Abstract
Senescence affect different traits of plants, such as the ripening of fruit, number, quality and timing of seed maturation. While senescence is induced by age, growth hormones and different environmental stresses, a highly organized genetic mechanism related to substantial changes in gene expression regulates the process. Only a few genes associated to senescence have been identified in crop plants despite the vital significance of senescence for crop yield. The S40 gene family has been shown to play a role in leaf senescence. The barley HvS40 gene is one of the senescence marker genes which shows expression during age-dependent as well as dark-induced senescence. Like barley HvS40, the Arabidopsis AtS40-3 gene is also induced during natural senescence as well as in response to treatment with abscisic acid, salicylic acid, darkness and pathogen attack. It is speculated that rice OsS40 has a similar function in the leaf senescence of rice.
Keywords: S40 protein family, senescence, environmental cues
1. Introduction
The last phase of leaf development prior to cell death is leaf senescence. The degradation of cellular materials and their remobilization to develop leaves and emerging seeds characterizes the process [1,2]. In cereal crops, a close relationship exists between leaf senescence and the seed maturation induction process [3,4]. While early senescence caused substantial production loss [5,6], increased yield was shown by stay-green phenotypes in different crops [7]. In the USA and Canada, the yield of Zea mays L. (maize) increased significantly due to delayed leaf senescence [8,9] while the yield of soybean decreased due to precocious leaf senescence [10]. Similarly, the weight of seeds in the maize plants increased by 15–30% and the yield of grains in rice increased by 24% [11] and 10.3% [12] due to delayed senescence. Leaf senescence directly influences the quality and yield of a crop, and therefore can be manipulated for improving crop quality and increasing the yield [11].
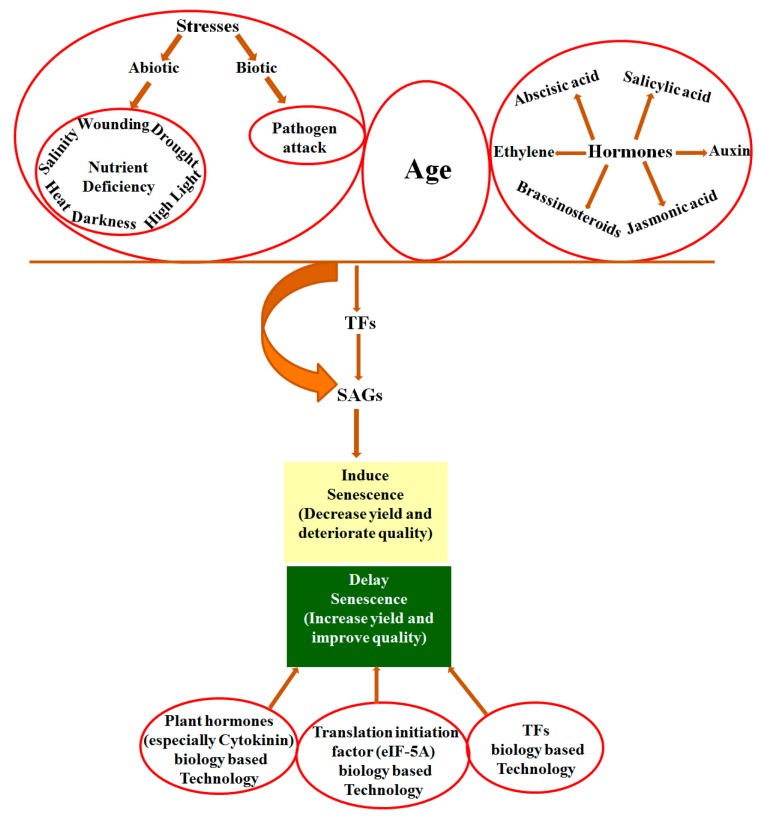
Leaf senescence is an age-dependent process, yet it is induced by various external and internal factors. External factors include biotic factors such as pathogen attack, and abiotic stresses such as salt stress, drought stress, wounding, heat, nutrient stresses, high light and darkness [13,14,15,16,17,18,19]. Internal factors include plant growth hormones and aging. Plant hormones include abscisic acid (ABA), salicylic acid (SA), jasmonic acid (JA), auxin, ethylene, and brassinosteroids (BR) that positively regulate senescence, while cytokinins and polyamines negatively regulate senescence [20,21]. In this sense, the suppression of plant hormones that promote senescence such as ethylene or overproduction of senescence-inhibiting hormones such as cytokinins could delay plant senescence [20,21].
Senescence is a highly organized process that requires the expression of specific genes [22,23], referred to as SAGs (senescence-associated genes), that account for 10% of a plant genome [24,25] during aging. Thousands of genes show differential expression patterns during the onset and development of senescence [25,26]. In Arabidopsis, around 20% of all the genes showed differential expression during leaf senescence [27] and 827 SAGs were found to be upregulated at least three times upon senescence [16]. Transcriptomic studies have revealed a wide range of change in gene-expression during senescence in many plants such as Arabidopsis thaliana [26,28], Triticum aestivum [29], Hordeum vulgare [30], Medicago truncatula [31], Oryza sativa [32] and Aspen [33]. Different transcription factors (TFs) among which transcription factor NAM (no apical meristem), ATAF, CUC (cup-shaped cotyledon) (NAC) and transcription factor in which N-terminal ends contain a conserved WRKYGQR amino acids sequences (WRKY) are the two major groups [25] that activate these SAGs, hence the regulation of a gene expression that encodes a specific, important transcription factor can influence the expression of numerous genes that are involved in plant senescence [34].
Arabidopsis NAP (AtNAP), one of the members of the NAC family, is a key transcription factor that plays a regulatory role in leaf senescence. The counterpart genes in other crops such as morning glory [35], Festuca arundinacea [36], Phaseolus vulgaris, Oryza sativa [37] and Bambusa vulgaris [38] also showed expression only in non-senescent leaves, but not in young leaves. Moreover, the expression pattern of these homologues was similar to AtNAP [36,37,38], which indicates their regulatory role in leaf senescence. The corresponding genes of AtNAP showed delayed leaf senescence in rice [39] and maize [40]. Similarly, WRKY transcription factors such as WRKY70, WRKY54 [41], WRKY53 [42], WRKY22 [43], WRKY23 [44], WRKY42 [45], WRKY80 [46] and WRKY6 [47] also play a role in regulating leaf senescence. Genomic pull-down assays identified sixty-three genes as direct targets of WRKY53, including at least six other members of the WRKY gene family. This indicates that WRKY53 transcription factors regulate SAGs and act as an upstream control element in a WRKY signaling cascade [42] (Figure 1).
Figure 1.
Transcriptional regulation and manipulation of leaf senescence.
Generally, in cereal crops, an inverse proportion exists between leaf senescence and seed production yields; that is, early senescence causes substantial production loss, and vice versa. It is important to manipulate plant senescence by biotechnology methods to increase crop yields. The S40 gene family is one of few genes identified as being associated with plant leaf senescence. This review deals with the role of the S40 gene family in, mainly, barley and rice as well as Arabidopsis. It describes (1) phylogenetic trees; (2) the alteration of gene expression; (3) the subcellular localization of S40 proteins; (4) the nature of S40 mutants; (5) regulation at the transcriptional level; (6) cis elements comparison; and (7) WHIRLY1 protein bindings to the HvS40 promoter. This compilation of information is thought to be timely and useful for people working with similar kinds of subjects.
