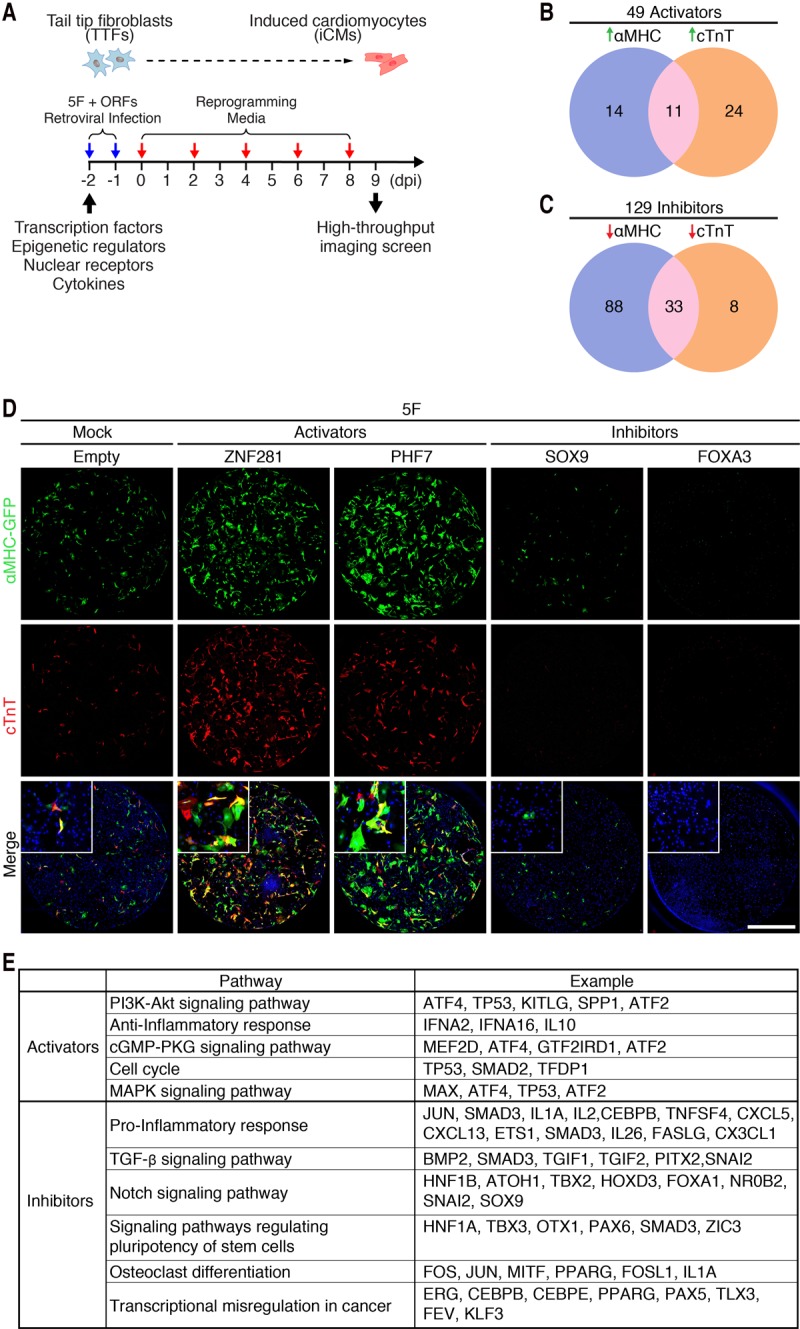
Identification of activators and inhibitors of 5F-mediated cardiac reprogramming from a human ORF cDNA screen. (A) Schematic diagram of the human ORF cDNA library screen strategy for cardiac reprogramming in adult TTFs. (B) Venn diagram showing the number of activators identified from the screen. Genes with Z-scores of αMHC-GFP or cTnT expression ≥2 were defined as activators. Twenty-five genes induced αMHC-GFP expression only, 35 genes induced cTnT expression only, and 11 genes induced expression of both markers. (C) Venn diagram showing the number of inhibitors identified from the screen. Genes with Z-scores of αMHC-GFP or cTnT expression −2 or lower were defined as inhibitors. One-hundred-twenty-one genes repressed αMHC expression only, 41 genes repressed cTnT expression only, and 33 genes repressed both αMHC-GFP and cTnT expression. (D) Representative immunocytochemistry images of TTFs from adult αMHC-GFP transgenic mice treated with 5F and either empty virus or viruses encoding activators (ZNF281 or PHF7) and inhibitors (FOXA3 or SOX9). Cells were fixed and stained for αMHC-GFP (green), cTnT (red), and Hoechst (blue) 9 d after infection. Bars, 2 mm. (E) Pathways enriched in activators (n = 49) and inhibitors (n = 129), respectively, by DAVID pathway analysis.
Among the 49 activators, 25 enhanced αMHC-GFP expression, 35 enhanced cTnT expression, and 11 enhanced expression of both cardiac markers (Fig. 1B). The two strongest activators were PHD finger protein 7 (PHF7), a histone H3-binding protein expressed only in the male germline (Yang et al. 2012), and the ZNF281 protein, about which little is known (Fig. 1D; Supplemental Table S2). Among the 129 inhibitors, 121 inhibited αMHC-GFP expression, 41 inhibited cTnT expression, and 33 inhibited both cardiac markers (Fig. 1C). Some of the repressors, such as forkhead box protein A3 (FOXA3), nearly abolished 5F-mediated cardiac reprogramming (Fig. 1D; Supplemental Table S2). Cell numbers were unaffected by the inhibitors, suggesting that they acted directly on the reprogramming process rather than through indirect mechanisms, such as causing cell death.
Pathway analysis of regulators of cardiac reprogramming
To identify key pathways that regulate cardiac reprogramming, we performed pathway enrichment analysis for activator and inhibitor genes. Given that this was a genome-wide screen, we expected that this analysis would identify pathways known to regulate cardiac reprogramming. Indeed, the PI3K–AKT signaling pathway, which has been shown to enhance cardiac reprogramming (Zhou et al. 2015), was among the most enriched pathways associated with the activators. Other enriched pathways associated with the activators were the anti-inflammatory pathway, the cGMP–PKG signaling pathway, the cell cycle pathway, and the MAPK signaling pathway (Fig. 1E). It is noteworthy that the TGF-β and Notch signaling pathways, which negatively regulate cardiac reprogramming (Ifkovits et al. 2014; Abad et al. 2017), were associated with the inhibitors. Other pathways associated with the inhibitors were the proinflammatory pathway and signaling pathways regulating pluripotency of stem cells, osteoclast differentiation, and transcriptional misregulation in cancer (Fig. 1E).
Because inflammatory signaling pathways were associated with both activators and inhibitors, we examined the functions of each individual gene within these pathways. Interestingly, we found that most of the identified activators possessed anti-inflammatory functions, including several anti-inflammatory cytokines, such as IFNA2, IFNA16, and IL10. Consistent with these findings, most identified inhibitors were proinflammatory, including several proinflammatory cytokines, such as IL1A, IL2, and IL26, and the inflammatory response transcription factor CEBPβ (Fig. 1E).
ZNF281 enhances cardiac reprogramming of adult fibroblasts
PHF7 and ZNF281 were the two strongest activators identified from our retroviral cDNA expression screen with 5F (Fig. 1D; Supplemental Table S2). We focused our initial attention on ZNF281, which has a broad expression pattern with enriched expression in the heart (Supplemental Fig. S1), and explored the mechanistic basis of its cardiac-inducing activity. Previous reports described the influence of ZNF281 on pluripotency, stemness, and epithelial–mesenchymal transition (EMT) (Hahn and Hermeking 2014). However, the potential involvement of ZNF281 in cardiac development has not been explored previously.
