Abstract
The circadian timing system (CTS) controls various biological functions in mammals including xenobiotic metabolism and detoxification, immune functions, cell cycle events, apoptosis and angiogenesis. Although the importance of the CTS is well known in the pharmacology of drugs, it is less appreciated at the clinical level. Genome-wide studies highlighted that the majority of drug target genes are controlled by CTS. This suggests that chronotherapeutic approaches should be taken for many drugs to enhance their effectiveness. Currently chronotherapeutic approaches are successfully applied in the treatment of different types of cancers. The chronotherapy approach has improved the tolerability and antitumor efficacy of anticancer drugs both in experimental animals and in cancer patients. Thus, chronobiological studies have been of importance in determining the most appropriate time of administration of anticancer agents to minimize their side effects or toxicity and enhance treatment efficacy, so as to optimize the therapeutic ratio. This review focuses on the underlying mechanisms of the circadian pharmacology i.e., chronopharmacokinetics and chronopharmacodynamics of anticancer agents with the molecular aspects, and provides an overview of chronotherapy in cancer and some of the recent advances in the development of chronopharmaceutics.
Keywords: biological rhythms, circadian timing system (CTS), chronopharmacodynamics, chronopharmacokinetics, cancer chronotherapy
1. Introduction
Circadian rhythms are oscillations in the behavior and biochemical reactions of organisms, and occur with a periodicity of approximately twenty-four hours (cf Latin circa, or “about”, and dies, or “day”) [1,2]. Three important properties of the circadian rhythm are: (i) its innate nature, that is, the intrinsic rhythm that exists without any sensory input from the environment; (ii) its capacity for temperature compensation, that is, the maintenance of the intrinsic period, phase, and amplitude of the rhythm despite external fluctuations in temperature, so long as these fluctuations do not interfere with physiological thermoregulation; and (iii) its photoentrainment, that is, the synchronization of the phases of the rhythm with the external light-dark cycles of the solar day [3,4,5,6]. Studies in the field of biological rhythms over the last decade have shown that many aspects of human biology have a circadian rhythm. We wake up, we are active, and we sleep at very similar times each day. Because the environmental changes that accompany the day are so predictable, nature has devised an ingenious system, the circadian clock, to anticipate these changes and regulate physiology and behavior accordingly [3,4,5]. For example, before we wake up, our heart rate and blood pressure increase in anticipation of this stressful event [6]. Likewise, our mental state and capacity for memory and learning, as well as our state of arousal and onset of sleep, are also clock-regulated [7,8,9]. The circadian clock regulates the timing of sleep and wakefulness, and therefore all dependent behavioral and physiological processes. In humans, a defect in the clock gene Period (PER2) produces familial advanced sleep phase syndrome (FASPS) [10,11], and an analogous mutation causes the same phenotype in mice [12]. People with a causal mutation in casein kinase, CSNK1D, and an associated variant, CSNK1E, display advanced sleep phase syndrome (ASPS) and delayed sleep phase syndrome (DSPS), respectively [13,14]. Finally, a human Circadian Locomotor Output Cycles Kaput (CLOCK) variant is associated with diurnal sleep preference [15]. Circadian clock genes are also associated with a host of neurological disorders including schizophrenia [16,17,18,19], unipolar major depression [20], and bipolar disorder [21,22]. Abnormalities in the circadian rhythms are also closely related with the pathophysiology of cancer [23,24,25]. Impairment of the CTS in cancer patients is concerned with many various systemic symptoms with the inclusion of fatigue, sleep disorders, body weight loss due to appetite loss in addition to poor therapeutic outcomes [23,26,27,28].
The toxicity of anticancer agents in healthy cells and the efficacy in malignant tumors may be modified depending on drug administration time in both experimental models and in cancer patients [27,28,29,30,31,32]. When drugs are given at their respective times of best tolerability, improved efficacy is seen [29]. Circadian rhythms of biological functions may affect pharmacokinetics, target organ toxicity and antitumor activity of anticancer agents as a function of dosing time [29,33,34,35]. Several physiological and molecular mechanisms involved in the regulations of pharmacological processes under the control of circadian clock, beginning with the entry of the anticancer drug to the organism and ending with the clinical response and/or toxic effects, have started to emerge only recently [29,32,35,36,37,38]. Today, several chronotherapeutic approaches are successfully applied in the treatment of many cancers [27,32,36,38]. Chronotherapeutics consists in the delivery of medications as a function of circadian rhythms in order to minimize adverse effects and/or to enhance efficacy. Randomised clinical trials revealed that chrono-modulated delivery of anticancer chemotherapy protocol was 5-fold less toxic and nearly twice as effective as compared with constant rate infusion in male patients [39,40]. This review focuses on the underlying mechanisms of the circadian pharmacology of anticancer agents and provides an overview of chronotherapy in cancer and some of the recent advances in the development of chronopharmaceutics.
2. The Circadian Timing System (CTS) and Molecular Mechanisms of Circadian Clock
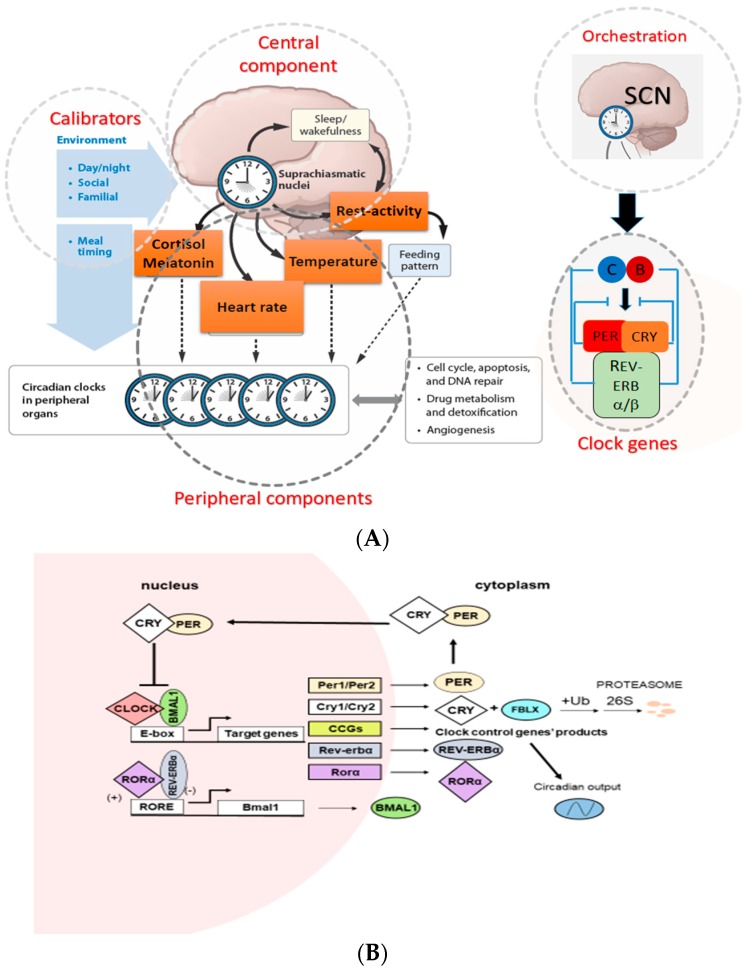
In mammals, the CTS are organized hierarchically, with light-responsive “master” pacemaker clocks located within the suprachiasmatic nucleus (SCN) of the hypothalamus that drive rhythmic cycles within extra-SCN neurons and peripheral tissues. The SCN aligns peripheral tissue clocks with the environmental light cycle through a combination of direct autonomic nervous system efferents and neuroendocrine signals (Figure 1A). At the molecular level, the clockwork of the cell involves several proteins that participate in positive and negative transcriptional feedback loops (Figure 1B). Brain and muscle aryl hydrocarbon receptor nuclear translocator 1 (BMAL1) and CLOCK are transcription factors that contain two basic helix-loop-helix domains and bind E-box elements (CACGTG) in the Period (Per) and Cryptochrome (Cry) genes and thereby exert a positive effect on circadian transcription [41,42,43]. The mammalian PER and CRY proteins act as negative regulators of transcription driven by the BMAL1/CLOCK heterodimer [44,45,46]. PER and CRY form heterodimers that interact with casein kinase Iε (CKIε) and then translocate into nucleus where CRY acts as a negative regulator of BMAL1/CLOCK–driven transcription [47]. In addition to the primary biochemical feedback loop that regulates cycling at the E-box, circadian gene expression is mediated by the transcription at the ROR/REV-ERB and the DBP-E4BP4 (D-box) binding elements NR1Ds and RORs (subfamilies of nuclear hormone receptors) which either activate or repress gene transcription form the ROR components in several clock genes [48]. Microarray studies have addressed the role of specific clock genes in peripheral oscillator function. For example, oscillations were lost in 90% of transcripts when the circadian clock was selectively disrupted in mouse hepatocytes, suggesting that the remaining 10% of rhythms were driven by systemic cues originating outside the liver but impinging on its function [49]. Genetic and bioinformatics studies suggested that there are more clock genes in mammals, which directly regulate core clock mechanisms or posttranslationally regulate core clock protein activity [50,51].
Figure 1.
Hierarchical organization of the CTS and its calibrators in mammals. (A) The CTS consists of three components: an input component, a clockwork and output component. The input component mediated at the macroscopic level by the visual perception of light. Additionally, timing of the meal and social interaction are considered as the input, which they act as Zeitgeber. The second one, the master circadian clock is located in the hypothalamus as a pair of neuron clusters known as the suprachiasmatic nuclei (SCN). The circadian clock in SCN is entrained at precisely 24-h by environmental factors. The output component where SCN synchronizes and coordinates the molecular clocks in the peripheral tissues with a complex interplay between behavioral, neuroendocrine and neuronal pathways as well as SCN-generated physiological rhythms such as body temperature and rest-activity rhythms, heart rate, cortisol and melatonin secretion called as circadian biomarkers. Molecular clocks rhythmically control xenobiotic metabolism and detoxification, cell cycle, apoptosis, DNA repair, and angiogenesis over a 24-h period. Adapted from [30]. (B) Circadian oscillator is composed of interlocked positive and negative transcriptional feedback loops. In positive transcriptional feedback loops this CLOCK or NPAS4 and BMAL1 form a dimer and bind E-box of the clock controlled genes (CCGs) including Per and Cry. Then CRY and PER along with casein kinase ε (CKIε) form a trimeric complex and moves into nucleus, where it suppresses the activity of BMAL:CLOCK activity, which forms negative transcriptional feedback loop. A second feedback loop is formed by the action of orphan receptors, RORα and REV-ERBα proteins, on Bmal1 transcription. RORα and REV-ERB activate and repress transcription of Bmal1 through their competitive action on response elements (ROREs) on the Bmal1 promoter, respectively. Adapted from [52].
3. Experimental Chronopharmacology of Anticancer Drugs
Chronopharmacology is a branch of pharmacology that deal with the biological rhythm dependencies of drugs and examines circadian variations of drug PK (chronoPK), drug PD (chronoPD) and toxicity (chronotoxicity) [1,29,53,54]. The CTS determines the optimal dosing times of several drugs including anticancer drugs via controlling drug metabolism and detoxification, drug transport, bioactivation, elimination, and molecular drug targets that account for the chronopharmacology (chronoPK and chronoPD) (Figure 2) [29,32,35,53]. We review the in vitro and in vivo studies and present possible molecular mechanisms behind the circadian time-dependent PK and PD of anticancer drugs.
Figure 2.
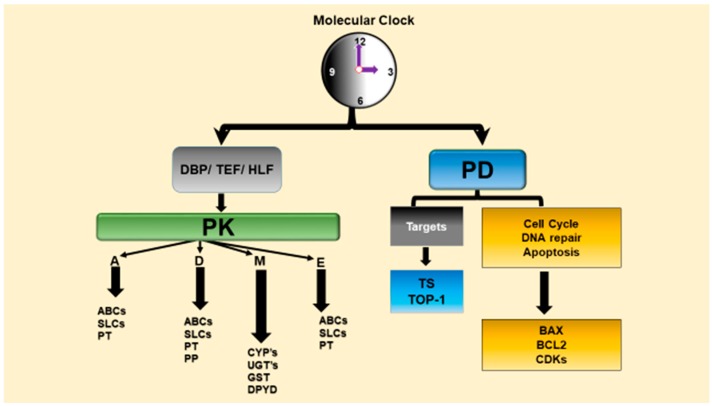
Molecular clock controls PK and PD of anticancer agents. The clock controls main pathways responsible for anticancer drug PK during the 24 h. Circadian clocks also controls several drug targets including cell cycle, DNA repair and apoptosis genes which are contributed to anticancer drugs’ PD. ABC: ATP-Binding Cassette, SLC: Solute Carriers, CYP: Cytochrome, UGT: Uridine diphosphate (UDP) glucronyl transferase, GST: Glutation S-transferase, DPYD: Dihydropyrimidine dehydrogenase, PT: Passive Transport, PP: plasma protein binding, TOP-1: topoisomerase 1, TS: thymidylate synthase, CDK: Cyclin-dependent kinase.
The circadian physiology and the genes involved in the metabolism (e.g., CYPs), transport (e.g., ABC carriers) and elimination of the anticancer drugs, as well as those that are involved in the cell cycle events and apoptosis and other molecular drug targets (e.g., thymidylate synthase for fluoropyrimidines) are controlled by the CTS [23,34,35,55,56,57]. Indeed, diurnal oscillations in drug absorption, distribution, metabolism, and excretion (ADME), as well as daily variations in the sensitivity of molecular drug targets lead to the dosing-time dependent changes in the anticancer drug efficacy and safety [32,34,35,58].
The cell cycle events and apoptosis process in bone marrow and tumor such as WEE1, cyclin-dependent kinase 2 (CDC2), P21, BCL-2, BCL-2-associated X protein (BAX) and/or other molecular drug targets that are controlled by the molecular circadian clocks (Figure 2), and therefore transcriptions of these genes show circadian variations [23,29,32,34,55,56,57]. In fact, a recent genomics studies revealed that 56 of the top 100 best-selling drugs are the target product of a circadian gene in the United States. Moreover, they also showed that the half-lives ≈ 50% of these drugs are less than 6 h [59]. These suggest that PK and PD of these drugs with short half-life should be evaluated according to CTS to find their most effective dosing time. Short elimination half-life drugs will reach the protein in the correct time i.e., the highest expression level of protein, and shortly interact with them. Diurnal oscillations in drug absorption, distribution, metabolism, and excretion, as well as daily variations in the sensitivity of molecular drug targets lead to the dosing-time dependent changes in the drug effectiveness and safety [29,32,58].
3.1. Interaction of Circadian Clock Network with Drug Metabolism, Detoxification and Transport
The CTS modifies ADME of drugs over a 24-h period, which provides the molecular basis for dosing time-dependency of drug toxicity and efficacy [29,32,35]. Biological rhythms in drug metabolism, detoxification and drug transport manifest itself in circadian PK and PD of drugs, hence produce circadian changes in drug effectiveness and toxicity [28]. In mammals, biotransformation/detoxification processes involve chemical changes of xenobiotics/drugs in various organs such as liver, intestine, and kidney. The main results of such metabolism are the increase in xenobiotics/drugs’ water solubility and the facilitation of their excretion into bile, feces and/or urine by the transporters [53]. Phase I metabolism reactions involve enzymes from 27 different genetic families, of which 150 members are responsible for oxidation, reduction and hydrolysis (Class I). In particular, cytochrome P-450 (CYP450) microsomal enzyme superfamily plays a crucial role through its high expression in liver and intestine. Phase II metabolizing or conjugating enzymes such as UDP-glucuronosyl transferases (UGTs), glutathione-S-transferases (GST), sulfotransferases (SULT), and N-acetyl transferases (NAT) are responsible for conjugation (Class II). Phase III components consist of ATP-Binding Cassette (ABC) transporters and solute carrier (SLC) super family which are responsible for cellular efflux and influx of xenobiotics/drugs, respectively (Class III) [29,35,60,61]. In addition, P450 oxidoreductase (POR) and aminolevulinic acid synthase (ALAS1) enzymes take role in the regulation of the activity of most of the Phase I enzymes [35].
Circadian expression of the Phase I, II, and III detoxification proteins are regulated by DBP, HLF, and TEF proteins which are the three members of the PARbZip transcription factor family [62,63,64,65]. PARbZip transcription factors regulate the transcription of numerous key genes involving in the Phase I and Phase II drug metabolism, and drug transport by rhythmically binding to D-box-containing promoters of these genes [64]. Gachon et al. [66] showed that PARbZip-triple knockout mice were highly vulnerable to harmful effects of anticancer drugs. These knockout mice had highly reduced capacity to metabolize xenobiotics [66]. The molecular clocks in liver, kidney and small intestine which are the three pivotal tissue for xenobiotic detoxification, control the circadian expression of these PARbZip transcription factors. In turn, PARbZip proteins govern the circadian expression of three well-known nuclear receptors involved in the control of drug metabolism including constitutive androstan receptor (CAR), peroxisome proliferator activated receptor-alpha (PPAR-α), and aryl hydrocarbon receptor (Ahr) [62,64,65,66,67]. However, how the PARbZip proteins drive the rhythmic expression of pregnane X receptor (PXR) is not known precisely, i.e., mechanisms facilitating rhythmic transcription of PXR targets generally are not well-understood. A recent study by Kriebs et al. [68] revealed that CRY1 interacts with more than half of all mouse nuclear receptors directly, suggesting that direct repression of PXR by CRY1/2 enables circadian modulation of this nuclear receptor, and also pose an additional avenue for circadian clock regulation of rhythmically transcribed other nuclear receptors, including CAR. Gorbacheva et al. [69] indicated that mice with a Bmal1 null allele or a mutation of the Clock gene exhibited enhanced sensibility to the toxic effect of the anticancer drug cyclophosphamide, however mice lacking of two Cry genes were more resistant to the toxic effect. These findings suggest that the circadian clock is directly involved in the chronotoxicity of drugs, via regulating genes involved in drug metabolism directly or through the circadian clock-regulated transcription factors [35]. PARbZip transcription factors also control the rhythmic expression of ALAS1 and POR in the mRNA and/or protein level [66].
Nuclear receptors function as xenobiotic sensors. In response to xenobiotic binding or activation signals induced by xenobiotics, they accumulate in the nucleus and activate the transcription of Phase I, II and III genes involving in the drug metabolism and transport [35,61,70]. The nuclear receptors REV-ERBα/β and RORα/γ are key regulators of core clock function, and many other nuclear receptors are rhythmically expressed under the control of core clock components [64,65,67]. In the RORα/γ double-knock out mice, major effects on gene regulation were observed as activation of Cyp2b9/10, Cyp4a10 and Cyp4a14, Sult1e1/Est and Sult2a9/2a, and suppression of Cyp7b1, Cyp8b1, Hsd3b4 (3β-hydroxysteroid dehydrogenase) and Hsd3b5 [71]. Diurnal mRNA expression profiles of 49 nuclear receptors in the liver, skeletal muscle, white fat and brown fat tissues of mice were investigated in a detailed analysis with High-Throughput Real-Time RT-PCR (Reverse Transcriptase-Polymerase Chain Reaction). More than 50% of nuclear receptors analyzed in the study exhibited rhythmic variations with tissue-specific oscillation in these metabolically active tissues [72]. Two key nuclear receptors, CAR and PPAR-α are responsible for modulating rhythmic expressions of CYP450 and ABC transporters involving in the xenobiotic detoxification [29,65,67,73]. The temporal mRNA oscillation of the CAR is resemble to temporal changes of Cyp1a and Cyp1b mRNA expressions in rat liver [74]. Cyp2b10 activity is highest at night and lowest at daytime in the mouse liver, which is a direct target of CAR [66]. Cyp2a5, Cyp2b10, and Cyp3a11 which show circadian rhythms in their expressions are the best-known targets of CAR nuclear receptor [75].