2. Phylogenetic Tree Construction
S40 belongs to the DUF584 family, containing the DUF584 domain. All members of the DUF584 group shared the sequence GRXLKGR(D/E)(L/M)XXXR(D/N/T)X(I/V)XXXXG(F/I) [48]. This sequence belongs to the C-terminal domain. In barley, only two proteins belong to DUF584 family, of which one is encoded by HvS40 gene, while in Arabidopsis, fifteen proteins belong to this family [49]. An alignment by Clustal W [50] showed that the proteins can be further divided into five groups on the basis of similarities among the amino acid sequences. Four DUF584 proteins of Arabidopsis and the HvS40 protein belong to the same group, group I. In the remaining eleven of the Arabidopsis DUF584 protein family, three proteins lie in group II, four proteins lie in group III, three proteins lie in group IV, while only one Arabidopsis protein belongs to Group V, which also has the second barley DUF584 protein [49].
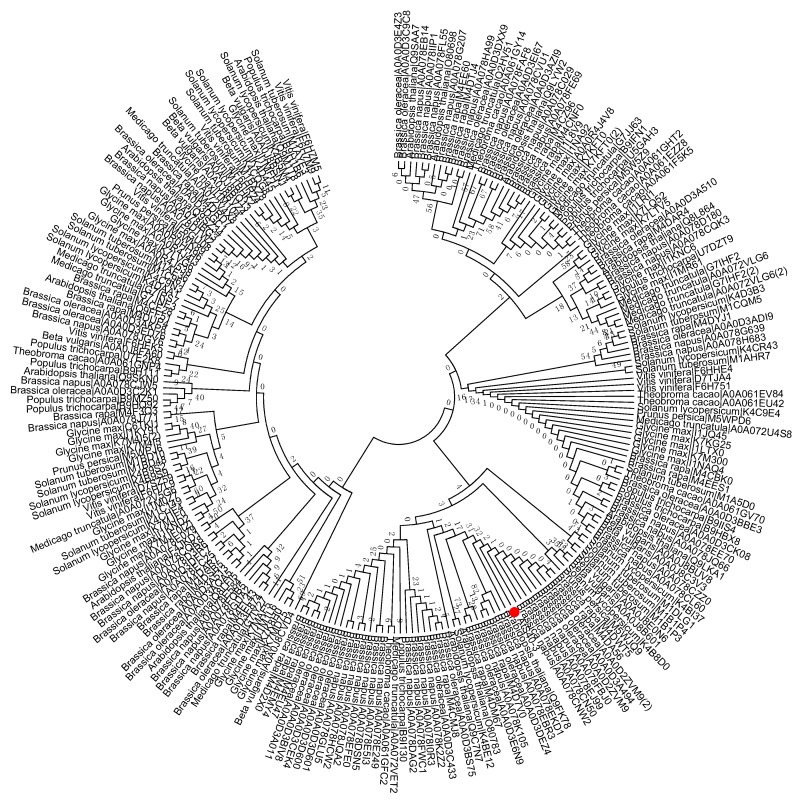
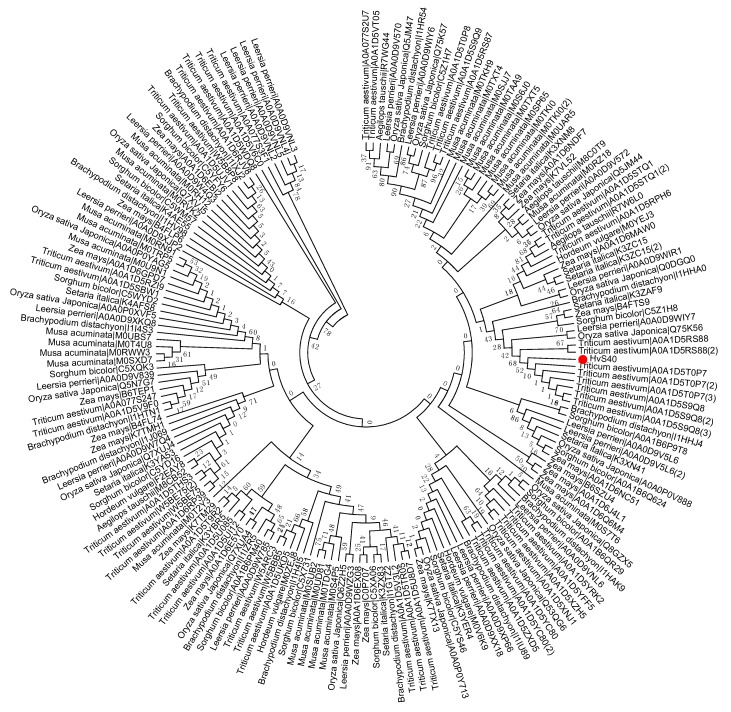
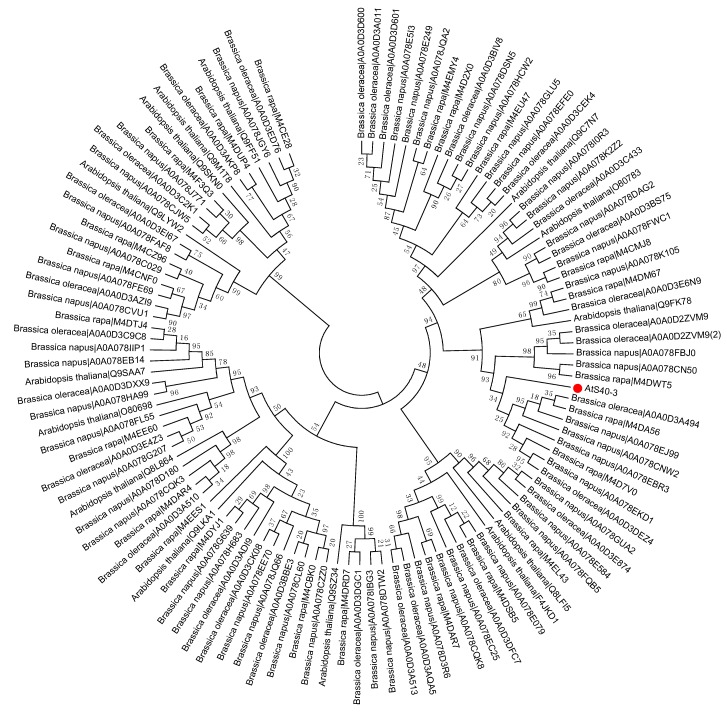
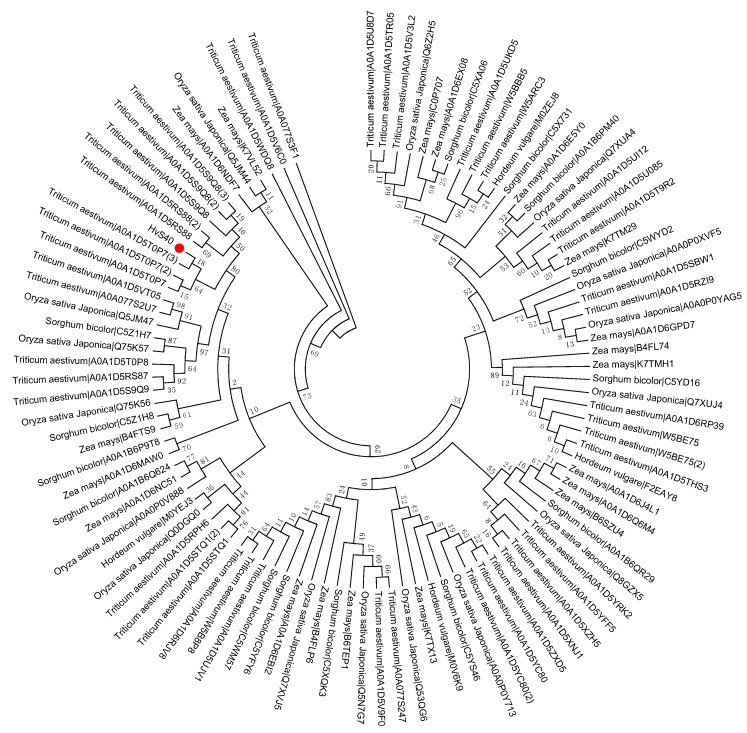
To study the evolutionary history of S40 proteins, a HvS40 protein sequence was used as a Basic Local Alignment Search Tool (BLAST) query against the available sequenced genome of plants using http://plants.ensembl.org/Multi/Tools/database. The aligned sequences were obtained from dicot plants including Arabidopsis thaliana, Beta vulgaris, Brassica napus, Brassica oleracea, Brassica rapa, Glycine max, Medicago truncatula, Populus trichocarpa, Prunus persica, Solanum lycopersicum, Solanum tuberosum, Theobroma cacao and Vitis vinifera, while monocot plants included Aegilops tauschii, Brachypodium distachyon, Hordeum vulgare, Leersia perrieri, Musa acuminate, Oryza sativa Japonica, Setaria italica, Sorghum bicolor, Triticum aestivum and Zea mays. When the HvS40 protein sequence was blasted against the genomes of Chlamydomonas reinhardtii, Chondrus crispus, Cyanidioschyzon merolae strain 10D, Ectocarpus siliculosus, Galdieria sulphuraria, Ostreococcus lucimarinus CCE9901, Picea sitchensis and Thalassiosira pseudonana, no similar hit was found. The MEGA 6.0 program was employed to construct a neighbor-joining phylogenetic tree using a JTT model with 1000 bootstrap replicates. The full length amino acid sequences were aligned by using Clustal Omega. To identify the HvS40 protein aligned members, four separate trees were constructed in which the first tree includes dicot plants (Figure 2), the second includes monocot plants (Figure 3), the third includes Brassicaceae family plants (Figure 4), and the fourth includes cereal crop plants (Figure 5). Among the dicot plants, several members from Brassica napus, Brassica oleracea and Brassica rapa were identified close to AtS40-3, while in monocot, several members from Triticum aestivum were identified close to HvS40 that may have similar functions in these plants. Plenty of HvS40 protein-aligned members were found in cereal crops as well as Brassicaceae family plants. Sixteen members of the S40 gene family were found in the rice (Oryza sativa Japonica) genome. It seems that the S40 gene family does not exist in lower organisms such as Thallophytic, and does also not exist in Gymnosperms plants.
Figure 2.
Phylogenetic tree of the S40 gene family in sequenced genome dicot plants including Arabidopsis thaliana, Beta vulgaris, Brassica napus, Brassica oleracea, Brassica rapa, Glycine max, Medicago truncatula, Populus trichocarpa, Prunus persica, Solanum lycopersicum, Solanum tuberosum, Theobroma cacao, and Vitis vinifera. Red dot indicates the S40 senescence reference gene, AtS40-3. The MEGA 6.0 program was employed to construct a neighbor-joining phylogenetic tree using the JTT model with 1000 bootstrap replicates. The full-length amino acid sequences were aligned using Clustal Omega.
Figure 3.
