Abstract
EvoDesign is a computational algorithm that allows the rapid creation of new protein sequences that are compatible with specific protein structures. As such, it can be used to optimize protein stability, to resculpt the protein surface to eliminate undesired protein-protein interactions, and to optimize protein-protein binding. A major distinguishing feature of EvoDesign in comparison to other protein design programs is the use of evolutionary information in the design process to guide the sequence search toward native-like sequences known to adopt structurally similar folds as the target. The observed frequencies of amino acids in specific positions in the structure in the form of structural profiles collected from proteins with similar folds and complexes with similar interfaces can implicitly capture many subtle effects that are essential for correct folding and protein-binding interactions. As a result of the inclusion of evolutionary information, the sequences designed by EvoDesign have native-like folding and binding properties not seen by other physics-based design methods. In this chapter, we describe how EvoDesign can be used to redesign proteins with a focus on the computational and experimental procedures that can be used to validate the designs.
Keywords: Protein design, Evolutionary profile, Protein structure modeling, Experimental protein validation, Recombinant expression, Circular dichroism, Nuclear magnetic resonance
1 Introduction
Computational protein design has expanded in recent years from the prediction of the effects of single site mutations to the complete redesign of entire proteins, including the alteration of protein-protein binding affinity and specificity [1–4], enzymatic activity [5, 6], and even the creation of new folds [7] and functions [8] that are not seen in nature. On the theoretical side, protein design has been used to find the sequence constraints necessary to generate specific folds or functions [9–11]. Through the use of these constraints, fundamental questions in protein evolution have been addressed by distinguishing what is physically possible from what is actually observed in evolution [10, 12].
However, full protein redesign beyond the mutation of a few hot spot residues, called de novo design, is computationally difficult, which is reflected in the relatively low successful percentage of successful designs. Most algorithms for de novo protein design approach the problem as reverse ab initio protein folding, evaluating the energy of the sequence according to all-atom physical potentials. Several problems become apparent in the naïve application of this approach: (1) A very large number of sequences must be considered, which limits the force field to only approximate energy terms that can be rapidly calculated; (2) there is a mismatch between the low-resolution models generated in the sequence search and the all-atom physical potentials used for evaluation. To make the design simulation computationally tractable, the possible conformations of the side-chains of the protein are restricted to a limited set of discrete rotamer conformations. The small steric clashes that necessarily result from this approximation force the use of dampened potentials that may miss subtle interactions that exist in the native protein [13, 14]; (3) the sequence search is considered only with the protein in isolation, not as the protein actually exists in the cellular context. This causes subtle problems in the real-life application of the designed proteins, particularly with respect to aggregation, as the highly hydrophobic sequences favored by folding energetics generally adopt highly compact sequences in silico but tend to aggregate in reality when actually expressed [15].
One approach to handle these challenges is to increase the accuracy of the design process by attempting to model physical reality at a higher resolution. In this spirit, design methodologies have been created that explicitly consider multiple conformations of the folded protein using ensemble techniques for multistate design [16–18] or that explicitly consider the unfolded state during the design process [18]. Alternatively, other design methodologies have been created that recognize the inherent inaccuracy of the force fields and attempt to diminish the effects of known inaccuracies. One example is the use of soft-core potentials that lessen repulsive interactions, preventing strongly unfavorable interactions that can be alleviated by small backbone motions from overriding the other terms [19]. Another example of this approach is the inclusion of additional terms in the force field that consider factors relevant to real proteins that are missing in the simulation, for example, the explicit consideration of inappropriate hydrophobic surfaces to limit aggregation in the designed sequences [18, 20]. The ongoing development of these methods has contributed greatly to the field and has led to some spectacular successes. However, complete de novo protein design is still a difficult process with routine application still in the future.
An alternative approach, based on hard-won knowledge from protein fold-recognition and structure prediction [21–24], is to recognize that evolution implicitly encodes information on protein folds and binding interactions that greatly exceeds our ability to describe it through reductionist, physics-based methods. This evolution-based method approach to protein design differs from the physics-based methods in that most energy terms are not dependent on the full-atom representation of each tested sequence, whose inaccuracy is a significant source of error. Instead, the sequence space search is constrained by the sequence and structural profiles collected from structurally analogous families, assisted by neural network predictions of local structural features, including secondary structure, backbone torsion angle, and solvation [25, 26].
2 Methods
2.1 EvoDesign: Evolution-Based Method to Design Protein Folds and Interactions
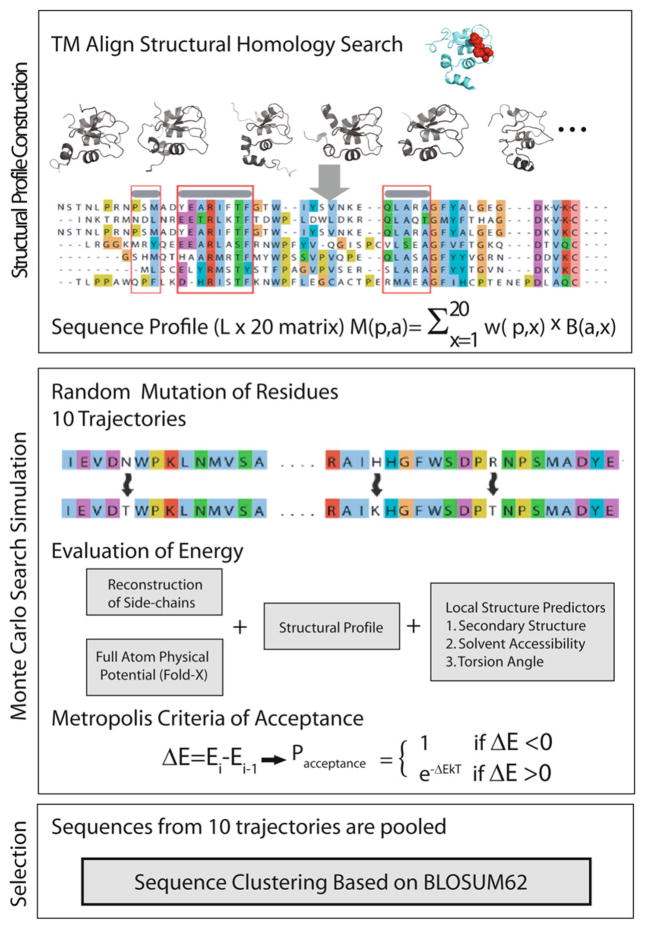
The principle of EvoDesign follows the critical lessons learned from threading-based protein structure prediction methods, i.e., to use the reliable “finger-print” of nature of multiple proteins from the same family in the form of structural profile information to guide the simulation to the sequence search. It first collects a set of proteins with similar folds to the target scaffold structure from the PDB library by the structural alignment program TM-align [27], using a TM-score cutoff value to define structural similarity (Fig. 1) [28]. In the second step, this set of structurally similar folds is used to create a position specific scoring matrix M(p, a) for evaluating potential sequences [29, 30].
Fig. 1.