Transcriptome profiling elicited that PARbZip-deficient mice display a general down-regulation of genes coding enzymes involved in xenobiotic detoxification in the liver and kidney [66]. These genes include the Phase I (Cyp2b, Cyp2c, Cyp2a and Cyp3a), Phase II (Ces3, GSTt1, and GSTa3) and Phase III (Abcg2) group of detoxification components. The expression of POR and ALAS1 is also decreased [66]. Recent findings suggested that PARbZip proteins directly regulate the expression of some of these enzymes by rhythmic binding to their promoter regions, for instance, in the case of Cyp3a4 [76] and Mdr1a (multi-drug resistance 1a or Abcb1a) genes [67]. DBP activates the transcription of the human Cyp3a4, its mouse homolog Cyp3a11, and rat Cyp2c6 genes through binding to their transcriptional start sites [77]. Lavery et al. [78] showed that DBP controls circadian regulation of Cyp2a4 and Cyp2a5 in mouse liver, where in Dbp null mice both genes showed significantly impaired expression. The mammalian CYP450 superfamily is mainly involved in xenobiotic metabolism, eicosanoid, cholesterol and bile acid biosynthesis, and arachidonic acid metabolism [77]. CYP450 is the major enzyme superfamily able to catalyze the oxidative biotransformation of most drugs, accounting for about ~75% of the total metabolism [79,80]. In human hepatoma (HepG2) cell line synchronized with serum shock, DBP activity and CYP3A4 transcription were highest in daytime. Conversely, the CYP3A4 suppressor E4BP4 expression was highest at night [76]. Most of the anticancer drugs that undergo oxidation, reduction, or hydrolysis reactions under Phase I metabolism in the liver, and to some extent, in the intestine, show circadian tolerability [29]. Cyp3a enzyme shows the chronotolerance patterns of seliciclib, docetaxel, irinotecan, mitoxantrone, and vinorelbine by oxidative metabolism [29]. PARbZip-deficient mice are found to be extremely susceptible to anticancer drug-induced toxicity such as cyclophosphamide and mitoxantrone [66]. On the other hand, non-rhythmic Cyp3a13, rhythmic Cyp2b10, and possibly Cyp2c29 participate in the circadian tolerability of cyclophosphamide [66]. The temporal microsomal oxidase and reductase expressions and activities of most of the CYP450 enzymes including Cyp1a, Cyp2a, Cyp3a11, Cyp4a14, Cyp2b10 mRNA levels and activity of Cyp2e1 are higher during the dark (activity) span and lower during the light (rest) span in the liver and kidney of rats or mice [29]. In vitro and in vivo studies involving the circadian rhythms in mRNA expressions, protein levels and activities of CYP450 enzymes are reviewed in Table 1.
Table 1.
This table summarizes the amount and activities of CYP 450 level at both molecular and biochemical levels in various circadian times in mammals along with rhythm parameters from different studies.
| Enzyme | Parameter | Method | Tissue/Cell Line | Species | Strain | Sex | Age | Peak | Trough | Cosinor | ANOVA | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cyp11a1 | mRNA | RT-PCR | Adrenal gland | Mouse | WT | - | - | ZT0 | ZT12 | - | - | [81] |
| Cyp11b1 | mRNA | RT-PCR | Adrenal gland | Mouse | Crem KO | - | - | ZT0 | ZT12 | - | - | [81] |
| Cyp17 | mRNA | Microarray | Liver | Mouse | CD-1 | Male | Mature | ZT14-18 | ZT2 | - | - | [82] |
| Cyp17a1 | mRNA | RT-PCR | Adrenal gland | Mouse | Crem KO | - | - | ZT12 | ZT0 | - | - | [81] |
| Cyp1a1 | mRNA | RT-PCR | Liver | Mouse | Kunming | Male | 8 W | ZT6 | ZT18-22 | p < 0.05 | - | [75] |
| Cyp1a1 | mRNA | RT-PCR | Liver | Mouse | Kunming | Female | 8 W | ZT6 | ZT22 | p < 0.05 | - | [75] |
| Cyp1a1 | mRNA | RT-PCR | Lung | Mouse | WT | - | - | ZT14 | ZT6 | - | p < 0.05 | [83] |
| Cyp1a1 | Protein | Western blot | Lung | Mouse | WT | - | - | ZT18 | ZT6 | - | - | [83] |
| Cyp1a1 | Activity | P450-Glo kit | Lung | Mouse | WT | - | - | ZT18 | ZT2 | - | p < 0.05 | [83] |
| Cyp1a2 | mRNA | RT-PCR | Liver | Mouse | C57BL/6 | Male | 8 W | ZT13 | ZT1 | - | - | [84] |
| Cyp1a2 | mRNA | RT-PCR | Liver | Mouse | Kunming | Male | 8 W | ZT6 | ZT18-22 | p < 0.05 | - | [75] |
| Cyp1a2 | mRNA | RT-PCR | Liver | Mouse | Kunming | Female | 8 W | ZT6 | ZT22 | p < 0.05 | - | [75] |
| Cyp21a1 | mRNA | RT-PCR | Adrenal gland | Mouse | WT | - | - | ZT12 | ZT0 | - | - | [81] |
| Cyp26a1 | mRNA | RT-PCR | Liver | Mouse | ICR | Male | 6 W | ZT6-10 | ZT2 | - | - | [85] |
| Cyp27a1 | mRNA | RT-PCR | Liver | Mouse | Kunming | Male | 8 W | ZT10 | ZT22 | p < 0.05 | - | [75] |
| Cyp27a1 | mRNA | RT-PCR | Liver | Mouse | Kunming | Female | 8 W | ZT10 | ZT22 | p < 0.05 | - | [75] |
| Cyp2a1 | Activity | THA | Testis | Rat | Fischer 344 | Male | 18 W | ZT8 | ZT20 | - | - | [86] |
| Cyp2a4 | mRNA | Microarray | Liver | Mouse | CD-1 | Male | Mature | ZT14-18 | ZT2 | - | - | [82] |
| Cyp2a4 | mRNA | RPA | Liver | Mouse | 129/Ola | Male | 10–16 W | ZT11-15 | ZT23 | - | - | [78] |
| Cyp2a4 | mRNA | RT-PCR | Liver | Mouse | Kunming | Male | 8 W | ZT10 | ZT22 | p < 0.05 | - | [75] |
| Cyp2a5 | mRNA | RPA | Liver | Mouse-NTF | - | - | 10–16 W | ZT13 | ZT0 | - | - | [87] |
| Cyp2a5 | mRNA | RPA | Liver | Mouse-DTF | - | - | 10–16 W | ZT0 | ZT12 | - | - | [87] |
| Cyp2a5 | mRNA | ADDER | Liver | Mouse-NTF | - | - | 10–16 W | ZT16 | ZT4 | - | - | [87] |
| Cyp2a5 | mRNA | ADDER | Liver | Mouse-DTF | - | - | 10–16 W | ZT4 | ZT16 | - | - | [87] |
| Cyp2a5 | mRNA | RBA | Liver | Mouse | 129/Ola | Male | 10–16 W | ZT11-15 | ZT23-3 | - | - | [78] |
| Cyp2b10 | mRNA | Northern blot (PTB induced) |
Liver | Mouse | - | - | - | ZT16 | ZT4 | - | - | [66] |
| Cyp2b10 | mRNA | Northern blot (PTB induced) |
Intestine | Mouse | - | - | - | ZT16 | ZT4 | - | - | [66] |
| Cyp2b10 | mRNA | RT-PCR | Liver | Mouse | Kunming | Male | 8 W | ZT14-18 | ZT22-2 | p < 0.05 | - | [75] |
| Cyp2b10 | mRNA | RT-PCR | Liver | Mouse | Kunming | Female | 8 W | ZT18 | ZT6 | p < 0.05 | - | [75] |
| Cyp2b10 | mRNA | Branched DNA assay | Liver | Mouse | C57BL/6 | Male | 9 W | ZT21 | ZT9 | - | - | [60] |
| Cyp2c11 | mRNA | RT-PCR | Hippocampus | Rat | Wistar | Male | - | ZT2.58 | - | - | p = 0.0017 | [88] |
| Cyp2c11 | mRNA | RT-PCR | Middle cerebral artery | Rat | Wistar | Male | - | ZT5.11 | - | - | p = 0.025 | [88] |
| Cyp2c11 | mRNA | RT-PCR | Iinferior vena cava | Rat | Wistar | Male | - | ZT4.20 | - | - | p = 0.011 | [88] |
| Cyp2c50 | pre-mRNA | RT-PCR | Liver | Mouse | - | - | - | ZT20 | ZT4 | - | - | [66] |
| Cyp2d10 | mRNA | RT-PCR | Liver | Mouse | FVB | Female | 8–12 W | ZT20 | ZT8 | p < 0.0413 | - | [89] |
| Cyp2d22 | mRNA | RT-PCR | Liver | Mouse | FVB | Female | 8–12 W | ZT20 | ZT8 | p < 0.0413 | - | [89] |
| Cyp2e1 | mRNA | Microarray | Liver | Mouse | CD-1 | Male | Mature | ZT14-18 | ZT2 | - | - | [82] |
| Cyp2e1 | Activity | p-Nitrophenol hydroxylation | Liver | Rat | Sprague Dawley | Male | - | ZT12 | ZT0 | - | - | [90] |
| Cyp2e1 | mRNA | RT-PCR | Liver | Rat | Wistar | Male | 8 W | ZT12 | ZT21 | - | p < 0.05 | [91] |
| Cyp2e1 | Protein | Western blot | Liver | Rat | Wistar | Male | 8 W | ZT18 | ZT3 | - | p < 0.01 | [91] |
| Cyp2e1 | Activity | Hydroxylation/HPLC | Liver | Rat | Wistar | Male | 8 W | ZT21 | ZT3 | - | p < 0.05 | [91] |
| Cyp2e1 | mRNA | RT-PCR | Kidney | Rat | Wistar | Male | 8 W | ZT12 | - | - | - | [91] |
| Cyp2e1 | Protein | Western blot | Kidney | Rat | Wistar | Male | 8 W | ZT21 | ZT6 | - | p < 0.05 | [91] |
| Cyp2e1 | Activity | Hydroxylation/HPLC | Kidney | Rat | Wistar | Male | 8 W | ZT21 | ZT6 | - | p < 0.05 | [91] |
| Cyp2e1 | mRNA | RT-PCR | Liver | Mouse | Kunming | Male | 8 W | ZT10 | ZT22 | p < 0.05 | - | [75] |
| Cyp2e1 | mRNA | RT-PCR | Liver | Mouse | Kunming | Female | 8 W | ZT10 | ZT22 | p < 0.05 | - | [75] |
| Cyp2e1 | Activity | p-Nitrophenol hydroxylation | Liver | Mouse-AD | ICR | Male | 7 W | ZT14-18 | ZT2-6 | - | p < 0.01 | [92] |
| Cyp2e1 | Activity | p-Nitrophenol hydroxylation | Liver | Mouse-TRF | ICR | Male | 7 W | ZT22-2 | ZT14 | - | p < 0.01 | [92] |
| Cyp2e1 | mRNA | Branched DNA assay | Liver | Mouse | C57BL/6 | Male | 9 W | ZT17 | ZT5 | - | - | [60] |
| Cyp3a | Activity | EDA | Liver | Rat | Wistar | Male | 10 W | ZT19 | ZT6 | - | - | [93] |
| Cyp3a | Activity | EDA | Liver | Rat | Wistar | Male | 22 W | ZT19 | ZT8 | - | - | [93] |
| Cyp3a11 | mRNA | RT-PCR | Small intestine | Mouse | FVB | Female | 8–12 W | ZT16 | ZT4 | p < 0.0172 | - | [89] |
| Cyp3a11 | mRNA | RT-PCR | Liver | Mouse | ICR | Male | 6 W | ZT10 | ZT22 | - | - | [85] |
| Cyp3a11 | mRNA | RT-PCR | Liver | Mouse | Kunming | Male | 8 W | ZT2 | ZT14-16 | p < 0.05 | - | [75] |
| Cyp3a11 | mRNA | RT-PCR | Liver | Mouse | Kunming | Female | 8 W | ZT2 | ZT14-22 | p < 0.05 | - | [75] |
| Cyp3a11 | mRNA | Branched DNA assay | Liver | Mouse | C57BL/6 | Male | 9 W | ZT21 | ZT1 | - | - | [60] |
| Cyp3a25 | mRNA | RT-PCR | Liver | Mouse | Kunming | Male | 8 W | ZT2 | ZT14-16 | p < 0.05 | - | [75] |
| Cyp3a25 | mRNA | RT-PCR | Liver | Mouse | Kunming | Female | 8 W | ZT2 | ZT14-22 | p < 0.05 | - | [75] |
| Cyp4a10 | mRNA | RT-PCR | Liver | Mouse | Kunming | Male | 8 W | ZT10 | ZT22 | p < 0.05 | - | [75] |
| Cyp4a10 | mRNA | RT-PCR | Liver | Mouse | Kunming | Female | 8 W | ZT18 | ZT6-10 | p < 0.05 | - | [75] |
| Cyp4a14 | mRNA | RT-PCR | Liver | Mouse | Kunming | Male | 8 W | ZT10 | ZT22 | p < 0.05 | - | [75] |
| Cyp4a14 | mRNA | RT-PCR | Liver | Mouse | Kunming | Female | 8 W | ZT10 | ZT22 | p < 0.05 | - | [75] |
| Cyp4a14 | mRNA | Branched DNA assay | Liver | Mouse | C57BL/6 | Male | 9 W | ZT13 | ZT21 | - | - | [60] |
| Cyp4x1 | mRNA | RT-PCR | Hippocampus | Rat | Wistar | Male | - | ZT8.24 | - | p = 0.0016 | [88] | |
| Cyp4x1 | mRNA | RT-PCR | Inferior vena cava | Rat | Wistar | Male | - | ZT18.58 | - | p = 0.0003 | [88] | |
| Cyp51 | mRNA | RT-PCR | Adrenal gland | Mouse | WT | - | ZT16 | ZT4 | - | - | [81] | |
| Cyp51 | mRNA | Northern blot | Liver | Rat | Wistar | Male | 8 W | ZT14-22 | ZT10 | - | - | [94] |
| Cyp7a | mRNA | in situ hybridization | Liver | Rat | Fischer | Male | - | ZT16 | ZT4 | - | - | [95] |
| Cyp7a | mRNA | RT-PCR | Liver | Rat | Wistar | Male | 8 W | ZT16 | ZT4 | - | - | [96] |
| Cyp7a | mRNA | Northern blot | Liver | Rat | Wistar | Male | 8 W | ZT18 | ZT6 | - | - | [94] |
| Cyp7a | Activity | 7α-hydroxylase | Liver | Rat-AD | Wistar | Male | - | ZT14 | ZT6 | - | p < 0.05 | [97] |
| Cyp7a | Activity | 7α-hydroxylase | Liver | Rat-TRF | Wistar | Male | - | ZT7 | ZT19 | - | p < 0.05 | [97] |
| Cyp7a | mRNA | Northern blot | Liver | Rat-AD | Wistar | Male | - | ZT14 | ZT7 | - | p < 0.01 | [97] |
| Cyp7a | mRNA | Northern blot | Liver | Rat-TRF | Wistar | Male | - | ZT7 | ZT14 | - | p < 0.01 | [97] |
| Cyp7a1 | Activity | Enzymatic | Liver | Rat | Wistar | Male | - | ZT16 | ZT0-4 | - | - | [98] |
| Cyp7a1 | mRNA | RT-PCR | Liver | Rat-AD | F344/DuCrj | Male | 7 W | ZT18 | ZT6 | - | p < 0.01 | [99] |
| Cyp7a1 | mRNA | RT-PCR | Liver | Rat-TRF | F344/DuCrj | Male | 7 W | ZT6 | ZT18 | - | p < 0.01 | [99] |
| Cyp7a1 | mRNA | RPA | Liver | Rat | Lewis | - | - | ZT16 | ZT4 | - | - | [100] |
| Cyp7a1 | mRNA | RT-PCR | Liver | Mouse | Kunming | Male | 8 W | ZT10 | ZT22 | p < 0.05 | - | [75] |
| Cyp7a1 | mRNA | RT-PCR | Liver | Mouse | Kunming | Female | 8 W | ZT10 | ZT22 | p < 0.05 | - | [75] |
| Cyp7a1 | mRNA | Northern blot | Liver | Rat | Wistar | Male | - | 10 p.m. | 10 a.m. | - | - | [101] |
| Cyp7a1 | Protein | Western blot | Liver | Rat | Wistar | Male | - | 10 p.m. | 10 a.m. | - | - | [101] |
| Cyp7a1 | Activity | Hydroxylase | Liver | Rat | Wistar | Male | - | 10 p.m. | 10 a.m. | - | - | [101] |
| Cyp7b1 | mRNA | RT-PCR | Liver | Mouse | Kunming | Female | 8 W | ZT6 | ZT22 | p < 0.05 | - | [75] |
| Cyp8b | mRNA | RT-PCR | Liver | Rat | Wistar | Male | 8 W | ZT7-10 | ZT19 | - | - | [96] |
| Cyp8b | mRNA | Northern blot | Liver | Rat | Wistar | Male | 8 W | ZT10 | ZT22 | - | - | [94] |
| Cyp8b | Activity | 12α-hydroxylase | Liver | Rat-AD | Wistar | Male | - | ZT14 | ZT6 | - | p < 0.05 | [97] |
| Cyp8b | Activity | 12α-hydroxylase | Liver | Rat-TRF | Wistar | Male | - | ZT7 | ZT19 | - | p < 0.05 | [97] |
| Cyp8b | mRNA | Northern blot | Liver | Rat-AD | Wistar | Male | - | ZT7 | ZT19 | - | p < 0.01 | [97] |
| Cyp8b | mRNA | Northern blot | Liver | Rat-TRF | Wistar | Male | - | ZT19 | ZT14 | - | p < 0.05 | [97] |
| P450 | Protein | from [102] | Liver | Rat-AD | F344 | - | 10 W | ZT3 | ZT19 | - | p < 0.01 | [103] |
| P450 | Protein | from [102] | Liver | Rat-fasted | F344 | - | 10 W | ZT19-23 | ZT7 | - | p < 0.01 | [103] |
| P450 | Activity | 7ACoD | Liver | Rat | F344 | - | 10 W | ZT19-23 | ZT7 | - | p < 0.01 | [103] |
| P450 | Activity | 7ACoD | Liver | Rat-AD | F344/DuCrj | Male | 7 W | ZT18 | ZT6 | - | p < 0.01 | [99] |
| P450 | Activity | 7ACoD | Liver | Rat-TRF | F344/DuCrj | Male | 7 W | ZT6 | ZT18 | - | p < 0.01 | [99] |
ZT, Zeitgeber Time; RT-PCR, reverse transcription-polymerase chain reaction; WT, wild type; KO, knock-out; THA, Testosterone hydroxylase activity; ADDER, amplification of double-stranded cDNA end restriction fragments; RPA, Ribonuclease Protection Assay; AD, ad libitum; TRF, time-restricted feeding; NTF, night time feeding; DTF, day time feeding; 7ACoD, 7-alkoxycoumarin o-dealkylase; EDA, Erythromycin N-demethylase Activity; PTB, pentobarbital; W, week; Y, year. (-), means “not mentioned”.
