Introduction
Diffuse Intrinsic Pontine Glioma (DIPG) is a very aggressive pediatric brainstem tumor characterized by rapid and uniform patient demise1. A heterozygous point mutation of histone H3 occurs in more than 80% of these tumors, and results in a lysine-to-methionine substitution (H3K27M)2,3. Expression of the histone mutant is accompanied by a reduction in the levels of Polycomb Repressive Complex 2 (PRC2) mediated H3K27 trimethylation (H3K27me3) and this is purported to be a driving event of DIPG oncogenesis4,5. Despite a major loss of this histone mark, a not insignificant amount of PRC2 activity can still be detected in DIPG cells positive for H3K27M histone mutant6,7. Along with the PRC2 role, the function of the H3K27M histone mutant in DIPG cells is not characterized. To investigate these issues, we profile the epigenome of H3K27M mutant DIPG cells and demonstrate that H3K27M co-localizes with H3K27 acetylation (H3K27ac). Heterotypic H3K27M-K27ac nucleosomes co-localize with bromodomain proteins at actively transcribed genes, whereas PRC2 is excluded from the chromatin of the H3K27M occupied regions. Residual PRC2 activity is required for maintaining DIPG proliferative potential by repressing neuronal differentiation and function. Moreover, we demonstrate that pharmacologic bromodomain protein inhibition suppresses tumor growth in vivo.
To advance understanding of the molecular function of H3K27M in oncogenesis, we mapped genome-wide occupancy of the mutant histone in DIPG cell lines by ChIP-seq, using an antibody directed against H3K27M (Supplementary Fig. 1). To confirm the specificity of the H3K27M antibody, we expressed wildtype or H3K27M mutant histone at equivalent levels in HCT-116 cells (Supplementary Fig. 1A) and performed ChIP-seq. ChIP-seq signal with the H3K27M antibody was detected exclusively in H3K27M expressing cells, whereas no enrichment was observed in cells expressing the wildtype counterpart (Supplementary Fig. 1B–D). ChIP-seq results from SF8628 human DIPG cells, containing a mutant H3F3A gene encoding H3.3K27M8, reveal a genome wide distribution of H3.3K27M that is highly correlated with active transcription, as indicated by the co-localization of acetylated H3K27 (H3K27ac) and RNA polymerase II (RNA pol II) (Fig. 1A–C and Supplementary Fig. 2A–B). Intriguingly, this observation is recapitulated in SU-DIPG-IV (hereafter called DIPG IV), a DIPG cell line carrying a point mutation in the HIST1H3B gene that results in the expression of H3.1K27M9, and is further supported by results obtained in SF7761, another H3F3A mutant DIPG line8 (Supplementary Figs. 3–4).
Figure 1. H3K27M correlates with H3K27ac and is excluded from PRC2 targets.
(A) Representative UCSC genome browser views of genomic regions enriched after chromatin immunoprecipitation (ChIP) for histones, histone post-translational modifications and proteins listed on the left of each track in SF8628. Input served as background control. (B) Heat map analysis of the most highly H3.3K27M enriched genomic loci (first 5000 in descending order top to bottom) in SF8628. The center of each vertical lane represents the summit of the peak while the left and right borders represent −5 kb and +5 kb, respectively. All the other antigens occupancy at those regions are plotted accordingly. Input served as background control. Scale bar represents relative intensities. (C) Metaplot showing average signal accumulation (RPM, reads per million) of the top 5000 regions bound by H3.3K27M in SF8628. All the other antigens average occupancy at those regions are plotted accordingly. Each plot is centered on the summit of the average occupancy and extended 5 kb upstream and downstream (−5kb and +5kb, respectively). Input served as background control. (D) Western blot analysis of immunoprecipitated (IP) histone H3K27ac from mononucleosomes containing C-terminal FLAG-tagged wild type H3.3 or mutant H3.3K27M purified from 293T cells (image representative of at least 2 independent experiments. Uncropped images in Supplementary Figure 18). (E) Same as (A). (F) As described in (B) showing 5000 most enriched regions bound by H3K27me3.
As shown by our group and others, H3K27M expression is associated with increased levels of H3K27ac4,5 (Fig. 1D). To directly demonstrate a functional link between the expression of H3K27M and H3K27ac accumulation, we used a doxycycline-inducible system and found that H3K27M induction of H3K27ac is a reversible process (Supplementary Fig. 5A). H3K27M localizes to H3K27ac sites that are present in wildtype H3.3 expressing cells, but importantly it can induce a marked increase of H3K27ac at regions showing only modest H3K27ac enrichment in control cells (Supplementary Fig. 5B–C). These changes in H3K27ac are reversible and return to distributions almost indistinguishable to those present in control H3.3 expressing cells after terminating H3K27M transgene expression (Supplementary Fig. 5B–C). Importantly, H3K27ac is the only significantly increased histone acetylation mark induced by H3K27M as determined by mass spectrometry analysis (Supplementary Fig. 6–7 and Supplementary Table 1). Thus H3K27M expression directly increases the levels of H3K27ac at specific genomic regions.
