Abstract
Neurofibromatosis type I, a genetic disorder due to mutations in the NF1 gene, is characterized by a high mutation rate (about 50% of the cases are de novo) but, with the exception of whole gene deletions associated with a more severe phenotype, no specific hotspots and few solid genotype/phenotype correlations. After retrospectively re-evaluating all NF1 gene variants found in the diagnostic activity, we studied 108 patients affected by neurofibromatosis type I who harbored mutations that had not been previously reported in the international databases, with the aim of analyzing their type and distribution along the gene and of correlating them with the phenotypic features of the affected patients. Out of the 108 previously unreported variants, 14 were inherited by one of the affected parents and 94 were de novo. Twenty-nine (26.9%) mutations were of uncertain significance, whereas 79 (73.2%) were predicted as pathogenic or probably pathogenic. No differential distribution in the exons or in the protein domains was observed and no statistically significant genotype/phenotype correlation was found, confirming previous evidences.
Keywords: neurofibromatosis type I, NF1 gene, genotype/phenotype correlation
1. Introduction
The rapid evolution of the high-throughput sequencing technologies presents significant challenges in the acquisition, analysis and interpretation of large data sets. In particular, the detection of several novel alleles in multiple-gene panels or clinical exomes brings about difficulties in the understanding of the significance of the variants found and finally in the interpretation and clinical use of the genetic result in settings like pre-symptomatic or prenatal diagnosis [1,2]. Disease-specific databases, knowledge of the allele frequencies and the type of nucleotide change help to sort single variants into benign or pathogenic, but some of them remain uncertain about their clinical significance [3], especially when they are found in genes causing very rare diseases or those with a reduced penetrance or with a high mutation rate [4]. The approach to the interpretation of single variants used for genes such as NF1 or BRCA1 [1,5], based on a distribution into classes of growing evidence of pathogenicity (five, at present), has then been translated to the interpretation of the results of clinical exomes or large gene-panels.
In addition to providing a strategy for the clinical use of new gene variants, NF1 (MIM*613113) is a model gene for the high rate of new alleles (about 50% of the total burden of mutations [6]) and for their fully penetrant mendelian behavior, which makes them detectable in specific phenotypes belonging to neurofibromatosis type I (NF1) (MIM#162200) (various combinations of multiple café-au-lait spots, axillary and inguinal freckling, multiple neurofibromas, and iris Lisch nodules) [7]. Understanding the features of the NF1 novel variants can shed light on the patterns of genome variability, natural selection, and evolution.
Moreover, the clinical phenotypic expression characterized by marked intra- and inter-familial variability with multisystemic complications including neurological, cardiovascular, gastrointestinal, endocrine, neoplastic, and orthopedic features [8] has long puzzled physicians and researchers in the attempt to predict which genotypes would harbor the highest risk of complications. Very few, however, are the established genotype–phenotype correlations, whose best example is represented by the NF1 extended deletions, which have been linked to a more severe phenotype than point mutations [9,10,11]. Finally, no evidence exists demonstrating that mutations cluster in specific gene regions or protein domains of which the NF1 protein, neurofibromin, is composed, including a cysteine–serine-rich domain (CSRD), a tubulin-binding domain (TBD), a central GTPase-activating protein-related (GRD), a SEC14 domain, a syndecan binding 1 (SH1) domain, a pleckstrin homology (PH) domain, a syndecan binding 2 (SH2) domain, and a carboxy-terminal domain (CTD) [12].
In the present study, we retrospectively re-evaluated all NF1 gene variants found in 17 years of diagnostic activity and selected all the mutations not reported in the international databases or in the medical literature. Those latter were stratified according to the five pathogenetic classes, analyzed for their type and their distribution in the exons of the NF1 gene and in the domains of the relative protein. Finally, after dividing the phenotypic features of the disease into cardinal signs and complications, a genotype/phenotype correlation was attempted according to the type and site of the variant found.
2. Results
Out of the 502 patients with a clinical diagnosis of neurofibromatosis type I, 108 harbored mutations that had never been previously reported in the international disease databases (Table S1). Among those, 45 were single-base substitutions (26 missense, 10 truncating, 6 splice-site, and 3 synonymous), 44 small deletions, (41 truncating, 2 in-frame, and 1 splice-site), 12 duplications (all producing truncating protein), 5 indels (4 truncating and 1 in-frame), 1 insertion (truncating), and 1 large deletion spanning Exons 2–19 (Table 1).
Table 1.
