Abstract
Antibiotic-resistant bacterial infections are increasingly prevalent worldwide and there is an urgent need for novel classes of antibiotics capable of overcoming existing resistance mechanisms. One potential antibiotic target is the bacterial single-stranded DNA binding protein (SSB) which serves as a hub for DNA repair, recombination and replication. Eight highly conserved residues at the C-terminus of SSB use direct protein-protein interactions (PPIs) to recruit more than a dozen important genome maintenance proteins to single-stranded DNA. Mutations that disrupt PPIs with the C-terminal tail of SSB are lethal, suggesting that small molecule inhibitors of these critical SSB PPIs could be effective antibacterial agents. As a first step toward implementing this strategy, we have developed orthogonal high-throughput screening assays to identify small molecule inhibitors of the Klebsiella pneumonia SSB-PriA interaction. Hits were identified from an initial screen of 72,474 compounds using an AlphaScreen (AS) primary screen and their activity was subsequently confirmed in an orthogonal fluorescence polarization (FP) assay. As an additional control, an FP assay targeted against an unrelated eukaryotic PPI was used to confirm specificity for the SSB-PriA interaction. Nine potent and selective inhibitors produced concentration-response curves with IC50 values <40 µM and the direct binding of two compounds were observed to bind to PriA, demonstrating the success of this screen strategy.
Antibiotic resistance among bacterial pathogens has become an increasing worldwide healthcare crisis. In the United States alone, an estimated 2 million people are infected with antibiotic-resistant bacteria annually, with over 23,000 attributed deaths each year.1 The emergence and spread of bacterial strains with resistance to antibiotics of last resort, such as carbapenems2 and colistin,3 have raised fears of a post-antibiotic world in which routine infections are untreatable.4, 5 While improved prescribing practices and limitations in the use of antibiotics in livestock production are essential parts of efforts to combat antibiotic resistance,6 novel classes of antibiotics that circumvent existing resistance mechanisms are also urgently needed.5
The molecular machinery essential for bacterial replication and genome maintenance is surprisingly dissimilar from analogous proteins in eukaryotes, so much so that it is thought that processes such as replication could have evolved independently.7 From a therapeutic perspective, this difference makes genome maintenance proteins excellent antibiotic targets since the possibility of cross-reaction with functionally analogous eukaryotic proteins is minimized.8, 9 Because DNA replication, recombination and repair pathways are essential for bacterial cell viability, antibacterial drugs that target these processes have the potential to be highly effective. Fluoroquinolone topoisomerase inhibitors such as ciprofloxacin, levofloxacin, and trovafloxacin, which are commonly used to treat hospital-acquired pneumonia, urinary tract infections and other antibiotic-resistant infections,10, 11 comprise the only current antibiotics that act on bacterial genomic targets. These therapeutics inhibit type-II topoisomerases creating DNA breaks that block DNA replication fork progression12 and are lethal unless repaired by homologous recombination and DNA replication restart processes that reload the DNA replication machinery. While the success of this class of antibiotics validates genome maintenance proteins as targets for antibacterial drug development, the therapeutic potential of numerous direct DNA replication, recombination and repair proteins remain untapped.8, 9
In genome maintenance reactions, duplex DNA must often be separated to allow enzymes access to genomic information. However, single-stranded (ss) DNA is sensitive to chemical and nucleolytic attack and can form secondary structures that block genome maintenance reactions. To avoid these potential problems, bacteria have evolved specialized ssDNA binding proteins (SSBs) that bind and protect ssDNA as it is formed in cells. For the majority of bacteria examined to date, the functional ssDNA binding unit is a homotetrameric core of individual oligonucleotide-binding (OB) folds that are responsible for high-affinity ssDNA binding. Extending from the C-terminus of each of the monomeric constituents are intrinsically disordered linkers that terminate with an acidic and highly conserved 8 amino acid sequence called the SSBct. The SSBct interacts with a variety of critical genome maintenance proteins, positioning the SSB-DNA complex as a central hub for repair, replication and recombination. The structures of multiple SSB-interacting proteins (IPs) bound to peptides mimicking the SSBct have been determined and the SSB binding pocket of each of these proteins show striking electrostatic similarities.13–18 Deletion or mutation of conserved residues within the SSBct disrupts its protein interactions and is lethal in Escherichia coli.19–25 Given the essential nature of SSB protein interactions, we have hypothesized that small molecules capable of disrupting SSB protein interfaces could prove to be valuable antibiotic lead compounds. Previously identified inhibitors that block E. coli SSBct interaction with Exonuclease I support this hypothesis; however, their potential as antibiotics is limited by the fact that the activity of Exonuclease I is not essential to bacterial viability.26, 27
As a step toward targeting SSB-protein interactions with molecules that could be used as antibacterial therapeutics, we have developed a high-throughput screening (HTS) platform to identify inhibitors that block interaction between the SSBct and the essential PriA DNA helicase from Klebsiella pneumonia, an important human pathogen.28 The HTS strategy was designed with cost savings considerations as a major factor. These savings are derived from both the use of 1536-well plates to minimize reagent use and by using tip-free liquid handling. In a pilot screen of 72,474 compounds, 9 potent inhibitors that selectively disrupt the Klebsiella SSBct-PriA interaction were identified and 2 of these compounds were observed to bind PriA. Identification of these compounds validates this screening strategy and lays the foundation for the optimization and antibacterial activity of these inhibitors.
