Abstract
Background
Analyses of multicenter studies often need to account for center clustering to ensure valid inference. For binary outcomes, it is particularly challenging to properly adjust for center when the number of centers or total sample size is small, or when there are few events per center. Our objective was to evaluate the performance of generalized estimating equation (GEE) log-binomial and Poisson models, generalized linear mixed models (GLMMs) assuming binomial and Poisson distributions, and a Bayesian binomial GLMM to account for center effect in these scenarios.
Methods
We conducted a simulation study with few centers (≤30) and 50 or fewer subjects per center, using both a randomized controlled trial and an observational study design to estimate relative risk. We compared the GEE and GLMM models with a log-binomial model without adjustment for clustering in terms of bias, root mean square error (RMSE), and coverage. For the Bayesian GLMM, we used informative neutral priors that are skeptical of large treatment effects that are almost never observed in studies of medical interventions.
Results
All frequentist methods exhibited little bias, and the RMSE was very similar across the models. The binomial GLMM had poor convergence rates, ranging from 27% to 85%, but performed well otherwise. The results show that both GEE models need to use small sample corrections for robust SEs to achieve proper coverage of 95% CIs. The Bayesian GLMM had similar convergence rates but resulted in slightly more biased estimates for the smallest sample sizes. However, it had the smallest RMSE and good coverage across all scenarios. These results were very similar for both study designs.
Conclusions
For the analyses of multicenter studies with a binary outcome and few centers, we recommend adjustment for center with either a GEE log-binomial or Poisson model with appropriate small sample corrections or a Bayesian binomial GLMM with informative priors.
Electronic supplementary material
The online version of this article (doi:10.1186/s13063-017-2248-1) contains supplementary material, which is available to authorized users.
Keywords: Multicenter studies, Relative risk, Generalized linear mixed models, Generalized estimating equations, Correlated binary data, Bayesian log binomial
Background
In multicenter studies, outcomes from the same center cannot be assumed to be independent, and analyses often need to account for center clustering. Neglecting to account for center may lead to erroneous conclusions, particularly when randomization is stratified by center [1–4]. Yet, authors of a recent review of multicenter studies published in four major medical journals (BMJ, New England Journal of Medicine, JAMA, and The Lancet) found that only 22% of randomized controlled trials (RCTs) with a binary outcome reported accounting for a center effect, a rate similar to past reviews [5, 6]. This result may be due to the fact that it is challenging to properly adjust for center when there are few centers, total sample size is small, or there are few events per center. Clear practical guidelines for the statistical analyses and reporting of multicenter studies are needed to assist investigators and data analysts in conducting appropriate multicenter analyses.
The best-suited methods to adjust for center include generalized estimating equations (GEEs) and generalized linear mixed models (GLMMs; also referred to as random effects, multilevel, or mixed effects models). However, careful application of these methods is needed for studies with few centers. For example, the robust SEs typically reported for GEEs are downward-biased when the number of centers is < 50 [7]. For GLMMs, the approximate Wald test and CIs may have inflated type I error rates [8].
Our objective in this article is to review and evaluate both frequentist and Bayesian state-of-the-art statistical methods for estimating relative risk (RR) for multicenter studies. We focus on RRs rather than ORs because RRs are considered a more meaningful and interpretable treatment measure [9, 10], and few studies have evaluated methods for estimating adjusted RRs. We provide recommendations and practical guidelines for analyzing both RCTs and observational multicenter studies.
Methods
We review three methods for estimating RRs: log-binomial regression, the widely used GEE method, and GLMMs. Specifically, we evaluate a total of ten regression models:
Log binomial
GEE binomial
GEE binomial with small-sample correction of SEs
GEE Poisson
GEE Poisson with small-sample correction of SEs
GLMM binomial
GLMM binomial with bootstrapped SEs
GLMM Poisson
GLMM Poisson with bootstrapped SEs
Bayesian GLMM binomial
We assume a study design with ≤ 30 centers, binary outcome, binary treatment/exposure variable, and a binary baseline covariate that could be a stratifying variable or a potential confounder. We also assume the center size variation is not very large (i.e., coefficient of variation < 0.40). Table 1 provides a summary of all the models and details of their specification evaluated in the simulation study.
Table 1.
Summary of details of ten regression models evaluated in the simulation study
| Model | SEs | 95% CI or posterior interval | Other assumptions |
|---|---|---|---|
| GLM-Bin | Model-based, unadjusted for center correlation | Wald | |
| GEE binomial | Robust sandwich | t-based | Exchangeable working correlation |
| GEE binomial KC-correcteda | Robust sandwich with small-sample correction | t-based | Exchangeable working correlation |
| GEE Poisson | Robust sandwich | t-based | Exchangeable working correlation |
| GEE Poisson KC-correcteda | Robust sandwich with small-sample correction | t-based | Exchangeable working correlation |
| GLMM binomial | Model-based | Wald | Adaptive quadrature with 10 points |
| GLMM binomial bootstrapa | Parametric bootstrap | Parametric bootstrap, quantile-based | Laplace for fitting bootstrap samples |
| GLMM Poisson | Model-based | Wald | Adaptive quadrature with 10 points |
| GLMM Poisson bootstrapa | Parametric bootstrap | Parametric bootstrap, quantile-based | Laplace for fitting bootstrap samples |
| Bayesian binomial GLMM | Posterior SD | Quantile-based posterior interval | Priors β0 ~ Normal(0,102); β1, β2 ~ Normal(0,1); σ ~ half-Normal(0,1) |
Abbreviations: GEE Generalized estimating equation, GLM-Bin Log-binomial regression model, GLMM Generalized linear mixed model, KC Kauermann and Carroll
aThe small sample KC correction or bootstrap samples correct only the SEs and 95% CIs and do not affect the point estimates of the risk ratio
We do not investigate methods treating center as a fixed effect, because problems with this approach (including exclusion of patients or centers, biased treatment effect estimate, and increased type I error) have been noted before [2–4].