Overview of the EvoDesign method showing the construction of the structural profile, the Monte Carlo search in sequence space, and the final selection of the sequences by sequence clustering
To create the position specific scoring matrix, first a multiple sequence alignment (MSA) is generated according to the pair-wise structural alignments between the structural analogs identified in the first step and the target structure (Fig. 1). An L × 20 matrix (where L = length of the protein) is then created according to
| (1) |
where x represents a particular amino acid, B(a, x) is the BLO-SUM62 substitution matrix [31] for amino acid x to amino acid a, and w(p, x) is the frequency of the amino acid x appearing at position p in the MSA created by TM-align. The matrix M(p, a) serves as a structural profile to guide the sequences toward native-like sequences known to adopt structurally similar folds as the target (Fig. 1).
While the structural profile as given by the position specific scoring matrix M(p, a) is efficient in guiding the global fold, optimization on the profile alone can result in singularities (i.e., disjointed “islands”) in local sequences. To smoothen these singularities, back propagation neural network predictors are used to estimate the secondary structure (SS), solvent accessibility (SA), and torsion angles (φ/ψ) of the sequence. Unlike other predictors for these properties [32–34], these single-sequence-based predictors do not require a computationally expensive PSI-BLAST search, which considerably speeds up prediction at little cost in accuracy [25].
The evolutionary potential in EvoDesign is defined as the maximum score of the optimal alignment path between the decoy and target structure obtained by Needleman-Wunsch dynamic programming, giving the energy function:
| (2) |
where ΔSS, ΔSA, Δφ, and Δψ are the difference in secondary structure, solvent accessibility and torsion angles between the target assignments, and the predictions from the decoy sequences. The weighting factors (wi) are decided by the relative accuracy of the single-sequence-based predictions for each term on a training set [25].
A physics-based potential can be used to predict potential favorable and unfavorable interactions among side-chains, such as steric interactions, which may be missed by the evolutionary-based terms defined above. While our computational benchmark results indicate the evolution-based energy function alone is sufficient to design protein sequences, adding a physics-based energy term from FoldX [35] improved the atomic packing of the local structures based on both computational structure prediction and experimental structure validations [25]. In this case, a full-atom representation of the sequence is needed which is created by SCWRL [36].
The final force field for single-chain protein design in EvoDesign is given by the weighted Z-scores of the evolution and physics-based terms:
| (3) |
where 〈…〉 and δ indicate the average and standard deviation of the energy terms.
To actually generate the designed sequences, Monte Carlo searches are performed starting from 10 random sequences that are updated by random residue mutations (Fig. 1). Due to the imprecision of the force field, the lowest energy states do not always correspond to the best sequence design. Instead of simply focusing on the lowest energy sequence, the sequences from all 10 runs are pooled and the sequence with the maximum number of neighbors is identified using the SPICKER clustering algorithm [37] where the pair-wise distance between sequences is measured by the sum of the BLOSUM62 substitution scores [38].
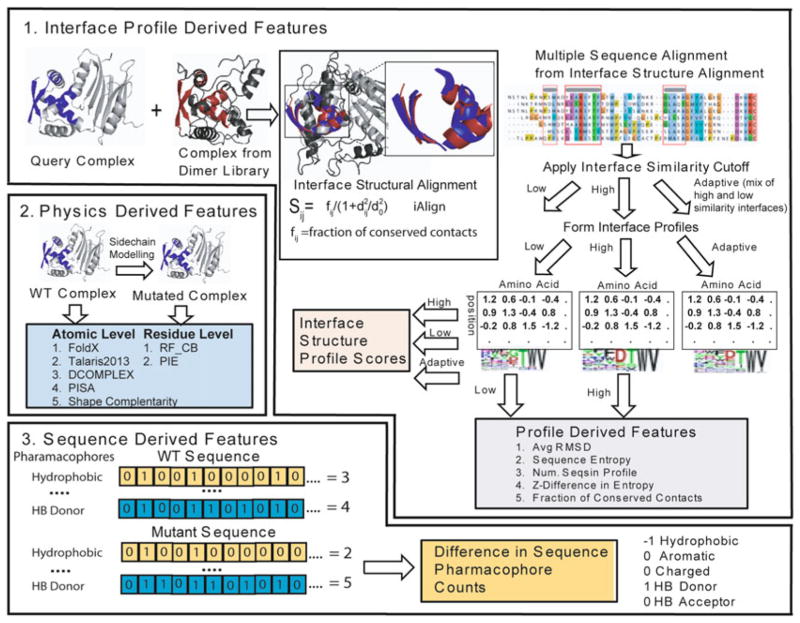
The above procedure finds sequences compatible with the target structure. To introduce new or altered functionality into the protein, the affinity of existing protein-protein interfaces can be improved by EvoDesign or new interfaces created though the optimization of non-native complexes created by docking. To modify interfaces, EvoDesign uses a multiscale approach incorporating a variety of features at different levels of structural resolution (Fig. 2).
Fig. 2.
Multiscale approach to predicting protein binding affinity using features derived from interface structural profiles, WT and mutant sequences, and physics-based scoring of the structures of the wild-type and mutant complexes. (1) Interface profile scores derived by structural alignment of structurally similar interfaces using an interface similarity cutoff to define the aligned sequences that are used to build the profile. (2) Physics-based scores are formed at the residue or atomic level formed by modeling the mutant monomeric protein and complex and evaluating the difference in energy. (3) Sequence features are formed by the difference between the WT and mutant sequences in the number of hydrophobic (V, I, L, M, F, W, or C), aromatic (Y, F, or W), charged (R, K, D, or E), hydrogen bond acceptors (D, E, N, H, Q, S, T, or Y), and hydrogen bond donating residues (R, K, W, N, Q, H, S, T, or Y) along with difference in amino acid volume calculated from the sequence
Similar to the design of protein folds with EvoDesign, a key feature of the binding potential is the mixture of physics-based and evolutionary terms in the energy function [39]. For interface modification, the evolutionary terms are created from the structural alignment of similar interfaces from the nonredundant COTH structural library of dimeric proteins [40] by the IAlign program [41]. A series of interface similarity cutoffs has been used to define three separate interface structure profiles along with different metrics designed to assess the accuracy of the profiles relative to the other terms [39]. The interface profiles scores are then combined with physics-based all-atom and residue level docking scores. Finally, sequence-based scores based on phamacophore count differences between the native and designed sequences are calculated to complete the multiscale approach. A random forest method trained to predict the experimental affinity changes (ΔΔG) associated with single and multiple mutations at the interface is used for the final interface energy score. This energy score has a correlation to the experimental ΔΔG values equivalent or superior to the best state-of-the-art mutation prediction programs (Pearson’s correlation coefficient = 0.83 for a 5 fold cross validated set) but is fast enough to calculate the thousands of potential mutations necessary for protein design. The interface energy is then added to the regular EvoDesign scoring potential, using a user-defined weighting function to balance fold stability and protein-protein affinity.
2.2 Using the EvoDesign Server Design Program
The EvoDesign program can be used as a server at http://zhanglab.ccmb.med.umich.edu/EvoDesign. The only input to the server is a PDB format file of the target structure, which can be either a full-atomic or backbone only model. In either case, the backbone of the protein structure should be complete without breaks in the chain. Currently, the server is limited to design of one protein chain only.