The transcriptional level of the major exsorptive ABC transporters and P-gp (P-glycoprotein, MDR1, ABCB1), which play an important role in the excretion of xenobiotics and expels drugs from blood to the intestinal lumen, respectively, are under the control of the CTS. P-gp is indirectly involved into xenobiotic metabolism resulting in the detoxification of potentially harmful substances by such “P-gp-CYP alliance”. Recent studies at the cellular level show that gene expression of ABC transporters including abcb1a (mdr1a), abcb1b (mdr1b), abcc1 (Multidrug resistance-associated protein 1; mrp1), abcc2 (mrp2), abcb4 (mdr2), bcrp1 are clock-controlled in mouse total ileum, ileal mucosa and liver [31,60,104], total jejunum of rats [105,106,107], and liver of monkeys [108]. Abcb1a transcription was shown to be rhythmic in mouse total ileum as a result of its alternative activation by HLF and repression by E4BP4. The role of clock on abcb1a circadian transcription was further demonstrated, since the abcb1a rhythm was disrupted in Clock/Clock mutant mice as compared to wild-type ones [67]. Ando et al. [109] found a clear 24 h rhythmicity in the abcb1a mRNA expression with a peak at ZT12-16 in the liver and intestine of C57BL/6J mice. Additionally, they also showed P-gp activity is circadian dependent where its substrate digoxin concentration was lower at ZT12. Studies involving the circadian rhythms in mRNA expressions, protein levels and activities of ABC transporters are summarized in Table 2.
Table 2.
This table summarizes the amount and activities of ABC transporter level at both molecular and biochemical levels in various circadian times in mammals along with rhythm parameters from different studies.
| Transporter | Parameter | Method | Tissue/Cell Line | Species | Strain | Sex | Age | Peak | Trough | Cosinor | ANOVA | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Abcb1 | mRNA | RT-PCR | Small intestine | Monkey | Cynomolgus | Male | 4–7 Y | ZT3 | ZT9 | - | NS | [108] |
| Abcb1 | mRNA | RT-PCR | Liver | Monkey | Cynomolgus | Male | 4–7 Y | ZT21 | ZT15 | - | NS | [108] |
| Abcb1a | mRNA | RT-PCR | Liver | Mouse | C57BL/6 | Male | 10 W | ZT12-16 | ZT0 | - | p < 0.01 | [109] |
| Abcb1a | mRNA | RT-PCR | Jejunum | Mouse | C57BL/6 | Male | 10 W | ZT12 | ZT0 | - | p < 0.01 | [109] |
| Abcb1a | mRNA | RT-PCR | Kidney | Mouse | C57BL/6 | Male | 10 W | ZT12 | ZT0 | - | NS | [109] |
| Abcb1a | mRNA | RT-PCR | Ileum | Mouse | B6D2F1 | Female | 10 W | ZT15 | ZT3 | - | NS | [33] |
| Abcb1a | mRNA | RT-PCR | Jejunum (proximal) | Rat-AD | Wistar | Male | 8 W | ZT12 | ZT0 | - | p < 0.05 | [105] |
| Abcb1a | mRNA | RT-PCR | Jejunum (proximal) | Rat-TRF | Wistar | Male | 8 W | ZT0 | ZT12 | - | p < 0.01 | [105] |
| Abcb1a | Activity | Digoxin conc./intestinal perfusion | Jejunum (proximal) | Rat-AD | Wistar | Male | 8 W | ZT18 | ZT6 | - | p < 0.05 | [105] |
| Abcb1a | Activity | Digoxin AUC/intestinal perfusion | Jejunum (proximal) | Rat-AD | Wistar | Male | 8 W | ZT18 | ZT6 | - | p < 0.01 | [105] |
| Abcb1a | Activity | Digoxin conc. | Jejunum (proximal) | Rat-TRF | Wistar | Male | 8 W | ZT6 | ZT18 | - | p < 0.05 | [105] |
| Abcb1a | Activity | Digoxin AUC | Jejunum (proximal) | Rat-TRF | Wistar | Male | 8 W | ZT6 | ZT18 | - | p < 0.05 | [105] |
| Abcb1a | mRNA | RT-PCR | Ileum | Mouse | C57BL/6 | Male | - | ZT10 | ZT2 | - | - | [67] |
| Abcb1a | mRNA | RT-PCR | Liver | Mouse | C57BL/6 | - | 8 W | ZT13 | ZT1 | p = 0.016 | p = 0.029 | [111] |
| Abcb1a/1b | mRNA | RT-PCR | Jejunal mucosa | Rat | SD | Male | - | ZT6 | ZT18 | p < 0.05 | p < 0.0001 | [107] |
| Abcb1a/1b | mRNA | Branched DNA assay | Liver | Mouse | C57BL76 | Male | 9 W | ZT16 | ZT0 | - | - | [60] |
| Abcb1b | mRNA | RT-PCR | Liver | Mouse | C57BL/6 | Male | 10 W | ZT16 | ZT0 | - | NS | [109] |
| Abcb1b | mRNA | RT-PCR | Jejunum | Mouse | C57BL/6 | Male | 10 W | ZT20 | ZT0 | - | NS | [109] |
| Abcb1b | mRNA | RT-PCR | Kidney | Mouse | C57BL/6 | Male | 10 W | ZT20 | ZT0 | - | NS | [109] |
| Abcb1b | mRNA | RT-PCR | Ileum | Mouse | B6D2F1 | Female | 10 W | ZT15 | ZT3 | - | p = 0.03 | [33] |
| ABCB1 | mRNA | RT-PCR | Caco-2 | Human | - | - | - | ZT16 | ZT4 | p = 0.013 | - | [112] |
| ABCB1 | mRNA | RT-PCR | Caco-2 | Human | - | - | - | ZT16 | ZT4 | - | - | [113] |
| abcb1a | mRNA | RT-PCR | AdenoCA colon 26 | Mouse | - | - | - | ZT0 | ZT12 | - | - | [67] |
| P-gp | Protein | Western blot | Liver | Mouse | C57BL/6 | - | 8 W | ZT20 | ZT0 | NS | - | [111] |
| P-gp | Protein | Western blot | Liver | Mouse | C57BL/6 | Male | 10 W | ZT8 | ZT20 | - | NS | [109] |
| P-gp | Protein | Western blot | Kidney | Mouse | C57BL/6 | Male | 10 W | ZT0 | ZT16 | - | NS | [109] |
| P-gp | Protein | Western blot | Jejunum | Mouse | C57BL/6 | Male | 10 W | ZT8 | ZT0 | - | p = 0.04 | [109] |
| P-gp | Activity | Digoxin accumulation | Jejunum | Mouse | C57BL/6 | Male | 10 W | ZT12 | ZT0 | - | p < 0.05 | [109] |
| P-gp | Protein | Western blot | Jejunum | Monkey | Cynomolgus | Male | 4–7 Y | ZT21 | ZT9 | - | NS | [108] |
| P-gp | Activity | Quinidine conc. | Brain homogenate | Rat | Wistar | Male | - | ZT8 | ZT20 | - | p < 0.05 | [114] |
| P-gp | Activity | Unbound quinidine in CSF | Microdialysis | Rat | Wistar | Male | - | ZT8 | ZT20 | - | p < 0.05 | [114] |
| P-gp | Protein | Western blot | Ileum | Mouse | C57BL/6 | Male | - | ZT10 | ZT2 | - | [67] | |
| P-gp | Activity | Talinolol intestinal perfusion | Jejunum | Rat-fasted | Wistar | Male | - | ZT13-15 | ZT1-3 | - | p < 0.05 | [115] |
| P-gp | Activity | Talinolol intestinal perfusion | Ileum | Rat-fasted | Wistar | Male | - | ZT13-15 | ZT1-3 | - | p < 0.05 | [115] |
| P-gp | Activity | Losartan intestinal perfusion | Jejunum | Rat-fasted | Wistar | Male | - | ZT13-15 | ZT1-3 | - | p < 0.05 | [115] |
| P-gp | Activity | Losartan intestinal perfusion | Ileum | Rat-fasted | Wistar | Male | - | ZT13-15 | ZT1-3 | - | p < 0.05 | [115] |
| P-gp | Activity | [18F]MC225 PET | Whole brain | Rat | SD | Male | 14–16 W | ZT3-9 | ZT15 | - | p < 0.001 | [116] |
| P-gp | Activity | [18F]MC225 PET | Cortex | Rat | SD | Male | 14–16 W | ZT3-9 | ZT15 | - | p < 0.001 | [116] |
| P-gp | Activity | [18F]MC225 PET | Striatum | Rat | SD | Male | 14–16 W | ZT3 | ZT15 | - | p < 0.01 | [116] |
| P-gp | Activity | [18F]MC225 PET | Hippocampus | Rat | SD | Male | 14–16 W | ZT3 | ZT15 | - | p < 0.01 | [116] |
| P-gp | Activity | [18F]MC225 PET | Cerebellum | Rat | SD | Male | 14–16 W | ZT3-9 | ZT15 | - | p < 0.01 | [116] |
| P-gp | Activity | [18F]MC225 PET | Pons | Rat | SD | Male | 14–16 W | ZT3-9 | ZT15 | - | p < 0.01 | [116] |
| Abcb4 | mRNA | RT-PCR | Liver | Mouse | C57BL/6 | Male | 10 W | ZT8 | ZT20 | - | p = 0.06 | [109] |
| Abcb4 | mRNA | RT-PCR | Jejunum | Mouse | C57BL/6 | Male | 10 W | ZT4 | ZT16 | - | NS | [109] |
| Abcb4 | mRNA | RT-PCR | Kidney | Mouse | C57BL/6 | Male | 10 W | ZT0 | ZT12 | - | NS | [109] |
| Abcb4 | mRNA | Northern blot | Liver | Mouse | Slc:ICR | Male | - | ZT0 | ZT16 | - | - | [106] |
| Abcb4 | mRNA | Branched DNA assay | Liver | Mouse | C57BL/6 | Male | 9 W | ZT4 | ZT16 | - | - | [60] |
| ABCC1 | mRNA | RT-PCR | Caco-2 | Human | - | - | - | ZT10 | ZT0 | - | - | [113] |
| ABCC2 | mRNA | RT-PCR | Caco-2 | Human | - | - | - | ZT12 | ZT0 | - | - | [113] |
| Abcc2 | mRNA | RT-PCR | Liver | Mouse | C57BL/6 | Male | 10 W | ZT12 | ZT0 | - | p < 0.01 | [109] |
| Abcc2 | mRNA | RT-PCR | Jejunum | Mouse | C57BL/6 | Male | 10 W | ZT8 | ZT20 | - | p < 0.01 | [109] |
| Abcc2 | mRNA | RT-PCR | Kidney | Mouse | C57BL/6 | Male | 10 W | ZT12 | ZT0 | - | p = 0.02 | [109] |
| Abcc2 | mRNA | RT-PCR | Liver | Mouse | C57BL/6 | - | 8 W | ZT7 | ZT19 | p = 0.032 | p = 0.045 | [111] |
| Abcc2 | Protein | Western blot | Liver | Mouse | C57BL/6 | - | 8 W | ZT16 | ZT4 | p < 0.05 | [111] | |
| Abcc2 | mRNA | RT-PCR | Ileum mucosa | Mouse | B6D2F1 | Male | - | ZT12 | ZT0 | p = 0.0023 | p = 0.04 | [31] |
| Abcc2 | mRNA | RT-PCR | Ileum mucosa | Mouse | B6D2F1 | Female | - | ZT9 | ZT0 | p = 0.0023 | p = 0.008 | [31] |
| Abcc2 | mRNA | RT-PCR | Ileum mucosa | Mouse | B6CBAF1 | Male | - | ZT12 | ZT0 | p = 0.00026 | p = 0.004 | [31] |
| Abcc2 | mRNA | RT-PCR | Ileum mucosa | Mouse | B6CBAF1 | Female | - | ZT9 | ZT0 | p = 0.00012 | p = 0.004 | [31] |
| Abcc2 | mRNA | RT-PCR | Ileum serosa | Mouse | B6D2F1 | Male | - | ZT4 | ZT20 | NS | NS | [31] |
| Abcc2 | Protein | IHC | Ileum mucosa | Mouse | B6D2F1 | Male | - | ZT12 | ZT15 | - | p < 0.001 | [31] |
| Abcc2 | Protein | IHC | Ileum mucosa | Mouse | B6D2F1 | Female | - | ZT12 | ZT3 | - | p < 0.001 | [31] |
| Abcc2 | Protein | IHC | Ileum mucosa | Mouse | B6CBAF1 | Male | - | ZT15 | ZT12 | - | p < 0.001 | [31] |
| Abcc2 | Protein | IHC | Ileum mucosa | Mouse | B6CBAF1 | Female | - | ZT0 | ZT15 | - | p < 0.001 | [31] |
| Abcc2 | mRNA | RT-PCR | Jejunal mucosa | Rat | SD | Male | - | ZT12 | ZT3 | p < 0.05 | p = 0.001 | [107] |
| Abcc2 | mRNA | Branched DNA assay | Liver | Mouse | C57BL/6 | Male | 9 W | ZT4 | ZT16 | - | - | [60] |
| MRP-2 | Protein | Western blot | Jejunum | Monkey | Cynomolgus | Male | 4–7 Y | ZT21 | ZT9 | - | NS | [108] |
| Abcg2 | mRNA | RT-PCR | Liver | Mouse | Per1 and Per2 knockout | - | 8 W | ZT7 | ZT19 | p = 0.023 | p = 0,049 | [111] |
| Abcg2 | mRNA | RT-PCR | Jejunal mucosa | Rat | SD | Male | - | ZT3 | ZT15 | p < 0.05 | p = 0.04 | [107] |
| Abcg2 | mRNA | Branched DNA assay | Liver | Mouse | C57BL/6 | Male | 9 W | ZT16 | ZT4 | - | - | [60] |
| Abcg2 isoform B | mRNA | RT-PCR | Liver | Mouse | ICR | - | - | ZT6 | ZT18 | - | p < 0.05 | [104] |
| Abcg2 isoform B | mRNA | RT-PCR | Kidney | Mouse | ICR | - | - | ZT10 | ZT18 | - | p < 0.05 | [104] |
| Abcg2 isoform B | mRNA | RT-PCR | Small intestine | Mouse | ICR | - | - | ZT6 | ZT22 | - | p < 0.05 | [104] |
| ABCG2 | Protein | Western blot | Jejunum | Monkey | Cynomolgus | Male | 4–7 Y | ZT15-21 | ZT9 | - | NS | [108] |
| ABCG2 | mRNA | RT-PCR | Caco-2 | Human | - | - | - | ZT12 | ZT0 | - | - | [113] |
| abcg2 | mRNA | RT-PCR | aMoS7 | Mouse | - | - | - | ZT12 | ZT0 | - | - | [104] |
ZT, Zeitgeber Time; NS, not significant; RT-PCR, reverse transcription-polymerase chain reaction; AUC, area under the curve; CSF, cerebrospinal fluid; PET, positron emission tomography; Caco-2, human colon carcinoma cell line; aMoS7, Immortalized small intestine epithelial cells; AdenoCA, adenocarcinoma; AD, ad libitum; TRF, Time restricted feeding; IHC, immunohistochemistry; SD, Sprague Dawley; W, week; Y, year. (-), means “not mentioned”.
SLC transporters are responsible for the influx of drugs in the intestine (absorption), cellular import of xenobiotics to hepatocytes, and reabsorption and excretion of xenobiotics in kidney. Import of xenobiotics to the liver possess particular importance since they metabolize in the hepatocytes. Influx of drugs via organic anion transporter polypeptide 1a2 (Oatp1a2) is important for liver metabolism of drugs. Zhang et al. [60] documented the daily mRNA expression rhythms of basolateral Oatp1a1 (Slco1a1), Oatp1a4 (Slco1a4), Oatp1b2 (Slco1b2), Oatp2b1 (Slco2b1), Oat2; (Slc22a7) and organic cation transporter-1 (Oct1; Slc22a1) in the mouse liver. Oct1 also display significant time dependent oscillation in the kidney, and clock rhythmically drives the renal expression of Oct1 protein through mediation PPARα. Likewise, daily intestinal mRNA expression of Oct novel type 1 (Octn1; Slc22a4) is governed by PPARα [110].
The carboxylesterases Ces1 and Ces2 are controlled rhythmically by CTS through PARbZip proteins in the liver and gastrointestinal tract [29,64]. Rhythmic expressions of Ces1 and Ces2 may be responsible for enhanced biotransformation of irinotecan into SN-38 during day time in male ICR mice [29,117]. Circadian detoxification of irinotecan was also observed in in vitro studies [112,113]. They showed in Caco-2 cells that the bioactivation (CES) and detoxification (UGT1A) enzymes of irinotecan were controlled by clock genes. Bioactivation of irinotecan by CES is maximum near the nadir of its detoxification enzyme UGT1A, which cause increased cytotoxicity. This led enhanced biotransformation to active metabolite SN38 and almost 4-fold change of irinotecan-induced apoptosis depending on the drug administration time [32]. Dihydropyrimidine dehydrogenase (DPYD) is another important circadian-controlled Phase I enzyme, responsible for detoxification of 5-fluorouracil (5-FU) and capecitabine [32]. Its transcription and activity are controlled by CTS in the liver of male B6D2F1 mice. DPYD activity peaks near the middle of the light span, when the animals rest [118]. In mononuclear cells of healthy subjects, and gastrointestinal or nasopharyngeal cancer patients, both DPYD mRNA and activity show circadian rhythms with a maximum at night (rest span), similar with mice [119,120,121]. DPYD activity was also highest at midnight in biopsies of oral mucosa in healthy subjects [122]. The opposite activity phases of DPYD and 5-FU target thymidylate synthase (TS) which is peaked at 1 p.m. in oral mucosa indicate an increased tolerability of 5-FU when administered at night [29]. Circadian rhythms in phase II detoxification by GST contribute to toxicities of platinum complexes [123]. GST catalyze the conjugation of reduced glutathione (GSH) to xenobiotics for detoxification [124]. GSH contents of liver and jejunum are nearly three-fold higher in the second half of the dark span in mice and rats as compared to mid-day, and suppression of GSH synthesis by buthionine sulfoximide greatly modifies the chronotolerance patterns of cisplatin and oxaliplatin in mice [125,126]. In summary, the circadian rhythmic changes in enzyme and transporter function may cause differences in absorption, metabolic degradation and excretion of drugs and might lead to daily bioavailability differences and unexpected outcomes. Furthermore, changing abundance of transporters and enzymes through 24-h may profoundly modify drug pharmacology/toxicology and this has potential implications for improving the tolerability of anticancer drugs. Aforementioned studies carried out by looking specific genes. To understand the effect of CTS at genome-wide level and identify biomarker genes further genomic studies require to be performed in both cancer patient and animal level.