We refer to our reprogramming mix of 5F plus ZNF281 as 6F. We validated the results of our screen by assessing GFP and cTnT expression in αMHC-GFP TTFs following 5F and 6F reprogramming after 7 d (Fig. 2A). Flow cytometry showed that addition of ZNF281 to 5F generated ∼33% αMHC-GFP+, ∼45% cTnT+, and ∼28% αMHC-GFP+/cTnT+ TTFs after 7 d of reprogramming (Fig. 2B,C). This TTF reprogramming efficiency using 6F is noteworthy when considering the relatively low statistical likelihood of each fibroblast taking up all five or six separate retroviruses encoding the reprogramming factors. We also examined the expression of cardiac and fibroblast transcripts by quantitative PCR (qPCR). Addition of ZNF281 to 5F increased the expression of cardiac marker genes Myh6 and Actc1 by ∼120-fold and ∼20-fold, respectively, and decreased expression of fibroblast marker genes Col1a2 and Sox9 by ∼30% and ∼60%, respectively (Fig. 2D). We also validated some of these results using αMHC-GFP CFs. The addition of ZNF281 to 5F increased the expression of GFP and cTnT in αMHC-GFP CFs evaluated by immunocytochemistry after 7 d of reprogramming (Supplemental Fig. S2A). These data were also corroborated by FACS analysis. The addition of ZNF281 to 5F generated ∼25% αMHC-GFP+, ∼45% cTnT+, and ∼21% αMHC-GFP+/cTnT+ iCMs (Supplemental Fig. S2B,C). Interestingly, ZNF281 in the presence of 5F also induced a twofold increase in the number of beating cells after 4 wk of reprogramming (Supplemental Fig. S2D).
Figure 2.
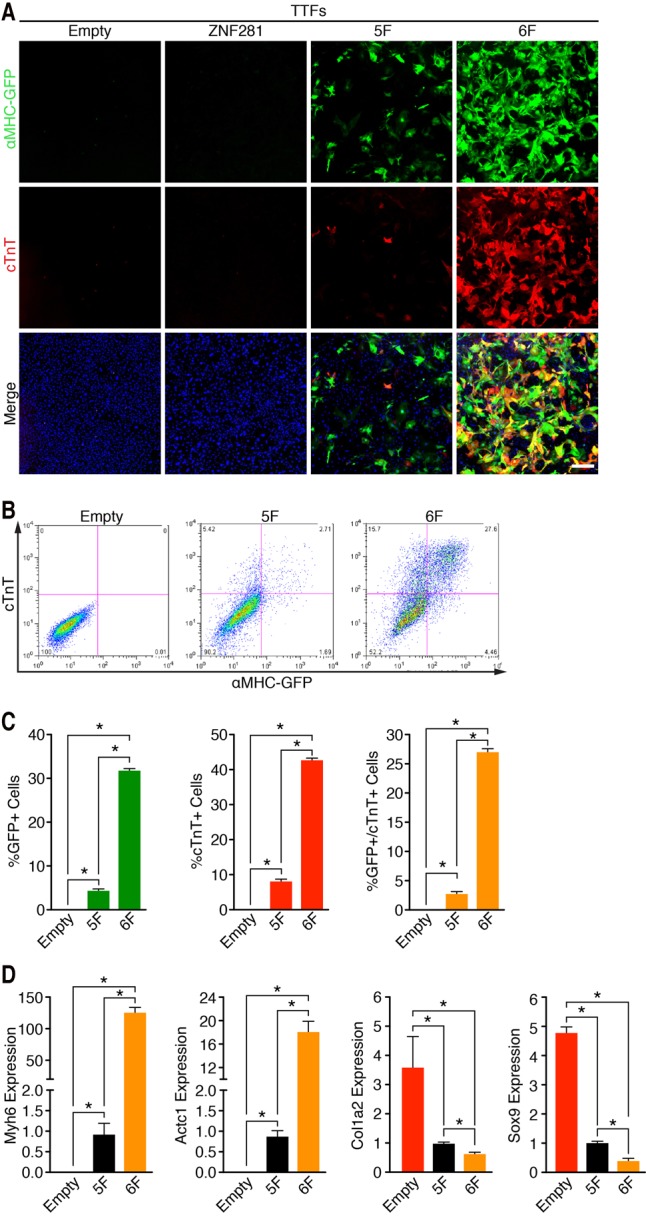
ZNF281 enhances cardiac reprogramming of adult fibroblasts. (A) Immunocytochemistry images of adult αMHC-GFP transgenic TTFs 7 d after infection with empty, ZNF281, 5F, or 6F retroviruses show that ZNF281 enhances expression of cardiac markers with 5F. (Green) αMHC-GFP; (red) cTnT; (blue) Hoechst. Bars, 500 µm. (B,C) Representative flow cytometry plot (B) and analyses (C) of αMHC-GFP+ and cTnT+ TTFs 7 d after infection with empty, 5F, or 6F. (D) TTFs were infected with empty, 5F plus empty, and ZNF281 for 7 d. Transcript levels of cardiac marker genes (Myh6 and Actc1) and fibroblast marker genes (Col1a2 and Sox9) were determined by qPCR. (*) P < 0.05.
Global reprogramming of the cardiac transcriptome by ZNF281
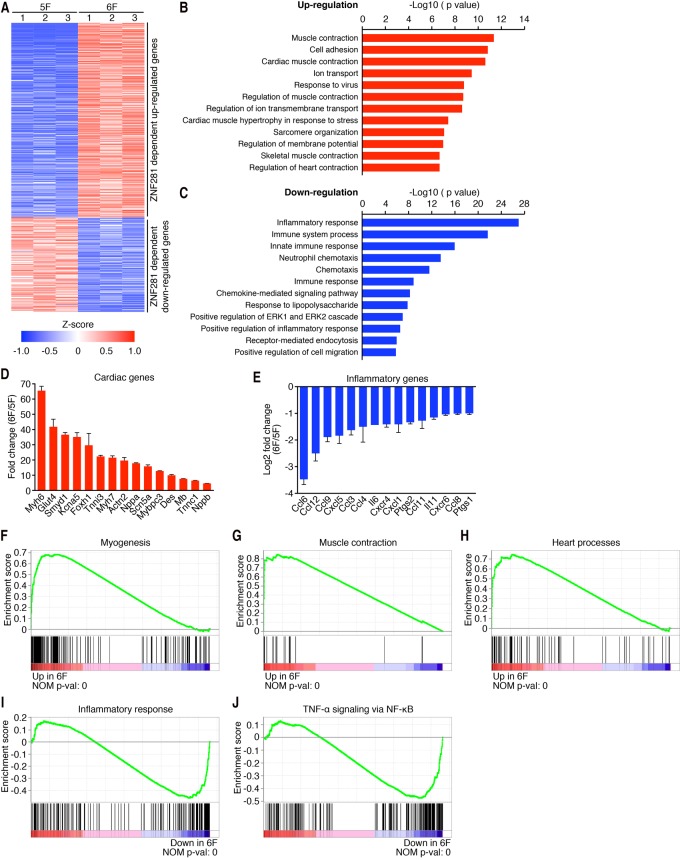
To further define the influence of ZNF281 on the reprogramming process, we performed RNA sequencing (RNA-seq) using adult TTFs reprogrammed for 7 d with 5F or 6F. Using a twofold cutoff and false discovery rate (FDR) <0.01 threshold for inclusion, we identified ∼1000 up-regulated genes and ∼500 down-regulated genes in 6F-treated compared with 5F-treated TTFs (Fig. 3A–C). Gene ontology (GO) enrichment analysis revealed that ZNF281 selectively up-regulated genes associated with muscle contractility (Fig. 3B,D), suggesting that ZNF281 globally enhanced cardiac reprogramming. Interestingly, the top GO terms enriched in the genes that were down-regulated by ZNF281 were all related to the inflammatory response (Fig. 3C,E). The gene set enrichment analysis (GSEA) also revealed that ZNF281 significantly up-regulated gene sets related to cardiac cells, such as myogenesis, muscle contraction, and heart processes (Fig. 3F–H). Among the most up-regulated genes in these three gene sets, we found structural genes such as troponin T and troponin I isoforms (Tnnt1 and Tnnt3 Tnni3), the major cardiac muscle isoform of myosin heavy and light chain (Myh6, Myh7, Myl4, and Myl2), specific muscle and cardiac actinin (Actc1 and Actn2), and the cardiac calcium channels (Ryr2), among other genes (Supplemental Fig. S3A–C). Similar to the GO analysis, ZNF281 in the presence of 5F significantly down-regulated gene sets involved in the inflammatory response and TNF-α signaling via NF-κB. (Fig. 3I,J) The majority of the down-regulated genes by ZNF281 in all of these three gene sets encoded cytokines and cytokine receptors involved in the immune response (e.g., IL6, Marco, TNF-α, and IL7r) (Supplemental Fig. S3D,E). These results suggest a dual role for ZNF281 in the activation of cardiac and the suppression of inflammatory gene programs.