Log-binomial regression
Binary regression models (i.e., logistic) without any adjustment for center correlation are the most often used methods for analyzing multicenter studies. To estimate RRs instead of ORs, we can use a log-binomial regression model (GLM-Bin) adjusting for covariates but with no adjustment for center correlation. Letting y ij be the observed binary outcome (yes/no) for subject i in center j, this model is specified as
| 1 |
where p ij is the probability of the outcome y ij, x 1ij is the treatment/exposure indicator, and x 2ij is a binary baseline covariate. A Bernoulli distribution is assumed for y ij. The RR of the treatment/exposure is given by exp(β1). Model-based SEs (obtained from the regression model and unadjusted for center clustering) and Wald-type 95% CIs [ 1 ± 1.96 × SE( 1)] are usually reported for this model. In addition to ignoring within-center correlation, this model also has the disadvantage of convergence problems at the parameter boundary and can lead to probability estimates > 1.
GEE models
The GEE log-binomial and log Poisson models take the same form as model (1), assuming either a binomial or Poisson distribution for y. However, the SEs are corrected by using GEEs with an exchangeable correlation matrix, which assumes that patient outcomes from the same center are correlated but are independent from patient outcomes in different centers. GEE Poisson (also referred to as modified Poisson) regression is widely used to estimate RRs because it provides consistent estimates of the RR and is more stable than the GEE binomial model [9, 11]. Similarly to the GLM-Bin, probability estimates from both of these GEE models can be > 1.
Robust sandwich SEs for possible misspecification of the covariance structure (and misspecification of the distribution for Poisson regression) are typically used with GEE methods [12]. When the number of centers is small, a bias-corrected variance sandwich estimator is needed to provide correct inference [13]. We use the Kauermann and Carroll (KC) [14] correction of robust SEs because it has been shown to perform well with small numbers of centers [15].
Similarly, the Wald test and CIs typically reported for GEEs have been noted to have inflated type I errors with few centers [15, 16]. An approximate t statistic that accounts for the large variation in the sandwich estimator often present with small samples has been shown to perform better than the Wald test in this setting [17].
For both the binomial and Poisson GEE models, we assess the performance of (1) robust SEs coupled with approximate t-based 95% CIs [ 1 ± t d × SErobust( 1)] and (2) KC-corrected robust SEs with t-based 95% CIs for the RR.
GLMMs
Random effects models account for within-center correlation by including a random center intercept. Estimates derived from GLMMs are interpreted as center-specific (or conditional) as opposed to the marginal interpretation of GEE estimates. However, we note that under a log link, the estimated treatment/exposure effect is the same for GEE and GLMMs [18]. Again letting y ij be the observed binary outcome for subject i in center j, a binomial GLMM is specified as
| 2 |
where u j is the random center effect and x 1ij and x 2ij are the treatment/exposure indicator and baseline binary covariate, respectively.
We fit GLMMs using adaptive Gauss-Hermite quadrature with 10 quadrature points to provide a good compromise between accuracy and computation time in the simulation study [19]. We use two methods to compute SEs and 95% CIs. First, we calculate Wald 95% CIs for the RR. Although t statistics with various methods for calculating the degrees of freedom have been proposed as a better alternative to the Wald test, these are still approximations in GLMMs and may not perform well, particularly with few centers [8]. Hence, in the second method, we use a parametric bootstrap to calculate SEs and 95% CIs, which is a better alternative for computing CIs for GLMMs [20].
We also assess the performance of log Poisson GLMM using model-based SEs coupled with Wald 95% CIs and compare them with those obtained from a parametric bootstrap. We again note that probability estimates from these GLMMs may be > 1.
Bayesian binomial GLMM
A Bayesian approach provides several advantages, including the ability to give direct estimates of probability of benefit or harm from an intervention or exposure [21]. For the binomial GLMM, weakly informative prior distributions help stabilize the parametric estimates and hence address the convergence issues often seen with the frequentist approach [10, 22–24]. Constraints on the parameters are also easily implemented to avoid probability estimates > 1 [22]. In contrast to frequentist methods, Bayesian SEs and credible intervals (CrIs) for the RR account for all uncertainty in the model, including the between-center variation. Another advantage is that Bayesian inference does not rely on asymptotic results, which is an important issue when the number of centers is limited. A Bayesian approach also allows for the inclusion of informative priors derived from external information to exclude unrealistic RR values [25, 26].