Materials and Methods
Protein expression
SSBct (Trp-Met-Asp-Phe-Asp-Asp-Asp-Ile-Pro-Phe), N-terminally biotinylated SSBct (Bio-SSBct), N-terminally fluorescein labeled SSBct (F-SSBct) and an SSBct lacking the C-terminal phenylalanine (ΔFSSBct) peptides were purchased from the University of Wisconsin Biotechnology center as lyophilized powders and resuspended in either dimethyl sulfoxide (DMSO) or 10 mM HEPES-HCl, pH 7.0.
Protein Purification
Rosetta 2 competent E. coli cells were transformed with a pET28b plasmid encoding the Klebsiella pneumonia PriA protein fused to an N-terminal 6xHis purification tag.13 Single colonies were used to inoculate 100 mL of LB supplemented with kanamycin and chloramphenicol, and incubated with shacking overnight at 37 °C. The culture was used to inoculate 2 L of auto-induction media29 supplemented with kanamycin and chloramphenicol, and allowed to grow at 37 °C with shaking for 24 hours. Cells were pelleted by centrifugation and stored at −80 °C. Cell pellets were thawed, mixed with lysis buffer (10 mM HEPES-HCl, pH 7.0, 300 mM Na2SO4, 100 mM glucose, 10% (v/v) glycerol, 20 mM imidazole, 1 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride and a protease inhibitor tablet (ThermoFisher, Waltham, MA)) and lysed by sonication. Insoluble components were removed by centrifugation and the supernatant was passed through a 0.22 µm filter prior to being loaded onto Ni-NTA agarose resin (Quiagen, Hilden, Germany). The column was washed with 15 column volumes (CV) of lysis buffer, then eluted with a gradient of lysis buffer containing 20 to 300 mM imidazole over 10 CV. Fractions containing PriA were pooled, diluted to 90 mM Na2SO4 with dilution buffer (10 mM HEPES-HCl, pH 7.0, 10 mM dithiotheritol (DTT), 10% (v/v) glycerol) and then loaded onto an SPFF column (GE Healthcare Bio-Sciences, Marlborough, MA) equilibrated with buffer A (10 mM HEPES, 100 mM Na2SO4, 10% glycerol, 1 mM DTT, pH 7.0). PriA was eluted from the SPFF column by gradient elution of buffer A containing 100 to 500 mM Na2SO4. PriA-containing fractions were pooled, concentrated by centrifugation and then purified on a S-100 size exclusion column (GE Healthcare Bio-Sciences) equilibrated with buffer A containing 500 mM Na2SO4. Purified PriA was concentrated to approximately 150 µM, aliquoted and flash frozen in liquid nitrogen.
SSBct-PriA AlphaScreen assay
Screening took place at the University of Wisconsin Small Molecule Screening Facility (SMSF). Controls and small molecules (stored as 10 mM DMSO stocks) were dispensed into a 1536-well white plates (Nunc 253607, ThermoFisher) using an Echo 550 (Labcyte, Sunnyvale, CA) acoustic liquid handler. A Mantis liquid handler equipped with a high volume silicone chip (Formulatrix, Bedford, MA) was used to add 3.0 µL of a master mix containing 10 mM HEPES-HCl, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 10 mM DTT, 1 mg/mL BSA, 0.01% Triton X-100, 0.1 µM PriA, 0.1 µM Bio-SSBct and 5 µg/mL AlphaScreen (AS) of both donor and acceptor beads (PerkinElmer, Waltham, MA) to each well of the plate. Master mix was prepared under diminished lighting immediately prior to dispensing. Plates were centrifuged briefly, rocked at room temperature for an hour and then read using a PheraStar (BMG Labtech, Offenburg, Germany) plate reader using the following settings: 0.1 s settling time, 0.3 s excitation, 0.6 s integration time with a 0.04 s delay between excitation and integration. Final concentrations of the positive (SSBct) and negative (ΔFSSBct) controls were 25 µM and all compounds were tested at 33.3 µM final concentration. Each screening plate contained 32 positive and negative control wells, 43 DMSO-only control wells and a control SSBct concentration-response curve conducted in triplicate with SSBct concentrations of 0.5, 1, 2, 4, 8, 16 and 32 µM. For compound concentration-response curves, the requisite amount of each compound (at 10 mM in DMSO) was added to the wells and then backfilled with 100% DMSO so that each well contained a final DMSO concentration of 0.33% (v/v). AS master mix was then added to each well as before.