Phylogenetic tree of the S40 gene family in sequenced genome monocot plants including Aegilops tauschii, Brachypodium distachyon, Hordeum vulgare, Leersia perrieri, Musa acuminate, Oryza sativa Japonica, Setaria italica, Sorghum bicolor, Triticum aestivum, Zea mays. Red dot indicates the S40 senescence reference gene, HvS40. The MEGA 6.0 program was employed to construct a neighbor-joining phylogenetic tree using the JTT model with 1000 bootstrap replicates. The full-length amino acid sequences were aligned using Clustal Omega.
Figure 4.
Phylogenetic tree of the S40 gene family in the sequenced genome Brassicaceae family including Arabidopsis thaliana, Brassica napus, Brassica oleracea, and Brassica rapa. Red dot indicates the S40 senescence reference gene, AtS40-3. The MEGA 6.0 program was employed to construct a neighbor-joining phylogenetic tree using the JTT model with 1000 bootstrap replicates. The full-length amino acid sequences were aligned using Clustal Omega.
Figure 5.
Phylogenetic tree of the S40 gene family in sequenced genome cereal crop plants including Oryza sativa Japonica, Triticum aestivum, Zea mays, Hordeum vulgare, and Sorghum bicolor. Red dot indicates the S40 senescence reference gene, HvS40. The MEGA 6.0 program was employed to construct a neighbor-joining phylogenetic tree using the JTT model with 1000 bootstrap replicates. The full-length amino acid sequences were aligned using Clustal Omega.
3. Alterations in S40 Genes Correspond to Age-Dependent as Well as Artificially-Induced Leaf Senescence
In barley flag leaves, the expression level of HvS40 increased simultaneously with chlorophyll degradation and reached the highest level shortly before the last stage of senescence [14]. Thus, HvS40 gene expression can be used as a sign for age-dependent senescence [51]. In Arabidopsis, seven out of eleven genes, AtS40-1, AtS40-2, AtS40-3, AtS40-4, AtS40-5, AtS40-6 and AtS40-7 showed enhanced transcripts levels in senescent leaves compared to non-senescent leaves [49]. When barley leaf segments were exposed to drought stress and treated with hydrogen peroxide, the HvS40 gene showed a highly upregulated expression level [52], while treatment with mannitol to provoke osmotic stress induced its expression only slightly [53]. Although HvS40 expression was enhanced by treatment of barley leaf parts with abscisic acid (ABA), salicylic acid (SA), ethylene and jasmonic acid (JA) [54], the response to ABA was highly substantial [55]. Similarly, the expression level of three genes AtS40-2, AtS40-3 and AtS40-4 were shown to be slightly increased after the treatment of the Arabidopsis plants with salicylic acid (SA) and ABA [49]. HvS40 expression can be used as a sign for senescence induced by darkness [14] and the gene showed highly upregulated expression levels in barley [56]. The expression level of AtS40-1, AtS40-2 and AtS40-5 genes were induced after two days of dark incubation while AtS40-3 and At-S40-4 showed increased expression only one day after dark incubation [49].
When the leaves of a barley plant were incubated with the pathogen Pyrenophora teres, the HvS40 gene showed a highly induced expression level in the infected leaf segments at the necrotic lesion sites surrounded by chlorotic tissue [54]. Similarly, after the infection of the Arabidopsis plants with the pathogen Pseudomonas syringae for only one day, AtS40-2, AtS40-3 and AtS40-4 also showed increased expression [49].
By using the Bio-Analytic Resource for the plant biology database (http://bar.utoronto.ca/efprice/cgi-bin/efpWeb.cgi), we predicted the expression pattern of the OsS40 gene family in rice. The sequences for 16 putative S40 gene family members in rice (Oryza sativa) were obtained from the Rice Genome Annotation project (http://rice.plantbiology.msu.edu/) that were labeled as OsS40-1 to OsS40-16. During age-dependent senescence, the expression patterns of S40-1, S40-13 and S40-16 were shown to be slightly upregulated, while S40-4, S40-7, S40-14 and S40-15 showed significant upregulation. S40-1, S40-4, S40-6, S40-7, S40-9, S40-13, S40-15 and S40-16 showed significant upregulation to salt stress while S40-5, S40-6, S40-7, S40-9 and S40-14 showed significant upregulation to drought stress. None of the genes showed upregulation during cold stress (Table 1).