Classification of the previously undescribed NF1 mutations.
| Type of Mutation, n | Missense (%) | Truncating (%) | Splice (%) | Silent (%) | In-Frame (%) |
|---|---|---|---|---|---|
| Single Nucleotide Variants, 45 | 26 (57.8) | 10 (22.2) | 6 (13.3) | 3 (6.7) | 0 (0) |
| Small Deletions, 44 | 0 (0) | 41 (93.2) | 1 (2.3) | 0 (0) | 2 (4.5) |
| Small Duplications, 12 | 0 (0) | 12 (100) | 0 (0) | 0 (0) | 0 (0) |
| Indels, 5 | 0 (0) | 4 (80) | 0 (0) | 0 (0) | 1 (20) |
| Insertions, 1 | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) |
| Large Deletions, 1 | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) |
| Total, 108 | 26 (24) | 69 (63.9) | 7 (6.5) | 3 (2.8) | 3 (2.8) |
Truncating variants are the most frequently encountered alterations in our population (69/108; 63.9%), followed by missense (26/108; 24%), splice-site (7/108; 6.5%), synonymous (3/108; 2.8%), and in-frame variants (3/108; 2.8%) (Table 1). Out of the 108 previously undescribed NF1 alleles, 14 were familial (inherited by one of the two parents presenting signs and symptoms of NF1) and 94 de novo. Twenty-nine (26.9%) variants were of unknown significance (25 missense, 3 synonymous, and 1 in-frame), 16 (14.8%) were likely pathogenic (7 splice-site, 8 truncating, and 1 missense) and 63 (58.3%) variants were pathogenic (61 truncating and 2 in-frame) (Table 2).
Table 2.
Classification of the NF1 mutations according with pathogenicity score.
| Mutation Type, n (%) | Pathogenity Score, n (%) | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Missense, 26 (24) | 0 (0) | 0 (0) | 25 (96.1) | 1 (3.9) | 0 (0) |
| Truncating, 69 (63.9) | 0 (0) | 0 (0) | 0 (0) | 8 (11.6) | 61 (88.4) |
| Splicing, 7 (6.5) | 0 (0) | 0 (0) | 0 (0) | 7 (100) | 0 (0) |
| Silent, 3 (2.8) | 0 (0) | 0 (0) | 3 (100) | 0 (0) | 0 (0) |
| In-frame, 3 (2.8) | 0 (0) | 0 (0) | 1 (33.3) | 0 (0) | 2 (66.7) |
| Total, 108 | 0 (0) | 0 (0) | 29 (26.9) | 16 (14.8) | 63 (58.3) |
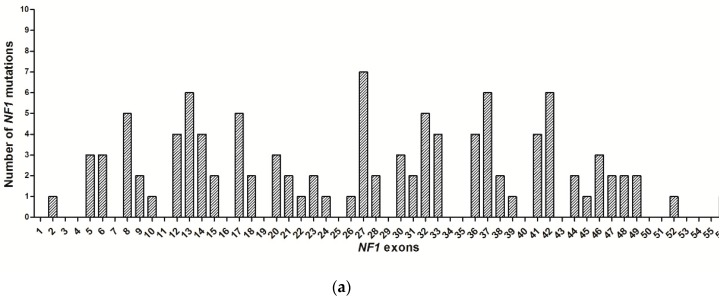
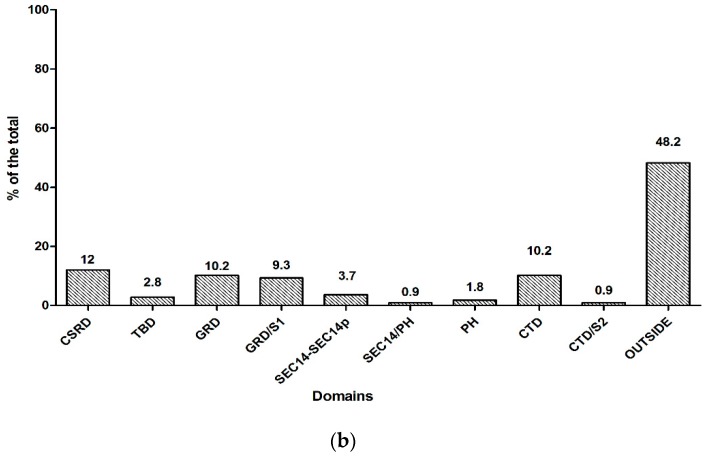
The distribution of the previously unreported variants in the NF1 gene showed that no mutational hotspots exist in the gene and that the most frequently hit exons (including the flanking introns, +/−2 bp) were Exons 13, 27, 37 (7 mutations), 8, 32, 42 (6 mutations), and 32 (5 mutations) (Figure 1a). Five out the 108 mutations (4.6%) were located within CpG dinucleotides. When the functional domains of the protein neurofibromin 1 were considered, 13 (12%) mutations were identified in the CSRD domain, 3 (2.8%) in TBD, 11 mutations (10.2%) in GRD, 10 (9.3%) in GRD/S1, 4 (3.7%) in SEC14-SEC14P, 1 (0.9%) in SEC14/PH, 2 (1.8%) in PH, 11 (10.2%) in CTD, and 1 (0.9) in CTD/S2. Furthermore, 52 (48.2%) mutations were localized outside the neurofibromin domains (Figure 1b).