Library Composition
The compound library comprised 72,474 unique small molecules originally purchased from Life Chemicals Inc. as part of their pre-plated diversity collection. The compounds were selected for diversity by the vendor, and cover a large number of distinct scaffolds.
Data analysis
To reduce the effect of plate and edge effects, two 1536-well plates containing AS master mix and 0.33% DMSO in every well were read. The mean intensity of each well from the DMSO only plates was used to normalize the matching well of the assay plates. Additionally, a vertical signal gradient was noted across each plate, likely resulting from incomplete temperature equilibration inside the plate reader. To compensate for this drift without slowing the screening, a normalization process was implemented.
First, the mean value of all samples in a given row, row i, was calculated (x̄row i). The mean of all row averages was then determined (x̄all rows) and a ratio of each of the individual row means to overall row mean was calculated. The reading of each well in row i was then multiplied by this row-specific ratio to calculate the corrected reading (xc,i) as in equation 1.
| (1) |
After the normalization process, Z’ scores for each plate were calculated and three plates with Z’ values lower than 0.5 were repeated.30 Small molecules producing more than a 35% inhibition in the SSBct-PriA AS were called hits. All hits were screened against Baell’s PAINS filters to remove small molecules containing motifs suggesting PAINS activity31 using the Drugs3 web service (fafdrugs3.mti.univ-paris-diderot.fr).32
Analysis of concentration-response curves was performed in Prism version 5.0c (GraphPad Software, La Jolla, CA) using a four-parameter logistic fit to determine IC50 values and errors.
Fluorescence polarization secondary and counter screens
To confirm the activity of compounds identified in the AS primary screen, hits were retested in duplicate at 33.3 µM in a fluorescence polarization (FP) assay. Compounds were added to black 384 shallow-well plates (ThermoFisher, 35 nL of a 10 mM DMSO stock) by an Echo acoustic liquid handler. FP master mix containing 10 mM HEPES-HCl, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM DTT, 4.85 µM PriA and 0.01 µM F-SSBct was prepared, and 10 µL of master mix was added with the Mantis liquid handler using a high volume silicone chip. After incubating for at least an hour at room temperature, plates were read using a PheraStar plate reader with the following setting: Excitation: 485 nm, Emission: 520 nm, settling time of 0.2 s and 200 flashes per well. Small molecules with an average inhibition of greater than 35% were tested in the FP counter screen, which was performed as previously described.33 Any compounds with an activity of > 25% inhibition in the RMI-MM2 FP assay were deemed promiscuous and not pursued.
Differential Scanning Fluorimetry
Differential scanning fluorimetry (DSF) was used to determine if any of the remaining compounds directly interact with PriA. In a 96-well, 0.2 µL thin-walled plate (Midsci, St. Louis, MO),10 µM PriA was incubated in 50 µL of DSF buffer containing 10 mM HEPES-HCl, 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM DTT, 2% DMSO (v/v) and 5X SYPRO orange (Sigma-Aldrich, St. Louis, MO). Compounds were tested at concentrations of 20 µM and 100 µM. In positive control wells, SSBct was included at a concentration of 20 µM. Using a CFX Connect real time PCR system (Bio-Rad, Hercules, CA), reactions were held at 25 °C for 10 minutes and then ramped to 90 °C at a rate of 0.5 °C/min. Fluorescence readings were taking using the FAM channel every 0.5 °C. For melting point (Tm) determinations, readings from a well containing no PriA were subtracted from each time point. To aid in analysis, the rate of change in fluorescence signal was calculated at every temperature and the Tm was determined to be the temperature with the maximum rate of change.