Table 1.
Predicted expression level of OsS40 genes under natural and stress-induced senescence (http://bar.utoronto.ca/efprice/cgi-bin/efpWeb.cgi).
| S40 Genes | Age Dependent Senescence | Salt Stress | Drought Stress | Cold Stress |
|---|---|---|---|---|
| OsS40-1 | Slightly increased | Increased | No change | No change |
| OsS40-2 | No change | No change | No change | No change |
| OsS40-3 | No change | No change | No change | No change |
| OsS40-4 | Increased | Increased | No change | No change |
| OsS40-5 | No change | No change | Increased | No change |
| OsS40-6 | No change | Increased | Increased | No change |
| OsS40-7 | Increased | Increased | Increased | No change |
| OsS40-8 | No change | No change | No change | No change |
| OsS40-9 | No change | Increased | Increased | No change |
| OsS40-10 | No change | No change | No change | No change |
| OsS40-11 | No change | No change | No change | No change |
| OsS40-12 | No change | No change | No change | No change |
| OsS40-13 | Slightly increased | Increased | No change | No change |
| OsS40-14 | Increased | No change | Increased | No change |
| OsS40-15 | Increased | Increased | No change | No change |
| OsS40-16 | Slightly increased | Increased | No change | No change |
4. Subcellular Localization
When the barley HvS40 15.4 kD small protein was fused to β-glucuronidase (GUS) [54] or to green fluorescent protein (GFP) [48], it was found to be located in the nucleus [54]. Due to the inclusion of two nuclear localization signals (NLS) in the sequence, this is expected. The theoretical molecular weights of the seven candidate proteins encoded by the AtS40 genes ranged from 12 to 25 kD. The six candidate proteins showed accumulation only in the cytoplasm while the AtS40-3-GUS fusion protein was localized both in the cytoplasm and nucleus. After transformation with the AtS40-3-GUS construct, GUS activity distribution in the cell [49] showed similarity to GUS activity distribution after transformation with the HvS40-GUS fusion construct [54].
The putative subcellular localization of S40 proteins in rice was predicted using the publicly available web services YLoc, WoLF PSORT, DISTILL 2.0, LOCALIZER 1.0, BaCelLo and Euk-mPLoc-2.0. A dominant number of prediction programs predicted the localization of OsS40-1, OsS40-2, OsS40-3, OsS40-5, OsS40-6, OsS40-7, OsS40-9, OsS40-14 and OsS40-16 in the nucleus while OsS40-12 and OsS40-13 localization were predicted to be in the chloroplast. The prediction programs predict different localizations of OsS40-4, OsS40-8, OsS40-11 and OsS40-15 as shown in the Table 2. Although the prediction tool shows a high ratio of accuracy, the exact localization must be tested via experimentation.
Table 2.
The putative subcellular localization of S40 protein in rice *.
| Loci Number in Genome | Gene Name | Yloc (http://abi.inf.uni-tuebingen.de/Services/YLoc/webloc.cgi) | WoLF PSORT (https://wolfpsort.hgc.jp/) | DISTILL 2.0 (http://distillf.ucd.ie/distill/) | LOCALI-ZER 1.0 (http://localizer.csiro.au/) | BaCelLo (http://gpcr.biocomp.unibo.it/bacello/pred.htm) | Euk-mPLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/euk-?multi-2/) |
|---|---|---|---|---|---|---|---|
| Loc_Hv FI496079 | HvS40—Nucleus (Krupinska et al. 2002) [54] | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Mitochondria. Nucleus |
| LOC_Os05g45450 | OsS40-1 | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus | Nucleus |
| LOC_Os05g44260 | OsS40-2 | Nucleus | Mitochondria | Nucleus | Nucleus | Nucleus | Extra cell |
| LOC_Os10g27350 | OsS40-3 | Nucleus | Chloroplast | Chloroplast | ------ | Nucleus | Nucleus |
| LOC_Os03g02280 | OsS40-4 | Nucleus | Chloroplast | Chloroplast | ------ | Secretory | Extra cell |
| LOC_Os04g45834 | OsS40-5 | Nucleus | Chloroplast | Chloroplast | Nucleus | Nucleus | Nucleus, Extra cell |
| LOC_Os04g33760 | OsS40-6 | Nucleus | Nucleus | Chloroplast | Nucleus | Nucleus | Nucleus, Extra cell |
| LOC_Os01g52730 | OsS40-7 | Nucleus | Nucleus | Chloroplast | ------- | Nucleus | Extra cell |
| LOC_Os10g33990 | OsS40-8 | Cytoplasm | Chloroplast | Chloroplast | Nucleus | Nucleus | Extra cell |
| LOC_Os01g64300 | OsS40-9 | Nucleus | Nucleus | Chloroplast | ------- | Nucleus | Extra cell |
| LOC_Os01g52740 | OsS40-10 | Cytoplasm | Nucleus | Chloroplast | Nucleus | Nucleus | Nucleus, Extra cell |
| LOC_Os12g05980 | OsS40-11 | Mitochondria | Chloroplast | Chloroplast | Nucleus | Nucleus | Extra cell |
| LOC_Os11g05600 | OsS40-12 | Chloroplast | Chloroplast | Chloroplast | Nucleus | Nucleus | Extra cell |
| LOC_Os04g43990 | OsS40-13 | Chloroplast | Mitochondria | Chloroplast | --------- | Chloroplast | Extra cell |
| LOC_Os05g45440 | OsS40-14 | Nucleus | Cytoplasm | Chloroplast | Nucleus | Nucleus | Nucleus |
| LOC_Os07g33270 | OsS40-15 | Nucleus | Mitochondria | Chloroplast | --------- | Nucleus | Extra cell |
| LOC_Os07g32810 | OsS40-16 | Nucleus | Nucleus | Chloroplast | ---------- | Nucleus | Extra cell |
* Publicly available web services YLoc, WoLF PSORT, DISTILL 2.0, LOCALI-ZER 1.0, BaCelLo and Euk-mPLoc-2.0 were used to predict the putative subcellular localization of S40 proteins in rice.
5. Natural Leaf Senescence Is Delayed and SAGs Are Repressed in the S40 Mutant
In Arabidopsis wildtype plants, the AtS40-3 gene showed 16 times higher expression levels in senescent leaves compared to non-senescent ones during natural senescence. While in the AtS40-3a mutant, the AtS40-3 gene expression level was similar in both non-senescent rosettes and senescent ones indicating that the gene was disabled for senescence-specific induction. The results showed a delayed senescence in the AtS40-3a mutant in comparison to the wildtype [49]. The expression level of the WRKY53 gene, which is a marker of early changes in gene expression during leaf senescence, was significantly down-regulated in the mutant in comparison to the wildtype. Similarly, the expression level of the SAG12 gene, which is a marker of late changes in gene expression, was clearly decreased in the AtS40-3a mutant in comparison to the wildtype at all stages of senescence [49]. The expression analyses in both the wildtype and mutants propose that the AtS40-3 gene acts as an activator of downstream SAGs, SAG12 and WRKY53.