Figure 3.
RNA-seq analysis shows that ZNF281 enhances cardiac genes and represses inflammatory genes. (A) Heat map of 1500 differentially expressed genes in 5F-treated versus 6F-treated TTFs identified by RNA-seq. (Red) Up-regulation; (blue) down-regulation. RNA-seq samples were prepared from adult TTFs reprogrammed for 7 d. (B,C) GO analysis showing biological processes associated with genes up-regulated (B) and down-regulated (C) by ZNF281. (D,E) Gene expression changes between 6F and 5F for selected cardiac markers (D) or inflammatory markers (E) as determined by RNA-seq. (F–H) Enrichment plots of the indicated gene sets and their nominal P-value of genes up-regulated by ZNF281. (I,J) Enrichment plots of the indicated gene sets and their nominal P-value of genes down-regulated by ZNF281.
ZNF281 broadly co-occupies cardiac enhancers with GATA4
To begin to define the molecular mechanism by which ZNF281 enhances the cardiac reprogramming activity of the 5F cocktail, we performed coimmunoprecipitation assays with ZNF281 and each reprogramming transcription factor in transfected HEK293 cells. We found that Flag-tagged ZNF281 interacted with Myc-tagged GATA4 but not with HAND2, MEF2C, or TBX5 (Fig. 4A). To determine whether ZNF281 could directly activate cardiac genes, we examined the ability of ZNF281 to activate a luciferase reporter controlled by the αMHC promoter (αMHC-Luciferase). Indeed, ZNF281 activated the αMHC-Luciferase reporter approximately sevenfold, and when ZNF281 was coexpressed with GATA4, αMHC-Luciferase was activated ∼15-fold (Supplemental Fig. S4), suggesting that ZNF281 and GATA4 cooperate to activate the αMHC promoter.
Figure 4.
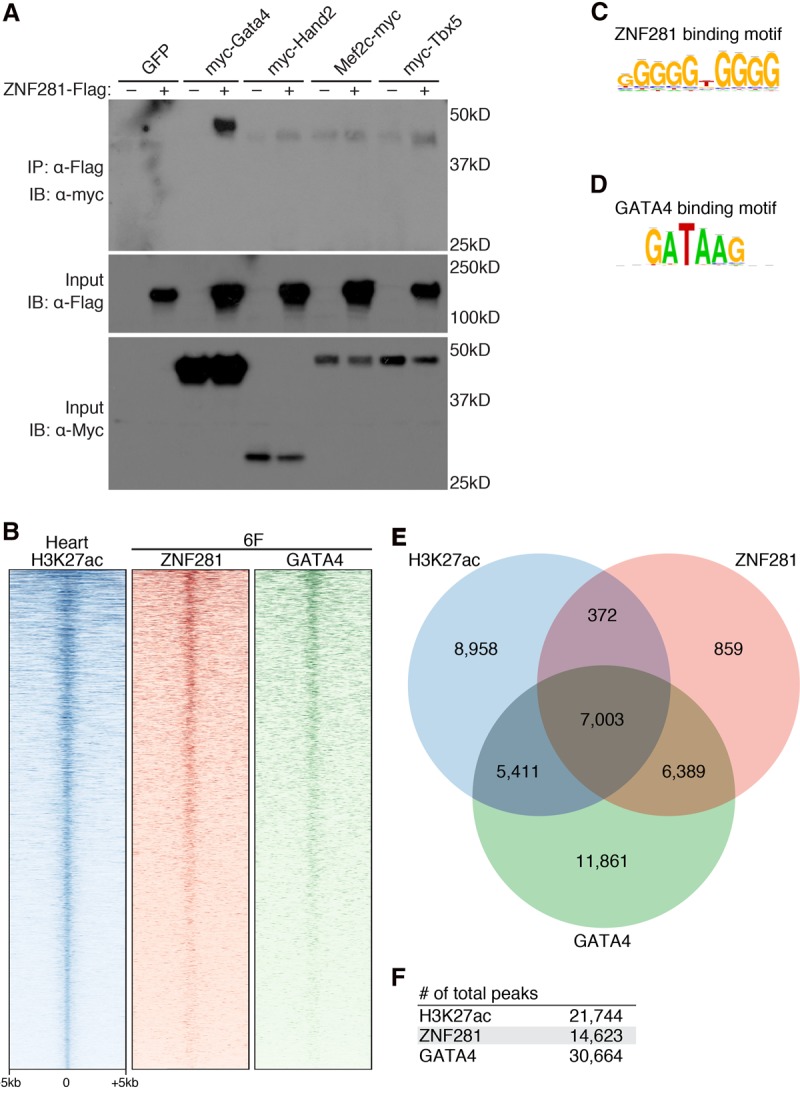
ZNF281 interacts with GATA4 to synergistically activate cardiac genes. (A) Coimmunoprecipitation assays were performed using HEK293 cells transfected with equal amounts of plasmid DNA encoding Myc-tagged GATA4, HAND2, MEF2C, or TBX5 and/or Flag-tagged ZNF281. (IP) Immunoprecipitation; (IB) immunoblot. (B) Heat maps showing ChIP-seq (chromatin immunoprecipitation [ChIP] followed by high-throughput sequencing) data for H3K27ac in adult mouse hearts and ZNF281 and GATA4 binding in reprogrammed TTFs at ±5 kb around the peak center. ChIP-seq experiments were performed using TTFs infected with 6F for 2 d. (C) ZNF281-binding motif enriched within ZNF281-binding peaks. (D) GATA4-binding motif enriched within GATA4-binding peaks. (E) Venn diagram showing the number of overlapping peaks between heart H3K27ac, ZNF281, and GATA4. (F) Total number of peaks identified by ChIP-seq.
To further understand the molecular relationship between ZNF281 and GATA4 in the cardiac reprogramming process, we examined the genomic locations of ZNF281 and GATA4 at an early stage of the reprogramming process (2 d after infection with 6F) by chromatin immunoprecipitation (ChIP) with antibodies to endogenous proteins followed by high-throughput sequencing (ChIP-seq) (Fig. 4B–E). De novo motif discovery on the binding peaks for ZNF281 and GATA4 revealed that the most significantly enriched motif associated with ZNF281 binding was GGGGTGGGG (Fig. 4C). For GATA4, the most enriched motif was GATAAG, which matches the consensus sequence for DNA binding of this transcription factor (Fig. 4D). ChIP-seq identified 14,623 peaks for ZNF281 and 30,664 peaks for GATA4 (Fig. 4E,F).