Results
Assay optimization in 1536-well plates
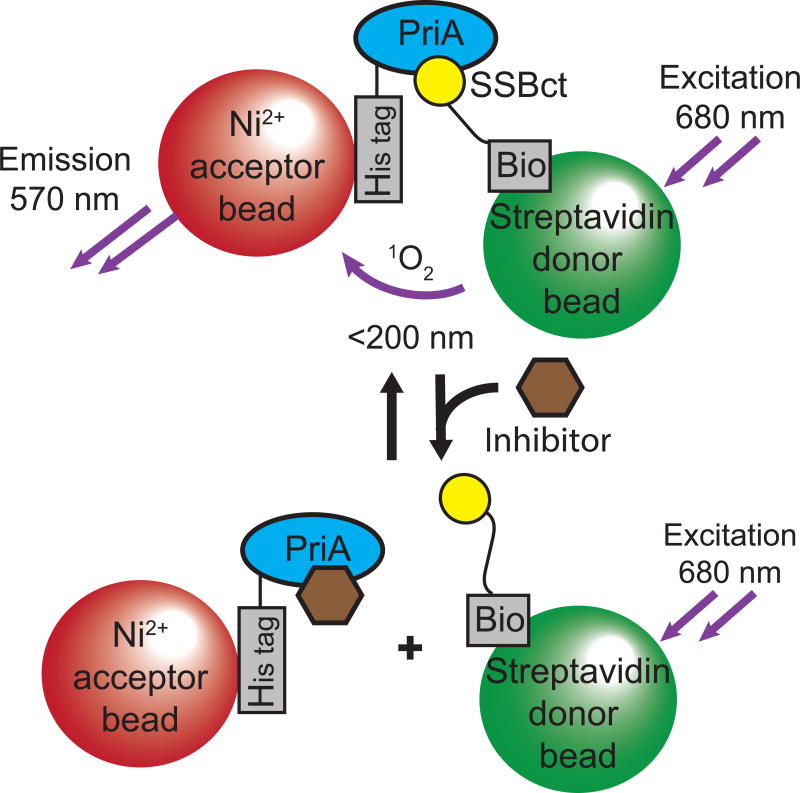
To identify inhibitors of SSB PPIs, we developed a high-throughput AS assay to quantify the interaction between SSBct and PriA helicase from Klebsiella pneumonia in a low volume, 1536-well plate format. In this bead-based proximity assay, Ni2+-coated acceptor beads are tethered to His-tagged PriA, while streptavidin-coated donor beads bind to Bio-SSBct peptides. If SSBct and PriA interact with one another, acceptor and donor beads will be held in close proximity and singlet oxygen released from the donor bead upon excitation with 680 nm light will diffuse to the acceptor bead and trigger the emission of a 570 nm wavelength signal. Disruption of the interaction greatly reduces the amount of the short-lived singlet oxygen that can reach acceptor beads, resulting in minimal 570 nm background emission (Figure 1). To measure the reproducibility of the SSBct-PriA AS assay and its suitability for use in high-throughput screening, the assay was tested in a 3-day validation study. Identical reactions (10 µL) containing His-tagged PriA and Bio-SSBct were challenged with unlabeled SSBct or ΔFSSBct competitor peptide in 384-well plates on three consecutive days. We observed robust separation between the positive (unlabeled SSBct competitor peptide) and negative (unlabeled ΔFSSBct peptide) control reactions across each day of the trial, resulting in a Z’ score of 0.81 and confirming its suitability for HTS. We have successfully used this assay as a counter screen in a previously published HTS.33
Figure 1. Schematic representation of the AS assay to identify inhibitors of the SSBct-PriA interaction.
SSBct and PriA are bound to donor and acceptor bead pairs that generate signal in close proximity. Disruption of the SSBct-PriA interaction by an inhibitor is detected by a loss of signal emission.
Given the difficulties associated with targeting PPIs, we anticipated a need to screen a large chemical library to successfully identify lead compounds. As protein production and AS beads are major contributors to the overall cost of the screen, we sought to minimize the assay volume without compromising assay performance. It was found that we could reduce our existing AS assay volume to 3 µL in a 1536-well format with only minimal losses in performance.