We investigate the performance of a Bayesian GLMM with the same form as that in model 2. For prior distributions, we use neutral priors for all parameters to represent equipoise: a Normal(0,102) for the intercept β0, Normal(0,1) for β1 and β2, and half-Normal(0,1) for σ. We use slightly informative priors on β1 and β2 with a 95% CrI of 0.14–7 in the RR scale to exclude unrealistic RR values that are almost never observed in studies of medical interventions. Similar priors skeptical of large treatment effects have been studied and shown to have good operating characteristics even with small sample sizes [26]. The half-Normal prior for the SD of the random center effect σ is a weakly informative prior that has been shown to perform well [27]. We constrain all p ij < 1 in the model (see Additional file 1 for sample code).
Simulation study
We conducted a simulation study assuming both a multicenter two-arm RCT and a multicenter observational study design. For each scenario, we simulated 1000 datasets from model 2 with 4, 10, or 30 centers. The number of subjects per center was sampled from a Poisson distribution with mean of 10, 20, and 50 to give average (expected) total samples sizes ranging from 40 to 1500. Under the RCT scenarios, randomization was stratified by center using permuted blocks of size 4. The covariate x 2ij was generated from Bernoulli(0.3), and the random center effect uj from Normal(0,0.4) to induce an intracluster correlation coefficient (ICC) of 0.08, where σ2 = ICC×(1−)/ and is the average probability in the sample [28]. The ICC represents the degree of dependence or correlation among observations from individuals within the same cluster or center [27]. The ICC value used in this simulation is within the range of values previously reported in cluster clinical trials [29].
For all RCT scenarios, we assumed a control outcome rate of 15% [i.e., exp(β0) = 0.15]. The treatment and covariate effects were both set to an RR of 1.5 [i.e., β1 = β2 = log(1.5)]. Whenever the simulated p ij was > 1, a new value of the random center effect u j was sampled until p ij < 1.
For the observational study scenarios, we assigned half of the subjects to exposure and the other half to a nonexposure group. To induce confounding, we generated the binary covariate x 2ij with prevalence of 0.4 in the exposure group and 0.2 in the nonexposed group using a discretized multivariate Normal method [30]. All other methods and parameters were the same as under the RCT scenarios.
Each dataset was analyzed using all the methods listed in Table 1. For the binomial and Poisson GLMMs, we used 3000 bootstrap samples for each dataset to calculate the bootstrap SEs and 95% CIs (from the quantiles). To speed up the calculation, we used Laplace approximation when fitting the models to the bootstrap samples.
The Bayesian GLMM was fitted via Markov chain Monte Carlo (MCMC) methods [31]. We used 3 MCMC chains, each with 10,000 iterations using the first 2000 as burn-in. Starting values were sampled from the estimated coefficients and SEs of the frequentist log Poisson model. We visually inspected the trace plots of all estimated parameters for the first 50 datasets of each scenario to monitor convergence of the chains. We additionally calculated the convergence diagnostic and deemed any datasets with an > 1.1 for any parameter as exhibiting nonconvergence (see below) [32]. We captured the posterior median of all four parameters and the 2.5% and 97.5% percentiles of β1 to calculate coverage of the 95% posterior interval. As a sensitivity analysis, for scenarios with ten subjects per center, we also fitted the Bayesian GLMM using vague Normal(0,104) priors for β0, β1, and β2 and half-Cauchy(0,1) [33] for σ.
For all models, we calculated the bias (βestimate − βtrue), root mean square error (RMSE), coverage of the 95% CI or posterior interval for the treatment/exposure effect β1, and convergence rate. We defined convergence as the percentage of simulated datasets where (1) the model converged (i.e., no error messages); (2) the absolute values of the point estimates for β0, β1, and β2 were < 5 (larger values would indicate unstable estimates); and (3) for the Bayesian models, the values for all parameters were < 1.10. For each model and scenario, we assessed bias, RMSE, and coverage only in datasets where convergence was achieved.
All simulations and analyses were conducted in R [34]. For the fitting of GEE models, we used the geepack [35] and geesmv [36] packages to calculate the degrees of freedom for the t statistic and the KC-corrected robust SEs. For GLMMs, we used the lme4 package [37] for the frequentist models and Stan [38] through the R interface rstan [39] to fit the Bayesian models. We provide sample code in Additional file 1.
Results
Convergence
For the Bayesian models, trace plots of the parameters showed the three chains mixing well after burn-in, except for a small percentage of the datasets, where one of the MCMC chains of σ failed to converge near 0 for a portion of the chain. (Other parameters also did not converge; an example of an RCT dataset is shown in Additional file 1: Figure S1.) These convergence issues were also detected by the diagnostic (>1.1), and these datasets were excluded from the results. Convergence rates for Bayesian models ranged from 92% for the RCT scenario with 4 centers and 10 subjects per center to 100% for some scenarios with 10 or 30 centers. Convergence rates for all scenarios are shown in Additional file 1: Table S1.
All frequentist models exhibit convergence problems for the smallest sample size for both designs, with convergence rates ranging from 45% for the binomial GLMM to 86% for the GEE Poisson model. For all other scenarios, convergence was not an issue, except for the binomial GLMM, which had poor convergence rates for all scenarios. Its lowest convergence rate was 27% for the scenario with 30 centers with 10 subjects/center (Additional file 1: Table S1).