During dark-induced senescence, the kinetics and expression level of SAGs changed in the AtS40-3a mutant in comparison to the wildtype. In the wildtype, the expression level of the SEN1 gene, SAG [57] was enhanced about 375-fold after one day of darkness and thereafter decreased. While in the AtS40-3a mutant, SEN1 expression increased only 100-fold after one day of dark incubation and increased further. In the wildtype, the expression level of the SAG12 gene was increased after dark incubation, which increased further with time in darkness. While in the AtS40-3a mutant, the SAG12 gene showed almost no expression after the first two days of dark incubation, and after three days of dark incubation it reached almost half of the expression level noticed in wildtype plants [49]. These results indicate that the AtS40-3 gene positively regulates senescence in natural light as well as dark conditions. The development of Hvs40 mutants for the phenotypic analysis as well as for the analysis of barley SAG expression in the mutants will improve the understanding of the senescence-associated role of HvS40.
6. Senescence-Related Expression of the S40 Gene Shows Regulation at the Transcriptional Level
Senescence processes were shown to be induced during dark incubation which is partially reverted after subsequent re-illumination. HvS40 was shown to accelerate after one day of dark incubation, which was decreased to unnoticeable levels after the exposure of plants to light for one day [56,58]. During dark treatment, the HvS40 gene was shown to be transcribed before the onset of senescence by the hybridization of nuclear run-on (NRO) transcripts. However, during dark-induced senescence, the transcription rates significantly increased and then declined when the senescence process was reversed by re-illumination [55]. The transcription rate of 19 barley SAGs, which showed increased levels of expression on exposure to darkness, were examined in parallel for comparison by NRO transcription assays. During the dark-induced senescence, five of 19 genes showed a higher rate of transcription [59]. By comparing the HvS40 gene promoter sequence with two other barley SAGs, one SAG (HvSD1) showed regulation at the transcriptional level only in leaves exposed to darkness while the other SAG (HvSD8) did not show regulation at the transcriptional level [55]. The HvS40 gene also showed increased transcription in the nuclei of senescent flag leaves [59], whereas HvSD1 transcription was not increased under the same conditions. Taken together these results illustrate that during senescence, the upstream regulatory components shared by various types of senescence regulate HvS40 gene expression at the transcriptional level. The regulation level of the senescence-related AtS40-3 gene in Arabidopsis is yet to be examined.
7. Cis Elements Comparison in the Promoters of S40 Genes in Barley, Arabidopsis and Rice
To reveal cis elements that might be involved in the initiation of HvS40 gene transcription during natural and dark-induced senescence and to gain insight into elements related to developmental senescence, the promoter of the HvS40 was compared with the promoters of S40 genes in barley, Arabidopsis and rice. Along with many myelocytomatosis (MYC, CANNTG) and myeloblastosis (MYB, ACGTG), several dehydration-responsive elements/C-repeats (DRE/CRT, A/GCCGAC) and abscisic acid responsive elements (ABRE, ACGTG) recognition sequences were found in HvS40 promoter sequence using in silico analyses, while only a few of the latter motifs were found in HvSD1 and HvSD8 promoters (Table 3). In the HvS40 gene upstream region, among the most abundant motifs, several DNA-binding with one finger (Dof) binding sites (AAAG) and many light-regulated elements (LREs) along with embryo-specific expression motifs and pathogen-responsive elements were also identified. Dof binding sites and pathogen-responsive elements were identified repeatedly in the HvSD1 gene promoter compared to the HvS40 gene promoter (Table 3). All the three analyzed promoters contained several W-box elements. W-boxes were shown to bind WRKY TFs [60,61].
Table 3.
Cis-elements in the promoters of S40 genes in barley, Arabidopsis, and rice.
| Cis-Elements | HvS40 | HvSDI | HvSD8 | At2g 34340 | At1g 29640 | At5g 45630 | AtS4-3 | OsS40-1 | OsS40-2 | OsS40-7 | OsS40-14 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| W-box | 4 | 4 | 5 | 5 | 6 | 5 | 2 | 2 | 2 | 5 | 3 |
| ERE | 2 | 1 | |||||||||
| MYB | 19 | 3 | 2 | 1 | 2 | 6 | 7 | 5 | 3 | 5 | 6 |
| LREs | 13 | 11 | 7 | 2 | 1 | ||||||
| MYC | 8 | 2 | 2 | 6 | 4 | 10 | 8 | 18 | 6 | 4 | 8 |
| ABRE | 6 | 5 | 4 | 4 | 15 | 15 | 6 | 4 | 8 | 4 | |
| Dof | 5 | 19 | 7 | 11 | 18 | 12 | 26 | 4 | 12 | 8 | 5 |
| PRE | 5 | 15 | 11 | ||||||||
| CGCG box | 3 | 3 | 3 | 8 | 26 | 14 | 20 | ||||
| SURE | 2 | 4 | 6 | 5 | 1 | 5 | 3 | 1 | 1 | ||
| DRE/CRT | 2 | 4 | 3 | 1 | 1 | 2 | 2 | 1 | |||
| LTR | 2 | 2 | 3 | 1 | 1 | 1 | 3 | 2 | 3 | ||
| ARF | 2 | 3 | 2 | 1 | 1 | ||||||
| DPBFCOREDCDC3 | 2 | 1 | 3 | 4 | 6 | 2 | 1 | 3 | 2 | ||
| Pyrimidine box | 1 | 3 | 2 | 1 | 2 | 1 | 2 | 1 | 3 | 1 | |
| G-box plus G | 1 |
Promoter regions of 840, 1000, and 708 bp upstream of S40 genes in barley, Arabidopsis, and rice respectively, were analyzed with the use of the PLACE program (Higo and others 1999). W-box: Binding site for WRKY TFs; ERE: Elicitor response element; MYB: Myeloblastosis; LREs: Light regulated elements; MYC: Myelocytomatosis; ABRE: Abscisic acid responsive elements; Dof: DNA-binding with one finger; PRE: Pathogen response elements; SURE: Sulfur response elements; DRE/CRT: Dehydration response elements/C-repeat; LTR: Low temperature response; ARF: Auxin response factor; DPBFCOREDCDC3: BZIP TFs binding core sequence; G-box plus G: TF OsIRO2-binding core sequence.
Two prominent motifs, W-box elements ([T]TGAC[C/T]) and inverted repeat motifs (IR2, TGTCA) were found only in the HvS40 promoter. The first motif (MI) that contained a W-box and an IR2 motif was situated 613–631-bp upstream of the transcription initiation site, while the second motif (MII), containing a W-box dimer motif, was located 401–420-bp upstream from the transcription initiation site [55]. The MI motif was shown to be highly identical to the elicitor response element (ERE) element found in the promoter of the PR10a gene in potato. In the potato, during pathogen attack, binding of StWHY1 TF to an ERE element that consists of a W-box and an IR2 element was observed [62,63]. The MII motif was found to be highly similar to a W-box dimer motif in the promoter of SIRK/FRK1 gene of Arabidopsis (At2g19190) that encodes a receptor kinase, which shows upregulated expression upon pathogen challenge, as well as during leaf senescence [64]. Moreover, the binding of WRKY11 and WRKY26 TFs to this W-box dimer motif was also found [61]. This suggests a multidimensional function of the W-box motifs that controls transcription during various developmental as well as environmental conditions by binding various TFs.
Using the phylogenetic analysis, the promoters of the S40 genes in rice and Arabidopsis that are closest to the HvS40 gene were analyzed by the plant cis-acting regulatory DNA elements (PLACE) database (http://www.dna.affrc.go.jp/PLACE/). Promoter regions of 1000 bp upstream of AtS40-3, At5g45630, At1g29640, At2g34340 and OsS40-1, OsS40-2, OsS40-7, OsS40-14 were analyzed to reveal cis regulatory elements. The prominent motif, a deoxyribonucleic acid (DNA) cis-regulatory element sequence, (T)TGAC)C/T), which is recognized by the family of WRKY transcription factors (W-box) along with many MYC, MYB, ABRE, and Dof binding sites and a few DRE/CRT recognition sequences were found in the promoter regions of Arabidopsis and rice S40 genes, as shown in Table 3. The W-box is an element known to be the binding site of WRKY transcription factors [65]; ABRE is an ABA-responsive cis-acting element found in many ABA-inducible genes [66,67,68]; and the Dof proteins are a family of plant-specific TFs that includes Dof1, Dof2, Dof3, and PBF [69] Similar function of the cis elements are speculated in the regulation of S40 genes in Arabidopsis and rice.