Pilot high-throughput screening campaign
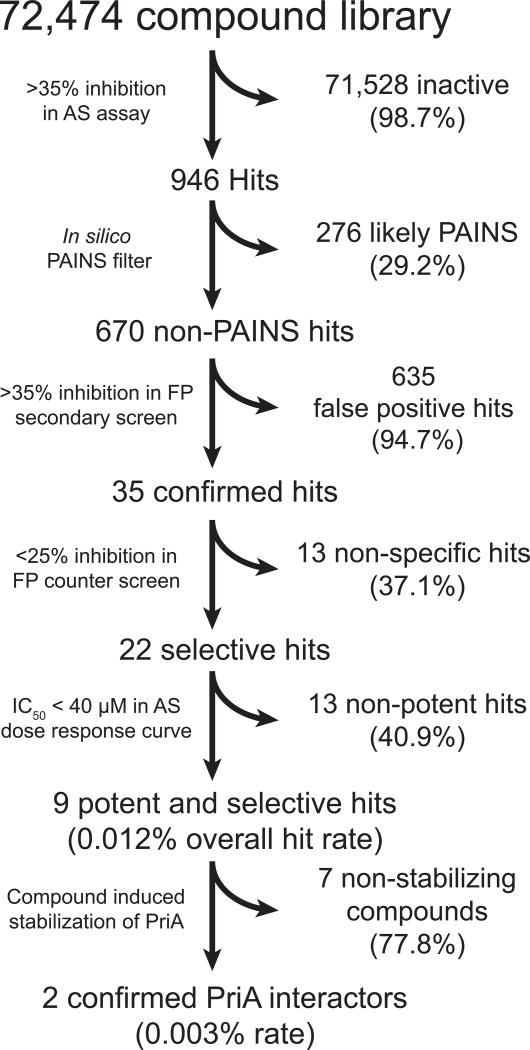
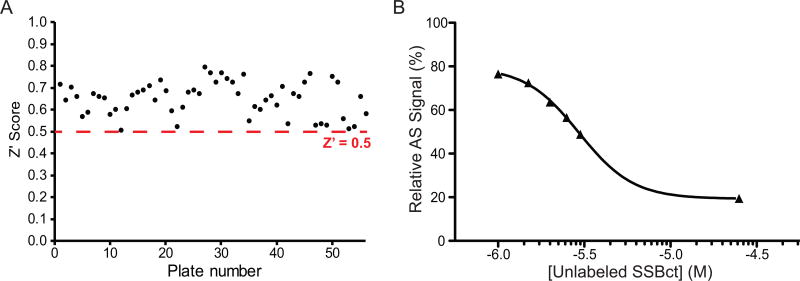
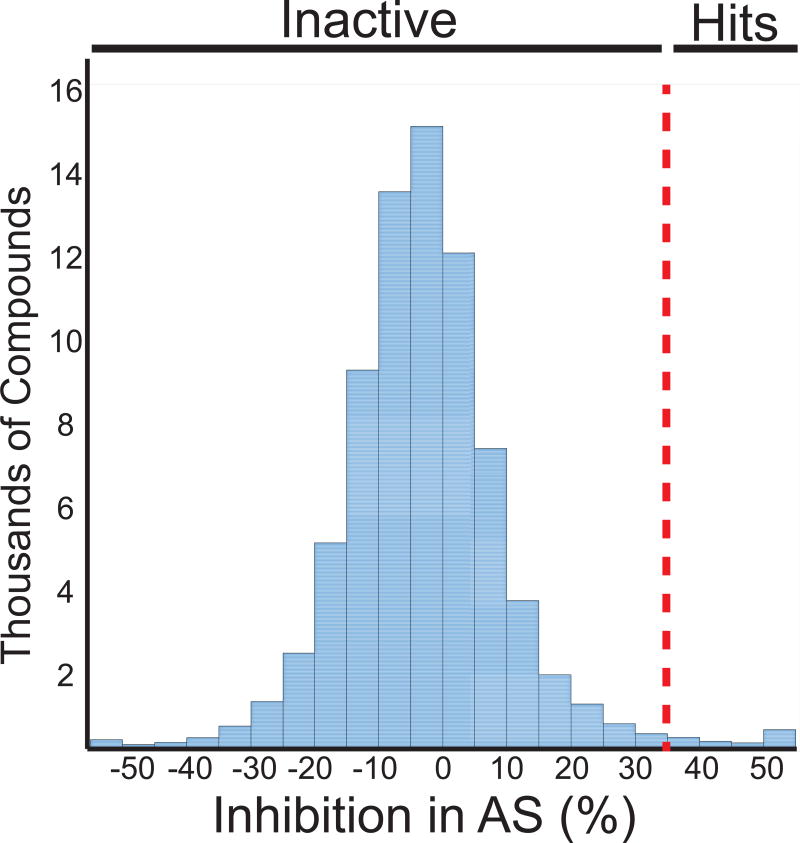
Having adapted an assay for use in high-throughput format, we screened a library of 72,474 unique compounds with our primary AS assay to identify inhibitors of the SSBct-PriA interaction (Figure 2). A mean Z’ score of 0.65 ± 0.08 was observed across 56 1536-well plates (Figure 3A). To allow for comparisons of potency across multiple plates, a concentration-response curve using unlabeled SSBct competitor peptide was included on each plate. Excellent consistency in the potency of the concentration-response curve was observed across plates and the IC50 determined from the concentration-response curve (3.0 ± 0.2 µM) was found to be in agreement with previously published values (2.4 ± 1.3 µM) of the E. coli SSBct-PriA interactions observed by surface plasmon resonance under similar salt concentrations (Figure 3B).34 In total, the mean inhibition from members of the compound library was -2.8% and normally distributed with a standard deviation of 13.5% (Figure 4). With a threshold of > 35% inhibition, corresponding to approximately 3 standard deviations from the mean, 946 small molecules were found to be active in the AS for a primary hit rate of 1.3%.
Figure 2. Screening and hit reduction workflow.
The selection criteria and number of compounds excluded are noted for each step.
Figure 3. The SSBct-PriA AS assay performs well under high-throughput conditions.
(A). Scatterplot of the Z’ scores observed for the each of the 56 plates used in the primary screen. The dashed red line indicates the minimum acceptable Z’ score of 0.5. Three plates were found to have Z’ scores below 0.5 and were repeated prior to data analysis. (B) Each plate contained a titration of unlabeled SSBct peptide in triplicate, the mean AS signal relative to the negative control wells on each plate is shown. High reproducibility of this concentration-response curve was observed between the plates. Error bars representing the SEM of all of the replicates at a given concentration of SSB are obscured behind data points.
Figure 4. Histogram showing the activity of the compounds tested in the SSBct-PriA primary AS assay.