There are three user-defined parameters to control the design simulation. The first parameter is the fold-similarity cutoff used for defining the structural profile (Eq. 1). By default, this is set to the relatively high value of a TM score of 0.7, which is relaxed if less than ten structural analogues are found in the PDB. This value can be adjusted to a higher or lower value; lower values incorporate more sequence and structural variability in constructing the profile while higher values incorporate less. The usual result is that higher cutoffs penalize deviations from the native sequence more strongly, which may or may not be desirable for the particular application. The second parameter controls whether the FoldX force field is used in the simulation or not. Inclusion of FoldX usually results in only a marginal improvement in the folding when validated by structure prediction (see the next section) [25], most likely due to the fact that the side-chains are modeled by a different force field from the SCWRL force field used for scoring. Including FoldX in the simulation requires that the full atomic model of each sequence be constructed, which is the most computationally demanding step in the simulation. For this reason, the FoldX force field is turned off by default. The last parameter does not affect the design simulation but controls whether structure prediction is performed for each of the designed sequences through the creation of I-TASSER models (see Subheading 2.3.1).
By default, the EvoDesign server operates without any residue restrictions on the design process. In many cases, it desirable to freeze certain residues in the design process, such as those involved in disulfide bond formation or in ligand binding. Taken further, in other cases, it is useful to redesign only the surface of the protein while keeping the inner core constant. An option is therefore provided to specify a set of residues (by residue number) which should be kept the same as in the input structure. It is also sometimes desirable to restrict the use of some residues completely or at certain positions. A prime example is cysteine residues on the surface, which can easily be oxidized to form intermolecular disulfide bonds that lead to a loss of activity through aggregation.
The output of the server is ten sequences in decreasing order of cluster size from the clusters generated by the SPICKER algorithm. For each sequence, the sequence identity to the native sequence is calculated along with the predicted normalized relative error for the secondary structure, solvent accessibility, and torsion angles. Each property is calculated by a high accuracy predictor using PSI-BLAST profiles along with neural network predictors (PSSPred for secondary structure prediction [42], ANGLOR for torsion angle prediction [32], and the method of SOLVE for solvent accessibility [43], respectively). The normalized relative error (NRE) is reported for each prediction, which is defined by [25].
| (4) |
where EDS refers to “error of designed sequence,” i.e., the mismatch between the predicted structure feature from the designed sequence and the target structure. ETS refers to “error of target sequence” that is defined similarly to EDS but for the target sequence. The NRE defined thus accounts for the uncertainty from the structure feature predictors. Finally, I-TASSER models of each of the designed sequences are provided if user selects the third option on I-TASSER modeling. The I-TASSER models represent a partial validation of the success of the design simulation as described below.
2.3 Computational Validation of Protein Designs
No computational design method is perfect, and validation remains an essential part of the design processes. Validating experimentally that the designed protein sequence successfully folds to the desired structure requires both successfully expressing the protein and successfully determining the structure. A full structure determination at the atomic level through either NMR spectroscopy or X-ray crystallography is a time-consuming and difficult task. Even simpler, less precise experimental methods for determining protein structure, such as comparing the secondary structure of the native and designed proteins through circular dichroism CD (see Subheading 2.4.7) and recognition of the presence of folded tertiary structure through 1D NMR (see Subheading 2.4.8), still require that the protein be successfully expressed. Compared to computational techniques, protein expression is relatively expensive, limited in throughput, and in some cases may be challenging to achieve. Before expression, it is therefore desirable to know which designed sequences are most likely to fold to the target structure. The first step is to visually confirm that the design sequences are compatible with the structure. Specifically, it is a good idea to look for buried charges without salt-bridges and buried side-chains without hydrogen bonding partners before proceeding. The EvoDesign program uses a fixed backbone approximation in its calculations. High energies from van der Waals clashes can usually be relieved by small changes in the backbone [44, 45]. However, buried charges and missing hydrogen bonds are much harder to compensate for by small structural movements. Since even one missed hydrogen bond or buried charge is enough to completely destabilize a structure, any designs possessing these features should be eliminated from consideration.
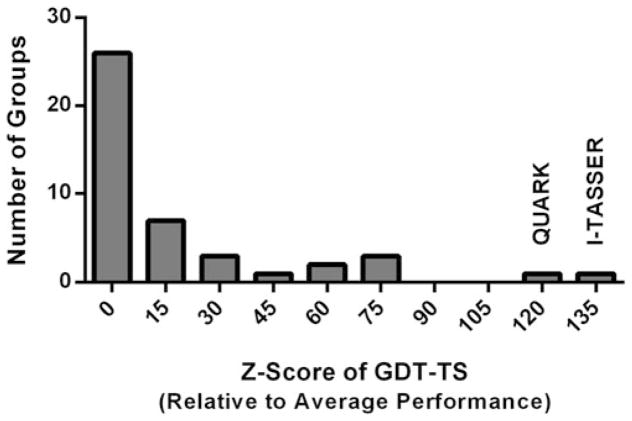
It is, however, not possible to tell reliably if a protein will fold correctly by simple visual analysis. Accurate structure prediction of designed sequences is therefore central to the EvoDesign methodology, as it allows a much larger number and variety of sequences to be tested for correct folding than can be experimentally checked. EvoDesign currently employs I-TASSER, which is a hierarchical approach to protein structure modeling that constructs protein 3D models by reassembling continuous fragments excised from the multiple threading templates [43, 46–48]. I-TASSER has been extensively tested in both benchmarking [46, 47, 49] and blind tests [50–53]. In particular, the community-wide CASP (Critical Assessment of protein Structure Prediction) experiment is designed to benchmark the state-of-the-art of protein structure predictions every two years since 1994 [54–56]. I-TASSER was tested (as “Zhang-server”) in the 7–11th CASP competitions in 2006–2015. Figure 3 shows the histogram of the Z-score of the GDT-score, which measures the significance of the model predictions by each group of automated structure predictors compared to the average performance, in the latest 11th CASP competition. The data shows the advantage of the I-TASSER in comparison to other state-of-the-art protein structure prediction methods, provided that the protein is already known to fold to a specific structure.
Fig. 3.
Histogram of the Z-scores of all automated protein structure predictors in the CASP11 experiment. The first bin contains groups that have Z-score below 0. Data are taken from official CASP webpage at URL http://www.predictioncenter.org/casp11/zscores_final.cgi?model_type=first&gr_type=server_only
2.3.1 Estimating Structural Fidelity and Foldability of Designed Sequences Using I-TASSER
The I-TASSER-based structure prediction of designed sequences in EvoDesign seeks to answer two related but distinct questions. First, does the designed sequence fold to any structure at all or is it only partially or completely unfolded when expressed? Second, given that the protein folds, does it fold to the correct structure? If a designed sequence is known to fold, there is considerable evidence from the benchmark and blind tests described above that I-TASSER could, with some confidence, tell if it will fold to its target structure. However, the ability of template-based protein structure programs to determine whether or not a given sequence can fold correctly to any structure at all has been tested much less extensively (see Note 1).