8. The WHIRLY1 Protein Binds to HvS40 Promoters at the MII Motif Region
The MI and MII motifs (W-box dimer motif) of HvS40 are separated by a 200 bp sequence stretch. Three DNA–protein complexes with similar mobility were identified (complexes I, II, III) with motifs as well as nuclear protein extracts from young and senescent leaves of barley using electrophoretic mobility shift assays. The complex of intermediate mobility among the three complexes that were formed at MII, which is close to the start codon, was derived solely with nuclear proteins obtained from young leaves. While the complex of intermediate mobility among the three complexes formed at MI was derived solely with nuclear proteins from senescent leaves. By using a biotinylated DNA fragment that contains the MII motif to bind the nuclear proteins from nonsenescent leaves, a protein of 24-kDa was extracted, which was recognized as WHIRLY1 on the basis of the amino acid sequences of three peptides [55].
Recombinant WHIRLY1 was shown to bind with MII as well as the MI motif. A complex of mobility formed at MI with nuclear proteins derived from senescent leaves showed similarity to the one that contained WHIRLY1 at MII in young leaves. This suggests that WHIRLY1 might negatively regulate the HvS40 gene, which represses the expression of the HvS40 gene prior to the initiation of senescence, because WHIRLY1 has already been shown as a factor binding to the MII of the HvS40 promoter only in young leaves. This also suggests that before the onset of senescence, WHIRLY1 might bind to the HvS40 promoter at different positions, as happens during senescence. It is possible that this alteration in position changes the binding efficiency of the promoter for various TFs. These different TFs that bind to the W-box motifs of MII may include WRKY TFs. As in the promoter of the SIRK/FRK1 gene of Arabidopsis, five nominated WRKY TFs that represent the three main WRKY protein groups were found to bind to the W-box dimer motif [61], which has high similarity to the HvS40 MII motif. Furthermore, various adjacent W-box elements have been reported in various gene promoters. In the case of HvWRKY38 TF in barley, two closely adjacent W-box motifs are essential for efficient DNA binding, while PcWRKY1 tends to have synergistic effects on DNA transcription [70,71]. However, most of the WRKY TFs, might bind as a monomer to only one of the two W-box regions contained within the dimer motifs [61]. For example, it is reported that for the effective DNA binding of HvWHIRLY1 on motif II (MIII) at the HvS40 promoter region, two unaltered W-box motifs are required. These findings clearly show that HvWHIRLY1 TF has a lower binding capability to the W-box motifs as compared to the WRKY TFs [55]. It is reported that there is a higher expression level of WRKY TFs during pathogen attack and during senescence [65], this may be due to altered gene expression resulting from the replacement of the WHIRLY1 TF from the promoter of HvS40. However, a chromatin immunoprecipitation comprised of HvWHIRLY1-specific antibody is needed to investigate whether the change of the position of WHIRLY1 at the promoter region of HvS40 during development alters its expression. Furthermore, the development of transgenic plants with different levels of WHIRLY1 TF is also needed to explore the effect of WHIRLY1 TF on HvS40 gene expression.
9. Conclusions
Regarding the potential use of senescence for improving yield, much of the fundamental knowledge of the regulation of senescence has been tested in crop species such as those with stay-green traits [72] and pSAG12:IPT technology [73]. Further elucidating the genes related to senescence will expand the understanding of leaf senescence regulation at the molecular level. The S40 gene family showed enhanced expression levels during natural as well as stress-induced senescence in barley and Arabidopsis. The AtS40-3 gene positively regulates senescence in natural light as well as in dark conditions. Upstream regulatory components shared by the various types of senescence regulate HvS40 gene expression at the transcriptional level during senescence. The single-stranded DNA-binding protein WHIRLY1 binds to the HvS40 promoter at the MII motif region. The S40 gene family plays a regulatory role in leaf senescence in Arabidopsis and barley plants. The predicted upregulated expression pattern of some members of the S40 gene family in rice insinuate its regulatory role in rice leaf senescence. Different parameters still need to be studied to gain insight into the complex regulation of senescence by the S40 gene family, particularly in the model plant, Arabidopsis. The development of transgenic plants and further efforts for detailed functional analyses of the S40 gene family in rice, wheat and other crops will explore the precise mechanism of function of the S40 gene family related to senescence.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Noodén L. The Phenomena of Senescence and Aging. Food and Agriculture Organization; Rome, Italy: 1988. [Google Scholar]
- 2.Matile P. Crop Photosynthesis: Spatial and Temporal Determinants. Elsevier; Amsterdam, The Netherlands: 1992. Chloroplast senescence; pp. 413–440. [Google Scholar]
- 3.Kohl S., Hollmann J., Blattner F.R., Radchuk V., Andersch F., Steuernagel B., Schmutzer T., Scholz U., Krupinska K., Weber H. A putative role for amino acid permeases in sink-source communication of barley tissues uncovered by RNA-seq. BMC Plant Biol. 2012;12:154. doi: 10.1186/1471-2229-12-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollmann J., Gregersen P.L., Krupinska K. Identification of predominant genes involved in regulation and execution of senescence-associated nitrogen remobilization in flag leaves of field grown barley. J. Exp. Bot. 2014;65:3963–3973. doi: 10.1093/jxb/eru094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pleuel H., Ojanpera K., Danielsson H., Sild E., Gelang J., Wallin G. Effects of Ozone on Leaf Senescence in Spring Wheat—Possible Consequences for Grain Yield. Food and Agriculture Organization; Rome, Italy: 1997. [Google Scholar]
- 6.Li H., Wang S., Li B., Tong Y., Yang X., Li Z. Comparative study on physiological traits related with grain filling and photosynthesis of flag leaf in early aging and normal near—Isogenic lines of common wheat. Zuo Wu Xue Bao. 2005;32:1642–1648. [Google Scholar]
- 7.Gregersen P.L., Culetic A., Boschian L., Krupinska K. Plant senescence and crop productivity. Plant Mol. Biol. 2013;82:603–622. doi: 10.1007/s11103-013-0013-8. [DOI] [PubMed] [Google Scholar]
- 8.Duvick D.N. Genetic Contributions to Yield Gains of Five Major Crop Plants. Crop Science Society of America and American Society of Agronomy; Madison, WI, USA: 1984. Genetic contributions to yield gains of US hybrid maize, 1930 to 1980; pp. 15–47. [Google Scholar]