The dashed line represents 35% inhibition in the assay. Compounds with more than 35% activity were advanced for further characterization.
Elimination of false positive hits
Others have noted that a subset of small molecules present in screening libraries have activity against a broad range of diverse assay targets, particularly in AS assays. We used an in silico filter to identify and remove 276 PAINS and other compounds with suspect chemical structures from our initial list of 946 hits.31, 32
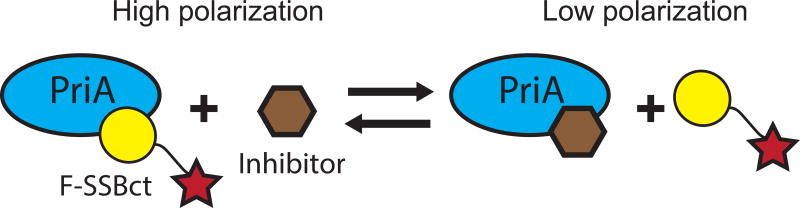
In order to further cull false positives not detected by the in silico PAINS filter from our initial AS HTS, we adapted an FP assay monitoring the SSBct-PriA interaction for use in 384-well plate format as an orthogonal secondary screen.20 In this assay, PriA is mixed with an SSBct peptide labelled with an N-terminal fluorescein (F-SSBct). The proportion of F-SSBct bound to PriA is determined by exciting the F-SSBct with polarized light and measuring the polarization of emitted fluorescence. Free F-SSBct tumbles rapidly before emission, resulting in a loss of polarization whereas the higher molecular weight PriA-F-SSBct complex tumbles more slowly and a greater proportion of the emitted light remains polarized (Figure 5).
Figure 5. Scheme depicting the SSBct-PriA fluorescence polarization assay.
Displacement of the F-SSBct from PriA by an inhibitor is detected as a decrease in polarization of light emitted from F-SSBct.
To confirm the hits selected by our primary AS assay were specific for the SSBct-PriA interaction, each of the remaining 670 compounds were tested in duplicate using the orthogonal SSBct-PriA FP assay. We expected compounds interfering with the AS chemistry rather than blocking SSBct would be inactive in the FP assay. Greater than 35% inhibition in the FP assay was observed for 35 hits (5.2%). To exclude any remaining non-specific inhibitors, each compound was tested for activity in an FP assay targeting an unrelated eukaryotic protein-protein interaction.33 Twenty-two of the 35 small molecules had <25% inhibitory activity at a concentration of 33.3 µM in this counter screen and were deemed specific for the SSBct-PriA interaction. Concentration-response curves were obtained using the AS assay and 9 potent and selective lead compounds were identified with IC50 values less than 40 µM.
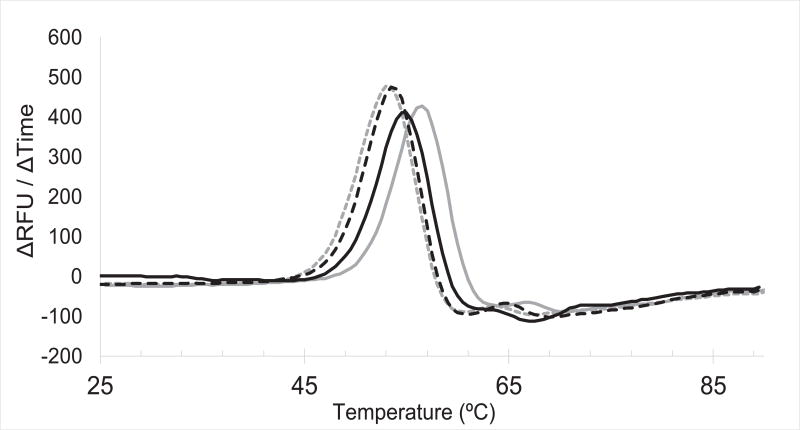
To assess if these compounds bind PriA, DSF was used to measure the Tm of PriA in the presence or absence of each of the 9 lead compounds. Small molecules are frequently observed to stabilize interacting proteins, thus an increase in the Tm of PriA indicates compound binding.35 PriA was thermally denatured in the presence of a dye which fluoresces only when bound to hydrophobic patches of PriA that are exposed as the protein unfolds. Concentration-dependent stabilization of PriA was observed for 2 compounds (Figure 6).
Figure 6. Small molecule stabilization of PriA during thermal denaturation indicates direct binding.
PriA was heated in the absence of ligand (gray dashed line), or in the presence of SSBct (solid gray line), 20 µM inhibitor (dashed black line) or 100 µM inhibitor (solid black line). The Tm for each condition is the temperature corresponding to the maximum change in fluorescence.