Figure 1.
(a) Distribution of the NF1 variants by exons. (b) Distribution of the variants in neurofibromin domains. CSRD: cysteine–serine-rich domain; TBD: tubulin-binding domain; GRD: GTPase-activating protein-related domain; S1: syndecan binding domain 1; PH: pleckstrin homology domain; CTD: carboxy-terminal domain; S2: syndecan binding domain 2; SEC14/ SEC14p: Sec14-like lipid binding module.
Twenty-nine variants (26.9% of the total) were classified as variants of unknown significance (VUS, Score 3), 16 (14.8%) as likely pathogenetic (Score 4) and 63 (58.3%) as pathogenetic (Score 5). De novo variants were assigned to Classes 4 and 5 in 73.4% (69 out of 94) of cases, whereas the familial ones were attributed to Class 4 or Class 5 in 71.4% (10 out of 14) of cases, χ-square = 0.004, p = 1) (Table 2 and Table S2).
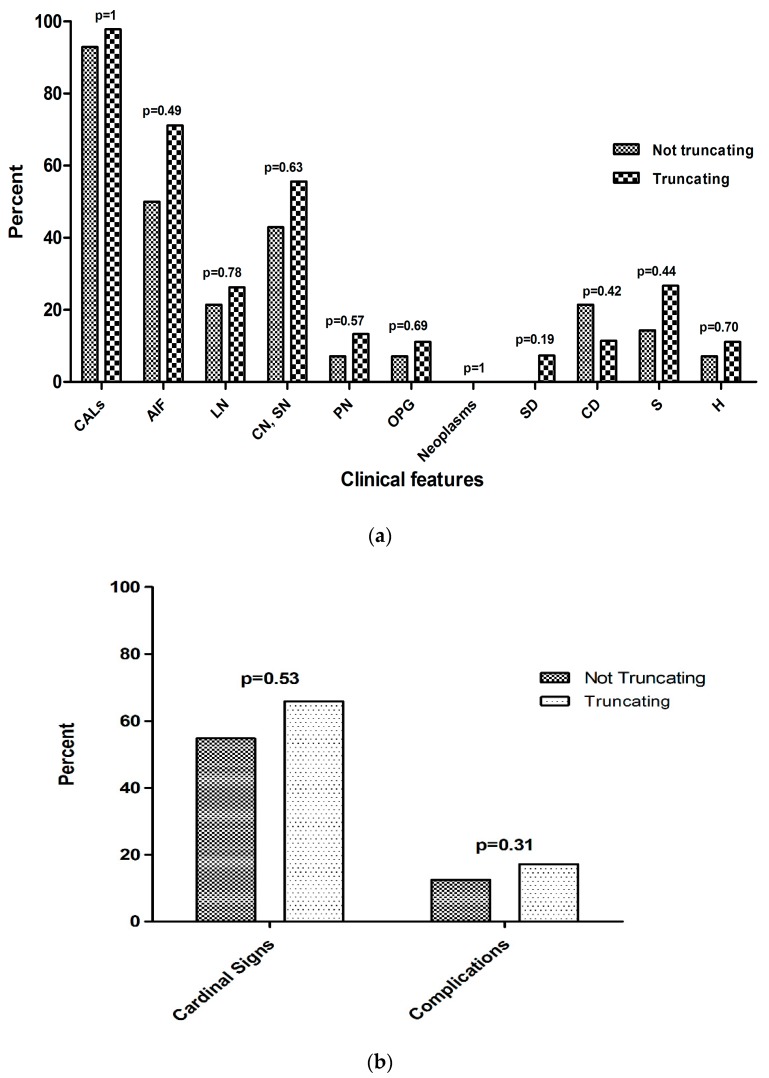
As far as the phenotypic features were concerned (details of each patient are reported in Table S2), a higher proportion of specific NF1 features was found in patients with truncating mutations, compared to patients with non-truncating ones (the 7 splice-site variants were excluded from this analysis due the lack of functional data about their effect on the protein), but none of the differences found reached statistical significance (cafe-au-lait patches (CALs): 97.8% vs. 92.9% for truncating mutations vs. non-truncating mutations, respectively, p = 1; axillary or groin freckling (AIF): 71.1% vs. 50%, p = 0.49; Lisch nodules (LN): 26.2% vs. 21.4%, p = 0.78; cutaneous/subcutaneous neurofibromas (CN/SN): 55.6% vs. 42.9%, p = 0.63; plexiform neurofibromas (PN): 13.3% vs. 7.1%, p = 0.57; optic pathway glioma (OPG): 11.1% vs. 7.1%, p = 0.69; neoplasms: 0% vs. 0%, p = 1; sphenoid wing dysplasia (SD): 7.3% vs. 0%, p = 0.19; cognitive deficit (CD): 11.4% vs. 21.4%, p = 0.42; scoliosis (S): 26.7% vs. 14.3%, p = 0.44, hypertension (H): 11.1% vs. 7.1%, p = 0.70) (Figure 2a). After the phenotypic features were grouped into cardinal signs (CALs, AIF, and LN) and complications (all the other signs), no significant differences were found between patients with truncating mutations vs. those with non-truncating mutations (17.2% vs. 12.5%, p = 0.31) (Figure 2b). The average paternal age in de novo mutations (36 years and 4 months) was higher than that in familial mutations (35 years), but, again, no statistically significant differences were found.
Figure 2.