3.2. Relevance of Circadian Rhythms for Chronopharmacodynamics of Anticancer Agents
ChronoPD examines the mechanisms of drug actions in relation to circadian clock and mainly deals with rhythm-dependent differences in the effects of drugs on the body, and rhythmic differences in the susceptibility or sensitivity of a biological target to drugs [53,54]. These administration time differences in the drug effects in the view of PD are due to the biological rhythms in the number and conformation of drug-specific receptors, membrane permeability, free-to-bound plasma fraction of medications, ion channel dynamics, second messengers, rate limiting steps in metabolic pathways, binding enzymatic activities and other drug targets in several molecular signal transduction pathways as well as cell cycle process [29,34,57,127,128]. For instance, dosing time-dependent anti-tumor effect of Interferon-β (IFN-β) is closely related to that of IFN receptors and interferon-stimulated gene factor (ISGF) expressions in tumor cells [129].
The CTS profoundly modifies the PD of anticancer medications in that it controls several molecular drug targets in many biological functions including cellular proliferation, cell cycle events, angiogenesis, DNA repair and apoptosis, and many signal transduction pathways which account for chronoPD in particular for anticancer drugs (see Table 3) [28,29,53,57,127]. Biological rhythms at the cellular level cause significant dosing time-dependent differences in the PD of medications, and thus may modify the efficacy and toxicity of drugs depending on the dosing time [29,112]. Mechanisms of chronotolerance and chronoefficacy involve circadian clock dependencies in anticancer drug PK, as well as circadian rhythms in relevant cellular targets in host and tumor cells. The CTS controls cyclins (CycB1, CycD, CycE), cyclin dependent kinases (CDK1, CDK2), cell cycle genes that gate G1/S or G2/M transitions such as p21, Wee-1, proto-oncogene c-Myc, DNA repair (ATM, AGT, p53), and apoptosis e.g., proapoptotic Bax and antiapoptotic Bcl2 genes (Figure 2) [23,37,128,130,131,132].
Table 3.
The targets of the anticancer drugs that are under the control of circadian clock.
| Target | Parameter | Method | Tissue | Species | Strain | Sex | Peak | Trough | Cosinor | ANOVA | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BCL2 | Protein | Western blot | Bone marrow | Mouse | C3H/HeN | Male | ZT3 | ZT15 | p = 0.024 | - | [56] |
| BCL2 | Protein | Western blot | Bone marrow | MA13/C bearing mouse | C3H/HeN | Male | ZT7 | ZT23 | p = 0.001 | - | [56] |
| BCL2 | Protein | Western blot | Bone marrow | Mouse | B6D2F1 | Male | ZT4–7 | ZT1 | p = 0.025 | - | [56] |
| c-Myc | mRNA | Northern blot | Liver | Mouse | 129/C57BL6 | - | ZT14 | ZT10 | - | - | [133] |
| Cdk-4 | mRNA | Northern blot | Liver | Mouse | 129/C57BL6 | - | ZT18 | ZT10 | - | - | [133] |
| Cyclin D1 | mRNA | Northern blot | Liver | Mouse | 129/C57BL6 | - | ZT14 | ZT22 | - | - | [133] |
| Cyclin A (G2) | Protein | IHC | Oral mucosa | Human | - | Male | ZT20 | ZT4 | p < 0.001 | - | [134] |
| Cyclin B1 (M) | Protein | IHC | Oral mucosa | Human | - | Male | ZT20 | ZT12 | p = 0.016 | - | [134] |
| Cyclin E (G1/S) | Protein | IHC | Oral mucosa | Human | - | Male | ZT16 | ZT4 | p < 0.001 | - | [134] |
| p53 (G1) | Protein | IHC | Oral mucosa | Human | - | Male | ZT12 | ZT0 | p = 0.016 | - | [134] |
| TS | Activity | Tritium release assay | Oral mucosa | Human | - | Male | ZT16 | ZT4 | p = 0.008 | NS | [135] |
| TS | Activity | Tritium release assay | Bone marrow | Mouse | CD2F1 | Female | ZT18 | ZT2 | p < 0.001 | p < 0.001 | [136] |
| TS | Activity | Tritium release assay | Small intestine | Mouse | CD2F1 | Female | ZT6 | ZT18 | p < 0.001 | p < 0.001 | [136] |
| TS | Activity | Tritium release assay | Tumor | Mouse | CD2F1 | Female | ZT6 | ZT14 | p = 0.003 | p < 0.001 | [136] |
| TS | mRNA | PCR | Tumor | Mouse | CD2F1 | Female | ZT2 | ZT14 | NS | NS | [136] |
| TS | Protein | Western blot | Tumor | Mouse | CD2F1 | Female | ZT22 | ZT14 | p < 0.001 | p = 0.008 | [136] |
| Wee-1 | Protein | Western blot | Tumor | Mouse | CD2F1 | Female | ZT14 | ZT18 | p = 0.011 | p = 0.047 | [136] |
| Top-1 | mRNA | RT-PCR | Liver | Mouse | ICR | Male | ZT21 | ZT9 | - | - | [137] |
| Top-1 | mRNA | RT-PCR | Tumor | Mouse | ICR | Male | ZT21 | ZT9 | - | - | [137] |
| Top-1 | Activity | Relaxation of SCP DNA | Sarcoma 180 tumor | Tumor bearing mouse | ICR | Male | ZT5 | ZT13 | - | p < 0.05 | [137] |
ZT, Zeitgeber Time; Cdk-4, cyclin-dependent kinase-4; PCR, polymerase chain reaction; IHC, Immunohistochemistry; Top-1, Topoisomerase-1 enzyme; TS, Thymidylate synthase; MA13/C, Mammary adenocarcinoma; NS, not significant; SCP, Supercoiled plasmid. (-), means “not mentioned”.
Most of the chemotherapeutic agents are toxic for both malignant and healthy cells (bone marrow, oral mucosa, gut, and skin) in the process of cell division [34]. In addition, most of the anticancer agents show their cytotoxic effects at specific phases of the cell division cycle. For example, cells in the S-phase (during DNA synthesis) are more susceptible to anticancer drugs 5-FU [138] and irinotecan [139]. Proportions of cells in S phase and in G2/M phase increase by about 50% in the second half of the darkness period, and in contrast G0/G1 cells predominate during the light period in the bone marrow tissues in mice [56]. Best tolerated circadian time of 5-FU, irinotecan, docetaxel and gemcitabine in mice corresponds to the time when the antiapoptotic BCL2 expression is high, and proapoptotic BAX expression is low, at the early light span in mice [140,141,142,143,144]. Some of the anticancer drugs like oxaliplatin which produces DNA cross links does not have phase specificity [145]. For the optimal timing of successful cancer therapy, it is important to know the time of day when tumors reproducibly express proliferative targets relevant to cancer cell DNA synthesis or cancer cell division.
Pharmacology, and thus efficacy and toxicity of the anticancer drug 5-FU are affected by the circadian rhythms in TS which is the target enzyme of this drug and rhythms in DPYD, the rate limiting enzyme for the catabolism of 5-FU [136]. The enzyme TS catalyzes the synthesis of thymidylate, which is essential for DNA replication. The expression of TS gene is under the control of circadian clock, and TS activity varies throughout the day in normal human and mouse tissues in parallel with tissue S-phase fraction [135,146]. Circadian coordination of tumor TS content and activity result in daily variation in the toxic-therapeutic ratio of the TS-targeted drug 5-FU. In the study of Wood et al. [136], the best time of day (early activity, 2 h after daily arising) for 5-FU treatment to tumor-bearing mice (low host toxicity and high tumor response) was associated with the maximum daily tumor nuclear BMAL-1 protein content and total cell WEE-1 (gates cell mitosis) protein concentration, which was coincident with lowest daily average tumor size and vascular endothelial growth factor (VEGF) content as well as the lowest daily tumor (and also lowest TS activity in bone marrow and gut tissue) TS activity. Tumor TS protein content and TS activity have been used to successfully predict 5-FU tumor response in experimental and human cancers [147,148]. Wood et al. [136] used three established therapeutic targets, mitosis (cell division), TS (DNA synthesis), and VEGF (angiogenesis and growth) as pharmacological determinants of 5-FU. Cell cycle progression is tied to the circadian clock through clock-controlled WEE1, which modulates cyclin-dependent kinases. Inhibition of cyclin-dependent kinase cause cell cycle arrest and subsequent circadian stage-dependent gating of cells at G2-M interface of the cell cycle [55,149]. VEGF protein shows circadian pattern with a peak at 2–6 h after light onset [136,150] leading to variations in the in vivo response to several angiogenesis inhibitors according to the circadian time of administration.
Antitumor effect of IFN-β in nocturnally active mice was found to be more efficient during the early rest phase than during the early active phase [129]. The higher sensitivity to tumor cells was observed when specific binding of IFN receptor and the proportion of tumor cells in S phase increased. The circadian time-dependent effect of IFN-β was also supported by the rhythmic expression of the cell proliferation inhibitor i.e., cyclin-dependent kinase (cdk) inhibitor p21 wild-type p53 fragment 1 (p21WAF1) protein induced by IFN-β [129]. In another study, the dosing time-dependent effect of imatinib mesylate on the ability of tumor growth inhibition and underlying mechanism were investigated in mice from the viewpoint of sensitivity of tumor cells to the drug [151]. Imatinib is an anticancer agent which inhibits the function of various receptors with tyrosine kinase activity, including Abl, the bcr-abl chimeric product, KIT, and platelet-derived growth factor (PDGF) receptors [152,153,154,155]. Tumor growth inhibition was greater when imatinib mesylate (50 mg/kg, i.p.) administered in the early light phase then when it was administered in the early dark phase. The dosing time-dependency of the anti-tumor efficacy of the drug was found to be related with drug-induced inhibition of PDGF receptor activity, but not with KIT or Abl. Anti-tumor activity of imatinib was increased by the delivery of the drug when PDGF receptor activity was enhanced. In other words, rhythmic changes in the tyrosine kinase activity of PDGF receptors on tumor cells seem to influence the anti-tumor efficacy of imatinib [151].
Cao et al. [156] demonstrated that mammalian target of rapamycin (mTOR) pathway was under the control of the circadian clock in the SCN in mice. Using phosphorylated S6 ribosomal protein (pS6) as a marker of mTOR activity, they showed that the mTOR cascade exhibited maximal activity during the subjective day (at CT4; Circadian Time), and minimal activity during the late subjective night [156]. A study by Okazaki et al. [157] has also reported that activity of the mTOR-signaling pathway showed circadian rhythm in renal carcinoma tumors and the 24-h rhythm of mTOR-signaling activity was regulated by protein degradation via Fbxw7 that was controlled by circadian clock systems through DBP. mTOR protein levels in both renal tumor mass and in the normal liver tissue displayed 24-h rhythms, which were higher in the dark phase than in the light phase. Improved antitumor activity was observed when everolimus was administered to tumor-bearing mice during the time of elevated mTOR activity, suggesting that high levels of mTOR activation in renal tumors rendered them more sensitive to everolimus. The survival rate of tumor-bearing mice was higher when everolimus was administered at ZT12 than administered at ZT0 [157]. In a recent preclinical study, dosing time-dependent anti-tumor effect of erlotinib and the underlying molecular mechanisms through the PI3K/AKT (phosphatidylinositol-3-kinase/protein kinase B) and ERK/MAPK (extracellular signal-regulated kinase/mitogen-activated protein kinase) pathways were investigated in nude mice HCC827 tumor xenografts models [158]. Erlotinib is a tyrosine kinase inhibitor that suppresses intracellular phosphorylation of tyrosine kinase related epidermal growth factor receptor (EGFR, whose gene is oncogene) which is a critical member of receptor tyrosine kinase family and is involved in many cancer-related signal transduction pathways including proliferation, metastasis and angiogenesis [159,160]. Rat sarcoma/rapidly accelerated fibrosarcoma/mitogen-activated protein kinase (Ras/Raf/MAPK) and PI3K/AKT are two principal pathways of EGFR downstream, which may promote mitosis and prevent apoptosis [161]. Receptor tyrosine kinase activity of EGFR, and its downstream signal pathways AKT and MAPK showed obvious 24-h oscillation [158,162]. In this study, erlotinib displayed dosing time-dependent anti-tumor activity, which was more effective on the tumor growth inhibiton when administered in the early light phase than in the early dark phase, when the activities of EGFR and its downstream factors increased. Overall, the inhibitory effect of erlotinib on phosphorylation of EGFR, AKT and MAPK changed by the dosing time, providing a clue to optimize the dosing schedule of this drug [158].
In vitro studies has also exhibited the crucial role of cellular rhythms as a major determinants of pharmacological response. Topoisomerase 1 (TOP1) is an enzyme that catalyze the breaking and rejoining of DNA strands in a way which allows the strands to pass through one another, thus altering the topology of DNA. This enzyme is the molecular target of the anticancer agent irinotecan which is active against colorectal cancer. Inhibition of this enzyme by irinotecan results in the DNA breaks and apoptosis [163]. Dulong et al. [112] reported that TOP1 enzyme showed circadian rhythms in the level of mRNA and protein in synchronized colorectal cancer cell cultures. The molecular rhythms were also found in the bioactivation, transport, metabolism, and detoxification processes of the drug leading to profound changes in the pharmacokinetics and pharmacodynamics, and resulting in a four-fold difference in irinotecan induced apoptosis according the circadian drug timing [112,113]. In this study, irinotecan cytotoxicity was found to be positively correlated with the expression level of clock gene BMAL1, both in clock-proficient and clock-deficient cells. In contrast, BMAL1 overexpression inhibited colorectal cancer cell proliferation and enhanced colorectal cancer sensitivity to oxaliplatin [164]. The overall survival of patients with colorectal cancer with high BMAL1 levels in their primary tumors was profoundly longer than that of patients with low BMAL1 levels (27 vs. 19 months; p = 0.04) [164].
3.3. Relevance of Circadian Rhythms for Chronotoxicity of Anticancer Agents
Many anticancer agents show severe toxicities and adverse effects including myelosuppression and reduced immune function, gastrointestinal reactions, liver and kidney dysfunction [165]. Though, chronotherapy approach cannot provide avoidance of all these adverse effects, it can alleviate unfavorable effects for achieving better compliance of patient from the cancer therapy and better treatment response [29,32,127]. The circadian timing of drug administration might be very critical in eliminating the risk of toxicity of anticancer medications, since PK and PD are influenced by the circadian rhythms in the biological functions. Studies in rodents have revealed that the dose of the drug may be lethal when given at the certain times of the day, whereas the same dose may have only little adverse effects when given at other times of the day [29]. In other words, the lethal toxicity of a fixed dose of a drug may vary as a function of dosing time. 24-h variation in drug toxicities has been documented in rodents kept in regular alternation of 12 h of light and 12 h of darkness (LD12:12) [29,32]. The tolerability of more than 40 anticancer drug may vary up to 10-fold as a function of dosing time in rodents as shown with methotrexate [117], cyclophosphamide [69], docetaxel and doxorubicin [140,141,166], vinorelbine [167], gemcitabine and cisplatin [143,168], antimetabolites 5-FU [136,144] and L-alanosine [169], irinotecan [33,142], oxaliplatin [170], seliciclib [171] and etoposide [172]. In a recently performed chronopharmacological study, a second generation nucleotide analogue clofarabine which is used for the treatment of hematological malignancies such as acute leukemia was administered to mice, and time- and dose-dependent toxicity was evaluated by examining biochemical parameters, histological changes, and organ indexes of mice. Clofarabine toxicity in mice when administered in the rest phase was more severe than the active phase, demonstrated by more severe liver damage, immunosuppression, higher mortality rate, and lower LD50 [173].
For an important issue, drug chronopharmacology and chronotoxicity usually display opposite 24-h patterns in nocturnal rodents as compared to human (day active), whose circadian physiology and molecular clock gene expression differ by almost 12 h relative to the light-dark schedule [32,54]. In a clinical study, the relationship between dosing time of gemcitabine and development of hematologic toxicity in cancer patients were revealed with the finding that gemcitabine-induced hematologic toxicity can be decreased by treating cancer patients at 9:00 [174]. The study demonstrated that treatment with gemcitabine at 9:00 decreased the toxicity on white blood cells (WBC) and platelets by nearly 10% as compared to treatment at 15:00. These findings were accompanied with the previous study results of Li et al. [143] performed in nocturnal mice which indicated that tolerability of gemcitabine was better when administered during the early active phase as compared to the late active phase. Thus, the findings of many circadian functions in rodents could be applied to humans by changing the phase of circadian regulation to the anti-phase of it. Researches on the chronopharmacology of anticancer drugs, and chronotoxicity findings in preclinical studies will benefit clinical practice in future. Hence, the undesired effects of anticancer drugs, in particularly the ones with narrow therapeutic index and high potential for adverse effects may be controlled with circadian drug timing.
4. Chronotherapeutics in Cancer Chemotherapy
Chronotherapeutics aims at improving the tolerability and efficacy of treatments, and decreasing the toxicity and undesirable side effects of medications through their delivery considering the circadian rhythms of physiological and biochemical processes. Circadian rhythms in the biological functions, and 24-h rhythm dependencies of drug PK, PD, and drug safety constitutes the rational for chronotherapy. Both the desired/beneficial and undesired/adverse effects of drugs may vary significantly depending on the dosing time. Chronotherapy is especially relevant for anticancer agents [28,29,32,127]. The activity of anticancer drugs may be restricted by their side effects, and their toxicities to healthy host tissues. Most of the antineoplastic agents show their cytotoxic effects on the cells which are at the specific phases of cell division [29]. Circadian timing of anticancer agents often results in predictable changes in drug pharmacology which may translate into clinically relevant differences in treatment outcomes. An overview of administration route of cancer chronotherapeutics in phase II and III clinical trials is presented in Figure 3.
Figure 3.
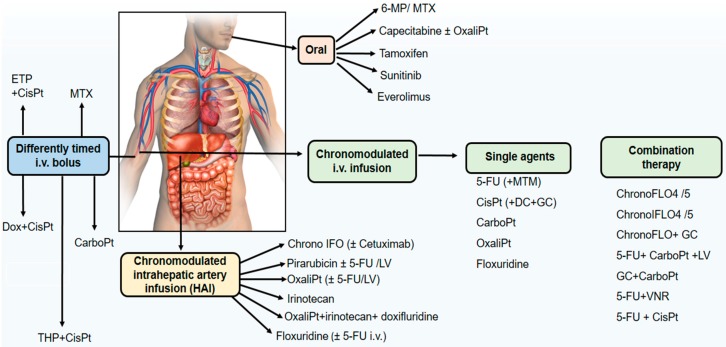
Administration route of cancer chronotherapeutics in phase II and III clinical trials. Anticancer drugs delivered by per oral, buccal, subcutaneous, intravenous (i.v.) and intrahepatic arterial (IHA) routes by using chronotherapeutic approaches. In i.v. administration routes, drugs are applied via chronomodulated fashion with different peak flow times or via i.v. bolus with different circadian time. HAI: Hepatic artery infusion; ETP: Etoposide; CisPT: Cisplatin; MTX: Methotrexate; Dox: Doxorubicin; THP: 4′-0-tetrahydropyranyl doxorubicin; CarboPT: Carboplatin; 6-MP: 6-Mercaptopurine; OxaliPt: Oxaliplatin; Chrono IFO: Chronomodulated irinotecan, 5-fluorouracil, leucovorin and oxaliplatin; LV; Leucovorin; GC: Gemcitabine; DC: Docetaxel; MTM: Mitomycin-C; ChronoFLO: Chronomodulated 5-FU, leucovorin, oxaliplatin; VNR: Vinorelbine.