Bias and RMSE
The bias was generally small for all frequentist models. It was larger for the smallest sample sizes and diminished as the number of centers and total sample size increased. The Bayesian estimates were more biased in the smallest sample sizes. The negative bias indicates that the posterior medians of the treatment effect are shrunk toward 0 because of the influence of the informative priors. The effect of the prior on the posterior estimates and the resulting bias from the Bayesian GLMM diminishes as the sample size increases and is smaller than the bias from frequentist models for some scenarios (Fig. 1). The Bayesian models have the smallest RMSE for scenarios with four or ten centers. All models give very similar RMSEs with 30 centers (Fig. 2). The bias and RMSE were very similar for both study designs.
Fig. 1.
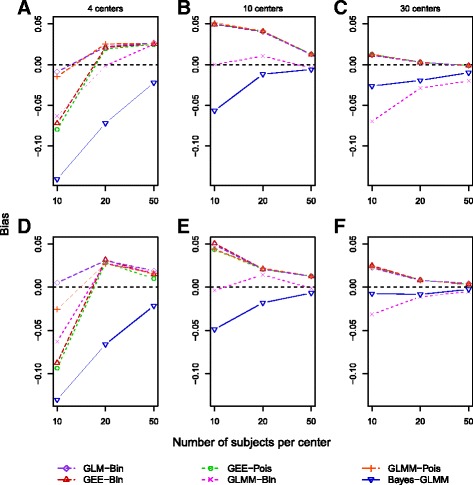
Bias of the estimates of β1 (calculated as βestimate − βtrue) for different scenarios under a multicenter randomized controlled trial study design (a–c) and an observational study design (d–f) based on 1000 simulations for each scenario. All scenarios used a β1 of log(1.5), a control outcome rate of 15%, and an intracluster correlation coefficient of 0.08. GEE Generalized estimating equation, GLM-Bin Log-binomial regression model, GLMM Generalized linear mixed model
Fig. 2.
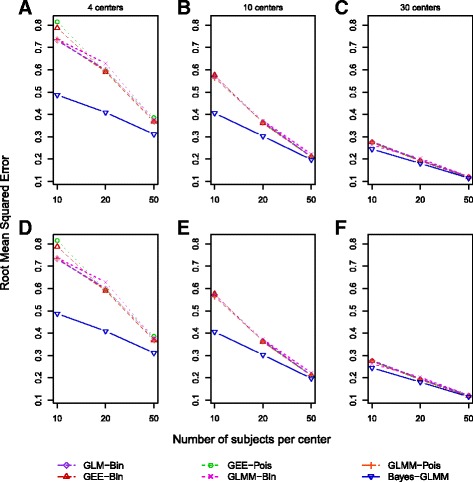
Root mean square error of β1 for scenarios under randomized controlled trial study designs (a–c) and observational study designs (d–f) based on 1000 simulations for each scenario. All scenarios used a β1 of log(1.5), a control outcome rate of 15%, and an intracluster correlation coefficient of 0.08. GEE Generalized estimating equation, GLM-Bin Log-binomial regression model, GLMM Generalized linear mixed model
Coverage
Figure 3 shows coverage of the 95% CIs and posterior interval for β1. The unadjusted GLM-Bin exhibits coverage above the nominal range (93.6–96.4% using 1000 datasets) with 4 or 10 centers and 10 or 20 subjects/center, but it has good coverage for all other scenarios. GEE CIs without the KC correction also have poor coverage with 4 centers (85–87%) and 10 centers with 50 subjects (~92%). KC-corrected GEE CIs have good coverage across the majority of scenarios, with the GEE Poisson having coverage closer to nominal than the GEE binomial. The binomial GLMM has coverage above the nominal value for scenarios with the three smallest total sample sizes; the coverage of the bootstrap CIs is more conservative than the Wald CIs for some scenarios. However, the GLMM Poisson model results in coverage well above the nominal value in all scenarios with both Wald-type and bootstrap CIs. The Bayesian model has conservative coverage for scenarios with total sample size ≤ 200; for all other scenarios, coverage is close to the nominal value. The study design had little effect on coverage.
Fig. 3.
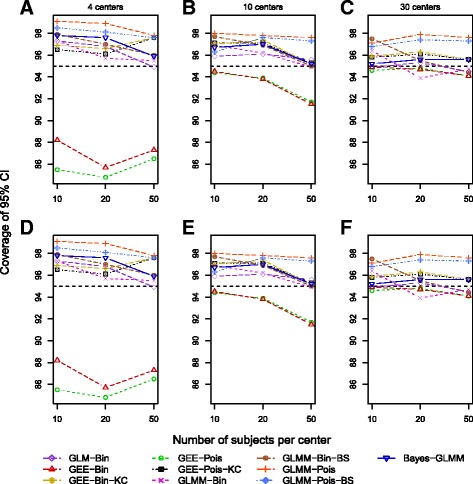
Coverage of 95% CI and posterior interval of β1 for scenarios under randomized controlled trial study designs (a–c) and observational study designs (d–f) based on 1000 simulations for each scenario. All scenarios used a β1 of log(1.5), control outcome rate of 15%, and an intracluster correlation coefficient of 0.08. GEE Generalized estimating equation, GLM-Bin Log-binomial regression model, GLMM Generalized linear mixed model, KC Kauermann and Carroll, BS Bootstrap
Sensitivity results
Under both study designs with four centers, Bayesian GLMMs with vague priors had a lower convergence rate, smaller bias, larger RMSE, and less conservative coverage than informative priors. For scenarios with 10 or 30 centers, vague priors resulted in similar convergence rate, bias, RMSE, and coverage compared with informative priors for both designs. In all scenarios, the estimates of the between-center SD were very similar under both vague and informative priors.