(a) Clinical features and (b) aggregation of cardinal signs and complications in patients with truncating and non-truncating mutations. CALs: café-au-lait patches; AIF: axillary or groin freckling; LN: Lisch nodules; CN/SN: cutaneous/subcutaneous; PN: plexiform neurofibromas; OPG: optic pathway glioma; SD: sphenoid wing dysplasia; CD: cognitive deficit; S: scoliosis; H: hypertension.
3. Discussion
Neurofibromatosis type I is a model genetic disorder due to the high mutation rate of its causative gene, NF1 [6]. However, despite the long-standing knowledge of the disease, studies of the genotype/phenotype correlation have failed to find clear associations with specific gene variations [13,14], probably due to the tumor suppressor mechanism of the NF1 gene, which acts in a recessive manner and requires a second random somatic, occult mutational event for its expression [15,16]. For these reasons, the majority of studies have yielded negative results [13,14,17], and very few have reported mutation-specific correlations: a 3 bp in-frame deletion, c.2970_2972delAAT, has been related to the absence of cutaneous neurofibromas [18,19], and missense mutations affecting codon 1809 (Arg1809Cys) have been associated with Noonan-like dysmorphisms and the absence of neurofibromas [20,21]. More general observations have indicated a poorer cognitive prognosis and a higher risk of tumors, neurofibromas, and MPNSTs for the whole NF1 gene deletions [9,10,11]; other reports have hypothesized an association of splice-site mutations with an increased tendency to develop neoplasms [22], of truncating mutations with the presence of Lisch nodules and CALs [23,24] and of non-truncating mutations with pulmonary stenosis [25]. The results of our study show that, starting from a population of affected patients, there are no hotspots for mutations in the NF1 gene, nor is there any preferential involvement of specific protein domains. We did not observe any significant correlation between the type of mutation and the phenotypic features, both taken individually (Figure 2a) and grouped together (Figure 2b). Moreover, no malignancies were registered (Figure 2a), possibly due to the young age of the population tested (median age 25 years and 5 months). In conclusion, our data confirm the majority of the previous studies about the weak predictive potential in clinical terms of the mutation found, which cannot be applied in individual cases, especially in predictive counselling settings like the prenatal diagnosis or when formulating the disease prognosis of an affected child.