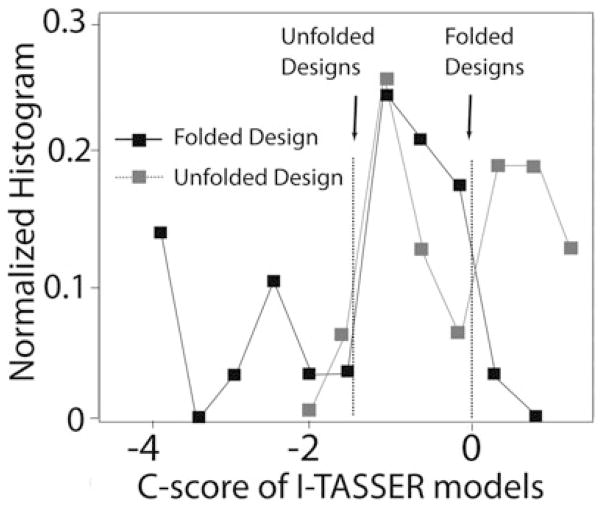
In an early test, I-TASSER was shown to cleanly distinguish native sequences from random sequences with similar sequence identity and secondary structural propensity [38]. For a more stringent benchmark test, we recently tested 16 successfully designed sequences that are known to match their target structure and 29 unsuccessful sequences that were known to either fold to a different structure or were unable to fold at all in the literature [25]. As shown in Fig. 4, I-TASSER successfully captured the deviation of the structures of the designed sequences from the target structure. Furthermore, the confidence level (C-score) [57] of the I-TASSER prediction is roughly correlated with the chance of success of the design: a C-score below −1.5 indicates an almost certain failure and a C-score above 0 indicates a very strong possibility of success. I-TASSER prediction on designed sequences can therefore allow a winnowing out of poorly designed sequences without resorting to the lengthy procedure of expressing and experimentally determining the structures of designed proteins at each step.
Fig. 4.
Divergence in the confidence score of the I-TASSER models for successfully and unsuccessfully designed sequences. Approximate cutoff values are indicated by the arrows. A C-score < −1.5 indicates a high probability that the design will not be folded correctly and a C-score > 0 indicates a high probability that the design will fold to the target structure
2.4 Experimental Validation of Designed Sequences
True validation of the designed protein requires that protein be characterized experimentally for structurally fidelity and activity. The processes listed below have been employed in the EvoDesign studies [25, 58], aiming to ensure that the designed proteins are thermodynamically stable, soluble, and adopt the desired fold. In all cases, the same tests should be performed with the wild-type protein as well for a control.
2.4.1 Expression and Purification of Designed Proteins
Before a protein can be characterized experimentally, the pure protein must be generated in sufficient quantities for the experiments. This is done through a process called recombinant expression, which involves incorporating the DNA sequence of the designed protein into the genome of another organism and using that organism’s protein production process to generate the target protein. Since there are many variations on the technique and the specifics of the process can vary with the protein being produced, a comprehensive description of the technique is not given here. Instead, key considerations are outlined in a basic manner for those unfamiliar with process. For further, more depth treatment readers are encouraged to consult several excellent reviews on this topic [59].
2.4.2 Choice of Host Cell
The first decision that must be made in setting up a recombinant protein expression system is the choice of the host cell whose protein synthesis machinery will produce the target protein. This choice is one of the most critical ones as the choice of the expression organism defines the scope of the project, the reagents and equipment needed, and the final outcome of the expression process [59]. Each protein expression has advantages and disadvantages. In most cases, bacterial expression systems are favored as they are low cost, easy to manipulate genetically, scale easily from small- to large-scale expression, and can easily incorporate isotopic labels for NMR studies. The main disadvantage of bacterial expression is that eukaryotic posttranslational modifications such as glycosylation and phosphorylation are not performed. In the case that these posttranslational modifications are essential, a eukaryotic host cell such as yeast or insect cells must usually be used and the process becomes considerably more complex.
Disulfide bond formation is also more difficult in bacteria, although this may be overcome in most cases by selecting a bacterial strain such as the Orgami cell line that have mutations in the thioredoxin reductase and glutathione reductase genes, which creates an oxidative environment that greatly enhances disulfide bond formation in the cytoplasm [60]. Expression can vary greatly for different bacterial strains. For this reason, different specialized strains of bacteria have been created to optimize the expression of recombinant proteins. Most specialized bacterial strains for expression start with the BL21 genetic background that is deficient in the Ion and ompT proteases that can lead to improper cleavage of the protein product. Other bacterial strains attempt to minimize the difference in codon usage between the natural codon usage of the bacteria and the codon usage required to express the protein.
Recombinant expression of proteins can lead to a high demand for specific tRNAs that are normally produced in only small amounts by the bacteria. Depletion of these low abundance tRNAs can cause translation to stall on the ribosome, leading to premature release from the ribosome and the generation of truncated versions of the protein [61]. From our studies [25, 58, 62], we recommend for routine use of the Rosetta 2 bacterial cell line that combines the protease mutations found in the BL21 strain along with additional modifications that allow the bacteria to generate low abundance tRNAs more efficiently and mutations that allow tunable expression through mutations in the Lac permease gene (see below). However, alternate strains may be considered in certain situations such as the Rosetta-gami strain, which adds the disulfide-bond promoting mutations of the Orgami strain to the Rosetta background.
2.4.3 Selection of Expression Vector
Once the host cell is selected, the next step is to create the vector that introduces the foreign DNA into host cell. This is typically a bacterial plasmid that contains several elements besides the DNA encoding the target protein. The first element is a gene for antibiotic resistance which provides a growth selection mechanism for discovery; only those bacteria that have incorporated the plasmid into their genome can grow in the presence of the antibiotic. The second is the promoter system, which ties the expression of the target protein to another protein whose expression is essential for the cell and whose expression can be readily induced at a specific time. Triggering expression at a specific time is essential as bacteria continue to grow during incubation and the time at which the protein is lysed determines the overall yield and final purity of the product. If the cell density is too low, the yield of expressed protein is naturally low. On the other hand, too high of cell density can also result in decreased yields and purity from loss of the plasmid from the bacteria [63], metabolism of the antibiotic within the medium, and death of the bacteria from lack of dissolved oxygen [64]. Typically, this is done through the use of the Lac operon, in which protein expression can be induced at a specific time period during growth with the lactose analog isopropyl β-D-1-thiogalacto-pyranoside (IPTG).
2.4.4 Purification of Expressed Protein
Once expressed, the expressed protein still needs to be purified from the other proteins in the bacterial cell. Although this may be accomplished using the sequence of the designed protein without modification using multiple steps of column chromatography, it is easier to fuse the designed sequence to other protein domains to make purification easier. In many cases, the expressed protein is not soluble at the very high concentrations generated during expression. In this situation, the expressed protein accumulates in an insoluble form in the bacteria as particles known as inclusion bodies. The formation of inclusion bodies can make purification easier or more difficult. The inclusion bodies generally contain the expressed protein in highly pure form with only a small amount of the other proteins of the host cell mixed in, a clear advantage for the purification process. On the other hand, proteins within inclusion bodies must be first disaggregated and then refolded with urea, which may prove a difficult process [65]. If the stability of the protein is unknown, such as the case with designed proteins, it is often easier to try to purify already folded, soluble proteins.