Administration time of anticancer drugs carries importance because the CTS influences the PK of many anticancer agents and affects clinical outcomes in cancer patients [29]. Strikingly, time course of plasma concentrations of 5-fluorouracil (5-FU) in cancer patients receiving constant rate i.v. infusion for 5 days. The highest plasma concentrations of 5-FU were found around 4:00 a.m. despite constant-rate infusion [34]. Moreover, the best-tolerated time of 5-FU was also found to be 4:00 a.m. in cancer patients with a large regular circadian variation in the 5-FU plasma concentrations and a maximum plasma concentration (Cmax) located at 4:00 a.m. [29,144]. In another study, irinotecan PK and toxicity were evaluated in 31 cancer patients. Irinotecan was administered as a conventional 30-min infusion in the morning or as a chronomodulated infusion from 2:00 a.m. to 8:00 a.m. Patients receiving the chronomodulated infusion, suffered less severe diarrhea and less interpatient variability of irinotecan and SN-38 as compared to conventional therapy [175]. Recent studies also showed that daily pharmacokinetic variations were seen in estrogen receptor modulators and tyrosine kinase inhibitors in mice and cancer patients [89,176,177].
Since antitumor drugs may kill tumor cells as well as normal cells, the maximization of the antitumor effects and the minimization of the toxicity of anticancer agents to normal tissues are substantial in cancer chemotherapy. Chronotherapy may play a vital role in the quality of life and survival rate for cancer patients. A significant improvement in the tolerability and efficacy was demonstrated in the international randomized trials in which cancer patients were treated with the same sinusoidal chronotherapy schedule over 24-h as compared to constant rate infusion [27]. In a randomised multicentre phase III trial which enrolled 186 patients with metastatic colorectal cancer, chronomodulated infusion of oxaliplatin, 5-FU and folinic acid (ChronoFLO) administered to coincide with relevant circadian rhythm in comparison to a constant rate method of infusion. Chronomodulated delivery decreased five-fold the rate of severe mucosal toxicity as compared to constant drug delivery (14% vs. 76%), and halved that of functional impairment from the peripheral sensitive neuropathy (16% vs. 31%). Median time to treatment failure was found to be 6.4 months on chronotherapy and 4.9 months on constant rate infusion [178]. In another randomized multicenter trial, irinotecan was given as standard conventional 30-min infusion in the morning or as a chronomodulated infusion from 2:00 a.m. to 8:00 a.m., with peak delivery rate at 5:00 a.m. to 31 cancer patients with metastatic colorectal cancer. In this study, irinotecan-induced severe diarrhea episodes was fewer in chronomodulated therapy than conventional treatment [175]. Several other Phase I to Phase III clinical trials have been validated the relevance of circadian timing of anticancer medications and confirmed the efficacy of chronomodulated therapy in different kinds of tumors [179,180,181] Randomised clinical trials revealed that the gender has a prominent role for tolerability and efficacy of chronotherapeutics. In three clinical trials involving advanced or metastatic colorectal or lung cancer, chronoFLO or chronomodulated 5-FU-LV-carboplatin produced significantly more adverse events in women as compared to men [40,178]. Recent clinical studies evaluating the role of circadian timing on anticancer drug pharmacology and treatment outcomes are exemplified in Table 4.
Table 4.
Examples from the literature on relationship between circadian clock and cancer treatment at clinical level.
| Anticancer Drug(s) | Cancer Type | Study Design | Dose/Chronomodulated Schedule | Main Pharmacological Findings | Main Clinical Findings | Reference |
|---|---|---|---|---|---|---|
| Pharmacokinetics/Pharmacodynamics | Efficacy/Toxicity/Adverse Effects | |||||
| Cisplatin (combined with Gemcitabine and Docetaxel) | Non-small cell lung cancer | Randomized controlled study, pharmacokinetic analysis | Cisplatin 30-min i.v. infusion at 06:00 (morning) and 18:00 (evening) | Total and unbound platin CL 18:00 > 06:00 |
Leucopenia, neutropenia and nausea symptoms lower at 18:00 | [187] |
| Irinotecan + Oxaliplatin + 5-FU (combined with cetuximab) | Colorectal cancer with liver metastases | Pharmacokinetic study | Hepatic artery infusion (chronomudulated) Irinotecan (180 mg/m2)-6 h sinusidal infusion peak at 05:00 h, Oxaliplatin (85 mg/m2)-12 h sinusoidal infusion peak at 16:00, 5-FU (2800 mg/m2)-12 h sinusoidal infusion peak at 04:00 h | Good correlation between AUC of irinotecan, SN-38, ultrafiltrated Pt and leukopenia. The AUC and Cmax of ultrafiltrated Pt were significantly correlated with the severity of diarrhea | - | [183] |
| 5-FU, LV and oxaliplatin | Colon or rectum cancers with metastases | Meta-analysis of three Phase III trials | ChronoFLO = 5-FU-LV from 2215 to 09:45 h with a peak at 04:00 h, and oxaliplatin from 10:15 to 21:45 h with a peak at 1600 h. | - | Overall survival was higher in males on chronoFLO when compared with CONV (p = 0.009) |
[188] |
| Capecitabine (combined with radiotherapy) | Rectal cancer | Prospective single-center, single-arm Phase II study | Oral Capecitabine (1650 mg/m2) 50% dose at 8:00 (morning) and 50% dose at 12:00 (noon)-BRUNCH | - | No Grade 2–3 toxicity of hand-foot syndrome, thrombocytopenia, diarrhea and mucositis. There were no grade IV toxicities | [189] |
| Capecitabine + Oxaliplatin | Treatment-naïve colorectal cancer patients with metastatic disease | Prospective single-center, single-arm, Phase II study | Oral Capecitabine (2000 mg/m2) 50% dose at 8:00 (morning) and 50% dose at 12:00 (noon)-BRUNCH | AUC0–4 h of Capecitabine 08:00 > 12:00 Cmax of Capecitabine 08:00 > 12:00 | Lower incidence of hand-foot syndrome | [190] |
| Tamoxifen | Breast cancer | Pharmacokinetic cross-over study | Oral 20 or 40 mg once a day at 8:00 or 13:00 or 20:00 | AUC0–8 h and Cmax of tamoxifen 08:00 > 20:00 (20%) tamoxifen tmax 08:00 < 20:00 Systemic exposure (AUC0–24 h) to endoxifen 08:00 > 20:00 (15%) |
- | [89] |
| Everolimus (combined with exemastan and tamoxifen) | Metastatic breast cancers | - | Oral morning or evening administration | - | Morning administration of everolimus minimize metabolic alteration and fatigues. No pneumonitis after morning administration | [191] |
| Sunitinib | Advanced clear cell renal cell carcinoma, Pancreatic neuro-endocrine tumors | Prospective randomized crossover study | 8:00. (morning), 13:00 (noon) and 18:00. (evening) | Ctrough 13:00. or 18:00 p.m. > Ctrough 8:00 p = 0.006 |
- | [176] |
| Linifanib | Advanced or metastatic solid tumors and refractory to standard therapy | Phase I, open-label, randomized, crossover study | 0.25 mg/kg (maximum 17.5 mg) morning or evening | Cmax morning > evening (p < 0.01) | - | [192] |
| Radiotherapy | Bone metastases | Cohort study | Treatment times are 08:00–11:00, 11:01–14:00 or 14:01–17:00 | - | Females in the 11:01 to 14:00 cohort exhibited higher response rate | [193] |
i.v., intravenous; CL, clearance; 5-FU, 5-fluorouracil; LV, leucovorin; CONV, conventional therapy; Cmax, maximum plasma concentration; tmax, time to reach Cmax; AUC, Area under curve; Cthrough, Trough plasma level after reaching steady-state concentration of the drug in repetitive administration; (-), means “not mentioned”.
5. Chronopharmaceutics and Innovative Chrono-Drug Delivery Systems in Cancer Treatment
The CTS may substantially influence the PK and PD of medications when injected, infused, inhaled or applied orally and dermally at the different circadian times of the day [38,182]. It has become indispensable to develop a drug delivery system which release the anticancer drug at the target site and appropriate time to achieve an improved progress in treatment outcomes. “Chronopharmaceutics” are new drug delivery systems which are used to synchronize drug concentrations according to circadian rhythms of the biological functions and drug pharmacology in the organism [182]. The fundamental aim of chronopharmaceutics and chrono-drug delivery systems (ChronoDDS) are to deliver drugs in the time of greatest need with the dose providing higher concentration and when the need is less providing lesser concentration for minimizing side effects and optimizing treatment outcomes of medications [29,182]. Today, the technologies available for chronopharmaceutics are programmable-in-time infusion pumps (chronomodulating infusion pumps), modified release (MR) of oral drugs, toward rhythm-sensing drug-releasing nanoparticles [38,127]. More attempts are being made for adjusting DDSs accurately to cancer patient needs, in terms of both treatment efficacy and compliance.
Conventional infusion protocols and drug formulations in the cancer treatment only consider drug doses, duration and frequency of the infusions keeping the in vivo drug concentration in the therapeutic level for a prolonged period of time. In contrast, circadian chronomodulated pulsatile drug delivery systems are characterized by a programmable lag phase after which the drug release is triggered automatically as time-controlled taking into account the rhythmic determinants in disease pathophysiology, chronopharmacology of drugs, and the CTS to identify the drug delivery pattern, dose and dosing time [38,182]. Pulsatile DDSs have the capability of releasing the drug after a predetermined lag period in pulsed or releasing in a controlled manner offering solutions for delivery of drugs which require nocturnal dosing. These systems have drawn the attention of both industry and academic research recently. Chronomodulating infusion pumps present a new infusion parameter which is called “the time of peak-flow rate” since the drug delivery pattern is not constant, i.e., it is rather semi-sinusoidal, with an increasing flow rate, a peak administration rate at a time specification and a gradual symmetric decrease in flow rate [53].
Chronotherapeutic approaches to drug delivery have triggered the industrial development of non-implantable multichannel programmable-in-time pumps for the treatment of cancer. Invention of the IntelliJectTM device with four 30-mL reservoirs which was approved in the European Union and in North America enabled the development of first circadian chronomodulated combination delivery schedule of 5-FU, leucovorin, and oxaliplatin used safely and with increased efficacy [29,38]. MélodieTM, a four-channel programmable pumps with the advantages of increased energy autonomy, flexible reservoir capacity, rapid programming of any delivery schedule and computer storage of treatment protocols and patient data. The direct administration of anticancer drugs into the hepatic artery has been shown to be beneficial in terms of both tolerability and antitumor activity on liver metastases from colorectal cancer. Since liver is one of the most rhythmic organs in the body, high differential cytotoxicity is expected from the direct chronomodulated infusion of chemotherapy into the hepatic artery that vascularizes liver metastases. Thus, liver metabolism and susceptibility are expected to be rhythmic, while liver metastases are expected to display no such robust circadian organization any more. An international trial (OPTILIV: Optimal control of liver metastases with hepatic artery infusion of 5-FU, oxaliplatin and irinotecan chemotherapy and systemic cetuximab) revealed that combination of agents by intrahepatic artery infusion elicited safe and effective therapy [183,184].
Chronomodulated infusions of chemotherapy have been given to non-hospitalized patients using a more recent developed elastomeric pump with programmed rhythmic electronic control of infusion flow rate, which is named CIPTM. These innovative chrono-drug delivery systems provide a chronomodulated delivery of up to four drugs over one to several days in out-patients and with minimal nursing need [29,30].
Recently, novel drug loaded nanocarriers have been searched for chronotherapy. Chronotherapy with drug-containing nanoformulation appears to be a new therapeutic strategy which may improve cancer curability with decreased side effects, costs, and risks for the cancer patients [185]. Chronopharmaceuticals coupled with nanotechnology could be the future of DDS, and lead to safer and more efficient disease therapy in the future [182]. In a recent study, synergistic anti-tumor efficacy of paclitaxel (PTX)-loaded polymeric nanoparticles (PTX-NPs) combined with circadian chronomodulated chemotherapy in xenografted human lung cancer was examined to screen out to best time of the day for the drug delivery [186]. The drug in PTX-NPs displayed an initial fast release and subsequently a slower and sustained release. Chronomodulated delivery of PTX-NPs exhibited greatest anti-tumor activity than that of PTX injection, and showed a time-dependent effect which was the best at 15 h after light onset, suggesting a potential treatment for lung cancer [186]. The synergistic mechanism was suggested to be related to the inhibition of tumor cell proliferation through its action on Ki-67, and decreased microvessel density associated with CD31 and promoted cell apoptosis [186]. New chronopharmaceutical technologies are underway of development and testing for delivering anticancer agents precisely in a circadian time-dependent manner by bedside or ambulatory pumps for cancer treatment.
6. Conclusions and Perspectives
Chronotherapy has become an intriguing research subject in recent years particularly in cancer, rheumatology, neurological/psychiatric disorders, and cardiovascular diseases. However, chronotherapeutic approaches do not reach to the desired level in the pharmacotherapy. Pharmaceutical industry, local drug authorities and clinicians should consider the effects of circadian rhythms on the efficacy and/or tolerability of drugs, and drug dosing time which is a critical factor should not be underestimated. The clinicians should more focus on this issue and perform new clinical trials to show benefit of chronotherapy in cancer, cardiovascular and other diseases.
The circadian rhythms in the biological functions and circadian physiology, clock genes as well as cell cycle events may substantially affect the cancer chemotherapy depending on the drug delivery schedule. Yet, many factors including gender, age, physical activity, disease states, genetic polymorphism and phenotypic properties of an individual result in the inter-individual and intra-individual differences in drug exposure and pharmacokinetic parameters leading to a large variability in the therapeutic response [27,29,194].
To establish a rational chronotherapeutic strategy for choosing the appropriate dosing time, it is compulsory to identify the CTS status and predict optimal treatment timing in cancer patients. With respect to this translational point, Ortiz-Tudela and co-workers proposed monitoring of temperature, activity and position of metastatic gastrointestinal cancer patients by employing inner wrist surface Temperature, arm Activity and Position (TAP) [195]. Pursuing of circadian biomarkers seems smart approach but clinical validation is necessary in that inter- and intra-individual changes for these parameters. Identification of the potential rhythmic key determinants of anticancer drug chronopharmacology associated with the drug metabolism and detoxification, molecular drug targets and their molecular clock control are necessary for optimally shaping the circadian drug delivery patterns to individual CTS in cancer patients. CYCLOPS i.e., cyclic ordering by periodic structure is very recently proposed algorithm by Anafi and coworkers [196]. To develop the algorithm, large human liver and lung biopsy as well as mouse tissue samples were collected and rhythmic transcripts of many drug targets and disease gene were identified. In vivo validation of the method was carried out by using streptozocin which destroys pancreatic beta cells. The authors showed that the dosage time can temporally segregate efficacy from dose-limiting toxicity of streptozocin. The TAP approach and CYCLOPS algorithm will enable us to assess the effectiveness and safety of the chemotherapy for individual patients, which bring a personalized chrono-chemotherapy.
We may claim that randomized clinical trials employed chronotherapy for cancer are limited as compared to other clinical studies. Further studies should be performed for better understanding of clinical outcomes of cancer chronotherapy in particular women cancer patients. Non-invasive and clinically validated techniques for determining the CTS status, data based mathematical modelling, system chronopharmacological and data based translational approaches will help achieving better efficacy and acceptable adverse effect of drugs in particular anticancer agents. New chronopharmaceutical and chrono-drug delivery products enable to clinicians for chronotherapeutic applications in cancer and other diseases.