Examples
Infection treatment multicenter trial
We analyzed the data presented by Beitler and Landis [40] arising from an eight-center RCT investigating the efficacy of an active drug compared with control for treatment of an infection. The primary outcome was favorable response to the drug. In the eight centers, the rate of success in the active drug group (n = 130) varied from 9% to 80%, whereas the control group (n = 143) had a rate of success ranging from 0 to 86%. We used the same methods as described for the simulation study (excluding Poisson GLMM, which did not perform well in the simulation study). Table 2 shows the estimated RRs derived from the different methods. The binomial GLMM model did not converge. The RR estimates differ across the models, with the binomial GEE resulting in the largest RR of 1.43 and the Bayesian GLMM resulting in the smallest RR of 1.27. The 95% CIs from the GEE models without the KC correction do not include 1.0. In comparison, the 95% CI from the unadjusted GLM and KC-corrected GEE models include 1.0. The Bayesian 95% CrI is the narrowest, despite properly accounting for center variation (estimated as 0.81) and might lead to a conclusion that the active drug is effective.
Table 2.
Estimated relative risk and 95% CI or credible interval for multicenter randomized controlled trial example
| Modela | RR | 95% CI or CrI |
|---|---|---|
| GLM-Bin | 1.29 | 0.95–1.75 |
| GEE binomial | 1.43 | 1.01–2.02 |
| GEE binomial, KC-corrected | 0.96–2.14 | |
| GEE Poisson | 1.42 | 1.01–2.01 |
| GEE Poisson, KC-corrected | 0.95–2.12 | |
| GLMM binomialb | – | – |
| Bayesian binomial GLMM | 1.27 | 1.00–1.65 |
Abbreviations: CrI Credible interval, GEE Generalized estimating equation, GLM-Bin Log-binomial regression model, GLMM Generalized linear mixed model, KC Kauermann and Carroll, RR Relative risk
Except for the GLM-Bin model, all models are adjusted for center correlation. The KC bias correction in the GEE models adjusts the robust SE for the small number of centers (estimate of RR does not change)
aGLMM Poisson models are excluded because of their poor performance in the simulation study
bModel did not converge
Pediatric appendicitis observational study
In this study, we compared cohorts of children before and after implementation of a clinical practice guideline for treatment of perforated appendicitis in children [41]. The study was conducted in a children’s hospital with 11 surgeons providing care. The primary outcome was the occurrence of any adverse event, such as readmission or surgical site infection. Totals of 191 and 122 patients were included in the pre- and postimplementation cohorts, respectively. We compared the analytical methods including identification of intra-abdominal abscess as a covariate, with surgeon as the clustering variable. Estimated RRs and 95% CIs are shown in Table 3. Compared with the other models, the RR estimates from both GEE models are closer to 1 and their CIs without the KC bias correction are narrower than all other intervals. However, the main conclusion of the intervention being associated with reduced adverse outcomes (although not statistically significant) would not differ on the basis of the analytical method chosen.
Table 3.
Results for pediatric appendicitis study
| Model | RR | 95% CI or CrI |
|---|---|---|
| GLM-Bin | 0.76 | 0.51–1.11 |
| GEE binomial | 0.79 | 0.56–1.13 |
| GEE binomial, KC-corrected | 0.53–1.19 | |
| GEE Poisson | 0.80 | 0.56–1.14 |
| GEE Poisson, KC-corrected | 0.53–1.21 | |
| GLMM binomial | 0.76 | 0.51–1.11 |
| GLMM binomial bootstrap | 0.51–1.10 | |
| Bayesian binomial GLMM | 0.73 | 0.49–1.07 |
Abbreviations: CrI Credible interval, GEE Generalized estimating equation, GLM-Bin Log-binomial regression model, GLMM Generalized linear mixed model, KC Kauermann and Carroll, RR Relative risk
The KC bias correction in the GEE models adjusts the robust SE for the small number of centers (estimate of RR does not change)
Discussion
For multicenter studies, it is important to adjust for possible center correlation when computing treatment effects, particularly when the proportion of center variability over total variability is large or when randomization is stratified by center to ensure correct SEs and CIs [1, 4]. However, no clear guidelines exist for the appropriate analyses, and it can be challenging for data analysts to perform a properly adjusted analysis when the outcome is binary and there are few centers. In this paper, we have reviewed and evaluated methods available for analyses of multicenter studies.
Summary of simulation results
For all but the smallest sample size, convergence rates were ≥ 96% for all models except the binomial GLMM. This model had convergence problems in all scenarios investigated, and its use may be limited. All frequentist estimates of the treatment effect had small and very similar bias. The Bayesian estimates were more biased in the smallest sample sizes. Although the binomial GLM with unadjusted SEs had very little bias, it had conservative coverage for the smallest sample sizes in both the RCT and observational designs.
The GEE models without a small sample correction for the sandwich SEs had very poor coverage with four or ten centers, even when coupled with approximate t-based CIs. This poor performance of sandwich SEs has been noted before in estimating ORs [3]. Using the KC bias correction greatly improved the performance of these models across all scenarios. These results are similar to those obtained by Yelland et al. [9] and Zou and Donner [11], although they used different corrections for the variance estimates.
