Genes & Development 30: 2286–2296 (2016)
We have realized that there is an error in Figure 4 of the above-mentioned article. By mistake, the images in Figure 4, A and D, originate from the same sample. This image is from the untreated control experiment (Fig. 4A) but was mistakenly also used in Figure 4D (2,4-D treatment). We have built a revised Figure 4 in which we corrected this error.
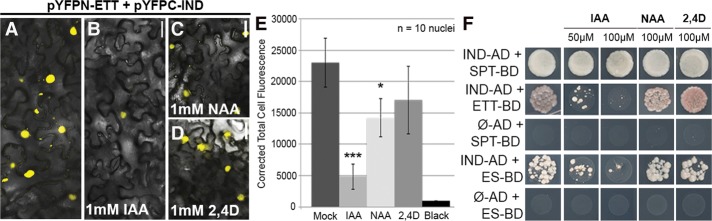
Figure 4.
ETT and IND protein interact in an IAA-sensitive manner. (A–D) BiFC with pYFPN-ETT and pYFPC-IND in the absence (A) and presence of IAA (B), NAA (C), and 2,4-D (D). All hormonal treatments were with 1 mM in lanolin. (E) Fluorescence quantification (CTCF) of split YFP signal between ETT and IND without treatment and with IAA, NAA, and 2,4-D. (*) P < 0.01; (***) P < 0.0001. Error bars show the standard deviation. (F) Y2H assays with increasing concentration of IAA, NAA, and 2,4D. Bars: A–D, 50 μm.
Moreover, we have added a raw data file as an Additional Supplemental Material link associated with the original article with the raw images of three independent experiments that were used for the quantification in Figure 4E (Figure_4_raw_data.pdf). For each condition, we have included a picture taken with a low magnification (a 10× objective). In addition, we have included four more images at higher magnification (a 20× objective) and have repeated the intensity quantification using the nuclei that are labeled with a number. The values are summarized below the images, and a new chart has been built with these data, including a new ANOVA analysis. These data show the same result as in the original Figure 4.
The authors apologize for this error.
doi: 10.1101/gad.306985.117