- 9.Tollenaar M., Wu J. Yield improvement in temperate maize is attributable to greater stress tolerance. Crop Sci. 1999;39:1597–1604. doi: 10.2135/cropsci1999.3961597x. [DOI] [Google Scholar]
- 10.Egli D.B. Seed-fill duration and yield of grain crops. Adv. Agron. 2004;83:243–279. [Google Scholar]
- 11.Guo Y., Gan S.-S. Translational researches on leaf senescence for enhancing plant productivity and quality. J. Exp. Bot. 2014;65:3901–3913. doi: 10.1093/jxb/eru248. [DOI] [PubMed] [Google Scholar]
- 12.Liang C., Wang Y., Zhu Y., Tang J., Hu B., Liu L., Ou S., Wu H., Sun X., Chu J. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Porc. Natl. Acad. Sic. USA. 2014;111:10013–10018. doi: 10.1073/pnas.1321568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitehead C.S., Halevy A.H., Reid M.S. Roles of ethylene and 1-aminocyclopropane-1-carboxylic acid in pollination and wound-induced senescence of Petunia hybrida flowers. Physiol. Plant. 1984;61:643–648. doi: 10.1111/j.1399-3054.1984.tb05184.x. [DOI] [Google Scholar]
- 14.Becker W., Apel K. Differences in gene expression between natural and artificially induced leaf senescence. Planta. 1993;189:74–79. doi: 10.1007/BF00201346. [DOI] [Google Scholar]
- 15.Lutts S., Kinet J., Bouharmont J. NaCl-induced senescence in leaves of rice (Oryza sativa L.) cultivars differing in salinity resistance. Ann. Bot. 1996;78:389–398. doi: 10.1006/anbo.1996.0134. [DOI] [Google Scholar]
- 16.Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P.O., Nam H.G., Lin J.F., Wu S.H., Swidzinski J., Ishizaki K. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005;42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- 17.Munns R. Genes and salt tolerance: Bringing them together. New Phytol. 2005;167:645–663. doi: 10.1111/j.1469-8137.2005.01487.x. [DOI] [PubMed] [Google Scholar]
- 18.Guo Y., GAN S.S. Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 2012;35:644–655. doi: 10.1111/j.1365-3040.2011.02442.x. [DOI] [PubMed] [Google Scholar]
- 19.He Y., Gan S. Molecular characteristics of leaf senescence. Plant Mol. Biol. 2003;1:1–17. [Google Scholar]
- 20.Gan S. Mitotic and postmitotic senescence in plants. Sci. Ageing Knowl. Environ. 2003 doi: 10.1126/sageke.2003.38.re7. [DOI] [PubMed] [Google Scholar]
- 21.Jibran R., Hunter D.A., Dijkwel P.P. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 2013;82:547–561. doi: 10.1007/s11103-013-0043-2. [DOI] [PubMed] [Google Scholar]
- 22.Buchanan-Wollaston V., Ainsworth C. Leaf senescence in Brassica napus: Cloning of senescence related genes by subtractive hybridisation. Plant Mol. Biol. 1997;33:821–834. doi: 10.1023/A:1005774212410. [DOI] [PubMed] [Google Scholar]
- 23.Lim P.O., Kim Y., Breeze E., Koo J.C., Woo H.R., Ryu J.S., Park D.H., Beynon J., Tabrett A., Buchanan-Wollaston V. Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J. 2007;52:1140–1153. doi: 10.1111/j.1365-313X.2007.03317.x. [DOI] [PubMed] [Google Scholar]
- 24.He Y., Tang W., Swain J.D., Green A.L., Jack T.P., Gan S. Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiol. 2001;126:707–716. doi: 10.1104/pp.126.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Y., Cai Z., Gan S. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 2004;27:521–549. doi: 10.1111/j.1365-3040.2003.01158.x. [DOI] [Google Scholar]
- 26.Breeze E., Harrison E., McHattie S., Hughes L., Hickman R., Hill C., Kiddle S., Kim Y.-S., Penfold C.A., Jenkins D. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell. 2011;23:873–894. doi: 10.1105/tpc.111.083345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zentgraf U., Jobst J., Kolb D., Rentsch D. Senescence-related gene expression profiles of rosette leaves of Arabidopsis thaliana: Leaf age versus plant age. Plant Biol. 2004;6:178–183. doi: 10.1055/s-2004-815735. [DOI] [PubMed] [Google Scholar]
- 28.Van der Graaff E., Schwacke R., Schneider A., Desimone M., Flügge U.-I., Kunze R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 2006;141:776–792. doi: 10.1104/pp.106.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregersen P.L., Holm P.B. Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.) Plant Biotechnol. J. 2007;5:192–206. doi: 10.1111/j.1467-7652.2006.00232.x. [DOI] [PubMed] [Google Scholar]
- 30.Parrott D.L., McInnerney K., Feller U., Fischer A.M. Steam-girdling of barley (Hordeum vulgare) leaves leads to carbohydrate accumulation and accelerated leaf senescence, facilitating transcriptomic analysis of senescence-associated genes. New Phytol. 2007;176:56–69. doi: 10.1111/j.1469-8137.2007.02158.x. [DOI] [PubMed] [Google Scholar]
- 31.De Michele R., Formentin E., Todesco M., Toppo S., Carimi F., Zottini M., Barizza E., Ferrarini A., Delledonne M., Fontana P. Transcriptome analysis of Medicago truncatula leaf senescence: Similarities and differences in metabolic and transcriptional regulations as compared with Arabidopsis, nodule senescence and nitric oxide signalling. New Phytol. 2009;181:563–575. doi: 10.1111/j.1469-8137.2008.02684.x. [DOI] [PubMed] [Google Scholar]
- 32.Liu L., Zhou Y., Zhou G., Ye R., Zhao L., Li X., Lin Y. Identification of early senescence-associated genes in rice flag leaves. Plant Mol. Biol. 2008;67:37–55. doi: 10.1007/s11103-008-9300-1. [DOI] [PubMed] [Google Scholar]
- 33.Andersson A., Keskitalo J., Sjödin A., Bhalerao R., Sterky F., Wissel K., Tandre K., Aspeborg H., Moyle R., Ohmiya Y. A transcriptional timetable of autumn senescence. Genome Biol. 2004;5:R24. doi: 10.1186/gb-2004-5-4-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo Y. Towards systems biological understanding of leaf senescence. Plant Mol. Biol. 2013;82:519–528. doi: 10.1007/s11103-012-9974-2. [DOI] [PubMed] [Google Scholar]
- 35.Shinozaki Y., Tanaka T., Ogiwara I., Kanekatsu M., van Doorn W.G., Yamada T. Expression of an AtNAP gene homolog in senescing morning glory (Ipomoea nil) petals of two cultivars with a different flower life span. J. Plant Physiol. 2014;171:633–638. doi: 10.1016/j.jplph.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Guo Y.-J., Wei Q., Kuai B.-K. Cloning and characterization of the AtNAP orthologous in Festuca arundinacea. J. Fudan Univ. (Nat. Sci.) 2010;49:544–551. [Google Scholar]
- 37.Guo Y., Gan S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006;46:601–612. doi: 10.1111/j.1365-313X.2006.02723.x. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y., Qiu K., Kuai B., Ding Y. Identification of an NAP-like transcription factor BeNAC1 regulating leaf senescence in bamboo (Bambusa emeiensis ‘Viridiflavus’) Physiol. Plant. 2011;142:361–371. doi: 10.1111/j.1399-3054.2011.01472.x. [DOI] [PubMed] [Google Scholar]
- 39.Tang X.-R., Zhang R.-K., Chen X., Wu X.-J., Ming F. Characterization of OsNAP from Oryza sativa L. and its application in molecular breeding. J. Fudan Univ. (Nat. Sci.) 2012;51:507–514. [Google Scholar]
- 40.Zhang Y., Cao Y., Shao Q., Wang L., Wang H., Li J., Li H. Regulating Effect of ZmNAP Gene on Anti-senescence and Yield Traits of Maize. J. Henan Agric. Sci. 2012;41:19–24. [Google Scholar]