When the binomial GLMM achieved convergence, it had good overall performance except for sample sizes < 100, where it had conservative coverage. Across all scenarios, the Poisson GLMM also had coverage above the nominal value even with bootstrapped CIs, which would lead to diminished power.
The Bayesian GLMM had good coverage across all scenarios, and the bias exhibited in the smallest sample sizes was only slightly larger than the other models evaluated. Its higher convergence rate for the smallest sample size is due to the use of informative priors that help stabilize the estimates, particularly in cases of complete separation [24, 26].
Although we do not report the estimate of the SD of the random effects (or ICC), the Bayesian GLMM outperformed the frequentist models with estimates that were very close to the true parameter value. In contrast, the frequentist models consistently underestimated this parameter even in the larger sample sizes. This downward bias of estimates of the variance components in GLMMs is well known [19, 42]. Here the Bayesian approach has a clear advantage over frequentist methods because it provides less biased variance estimates and automatically produces CIs for these parameters. More importantly, the point estimates and CIs of the treatment/exposure effect appropriately account for the uncertainty in the variance parameter.
The sensitivity analysis for the Bayesian GLMM using vague priors produced results very similar to those under the informative priors. However, the range of effect sizes supported by the vague priors is unrealistic, and we strongly recommend against using these priors.
Recommendations
On the basis of our simulation results and other studies [3, 43], we recommend that both RCT and observational multicenter studies adjust for center in the analysis. Although adjusting for center when the ICC is small may not provide a great advantage, it also does not adversely affect the point estimates, SEs, and type I error rates. Furthermore, methods that properly adjust for center clustering are easy to implement in most statistical software.
We do not recommend the use of a Poisson GLMM to estimate adjusted RR. We do recommend use of a binomial GLMM, except when the number of centers is < 5 or the total sample size is < 100, although convergence may be a problem. The most robust frequentist methods appear to be either a GEE log-binomial model or a Poisson model with an exchangeable correlation. When the number of centers or clusters is < 50, a sandwich variance estimator needs to include a small sample correction such as the KC correction used here. Kahan et al. [3] reported that model-based SEs may be another option for GEE models, but we did not evaluate them here.
A Bayesian GLMM is a robust alternative. This method performed the best in terms of all measures of convergence, bias, RMSE, and coverage. Another advantage of a Bayesian approach is that exterior information about treatment/exposure effect can be formally incorporated into the prior distributions. As we did here, the priors can explicitly exclude large effects, which are unlikely in clinical studies. Probabilities of benefit or harm are easily obtained and can be more informative for investigators than the traditional p value. Although our focus was not on the estimation of the between-center variance, the Bayesian model outperformed all other methods in estimating this parameter. Implementation of the Bayesian model can be done in OpenBUGS or in Stan through R as was done here.
Limitations
Our simulation study was limited to one treatment/exposure effect size and control rate. However, others have noted similar performance when the effect size or control outcome rate was varied [3, 9, 11, 15]. We also did not investigate fixed effects models, because their limitations have been noted before [2–4]. However, these methods could be an alternative method of analysis for studies with three or fewer centers. We also note computational limitations faced in most simulation studies. In particular, results from the Bayesian models would have benefited from running longer chains. Increasing the number of bootstrap samples for the GLMMs could also potentially improve their performance. Our simulation study did not include scenarios with an ICC of 0. However, others have found that the methods recommended here perform well even in cases where the ICC is very close to 0 [8, 15, 43]. We investigated the performance of only an exchangeable correlation matrix for the GEE models, which is a plausible assumption for multicenter data. However, other correlation structures can be used, and the performance of GEE models has been shown to be robust to the choice of the correlation structure [11].
We chose to focus on binary outcome because it is the most common type of outcome reported in medical research, but it would be important to investigate methods for other types of outcomes (i.e., time-to-event data). Last, we did not investigate treatment by center interactions, and this is an important issue that needs to be investigated in future studies.
Conclusions
For the analysis of multicenter studies with a binary outcome, we recommend adjustment for center with either a GEE log-binomial or Poisson model or a Bayesian binomial GLMM with informative priors. The GEE models should include a small sample variance correction for sandwich estimators when the number of center is < 30. The Bayesian model with informative priors provides stable estimates, greater flexibility, and good performance even with very small sample sizes.
Acknowledgements
The authors thank Dr. Martin Blakely for providing access to the pediatric appendicitis study data.
Funding
This research was supported by no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Availability of data and materials
R code for implementing all regression models is given in the supplemental material (Additional file 1). The multicenter trial data from the second example are available from the authors upon reasonable request and with permission of Dr. Martin Blakely.
Abbreviations
- CrI
Credible interval
- GEE
Generalized estimating equation
- GLM-Bin
Log-binomial regression model
- GLMM
Generalized linear mixed model
- ICC
Intracluster correlation coefficient
- KC
Kauermann and Carroll
- MCMC
Markov chain Monte Carlo
- RCT
Randomized controlled trial
- RMSE
Root mean squared error
- RR
Relative risk
Additional file
Figure S1. Trace plots of parameters for an example dataset exhibiting convergence issues. Table S1. Convergence rates for regression models based on 1000 simulated datasets. Sample R code for fitting models used in the simulation study. (PDF 431 kb)
Authors’ contributions
CP conceived the research question, initiated the simulation code, and supervised the implementation of the simulation study. VTTT helped develop and implement the code. Both authors drafted the manuscript and critically reviewed it. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13063-017-2248-1) contains supplementary material, which is available to authorized users.
