Abstract
The common p.D358A variant (rs2228145) in IL-6R is associated with risk for multiple diseases and with increased levels of soluble IL-6R in the periphery and central nervous system (CNS). Here, we show that the p.D358A allele leads to increased proteolysis of membrane bound IL-6R and demonstrate that IL-6R peptides with A358 are more susceptible to cleavage by ADAM10 and ADAM17. IL-6 responsive genes were identified in primary astrocytes and microglia and an IL-6 gene signature was increased in the CNS of late onset Alzheimer’s disease subjects in an IL6R allele dependent manner. We conducted a screen to identify variants associated with the age of AD onset in APOE ε4 carriers. Across 5 datasets, p.D358A had a meta P=3×10−4 and an odds ratio (OR) = 1.3, 95% confidence interval (CI) 1.12 – 1.48. Our study suggests that a common coding region variant of the IL-6 receptor results in neuroinflammatory changes that may influence the age of onset in AD in APOE ε4 carriers.
Keywords: Alzheimer Disease, IL-6, Metalloproteases, Astrocytes, Microglia
Introduction
IL-6R is a receptor for IL-6, a cytokine that can trigger a range of cellular responses including inflammation [1]. IL-6R is primarily expressed in myeloid lineage cells in both transmembrane and soluble (sIL-6R) versions. On cells expressing transmembrane IL-6R, IL-6 binds to a complex of IL-6R and gp130, inducing cis-signaling. The transmembrane IL6-R can be cleaved and shed as sIL-6R. Unlike many cytokines, sIL-6R is not a decoy receptor. On cells that express gp130 but not transmembrane IL-6R, IL-6 and soluble IL-6R form a complex with membrane bound gp130 to induce trans-signaling[1].
A common variant in IL-6R, p.D358A (rs2228145) has been associated with multiple phenotypes, with A358 increasing risk of asthma[2], higher sIL-6R serum and cerebrospinal fluid levels[3,4], lower serum C-reactive protein levels[5], lower serum fibrinogen levels[6], and reduced risk of coronary heart disease[7] and rheumatoid arthritis[8,9]. Amino acid 358 is within a predicted metalloprotease cleavage site of the IL-6R receptor and the A358 variant is thought to play a role in increased shedding of the extracellular portion of the receptor [10–15].
In the central nervous system (CNS), IL6R is primarily expressed by microglia. We hypothesized that alterations in the ratio of transmembrane to soluble IL-6R, conferred by the D358A variant, may result in detectable gene expression changes in neurodegenerative diseases, including late onset Alzheimer’s disease (LOAD). Neuroinflammation is increasingly recognized as a key pathway in the development and severity of LOAD [16]. LOAD is the most common cause of dementia and is characterized by progressive cognitive decline[17]. Individuals with LOAD have regional neuronal loss, neuroinflammation, plaques containing amyloid β, and neurofibrillary tangles composed of tau[17]. Genome wide association studies (GWAS) in European and African American populations have identified >20 loci associated with LOAD risk[18–25], and several of the risk genes are likely to influence inflammation, including TREM2, CR1, CD33, and INPP5D. The APOE ε4 (APOE4) allele accounts for the largest fraction of the heritable risk of AD (20–40%)[26]. There are three forms of APOE that are defined by haplotypes of 2 coding variants (ε2, ε3, ε4). By age 85, 11% of the general population will develop LOAD, compared to 55% of APOE4 homozygotes and ~25% of APOE4 heterozygotes[27]. Although APOE4 is the strongest common variant associated with LOAD, there is a great deal of heterogeneity in the age of disease onset, even among APOE4 carriers.
We investigated the mechanism by which the IL-6R D358A allele leads to increased sIL6R, and looked for evidence that this functional variant contributes to the pathophysiology of Alzheimer’s disease.
Materials and Methods
Analysis of CSF
CSF data for IL-6R levels and genotype was obtained from the ADNI study and individual data was downloaded from the LONI website.
Detection of soluble and membrane bound IL-6R in cultured human CD4+ T cells
Healthy human volunteers were genotyped for IL-6R SNP rs2228145 by ABI C_16170664_10 assay. PBMC’s were obtained by Ficol gradient from five pairs of homozygous donors (one with each genotype AA/CC) age, gender and ethnicity matched. CD4+T cells were purified from PBMCs by negative selection using EasySep CD4+T cells enrichment kit (STEMCELL Technologies-19052) as recommended by the manufacturer.
CD4+T cells were then cultured for 72 hours in RPMI 1640+10% FBS + 2-Mercaptoethanol treated with and without 100nM PMA for 60 minutes. Cells were harvested soon after the treatment and stained with IL-6R-PE antibody (BD-Cat. No-551850). Membrane bound IL-6R was analyzed by FACS.
Supernatant was also collected after 24, 48 and 72 hours of CD4+T cell culture to measure sIL-6R concentration by ELISA (Human IL-6 R alpha Quantikine ELISA Kit, R&D Systems Cat-No DR600).
In vitro IL-6R shedding in 293T cells
293T cells were transfected with D358 (WT) or A358 (Mut) constructs of IL-6R along with GFP as transfection control. To study the effect over time cells were treated 48 hours after transfection with 100nM PMA for 0, 30, 60 and 120 minutes or to check the effect of MMP inhibitors, cells were treated with/without TAPI-2 (adam 17 Inhibitor) 20uM or with/without GM 6001 (pan MMP inhibitor) 100nM for 60 min prior to treatment with/without 100nM PMA for 60 min. Cell were harvested after the treatment and stained with IL-6R-PE antibody (BD-Cat. No-551850). Membrane bound IL-6R was analyzed by FACS by gating double positive cells for GFP and IL-6R.
Adam 10 and Adam 17 in-vitro enzymatic assay
The effect of recombinant MMP (Adam10 and Adam17) on the IL-6R synthetic peptide was checked by in-vitro enzymatic assay. Synthetic IL-6R peptides with a difference of one amino acid aspartate (D) to alanine (A) were synthesized.
| IL-6R D358 | -DSANATSLPVQDSSSVPLPTFL |
| IL-6R A358 | -DSANATSLPVQASSSVPLPTFL |
Adam 17 or 10 enzymes (R&D system) at 2.5ug/ml and substrate (peptides) at 10 uM concentration were added in an assay buffer (25mM Tris, 2.5 uM ZnCl2, 0.005 Brij-35 (v/v) pH 9.0) and incubated at 37°C for indicated time. Quantitation of cleaved vs. uncleaved peptide was done by mass-spectrometry.
Quantification of peptides by Mass-spectrometry
Selected reaction monitoring quantitation for the IL-6R reaction mixtures was performed on a triple quadrupole/linear ion trap mass spectrometer, 4000 QTRAP (Applied Biosystems, Foster City, CA) coupled to a Tempo Autosampler (Applied Biosystems), and a 1200 capillary LC system (Agilent, Santa Clara, CA). Samples were loaded via full (2uL) loop injection directly onto a MetaSil AQ 3 C18 150 × 2.0mm column (Varian, Lake Forrest, CA) and separated by reverse phase chromatography at a flow rate of 200 μL/min where solvent A was 98% H2O/2% Acetonitrile/0.1% formic acid and solvent B was 98% Acetonitrile/2% H2O/0.1% formic acid. Samples were loaded in 98% water/2% Acetonitrile/0.1% FA and eluted with a gradient of 5–30% solvent B for 1 min, 30–65% solvent B for 1min, and 65–90% solvent B for 4min; with a total run time of 15min. Samples were ionized via a Turbo Ionspray source with the spray voltage set at 5200V, curtain gas of 20 psi, and an interface heating temperature of 550°C. The declustering potential (DP) was 60V, collision energy was 35V, dwell time was 100ms per transition, for a total cycle time of 0.420s. Q1 and Q3 resolution settings were set at low and unit, respectively. The most abundant charge states of each intact and Adam10 or 17 processed peptides were determined empirically and used for SRM transition development. SRM transitions were selected to monitor the fragment ion with the highest intensity and uniqueness to the peptide. The following transitions were monitored for the IL-6R A358 reaction: the [M+3H]3+ precursor of DSANATSLPVQASSSVPLPTFL at 802.4 to 477.5 (y4), the [M+2H]2+ precursor of VQASSSVPLPTFL at 673.4 to 477.5 (y4), the [M+2H] 2+ precursor of ASSSVPLPTFL at 559.8 to 477.5 (y4), and the [M+2H] 2+ precursor of PVQASSSVPLPTFL at 721.8 to 477.5 (y4). The following transitions were monitored for the IL-6R D358 reaction: the [M+3H] 3+ precursor of DSANATSLPVQDSSSVPLPTFL at 817.1 to 477.5 (y4), the [M+2H] 2+ precursor of VQDSSSVPLPTFL at 695.4 to 477.5 (y4), the [M+2H] 2+ precursor of DSSSVPLPTFL at 581.8 to 477.5 (y4), and the [M+2H] 2+ precursor of PVQDSSSVPLPTFL at 743.9 to 477.5 (y4). Transitions corresponding to the N-terminal peptide fragments were not monitored due to the hydrophilicity and suboptimal chromatographic peak shape observed (data not shown). Peak integration was performed using MultiQuant 1.1 software (Applied Biosystems), and area under the curve (AUC) was used to determine the abundance of each peptide fragment relative to intact peptide.