Based on the interaction of ZNF281 with GATA4, we predicted that ZNF281 co-occupies genomic sites with GATA4 on cardiac enhancers. Indeed, we found a high degree of overlap of ZNF281 and GATA4 genomic binding sites. Among the 14,623 peaks of ZNF281, we found that 91.6% (13,392 of 14,623) overlapped with peaks of GATA4 binding, and only 8.4% of ZNF281 peaks did not overlap with GATA4, indicating the co-occupancy of ZNF281 and GATA4 on endogenous genomic sites at an early stage of the cardiac reprogramming process (Fig. 4E). In contrast, the overlapping peaks of ZNF281 and GATA4 accounted for only 43.7% of the total GATA4 peaks (13,392 of 30,664) (Fig. 4E), suggesting that GATA4 likely co-occupies additional sites with other reprogramming factors besides ZNF281.
To assess genome-wide localization of ZNF281 on cardiac enhancers, we established a chromatin landscape for heart tissue using H3K27ac ChIP data from The Mouse ENCODE Consortium (Mouse ENCODE Consortium et al. 2012) to reveal cardiac enhancers. Rank ordering of all cardiac enhancers based on H3K27ac enrichment revealed that GATA4 bound to the majority (57.1%; 12,414 of 21,744) of active cardiac enhancers, consistent with the established role of GATA4 in heart development. ZNF281 bound to 33.9% (7375 of 21,744) of active cardiac enhancers, while ZNF281 and GATA4 co-occupied 32.2% (7003 of 21,744) of cardiac enhancers (Fig. 4B,E). Genome-wide binding data for ZNF281 and GATA4 suggested that ZNF281 is most commonly associated with GATA4 on cardiac enhancers, defined by the presence of H3K27ac (Fig. 4B,E).
GATA4 recruits ZNF281 to cardiac enhancers
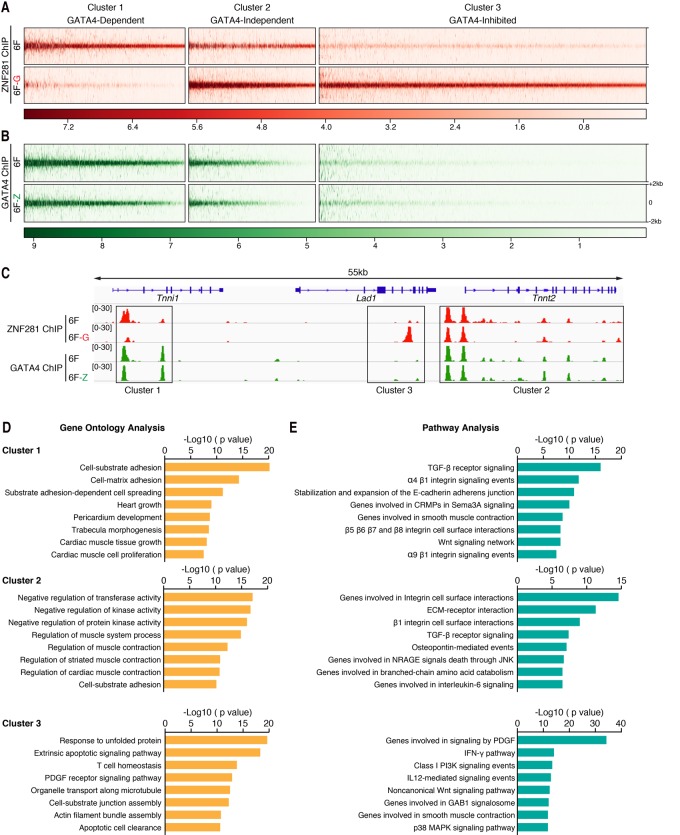
We compared the occupancy patterns of ZNF281 in the presence (referred to as “6F”) or absence of GATA4 (referred to as “6F − G”) (Fig. 5A). Additionally, we examined the genomic occupancy pattern of GATA4 in the absence of ZNF281 (referred to as “6F − Z”) following reprogramming (Fig. 5B). We found that the occupancy pattern of GATA4 throughout the genome was not dramatically affected by the presence or absence of ZNF281 (Fig. 5B). However, the presence of GATA4 strongly impacted the genomic occupancy pattern of ZNF281 (Fig. 5A). We assigned 33,934 ZNF281-binding peaks into three clusters based on how GATA4 influenced ZNF281 occupancy: a GATA4-dependent cluster (cluster 1, 8882 of 33,934 peaks), a GATA4-independent cluster (cluster 2, 7003 of 33,934 peaks), and a GATA4-inhibited cluster (cluster 3, 18,049 of 33,934 peaks) (Fig. 5A,B). Interestingly, cluster 3, in which the occupancy of ZNF281 was inhibited by the presence of GATA4, also correlated with lower binding affinity of GATA4 compared with clusters 1 and 2 (Fig. 5A,B). It seems that the higher-affinity GATA4-binding sites in cluster 1 competed ZNF281 away from the lower-affinity GATA4-binding sites in cluster 3. For example, a region that is located at chromosome 1 (137,694,960–137,749,970; mm9) shows binding peaks that belong to three different clusters (Fig. 5C). Binding peaks that associated with the sarcomere genes Tnni1 and Tnnt2, which encode subunits of the troponin complex involved in regulating muscle contraction, were assigned to cluster 1 and cluster 2, respectively. Binding peaks that associated with a nonmuscle gene, ladinin-1 (Lad1), which encodes a basement membrane protein, were assigned to cluster 3.
Figure 5.
GATA4 recruits ZNF281 to cardiac enhancers. (A,B) Heat map for ZNF281 or GATA4 genomic binding at ±2 kb around the peak center in each cluster. ChIP-seq experiments were performed using adult TTFs reprogrammed for 2 d with 6F, 6F − G, or 6F − Z. (C) Integrative Genomics Viewer (IGV) browser tracks at chromosome 1 (137,694,960–137,749,970; mm9) show an example of peaks that belong to each indicated cluster. (D,E) GOs (D) and pathways (E) enriched in genes that associate with each indicated cluster identified by GO enrichment analysis and pathway analysis.
To investigate the functional significance of the GATA4- and ZNF281-binding peaks in the three clusters, we performed GO and pathway enrichment analysis using the Genomic Regions Enrichment of Annotations Tool (GREAT) (McLean et al. 2010). For GO enrichment analysis, we found that most of the GO terms enriched in clusters 1 and 2 were heart- or muscle-related (Fig. 5D), whereas most of the GO terms enriched in cluster 3 were related to stress and inflammatory responses. For pathway enrichment analysis, several pathways, such as the TGF-β and Wnt signaling pathways, which are important for cardiogenesis, heart repair, and cardiac reprogramming (Ifkovits et al. 2014), were enriched in clusters 1 and 2, whereas inflammatory response pathways were enriched in cluster 3 (Fig. 5E). We conclude that GATA4 directly recruits ZNF281 to cardiac enhancers to activate cardiac gene expression.
In contrast to the effect on cardiac enhancers, GATA4 did not attenuate ZNF281 binding to inflammatory enhancers (Fig. 5A,D,E). Additionally, ZNF281 served as a repressor instead of an activator of inflammatory genes, suggesting that ZNF281 acts through distinct mechanisms to regulate expression of cardiac and inflammatory genes.