Abbreviations
| 5-FU | 5-Fluorouracil |
| ABC | ATP-Binding Cassette |
| AGT | O6-alkylguanine transferase |
| Ahr | Aryl hydrocarbon receptor |
| ALAS1 | Aminolevulinic acid synthase |
| ASPS | Advanced sleep phase syndrome |
| ATM | Ataxia telangiectasia mutated |
| BAX | BCL-2-associated X protein |
| BMAL1 | Brain and muscle aryl hydrocarbon receptor nuclear translocator 1 |
| CAR | Constitutive androstan receptor |
| CCGs | Clock controlled genes |
| CDC2 | Cyclin-dependent kinase 2 |
| CES | Carboxylesterase |
| ChronoDDS | Chrono-drug delivery system |
| ChronoPD | Chronopharmacodynamics |
| ChronoPK | Chronopharmacokinetics |
| CKIε | Casein kinase Iε |
| CLOCK | Circadian locomotor output cycles kaput |
| Cmax | Maximum plasma concentration |
| Cry | Cryptochrome |
| CTS | Circadian timing system |
| Cyc | Cyclin |
| CYCLOPS | Cyclic ordering by periodic structure |
| CYP450 | Cytochrome P-450 |
| DBP | Albumin D-site-binding protein |
| DDS | Drug delivery systems |
| DPYD | Dihydropyrimidine dehydrogenase |
| DSPS | Delayed sleep phase syndrome |
| EGFR | Epidermal growth factor receptor |
| ERK/MAPK | Extracellular signal-regulated kinase/mitogen-activated protein kinase |
| FASPS | Familial advanced sleep phase syndrome |
| GSH | Reduced glutathione |
| GST | Glutathione-S-transferases |
| HLF | Hepatic leukemia factor |
| Hsd3b | 3β-hydroxysteroid dehydrogenase |
| IFN-β | Interferon-β |
| ISGF | Interferon-stimulated gene factor |
| MDR1 | Multidrug resistance protein 1 (ABCB1, P-glycoprotein, P-gp in human) |
| Mdr1 | Multidrug resistance protein 1 (abcb1a/b mdr1a/b, P-gp in rodents) |
| MR | Modified Release |
| Mrp | Multidrug resistance-associated protein (Abcc) |
| mTOR | Mammalian target of rapamycin |
| NAT | N-acetyl transferases |
| NFIL3/E4BP4 | Nuclear factor, interleukin 3 regulated |
| NPAS2 | Neuronal PAS-domain protein 2 |
| Oatp | Organic anion transporter polypeptide |
| Oct | Organic cation transporter |
| Octn1 | Oct novel type 1 |
| p21WAF1 | p21 wild-type p53 fragment 1 |
| PARbZip | Proline-acidic amino acid-rich basic leucine zipper |
| PD | Pharmacodynamics |
| PDGF | Platelet-derived growth factor |
| Per | Period |
| P-gp | P-glycoprotein |
| PI3K/AKT | Phosphatidylinositol-3-kinase/protein kinase B |
| PK | Pharmacokinetics |
| POR | P450 oxidoreductase |
| PPAR-α | Peroxisome proliferator activated receptor-alpha |
| PTX | Paclitaxel |
| PTX-NPs | PTX-loaded polymeric nanoparticles |
| PXR | Pregnane X receptor |
| REV-ERB/NR1D | Reverse strand of ERB |
| RORs | Retinoic acid-receptor-related orphan receptor |
| SCN | Suprachiasmatic nuclei |
| SLC | Solute carrier |
| SULT | Sulfotransferases |
| TEF | Thyrotroph embryonic factor |
| Top1 | Topoisomerase 1 |
| TS | Thymidylate synthase |
| UDP | Uridine diphosphate |
| UGT | Glucuronosyl transferase |
| VEGF | Vascular endothelial growth factor |
| WBC | White blood cells |
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Smolensky M.H., Peppas N.A. Chronobiology, drug delivery, and chronotherapeutics. Adv. Drug Deliv. Rev. 2007;59:828–851. doi: 10.1016/j.addr.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Schulz P., Steimer T. Neurobiology of circadian systems. CNS Drugs. 2009;23(Suppl S2):3–13. doi: 10.2165/11318620-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 3.Baggs J.E., Hogenesch J.B. Genomics and systems approaches in the mammalian circadian clock. Curr. Opin. Genet. Dev. 2010;20:581–587. doi: 10.1016/j.gde.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gachon F., Nagoshi E., Brown S.A., Ripperger J., Schibler U. The mammalian circadian timing system: From gene expression to physiology. Chromosoma. 2004;113:103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- 5.Green C.B., Takahashi J.S., Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kovac J., Husse J., Oster H. A time to fast, a time to feast: The crosstalk between metabolism and the circadian clock. Mol. Cells. 2009;28:75–80. doi: 10.1007/s10059-009-0113-0. [DOI] [PubMed] [Google Scholar]
- 7.Eckel-Mahan K.L., Storm D.R. Circadian rhythms and memory: Not so simple as cogs and gears. EMBO Rep. 2009;10:584–591. doi: 10.1038/embor.2009.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franken P., Dijk D.J. Circadian clock genes and sleep homeostasis. Eur. J. Neurosci. 2009;29:1820–1829. doi: 10.1111/j.1460-9568.2009.06723.x. [DOI] [PubMed] [Google Scholar]
- 9.Kyriacou C.P., Hastings M.H. Circadian clocks: Genes, sleep, and cognition. Trends Cogn. Sci. 2010;14:259–267. doi: 10.1016/j.tics.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Toh K.L., Jones C.R., He Y., Eide E.J., Hinz W.A., Virshup D.M., Ptacek L.J., Fu Y.H. An hper2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 11.Vanselow K., Vanselow J.T., Westermark P.O., Reischl S., Maier B., Korte T., Herrmann A., Herzel H., Schlosser A., Kramer A. Differential effects of per2 phosphorylation: Molecular basis for the human familial advanced sleep phase syndrome (fasps) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y., Toh K.L., Jones C.R., Shin J.Y., Fu Y.H., Ptacek L.J. Modeling of a human circadian mutation yields insights into clock regulation by per2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weitzman E.D., Czeisler C.A., Coleman R.M., Spielman A.J., Zimmerman J.C., Dement W., Richardson G., Pollak C.P. Delayed sleep phase syndrome. A chronobiological disorder with sleep-onset insomnia. Arch. Gen. Psychiatry. 1981;38:737–746. doi: 10.1001/archpsyc.1981.01780320017001. [DOI] [PubMed] [Google Scholar]
- 14.Takano A., Uchiyama M., Kajimura N., Mishima K., Inoue Y., Kamei Y., Kitajima T., Shibui K., Katoh M., Watanabe T., et al. A missense variation in human casein kinase i epsilon gene that induces functional alteration and shows an inverse association with circadian rhythm sleep disorders. Neuropsychopharmacology. 2004;29:1901–1909. doi: 10.1038/sj.npp.1300503. [DOI] [PubMed] [Google Scholar]
- 15.Katzenberg D., Young T., Finn L., Lin L., King D.P., Takahashi J.S., Mignot E. A clock polymorphism associated with human diurnal preference. Sleep. 1998;21:569–576. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- 16.Lamont E.W., Coutu D.L., Cermakian N., Boivin D.B. Circadian rhythms and clock genes in psychotic disorders. Isr. J. Psychiatry Relat. Sci. 2010;47:27–35. [PubMed] [Google Scholar]
- 17.Lamont E.W., Legault-Coutu D., Cermakian N., Boivin D.B. The role of circadian clock genes in mental disorders. Dialogues Clin. Neurosci. 2007;9:333–342. doi: 10.31887/DCNS.2007.9.3/elamont. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mansour H.A., Wood J., Logue T., Chowdari K.V., Dayal M., Kupfer D.J., Monk T.H., Devlin B., Nimgaonkar V.L. Association study of eight circadian genes with bipolar i disorder, schizoaffective disorder and schizophrenia. Genes Brain Behav. 2006;5:150–157. doi: 10.1111/j.1601-183X.2005.00147.x. [DOI] [PubMed] [Google Scholar]
- 19.Moons T., Claes S., Martens G.J., Peuskens J., Van Loo K.M., Van Schijndel J.E., De Hert M., van Winkel R. Clock genes and body composition in patients with schizophrenia under treatment with antipsychotic drugs. Schizophr. Res. 2011;125:187–193. doi: 10.1016/j.schres.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Soria V., Martinez-Amoros E., Escaramis G., Valero J., Perez-Egea R., Garcia C., Gutierrez-Zotes A., Puigdemont D., Bayes M., Crespo J.M., et al. Differential association of circadian genes with mood disorders: Cry1 and npas2 are associated with unipolar major depression and clock and vip with bipolar disorder. Neuropsychopharmacology. 2010;35:1279–1289. doi: 10.1038/npp.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benedetti F., Dallaspezia S., Colombo C., Pirovano A., Marino E., Smeraldi E. A length polymorphism in the circadian clock gene per3 influences age at onset of bipolar disorder. Neurosci. Lett. 2008;445:184–187. doi: 10.1016/j.neulet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 22.McCarthy M.J., Welsh D.K. Cellular circadian clocks in mood disorders. J. Biol. Rhythms. 2012;27:339–352. doi: 10.1177/0748730412456367. [DOI] [PubMed] [Google Scholar]
- 23.Fu L., Kettner N.M. The circadian clock in cancer development and therapy. Prog. Mol. Biol. Transl. Sci. 2013;119:221–282. doi: 10.1016/B978-0-12-396971-2.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zee P.C., Attarian H., Videnovic A. Circadian rhythm abnormalities. Continuum (Minneap Minn) 2013;19:132–147. doi: 10.1212/01.CON.0000427209.21177.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuhr L., Abreu M., Pett P., Relogio A. Circadian systems biology: When time matters. Comput. Struct. Biotechnol. J. 2015;13:417–426. doi: 10.1016/j.csbj.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mormont M.C., Waterhouse J., Bleuzen P., Giacchetti S., Jami A., Bogdan A., Lellouch J., Misset J.L., Touitou Y., Levi F. Marked 24-h rest/activity rhythms are associated with better quality of life, better response, and longer survival in patients with metastatic colorectal cancer and good performance status. Clin. Cancer Res. 2000;6:3038–3045. [PubMed] [Google Scholar]
- 27.Ortiz-Tudela E., Iurisci I., Beau J., Karaboue A., Moreau T., Rol M.A., Madrid J.A., Levi F., Innominato P.F. The circadian rest-activity rhythm, a potential safety pharmacology endpoint of cancer chemotherapy. Int. J. Cancer. 2014;134:2717–2725. doi: 10.1002/ijc.28587. [DOI] [PubMed] [Google Scholar]
- 28.Innominato P.F., Roche V.P., Palesh O.G., Ulusakarya A., Spiegel D., Levi F.A. The circadian timing system in clinical oncology. Ann. Med. 2014;46:191–207. doi: 10.3109/07853890.2014.916990. [DOI] [PubMed] [Google Scholar]
- 29.Levi F., Okyar A., Dulong S., Innominato P.F., Clairambault J. Circadian timing in cancer treatments. Annu. Rev. Pharmacol. Toxicol. 2010;50:377–421. doi: 10.1146/annurev.pharmtox.48.113006.094626. [DOI] [PubMed] [Google Scholar]
- 30.Levi F., Okyar A. Circadian clocks and drug delivery systems: Impact and opportunities in chronotherapeutics. Expert Opin. Drug Deliv. 2011;8:1535–1541. doi: 10.1517/17425247.2011.618184. [DOI] [PubMed] [Google Scholar]
- 31.Okyar A., Piccolo E., Ahowesso C., Filipski E., Hossard V., Guettier C., La Sorda R., Tinari N., Iacobelli S., Levi F. Strain- and sex-dependent circadian changes in abcc2 transporter expression: Implications for irinotecan chronotolerance in mouse ileum. PLoS ONE. 2011;6:e20393. doi: 10.1371/journal.pone.0020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dallmann R., Okyar A., Levi F. Dosing-time makes the poison: Circadian regulation and pharmacotherapy. Trends Mol. Med. 2016;22:430–445. doi: 10.1016/j.molmed.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Filipski E., Berland E., Ozturk N., Guettier C., van der Horst G.T., Levi F., Okyar A. Optimization of irinotecan chronotherapy with p-glycoprotein inhibition. Toxicol. Appl. Pharmacol. 2014;274:471–479. doi: 10.1016/j.taap.2013.12.018. [DOI] [PubMed] [Google Scholar]
- 34.Levi F., Schibler U. Circadian rhythms: Mechanisms and therapeutic implications. Annu. Rev. Pharmacol. Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 35.Gachon F., Firsov D. The role of circadian timing system on drug metabolism and detoxification. Expert Opin. Drug Metab. Toxicol. 2011;7:147–158. doi: 10.1517/17425255.2011.544251. [DOI] [PubMed] [Google Scholar]
- 36.Zarogoulidis P., Trakada G., Zarogoulidis K. A chrono-target chemotherapy treatment model for lung cancer treatment. Ther. Deliv. 2013;4:5–8. doi: 10.4155/tde.12.137. [DOI] [PubMed] [Google Scholar]
- 37.Sancar A., Lindsey-Boltz L.A., Gaddameedhi S., Selby C.P., Ye R., Chiou Y.Y., Kemp M.G., Hu J., Lee J.H., Ozturk N. Circadian clock, cancer, and chemotherapy. Biochemistry. 2015;54:110–123. doi: 10.1021/bi5007354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ballesta A., Innominato P.F., Dallmann R., Rand D.A., Levi F.A. Systems chronotherapeutics. Pharmacol. Rev. 2017;69:161–199. doi: 10.1124/pr.116.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouchahda M., Macarulla T., Liedo G., Levi F., Elez M.E., Paule B., Karaboue A., Artru P., Tabernero J., Machover D., et al. Feasibility of cetuximab given with a simplified schedule every 2 weeks in advanced colorectal cancer: A multicenter, retrospective analysis. Med. Oncol. 2011;28(Suppl S1):S253–S258. doi: 10.1007/s12032-010-9716-8. [DOI] [PubMed] [Google Scholar]
- 40.Levi F., Focan C., Karaboue A., de la Valette V., Focan-Henrard D., Baron B., Kreutz F., Giacchetti S. Implications of circadian clocks for the rhythmic delivery of cancer therapeutics. Adv. Drug Deliv. Rev. 2007;59:1015–1035. doi: 10.1016/j.addr.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Gekakis N., Staknis D., Nguyen H.B., Davis F.C., Wilsbacher L.D., King D.P., Takahashi J.S., Weitz C.J. Role of the clock protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 42.Hogenesch J.B., Gu Y.Z., Jain S., Bradfield C.A. The basic-helix-loop-helix-pas orphan mop3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.King D.P., Zhao Y., Sangoram A.M., Wilsbacher L.D., Tanaka M., Antoch M.P., Steeves T.D., Vitaterna M.H., Kornhauser J.M., Lowrey P.L., et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/S0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kume K., Zylka M.J., Sriram S., Shearman L.P., Weaver D.R., Jin X., Maywood E.S., Hastings M.H., Reppert S.M. Mcry1 and mcry2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/S0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 45.Shearman L.P., Sriram S., Weaver D.R., Maywood E.S., Chaves I., Zheng B., Kume K., Lee C.C., van der Horst G.T., Hastings M.H., et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 46.Van der Horst G.T., Muijtjens M., Kobayashi K., Takano R., Kanno S., Takao M., de Wit J., Verkerk A., Eker A.P., van Leenen D., et al. Mammalian cry1 and cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 47.Vielhaber E.L., Duricka D., Ullman K.S., Virshup D.M. Nuclear export of mammalian period proteins. J. Biol. Chem. 2001;276:45921–45927. doi: 10.1074/jbc.M107726200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ukai-Tadenuma M., Kasukawa T., Ueda H.R. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat. Cell Biol. 2008;10:1154–1163. doi: 10.1038/ncb1775. [DOI] [PubMed] [Google Scholar]
- 49.Kornmann B., Schaad O., Bujard H., Takahashi J.S., Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi J.S. Molecular architecture of the circadian clock in mammals. In: Sassone-Corsi P., Christen Y., editors. A time for Metabolism and Hormones. Springer; Cham, Switzerland: 2016. [PubMed] [Google Scholar]
- 51.Anafi R.C., Lee Y., Sato T.K., Venkataraman A., Ramanathan C., Kavakli I.H., Hughes M.E., Baggs J.E., Growe J., Liu A.C., et al. Machine learning helps identify chrono as a circadian clock component. PLoS Biol. 2014;12:e1001840. doi: 10.1371/journal.pbio.1001840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kavakli I.H., Baris I., Tardu M., Gul S., Oner H., Cal S., Bulut S., Yarparvar D., Berkel C., Ustaoglu P., et al. The photolyase/cryptochrome family of proteins as DNA repair enzymes and transcriptional repressors. Photochem. Photobiol. 2017;93:93–103. doi: 10.1111/php.12669. [DOI] [PubMed] [Google Scholar]
- 53.Erkekoglu P., Baydar T. Chronopharmacodynamics of drugs in toxicological aspects: A short review for clinical pharmacists and pharmacy practitioners. J. Res. Pharm. Pract. 2012;1:41–47. doi: 10.4103/2279-042X.108369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tahara Y., Shibata S. Chrono-biology, chrono-pharmacology, and chrono-nutrition. J. Pharmacol. Sci. 2014;124:320–335. doi: 10.1254/jphs.13R06CR. [DOI] [PubMed] [Google Scholar]
- 55.Matsuo T., Yamaguchi S., Mitsui S., Emi A., Shimoda F., Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 56.Granda T.G., Liu X.H., Smaaland R., Cermakian N., Filipski E., Sassone-Corsi P., Levi F. Circadian regulation of cell cycle and apoptosis proteins in mouse bone marrow and tumor. FASEB J. 2005;19:304–306. doi: 10.1096/fj.04-2665fje. [DOI] [PubMed] [Google Scholar]
- 57.Kelleher F.C., Rao A., Maguire A. Circadian molecular clocks and cancer. Cancer Lett. 2014;342:9–18. doi: 10.1016/j.canlet.2013.09.040. [DOI] [PubMed] [Google Scholar]
- 58.Paschos G.K., Baggs J.E., Hogenesch J.B., FitzGerald G.A. The role of clock genes in pharmacology. Annu. Rev. Pharmacol. Toxicol. 2010;50:187–214. doi: 10.1146/annurev.pharmtox.010909.105621. [DOI] [PubMed] [Google Scholar]
- 59.Zhang R., Lahens N.F., Ballance H.I., Hughes M.E., Hogenesch J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y.K., Yeager R.L., Klaassen C.D. Circadian expression profiles of drug-processing genes and transcription factors in mouse liver. Drug Metab. Dispos. 2009;37:106–115. doi: 10.1124/dmd.108.024174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zmrzljak U.P., Rozman D. Circadian regulation of the hepatic endobiotic and xenobitoic detoxification pathways: The time matters. Chem. Res. Toxicol. 2012;25:811–824. doi: 10.1021/tx200538r. [DOI] [PubMed] [Google Scholar]
- 62.Handschin C., Meyer U.A. Induction of drug metabolism: The role of nuclear receptors. Pharmacol. Rev. 2003;55:649–673. doi: 10.1124/pr.55.4.2. [DOI] [PubMed] [Google Scholar]
- 63.Xu C., Li C.Y., Kong A.N. Induction of phase i, ii and iii drug metabolism/transport by xenobiotics. Arch. Pharm. Res. 2005;28:249–268. doi: 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]
- 64.Gachon F. Physiological function of parbzip circadian clock-controlled transcription factors. Ann. Med. 2007;39:562–571. doi: 10.1080/07853890701491034. [DOI] [PubMed] [Google Scholar]