Astrocyte and microglia cell culture
Primary mouse astrocyte and microglia co-culture and RNA extraction was performed as previously described[28]. We determined that 1.25 nM of both IL-6 (R&D 406-ML-005/CF) and sIL-6R was an optimal dose, and treated a new set of co-cultures at these levels for 24 hours prior to RNA extraction. Treated co-cultures were shaken to separate the culture into a microglia-enriched sample of detached cells in the media and an astrocyte-enriched sample adhering to the flask.
Western blotting of IL-6 in astrocytes and microglia
Primary mouse astrocyte and microglia co-culture were treated with 1.25 nM IL-6 or control media for 15 minutes, washed once with cold PBS, and lysed in RIPA buffer (Sigma R0278) with PhosStop phosphatase inhibitors (Roche 04906837001) and cOmplete protease inhibitors (Roche 11836170001) at 4C on a rotator for 30 minutes. The lysate was spun at 20,000g at 4C and the supernatant was transferred to a new tube to remove insoluble material and stored at −80C. Protein lysate concentrations were measured by a BCA protein assay (Thermo Scientific 23227) following the kit provided protocol. 25ug of lysate was loaded per well of a 10% bis-tris gel (Invitrogen WG1202) and immunoblotted using standard procedures using 1:1,000 anti-rabbit pT705 STAT3 (Cell signaling 9131) and 1:20,000 anti-mouse beta-actin (Sigma A3854) antibodies. Proteins on blots were visualized using the LI-COR Odyssey system.
Quantitative PCR and RT-PCR
RNA sample quality was assessed using a RNA 6000 Pico kit (Agilent Technologies 5067-1513) and 2100 Bioanalyzer (Agilent Technologies G2939AA). Technical triplicate quantitative PCR reactions were performed on a Biomark HD System (Fluidigm) using iScript cDNA generated from microglia or astrocyte enriched RNA as a template (Bio-rad 170-8891) and the following Taqman primer sets: Actb (Mm00607939_s1), BC055004 (Mm01290815_m1), Bcl3 (Mm00504306_m1), C4b (Mm00437893_g1), Ccl12 (Mm01617100_m1), Ccl8 (Mm01297183_m1), Cdsn (Mm01275230_m1), Cebpd (Mm00786711_s1), Fn1 (Mm01256744_m1), Gapdh (Mm99999915_g1), Gda (Mm00515820_m1), Hp (Mm00516884_m1), Hprt (Mm01545399_m1), Hspb1 (Mm00834384_g1), Lcn2 (Mm01324470_m1), Saa1 (Mm00656927_g1), Saa3 (Mm00441203_m1), Serpina3n (Mm00776439_m1), Socs3 (Mm00545913_s1), Tgm1 (Mm00498375_m1), Timp1 (Mm00441818_m1) (Life Technologies). Gene expression was normalized to the geometric mean of Gapdh, Actb, and Hprt expression.
Microarray analysis of astrocyte and microglia cultures treated with IL-6 + sIL-6R
Quantity and quality of total RNA samples was determined using ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE) and Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA), respectively. We used Agilent’s method for preparation of Cy-dye labeled cRNA and array hybridization (Santa Clara, CA). Briefly, total RNA was converted to double-stranded cDNA and then to Cy-dye labeled cRNA using Agilent’s Quick Amp Labeling Kit. The labeled cRNA was purified using an RNeasy mini kit (Qiagen, San Diego, CA). cRNA yield and Cy-dye incorporation was determined using an ND-1000 spectrophotometer (Thermo Scientific). 750 ng of the labeled cRNA was fragmented and hybridized to the Agilent’s Whole Mouse Genome 4×44Kv2 arrays as described in the manufacturer’s hybridization kit. All samples were labeled with Cy5 and hybridized against Cy3 labeled universal mouse reference (Stratagene, La Jolla, CA). Following hybridization, the arrays were washed, dried and scanned on Agilent’s microarray scanner. Agilent’s Feature Extraction software 11.0 was used to analyze acquired array images.
Bioinformatics analyses were completed with the Bioconductor (v2.13)[29] software and the R (v3.0.2) programming language. Background correction of acquired images was performed using the normexp function in the limma package using an offset of 50[30]. Within array normalization was performed using the normalizeWithinArrays function with the loess method. Lastly, arrays were normalized with the normalizeBetweenArrays function using the aquantile method. Control probes were removed from the analysis. Data for duplicate probes on the Agilent array were averaged using the avereps function. Prior to comparison between groups, probes were filtered to ensure only a single probe was represented for each gene using the featureFilter function with default parameters. QC was performed using the arrayQualityMetrics package.
Differential expression of genes was determined using the limma package, and p-values reported in the text were corrected for multiple testing using Benjamini and Hochberg’s method. Genes that were not also differentially expressed by PCR were excluded from further analyses.
Creation of IL-6 + sIL-6R responsive gene sets in astrocytes and microglia and pathway analysis
Microarray gene sets of IL-6 + sIL-6R microglia or astrocyte response were determined by selecting the top differentially expressed genes (adjusted p-value < 0.01), ranked by fold change (Table S1). Gene Ontology analysis (Table S2) was performed using the GOstats and ReportingTools packages[31]. We used the conditional setting for the GOstats hypergeometric test to take into account the nesting structure of the GO categories. We considered all ‘Biological Process’ categories with at least 10 gene members.
Public IL-6 responsive gene sets were selected from mSigDB v4.0 and their enrichment in the astrocyte and microglia cultures was determined via the same gene set enrichment methods discussed below. As these publicly available IL-6 responsive gene sets were not generated using CNS cell types they may contain significant numbers of genes that are not expressed in the CNS. A notable example is CRP which is not expressed in our culture system or in our human cortical tissue sets. From this analysis, only a fibroblast enriched gene set from Dasu et al (2004) was significantly enriched in LOAD samples with the risk allele (Table S4)[32].
Analysis of human cortex gene expression and genotype data
The top differentially expressed genes ranked by microarray fold change were explored in human microarray data. Publicly available expression arrays for the temporal region of human cortex samples were downloaded from the Gene Expression Omnibus[33] (GEO, GSE 15222). Raw data was normalized via the variance stabilizing normalization and non-specific filtering was performed with nsFilter to remove duplicate probes. Covariates and genotype data were downloaded from the author’s website (http://labs.med.miami.edu/myers/LFuN/data.html). We discovered a strong batch and sex effect in this data. All subsequent analyses corrected for these covariates. Because our discovered IL6R variant was associated with the age of onset of LOAD patients, we did not adjust for age in our models. We removed samples that were APOE 2/4. After removing SNPs with missing genotype rate > 1% we performed genotype imputation using Shapeit, IMPUTE2, and 1000 Genomes haplotypes as described above.
Gene sets and gene set enrichment analysis
Gene sets generated from IL-6 and sIL-6R stimulated microglia- or astrocyte-enriched cultures were identified as described above. Mouse Entrez IDs were mapped to Human Entrez IDs using the getLDS function of the biomaRt package[34]. Several mouse genes mapped to multiple human homologs; homologene (http://www.ncbi.nlm.nih.gov/homologene) was used to determine a unique human homolog for these genes. Genes that did not map to human or did not map to a probe in the Webster et al. (2009) data set were excluded[35].
To determine the level of enrichment of our IL-6 responsive gene sets in human data, we performed gene set enrichment analysis (GSEA)[36]. GSEA determines the level of the association of a set of related genes with a particular variable of interest. GSEA was performed using the permutation-based JG-Score[37] and rotation-based ROAST[38] methods in corresponding Bioconductor packages (GSEAlm and limma, respectively). The JG-score is a weighted sum of the individual differential expression statistics, and is thus sensitive to outliers. 9,999 permutations were used to calculate the p-value for observed JG-score statistic. Due to the granularity of the permutation method, minimum permutation p-values are 0.0001. For ROAST, the ‘floormean’ method was used; this method is less sensitive than the JG-score to outliers. 9,999 rotations were used to calculate the ROAST p-values. Genes within a set are considered up-regulated if their squared z-statistic for differential expression is greater than 2.
We considered four null hypotheses for the IL-6 responsive gene set in the AD patients: (1) the set is not enriched in A358 carriers versus D358 carriers; (2) the set is not enriched in A358 + APOE4 carriers versus D358 + APOE4 carriers; (3) the set is not enriched in A358 + APOE3 carriers versus D358 + APOE3 carriers; (4) the set is not enriched in A358 + APOE4 carriers versus A358 + APOE3 carriers. For each analysis, we adjusted for sex and batch effects. In the first comparison, we also adjusted for APOE status.
IL-6 score in clinical samples
Expression data from the human AD and control samples was adjusted for batch and sex by fitting the following linear model for the ith gene:
We then calculated the corresponding residuals for each sample with the lmPerGene and getResidPerGene functions in the GSEAlm package.
We normalized the adjusted expression in each gene (Yij) by subtracting its mean and dividing by its standard deviation in all samples (si):
For the jth subject, the IL-6 responsive score is the sum of K normalized adjusted expression values of each gene in the gene set divided by the size of the set:
Two-sample t-tests for both the AD and control samples were performed to determine whether or not the A358 and D358 samples have the same score.