Corepression of inflammatory genes by ZNF281 and nucleosome remodeling deacetylase (NuRD) complex components
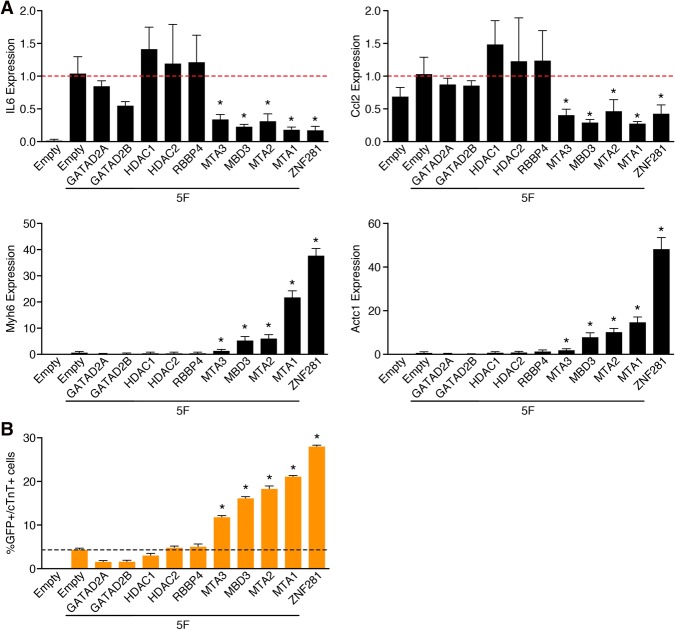
Previously, it was reported that ZNF281 associates with the NuRD complex to repress transcription (Fidalgo et al. 2012, 2016). The NuRD complex is an ATP-dependent chromatin remodeling complex that contains multiple subunits. Notably, we identified the NuRD complex component MTA1 as an activator of cardiac reprogramming in our screen (Supplemental Table S2). Because the NuRD complex has also been shown to repress inflammatory signaling (Ramirez-Carrozzi et al. 2006), we tested other NuRD complex subunits, including MTA2, MTA3, MBD3, GATAD2A, GATAD2B, RBBP4, HDAC1, and HDAC2, in the reprogramming assay with 5F in adult TTFs. Indeed, addition of four of the NuRD complex subunits (MTA1, MTA2, MTA3, and MBD3) to 5F substantially decreased expression of the inflammatory markers (IL6 and Ccl2) and increased expression of cardiac markers (Myh6 and Actc1) (Fig. 6A). Addition of NuRD complex subunits also promoted cardiac reprogramming, as measured by flow cytometry using the cardiac markers αMHC-GFP and cTnT (Fig. 6B; Supplemental Fig. S5A,B).
Figure 6.
ZNF281 represses the inflammatory response through the NuRD complex. (A) TTFs were infected with empty, 5F plus empty, ZNF281, or each individual NuRD complex subunit retrovirus for 7 d. Transcript levels of inflammatory (IL6 and Ccl2) and cardiac (Myh6 and Actc1) marker genes were determined by qPCR. (B) Representative flow cytometry plot of αMHC-GFP+ and cTnT+ TTFs 7 d after infection with empty, 5F plus empty, ZNF281, or each individual NuRD complex subunit retroviruses. The dashed line indicates control. (*) P < 0.05.
Because the NuRD complex is composed of several regulatory subunits, including HDAC1 and HDAC2, we tested an inhibitor of these enzymes (FK228) for its potential impact on reprogramming. Consistent with our model, we observed that inhibition of HDAC1/2 activity by FK228 decreased cardiac gene expression in the presence of 5F to a level similar to control cells without 5F (Supplemental Fig. S5C,D).
Anti-inflammatory drugs promote cardiac reprogramming
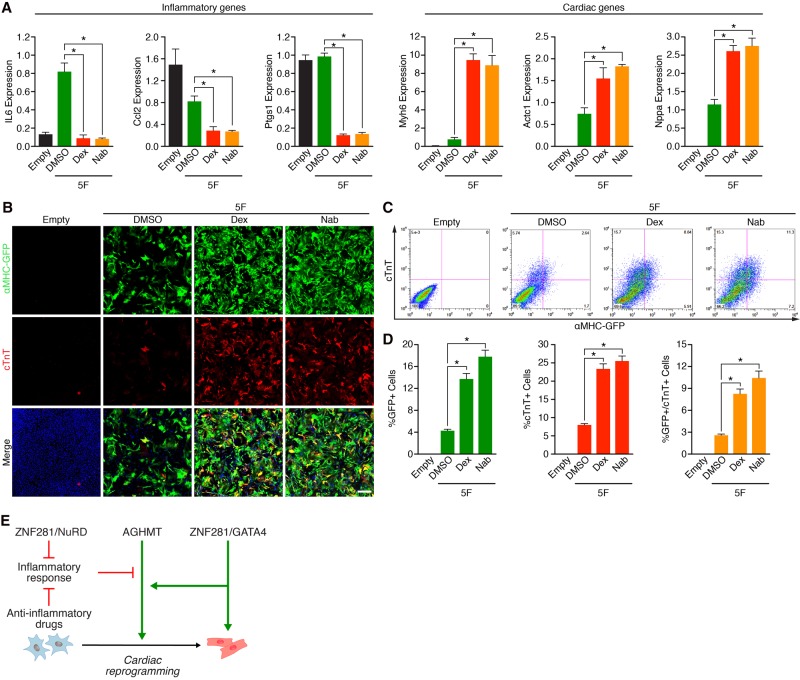
Given the influence of ZNF281 on inflammatory gene expression and the finding that numerous activators and inhibitors identified in our screen were also implicated in inflammatory pathways, we postulated that inhibition of the inflammatory response by anti-inflammatory drugs would also enhance reprogramming. To test this hypothesis, we evaluated the effects of two anti-inflammatory drugs—dexamethasone (Dex), a steroidal anti-inflammatory drug, and the cyclooxygenase enzyme inhibitor nabumetone (Nab), a nonsteroidal anti-inflammatory drug—on the reprogramming process. Anti-inflammatory drugs were added to 5F reprogrammed TTFs after viral infection. After 7 d of drug treatment, the transcript levels of inflammatory (IL6, Ccl2, and Ptgs1) and cardiac (Myh6, Actc1, and Nppa) markers were examined by qPCR. Addition of anti-inflammatory drugs to 5F reprogrammed TTFs decreased expression of inflammatory markers, as expected, but increased the expression of cardiac markers from twofold to 10-fold, indicating enhanced reprogramming efficiency (Fig. 7A). Anti-inflammatory drugs also enhanced the reprogramming process as assayed by immunocytochemistry (Fig. 7B) and flow cytometry (Fig. 7C,D) for the cardiac markers αMHC-GFP and cTnT, respectively. Thus, we conclude that inflammatory signaling imposes an inhibitory influence on the cardiac reprogramming process.
Figure 7.
Anti-inflammatory drugs promote cardiac reprogramming. (A) 5F reprogrammed TTFs treated with either DMSO or the indicated anti-inflammatory drugs for 7 d after infection. Expression of inflammatory genes (IL6, Ccl2, and Ptgs1), and cardiac genes (Myh6, Actc1, and Nppa) was determined by qPCR. (Dex) 10 µM Dex; (Nab) 10 µM Nab. (B) Immunocytochemistry images of 5F reprogrammed adult αMHC-GFP transgenic TTFs treated with DMSO or the indicated anti-inflammatory drugs for 7 d. (Green) αMHC-GFP; (red) cTnT; (blue) Hoechst. Bars, 500 µm. (C,D) Representative flow cytometry plot (C) and analyses (D) of αMHC-GFP+ and cTnT+ cells in 5F reprogrammed adult αMHC-GFP transgenic TTFs treated with DMSO or the indicated anti-inflammatory drugs for 7 d. (*) P < 0.05. (E) Model showing the mechanism of action of ZNF281 on 5F-mediated direct cardiac reprogramming. ZNF281 is a cardiac transcription coactivator recruited by GATA4 to cardiac enhancers to activate cardiac gene expression. ZNF281 also represses the inflammatory response, which acts as a barrier pathway to cardiac reprogramming.