Contributor Information
Claudia Pedroza, Phone: 713-500-5721, Email: claudia.pedroza@uth.tmc.edu.
Van Thi Thanh Truong, Email: van.t.truong@uth.tmc.edu.
References
- 1.Parzen M, Lipsitz SR, Dear KBG. Does clustering affect the usual test statistics of no treatment effect in a randomized clinical trial? Biom J. 1998;40:385–402. doi: 10.1002/(SICI)1521-4036(199808)40:4<385::AID-BIMJ385>3.0.CO;2-#. [DOI] [Google Scholar]
- 2.Agresti A, Hartzel J. Strategies for comparing treatments on a binary response with multi-centre data. Stat Med. 2000;19:1115–39. doi: 10.1002/(SICI)1097-0258(20000430)19:8<1115::AID-SIM408>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 3.Kahan BC. Accounting for centre-effects in multicentre trials with a binary outcome - when, why, and how? BMC Med Res Methodol. 2014;14:20. doi: 10.1186/1471-2288-14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kahan BC, Morris TP. Improper analysis of trials randomised using stratified blocks or minimisation. Stat Med. 2012;31:328–40. doi: 10.1002/sim.4431. [DOI] [PubMed] [Google Scholar]
- 5.Kahan BC, Harhay MO. Many multicenter trials had few events per center, requiring analysis via random-effects models or GEEs. J Clin Epidemiol. 2015;68:1504–11. doi: 10.1016/j.jclinepi.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tangri N, Kitsios GD, Su SH, Kent DM. Accounting for center effects in multicenter trials. Epidemiology. 2010;21:912–3. doi: 10.1097/EDE.0b013e3181f56fc0. [DOI] [PubMed] [Google Scholar]
- 7.Mancl LA, DeRouen TA. A covariance estimator for GEE with improved small-sample properties. Biometrics. 2001;57:126–34. doi: 10.1111/j.0006-341X.2001.00126.x. [DOI] [PubMed] [Google Scholar]
- 8.Li P, Redden DT. Comparing denominator degrees of freedom approximations for the generalized linear mixed model in analyzing binary outcome in small sample cluster-randomized trials. BMC Med Res Methodol. 2015;15:38. doi: 10.1186/s12874-015-0026-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yelland LN, Salter AB, Ryan P. Performance of the modified Poisson regression approach for estimating relative risks from clustered prospective data. Am J Epidemiol. 2011;174:984–92. doi: 10.1093/aje/kwr183. [DOI] [PubMed] [Google Scholar]
- 10.Chu H, Cole SR. Estimation of risk ratios in cohort studies with common outcomes: a Bayesian approach. Epidemiology. 2010;21:855–62. doi: 10.1097/EDE.0b013e3181f2012b. [DOI] [PubMed] [Google Scholar]
- 11.Zou GY, Donner A. Extension of the modified Poisson regression model to prospective studies with correlated binary data. Stat Methods Med Res. 2013;22:661–70. doi: 10.1177/0962280211427759. [DOI] [PubMed] [Google Scholar]
- 12.Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. doi: 10.1093/biomet/73.1.13. [DOI] [Google Scholar]
- 13.Lu B, Preisser JS, Qaqish BF, Suchindran C, Bangdiwala SI, Wolfson M. A comparison of two bias-corrected covariance estimators for generalized estimating equations. Biometrics. 2007;63:935–41. doi: 10.1111/j.1541-0420.2007.00764.x. [DOI] [PubMed] [Google Scholar]
- 14.Kauermann G, Carroll RJ. A note on the efficiency of sandwich covariance matrix estimation. J Am Stat Assoc. 2001;96:1387–96. doi: 10.1198/016214501753382309. [DOI] [Google Scholar]
- 15.Li P, Redden DT. Small sample performance of bias-corrected sandwich estimators for cluster-randomized trials with binary outcomes. Stat Med. 2015;34:281–96. doi: 10.1002/sim.6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M, Long Q. Modified robust variance estimator for generalized estimating equations with improved small-sample performance. Stat Med. 2011;30:1278–91. doi: 10.1002/sim.4150. [DOI] [PubMed] [Google Scholar]
- 17.Wang M, Kong L, Li Z, Zhang L. Covariance estimators for generalized estimating equations (GEE) in longitudinal analysis with small samples. Stat Med. 2016;35:1706–21. doi: 10.1002/sim.6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritz J, Spiegelman D. Equivalence of conditional and marginal regression models for clustered and longitudinal data. Stat Methods Med Res. 2004;13:309–23. [Google Scholar]
- 19.Pinheiro JC, Chao EC. Efficient Laplacian and adaptive Gaussian quadrature algorithms for multilevel generalized linear mixed models. J Comput Graph Stat. 2006;15:58–81. doi: 10.1198/106186006X96962. [DOI] [Google Scholar]
- 20.Faraway JJ. Extending the linear model with R: generalized linear, mixed effects and nonparametric regression models. Boca Raton, FL: Chapman & Hall/CRC; 2006. [Google Scholar]
- 21.Spiegelhalter DJ, Abrams KR, Myles JP. Bayesian approaches to clinical trials and health care evaluation. Chichester, UK: John Wiley & Sons, Ltd; 2004. [Google Scholar]