Subjects and genotyping
For all of the cohorts, we selected individuals of self reported European ancestry that matched the following criteria: APOE ε4 carriers (one or two copies) Alzheimer’s cases with age ≤ 65 yrs, or controls with age ≥ 80 yrs and one copy of ε4, or controls with age ≥ 75 yrs and two copies of ε4 (Table S7).
The primary study included 103 total cases and 109 that were genotyped using Illumina Omni 1M Quad SNP array, which included 1,140,419 SNPs. All samples from this primary study were from NIA ADC cohort (see below for description).
The first US replication cohort included 58 total cases and 275 controls genotyped using Illumina Omni2.5M SNP array (2,379,855 SNPs). The analysis included 215 samples from Cache County[39] and 55 samples from 23andMe.
The second US replication cohort included data obtained from dbGap phs000168.v1.p1 which included 207 cases and 386 controls from the National Institute on Aging – Late Onset Alzheimer’s Disease Study, that were genotyped using Illumina Human610-Quad SNP array (620,901 SNPs).
The third US replication study included 702 cases and 258 controls from the NIA ADC cohort genotyped (see description below) using Illumina HumanExome-12v1 SNP array (247,870 SNPs).
The NIA ADC cohort, assembled by the Alzheimer’s Disease Genetics Consortium (ADGC), included subjects ascertained and evaluated by the clinical and neuropathology cores of the 29 NIA funded ADCs. Data collection is coordinated by the National Alzheimer’s Coordinating Center (NACC). NACC coordinates collection of phenotype data from the 29 ADCs, cleans all data, coordinates implementation of definitions of AD cases and controls, and coordinates collection of samples. The ADC cohort consists of 2,499 autopsy-confirmed and 1,748 clinically confirmed AD cases, 175 cognitively normal elders (CNEs) with complete neuropathology data who were older than 60 years at age of death, and 2,669 living CNEs evaluated using the Uniform data set (UDS) protocol40,41 who were documented to not have mild cognitive impairment (MCI) and were between 60 and 100 years of age at assessment. Among cases, the average age at onset was 71.6 years (±9.0 years), and APOE genotypes were 0.1% e22, 3.5% e23, 31.6% e33, 2.8% e24, 44.3% e34, and 14.7% e44. Among controls, the average age at last exam was 76.6 years (±9.4 years), and APOE genotypes were 0.7% e22, 13.0% e23, 56.8% e33, 2.1% e24, 21.4% e34, and 2.2% e44. Based on the data collected by NACC, the ADGC Neuropathology Core Leaders Subcommittee derived inclusion and exclusion criteria for AD and control samples. All autopsied subjects were age ≥60 years at death. AD cases had dementia according to DSM-IV criteria or Clinical Dementia Rating (CDR) ≥1. Neuropathologic stratification of cases followed NIA/Reagan criteria explicitly or used a similar approach when NIA/Reagan criteria42 were coded as not done, missing, or unknown. Cases were intermediate or high likelihood by NIA/Reagan criteria with moderate to frequent amyloid plaques and neurofibrillary tangle (NFT) Braak stage of III–VI[40,41]. Persons with Down’s syndrome, non-AD tauopathies and synucleinopathies were excluded. All autopsied controls had a clinical evaluation within 2 years of death. Controls did not meet DSM-IV criteria for dementia, did not have a diagnosis of mild cognitive impairment (MCI), and had a CDR of 0, if performed. Controls did not meet or were low-likelihood AD by NIA/Reagan criteria, had sparse or no amyloid plaques, and a Braak NFT stage of 0–II. ADCs sent frozen tissue from autopsied subjects and DNA samples from some autopsied subjects and from living subjects to the ADCs to the National Cell Repository for Alzheimer’s Disease (NCRAD). DNA was prepared by NCRAD and sent either by NCRAD or by the University of Pennsylvania and sent to the North Shore Medical Center for genotyping.
The Icelandic study (Decode Genetics) included whole genome sequencing of 75 cases and 75 controls using Illumina Hi-Seq at an average read coverage of 30× using 100bp paired end reads. The reads were aligned to the reference genome using BWA[42]. QC of the alignment data was performed using GATK[43] and involved removal of PCR duplicates, base quality recalibration, and realignment around short indels. Variant calling was also performed using GATK[43].
Quality control, genotype imputation, and association analysis
In the analysis of the primary study data, we excluded SNPs with >5% missing genotypes, Hardy-Weinberg test p-value < 10−4, minor allele frequency < 0.01. We performed identity-by-descent analysis to rule out any closely related individuals and PCA to rule out any population outliers and to identify eigenvectors that were significantly associated with case-control status. We then performed logistic regression of the case-control status on the genotype (coded additively 0,1,2) at every SNP, including the above eigenvectors as covariates to account for possible population stratification. All association analysis was performed using SNPTEST[44].
The SNP of interest, rs2228145, was directly genotyped in all but the US replication 2 study. Genotype imputation was performed for those samples using a workflow that included pre-phasing using Shapeit[45] followed by imputation using IMPUTE2[46] and reference haplotypes from the 1000 Genomes project. Meta-analysis of results was performed using METASOFT[47].
Results
A358 IL-6R is associated with increased shedding of membrane bound IL-6R
sIL-6R concentrations have been shown to be higher in A358 IL6R carriers compared to those with the D358 IL6R variant in both serum[2,11,48,49] and the CSF[4]. We confirmed the previous observations that the minor allele (A358) is associated with increased circulating levels of sIL-6R in cerebrospinal fluid (CSF, Fig. S1, each two-way comparison p-value ≤ 5.31 × 10−5) and serum (p-value = 0.022; Fig. S2A) of Alzheimer’s Disease Neuroimaging Initiative (ADNI) study participants
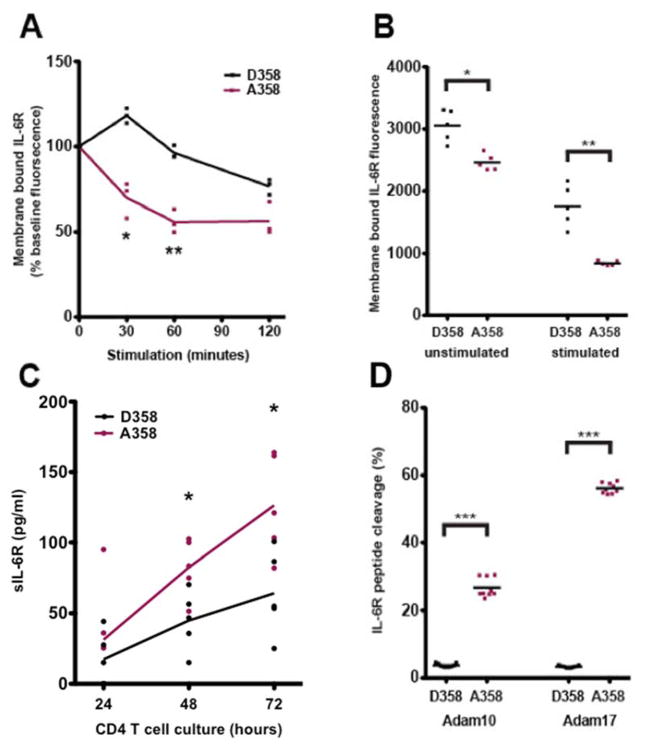
ADAM17 is a metalloprotease that is activated in cells by phorbol 12-myristate 13-acetate (PMA) and generates sIL-6R by cleavage of IL-6R between amino acids 357 and 358[9,50]. In transfected 293T cells, the A358 variant of IL-6R showed 45% lower cell surface levels of IL-6R than p.D358 after 60 minutes of PMA treatment (p-value = 0.0025; Fig. 1A).
Fig. 1. A358 is associated with increased ectodomain shedding of IL-6R by Adam 17.
(A) 293T cells transfected with D358 or A358 IL-6R were treated with 100nM PMA for 0, 30, 60 and 120 minutes. Membrane bound IL-6R was analyzed by FACS. The mean fluorescence intensity relative to the initial time point of three experiments is shown. At each time point, the A358 samples have decreased levels of mIL-6R compared to the D358 samples (p30minutes=0.0079; p60minutes=0.0025; p120minutes=0.050). (B) CD4+ T cells cultured for 72 hours in RPMI 1640+10% FBS + 2-Mercaptoethanol treated without (unstimulated) or with (stimulated) 100nM PMA for 60 minutes. Membrane bound IL-6R was analyzed by FACS. The mean fluorescence intensity from five pairs of genotyped donors is shown. Both the unstimulated (p-value = 0.014) and stimulated (p-value = 0.0030) A358 samples have decreased mIL-6R compared to their D358 counterparts. (C) Supernatant was collected after 24, 48 and 72 hours of CD4+T cell culture and sIL-6R was measured by ELISA (D) ADAM10 and ADAM17 activity on the IL-6R peptides after 6 hours. Quantification of cleaved and uncleaved product by single reaction monitoring mass spectrometry. Area under the curve of transition ion peaks were used to calculate percent cleaved product considering total (cleaved + uncleaved area under the curve) as 100% is shown. P-value <0.05= *, <0.005=**, <0.0005=***.