Discussion
Here we performed an unbiased screen for regulators of adult cardiac reprogramming and identified 178 new activators and inhibitors that belong to various biological pathways. These different regulators revealed that anti-inflammatory and proinflammatory factors evoke opposing effects on cardiac reprogramming. We found that proinflammatory molecules prevented reprogramming, whereas anti-inflammatory drugs enhanced cardiac reprogramming. Among the identified activators, ZNF281 showed the most potent stimulatory activity. The effect of ZNF281 on cardiac reprogramming appears to be mediated by association with GATA4 on cardiac enhancers and by inhibition of inflammatory signaling, which antagonizes cardiac reprogramming (Fig. 7E). We conclude that ZNF281 acts at the nexus of the opposing transcriptional programs for cardiac and inflammatory gene expression.
Stimulation of cardiac reprogramming by ZNF281
The potential involvement of ZNF281 in cardiac gene regulation has not been recognized previously. Our results indicate that ZNF281 functions as a positive regulator of cardiogenesis by associating with GATA4 on cardiac enhancers, resulting in synergistic activation of a broad collection of cardiac genes, including those encoding cardiac transcription factors, calcium-handling proteins, cardiac metabolic enzymes, and components of the sarcomere. ZNF281 is expressed in a variety of tissues during various developmental stages, and its absence in mice results in embryonic lethality between embryonic days 7.5 and 8.5, prior to the formation of primitive ventricles and atria (Fidalgo et al. 2011; Xin et al. 2013). We speculate that ZNF281 may also have important functions in the early stage of cardiac development.
It has been suggested that reprogramming requires pioneer factors to first engage and open target sites in chromatin and confer competency for other factors to bind (Soufi et al. 2015). Previously, GATA4 was shown to serve as a pioneer factor for the albumin gene enhancer in liver precursor cells by initiating chromatin opening (Cirillo et al. 2002). Cardiac gene expression during heart development requires regulated interactions among the transcription factors GATA4, TBX5, and NKX2-5. The combined cooperativity of these factors is critical for the expansion of cardiac progenitors and the regulation of the cardiac differentiation program (Luna-Zurita et al. 2016). Moreover, it has been shown in human cardiomyocytes that GATA4 broadly co-occupies cardiac enhancers with TBX5 (Ang et al. 2016). This association is physiologically relevant because a human mutation in GATA4 that disrupts its association with TBX5 leads to aberrant chromatin states and impaired contractility, calcium handling, and metabolic activity (Ang et al. 2016).
We show that ZNF281 does not bind to cardiac enhancers in fibroblasts without the presence of GATA4, indicating that ZNF281 is unlikely to be a pioneer factor that opens chromatin. However, we show that ZNF281 requires the presence of the pioneer factor GATA4 to bind cardiac enhancers. Understanding the mechanism by which pioneer factors and other transcription factors interact and induce conformational changes in chromatin structure will be necessary to decipher the molecular mechanism involved in cardiac reprogramming.
The influence of inflammation and the NuRD complex on cardiac reprogramming
Following injury, inflammatory infiltration is among the earliest responses, which is necessary to clear cellular debris and promote scar formation (Aurora and Olson 2014). Recent studies revealed both positive and negative roles for the inflammatory response in cardiac repair and regeneration. Inhibition of C/EBP signaling in the adult epicardium reduces injury-induced neutrophil influx, leading to a reduction of fibrosis and maintenance of cardiac function (Huang et al. 2012). In contrast, the injury-induced macrophage response is necessary for heart regeneration in neonatal mice and zebrafish (Aurora et al. 2014; Lai et al. 2017). Our results show that the inflammatory response acts as a barrier for cardiac reprogramming, and ZNF281 appears to impose an anti-inflammatory influence on cardiac reprogramming.
The NuRD chromatin remodeling complex has been shown to repress proinflammatory gene expression in macrophages (Ramirez-Carrozzi et al. 2006). We show that in addition to direct cardiac gene activation, ZNF281 and some components of the NuRD complex also repress inflammatory signaling and activate cardiac reprogramming. Unlike cardiac reprogramming, ZNF281 has been shown to repress iPSC reprogramming through the NuRD complex (Fidalgo et al. 2011). Given that the inflammatory response is critical for iPSC formation (Lee et al. 2012), we surmise that in addition to repression of embryonic stem cell-specific genes, the anti-inflammatory effect of the ZNF281/NuRD complex also contributes to the repression of iPSC reprogramming. We cannot exclude the possibility that the NuRD complex has a more direct role in the activation of the cardiac gene program. In this regard, preservation of the identity of heart or skeletal muscle cell types has also been shown to depend on NuRD-mediated epigenetic repression of alternate lineage gene expression. Loss of the NuRD subunit Chd4 in the heart triggers aberrant expression of skeletal muscle gene expression and leads to hybrid striated muscle tissue (Gomez-Del Arco et al. 2016).
The opposing effects of inflammation on cardiac versus iPSC reprogramming suggest that these two types of reprogramming use distinct regulatory mechanisms. Given that embryonic stem cells are pluripotent and able to generate different cell types, whereas cardiomyocytes are terminally differentiated cells, it is possible that inflammation promotes epigenetic remodeling that is favorable for iPSC but not cardiac reprogramming.
Clinical implications
Direct reprogramming of adult fibroblasts to cardiomyocytes represents a potential approach for repair of the heart following injury. As an activator of cardiac reprogramming, ZNF281 offers new insights into the molecular basis of this process. Additionally, our finding that ZNF281 represses the inflammatory response reveals a role for anti-inflammatory signaling in cardiac reprogramming and highlights the potential clinical application of commonly used anti-inflammatory drugs in cardiac repair.
Materials and methods
Isolation of adult mouse fibroblasts
For isolation of adult mouse TTFs, tails were cut from 8- to 12-wk-old adult wild-type or αMHC-GFP mice and minced into 1-cm pieces with razor blades after peeling off the superficial dermis. The minced tails were placed in fibroblast growth medium (DMEM supplemented with 10% FBS and 1% [v/v] penicillin/streptomycin). TTFs migrated out from the explants within 1 wk and were passaged once before use. For the adult mouse CF isolation, hearts from 8- to 12-wk-old adult wild-type or αMHC-GFP mice were chopped with razor blades into small pieces. The chopped hearts were digested for 1 h at 37°C in a digestion medium containing Liberase TH and Liberase TM (Roche), DNase I (New England Biolabs), and polaxomer (Sigma). After digestion, cell suspensions were filtered through a 70-µm strainer and placed in fibroblast growth medium (DMEM supplemented with 10% FBS and 1% [v/v] penicillin/streptomycin). All animal experiments described in this study were approved and conducted under the oversight of the University of Texas Southwestern Institutional Animal Care and Use Committee.
Construction of the human retroviral ORF cDNA library
Gateway-compatible human ORF pEntry vectors were purchased from Thermo Fisher Scientific. Gateway-compatible retroviral destination vector pMXs-gw was a gift from Shinya Yamanaka (Addgene, plasmid no. 18656) (Takahashi and Yamanaka 2006). We transferred each ORF individually into pMXs-gw by performing site-specific LR recombination using Gateway LR Clonase II enzyme mix kit (Thermo Fisher Scientific).
Retrovirus production, cellular reprogramming, and treatment
For retroviruses production, Platinum E cells were seeded into culture dishes (1 × 105 cells per square centimeter) 1 d before transfection in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. Cells reached ∼80% confluency on the day of transfection. DNA plasmids were transfected into Platinum E cells using FuGENE 6 transfection reagent. Twenty-four hours after transfection, wild-type or αMHC-GFP TTFs were seeded into culture dishes or plates that were precoated with SureCoat (Cellutron) for 1 h at a density of 6 × 103 per square centimeter. Forty-eight hours after transfection, polybrene was added to viral medium that was filtered through a 0.45-µm filter at a concentration of 8 µg/mL. The mixture replaced the growth medium in the cell culture plate with TTFs. The viral infection was serially repeated twice. Twenty-four hours after the second infection, the viral medium was replaced with induction medium, composed of DMEM/199 (4:1), 10% FBS, 5% horse serum, 1% penicillin/streptomycin, 1% nonessential amino acids, 1% essential amino acids, 1% B-27, 1% insulin–selenium–transferrin, 1% vitamin mixture, and 1% sodium pyruvate (Invitrogen). Beating cell analysis was performed as described by Zhou et al. (2015). Small molecule treatments throughout the reprogramming process used 10 µM Dex, 10 µM Nab (Sigma), and 10 nM FK228 (Selleckchem).