- 22.Warn DE, Thompson SG, Spiegelhalter DJ. Bayesian random effects meta-analysis of trials with binary outcomes: methods for the absolute risk difference and relative risk scales. Stat Med. 2002;21:1601–23. doi: 10.1002/sim.1189. [DOI] [PubMed] [Google Scholar]
- 23.Torman VB, Camey SA. Bayesian models as a unified approach to estimate relative risk (or prevalence ratio) in binary and polytomous outcomes. Emerg Themes Epidemiol. 2015;12:8. doi: 10.1186/s12982-015-0030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gelman A, Jakulin A, Pittau MG, Su YS. A weakly informative default prior distribution for logistic and other regression models. Ann Appl Stat. 2008;2:1360–83. doi: 10.1214/08-AOAS191. [DOI] [Google Scholar]
- 25.Greenland S. Putting background information about relative risks into conjugate prior distributions. Biometrics. 2001;57:663–70. doi: 10.1111/j.0006-341X.2001.00663.x. [DOI] [PubMed] [Google Scholar]
- 26.Pedroza C, Han W, Truong VT, Green C, Tyson JE. Performance of informative priors skeptical of large treatment effects in clinical trials: A simulation study. Stat Methods Med Res. 2015:0962280215620828. Epub ahead of print. [DOI] [PubMed]
- 27.Lambert PC, Sutton AJ, Burton PR, Abrams KR, Jones DR. How vague is vague? A simulation study of the impact of the use of vague prior distributions in MCMC using WinBUGS. Stat Med. 2005;24:2401–28. doi: 10.1002/sim.2112. [DOI] [PubMed] [Google Scholar]
- 28.Thompson SG, Warn DE, Turner RM. Bayesian methods for analysis of binary outcome data in cluster randomized trials on the absolute risk scale. Stat Med. 2004;23:389–410. doi: 10.1002/sim.1567. [DOI] [PubMed] [Google Scholar]
- 29.Eldridge SM, Ashby D, Feder GS, Rudnicka AR, Ukoumunne OC. Lessons for cluster randomized trials in the twenty-first century: a systematic review of trials in primary care. Clin Trials. 2004;1:80–90. doi: 10.1191/1740774504cn006rr. [DOI] [PubMed] [Google Scholar]
- 30.Emrich LJ, Piedmonte MR. A method for generating high-dimensional multivariate binary variates. Am Stat. 1991;45:302–4. [Google Scholar]
- 31.Zeger SL, Karim MR. Generalized linear models with random effects; a Gibbs sampling approach. J Am Stat Assoc. 1991;86:79–86. doi: 10.1080/01621459.1991.10475006. [DOI] [Google Scholar]
- 32.Brooks SP, Gelman A. General methods for monitoring convergence of iterative simulations. J Comput Graph Stat. 1998;7:434–55. [Google Scholar]
- 33.Gelman A. Prior distributions for variance parameters in hierarchical models (comment on article by Browne and Draper) Bayesian Anal. 2006;1:515–34. doi: 10.1214/06-BA117A. [DOI] [Google Scholar]
- 34.R Core Team. R: a language and environment for statistical computing [Internet]. Vienna: R Foundation for Statistical Computing; 2015. http://www.R-project.org/.
- 35.Halekoh U, Højsgaard S, Yan J. The R package geepack for generalized estimating equations. J Stat Softw. 2006;15(2):1–11. doi: 10.18637/jss.v015.i02. [DOI] [Google Scholar]
- 36.Wang M. geesmv: Modified variance estimators for generalized estimating equations [Internet]. R package version 1.3; 2015. http://cran.r-project.org/package=geesmv/.
- 37.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 38.Carpenter B, Gelman A, Hoffman M, Lee D, Goodrich B, Betancourt M, et al. Stan: a probabilistic programming language. J Stat Softw. 2016. In Press. [DOI] [PMC free article] [PubMed]
- 39.Stan Development Team. RStan: the R interface to Stan [Internet]. Version 2.9.0. http://mc-stan.org/.
- 40.Beitler PJ, Landis JR. A mixed-effects model for categorical data. Biometrics. 1985;41:991–1000. doi: 10.2307/2530970. [DOI] [PubMed] [Google Scholar]
- 41.Willis ZI, Duggan EM, Bucher BT, Pietsch JB, Milovancev M, Wharton W, et al. Effect of a clinical practice guideline for pediatric complicated appendicitis. JAMA Surg. 2016;151:e160194. doi: 10.1001/jamasurg.2016.0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breslow NE, Clayton DG. Approximate inference in generalized linear mixed models. J Am Stat Assoc. 1993;88:9–25. [Google Scholar]
- 43.Kahan BC, Morris TP. Analysis of multicentre trials with continuous outcomes: when and how should we account for centre effects? Stat Med. 2013;32:1136–49. doi: 10.1002/sim.5667. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
R code for implementing all regression models is given in the supplemental material (Additional file 1). The multicenter trial data from the second example are available from the authors upon reasonable request and with permission of Dr. Martin Blakely.