Steady state and PMA treated CD4+T cells isolated from healthy human donors revealed a similar in-vivo genotype-dependent effect on cleavage showing decreased cell surface IL6R (paired t-test p-value = 0.0030; Fig. 1B) and increased sIL6R (Fig. 1C). These data are consistent with a model where A358 IL6R is associated with an increased rate of proteolytic cleavage of membrane bound IL-6R in A358 carriers at steady state and following PMA stimulation.
ADAM17 and ADAM10 preferentially cleave IL6R A358
We next tested the effect of the probable IL-6R protease ADAM17 and pan-metalloprotease inhibitors on WT (D358), minor allele (A358) and an ADAM cleavage insensitive (Del 353-362) IL-6R constructs in 293T cells. Consistent with the above experiments, A358 showed decreased cell surface staining (p-value = 0.0029) while, Del 353-362 had no increase in shedding upon PMA induction (p-value = 0.580; Fig. S2B). Decreased cell surface staining of IL-6R A358 was completely abrogated by incubating the cells with pan MMP inhibitor (p-value = 0.146), or with an ADAM17 inhibitor prior to PMA treatment (p-value = 0.858; Fig. S2B). Secretion of sIL-6R by ectodomain shedding in CD4+ T cells is most likely mediated by ADAM17[51], though ADAM10 may contribute in other cell types. The cleavage site of ADAM10 on IL-6R has not previously been characterized. ADAM10 and ADAM17 have over 100 known substrates that only partially overlap[52], and no consensus cleavage sequence exists at this time[53]. To determine ADAM10 and ADAM17 activity on IL-6R, we generated synthetic peptides that contained either D358 or A358, and performed an in-vitro cleavage assay. Both ADAM10 and ADAM17 were more active on the A358 compared to D358 peptide (p-value = 4.07 × 10−9), with ADAM17 having more overall activity after 6 hours (p-value = 1.08 × 10−14; Fig. 1D, Fig. S2C). These data support the conclusion that increased circulating levels of IL-6R in A358 carriers are likely a result of increased proteolytic cleavage of membrane bound IL-6R by ADAM17 and ADAM10.
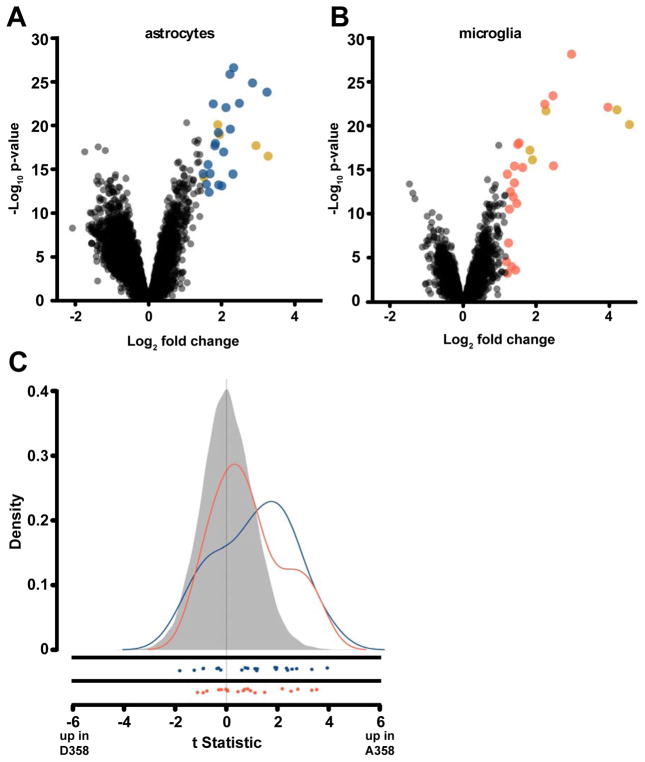
Identification of astrocyte and microglia IL-6 + sIL-6R responsive gene signatures
We next investigated whether there was evidence for differences in IL-6 signaling in Alzheimer’s disease brains conferred by p.D358A IL-6R. To address this, we first measured transcriptional responses to IL-6 + sIL-6R in co-cultures of mouse astrocytes and microglia. Activation of IL-6 signaling by treatment with IL-6 and sIL-6R was confirmed by detection of STAT3 phosphorylation (Fig. S3A). Treated co-cultures were separated into microglia-enriched and astrocyte-enriched samples and RNA was isolated for microarray analysis (see Methods). In the astrocyte-enriched samples, 68 genes were up-regulated and 246 genes were down-regulated following IL-6 + sIL-6R treatment (adjusted p-value < 0.01 and log2 fold change > 2; Fig. 2A). In the microglia-enriched cultures, 33 genes were up-regulated and 6 genes were down-regulated after IL-6 + sIL-6R treatment compared to controls (Fig. 2B) using the same significance and fold change filters. Differential expression of the top IL-6 + sIL-6R responsive genes was confirmed by qPCR (Table S1).
Fig. 2. IL-6 induced gene expression in astrocytes and microglia.
(A,B) Volcano plot for IL-6 and sIL-6R treated astrocytes and microglia. Average log2 (fold change) versus −log10 (p-value) for all genes in the (A) astrocyte and (B) microglia samples. The top 25 genes up-regulated by IL-6 and sIL-6R treatment by fold change are noted by color (blue : astrocytes, orange : microglia, and gold : both). (C) IL-6 set enriched in patients with AD and IL6R risk variant. Density of the t-statistics is shown for all genes in human AD patients (grey), astrocyte set (blue), and microglia set (orange). Each t-statistic is measuring the relative difference between the average expression in patients who carry A358 and those that do not (after adjusting for APOE, sex and batch). The individual gene statistics of the set are plotted below the density plots. IL-6 responsive gene set densities are shifted to the right, indicating higher expression of these genes in A358 carriers compared to non-carriers. The astrocyte set has the highest level of enrichment among A358 carriers with 40.9% of genes up-regulated in this set (p-value = 0.003) and the microglia gene set is also enriched (p-value = 0.0267).
As expected, genes significantly up-regulated in astrocytes and microglia were enriched in immune related Gene Ontology categories, including “inflammatory response”, “response to stress”, and “acute-phase response” (Table S2). Gene set enrichment analysis[36,54] (GSEA, see Methods) also identified previously identified IL-6 responsive gene sets derived from non-CNS cell types (Table S3). The top 25 up-regulated genes in each cell type were selected for further analysis and were used to generate IL-6 responsive gene sets (Table S1, Fig. S3B,C).
IL-6 responsive genes are enriched in brains of A358 carriers with AD
Matched genotype and gene expression data were obtained from a recent study of the temporal cortex of LOAD patients (n = 94) and normal healthy controls (n = 124)[35]. Combining the genotype and expression data, we analyzed the effect of the A358 and D358 IL6R variants on the expression of genes derived from the astrocyte and microglia experiments. First, we used GSEA as a sensitive method to examine the behavior of the IL-6 responsive gene sets in the LOAD expression data. Astrocyte-specific IL-6 responsive genes were significantly enriched in LOAD patients with the A358 allele (p-value = 0.003; Fig. 2C; Table S4). The microglia-derived gene set was also enriched among A358 carriers though to a lesser degree (p-value = 0.01, Table S4).
To assess the impact of APOE4 status on the IL-6 responsive gene sets in IL6R A358 carriers, LOAD patients were divided into two groups; APOE4 carriers and APOE4 non-carriers. Within each of these groups, IL6R A358 carriers were compared to D358 carriers and the behavior of the IL-6 responsive gene sets were examined in the resulting data using GSEA. In the APOE4 carrier group, the astrocyte gene set remained significantly enriched in IL6R A358 carriers (p-value = 0.019), but the microglia gene set did not (p-value = 0.107) (Fig. S4A; Table S6). The astrocyte IL-6 responsive gene set was also significantly enriched in APOE4 non-carriers with the A358 allele (p-value = 0.040) with the microglia gene sets again not reaching significance (p-value = 0.065) (Fig. S4B; Table S4). These data show that in both APOE4+ and APOE4− AD subjects, astrocyte derived IL-6 responsive genes were significantly enriched in carriers of the IL6R A358 allele. APOE4 carriers were also compared to APOE4 non-carriers regardless of A358 status to rule out enrichment of the IL-6 signature due to APOE mediated inflammatory effects. There was no evidence of enrichment of any of the gene sets in this comparison (Fig. S4C; Table S4), consistent with a model in which differential expression of IL-6R responsive gene sets is driven primarily by the IL-6R genotype.
We next examined individual IL-6 responsive genes in an attempt to increase sensitivity and specificity. We pre-specified genes to combine into a summary response score based on the highest observed summed fold change in the IL-6 treated astrocyte and microglia arrays that also had validation by qRT-PCR. The top pre-specified and validated genes (SERPINA3, TIMP1, TGM1 and FN1) were then tested in LOAD patients with the A358 genotype and compared to those with the D358 genotype. We found significant enrichment of these genes in A358 carrier brains, with SERPINA3 (p-value = 0.0016) and FN1 (p-value = 0.046) driving the association and TIMP1 and TGM1 also showing enrichment but to a lesser extent (Fig. S5A–D).
