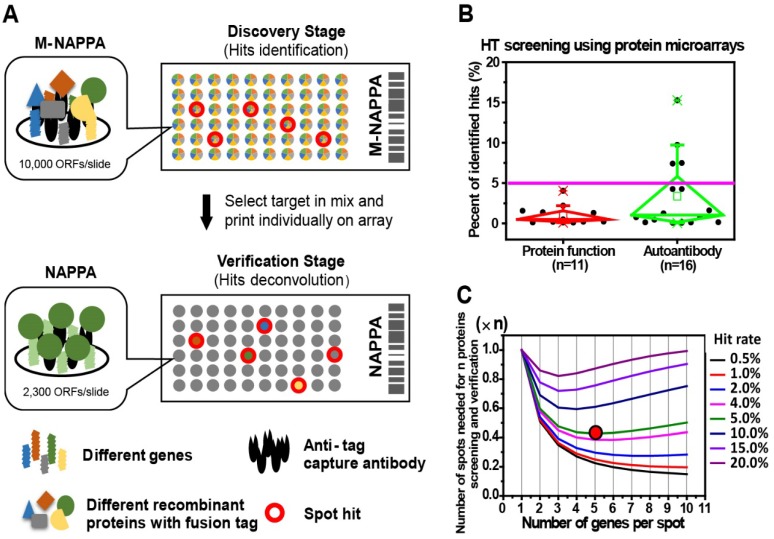
Figure 1.
Multiplexed proteins per feature is made possible by low hit rates in HT screening studies. (A) Schematic illustration of unbiased target discovery using M-NAPPA protein microarrays where each spot contains five gene plasmids encoding for different proteins. The hits identified from high-density M-NAPPA protein microarrays could be deconvoluted by non-multiplexed NAPPA arrays. (B) Percentage of hits identified in high throughput protein function and autoantibody biomarker studies in 37 publications within the last five years. (C) Plot of the total number of features needed to evaluate hits in a primary screen followed by verification study at different multiplicities and hit rates. The red arrow points to the optimal number of genes printed within each spot for 10k proteins with an estimated hit rate of 5%.