To enhance the solubility of proteins during purification, a solubility tag such as the Mocr domain [66] can be fused to the target protein. This domain is usually fused N-terminal to the designed sequence. Since it is localized to the N-terminus, the Mocr domain is therefore synthesized first and folds into its native form before the translation of the designed sequence, stabilizing the designed domain’s folding process. Moreover, the high negative charge on the Mocr domain increases the solubility during the purification process by preventing self-association by electrostatic repulsion. Along with the solubility tag, another sequence that specifically binds a particular column can be incorporated to assist purification. A common choice is the His tag, six consecutive histidine residues that strongly bind nickel (Ni) columns. A protease cleavage site is often placed between the Mocr domain with the His tag and the sequence of the designed protein so that the two domains can be separated. The expressed protein with the Mocr/His tag will bind the Ni column; most other bacterial proteins will not. The Mocr/His domain is then cleaved from the target sequence by the addition of a protease specific to the cleavage site and passed through the Ni column again. This time, the target protein does not bind the Ni column but all other nickel-binding proteins will remain bound to the column. The end result of this process is a highly pure protein in a soluble form.
2.4.5 Confirmation of Protein Solubility
In addition to adopting a stable folded conformation, many proteins must be soluble in water to perform their biological function. This requirement constrains the design process, as sequences that are optimized only for stability of the folded conformation may not be optimized for solubility. A key advantage of the EvoDesign method is that the structural profiles implicitly include all the constraints involved in determining the sequences that are compatible with a specific fold, not just those concerned with fold stability. As a result, sequences designed by EvoDesign are significantly more native-like in composition than those designed by physics only methods [25], which tend to overemphasize hydrophobic residues on the surface more than is found in native proteins [20, 38, 67]. Consequently, aggregation by the coalescence of exposed hydrophobic patches is a common source of failure in physics-based design [20].
As aggregation generally makes a protein useless for most applications, the oligomeric state of the protein should be determined before proceeding at the highest concentration used for the other biophysical experiments. Typically, this is around 100 μM for a 100-residue domain. The limiting factor is usually sensitivity of the 1D NMR experiment for tertiary structure estimation and sensitivity of the urea denaturation experiment used for the determination of protein stability (see Note 2). An approximate concentration range may be established by measuring the signal-to-noise ratio at different concentrations of the native protein. The signal of both experiments is actually more sensitive to the total concentration by weight than the molar concentration. The 100 μM value may need to be adjusted upward or downward for proteins significantly shorter or longer than 100 residues.
The presence of aggregation is most readily determined quantitatively by dynamic light scattering, which measures the hydrodynamic radius of proteins in solution, or from a correctly calibrated analytical size exclusion column. In the absence of either instrument, aggregation may be measured semiquantitatively by the absorbance at 400 nm. At this wavelength range, the protein does not absorb light and increases in absorbance are due to Raleigh scattering, which is proportional to the sixth power of the particle radius. A comparison to the corresponding absorbance at 400 nm of the native protein provides a qualitative estimate of the amount of aggregation in the sample (see Note 3).
2.4.6 Confirmation of Structural Fidelity
X-ray crystallography remains the gold standard for confirming whether a protein design has the desired structure. However, not all well-folded proteins crystallize and the expense of X-ray crystallography severely restricts the number of designs that can be studied. From a functional perspective, absolute structural fidelity is not necessary in many cases and small changes on the atomic scale are tolerated if the protein is stable, soluble, and functional. To test a larger number of sequences, faster low-resolution biophysical techniques can be used to eliminate obviously badly designed sequences [68, 69].
2.4.7 Confirmation of Secondary Structure
Secondary structure is the most basic building block of protein structure. The existence of severely incorrect secondary structure in the designed protein therefore very strongly implicates a failed design. Since each secondary structure element (α-helix, β-sheet, and random coil) has a distinct circular dichroism (CD) spectra, the relative fractions of each in a protein can be estimated from a CD spectra by fitting to a reference set of proteins with known CD spectra and secondary structure [70]. The accuracy of this procedure is typically around ±5 %, with α-helical content determined more precisely than either random coil or beta sheet content. If available, infrared (IR) spectra can also be used in a similar manner to characterize the secondary structure, as it has been shown that IR and CD are largely complementary and a combination of the two techniques gives a more accurate picture of the secondary structure than either technique alone [71].
2.4.8 Confirmation of Existence of Tertiary Structure
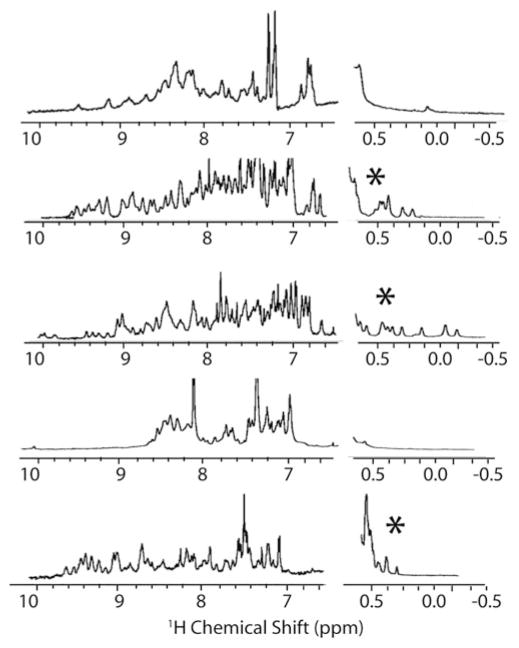
The existence of tertiary structure has traditionally been defined in a qualitative way from the appearance of the 1D 1H NMR spectra of the protein. A protein that is poorly folded, without extensive contacts within the protein core, has a distinctive 1D NMR spectra characterized by the lack of highly shielded peaks in the region of the spectra from −1 to 0.5 ppm and poor dispersion of the signal within the amide region (see Fig. 5) [72, 73]. While this method is standard in the protein design field [68, 69], it is subjective and qualitative. A more objective and quantitative method is to use the autocorrelation of a 1D 1H [74] or unassigned 3D 15N NOESY-HSQC NMR spectrum [75], which have been shown to accurately distinguish folded and unfolded proteins. A comparison of the binding of the dye SYPRO Orange, which binds to exposed hydrophobic surfaces, to the native sequence can provide an additional test for a misfolded protein structure [76].
Fig. 5.
NMR spectra of folded (with asterisk) and unfolded designed proteins. The folded designs have a wider range of chemical shift values in the amide region of the spectrum (7–10 ppm) and have chemical shift values below 0.5 ppm indicating side-chains strongly shielded from solvent, as would be expected in a well-packed protein core
2.4.9 Confirmation of Fold Stability
The free energy of folding can be measured using chemical denaturation with urea, with denaturation measured by the decrease in secondary structure as determined by CD [25]. As the concentration of urea is increased, the protein unfolds, in most cases by a two-step process without a significant population of partially unfolded intermediates. The first step of determining the stability is to measure the CD signal without denaturant (CDfolded), where it is assumed to be completely folded, and at a high concentration of denaturant, where it is assumed to be completely folded (CDunfolded). If unfolding is a two-step process, the CD signal as a function of the urea concentration is [77]:
| (5) |
where ffolded(urea) and funfolded(urea) refer to the fractions of folded and unfolded proteins respectively, at a given urea concentration. Since the equilibrium constant can be calculated directly from fraction of folded and unfolded proteins, the Gibbs free energy of unfolding can be calculated for each urea concentration [77]:
| (6) |
| (7) |
The relevant free energy is the free energy of unfolding in the absence of denaturant, which can be obtained by linear extrapolation of the free energy to zero urea concentration.