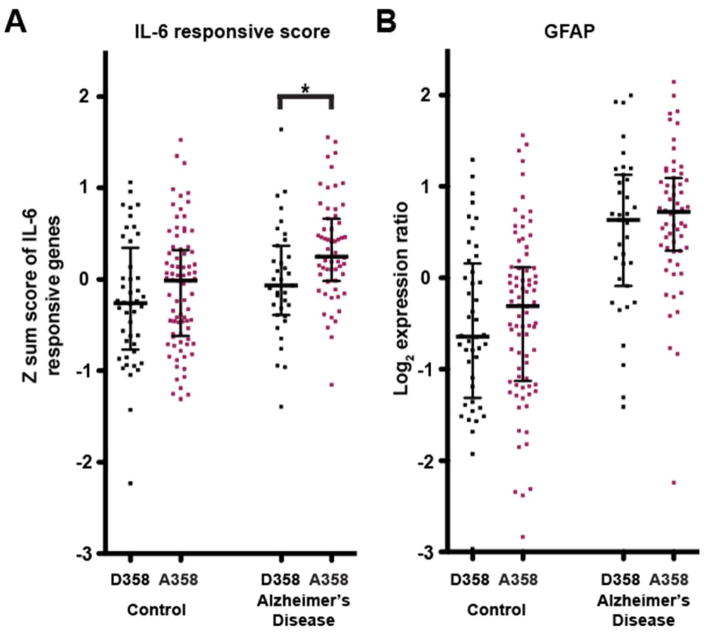
For each patient, we calculated a score representing the weighted sum of the age-, sex-, and batch-adjusted expression of the IL-6 responsive genes (see Methods). This score is significantly higher among LOAD patients who carry the A358 allele versus those who are homozygous for the D358 allele (two-sided t-test p-value = 0.0080, Fig. 3A). There was no evidence of an association between IL6R genotype and the IL-6 responsive score in the normal healthy control subjects.
Fig. 3. Elevated IL-6 responsive score in IL6R A358 carriers.
(A) IL-6 responsive scores for each subject. The score is calculated as the normalized sum of the modified expression (adjusted for batch and sex) of four genes (TIMP1, TGM1, FN1 and SERPINA3). The score is significantly higher among AD patients with A358 versus D358 (p-value = 0.0080) IL6R variant. Whiskers represent interquartile range. (B) Normalized GFAP expression in all patients stratified by disease and IL6R genotype. Note that GFAP expression is higher in AD patients versus controls (p-value < 1 × 10−15), but that this effect is not dependent on IL6R genotype. Whiskers represent interquartile range.
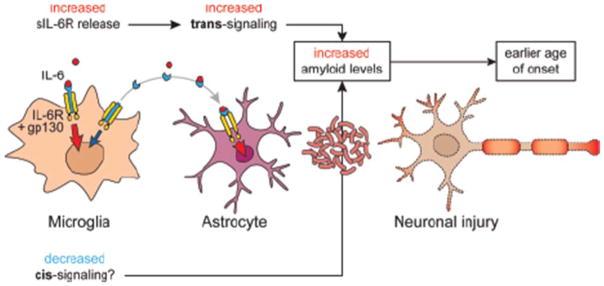
As the individual top responsive genes were induced in astrocytes, and the larger astrocyte gene set was consistently enriched, we examined the abundance of IL-6R signaling components in isolated microglia and astrocytes. In our primary cultures, Il6ra (the IL6R mouse homolog) was highly expressed in microglia compared to astrocytes, and Il6st (encoding GP130) was expressed in both astrocytes and microglia. This suggests that increased shedding of IL-6R from the cell surface of microglia may result in reduced IL-6 cis-responsiveness concomitant with an increase in trans-signaling through gp130 on astrocytes.
As the astrocyte derived IL-6 responsive gene set and individual genes in the gene score were enriched in A358 LOAD cases, we examined the expression of GFAP, a marker of astrocytes which can also be elevated under inflammatory conditions. GFAP expression was significantly higher in LOAD versus control subjects (p-value < 1 ×10−15), but there was no association between GFAP expression and IL6R genotype in LOAD subjects (p- value = 0.378, Fig. 3B). This is consistent with a model in which the change in expression of the IL-6 responsive gene signatures is A358 dependent, and not driven by a difference in astrocyte number or activity.
An IL-6R coding region SNP is a candidate modifier of age of onset
In large genome-wide association studies of LOAD, D358A IL-6R has not been associated with the risk of developing Alzheimer’s[18–25]. However, we hypothesized that the changes in neuroinflammatory signaling conferred by D358A IL-6R might be enriched in individuals with an earlier age of onset and reduced in cognitively intact elderly individuals that carry the major LOAD risk allele APO ε4. We performed an initial GWAS (primary study) aimed at identifying common variants that modify the effect of APOE risk by comparing 100 LOAD cases and 102 elderly controls that were homozygous or heterozygous for the ε4 allele and lacked ε2. To augment statistical power, we selected individuals from the extremes of the age distribution and identified cases with early age of onset LOAD (<65 years) and elderly controls (>75 years ε4/ε4, >80 ε3/ε4) with no evidence of cognitive decline. The strongest association was found at rs4474240 (OR = 3.42, p-value = 5.01 × 10−6) in the IL-6R region (Fig. S6A,B). rs4474240 is on the same haplotype (D′ = 0.88, r2 = 0.1) as a missense variant in IL6R (rs2228145, p.D358A) that has been reported to be associated with LOAD in Han Chinese[55]. Amino acid 358 is a site of cleavage of the IL-6R receptor by ADAM17 (Fig. 4A) and the A358 variant is thought to play a role in increased shedding of the extracellular portion of the receptor (as discussed above).
Fig. 4. Association of rs2228145 in IL6R with earlier age of AD onset in APOE4 carriers.
(A) The IL6R risk allele discovered in our modifier screen changes the 358th amino acid in IL-6R from D to A. This amino acid lies at the cleavage site for IL-6R. Box = transmembrane spanning residues. (B) Summary of association at rs2228145 in the five datasets. The case and controls frequencies are those of the minor allele “C”.
We examined four additional case/control cohorts (total of 1145 cases and 889 controls) matching the criteria in the primary analysis for APOE4 status and age, for association to rs4474240 and rs2228145 (IL6R p.D358A). rs4474240 showed significant, but modest, association with early onset Alzheimer’s in the replication cohorts (p-value = 0.037). In two of the four replication cohorts, IL6R p.D358A was nominally associated: US replication 1 (OR = 1.4, p-value = 0.086) and US replication 2 (OR = 1.6, p-value = 0.00062), both cohorts are from single site centers. In two other studies, we observed no significant association for IL6R p.D358A: US replication 3 (OR = 1.01, p-value = 0.72) and Icelandic (OR = 1.2, p-value = 0.51). A meta-analysis of IL6R p.D358A in all 5 cohorts resulted in a combined OR= 1.3 and p-value = 0.0003, using fixed effect meta- analysis (Fig. 4B). We conclude that IL6R D358A is a candidate modifier of age of onset in APOE4 carriers, specifically associated with early disease onset.
The analysis was then extended to an Alzheimer’s case/control cohort of totaling 7102 individuals (4121 AD cases and 2981 controls) without the age and APOE status filters used in the modifier screen above. When the whole cohort was examined for an association to IL6R A358, no significant enrichment is observed, with the frequency of IL6R A358 of 41.0% in AD cases and 40.7% in controls (Table S5). In order to test whether IL6R A358 was enriched in APOE4 carriers regardless of age of onset, we stratified the cohort by APOE4 status and observed a modest dose effect, with a 40.2% frequency of IL6R A358 in APOE4− carriers (OR all controls = 0.98), a 41.4% frequency in APOE3/4 carriers (OR relative to controls = 1.03) and a frequency of 42.0% in APOE4/4 cases (OR relative to controls = 1.05, Table S1). The age of onset in APOE3/4 and APOE4/4 carriers was tested for a correlation with IL6R A358 genotype (Table S6). Among AD cases carrying APOE3/4 and APOE4/4, a frequency of 43.1% (OR relative to APOE4+ controls = 1.17) was observed in cases with an age of onset <65 years of age, 41.3% (OR relative to APOE4+ controls = 1.09) in those with an age of onset between 66 and 74 years of age, and 40.6% (OR relative to APOE4+ controls = 1.06) in AD cases with an age of onset >75 years of age. Our data suggests that IL6R A358 is modestly enriched in all AD cases carrying the APOE4 allele, and is further enriched in cases with an age of onset <65 years of age.
Discussion
The 358A variant has been associated with elevated plasma sIL-6R levels in an allelic dose dependent manner[11,12,49] and this effect is also true in Alzheimer’s CSF with genotype being a major driver of sIL-6R concentration[4]. In the CSF, heterozygote carriers of 358A had ~45% higher levels of sIL-6R than non-carriers, however disease status (AD, mild cognitive impairment or control) had no significant effect on sIL-6R levels.