Another focus of our study was to highlight potential factors involved in the high mutation rate in the NF1 gene, for which two main risk factors have been advocated: the paternal age of the affected [26] and the genomic context [27], which are the common determinants of the genetic variability at the nucleotide level [28]. Considering our population, the paternal age at birth was slightly higher in de novo mutations (mean age 36 years and 4 months) than in familial ones (mean 35 age), although no statistically significant association was found, also probably due to a type 2 statistical error for the limited number of observations in the familial group (14 cases). The context, i.e., the identity of the nucleotides surrounding the mutation, had clearly no impact on the mutational load, since only 5 of 108 fell into the CpG dinucleotides, the most mutable sequences in humans [27]. Regardless of the cause, the high variability of the gene is evident when querying the ClinVar database [3] for all known NF1 exonic variants (including splice-site +/− 2 bp): the reported mutations occur, on average, in one out of every eight nucleotides—mostly missense (74%), followed by truncating (20%), and splice-site (6%). They present an inverted ratio compared to our NF1 cohort, in which the most common mutation group is represented by the truncating (63.9%) variations, followed by missense (24%) and splice-site (6.5%) variations. This difference is reflected in the clinical significance of the variant found (for truncating mutations, it is easier to foresee a causal role for the disease), which in the ClinVar database was in 44% of the cases considered as VUS, a ratio that in our cohort was 26.9% (29/108), a lower but still relevant percentage of tests with inconclusive results about pathogenesis.
In conclusion, the present genotype/phenotype correlation study based on 108 previously undescribed NF1 variants failed to show any gene hotspot for mutation and any significant association of the clinical presentation of the disease with the type of gene mutation found. In at least one fourth of the cases, the clinical significance of the variant found at the genetic testing remains uncertain, with important reflections on the genetic counseling of the patients and their families.
4. Materials and Methods
4.1. Patients
The analysis is based on the records of Parma University Hospital’s Unit of Medical Genetics, covering the years 2000 to 2017. The laboratory functions as a hub for the entire Emilia Romagna Region (4.5 million inhabitants) and attracts patients from other Italian regions as well. Genetic testing was performed on patients having a clinical suspicion of neurofibromatosis type I based on the presence of at least two of the clinical manifestations proposed by the National Institute of Health Consensus Development Conference, i.e., the presence of six or more CALs >15 mm in adults and >5 mm in children, two or more CN/SN or PN, AIF, LN, OPG, MPNST, PCC, CD, and distinctive osseous lesions such as SD or S [29]. Each sample given to our laboratory was accompanied by a clinical chart in which a clinical description of the patient was reported. Patient’s clinical records, genealogical trees, and genetic test results were all collected and archived in a specifically dedicated Excel file. In the 2000–2017 period, 502 subjects with a clinical suspicion of neurofibromatosis type I were subjected to genetic testing as part of the diagnostic process.
4.2. NF1 Genetic Test
From April 2000 to June 2016 (397 out of the total 502 patients, 79.1%), genetic analyses on genomic DNA were conducted using denaturing high pressure liquid chromatography (DHPLC), which exploits the differential retention of homoduplex and heteroduplex DNA under conditions of partial thermal denaturation (reported detection rate for NF1 mutations: 72% [30]). Primer pairs were designed according to the published reference genomic NF1 sequence (GenBank accession number NG_009018.1). To characterize the samples with a profile different from a wild-type control, direct sequencing of the fragments was performed using M13 Universal primers and a GenomeLab DTCS Quick Start kit for Dye Cycle Sequencing on a CEQ 2000XL Sequence Analyzer (Beckman Coulter, CA, USA). Starting from June 2016 (105 out of the total 502 patients, 20.9%), the genetic analyses were performed via next-generation sequencing using the Illumina MiSeq (TruSeq Custom Amplicon v.1.5, San Diego, CA, USA) platform (reported detection rate for NF1 mutations: 88% [15]). For the description and nomenclature of sequence variations at the DNA and protein level, the Mutalyzer software version 2.0.22 [31] was used with the NM_000267.3 reference sequence. Multiplex ligation-dependent probe amplification (MLPA, MRC Holland Amsterdam, The Netherlands) as been used for the detection of deletions or duplications spanning one or more exons in those cases negative at the sequencing analysis. For predicting the pathogenicity of the missense mutations, the REVEL (Rare Exome Variant Ensemble Learner) tool was used, an Ensemble method based on the 13 most commonly used prediction software [32].