3 Conclusions
Using an evolution-based approach, we have successfully designed, expressed, and experimentally characterized a number of single domain proteins [25, 58]. In the first benchmark test, we used EvoDesign to redesign 87 globular proteins randomly collected from the PISCES server. I-TASSER was then used to test the fidelity of the predicted structure to the target. Although all homologous templates have been excluded from the I-TASSER template library, out of the 87 designed sequences, 80 % were predicted to fold to structure with an RMSD of <2.0 Å to the target scaffold, and 42.5 % were predicted to fold to an essentially identical structure with an RMSD < 1.0 Å. This was a clear difference from designed sequences created using only the FoldX force field, for which only 54 % of the predicted structures have an RMSD < 2.0 Å to the target structure, and only 31 % have an RMSD < 1.0 Å.
In a separate test, we redesigned five globular proteins by EvoDesign and used the experimental validation procedures described in Subheading 2.4 to confirm the success of the designs. All five proteins were successfully expressed using the expression system in Subheading 2.4.3 and were soluble to at least 70 μM. Further, all five designed proteins have secondary structure consistent with the target protein (<12 % difference). Three out of the five had a compact tertiary structure confirmed by NMR (Subheading 2.4.8, Fig. 5), for an overall success rate of 60 %. One of the three, the Phox homology domain of the cytokine-independent survival kinase (CISK-PX), could be crystallized and its structure compared to the native protein [78]. Despite having only 32 % sequence identity, the structure of the designed protein showed a very close similarity to the target with a RMSD of 1.54 Å and a TM score of 0.90 to the target template. The RMSD and TM score between the I-TASSER model and the X-ray crystal structure of CISK-PX are 1.32 Å and 0.91, respectively. Most of the difference between the two structures was in a loop that is disordered in the original structure.
Finally, we have shown that EvoDesign can be used to create functional complexes for the X-linked inhibitor of apoptosis proteins (XIAP) with improved properties by designing a peptide-protein complex involved in apoptosis inhibition [58]. The XIAP protein inhibits apotosis by binding caspase-9, an activity that is in turn regulated by the second mitochondria-derived activator of caspases (SMAC). The designed XIAP protein by EvoDesign binds SMAC but does not possess affinity for caspase-9. As such, the designed protein can serve as a SMAC sink, altering the normal protein-protein interaction network involved in cell death. The circular dichroism and isothermal calorimetry data showed that the designed XIAP domain was more stable than WT-XIAP and bound the SMAC derived peptide with a Kd of 167 ± 67 nM, which compares favorably with the 80 ± 25 nM Kd found for WT-XIAP. Interestingly, a designed version of XIAP with native interface residues actually showed worse binding (Kd of 352 ± 79 nM) and stability than the fully designed sequence, highlighting the efficiency of evolution-based full protein design.
Acknowledgments
The project is supported in part by the National Institute of General Medical Sciences (GM083107).
Footnotes
The distinction between these two questions becomes clear when the nature of the benchmarks is considered. Due to the experimental requirements of structure determination, the benchmark test largely consists of proteins that can be successfully expressed, successfully purified, and are stable for a prolonged period of time at high concentration. In addition, the protein also must be crystallized in the case of X-ray structures, which is a rather severe restriction for proteins with large unfolded regions as the disordered regions have poor crystal contacts which interferes with the crystallization process [79]. Even if the protein can be crystallized, the disordered regions will have poor electron density and will therefore not be resolved in the structure. Similarly, the structure of unfolded proteins is difficult to determine by NMR due to the lack of long-range NOE constraints and poor chemical shift dispersion [80]. These experimental constraints suggest that though the PDB library is largely complete with respect to the possible universe of monomeric folded domains [81, 82], it is still biased toward compact folded structures, as proteins that are intrinsically unstable or unfolded are difficult to observe. The PDB library should therefore not be considered as completely representative of the conformational ensembles, folded or not, that all protein sequences can adopt.
The signal-to-noise ratio in an NMR experiment depends on a number of factors including the field strength of the NMR spectrometer (higher magnetic fields give higher resolution spectra and hence higher signal-to-noise ratios), the size of the protein (larger proteins give rise to broader signals), and other factors such as conformational exchange (transitions between conformations under certain timescales give rise broader signals). The signal-to-noise ratio in a CD spectrum also depends on a variety of factors, including the transparency of the buffer in the far UV region of the spectrum (180–260 nm), the path-length of the cuvette, and the age of the xenon lamp used to acquire the spectrum. Of these factors, the transparency of the buffer usually has the most impact. A buffer strongly absorbing in the UV serves as an inner filter that attenuates the incoming light reaching the protein. Phosphate buffers are optimal for CD due to their transparency in the far UV region of the spectrum, although Tris buffers are nearly as good. Chloride ions absorb in this region and the proteins in NaCl solutions should be dialyzed against an equivalent of concentration of NaF. Finally, many additives used to stabilize proteins, such as glycerol, arginine, and Triton-X, absorb strongly in the UV and are incompatible with CD spectroscopy for this reason.
An alternative wavelength can be used if the protein possesses a cofactor such as FAD or FMN that absorbs in the visible light range.