Discussion
Here we describe a methodology for a low-volume AS HTS performed in 1536-well plates. While the manufacturer recommended 25 µL AS reaction volume provides for 10,000 data points per 5 mg of beads (at 20 µg/mL beads), our further miniaturization of the assay to a 3 µL final volume and reduction of bead concentration to 5 µg/mL allows for a 41-fold decrease in bead use. This allowed us to assay ~300,000 data points with an equivalent amount of reagents. Similarly, protein usage was reduced to less than 5 mg for the entire screen as compared to ~1 g estimated to be required for a comparable FP screen. Liquid handling via the Echo and Mantis instruments is both rapid (6 min per 1536-well plate ECHO and 13 min per plate Mantis) and independent of consumables. We found that the vacuum chips on the Mantis needed to be occasionally replaced, but this is only trivially added to the total cost. Of special note is the exceptionally low dead volume of AS master mix that is achieved with our liquid handling approach. On average the dead volume was only 30 µL per 1536-well plate.
During assay development we noted plate effects in the AS assay plates in the form of a signal loss gradient vertically down the plate and a significantly higher AS signal in the top row of each plate. Small temperature changes have been noted to alter AS signal and likely explain the signal gradient.36 However, we have consistently observed the top row phenomena across different incubation times, assay plates, assay targets and plate readers. While we have been unable to eliminate this effect, its impact can be mitigated by the normalization process described herein (Figure S1).
We observed a relatively low hit confirmation rate between the primary AS and secondary FP assay. The low confirmation rate likely resulted from using a relatively permissive hit threshold to determine which compounds were selected from the primary AS assay. This threshold was chosen to avoid prematurely eliminating inhibitors with more modest potency which could serve as lead compounds for the future development of more potent derivatives. One major limitation of such an approach is that the majority of these putative weak inhibitors are inactive and will need to be eliminated in downstream screening steps. However, given the historical difficulties in developing PPI inhibitors, we reasoned that the chance of identifying a bona fide, albeit weak, inhibitor would outweigh the additional screening burden.
Our HTS platform identified 9 selective inhibitors of the SSBct-PriA interaction with IC50 values < 40 µM. Of these 9 inhibitors, 2 were observed to interact with PriA by thermal stabilization in a DSF assay. While no stabilization was detected for 7 compounds, this does not exclude PriA binding or inhibition. Binding by non-stabilizing ligands or ligands that stabilize both folded and unfolded PriA to a similar extent will not be detected in this assay.37
Further biophysical and structural studies to characterize the interaction between these inhibitors and PriA are underway. These inhibitors are also being tested for activity against multiple SSB interacting partners to determine if they are active against other targets important for bacterial replication and genome maintenance. Compounds discriminating between specific SSB PPIs could serve as valuable chemical probes to dissect the role of individual interactions while those inhibiting SSB PPIs indiscriminately may represent a novel class of antibiotics capable of targeting pathogenic bacteria resistant to currently available antibiotics.
Supplementary Material
Acknowledgments
The authors thank Song Guo for assisting with the screening and members of the Keck lab for critical review of the manuscript.
Funding
The authors disclosed receipt of the following financial support for the research, authorship and/or publication of this article: The project was supported by NIH R33 AI111676 (J.L.K.). A.F.V. was supported by the University of Wisconsin-Madison Integrated Training for Physician-Scientists NIH Training Grant GM008692, the Clinical and translational Science award program, through the NIH National Center for Advancing Translational Sciences (NCATS), grant UL1TR000427, and NIH F30 CA210465. The UW Small Molecule Screening Facility Is supported by the University of Wisconsin Carbone Cancer Center Support Grant P30 CA014520.
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH). M.P.K designed, optimized and validated the AS and FP assay, A.F.V. and G.E.A. miniaturized and performed the AS HTS. A.F.V., M.P.K., S.A.W., G.E.A., F.M.H., and J.L.K. carried out data analysis and wrote the manuscript.