4.3. The Database
After the consultation of the dedicated international databases (Clinvar [3]; The Human Gene Mutation Database, HGMD [33]; Leiden Open Variation Database 3.0, LOVD [34]), the previously undescribed NF1 variants together with the genetic information were grouped and prepared in an Excel file. In particular, records have included, for each variant, the type, effect, aminoacid change, exon involved, protein domain, segregation (de novo/familial), and population frequency according to Ensembl [35] and ExAC [36]. All the information has been submitted to the ClinVar [3] gene variant database (Accession number: SUB2798650). Moreover, the main clinical features of the patients with previously undetected mutations were recorded according to the Human Phenotype Ontology (HPO) [37] (Table 3). Finally, the classical 5-tiered system, which classifies the variants on the basis of standard criteria [1] as 1 (benign), 2 (likely benign), 3 (uncertain significance), 4 (likely pathogenic), and 5 (pathogenic) was adopted.
Table 3.
Clinical features of patients with novel NF1 mutation.
| Clinical Features (HPO Code) | Number of Patients with Known Status | Number of Patients with Positive Status (%) |
|---|---|---|
| Café-au-lait spots (0000957) | 59 | 57/59 (96.6) |
| Axillary or inguinal freckles (0000997, 0030052) | 59 | 39/59 (66.1) |
| Lisch nodules (0009737) | 56 | 14/56 (25) |
| Cutaneous, subcutaneous neurofibromas (0100698, 0100698) | 59 | 31/59 (52.5) |
| Plexiform neurofibromas (0009732) | 59 | 7/59 (11.9) |
| Optic pathway glioma (0009734) | 59 | 6/59 (10.2) |
| Neoplasms (0002664) | 59 | 0/59 (0) |
| Sphenoid-shank dysplasia (0000924) | 55 | 3/55 (5.4) |
| Cognitive defects (0100543) | 58 | 8/58 (13.8) |
| Scoliosis (0002650) | 59 | 14/59 (23.7) |
| Hypertension (0000822) | 59 | 6/59 (10.2) |
The statistical analysis was performed using the SPSS software (version 10.0, SPSS Corp, Chicago, IL, USA), converting descriptive variables to numerical values and using the chi-square test to evaluate differences in mutation distribution among exons and protein domains of neurofibromin and for genotype–phenotype comparisons between groups. Statistical results were corrected for multiple testing using Holm’s test [38].
The local Institutional Review Board approval was requested and obtained for this study (Prot. N. 0051/2017).
Acknowledgments
This study was supported by “Fondazione Emma ed Ernesto Rulfo per la Genetica Medica” and by the Regione Emilia Romagna, “RARER”, Area 1 (E35E09000880002). Simonetta Rosato, Enrico Grosso, Andrea Zonta, Marcella Neri, Antonella Maffè, Mattia Gentile, Luca Rocchetti, Giovanni Ponti, Edoardo Caleffi are acknowledged for their clinical contribution to the study. Matteo Goldoni is acknowledged for his critical revision of the manuscript.
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/18/10/2071/s1.
Author Contributions
Antonio Percesepe and Francesco Bonatti conceived and designed the study; Davide Martorana, Alessia Adorni, and Annalisa Matichecchia performed the experiments; Francesco Bonatti, Alessia Adorni, Annalisa Matichecchia, Paola Mozzoni, and Davide Martorana analyzed the data; Vera Uliana, Francesco Pisani, Livia Garavelli, Claudio Graziano, Maria Gnoli, Diana Carli, Stefania Bigoni, Elena Boschi performed the phenotypic characterization of patients; Francesco Bonatti and Antonio Percesepe wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacArthur D.G., Manolio T.A., Dimmock D.P., Rehm H.L., Shendure J., Abecasis G.R., Adams D.R., Altman R.B., Antonarakis S.E., Ashley E.A., et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinvar. [(accessed on 28 August 2017)]; Available online: https://www.ncbi.nlm.nih.gov/clinvar/
- 4.Timmermans S., Tietbohl C., Skaperdas E. In: BioSocieties. Palgrave M., editor. Springer; Berlin, Germany: 2016. pp. 1–20. [Google Scholar]