- 65.Dallmann R., Brown S.A., Gachon F. Chronopharmacology: New insights and therapeutic implications. Annu. Rev. Pharmacol. Toxicol. 2014;54:339–361. doi: 10.1146/annurev-pharmtox-011613-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gachon F., Olela F.F., Schaad O., Descombes P., Schibler U. The circadian par-domain basic leucine zipper transcription factors dbp, tef, and hlf modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 67.Murakami Y., Higashi Y., Matsunaga N., Koyanagi S., Ohdo S. Circadian clock-controlled intestinal expression of the multidrug-resistance gene mdr1a in mice. Gastroenterology. 2008;135:1636–1644.e3. doi: 10.1053/j.gastro.2008.07.073. [DOI] [PubMed] [Google Scholar]
- 68.Kriebs A., Jordan S.D., Soto E., Henriksson E., Sandate C.R., Vaughan M.E., Chan A.B., Duglan D., Papp S.J., Huber A.L., et al. Circadian repressors cry1 and cry2 broadly interact with nuclear receptors and modulate transcriptional activity. Proc. Natl. Acad. Sci. USA. 2017;114:8776–8781. doi: 10.1073/pnas.1704955114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gorbacheva V.Y., Kondratov R.V., Zhang R., Cherukuri S., Gudkov A.V., Takahashi J.S., Antoch M.P. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the clock/bmal1 transactivation complex. Proc. Natl. Acad. Sci. USA. 2005;102:3407–3412. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao X., Cho H., Yu R.T., Atkins A.R., Downes M., Evans R.M. Nuclear receptors rock around the clock. EMBO Rep. 2014;15:518–528. doi: 10.1002/embr.201338271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wada T., Kang H.S., Jetten A.M., Xie W. The emerging role of nuclear receptor roralpha and its crosstalk with lxr in xeno- and endobiotic gene regulation. Exp. Biol. Med. (Maywood) 2008;233:1191–1201. doi: 10.3181/0802-MR-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang X., Downes M., Yu R.T., Bookout A.L., He W., Straume M., Mangelsdorf D.J., Evans R.M. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 73.Claudel T., Cretenet G., Saumet A., Gachon F. Crosstalk between xenobiotics metabolism and circadian clock. FEBS Lett. 2007;581:3626–3633. doi: 10.1016/j.febslet.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 74.Kanno Y., Otsuka S., Hiromasa T., Nakahama T., Inouye Y. Diurnal difference in car mrna expression. Nucl. Recept. 2004;2:6. doi: 10.1186/1478-1336-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu Y.F., Jin T., Xu Y., Zhang D., Wu Q., Zhang Y.K., Liu J. Sex differences in the circadian variation of cytochrome p450 genes and corresponding nuclear receptors in mouse liver. Chronobiol. Int. 2013;30:1135–1143. doi: 10.3109/07420528.2013.805762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takiguchi T., Tomita M., Matsunaga N., Nakagawa H., Koyanagi S., Ohdo S. Molecular basis for rhythmic expression of cyp3a4 in serum-shocked hepg2 cells. Pharmacogenet. Genom. 2007;17:1047–1056. doi: 10.1097/FPC.0b013e3282f12a61. [DOI] [PubMed] [Google Scholar]
- 77.Froy O. Cytochrome p450 and the biological clock in mammals. Curr. Drug Metab. 2009;10:104–115. doi: 10.2174/138920009787522179. [DOI] [PubMed] [Google Scholar]
- 78.Lavery D.J., Lopez-Molina L., Margueron R., Fleury-Olela F., Conquet F., Schibler U., Bonfils C. Circadian expression of the steroid 15 alpha-hydroxylase (cyp2a4) and coumarin 7-hydroxylase (cyp2a5) genes in mouse liver is regulated by the par leucine zipper transcription factor dbp. Mol. Cell Biol. 1999;19:6488–6499. doi: 10.1128/MCB.19.10.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guengerich F.P. Cytochrome p450 and chemical toxicology. Chem. Res. Toxicol. 2008;21:70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 80.Zanger U.M., Schwab M. Cytochrome p450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 81.Kosir R., Zmrzljak U.P., Bele T., Acimovic J., Perse M., Majdic G., Prehn C., Adamski J., Rozman D. Circadian expression of steroidogenic cytochromes p450 in the mouse adrenal gland—Involvement of camp-responsive element modulator in epigenetic regulation of cyp17a1. FEBS J. 2012;279:1584–1593. doi: 10.1111/j.1742-4658.2011.08317.x. [DOI] [PubMed] [Google Scholar]
- 82.Akhtar R.A., Reddy A.B., Maywood E.S., Clayton J.D., King V.M., Smith A.G., Gant T.W., Hastings M.H., Kyriacou C.P. Circadian cycling of the mouse liver transcriptome, as revealed by cdna microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 2002;12:540–550. doi: 10.1016/S0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 83.Tanimura N., Kusunose N., Matsunaga N., Koyanagi S., Ohdo S. Aryl hydrocarbon receptor-mediated cyp1a1 expression is modulated in a clock-dependent circadian manner. Toxicology. 2011;290:203–207. doi: 10.1016/j.tox.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 84.Kakan X., Chen P., Zhang J. Clock gene mper2 functions in diurnal variation of acetaminophen induced hepatotoxicity in mice. Exp. Toxicol. Pathol. 2011;63:581–585. doi: 10.1016/j.etp.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 85.Hamamura K., Matsunaga N., Ikeda E., Kondo H., Ikeyama H., Tokushige K., Itcho K., Furuichi Y., Yoshida Y., Matsuda M., et al. Alterations of hepatic metabolism in chronic kidney disease via d-box-binding protein aggravate the renal dysfunction. J. Biol. Chem. 2016;291:4913–4927. doi: 10.1074/jbc.M115.696930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seng J.E., Gandy J., Turturro A., Lipman R., Bronson R.T., Parkinson A., Johnson W., Hart R.W., Leakey J.E. Effects of caloric restriction on expression of testicular cytochrome p450 enzymes associated with the metabolic activation of carcinogens. Arch. Biochem. Biophys. 1996;335:42–52. doi: 10.1006/abbi.1996.0480. [DOI] [PubMed] [Google Scholar]
- 87.Damiola F. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carver K.A., Lourim D., Tryba A.K., Harder D.R. Rhythmic expression of cytochrome p450 epoxygenases cyp4x1 and cyp2c11 in the rat brain and vasculature. Am. J. Physiol. Cell Physiol. 2014;307:C989–C998. doi: 10.1152/ajpcell.00401.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Binkhorst L., Kloth J.S.L., de Wit A.S., de Bruijn P., Lam M.H., Chaves I., Burger H., van Alphen R.J., Hamberg P., van Schaik R.H.N., et al. Circadian variation in tamoxifen pharmacokinetics in mice and breast cancer patients. Breast Cancer Res. Treat. 2015;152:119–128. doi: 10.1007/s10549-015-3452-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bruckner J.V., Ramanathan R., Lee K.M., Muralidhara S. Mechanisms of circadian rhythmicity of carbon tetrachloride hepatotoxicity. J. Pharmacol. Exp. Ther. 2002;300:273–281. doi: 10.1124/jpet.300.1.273. [DOI] [PubMed] [Google Scholar]
- 91.Khemawoot P., Nishino K., Ishizaki J., Yokogawa K., Miyamoto K. Circadian rhythm of cytochrome p4502e1 and its effect on disposition kinetics of chlorzoxazone in rats. Eur. J. Pharmacol. 2007;574:71–76. doi: 10.1016/j.ejphar.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 92.Matsunaga N., Nakamura N., Yoneda N., Qin T., Terazono H., To H., Higuchi S., Ohdo S. Influence of feeding schedule on 24-h rhythm of hepatotoxicity induced by acetaminophen in mice. J. Pharmacol. Exp. Ther. 2004;311:594–600. doi: 10.1124/jpet.104.069062. [DOI] [PubMed] [Google Scholar]
- 93.Martin C., Dutertre-Catella H., Radionoff M., Debray M., Benstaali C., Rat P., Thevenin M., Touitou Y., Warnet J.M. Effect of age and photoperiodic conditions on metabolism and oxidative stress related markers at different circadian stages in rat liver and kidney. Life Sci. 2003;73:327–335. doi: 10.1016/S0024-3205(03)00271-6. [DOI] [PubMed] [Google Scholar]
- 94.Noshiro M., Kawamoto T., Furukawa M., Fujimoto K., Yoshida Y., Sasabe E., Tsutsumi S., Hamada T., Honma S., Honma K., et al. Rhythmic expression of dec1 and dec2 in peripheral tissues: Dec2 is a potent suppressor for hepatic cytochrome p450s opposing dbp. Genes Cells. 2004;9:317–329. doi: 10.1111/j.1356-9597.2004.00722.x. [DOI] [PubMed] [Google Scholar]
- 95.Berkowitz C.M., Shen C.S., Bilir B.M., Guibert E., Gumucio J.J. Different hepatocytes express the cholesterol 7 alpha-hydroxylase gene during its circadian modulation in vivo. Hepatology. 1995;21:1658–1667. doi: 10.1016/0270-9139(95)90472-7. [DOI] [PubMed] [Google Scholar]
- 96.Ishida H., Yamashita C., Kuruta Y., Yoshida Y., Noshiro M. Insulin is a dominant suppressor of sterol 12 alpha-hydroxylase p450 (cyp8b) expression in rat liver: Possible role of insulin in circadian rhythm of cyp8b. J. Biochem. 2000;127:57–64. doi: 10.1093/oxfordjournals.jbchem.a022584. [DOI] [PubMed] [Google Scholar]
- 97.Yamada M., Nagatomo J., Setoguchi Y., Kuroki N., Higashi S., Setoguchi T. Circadian rhythms of sterol 12alpha-hydroxylase, cholesterol 7alpha-hydroxylase and dbp involved in rat cholesterol catabolism. Biol. Chem. 2000;381:1149–1153. doi: 10.1515/BC.2000.142. [DOI] [PubMed] [Google Scholar]
- 98.Gielen J., Van Cantfort J., Robaye B., Renson J. Rat-liver cholesterol 7alpha-hydroxylase. 3. New results about its circadian rhythm. Eur. J. Biochem. 1975;55:41–48. doi: 10.1111/j.1432-1033.1975.tb02136.x. [DOI] [PubMed] [Google Scholar]
- 99.Hirao J., Arakawa S., Watanabe K., Ito K., Furukawa T. Effects of restricted feeding on daily fluctuations of hepatic functions including p450 monooxygenase activities in rats. J. Biol. Chem. 2006;281:3165–3171. doi: 10.1074/jbc.M511194200. [DOI] [PubMed] [Google Scholar]
- 100.Lavery D.J., Schibler U. Circadian transcription of the cholesterol 7 alpha hydroxylase gene may involve the liver-enriched bzip protein dbp. Genes Dev. 1993;7:1871–1884. doi: 10.1101/gad.7.10.1871. [DOI] [PubMed] [Google Scholar]
- 101.Noshiro M., Nishimoto M., Okuda K. Rat liver cholesterol 7 alpha-hydroxylase. Pretranslational regulation for circadian rhythm. J. Biol. Chem. 1990;265:10036–10041. [PubMed] [Google Scholar]
- 102.Omura T., Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J. Biol. Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 103.Furukawa T., Manabe S., Ohashi Y., Sharyo S., Kimura K., Mori Y. Daily fluctuation of 7-alkoxycoumarin o-dealkylase activities in the liver of male f344 rats under ad libitum-feeding or fasting condition. Toxicol. Lett. 1999;108:11–16. doi: 10.1016/S0378-4274(99)00065-X. [DOI] [PubMed] [Google Scholar]
- 104.Hamdan A.M., Koyanagi S., Wada E., Kusunose N., Murakami Y., Matsunaga N., Ohdo S. Intestinal expression of mouse abcg2/breast cancer resistance protein (bcrp) gene is under control of circadian clock-activating transcription factor-4 pathway. J. Biol. Chem. 2012;287:17224–17231. doi: 10.1074/jbc.M111.333377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hayashi Y., Ushijima K., Ando H., Yanagihara H., Ishikawa E., Tsuruoka S., Sugimoto K., Fujimura A. Influence of a time-restricted feeding schedule on the daily rhythm of abcb1a gene expression and its function in rat intestine. J. Pharmacol. Exp. Ther. 2010;335:418–423. doi: 10.1124/jpet.110.170837. [DOI] [PubMed] [Google Scholar]
- 106.Kotaka M., Onishi Y., Ohno T., Akaike T., Ishida N. Identification of negative transcriptional factor e4bp4-binding site in the mouse circadian-regulated gene mdr2. Neurosci. Res. 2008;60:307–313. doi: 10.1016/j.neures.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 107.Stearns A.T., Balakrishnan A., Rhoads D.B., Ashley S.W., Tavakkolizadeh A. Diurnal rhythmicity in the transcription of jejunal drug transporters. J. Pharmacol. Sci. 2008;108:144–148. doi: 10.1254/jphs.08100SC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Iwasaki M., Koyanagi S., Suzuki N., Katamune C., Matsunaga N., Watanabe N., Takahashi M., Izumi T., Ohdo S. Circadian modulation in the intestinal absorption of p-glycoprotein substrates in monkeys. Mol. Pharmacol. 2015;88:29–37. doi: 10.1124/mol.114.096735. [DOI] [PubMed] [Google Scholar]
- 109.Ando H., Yanagihara H., Sugimoto K., Hayashi Y., Tsuruoka S., Takamura T., Kaneko S., Fujimura A. Daily rhythms of p-glycoprotein expression in mice. Chronobiol. Int. 2005;22:655–665. doi: 10.1080/07420520500180231. [DOI] [PubMed] [Google Scholar]
- 110.Wada E., Koyanagi S., Kusunose N., Akamine T., Masui H., Hashimoto H., Matsunaga N., Ohdo S. Modulation of peroxisome proliferator-activated receptor-alpha activity by bile acids causes circadian changes in the intestinal expression of octn1/slc22a4 in mice. Mol. Pharmacol. 2015;87:314–322. doi: 10.1124/mol.114.094979. [DOI] [PubMed] [Google Scholar]
- 111.Oh J.H., Lee J.H., Han D.H., Cho S., Lee Y.J. Circadian clock is involved in regulation of hepatobiliary transport mediated by multidrug resistance-associated protein 2. J. Pharm. Sci. 2017;106:2491–2498. doi: 10.1016/j.xphs.2017.04.071. [DOI] [PubMed] [Google Scholar]
- 112.Dulong S., Ballesta A., Okyar A., Levi F. Identification of circadian determinants of cancer chronotherapy through in vitro chronopharmacology and mathematical modeling. Mol. Cancer Ther. 2015;14:2154–2164. doi: 10.1158/1535-7163.MCT-15-0129. [DOI] [PubMed] [Google Scholar]
- 113.Ballesta A., Dulong S., Abbara C., Cohen B., Okyar A., Clairambault J., Levi F. A combined experimental and mathematical approach for molecular-based optimization of irinotecan circadian delivery. PLoS Comput. Biol. 2011;7:e1002143. doi: 10.1371/journal.pcbi.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kervezee L., Hartman R., van den Berg D.J., Shimizu S., Emoto-Yamamoto Y., Meijer J.H., de Lange E.C. Diurnal variation in p-glycoprotein-mediated transport and cerebrospinal fluid turnover in the brain. AAPS J. 2014;16:1029–1037. doi: 10.1208/s12248-014-9625-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Okyar A., Dressler C., Hanafy A., Baktir G., Lemmer B., Spahn-Langguth H. Circadian variations in exsorptive transport: In situ intestinal perfusion data and in vivo relevance. Chronobiol. Int. 2012;29:443–453. doi: 10.3109/07420528.2012.668996. [DOI] [PubMed] [Google Scholar]
- 116.Savolainen H., Meerlo P., Elsinga P.H., Windhorst A.D., Dierckx R.A., Colabufo N.A., van Waarde A., Luurtsema G. P-glycoprotein function in the rodent brain displays a daily rhythm, a quantitative in vivo pet study. AAPS J. 2016;18:1524–1531. doi: 10.1208/s12248-016-9973-3. [DOI] [PubMed] [Google Scholar]
- 117.Ohdo S., Makinosumi T., Ishizaki T., Yukawa E., Higuchi S., Nakano S., Ogawa N. Cell cycle-dependent chronotoxicity of irinotecan hydrochloride in mice. J. Pharmacol. Exp. Ther. 1997;283:1383–1388. [PubMed] [Google Scholar]
- 118.Porsin B., Formento J.L., Filipski E., Etienne M.C., Francoual M., Renée N., Magné N., Lévi F., Milano G. Dihydropyrimidine dehydrogenase circadian rhythm in mouse liver. Eur. J. Cancer. 2003;39:822–828. doi: 10.1016/S0959-8049(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 119.Tuchman M., Roemeling R.V., Hrushesky W.A., Sothern R.B., O’Dea R.F. Dihydropyrimidine dehydrogenase activity in human blood mononuclear cells. Enzyme. 1989;42:15–24. doi: 10.1159/000469002. [DOI] [PubMed] [Google Scholar]
- 120.Harris B.E., Song R., Soong S.J., Diasio R.B. Relationship between dihydropyrimidine dehydrogenase activity and plasma 5-fluorouracil levels with evidence for circadian variation of enzyme activity and plasma drug levels in cancer patients receiving 5-fluorouracil by protracted continuous infusion. Cancer Res. 1990;50:197–201. [PubMed] [Google Scholar]
- 121.Zeng Z.L., Sun J., Guo L., Li S., Wu M.W., Qiu F., Jiang W.Q., Levi F., Xian L.J. Circadian rhythm in dihydropyrimidine dehydrogenase activity and reduced glutathione content in peripheral blood of nasopharyngeal carcinoma patients. Chronobiol. Int. 2005;22:741–754. doi: 10.1080/07420520500179969. [DOI] [PubMed] [Google Scholar]
- 122.Barrat M.A., Renee N., Mormont M.C., Milano G., Levi F. [circadian variations of dihydropyrimidine dehydrogenase (dpd) activity in oral mucosa of healthy volunteers] Pathol. Biol. (Paris) 2003;51:191–193. doi: 10.1016/S0369-8114(03)00035-X. [DOI] [PubMed] [Google Scholar]
- 123.Sawers L., Ferguson M.J., Ihrig B.R., Young H.C., Chakravarty P., Wolf C.R., Smith G. Glutathione s-transferase p1 (gstp1) directly influences platinum drug chemosensitivity in ovarian tumour cell lines. Br. J. Cancer. 2014;111:1150–1158. doi: 10.1038/bjc.2014.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Townsend D.M., Tew K.D. The role of glutathione-s-transferase in anti-cancer drug resistance. Oncogene. 2003;22:7369–7375. doi: 10.1038/sj.onc.1206940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li X.M., Metzger G., Filipski E., Boughattas N., Lemaigre G., Hecquet B., Filipski J., Levi F. Pharmacologic modulation of reduced glutathione circadian rhythms with buthionine sulfoximine: Relationship with cisplatin toxicity in mice. Toxicol. Appl. Pharmacol. 1997;143:281–290. doi: 10.1006/taap.1996.8088. [DOI] [PubMed] [Google Scholar]
- 126.Li X.M., Metzger G., Filipski E., Lemaigre G., Levi F. Modulation of nonprotein sulphydryl compounds rhythm with buthionine sulphoximine: Relationship with oxaliplatin toxicity in mice. Arch. Toxicol. 1998;72:574–579. doi: 10.1007/s002040050545. [DOI] [PubMed] [Google Scholar]
- 127.Ohdo S. Chronotherapeutic strategy: Rhythm monitoring, manipulation and disruption. Adv. Drug Deliv. Rev. 2010;62:859–875. doi: 10.1016/j.addr.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 128.Aleem E., Arceci R.J. Targeting cell cycle regulators in hematologic malignancies. Front. Cell Dev. Biol. 2015;3:16. doi: 10.3389/fcell.2015.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Takane H., Ohdo S., Yamada T., Yukawa E., Higuchi S. Chronopharmacology of antitumor effect induced by interferon-beta in tumor-bearing mice. J. Pharmacol. Exp. Ther. 2000;294:746–752. [PubMed] [Google Scholar]