Luciferase assay
Expression constructs (pBx-GATA4 and pMXs-ZNF281) and a reporter construct (p-αMHC-Luciferase) were transfected into HEK293T cells using Fugene HD (Promega) according to manufacturer's instructions. The total amount of DNA per well was kept constant by adding the corresponding amount of expression vector without a cDNA insert. Twenty hours after transfection, cell extracts were assayed for luciferase expression using the luciferase assay kit (Promega). Relative promoter activities were expressed as fluorescence relative units normalized to pBX-GFP expression in the cell.
Flow cytometry
For flow cytometry, cells were trypsinized and fixed with fixation buffer (BD Bioscience) for 15 min on ice. Fixed cells were washed three times with Perm/Wash buffer (BD Bioscience). Washed cells were incubated with mouse monoclonal anti-cTnT antibody (Thermo Scientific, MA5-12960) at 1:200 dilution and rabbit anti-GFP antibody (Thermo Scientific, A-11122) at 1:200 dilution in Perm/Wash buffer for 1 h on ice. Cells then were washed three times with Perm/Wash buffer followed by incubation with donkey anti-mouse Alexa fluor 647 (Invitrogen, A-31571) at 1:200 and goat anti-rabbit Alexa fluor 488 (Invitrogen, A-11008) at 1:200. Cells were washed with Perm/Wash buffer and then analyzed using FACS Caliber (BD Sciences) and FlowJo software.
Immunocytochemistry
For immunocytochemistry, cells were fixed in 4% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 at room temperature. Cells were washed three times with PBS followed by blocking with 10% goat serum for 1 h. Cells then were incubated with mouse monoclonal anti-cTnT antibody (Thermo Scientific, MA5-12960) at 1:500 dilutions and rabbit anti-GFP antibody (Thermo Scientific, A-11122) at 1:500 dilutions in 5% goat serum for 1 h. After three washes with PBS, cells then were incubated with donkey anti-mouse Alexa fluor 647 (Invitrogen, A31571) at 1:500 and goat anti-rabbit Alexa fluor 488 (Invitrogen, A-11008) at 1:500. Image acquisition was done on an INCell Analyzer 6000 (GE, Inc.) using a Nikon 4× objective. Samples were imaged using 488 and 561 laser lines for αMHC-GFP and cTnT, respectively. Four fields were used to capture the entire well. Image processing was performed in GE Developer Toolbox (version 1.9.3). The acquired images were segmented on the αMHC-GFP and cTnT channels separately. We then calculated the total area (square micrometers) and total signal intensity (arbitrary units) of each probe within their respective masks. Results for both channels were imported into the GeneData Analyzer module (version 13, GeneData, Inc.) and normalized against the robust median of the test population. From the normalized data, Z-scores were calculated as described previously (Wu et al. 2008).
Quantitative mRNA measurement
Total RNA was extracted using TRIzol (Invitrogen) according to the vender's protocol. RNAs were retrotranscribed to cDNA using iScript Supermix (Bio-Rad). qPCR was performed using KAPA SYBR Fast (Kapa Biosystems). mRNA levels were normalized by comparison with Gapdh mRNA. Primer sequences are listed in Supplemental Table S3.
Immunoprecipitation and Western blot analysis
Flag-epitope tag ZNF281 fusion proteins were coexpressed with myc-epitope tag fusion GATA4, HAND2, MEF2C, or TBX5 in HEK293 cells for 48 h. Cell lysates were incubated overnight with 1 µg of mouse monoclonal anti-Flag antibody (Sigma, F3165). The cell lysates were pulled down using magnetic Protein G Dynabeads (Invitrogen), and then the Flag-ZNF281 was eluted using 0.5 mg/mL free Flag peptide (Sigma). The final elution and the input obtained before the immunoprecipitation were analyzed by SDS-PAGE Western blot using a mouse monoclonal anti-Myc antibody (Novex, R950-25) or mouse monoclonal anti-Flag antibody (Sigma, F3165).
RNA-seq and GO analysis
For RNA-seq, total RNA was extracted from TTFs transduced with the indicated retroviruses using TRIzol (Invitrogen) according to the vender's protocol. Illumina RNA-seq was performed by the University of Texas Southwestern Microarray Core Facility. RNA-seq and transcriptome analysis were performed as described in Abad et al. (2017). Briefly, quality assessment of the RNA-seq data was done using NGS-QC-Toolkit. Reads with >30% nucleotides with Phred quality scores <20 were removed from further analysis. Quality-filtered reads were then aligned to the mouse reference genome GRCm38 (mm10) using the Hisat (version 2.0.0) aligner using default settings. Aligned reads were counted using featureCounts (version 1.4.6) per gene ID. Differential gene expression analysis was done using the R package edgeR (version 3.8.6). For each comparison, genes were required to have 10 cpm (counts per million) in at least three samples to be considered as expressed. They were used for normalization factor calculation. Gene differential expression analysis was done using the GLM approach following edgeR official documentation. Cutoff values of fold change >2 and FDR <0.01 were then used to select for differentially expressed genes between sample group comparisons. The DAVID gene functional annotation and classification tool was used to annotate the list of differentially expressed genes. GO enrichment analysis was performed to determine molecular and biological functional categories. The enrichment plots were performed using GSEA software. All RNA-seq data have been deposited in the Sequence Read Archive (SRA) with the accession numbers listed in Supplemental Table S4.
ChIP-seq, GO, and pathway analysis
For ChIP-seq, TTFs were transduced with the indicated retroviruses. Two days after retroviral transduction, TTFs were cross-linked with 1% formaldehyde in PBS for 30 min and neutralized by the addition of glycine to a final concentration of 0.125 M for 5 min. TTFs were harvested and washed with cold PBS, and then ChIP was performed using anti-GATA4 antibody (Santa Cruz Biotechnology, sc-1237), anti-ZNF281 antibody (Abcam, ab101218), and ChIP-IT Express ChIP kits (Active Motif) according to the vender's protocol. Subsequent library construction and massive parallel sequencing were performed at the University of Texas Southwestern Genomics and Microarray Core Facility. For ChIP-seq analysis, raw reads were mapped to GRCm38 (mm10) using Bowtie2 (version 2.2.8). An average of ∼50 million uniquely mapped (single end) reads was obtained per sample. Peak calling was performed using MACS2 (version 2.1.0). Resulting peak files were processed using bedTools (version 2.26) and deepTools (Ramirez et al. 2016) to generate coverage heat maps and obtain overlapping regions. The GREAT kits (McLean et al. 2010) were used for GO and pathway analysis. All ChIP-seq data have been deposited in the SRA with the accession numbers listed in Supplemental Table S4.
Quantification and statistical analysis
All data are presented as mean with SEM and have n = 3 per group (except Supplemental Fig. S4, with n = 6 per group). P-values were calculated with either unpaired/two-way t-test or one-way ANOVA. Statistical analyses were run using the GraphPad Prism 7 software package. P < 0.05 was considered significant in all cases after corrections were made for multiple pairwise comparisons.