- 5.Thompson B.A., Spurdle A.B., Plazzer J.P., Greenblatt M.S., Akagi K., Al-Mulla F., Bapat B., Bernstein I., Capellá G., den Dunnen J.T., et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat. Genet. 2014;46:107–115. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jouhilahti E.M., Peltonen S., Heape A.M., Peltonen J. The pathoetiology of neurofibromatosis 1. Am. J. Pathol. 2011;178:1932–1939. doi: 10.1016/j.ajpath.2010.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutmann D.H., Ferner R.E., Listernick R.H., Korf B.R., Wolters P.L., Johnson K.J. Neurofibromatosis type 1. Nat. Rev. Dis. Primers. 2017;3:17004. doi: 10.1038/nrdp.2017.4. [DOI] [PubMed] [Google Scholar]
- 8.Upadhyaya M. Neurofibromatosis type 1: Diagnosis and recent advances. Expert. Opin. Med. Diagn. 2010;4:307–322. doi: 10.1517/17530059.2010.494660. [DOI] [PubMed] [Google Scholar]
- 9.Pasmant E., Sabbagh A., Spurlock G., Laurendeau I., Grillo E., Hamel M.J., Martin L., Barbarot S., Leheup B., Rodriguez D., et al. NF1 microdeletions in Neurofibromatosis type 1: From genotype to phenotype. Hum. Mutat. 2010;31:1506–1518. doi: 10.1002/humu.21271. [DOI] [PubMed] [Google Scholar]
- 10.Kehrer-Sawatzki H., Mautner V.F., Cooper D.N. Emerging genotype-phenotype relationships in patients with large NF1 deletions. Hum. Genet. 2017;136:349–376. doi: 10.1007/s00439-017-1766-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cnossen M.H., van der Est M.N., Breuning M.H., van Asperen C.J., Breslau-Siderius E.J., van der Ploeg A.T., de Goede-Bolder A., van den Ouweland A.M., Halley D.J., Niermeijer M.F. Deletions spanning the Neurofibromatosis type 1 gene: Implications for genotype-phenotype correlations in Neurofibromatosis type 1? Hum. Mutat. 1997;9:458–464. doi: 10.1002/(SICI)1098-1004(1997)9:5<458::AID-HUMU13>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 12.Ratner N., Miller S.J. A RASopathy gene commonly mutated in cancer: The Neurofibromatosis type 1 tumour suppressor. Nat. Rev. Cancer. 2015;15:290–301. doi: 10.1038/nrc3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Luca A., Schirinzi A., Buccino A., Bottillo I., Sinibaldi L., Torrente I., Ciavarella A., Dottorini T., Porciello R., Giustini S., et al. Novel and recurrent mutations in the NF1 gene in Italian patients with Neurofibromatosis type 1. Hum. Mutat. 2004;23:629. doi: 10.1002/humu.9245. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J., Tong H., Fu X., Zhang Y., Liu J., Cheng R., Liang J., Peng J., Sun Z., Liu H., et al. Molecular characterization of NF1 and Neurofibromatosis type 1 genotype-phenotype correlations in a Chinese population. Sci. Rep. 2015;5:11291. doi: 10.1038/srep11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasmant E., Parfait B., Luscan A., Goussard P., Briand-Suleau A., Laurendeau I., Fouveaut C., Leroy C., Montadert A., Wolkenstein P., et al. Neurofibromatosis type 1 molecular diagnosis: What can NGS do for you when you have a large gene with loss of function mutations? Eur. J. Hum. Genet. 2015;23:596–601. doi: 10.1038/ejhg.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serra E., Puig S., Otero D., Gaona A., Kruyer H., Ars E., Estivill X., Lázaro C. Confirmation of a double-hit model for the NF1 gene in benign Neurofibromas. Am. J. Hum. Genet. 1997;61:512–519. doi: 10.1086/515504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bianchessi D., Morosini S., Saletti V., Ibba M.C., Natacci F., Esposito S., Cesaretti C., Riva D., Finocchiaro G., Eoli M. 126 novel mutations in Italian patients with Neurofibromatosis type 1. Mol. Genet. Genom. Med. 2015;3:513–525. doi: 10.1002/mgg3.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quintáns B., Pardo J., Campos B., Barros F., Volpini V., Carracedo A., Sobrido M.J. Neurofibromatosis without Neurofibromas: Confirmation of a genotype-phenotype correlation and implications for genetic testing. Case. Rep. Neurol. 2011;3:86–90. doi: 10.1159/000327557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Upadhyaya M., Huson S.M., Davies M., Thomas N., Chuzhanova N., Giovannini S., Evans D.G., Howard E., Kerr B., Griffiths S., et al. An absence of cutaneous neurofibromas associated with a 3-bp inframe deletion in exon 17 of the NF1 gene (c.2970-2972 delAAT): Evidence of a clinically significant NF1 genotype-phenotype correlation. Am. J. Hum. Genet. 