We sought to determine the functional consequences of increased cleavage and elevated levels of sIL-6R in the CNS of AD cases. We hypothesized from prior studies of IL-6 in peripheral cells expressing IL-6R that IL-6 trans-signaling may be enhanced concomitantly with cis-signaling being impaired in carriers of the rs2228145 variant, as evidenced by observations of reduced STAT3 signaling[10,56] and reduced CRP release[57]. By examination of gene expression patterns in primary cells we determined that in the CNS, microglia are the primary source of IL-6R and that astrocytes are competent for IL- 6 trans-signaling through gp130. In addition to our work, other published studies have also observed higher Il6ra (IL6R) expression in microglia compared to astrocytes and oligodendrocytes[58,59]. The expression of Il6st (gp130) however, is consistently higher in astrocytes versus neurons and oligodendrocytes[60]. We then identified IL-6 responsive genes in these cell types by stimulation with IL-6 and sIL-6R. While some gene expression differences were common between enriched cell types, the majority of differentially expressed genes were unique to either astrocytes or microglia. For example, prominent up regulation of SERPINA3 and TIMP1 was observed specifically in astrocytes. Enrichment of these individual genes in A358 carriers was observed in addition to the composite score analysis revealing a clear elevation of signal in A358 IL6R carriers with AD but little effect in control subjects. The possibility of enhanced trans-signaling and reduced cis-signaling would be congruent with our observation of a strong astrocyte derived IL-6 gene signal in LOAD subjects and a weaker IL-6 signal from the microglia derived gene set (Fig. 5).
Fig. 5.
Proposed model for IL-6 cis- and trans-signaling in the CNS.
Given the most prominently altered genes are predominantly expressed by astrocytes, we examined the distribution of GFAP expression as a way to assess the level of astrogliosis across the sample series. As has been previously observed[61,62] there was a significant increase in GFAP expression in AD cases relative to controls, but importantly this occured independently of IL6R genotype. This suggests that the observed increases in astrocyte IL-6 pathway activity are driven by IL6R genotype and not due to unequal astrocyte densities between groups.
Though APOE4 is a major risk factor for developing LOAD, it is incompletely penetrant and individuals display a wide-range of age of onset, suggesting that modifiers of APOE4 risk may exist. We designed a genetic modifier screen that compared APOE4 carriers with an early age of onset of LOAD (<65 years) to elderly cognitively intact APOE4 carriers. Our initial study identified a variant in the IL6R locus as a candidate APOE4 modifier, and subsequent studies supported this finding.
In conclusion, our data indicate that the IL6R D358A polymorphism leads to increased shedding of IL6R from the cell surface, which in AD cases leads to altered IL-6 pathway activity particularly in astrocytes, and may be associated with an earlier age of AD onset in APOE4+ cases. Therapeutics targeting IL-6 signaling for rheumatoid arthritis have been shown to be efficacious thus further work is warranted to consider inhibition of the IL-6 pathway in Alzheimer’s disease.
Supplementary Material
Acknowledgments
The authors would like to thank the patients who generously participated in this study. This work was supported by grants from the National Institutes of Health (R01-NS085419; R01-AG044546, P01-AG003991, U01 AG049508 and R01-AG035083). The recruitment and clinical characterization of research participants at Washington University were supported by NIH P50 AG05681, P01 AG03991, and P01 AG026276.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
We thank Jason Hackney, Melanie Huntley, Pascal Steiner, and Gerard Manning for their insightful comments in the design and analysis of this project. The authors would also like to thank the Genentech, Inc. microarray lab for their technical assistance with the microarray samples, GAP donation program at Genentech, and Allison Bruce for assistance with the schematic illustration.
Footnotes
Ethics Statement
The Genentech Institutional Animal Care and Use Committee approved all mouse studies.
ADNI
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California – San Francisco. ADNI is the result of efforts of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 subjects but ADNI has been followed by ADNI-GO and ADNI-2. To date these three protocols have recruited over 1500 adults, ages 55 to 90, to participate in the research, consisting of cognitively normal older individuals, people with early or late MCI, and people with early AD. The follow up duration of each group is specified in the protocols for ADNI-1, ADNI-2 and ADNI-GO. Subjects originally recruited for ADNI-1 and ADNI-GO had the option to be followed in ADNI-2. For up-to-date information, see www.adni-info.org.
Author contributions: R.R.G., J.S.K., P.C.G.H., and M.v.d.B. conceived the study and designed the experiments. T.R.B. performed the genetic analysis. N.R. performed all the experiments for functional characterization of the SNP rs2228145. P.C.G.H. implemented the cell culture and PCR experiments. J.L.L. performed the bioinformatics and statistical analyses. Q.T.P. and J.R.L. did the Mass-spec analysis. M.A.P-V, J.H, L.A.F, J.S.K, G.D.S, C.C., and A.M.G contributed genetic data and/or DNA samples. J.L.L., T.R.B, P.C.G.H., N.R, R.R.G, J.S.K., and M.v.d.B. wrote and reviewed the manuscript. All the authors read and approved the manuscript.
Competing interests: P.C.G.H., N.R., T.R.B., Q.T.P., K.S., D.V.H., J.R.L., T.W.B., J.S.K., R.R.G., and M.v.d.B. are paid employees of Genentech Inc.
References
- 1.Spooren A, Kolmus K, Laureys G, Clinckers R, De Keyser J, Haegeman G, Gerlo S. Interleukin-6, a mental cytokine. Brain Res Rev. 2011;67:157–183. doi: 10.1016/j.brainresrev.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Ferreira MAR, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souëf P, Danoy P, Baltic S, Nyholt DR, Jenkins M, Hayden C, Willemsen G, Ang W, Kuokkanen M, Beilby J, Cheah F, de Geus EJC, Ramasamy A, Vedantam S, Salomaa V, Madden PA, Heath AC, Hopper JL, Visscher PM, Musk B, Leeder SR, Jarvelin M-R, Pennell C, Boomsma DI, Hirschhorn JN, Walters H, Martin NG, James A, Jones G, Abramson MJ, Robertson CF, Dharmage SC, Brown MA, Montgomery GW, Thompson PJ. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet (London, England) 2011;378:1006–1014. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melzer D, Perry JRB, Hernandez D, Corsi A-M, Stevens K, Rafferty I, Lauretani F, Murray A, Gibbs JR, Paolisso G, Rafiq S, Simon-Sanchez J, Lango H, Scholz S, Weedon MN, Arepalli S, Rice N, Washecka N, Hurst A, Britton A, Henley W, van de Leemput J, Li R, Newman AB, Tranah G, Harris T, Panicker V, Dayan C, Bennett A, McCarthy MI, Ruokonen A, Jarvelin M-R, Guralnik J, Bandinelli S, Frayling TM, Singleton A, Ferrucci L. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kauwe JSK, Bailey MH, Ridge PG, Perry R, Wadsworth ME, Hoyt KL, Staley LA, Karch CM, Harari O, Cruchaga C, Ainscough BJ, Bales K, Pickering EH, Bertelsen S, Fagan AM, Holtzman DM, Morris JC, Goate AM. Genome-wide association study of CSF levels of 59 alzheimer’s disease candidate proteins: significant associations with proteins involved in amyloid processing and inflammation. PLoS Genet. 2014;10:e1004758. doi: 10.1371/journal.pgen.1004758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dehghan A, Dupuis J, Barbalic M, Bis JC, Eiriksdottir G, Lu C, Pellikka N, Wallaschofski H, Kettunen J, Henneman P, Baumert J, Strachan DP, Fuchsberger C, Vitart V, Wilson JF, Paré G, Naitza S, Rudock ME, Surakka I, de Geus EJC, Alizadeh BZ, Guralnik J, Shuldiner A, Tanaka T, Zee RYL, Schnabel RB, Nambi V, Kavousi M, Ripatti S, Nauck M, Smith NL, Smith AV, Sundvall J, Scheet P, Liu Y, Ruokonen A, Rose LM, Larson MG, Hoogeveen RC, Freimer NB, Teumer A, Tracy RP, Launer LJ, Buring JE, Yamamoto JF, Folsom AR, Sijbrands EJG, Pankow J, Elliott P, Keaney JF, Sun W, Sarin A-P, Fontes JD, Badola S, Astor BC, Hofman A, Pouta A, Werdan K, Greiser KH, Kuss O, Meyer zu Schwabedissen HE, Thiery J, Jamshidi Y, Nolte IM, Soranzo N, Spector TD, Völzke H, Parker AN, Aspelund T, Bates D, Young L, Tsui K, Siscovick DS, Guo X, Rotter JI, Uda M, Schlessinger D, Rudan I, Hicks AA, Penninx BW, Thorand B, Gieger C, Coresh J, Willemsen G, Harris TB, Uitterlinden AG, Järvelin M-R, Rice K, Radke D, Salomaa V, Willems van Dijk K, Boerwinkle E, Vasan RS, Ferrucci L, Gibson QD, Bandinelli S, Snieder H, Boomsma DI, Xiao X, Campbell H, Hayward C, Pramstaller PP, van Duijn CM, Peltonen L, Psaty BM, Gudnason V, Ridker PM, Homuth G, Koenig W, Ballantyne CM, Witteman JCM, Benjamin EJ, Perola M, Chasman DI. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123:731–738. doi: 10.1161/CIRCULATIONAHA.110.948570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabater-Lleal M, Huang J, Chasman D, Naitza S, Dehghan A, Johnson AD, Teumer A, Reiner AP, Folkersen L, Basu S, Rudnicka AR, Trompet S, Mälarstig A, Baumert J, Bis JC, Guo X, Hottenga JJ, Shin S-Y, Lopez LM, Lahti J, Tanaka T, Yanek LR, Oudot-Mellakh T, Wilson JF, Navarro P, Huffman JE, Zemunik T, Redline S, Mehra R, Pulanic D, Rudan I, Wright AF, Kolcic I, Polasek O, Wild SH, Campbell H, Curb JD, Wallace R, Liu S, Eaton CB, Becker DM, Becker LC, Bandinelli S, Räikkönen K, Widen E, Palotie A, Fornage M, Green D, Gross M, Davies G, Harris SE, Liewald DC, Starr JM, Williams FMK, Grant PJ, Spector TD, Strawbridge RJ, Silveira A, Sennblad B, Rivadeneira F, Uitterlinden AG, Franco OH, Hofman A, van Dongen J, Willemsen G, Boomsma DI, Yao J, Swords Jenny N, Haritunians T, McKnight B, Lumley T, Taylor KD, Rotter JI, Psaty BM, Peters A, Gieger C, Illig T, Grotevendt A, Homuth G, Völzke H, Kocher T, Goel A, Franzosi MG, Seedorf U, Clarke R, Steri M, Tarasov KV, Sanna S, Schlessinger D, Stott DJ, Sattar N, Buckley BM, Rumley A, Lowe GD, McArdle WL, Chen M-H, Tofler GH, Song J, Boerwinkle E, Folsom AR, Rose LM, Franco-Cereceda A, Teichert M, Ikram MA, Mosley TH, Bevan S, Dichgans M, Rothwell PM, Sudlow CLM, Hopewell JC, Chambers JC, Saleheen D, Kooner JS, Danesh J, Nelson CP, Erdmann J, Reilly MP, Kathiresan S, Schunkert H, Morange P-E, Ferrucci L, Eriksson JG, Jacobs D, Deary IJ, Soranzo N, Witteman JCM, de Geus EJC, Tracy RP, Hayward C, Koenig W, Cucca F, Jukema JW, Eriksson P, Seshadri S, Markus HS, Watkins H, Samani NJ, Wallaschofski H, Smith NL, Tregouet D, Ridker PM, Tang W, Strachan DP, Hamsten A, O’Donnell CJ. Multiethnic meta-analysis of genome-wide association studies in >100 000 subjects identifies 23 fibrinogen-associated Loci but no strong evidence of a causal association between circulating fibrinogen and cardiovascular disease. Circulation. 2013;128:1310–1324. doi: 10.1161/CIRCULATIONAHA.113.002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott P, Chambers JC, Zhang W, Clarke R, Hopewell JC, Peden JF, Erdmann J, Braund P, Engert JC, Bennett D, Coin L, Ashby D, Tzoulaki I, Brown IJ, Mt-Isa S, McCarthy MI, Peltonen L, Freimer NB, Farrall M, Ruokonen A, Hamsten A, Lim N, Froguel P, Waterworth DM, Vollenweider P, Waeber G, Jarvelin M-R, Mooser V, Scott J, Hall AS, Schunkert H, Anand SS, Collins R, Samani NJ, Watkins H, Kooner JS. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA. 2009;302:37–48. doi: 10.1001/jama.2009.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, Yoshida S, Graham RR, Manoharan A, Ortmann W, Bhangale T, Denny JC, Carroll RJ, Eyler AE, Greenberg JD, Kremer JM, Pappas DA, Jiang L, Yin J, Ye L, Su D-F, Yang J, Xie G, Keystone E, Westra H-J, Esko T, Metspalu A, Zhou X, Gupta N, Mirel D, Stahl EA, Diogo D, Cui J, Liao K, Guo MH, Myouzen K, Kawaguchi T, Coenen MJH, van Riel PLCM, van de Laar MAFJ, Guchelaar H-J, Huizinga TWJ, Dieudé P, Mariette X, Bridges SL, Zhernakova A, Toes REM, Tak PP, Miceli-Richard C, Bang S-Y, Lee H-S, Martin J, Gonzalez-Gay MA, Rodriguez-Rodriguez L, Rantapää-Dahlqvist S, Arlestig L, Choi HK, Kamatani Y, Galan P, Lathrop M, Eyre S, Bowes J, Barton A, de Vries N, Moreland LW, Criswell LA, Karlson EW, Taniguchi A, Yamada R, Kubo M, Liu JS, Bae S-C, Worthington J, Padyukov L, Klareskog L, Gregersen PK, Raychaudhuri S, Stranger BE, De Jager PL, Franke L, Visscher PM, Brown MA, Yamanaka H, Mimori T, Takahashi A, Xu H, Behrens TW, Siminovitch KA, Momohara S, Matsuda F, Yamamoto K, Plenge RM. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baran P, Nitz R, Grötzinger J, Scheller J, Garbers C. Minimal interleukin 6 (IL-6) receptor stalk composition for IL-6 receptor shedding and IL-6 classic signaling. J Biol Chem. 2013;288:14756–14768. doi: 10.1074/jbc.M113.466169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferreira RC, Freitag DF, Cutler AJ, Howson JMM, Rainbow DB, Smyth DJ, Kaptoge S, Clarke P, Boreham C, Coulson RM, Pekalski ML, Chen W-M, Onengut-Gumuscu S, Rich SS, Butterworth AS, Malarstig A, Danesh J, Todd JA. Functional IL6R 358Ala allele impairs classical IL-6 receptor signaling and influences risk of diverse inflammatory diseases. PLoS Genet. 2013;9:e1003444. doi: 10.1371/journal.pgen.1003444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galicia JC, Tai H, Komatsu Y, Shimada Y, Akazawa K, Yoshie H. Polymorphisms in the IL-6 receptor (IL-6R) gene: strong evidence that serum levels of soluble IL-6R are genetically influenced. Genes Immun. 2004;5:513–516. doi: 10.1038/sj.gene.6364120. [DOI] [PubMed] [Google Scholar]
- 12.Marinou I, Walters K, Winfield J, Bax DE, Wilson AG. A gain of function polymorphism in the interleukin 6 receptor influences RA susceptibility. Ann Rheum Dis. 2010;69:1191–1194. doi: 10.1136/ard.2008.100644. [DOI] [PubMed] [Google Scholar]
- 13.Garbers C, Jänner N, Chalaris A, Moss ML, Floss DM, Meyer D, Koch-Nolte F, Rose-John S, Scheller J. Species specificity of ADAM10 and ADAM17 proteins in interleukin-6 (IL-6) trans-signaling and novel role of ADAM10 in inducible IL-6 receptor shedding. J Biol Chem. 2011;286:14804–14811. doi: 10.1074/jbc.M111.229393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matthews V, Schuster B, Schütze S, Bussmeyer I, Ludwig A, Hundhausen C, Sadowski T, Saftig P, Hartmann D, Kallen K-J, Rose-John S. Cellular cholesterol depletion triggers shedding of the human interleukin-6 receptor by ADAM10 and ADAM17 (TACE) J Biol Chem. 2003;278:38829–38839. doi: 10.1074/jbc.M210584200. [DOI] [PubMed] [Google Scholar]
- 15.Garbers C, Monhasery N, Aparicio-Siegmund S, Lokau J, Baran P, Nowell MA, Jones SA, Rose-John S, Scheller J. The interleukin-6 receptor Asp358Ala single nucleotide polymorphism rs2228145 confers increased proteolytic conversion rates by ADAM proteases. Biochim Biophys Acta. 2014;1842:1485–1494. doi: 10.1016/j.bbadis.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 16.Villegas-Llerena C, Phillips A, Reitboeck PG, Hardy J, Pocock JM. Microglial genes regulating neuroinflammation in the progression of Alzheimer’s disease. Curr Opin Neurobiol. 2015;36:74–81. doi: 10.1016/j.conb.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Blennow K, de Leon MJ, Zetterberg H. Alzheimer’s disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- 18.Reitz C, Jun G, Naj A, Rajbhandary R, Vardarajan BN, Wang L-S, Valladares O, Lin C-F, Larson EB, Graff-Radford NR, Evans D, De Jager PL, Crane PK, Buxbaum JD, Murrell JR, Raj T, Ertekin-Taner N, Logue M, Baldwin CT, Green RC, Barnes LL, Cantwell LB, Fallin MD, Go RCP, Griffith P, Obisesan TO, Manly JJ, Lunetta KL, Kamboh MI, Lopez OL, Bennett DA, Hendrie H, Hall KS, Goate AM, Byrd GS, Kukull WA, Foroud TM, Haines JL, Farrer LA, Pericak-Vance MA, Schellenberg GD, Mayeux R. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E ε4, and the risk of late-onset Alzheimer disease in African Americans. JAMA. 2013;309:1483–1492. doi: 10.1001/jama.2013.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Morón FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fiévet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossù P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nöthen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JSK, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Lambert J-C, Amouyel P, Goate A, Rademakers R, Morgan K, Powell J, St George-Hyslop P, Singleton A, Hardy J. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J-C, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ERLC, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Rüther E, Schürmann B, Heun R, Kölsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frölich L, Hampel H, Gallacher J, Hüll M, Rujescu D, Giegling I, Goate AM, Kauwe JSK, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Mühleisen TW, Nöthen MM, Moebus S, Jöckel K-H, Klopp N, Wichmann H-E, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MMB, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues J-F, Tzourio C, Alpérovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Björnsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossù P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schürmann B, Heun R, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frölich L, Hampel H, Hüll M, Rujescu D, Goate AM, Kauwe JSK, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Mühleisen TW, Nöthen MM, Moebus S, Jöckel K-H, Klopp N, Wichmann H-E, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambert J-C, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fiévet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossù P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanché H, Dartigues J-F, Tzourio C, Gut I, Van Broeckhoven C, Alpérovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 24.Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JSK, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin L-W, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EMC, Ramirez-Lorca R, Debette S, Longstreth WT, Janssens ACJW, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MMB. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 27.Genin E, Hannequin D, Wallon D, Sleegers K, Hiltunen M, Combarros O, Bullido MJ, Engelborghs S, De Deyn P, Berr C, Pasquier F, Dubois B, Tognoni G, Fiévet N, Brouwers N, Bettens K, Arosio B, Coto E, Del Zompo M, Mateo I, Epelbaum J, Frank-Garcia A, Helisalmi S, Porcellini E, Pilotto A, Forti P, Ferri R, Scarpini E, Siciliano G, Solfrizzi V, Sorbi S, Spalletta G, Valdivieso F, Vepsäläinen S, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossù P, Hanon O, Piccardi P, Annoni G, Seripa D, Galimberti D, Licastro F, Soininen H, Dartigues J-F, Kamboh MI, Van Broeckhoven C, Lambert JC, Amouyel P, Campion D. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol Psychiatry. 2011;16:903–907. doi: 10.1038/mp.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benitez BA, Jin SC, Guerreiro R, Graham R, Lord J, Harold D, Sims R, Lambert J-C, Gibbs JR, Bras J, Sassi C, Harari O, Bertelsen S, Lupton MK, Powell J, Bellenguez C, Brown K, Medway C, Haddick PCG, van der Brug MP, Bhangale T, Ortmann W, Behrens T, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD, Haines JL, Turton J, Braae A, Barber I, Fagan AM, Holtzman DM, Morris JC, Williams J, Kauwe JSK, Amouyel P, Morgan K, Singleton A, Hardy J, Goate AM, Cruchaga C. Missense variant in TREML2 protects against Alzheimer’s disease. Neurobiol Aging. 2014;35:1510, e19–26. doi: 10.1016/j.neurobiolaging.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smyth G. limma: Linear Models for Microarray Data. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. 2005:397–420. [Google Scholar]
- 31.Huntley MA, Larson JL, Chaivorapol C, Becker G, Lawrence M, Hackney JA, Kaminker JS. ReportingTools: an automated result processing and presentation toolkit for high-throughput genomic analyses. Bioinformatics. 2013;29:3220–3221. doi: 10.1093/bioinformatics/btt551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dasu MRK, Hawkins HK, Barrow RE, Xue H, Herndon DN. Gene expression profiles from hypertrophic scar fibroblasts before and after IL-6 stimulation. J Pathol. 2004;202:476–485. doi: 10.1002/path.1539. [DOI] [PubMed] [Google Scholar]
- 33.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P, Rohrer K, Zhao A, Marlowe L, Kaleem M, McCorquodale DS, Cuello C, Leung D, Bryden L, Nath P, Zismann VL, Joshipura K, Huentelman MJ, Hu-Lince D, Coon KD, Craig DW, Pearson JV, Heward CB, Reiman EM, Stephan D, Hardy J, Myers AJ. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang Z, Gentleman R. Extensions to gene set enrichment. Bioinformatics. 2007;23:306–313. doi: 10.1093/bioinformatics/btl599. [DOI] [PubMed] [Google Scholar]
- 38.Wu D, Lim E, Vaillant F, Asselin-Labat M-L, Visvader JE, Smyth GK. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics. 2010;26:2176–2182. doi: 10.1093/bioinformatics/btq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Breitner JC, Wyse BW, Anthony JC, Welsh-Bohmer KA, Steffens DC, Norton MC, Tschanz JT, Plassman BL, Meyer MR, Skoog I, Khachaturian A. APOE-epsilon4 count predicts age when prevalence of AD increases, then declines: the Cache County Study. Neurology. 1999;53:321–331. doi: 10.1212/wnl.53.2.321. [DOI] [PubMed] [Google Scholar]
- 40.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 41.Nagy Z, Yilmazer-Hanke DM, Braak H, Braak E, Schultz C, Hanke J. Assessment of the pathological stages of Alzheimer’s disease in thin paraffin sections: a comparative study. Dement Geriatr Cogn Disord. 9:140–144. doi: 10.1159/000017038. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 45.Delaneau O, Zagury J-F, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 46.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marinou I, Maxwell JR, Wilson AG. Genetic influences modulating the radiological severity of rheumatoid arthritis. Ann Rheum Dis. 2010;69:476–482. doi: 10.1136/ard.2009.117721. [DOI] [PubMed] [Google Scholar]
- 49.Deming Y, Xia J, Cai Y, Lord J, Del-Aguila JL, Fernandez MV, Carrell D, Black K, Budde J, Ma S, Saef B, Howells B, Bertelsen S, Bailey M, Ridge PG, Holtzman D, Morris JC, Bales K, Pickering EH, Lee J-M, Heitsch L, Kauwe J, Goate A, Piccio L, Cruchaga C. Genetic studies of plasma analytes identify novel potential biomarkers for several complex traits. Sci Rep. 2016;6:18092. doi: 10.1038/srep18092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müllberg J, Oberthür W, Lottspeich F, Mehl E, Dittrich E, Graeve L, Heinrich PC, Rose-John S. The soluble human IL-6 receptor. Mutational characterization of the proteolytic cleavage site. J Immunol. 1994;152:4958–4968. [PubMed] [Google Scholar]
- 51.Briso EM, Dienz O, Rincon M. Cutting edge: soluble IL-6R is produced by IL-6R ectodomain shedding in activated CD4 T cells. J Immunol. 2008;180:7102–7106. doi: 10.4049/jimmunol.180.11.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheller J, Chalaris A, Garbers C, Rose-John S. ADAM17: a molecular switch to control inflammation and tissue regeneration. Trends Immunol. 2011;32:380–387. doi: 10.1016/j.it.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 53.Caescu CI, Jeschke GR, Turk BE. Active-site determinants of substrate recognition by the metalloproteinases TACE and ADAM10. Biochem J. 2009;424:79–88. doi: 10.1042/BJ20090549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 55.Wang M, Song H, Jia J. Interleukin-6 receptor gene polymorphisms were associated with sporadic Alzheimer’s disease in Chinese Han. Brain Res. 2010;1327:1–5. doi: 10.1016/j.brainres.2010.02.067. [DOI] [PubMed] [Google Scholar]
- 56.Harrison SC, Smith AJP, Jones GT, Swerdlow DI, Rampuri R, Bown MJ, Folkersen L, Baas AF, de Borst GJ, Blankensteijn JD, Price JF, van der Graaf Y, McLachlan S, Agu O, Hofman A, Uitterlinden AG, Franco-Cereceda A, Ruigrok YM, van’t Hof FN, Powell JT, van Rij AM, Casas JP, Eriksson P, Holmes MV, Asselbergs FW, Hingorani AD, Humphries SE. Interleukin-6 receptor pathways in abdominal aortic aneurysm. Eur Heart J. 2013;34:3707–3716. doi: 10.1093/eurheartj/ehs354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wypasek E, Potaczek DP, Lamplmayr M, Sadowski J, Undas A. Interleukin-6 receptor Asp358Ala gene polymorphism is associated with plasma C-reactive protein levels and severity of aortic valve stenosis. Clin Chem Lab Med. 2014;52:1049–1056. doi: 10.1515/cclm-2013-0606. [DOI] [PubMed] [Google Scholar]
- 58.Orre M, Kamphuis W, Osborn LM, Melief J, Kooijman L, Huitinga I, Klooster J, Bossers K, Hol EM. Acute isolation and transcriptome characterization of cortical astrocytes and microglia from young and aged mice. Neurobiol Aging. 2014;35:1–14. doi: 10.1016/j.neurobiolaging.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 59.Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang L, Means TK, El Khoury J. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Panter SS, McSwigan JD, Sheppard JR, Emory CR, Frey WH. Glial fibrillary acidic protein and Alzheimer’s disease. Neurochem Res. 1985;10:1567–1576. doi: 10.1007/BF00988599. [DOI] [PubMed] [Google Scholar]
- 62.Duffy PE, Rapport M, Graf L. Glial fibrillary acidic protein and Alzheimer-type senile dementia. Neurology. 1980;30:778–782. doi: 10.1212/wnl.30.7.778. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.