Declaration of Conflicting Interests
The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Antibiotic Resistance Threats in the United States. CDC; 2013. [PubMed] [Google Scholar]
- 2.Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo--lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y-Y, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 4.Nordmann P, Poirel L, Toleman MA, et al. Does broad-spectrum-lactam resistance due to NDM-1 herald the end of the antibiotic era for treatment of infections caused by Gram-negative bacteria? J. Antimicrob. Chemother. 2011;66:689–692. doi: 10.1093/jac/dkq520. [DOI] [PubMed] [Google Scholar]
- 5.Carlet J, Jarlier V, Harbarth S, et al. Ready for a world without antibiotics? The pensières antibiotic resistance call to action. Antimicrob. Resist. Infect. Control. 2012;1:1. doi: 10.1186/2047-2994-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laxminarayan R, Duse A, Wattal C, et al. Antibiotic resistance—the need for global solutions. Lancet Infect. Dis. 2013;13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 7.Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3390–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shereda RD, Kozlov AG, Lohman TM, et al. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson AJ, Causer RJ, Dixon NE. Architecture and conservation of the bacterial DNA replication machinery, an underexploited drug target. Curr. Drug Targets. 2012;13:352–372. doi: 10.2174/138945012799424598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated Pneumonia. Am. J. Respir. Crit. Care Med. 2005;171:388–416. doi: 10.1164/rccm.200405-644ST. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Mulholland SG. Appropriate antibiotic treatment of genitourinary infections in hospitalized patients. Am. J. Med. 2005;118:14–20. doi: 10.1016/j.amjmed.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharyya B, George NP, Thurmes TM, et al. Structural mechanisms of PriA-mediated DNA replication restart. Proc. Natl. Acad. Sci. 2014;111:1373–1378. doi: 10.1073/pnas.1318001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petzold C, Marceau AH, Miller KH, et al. Interaction with single-stranded DNA-binding protein stimulates Escherichia coli ribonuclease HI enzymatic activity. J. Biol. Chem. 2015;290:14626–14636. doi: 10.1074/jbc.M115.655134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu D, Keck JL. Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I. Proc. Natl. Acad. Sci. 2008;105:9169–9174. doi: 10.1073/pnas.0800741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marceau AH, Bahng S, Massoni SC, et al. Structure of the SSB-DNA polymerase III interface and its role in DNA replication. EMBO. J. 2011;30:4236–4247. doi: 10.1038/emboj.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryzhikov M, Koroleva O, Postnov D, et al. Mechanism of RecO recruitment to DNA by single-stranded DNA binding protein. Nucleic Acids Res. 2011;39:6305–6314. doi: 10.1093/nar/gkr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng K, Xu H, Chen X, et al. Structural basis for DNA 5'-end resection by RecJ. eLife. 2016;5:e14294. doi: 10.7554/eLife.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curth U, Genschel J, Urbanke C, et al. In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res. 1996;24:2706–2711. doi: 10.1093/nar/24.14.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu D, Windsor MA, Gellman SH, et al. Peptide inhibitors identify roles for SSB C-terminal residues in SSB/exonuclease I complex formation. Biochemistry (Mosc.) 2009;48:6764–6771. doi: 10.1021/bi900361r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelman Z, Yuzhakov A, Andjelkvoic J, et al. Devoted to the lagging Strand - the χ subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO. J. 1998;17:2436–2449. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbio. Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenberg J, Berends LJ, Donch J, et al. exrB: a malB-linked gene in Escherichia coli B involved in sensitivity to radiation and filament formation. Gen. Res. 1974;23:175–184. doi: 10.1017/s0016672300014798. [DOI] [PubMed] [Google Scholar]
- 24.Genschel J, Curth U, Urbanke C. Interaction of E. coli single-stranded DNA binding protein (SSB) with exonuclease I. The carboxy-terminus of SSB is the recognition site for the nuclease. Biol. Chem. 2000;381:183–192. doi: 10.1515/BC.2000.025. [DOI] [PubMed] [Google Scholar]
- 25.Wang TV, Smith KC. Effects of the ssb-1 and ssb-113 mutations on survival and DNA repair in UV-irradiated ΔuvrB strains of Escherichia coli K-12. J. Bact. 1982;151:186–192. doi: 10.1128/jb.151.1.186-192.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu D, Bernstein DA, Satyshur KA, et al. Small-molecule tools for dissecting the roles of SSB/protein interactions in genome maintenance. Proc. Natl. Acad. Sci. 2010;107:633–638. doi: 10.1073/pnas.0909191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marceau AH, Bernstein DA, Walsh BW, et al. Protein interactions in genome maintenance as novel antibacterial targets. PLoS ONE. 2013;8:e58765. doi: 10.1371/journal.pone.0058765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nordmann P, Cuzon G, Thierry N. The real threat of Klebsiella pneumoniae carbapenemase producing bacteria. Lancet Infect. Dis. 2009;9:228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 29.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 31.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 32.Lagorce D, Sperandio O, Baell JB, et al. FAF-Drugs3: A web server for compound property calculation and chemical library design. Nucleic Acids Res. 2015;43(W1):W200–W207. doi: 10.1093/nar/gkv353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voter AF, Manthei KA, Keck JL. A high-throughput screening strategy to identify protein-protein interaction inhibitors that block the Fanconi snemia DNA repair pathway. J. Biomol. Screen. 2016;21:626–633. doi: 10.1177/1087057116635503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cadman CJ, McGlynn P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 2004;32:6378–6387. doi: 10.1093/nar/gkh980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature Protocols. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 36.A Practical Guide to working with AlphaScreen. PerkinElmer; 2004. [accessed Feb 21, 2017]. https://www.urmc.rochester.edu/MediaLibraries/URMCMedia/hts/documents/AlphaScreenPracticalGuide.pdf. [Google Scholar]
- 37.Simeonov A. Recent Developments in the Use of Differential Scanning Fluorometry in Protein and Small Molecule Discovery and Characterization. Expert Opin. Drug Discov. 2013;8:1071–1082. doi: 10.1517/17460441.2013.806479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.