2007;80:140–151. doi: 10.1086/510781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinna V., Lanari V., Daniele P., Consoli F., Agolini E., Margiotti K., Bottillo I., Torrente I., Bruselles A., Fusilli C., et al. p.Arg1809Cys substitution in neurofibromin is associated with a distinctive NF1 phenotype without neurofibromas. Eur. J. Hum. Genet. 2015;23:1068–1071. doi: 10.1038/ejhg.2014.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rojnueangnit K., Xie J., Gomes A., Sharp A., Callens T., Chen Y., Liu Y., Cochran M., Abbott M.A., Atkin J., et al. High Incidence of Noonan syndrome features including short stature and pulmonic stenosis in patients carrying NF1 missense mutations affecting p.Arg1809: Genotype-phenotype correlation. Hum. Mutat. 2015;36:1052–1063. doi: 10.1002/humu.22832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alkindy A., Chuzhanova N., Kini U., Cooper D.N., Upadhyaya M. Genotype-phenotype associations in Neurofibromatosis type 1 (NF1): An increased risk of tumor complications in patients with NF1 splice-site mutations? Hum. Genom. 2012;6:12. doi: 10.1186/1479-7364-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castle B., Baser M.E., Huson S.M., Cooper D.N., Upadhyaya M. Evaluation of genotype-phenotype correlations in Neurofibromatosis type 1. J. Med. Genet. 2003;40:109. doi: 10.1136/jmg.40.10.e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabbagh A., Pasmant E., Imbard A., Luscan A., Soares M., Blanché H., Laurendeau I., Ferkal S., Vidaud M., Pinson S., et al. NF1 molecular characterization and Neurofibromatosis type I genotype-phenotype correlation: The French experience. Hum. Mutat. 2013;34:1510–1518. doi: 10.1002/humu.22392. [DOI] [PubMed] [Google Scholar]
- 25.Ben-Shachar S., Constantini S., Hallevi H., Sach EK., Upadhyaya M., Evans G.D., Huson S.M. Increased rate of missense/in-frame mutations in individuals with NF1-related pulmonary stenosis: A novel genotype-phenotype correlation. Eur. J. Hum. Genet. 2013;21:535–539. doi: 10.1038/ejhg.2012.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toriello H.V., Meck J.M. Statement on guidance for genetic counseling in advanced paternal age. Genet. Med. 2008;10:457–460. doi: 10.1097/GIM.0b013e318176fabb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francioli L.C., Polak P.P., Koren A., Menelaou A., Chun S., Renkens I., Genome of the Netherlands Consortium. van Duijn C.M., Swertz M., Wijmenga C., et al. Genome-wide patterns and properties of de novo mutations in humans. Nat. Genet. 2015;47:822–826. doi: 10.1038/ng.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veltman J.A., Brunner H.G. De novo mutations in human genetic disease. Nat. Rev. Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 29.Gutmann D.H., Aylsworth A., Carey J.C., Korf B., Marks J., Pyeritz R.E., Rubenstein A., Viskochil D. The diagnostic evaluation and multidisciplinary management of Neurofibromatosis 1 and Neurofibromatosis 2. JAMA. 1997;278:51–57. doi: 10.1001/jama.1997.03550010065042. [DOI] [PubMed] [Google Scholar]
- 30.De Luca A., Buccino A., Gianni D., Mangino M., Giustini S., Richetta A., Divona L., Calvieri S., Mingarelli R., Dallapiccola B. NF1 gene analysis based on DHPLC. Hum. Mutat. 2003;21:171–172. doi: 10.1002/humu.9111. [DOI] [PubMed] [Google Scholar]
- 31.Mutalyzer. [(accessed on 28 August 2017)]; Available online: https://mutalyzer.nl/
- 32.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., et al. REVEL: An ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Human Gene Mutation Database, HGMD. [(accessed on 14 August 2017)]; Available online: http://www.hgmd.cf.ac.uk/ac/all.php.
- 34.Leiden Open Variation Database 3.0, LOVD. [(accessed on 28 August 2017)]; Available online: http://www.lovd.nl/3.0/home.
- 35.Ensembl. [(accessed on 28 August 2017)]; Available online: http://www.ensembl.org/index.html.
- 36.ExAC Browser. [(accessed on 28 August 2017)]; Available online: http://exac.broadinstitute.org/
- 37.Human Phenotype Ontology (HPO) [(accessed on 28 July 2017)]; Available online: http://human-phenotype-ontology.github.io/
- 38.Roback P.J., Askins R.A. Judicious use of multiple hypotesis test. Conserv. Biol. 2005;19:261–267. doi: 10.1111/j.1523-1739.2005.00269.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.