Supplementary Material
Acknowledgments
We thank Jose Cabrera for assistance with graphics. This work was supported by grants from the National Institutes of Health (NIH; HL130253, HL-077439, DK-099653, and AR-067294), the Foundation Leducq Networks of Excellence (14CVD04 to E.N.O.), the Cancer Prevention and Research Institute of Texas (RP150596 to M.S.K.), and the Robert A. Welch Foundation (grant 1-0025 to E.N.O.). H.H. is supported by a Uehara Memorial Foundation Post-doctoral Fellowship and a Kanae Foreign Study Grant. M.G.M. is a Pew Latin American Fellow in the Biomedical Sciences, supported by the Pew Charitable Trusts. H.Z. was supported by a predoctoral fellowship (14PRE20030030) from the American Heart Association. We are also grateful for the support provided to the High-Throughput Screening Core Facility (B.A.P. and H.N.) by the Simmons Cancer Center Support Grant (P30 CA142543-06) and the NIH-sponsored S10 grant (1S10OD018005-01 to B.A.P.) awarded for the INCell Analyzer 6000.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.305482.117.
References
- Abad M, Hashimoto H, Zhou H, Morales MG, Chen B, Bassel-Duby R, Olson EN. 2017. Notch inhibition enhances cardiac reprogramming by increasing MEF2C transcriptional activity. Stem Cell Reports 8: 548–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang YS, Rivas RN, Ribeiro AJ, Srivas R, Rivera J, Stone NR, Pratt K, Mohamed TM, Fu JD, Spencer CI, et al. 2016. Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell 167: 1734–1749.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurora AB, Olson EN. 2014. Immune modulation of stem cells and regeneration. Cell Stem Cell 15: 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurora AB, Porrello ER, Tan W, Mahmoud AI, Hill JA, Bassel-Duby R, Sadek HA, Olson EN. 2014. Macrophages are required for neonatal heart regeneration. J Clin Invest 124: 1382–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill TJ, Choudhury RP, Riley PR. 2017. Heart regeneration and repair after myocardial infarction: translational opportunities for novel therapeutics. Nat Rev Drug Discov 10.1038/nrd.2017.106. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9: 279–289. [DOI] [PubMed] [Google Scholar]
- Fidalgo M, Shekar PC, Ang YS, Fujiwara Y, Orkin SH, Wang J. 2011. Zfp281 functions as a transcriptional repressor for pluripotency of mouse embryonic stem cells. Stem Cells 29: 1705–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo M, Faiola F, Pereira CF, Ding J, Saunders A, Gingold J, Schaniel C, Lemischka IR, Silva JC, Wang J. 2012. Zfp281 mediates Nanog autorepression through recruitment of the NuRD complex and inhibits somatic cell reprogramming. Proc Natl Acad Sci 109: 16202–16207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo M, Huang X, Guallar D, Sanchez-Priego C, Valdes VJ, Saunders A, Ding J, Wu WS, Clavel C, Wang J. 2016. Zfp281 coordinates opposing functions of Tet1 and Tet2 in pluripotent states. Cell Stem Cell 19: 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli BA, Milliar H, Toolan KP, Harkin J, Fuchs RK, Robling AG, Firulli AB. 2017. Defective Hand1 phosphoregulation uncovers essential roles for Hand1 in limb morphogenesis. Development 144: 2480–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu JD, Stone NR, Liu L, Spencer CI, Qian L, Hayashi Y, Delgado-Olguin P, Ding S, Bruneau BG, Srivastava D. 2013. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Reports 1: 235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Del Arco P, Perdiguero E, Yunes-Leites PS, Acin-Perez R, Zeini M, Garcia-Gomez A, Sreenivasan K, Jimenez-Alcazar M, Segales J, Lopez-Maderuelo D, et al. 2016. The chromatin remodeling complex Chd4/NuRD controls striated muscle identity and metabolic homeostasis. Cell Metab 23: 881–892. [DOI] [PubMed] [Google Scholar]
- Hahn S, Hermeking H. 2014. ZNF281/ZBP-99: a new player in epithelial–mesenchymal transition, stemness, and cancer. J Mol Med 92: 571–581. [DOI] [PubMed] [Google Scholar]
- Han Z, Yi P, Li X, Olson EN. 2006. Hand, an evolutionarily conserved bHLH transcription factor required for Drosophila cardiogenesis and hematopoiesis. Development 133: 1175–1182. [DOI] [PubMed] [Google Scholar]
- Huang GN, Thatcher JE, McAnally J, Kong Y, Qi X, Tan W, DiMaio JM, Amatruda JF, Gerard RD, Hill JA, et al. 2012. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science 338: 1599–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. 2010. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifkovits JL, Addis RC, Epstein JA, Gearhart JD. 2014. Inhibition of TGFβ signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS One 9: e89678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Ieda M. 2017. Discovery and progress of direct cardiac reprogramming. Cell Mol Life Sci 74: 2203–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Marin-Juez R, Moura PL, Kuenne C, Lai JKH, Tsedeke AT, Guenther S, Looso M, Stainier DY. 2017. Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration. Elife 6: e25605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Sayed N, Hunter A, Au KF, Wong WH, Mocarski ES, Pera RR, Yakubov E, Cooke JP. 2012. Activation of innate immunity is required for efficient nuclear reprogramming. Cell 151: 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Zurita L, Stirnimann CU, Glatt S, Kaynak BL, Thomas S, Baudin F, Samee MA, He D, Small EM, Mileikovsky M, et al. 2016. Complex interdependence regulates heterotypic transcription factor distribution and coordinates cardiogenesis. Cell 164: 999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed TM, Stone NR, Berry EC, Radzinsky E, Huang Y, Pratt K, Ang YS, Yu P, Wang H, Tang S, et al. 2017. Chemical enhancement of in vitro and in vivo direct cardiac reprogramming. Circulation 135: 978–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse ENCODE Consortium, Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, et al. 2012. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 13: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka N, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Isomi M, Nakashima H, Akiyama M, Wada R, Inagawa K, et al. 2014. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J 33: 1565–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R, et al. 2013. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci 110: 5588–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. 2012. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485: 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. 2006. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes Dev 20: 282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, et al. 2012. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485: 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. 2015. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161: 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D, DeWitt N. 2016. In vivo cellular reprogramming: the next generation. Cell 166: 1386–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. 1997. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet 16: 154–160. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Molkentin JD. 2017. Redefining the identity of cardiac fibroblasts. Nat Rev Cardiol 14: 484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseghi H, Liu J, Qian L. 2017. Molecular barriers to direct cardiac reprogramming. Protein Cell 10.1007/s13238-017-0402-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu Z, Yin C, Asfour H, Chen O, Li Y, Bursac N, Liu J, Qian L. 2015. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ Res 116: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Liu D, Sui Y. 2008. Quantitative assessment of hit detection and confirmation in single and duplicate high-throughput screenings. J Biomol Screen 13: 159–167. [DOI] [PubMed] [Google Scholar]
- Xin M, Olson EN, Bassel-Duby R. 2013. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol 14: 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa H, Muraoka N, Miyamoto K, Sadahiro T, Isomi M, Haginiwa S, Kojima H, Umei T, Akiyama M, Kuishi Y, et al. 2015. Fibroblast growth factors and vascular endothelial growth factor promote cardiac reprogramming under defined conditions. Stem Cell Reports 5: 1128–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SY, Baxter EM, Van Doren M. 2012. Phf7 controls male sex determination in the Drosophila germline. Dev Cell 22: 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Londono P, Cao Y, Sharpe EJ, Proenza C, O'Rourke R, Jones KL, Jeong MY, Walker LA, Buttrick PM, et al. 2015. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat Commun 6: 8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Dickson ME, Kim MS, Bassel-Duby R, Olson EN. 2015. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc Natl Acad Sci 112: 11864–11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wang L, Vaseghi HR, Liu Z, Lu R, Alimohamadi S, Yin C, Fu JD, Wang GG, Liu J, et al. 2016. Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell 18: 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.