- 41.Besseau S., Li J., Palva E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012;63:2667–2679. doi: 10.1093/jxb/err450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miao Y., Laun T., Zimmermann P., Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004;55:853–867. doi: 10.1007/s11103-005-2142-1. [DOI] [PubMed] [Google Scholar]
- 43.Zhou X., Jiang Y., Yu D. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells. 2011;31:303–313. doi: 10.1007/s10059-011-0047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jing S., Zhou X., Song Y., Yu D. Heterologous expression of OsWRKY23 gene enhances pathogen defense and dark-induced leaf senescence in Arabidopsis. Plant Growth Regul. 2009;58:181–190. doi: 10.1007/s10725-009-9366-z. [DOI] [Google Scholar]
- 45.Han M., Kim C.-Y., Lee J., Lee S.-K., Jeon J.-S. OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Mol. Cells. 2014;37:532. doi: 10.14348/molcells.2014.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ricachenevsky F.K., Sperotto R.A., Menguer P.K., Fett J.P. Identification of Fe-excess-induced genes in rice shoots reveals a WRKY transcription factor responsive to Fe, drought and senescence. Mol. Biol. Rep. 2010;37:3735–3745. doi: 10.1007/s11033-010-0027-0. [DOI] [PubMed] [Google Scholar]
- 47.Robatzek S., Somssich I.E. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence-and defence-related processes. Plant J. 2001;28:123–133. doi: 10.1046/j.1365-313X.2001.01131.x. [DOI] [PubMed] [Google Scholar]
- 48.Kilbienski I. Untersuchungen zur Funktion und Regulation des Seneszenzassoziierten HvS40-Genes der Gerste (Hordeum vulgare L.) und HvS40 Verwandter Gene von Arabidopsis Thaliana. Christian-Albrechts Universität Kiel; Kiel, Germany: 2007. [Google Scholar]
- 49.Fischer-Kilbienski I., Miao Y., Roitsch T., Zschiesche W., Humbeck K., Krupinska K. Nuclear targeted AtS40 modulates senescence associated gene expression in Arabidopsis thaliana during natural development and in darkness. Plant Mol. Biol. 2010;73:379–390. doi: 10.1007/s11103-010-9618-3. [DOI] [PubMed] [Google Scholar]
- 50.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Humbeck K., Quast S., Krupinska K. Functional and molecular changes in the photosynthetic apparatus during senescence of flag leaves from field-grown barley plants. Plant Cell Environ. 1996;19:337–344. doi: 10.1111/j.1365-3040.1996.tb00256.x. [DOI] [Google Scholar]
- 52.Falk J., Krauß N., Dähnhardt D., Krupinska K. The senescence associated gene of barley encoding 4-hydroxyphenylpyruvate dioxygenase is expressed during oxidative stress. J. Plant Physiol. 2002;159:1245–1253. doi: 10.1078/0176-1617-00804. [DOI] [Google Scholar]
- 53.Haussuhl K. Charakterisierung der Blattseneszenz von Hordeum vulgare L. Unter Besonderer Berucksichtigung des Gens HvS40. Christian-Albrechts-Universität zu Kiel; Kiel, Germany: 1998. [Google Scholar]
- 54.Krupinska K., Haussühl K., Schäfer A., van der Kooij T.A., Leckband G., Lörz H., Falk J. A novel nucleus-targeted protein is expressed in barley leaves during senescence and pathogen infection. Plant Physiol. 2002;130:1172–1180. doi: 10.1104/pp.008565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krupinska K., Dähnhardt D., Fischer-Kilbienski I., Kucharewicz W., Scharrenberg C., Trösch M., Buck F. Identification of WHIRLY1 as a factor binding to the promoter of the stress-and senescence-associated gene HvS40. Plant Growth Regul. 2014;33:91–105. doi: 10.1007/s00344-013-9378-9. [DOI] [Google Scholar]
- 56.Kleber-Janke T., Krupinska K. Isolation of cDNA clones for genes showing enhanced expression in barley leaves during dark-induced senescence as well as during senescence under field conditions. Planta. 1997;203:332–340. doi: 10.1007/s004250050199. [DOI] [PubMed] [Google Scholar]
- 57.Oh S.A., Lee S.Y., Chung I.K., Lee C.-H., Nam H.G. A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol. Biol. 1996;30:739–754. doi: 10.1007/BF00019008. [DOI] [PubMed] [Google Scholar]
- 58.Krause K., Falk J., Humbeck K., Krupinska K. Responses of the transcriptional apparatus of barley chloroplasts to a prolonged dark period and to subsequent reillumination. Physiol. Plant. 1998;104:143–152. doi: 10.1034/j.1399-3054.1998.1040201.x. [DOI] [Google Scholar]
- 59.Scharrenberg C. Untersuchungen zur Transkriptionellen Kontrolle der Genexpression Während der Blattseneszenz der Gerste (Hordeum vulgare L.) Christian-Albrechts Universität Kiel; Kiel, Germany: 2002. [Google Scholar]
- 60.Rushton P.J., Somssich I.E. Transcriptional control of plant genes responsive to pathogens. Curr. Opin. Plant Biol. 1998;1:311–315. doi: 10.1016/1369-5266(88)80052-9. [DOI] [PubMed] [Google Scholar]
- 61.Ciolkowski I., Wanke D., Birkenbihl R.P., Somssich I.E. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008;68:81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Desveaux D., Després C., Joyeux A., Subramaniam R., Brisson N. PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell. 2000;12:1477–1489. doi: 10.1105/tpc.12.8.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Desveaux D., Subramaniam R., Després C., Mess J.-N., Lévesque C., Fobert P.R., Dangl J.L., Brisson N. A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev. Cell. 2004;6:229–240. doi: 10.1016/S1534-5807(04)00028-0. [DOI] [PubMed] [Google Scholar]
- 64.Robatzek S., Somssich I.E. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002;16:1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 66.Bonetta D., McCourt P. Genetic analysis of ABA signal transduction pathways. Trends Plant Sci. 1998;3:231–235. doi: 10.1016/S1360-1385(98)01241-2. [DOI] [Google Scholar]
- 67.Grill E., Himmelbach A. ABA signal transduction. Curr. Opin. Plant Biol. 1998;1:412–418. doi: 10.1016/S1369-5266(98)80265-3. [DOI] [PubMed] [Google Scholar]
- 68.Guiltinan M.J., Marcotte W.R., Jr., Quatrano R.S. A plant leucine zipper protein that recognizes an abscisic acid response element. Science. 1990;250:267. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- 69.Yanagisawa S. Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J. 2000;21:281–288. doi: 10.1046/j.1365-313x.2000.00685.x. [DOI] [PubMed] [Google Scholar]
- 70.Eulgem T., Rushton P.J., Schmelzer E., Hahlbrock K., Somssich I.E. Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J. 1999;18:4689–4699. doi: 10.1093/emboj/18.17.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marè C., Mazzucotelli E., Crosatti C., Francia E., Stanca A.M., Cattivelli L. Hv-WRKY38: A new transcription factor involved in cold-and drought-response in barley. Plant Mol. Biol. 2004;55:399–416. doi: 10.1007/s11103-004-0906-7. [DOI] [PubMed] [Google Scholar]
- 72.Thomas H., Ougham H. The stay-green trait. J. Exp. Bot. 2014;65:3889–3900. doi: 10.1093/jxb/eru037. [DOI] [PubMed] [Google Scholar]
- 73.Gan S., Amasino R.M. Inhibition of leaf senescence by autoregulated production of cytokinin. Science. 1995;270:1986–1988. doi: 10.1126/science.270.5244.1986. [DOI] [PubMed] [Google Scholar]