- 130.Sancar A., Lindsey-Boltz L.A., Unsal-Kacmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 131.Williams A.B., Schumacher B. P53 in the DNA-damage-repair process. Cold. Spring Harb. Perspect. Med. 2016;6 doi: 10.1101/cshperspect.a026070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Banks P., Xu W., Murphy D., James P., Sandhu S. Relevance of DNA damage repair in the management of prostate cancer. Curr. Probl. Cancer. 2017;41:287–301. doi: 10.1016/j.currproblcancer.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 133.Fu L., Pelicano H., Liu J., Huang P., Lee C. The circadian gene period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/S0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 134.Bjarnason G.A., Jordan R.C., Sothern R.B. Circadian variation in the expression of cell-cycle proteins in human oral epithelium. Am. J. Pathol. 1999;154:613–622. doi: 10.1016/S0002-9440(10)65306-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bjarnason G.A., Jordan R.C., Wood P.A., Li Q., Lincoln D.W., Sothern R.B., Hrushesky W.J., Ben-David Y. Circadian expression of clock genes in human oral mucosa and skin: Association with specific cell-cycle phases. Am. J. Pathol. 2001;158:1793–1801. doi: 10.1016/S0002-9440(10)64135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wood P.A., Du-Quiton J., You S., Hrushesky W.J. Circadian clock coordinates cancer cell cycle progression, thymidylate synthase, and 5-fluorouracil therapeutic index. Mol. Cancer Ther. 2006;5:2023–2033. doi: 10.1158/1535-7163.MCT-06-0177. [DOI] [PubMed] [Google Scholar]
- 137.Kuramoto Y., Hata K., Koyanagi S., Ohdo S., Shimeno H., Soeda S. Circadian regulation of mouse topoisomerase i gene expression by glucocorticoid hormones. Biochem. Pharmacol. 2006;71:1155–1161. doi: 10.1016/j.bcp.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 138.Flatten K., Dai N.T., Vroman B.T., Loegering D., Erlichman C., Karnitz L.M., Kaufmann S.H. The role of checkpoint kinase 1 in sensitivity to topoisomerase i poisons. J. Biol. Chem. 2005;280:14349–14355. doi: 10.1074/jbc.M411890200. [DOI] [PubMed] [Google Scholar]
- 139.Sigmond J., Peters G.J. Pyrimidine and purine analogues, effects on cell cycle regulation and the role of cell cycle inhibitors to enhance their cytotoxicity. Nucleosides Nucleotides Nucleic Acids. 2005;24:1997–2022. doi: 10.1080/15257770500269556. [DOI] [PubMed] [Google Scholar]
- 140.Tampellini M., Filipski E., Liu X.H., Lemaigre G., Li X.M., Vrignaud P., Francois E., Bissery M.C., Levi F. Docetaxel chronopharmacology in mice. Cancer Res. 1998;58:3896–3904. [PubMed] [Google Scholar]
- 141.Granda T.G., Filipski E., D’Attino R.M., Vrignaud P., Anjo A., Bissery M.C., Levi F. Experimental chronotherapy of mouse mammary adenocarcinoma ma13/c with docetaxel and doxorubicin as single agents and in combination. Cancer Res. 2001;61:1996–2001. [PubMed] [Google Scholar]
- 142.Filipski E., Lemaigre G., Liu X.H., Mery-Mignard D., Mahjoubi M., Levi F. Circadian rhythm of irinotecan tolerability in mice. Chronobiol. Int. 2004;21:613–630. doi: 10.1081/CBI-120040183. [DOI] [PubMed] [Google Scholar]
- 143.Li X.M., Tanaka K., Sun J., Filipski E., Kayitalire L., Focan C., Levi F. Preclinical relevance of dosing time for the therapeutic index of gemcitabine-cisplatin. Br. J. Cancer. 2005;92:1684–1689. doi: 10.1038/sj.bjc.6602564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lévi F. Circadian rhythms in 5-fluorouracil pharmacology and therapeutic applications. In: Rustum Y.M., editor. Fluoropyrimidines in Cancer Therapy. Humana Press; Totowa, NJ, USA: 2003. pp. 107–128. [Google Scholar]
- 145.Wang D., Lippard S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 146.Lincoln D.W., 2nd, Hrushesky W.J., Wood P.A. Circadian organization of thymidylate synthase activity in normal tissues: A possible basis for 5-fluorouracil chronotherapeutic advantage. Int. J. Cancer. 2000;88:479–485. doi: 10.1002/1097-0215(20001101)88:3<479::AID-IJC23>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 147.Johnston P.G., Drake J.C., Trepel J., Allegra C.J. Immunological quantitation of thymidylate synthase using the monoclonal antibody ts 106 in 5-fluorouracil-sensitive and -resistant human cancer cell lines. Cancer Res. 1992;52:4306–4312. [PubMed] [Google Scholar]
- 148.Johnston P.G., Lenz H.J., Leichman C.G., Danenberg K.D., Allegra C.J., Danenberg P.V., Leichman L. Thymidylate synthase gene and protein expression correlate and are associated with response to 5-fluorouracil in human colorectal and gastric tumors. Cancer Res. 1995;55:1407–1412. [PubMed] [Google Scholar]
- 149.McGowan C.H., Russell P. Cell cycle regulation of human wee1. EMBO J. 1995;14:2166–2175. doi: 10.1002/j.1460-2075.1995.tb07210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Koyanagi S., Kuramoto Y., Nakagawa H., Aramaki H., Ohdo S., Soeda S., Shimeno H. A molecular mechanism regulating circadian expression of vascular endothelial growth factor in tumor cells. Cancer Res. 2003;63:7277–7283. [PubMed] [Google Scholar]
- 151.Nakagawa H., Takiguchi T., Nakamura M., Furuyama A., Koyanagi S., Aramaki H., Higuchi S., Ohdo S. Basis for dosing time-dependent change in the anti-tumor effect of imatinib in mice. Biochem. Pharmacol. 2006;72:1237–1245. doi: 10.1016/j.bcp.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 152.Buchdunger E., Cioffi C.L., Law N., Stover D., Ohno-Jones S., Druker B.J., Lydon N.B. Abl protein-tyrosine kinase inhibitor sti571 inhibits in vitro signal transduction mediated by c-kit and platelet-derived growth factor receptors. J. Pharmacol. Exp. Ther. 2000;295:139–145. [PubMed] [Google Scholar]
- 153.Druker B.J., Talpaz M., Resta D.J., Peng B., Buchdunger E., Ford J.M., Lydon N.B., Kantarjian H., Capdeville R., Ohno-Jones S., et al. Efficacy and safety of a specific inhibitor of the bcr-abl tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 154.Uehara H., Kim S.J., Karashima T., Shepherd D.L., Fan D., Tsan R., Killion J.J., Logothetis C., Mathew P., Fidler I.J. Effects of blocking platelet-derived growth factor-receptor signaling in a mouse model of experimental prostate cancer bone metastases. J. Natl. Cancer Inst. 2003;95:458–470. doi: 10.1093/jnci/95.6.458. [DOI] [PubMed] [Google Scholar]
- 155.Kim R., Emi M., Arihiro K., Tanabe K., Uchida Y., Toge T. Chemosensitization by sti571 targeting the platelet-derived growth factor/platelet-derived growth factor receptor-signaling pathway in the tumor progression and angiogenesis of gastric carcinoma. Cancer. 2005;103:1800–1809. doi: 10.1002/cncr.20973. [DOI] [PubMed] [Google Scholar]
- 156.Cao R., Anderson F.E., Jung Y.J., Dziema H., Obrietan K. Circadian regulation of mammalian target of rapamycin signaling in the mouse suprachiasmatic nucleus. Neuroscience. 2011;181:79–88. doi: 10.1016/j.neuroscience.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Okazaki H., Matsunaga N., Fujioka T., Okazaki F., Akagawa Y., Tsurudome Y., Ono M., Kuwano M., Koyanagi S., Ohdo S. Circadian regulation of mtor by the ubiquitin pathway in renal cell carcinoma. Cancer Res. 2014;74:543–551. doi: 10.1158/0008-5472.CAN-12-3241. [DOI] [PubMed] [Google Scholar]
- 158.Lin P., An F., Xu X., Zhao L., Liu L., Liu N., Wang P., Liu J., Wang L., Li M. Chronopharmacodynamics and mechanisms of antitumor effect induced by erlotinib in xenograft-bearing nude mice. Biochem. Biophys. Res. Commun. 2015;460:362–367. doi: 10.1016/j.bbrc.2015.03.039. [DOI] [PubMed] [Google Scholar]
- 159.Berg M., Soreide K. Egfr and downstream genetic alterations in kras/braf and pi3k/akt pathways in colorectal cancer: Implications for targeted therapy. Discov. Med. 2012;14:207–214. [PubMed] [Google Scholar]
- 160.Steins M., Thomas M., Geissler M. Erlotinib. Recent Res. Cancer Res. 2014;201:109–123. doi: 10.1007/978-3-642-54490-3_6. [DOI] [PubMed] [Google Scholar]
- 161.Goffin J.R., Zbuk K. Epidermal growth factor receptor: Pathway, therapies, and pipeline. Clin. Ther. 2013;35:1282–1303. doi: 10.1016/j.clinthera.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 162.Lauriola M., Enuka Y., Zeisel A., D’Uva G., Roth L., Sharon-Sevilla M., Lindzen M., Sharma K., Nevo N., Feldman M., et al. Diurnal suppression of egfr signalling by glucocorticoids and implications for tumour progression and treatment. Nat. Commun. 2014;5:5073. doi: 10.1038/ncomms6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gilbert D.C., Chalmers A.J., El-Khamisy S.F. Topoisomerase i inhibition in colorectal cancer: Biomarkers and therapeutic targets. Br. J. Cancer. 2012;106:18–24. doi: 10.1038/bjc.2011.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zeng Z.L., Luo H.Y., Yang J., Wu W.J., Chen D.L., Huang P., Xu R.H. Overexpression of the circadian clock gene bmal1 increases sensitivity to oxaliplatin in colorectal cancer. Clin. Cancer Res. 2014;20:1042–1052. doi: 10.1158/1078-0432.CCR-13-0171. [DOI] [PubMed] [Google Scholar]
- 165.Dy G.K., Adjei A.A. Understanding, recognizing, and managing toxicities of targeted anticancer therapies. CA Cancer J. Clin. 2013;63:249–279. doi: 10.3322/caac.21184. [DOI] [PubMed] [Google Scholar]
- 166.To H., Ohdo S., Shin M., Uchimaru H., Yukawa E., Higuchi S., Fujimura A., Kobayashi E. Dosing time dependency of doxorubicin-induced cardiotoxicity and bone marrow toxicity in rats. J. Pharm. Pharmacol. 2003;55:803–810. doi: 10.1211/002235703765951410. [DOI] [PubMed] [Google Scholar]
- 167.Filipski E., Amat S., Lemaigre G., Vincenti M., Breillout F., Levi F.A. Relationship between circadian rhythm of vinorelbine toxicity and efficacy in p388-bearing mice. J. Pharmacol. Exp. Ther. 1999;289:231–235. [PubMed] [Google Scholar]
- 168.Li X.M., Levi F. Circadian physiology is a toxicity target of the anticancer drug gemcitabine in mice. J. Biol. Rhythms. 2007;22:159–166. doi: 10.1177/0748730406298984. [DOI] [PubMed] [Google Scholar]
- 169.Li X.M., Kanekal S., Crepin D., Guettier C., Carriere J., Elliott G., Levi F. Circadian pharmacology of l-alanosine (sdx-102) in mice. Mol. Cancer Ther. 2006;5:337–346. doi: 10.1158/1535-7163.MCT-05-0332. [DOI] [PubMed] [Google Scholar]
- 170.Boughattas N.A., Levi F., Fournier C., Lemaigre G., Roulon A., Hecquet B., Mathe G., Reinberg A. Circadian rhythm in toxicities and tissue uptake of 1,2-diamminocyclohexane(trans-1)oxalatoplatinum(ii) in mice. Cancer Res. 1989;49:3362–3368. [PubMed] [Google Scholar]
- 171.Iurisci I., Filipski E., Reinhardt J., Bach S., Gianella-Borradori A., Iacobelli S., Meijer L., Levi F. Improved tumor control through circadian clock induction by seliciclib, a cyclin-dependent kinase inhibitor. Cancer Res. 2006;66:10720–10728. doi: 10.1158/0008-5472.CAN-06-2086. [DOI] [PubMed] [Google Scholar]
- 172.Levi F., Mechkouri M., Roulon A., Bailleul F., Horvath C., Reinberg A., Mathe G. Circadian rhythm in tolerance of mice for etoposide. Cancer Treat. Rep. 1985;69:1443–1445. [PubMed] [Google Scholar]
- 173.Luan J.J., Zhang Y.S., Liu X.Y., Wang Y.Q., Zuo J., Song J.G., Zhang W., Wang W.S. Dosing-time contributes to chronotoxicity of clofarabine in mice via means other than pharmacokinetics. Kaohsiung J. Med. Sci. 2016;32:227–234. doi: 10.1016/j.kjms.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 174.Iwata K., Aizawa K., Sakai S., Jingami S., Fukunaga E., Yoshida M., Hamada A., Saito H. The relationship between treatment time of gemcitabine and development of hematologic toxicity in cancer patients. Biol. Pharm. Bull. 2011;34:1765–1768. doi: 10.1248/bpb.34.1765. [DOI] [PubMed] [Google Scholar]
- 175.Giacchetti S., Cur´e H., Adenis A., Tubiana N., Vernillet L., Chedouba-Messali L., Chevalier V., Germa C., Chollet P., Levi F. Randomized multicenter trial of irinotecan (cpt) chronomodulated (chrono) versus standard (std) infusion in patients (pts) with metastatic colorectal cancer (mcc) Eur. J. Cancer. 2001;37:S309. doi: 10.1016/S0959-8049(01)81635-X. [DOI] [Google Scholar]
- 176.Kloth J.S., Binkhorst L., de Wit A.S., de Bruijn P., Hamberg P., Lam M.H., Burger H., Chaves I., Wiemer E.A., van der Horst G.T., et al. Relationship between sunitinib pharmacokinetics and administration time: Preclinical and clinical evidence. Clin. Pharmacokinet. 2015;54:851–858. doi: 10.1007/s40262-015-0239-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Liu J., Wang C.Y., Ji S.G., Xu X., Wang P.P., Zhang B., Zhao L.Y., Liu L., Lin P.P., Liu L.K., et al. Chronopharmacokinetics of erlotinib and circadian rhythms of related metabolic enzymes in lewis tumor-bearing mice. Eur. J. Drug Metab. Pharmacokinet. 2016;41:627–635. doi: 10.1007/s13318-015-0284-3. [DOI] [PubMed] [Google Scholar]
- 178.Levi F., Zidani R., Misset J.L. Randomised multicentre trial of chronotherapy with oxaliplatin, fluorouracil, and folinic acid in metastatic colorectal cancer. International organization for cancer chronotherapy. Lancet. 1997;350:681–686. doi: 10.1016/S0140-6736(97)03358-8. [DOI] [PubMed] [Google Scholar]
- 179.Chen D., Cheng J., Yang K., Ma Y., Yang F. Retrospective analysis of chronomodulated chemotherapy versus conventional chemotherapy with paclitaxel, carboplatin, and 5-fluorouracil in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Onco Targets Ther. 2013;6:1507–1514. doi: 10.2147/OTT.S53098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bajetta E., Pietrantonio F., Buzzoni R., Ferrario E., Valvo F., Mariani L., Dotti K.F., Biondani P., Formisano B., Gevorgyan A., et al. Chronomodulated capecitabine and adjuvant radiation in intermediate-risk to high-risk rectal cancer: A phase ii study. Am. J. Clin. Oncol. 2014;37:545–549. doi: 10.1097/COC.0b013e31827ecd1d. [DOI] [PubMed] [Google Scholar]
- 181.Tanaka K., Yabushita Y., Nakagawa K., Endo I. [circadian chronotherapy for metastatic liver tumor] Nihon Rinsho. 2013;71:2158–2164. [PubMed] [Google Scholar]
- 182.Patil S.S., Shahiwala A. Patented pulsatile drug delivery technologies for chronotherapy. Expert Opin. Ther. Pat. 2014;24:845–856. doi: 10.1517/13543776.2014.916281. [DOI] [PubMed] [Google Scholar]
- 183.Levi F., Karaboue A., Etienne-Grimaldi M.C., Paintaud G., Focan C., Innominato P., Bouchahda M., Milano G., Chatelut E. Pharmacokinetics of irinotecan, oxaliplatin and 5-fluorouracil during hepatic artery chronomodulated infusion: A translational european optiliv study. Clin. Pharmacokinet. 2017;56:165–177. doi: 10.1007/s40262-016-0431-2. [DOI] [PubMed] [Google Scholar]
- 184.Levi F.A., Boige V., Hebbar M., Smith D., Lepere C., Focan C., Karaboue A., Guimbaud R., Carvalho C., Tumolo S., et al. Conversion to resection of liver metastases from colorectal cancer with hepatic artery infusion of combined chemotherapy and systemic cetuximab in multicenter trial optiliv. Ann. Oncol. 2016;27:267–274. doi: 10.1093/annonc/mdv548. [DOI] [PubMed] [Google Scholar]
- 185.Youan B.B. Chronopharmaceutical drug delivery systems: Hurdles, hype or hope? Adv. Drug Deliv. Rev. 2010;62:898–903. doi: 10.1016/j.addr.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hu J., Fu S., Peng Q., Han Y., Xie J., Zan N., Chen Y., Fan J. Paclitaxel-loaded polymeric nanoparticles combined with chronomodulated chemotherapy on lung cancer: In vitro and in vivo evaluation. Int. J. Pharm. 2017;516:313–322. doi: 10.1016/j.ijpharm.2016.11.047. [DOI] [PubMed] [Google Scholar]
- 187.Li J., Chen R., Ji M., Zou S.L., Zhu L.N. Cisplatin-based chronotherapy for advanced non-small cell lung cancer patients: A randomized controlled study and its pharmacokinetics analysis. Cancer Chemother. Pharmacol. 2015;76:651–655. doi: 10.1007/s00280-015-2804-x. [DOI] [PubMed] [Google Scholar]
- 188.Giacchetti S., Dugue P.A., Innominato P.F., Bjarnason G.A., Focan C., Garufi C., Tumolo S., Coudert B., Iacobelli S., Smaaland R., et al. Sex moderates circadian chemotherapy effects on survival of patients with metastatic colorectal cancer: A meta-analysis. Ann. Oncol. 2012;23:3110–3116. doi: 10.1093/annonc/mds148. [DOI] [PubMed] [Google Scholar]
- 189.Akgun Z., Saglam S., Yucel S., Gural Z., Balik E., Cipe G., Yildiz S., Kilickap S., Okyar A., Kaytan-Saglam E. Neoadjuvant chronomodulated capecitabine with radiotherapy in rectal cancer: A phase ii brunch regimen study. Cancer Chemother. Pharmacol. 2014;74:751–756. doi: 10.1007/s00280-014-2558-x. [DOI] [PubMed] [Google Scholar]
- 190.Pilanci K.N., Saglam S., Okyar A., Yucel S., Pala-Kara Z., Ordu C., Namal E., Ciftci R., Iner-Koksal U., Kaytan-Saglam E. Chronomodulated oxaliplatin plus capecitabine (xelox) as a first line chemotherapy in metastatic colorectal cancer: A phase ii brunch regimen study. Cancer Chemother. Pharmacol. 2016;78:143–150. doi: 10.1007/s00280-016-3067-x. [DOI] [PubMed] [Google Scholar]
- 191.Giacchetti S., Li X.M., Ozturk N., Cuvier C., Machowiak J., Arrondeau J., Chang-Marchand Y., Espie M., Okyar A., Levi F. Consistent dosing-time dependent tolerability of everolimus (ev) in a pilot study in women with metastatic breast cancers (mbc) and in a mouse chronopharmacology investigation. Cancer Res. 2017;77:2. doi: 10.1158/1538-7445.SABCS16-P4-06-06. [DOI] [Google Scholar]
- 192.Xiong H., Chiu Y.L., Ricker J.L., LoRusso P. Results of a phase 1, randomized study evaluating the effects of food and diurnal variation on the pharmacokinetics of linifanib. Cancer Chemother. Pharmacol. 2014;74:55–61. doi: 10.1007/s00280-014-2475-z. [DOI] [PubMed] [Google Scholar]
- 193.Chan S., Zhang L., Rowbottom L., McDonald R., Bjarnason G.A., Tsao M., Barnes E., Danjoux C., Popovic M., Lam H., et al. Effects of circadian rhythms and treatment times on the response of radiotherapy for painful bone metastases. Ann. Palliat. Med. 2017;6:14–25. doi: 10.21037/apm.2016.09.07. [DOI] [PubMed] [Google Scholar]
- 194.Roche V.P., Mohamad-Djafari A., Innominato P.F., Karaboue A., Gorbach A., Levi F.A. Thoracic surface temperature rhythms as circadian biomarkers for cancer chronotherapy. Chronobiol. Int. 2014;31:409–420. doi: 10.3109/07420528.2013.864301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ortiz-Tudela E., Innominato P.F., Rol M.A., Levi F., Madrid J.A. Relevance of internal time and circadian robustness for cancer patients. BMC Cancer. 2016;16:285. doi: 10.1186/s12885-016-2319-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Anafi R.C., Francey L.J., Hogenesch J.B., Kim J. Cyclops reveals human transcriptional rhythms in health and disease. Proc. Natl. Acad. Sci. USA. 2017;114:5312–5317. doi: 10.1073/pnas.1619320114. [DOI] [PMC free article] [PubMed] [Google Scholar]