References
- 1.Karanicolas J, Kuhlman B. Computational design of affinity and specificity at protein-protein interfaces. Curr Opin Struct Biol. 2009;19(4):458–463. doi: 10.1016/j.sbi.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kortemme T, Joachimiak LA, Bullock AN, Schuler AD, Stoddard BL, Baker D. Computational redesign of protein-protein interaction specificity. Nat Struct Mol Biol. 2004;11(4):371–379. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- 3.Shifman JM, Mayo SL. Exploring the origins of binding specificity through the computational redesign of calmodulin. Proc Natl Acad Sci U S A. 2003;100(23):13274–13279. doi: 10.1073/pnas.2234277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopes A, Busch MSA, Simonson T. Computational design of protein-ligand binding: modifying the specificity of asparaginyl-tRNA synthetase. J Comput Chem. 2010;31(6):1273–1286. doi: 10.1002/jcc.21414. [DOI] [PubMed] [Google Scholar]
- 5.Procko E, Hedman R, Hamilton K, Seetharaman J, Fleishman SJ, Su M, Aramini J, Kornhaber G, Hunt JF, Tong L, Montelione GT, Baker D. Computational design of a protein-based enzyme inhibitor. J Mol Biol. 2013;425(18):3563–3575. doi: 10.1016/j.jmb.2013.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, 3rd, Hilvert D, Houk KN, Stoddard BL, Baker D. De novo computational design of retro-aldol enzymes. Science. 2008;319(5868):1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302(5649):1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 8.Siegel JB, Smith AL, Poust S, Wargacki AJ, Bar-Even A, Louw C, Shen BW, Eiben CB, Tran HM, Noor E, Gallaher JL, Bale J, Yoshikuni Y, Gelb MH, Keasling JD, Stoddard BL, Lidstrom ME, Baker D. Computational protein design enables a novel one-carbon assimilation pathway. Proc Natl Acad Sci U S A. 2015;112(12):3704–3709. doi: 10.1073/pnas.1500545112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ollikainen N, Kortemme T. Computational protein design quantifies structural constraints on amino acid covariation. PLoS Comput Biol. 2013;9(11):e1003313. doi: 10.1371/journal.pcbi.1003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fromer M, Linial M. Exposing the co-adaptive potential of protein-protein interfaces through computational sequence design. Bioinformatics. 2010;26(18):2266–2272. doi: 10.1093/bioinformatics/btq412. [DOI] [PubMed] [Google Scholar]
- 11.McLaughlin RN, Poelwijk FJ, Raman A, Gosal WS, Ranganathan R. The spatial architecture of protein function and adaptation. Nature. 2012;491(7422):138–142. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaefer C, Schlessinger A, Rost B. Protein secondary structure appears to be robust under in silico evolution while protein disorder appears not to be. Bioinformatics. 2010;26(5):625–631. doi: 10.1093/bioinformatics/btq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ollikainen N, Smith CA, Fraser JS, Kortemme T. Flexible backbone sampling methods to model and design protein alternative conformations. Methods Enzymol. 2013;523:61–85. doi: 10.1016/B978-0-12-394292-0.00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellogg EH, Leaver-Fay A, Baker D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins. 2011;79(3):830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424(6950):805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 16.Smith CA, Kortemme T. Predicting the tolerated sequences for proteins and protein interfaces using RosettaBackrub flexible backbone design. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0020451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harbury PB, Plecs JJ, Tidor B, Alber T, Kim PS. High-resolution protein design with backbone freedom. Science. 1998;282(5393):1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- 18.Pokala N, Handel TM. Energy functions for protein design: adjustment with protein-protein complex affinities, models for the unfolded state, and negative design of solubility and specificity. J Mol Biol. 2005;347(1):203–227. doi: 10.1016/j.jmb.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Yang Y, Zhan J, Dai L, Zhou Y. Energy functions in de novo protein design: current challenges and future prospects. Annu Rev Biophys. 2013;42:315–335. doi: 10.1146/annurev-biophys-083012-130315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacak R, Leaver-Fay A, Kuhlman B. Computational protein design with explicit consideration of surface hydrophobic patches. Proteins. 2012;80(3):825–838. doi: 10.1002/prot.23241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowie JU, Luthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253(5016):164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 22.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21(7):951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 23.Wu S, Zhang Y. MUSTER: improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins. 2008;72(2):547–556. doi: 10.1002/prot.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y. Progress and challenges in protein structure prediction. Curr Opin Struct Biol. 2008;18(3):342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitra P, Shultis D, Brender JR, Czajka J, Marsh D, Gray F, Cierpicki T, Zhang Y. An evolution-based approach to de novo protein design and case study on Mycobacterium tuberculosis. PLoS Comput Biol. 2013;9(10):e1003298. doi: 10.1371/journal.pcbi.1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitra P, Shultis D, Zhang Y. EvoDesign: de novo protein design based on structural and evolutionary profiles. Nucleic Acids Res. 2013;41(W1):W273–W280. doi: 10.1093/nar/gkt384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33(7):2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics. 2010;26(7):889–895. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gribskov M, Homyak M, Edenfield J, Eisenberg D. Profile scanning for 3-dimensional structural patterns in protein sequences. Comput Appl Biosci. 1988;4(1):61–66. doi: 10.1093/bioinformatics/4.1.61. [DOI] [PubMed] [Google Scholar]
- 30.Gribskov M, Mclachlan AD, Eisenberg D. Profile analysis – detection of distantly related proteins. Proc Natl Acad Sci U S A. 1987;84(13):4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu ST, Zhang Y. ANGLOR: a composite machine-learning algorithm for protein backbone torsion angle prediction. PLoS One. 2008;3(10) doi: 10.1371/journal.pone.0003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen HL, Zhou HX. Prediction of solvent accessibility and sites of deleterious mutations from protein sequence. Nucleic Acids Res. 2005;33(10):3193–3199. doi: 10.1093/nar/gki633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faraggi E, Zhang T, Yang YD, Kurgan L, Zhou YQ. SPINE X: improving protein secondary structure prediction by multistep learning coupled with prediction of solvent accessible surface area and backbone torsion angles. J Comput Chem. 2012;33(3):259–267. doi: 10.1002/jcc.21968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33(Web Server issue):382–388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krivov GG, Shapovalov MV, Dunbrack RL. Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77(4):778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Skolnick J. SPICKER: a clustering approach to identify near-native protein folds. J Comput Chem. 2004;25(6):865–871. doi: 10.1002/jcc.20011. [DOI] [PubMed] [Google Scholar]
- 38.Bazzoli A, Tettamanzi AGB, Zhang Y. Computational protein design and large-scale assessment by I-TASSER structure assembly simulations. J Mol Biol. 2011;407(5):764–776. doi: 10.1016/j.jmb.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brender JR, Zhang Y. Recognizing mutations on protein-protein binding interactions through structure-based interface profiles. PLoS Comput Biol. 2015 doi: 10.1371/journal.pcbi.1004494. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee S, Zhang Y. Protein-protein complex structure predictions by multimeric threading and template recombination. Structure. 2011;19(7):955–966. doi: 10.1016/j.str.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao M, Skolnick J. iAlign: a method for the structural comparison of protein-protein interfaces. Bioinformatics. 2010;26(18):2259–2265. doi: 10.1093/bioinformatics/btq404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y. 2012 http://zhanglab.ccmb.med.umich.edu/PSSpred.
- 43.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis IW, Arendall WB, Richardson DC, Richardson JS. The backrub motion: how protein backbone shrugs when a sidechain dances. Structure. 2006;14(2):265–274. doi: 10.1016/j.str.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 45.Smith CA, Kortemme T. Backrub-like backbone simulation recapitulates natural protein conformational variability and improves mutant side-chain prediction. J Mol Biol. 2008;380(4):742–756. doi: 10.1016/j.jmb.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu S, Skolnick J, Zhang Y. Ab initio modeling of small proteins by iterative TAS-SER simulations. BMC Biol. 2007;5:17. doi: 10.1186/1741-7007-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69(S8):108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kopp J, Bordoli L, Battey JN, Kiefer F, Schwede T. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69(Suppl 8):38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 51.Cozzetto D, Kryshtafovych A, Fidelis K, Moult J, Rost B, Tramontano A. Evaluation of template-based models in CASP8 with standard measures. Proteins. 2009;77(Suppl 9):18–28. doi: 10.1002/prot.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Montelione GT. Template based modeling assessment in CASP10. Paper presented at the 10th community wide experiment on the critical assessment of techniques for protein structure prediction; Gaeta, Italy. 9–12 Dec 2012.2012. [Google Scholar]
- 53.Lee BK. Template free modeling assessment in CASP10. Paper presented at the 10th community wide experiment on the critical assessment of techniques for protein structure prediction; Gaeta, Italy. 2012. [Google Scholar]
- 54.Moult J, Pedersen JT, Judson R, Fidelis K. A large-scale experiment to assess protein structure prediction methods. Proteins. 1995;23(3):2–5. doi: 10.1002/prot.340230303. [DOI] [PubMed] [Google Scholar]
- 55.Moult J, Fidelis K, Kryshtafovych A, Rost B, Tramontano A. Critical assessment of methods of protein structure prediction-round VIII. Proteins Struct Funct Bioinf. 2009;77:1–4. doi: 10.1002/prot.22589. [DOI] [PubMed] [Google Scholar]
- 56.Moult J. A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction. Curr Opin Struct Biol. 2005;15(3):285–289. doi: 10.1016/j.sbi.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008:9. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shultis D, Mitra P, Aslam N, Gray F, Piper C, Chinnaswamy K, Stuckey J, Cierpicki T, Wang S, Lei M, Zhang Y. Redesigning the fold and binding specificity of BIR3 domain of X-linked inhibitor of apoptosis proteins using evolutionary profiles. 2015 (submitted) [Google Scholar]
- 59.Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:172. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prinz WA, Aslund F, Holmgren A, Beckwith J. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J Biol Chem. 1997;272(25):15661–15667. doi: 10.1074/jbc.272.25.15661. [DOI] [PubMed] [Google Scholar]
- 61.Buchan JR, Stansfield I. Halting a cellular production line: responses to ribosomal pausing during translation. Biol Cell. 2007;99(9):475–487. doi: 10.1042/BC20070037. [DOI] [PubMed] [Google Scholar]
- 62.Shultis D, Czajka J, Marsh D, Gray F, Brender JR, Mitra P, Cierpicki T, Zhang Y. Structural validation of computational protein designed through evolutionary methods. (in preparation) [Google Scholar]
- 63.Baneyx F. Recombinant protein expression in Escherichia coli. Curr Opin Biotechnol. 1999;10(5):411–421. doi: 10.1016/s0958-1669(99)00003-8. [DOI] [PubMed] [Google Scholar]
- 64.Jana S, Deb JK. Strategies for efficient production of heterologous proteins in Escherichia coli. Appl Microbiol Biotechnol. 2005;67(3):289–298. doi: 10.1007/s00253-004-1814-0. [DOI] [PubMed] [Google Scholar]
- 65.Burgess RR. Refolding solubilized inclusion body proteins. Methods Enzymol. 2009;463:259–282. doi: 10.1016/S0076-6879(09)63017-2. [DOI] [PubMed] [Google Scholar]
- 66.DelProposto J, Majmudar CY, Smith JL, Brown WC. Mocr: a novel fusion tag for enhancing solubility that is compatible with structural biology applications. Protein Expr Purif. 2009;63(1):40–49. doi: 10.1016/j.pep.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dantas G, Kuhlman B, Callender D, Wong M, Baker D. A large scale test of computational protein design: folding and stability of nine completely redesigned globular proteins. J Mol Biol. 2003;332(2):449–460. doi: 10.1016/s0022-2836(03)00888-x. [DOI] [PubMed] [Google Scholar]
- 68.Koga N, Tatsumi-Koga R, Liu GH, Xiao R, Acton TB, Montelione GT, Baker D. Principles for designing ideal protein structures. Nature. 2012;491(7423):222. doi: 10.1038/nature11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, Ranganathan R. Evolutionary information for specifying a protein fold. Nature. 2005;437(7058):512–518. doi: 10.1038/nature03991. [DOI] [PubMed] [Google Scholar]
- 70.Sreerama N, Woody RW. Analysis of protein CD spectra: comparison of CONTIN, SELCON3, and CDSSTR methods in CDPro software. Biophys J. 2000;78(1):334. [Google Scholar]
- 71.Oberg KA, Ruysschaert JM, Goormaghtigh E. The optimization of protein secondary structure determination with infrared and circular dichroism spectra. Eur J Biochem. 2004;271(14):2937–2948. doi: 10.1111/j.1432-1033.2004.04220.x. [DOI] [PubMed] [Google Scholar]
- 72.Rehm T, Huber R, Holak TA. Application of NMR in structural proteomics: screening for proteins amenable to structural analysis. Structure. 2002;10(12):1613–1618. doi: 10.1016/s0969-2126(02)00894-8. [DOI] [PubMed] [Google Scholar]
- 73.Scheich C, Leitner D, Sievert V, Leidert M, Schlegel B, Simon B, Letunic I, Bussow K, Diehl A. Fast identification of folded human protein domains expressed in E. coli suitable for structural analysis. BMC Struct Biol. 2004;4:4. doi: 10.1186/1472-6807-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoffmann B, Eichmuller C, Steinhauser O, Konrat R. Rapid assessment of protein structural stability and fold validation via NMR. Methods Enzymol. 2005;394:142. doi: 10.1016/S0076-6879(05)94006-8. [DOI] [PubMed] [Google Scholar]
- 75.Schedlbauer A, Coudevylle N, Auer R, Kloiber K, Tollinger M, Konrat R. Autocorrelation analysis of NOESY data provides residue compactness for folded and unfolded proteins. J Am Chem Soc. 2009;131(17):6038. doi: 10.1021/ja8074067. [DOI] [PubMed] [Google Scholar]
- 76.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2(9):2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 77.Pace CN, Scholtz JM. Measuring the conformational stability of a protein. In: Creighton TE, editor. Protein structure: a practical approach. Oxford University Press; New York, NY: 1997. pp. 299–321. [Google Scholar]
- 78.Shultis D, Dodge G, Zhang Y. Crystal structure of designed PX domain from cytokine-independent survival kinase and implications on evolution-based protein engineering. 2015 doi: 10.1016/j.jsb.2015.06.009. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Price WN, 2nd, Chen Y, Handelman SK, Neely H, Manor P, Karlin R, Nair R, Liu J, Baran M, Everett J, Tong SN, Forouhar F, Swaminathan SS, Acton T, Xiao R, Luft JR, Lauricella A, DeTitta GT, Rost B, Montelione GT, Hunt JF. Understanding the physical properties that control protein crystallization by analysis of large-scale experimental data. Nat Biotechnol. 2009;27(1):51–57. doi: 10.1038/nbt.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O’Hare B, Benesi AJ, Showalter SA. Incorporating 1H chemical shift determination into 13C-direct detected spectroscopy of intrinsically disordered proteins in solution. J Magn Reson. 2009;200(2):354–358. doi: 10.1016/j.jmr.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Y, Hubner IA, Arakaki AK, Shakhnovich E, Skolnick J. On the origin and highly likely completeness of single-domain protein structures. Proc Natl Acad Sci U S A. 2006;103(8):2605–2610. doi: 10.1073/pnas.0509379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brylinski M, Gao M, Skolnick J. Why not consider a spherical protein? Implications of backbone hydrogen bonding for protein structure and function. Phys Chem Chem Phys. 2011;13(38):17044–17055. doi: 10.1039/c1cp21140d. [DOI] [PMC free article] [PubMed] [Google Scholar]