The H3K27M histone mutant is suggested to have increased binding affinity for EZH24,6,10, however, our previous biochemical purifications of mono-nucleosomes containing H3K27M did not show any preferential enrichment for PRC2 subunits5. To determine if PRC2 subunits co-localize with H3K27M in vivo on chromatin, we analyzed the genome-wide distribution of PRC2, H3K27me3 and H3K27M histone in DIPG cells. Strikingly, PRC2 subunits EZH2 and SUZ12, as well as their enzymatic product, H3K27me3, are largely excluded from chromatin on sites containing H3K27M (Fig. 1E–F; Supplementary Fig. 2 and Supplementary Fig. 8). Collectively, these results indicate that H3K27M is not involved in PRC2 recruitment or sequestration on chromatin in vivo, but on the contrary, suggest that PRC2 binding to chromatin is excluded by locus specific formation of H3K27M/K27ac heterotypic nucleosomes.
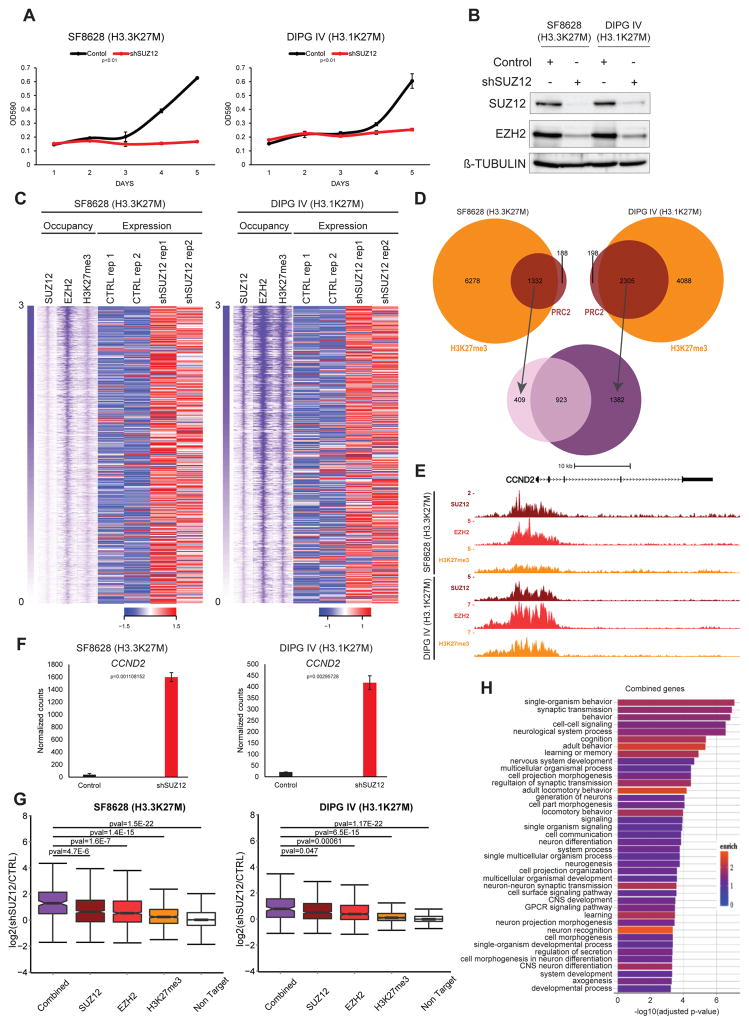
Histone H3K27M has been hypothesized to drive gliomagenesis through PRC2 catalytic inhibition4, suggesting that lack of PRC2 function is an important contributing factor to the development of H3K27M tumors. However, our results indicate both PRC2 and the product of its catalytic activity, H3K27me3, are present at several loci in DIPG cells6,7 (Fig. 1E–F and Supplementary Fig. 8 a–f). The functional consequence of this residual PRC2 activity in DIPG has yet to be examined. To address whether PRC2 function is required by H3K27M tumor cells, we used lentiviral shRNAs to suppress the expression of essential PRC2 subunits SUZ12 and EED11. The growth and colony forming ability of SUZ12 or EED knockdown cells are greatly reduced relative to control cells (Fig. 2A–B, Supplementary Fig. 9). Unexpectedly, these results demonstrate a role for PRC2 function in sustaining DIPG growth.
Figure 2. PRC2 is required for the oncogenic potential of H3K27M DIPG cells.
(A) (left) 5-days growth curve of SF8628 and (right) DIPG IV stably expressing a SUZ12 specific shRNA compared to control. The plots represent crystal violet absorbance quantification (λ=590 nm) measured at each day (one representative of 2 independent experiments. Each point is an average of cell culture replicates, n=3). Error bars indicate SD. P-values from a two-tailed unpaired t-test. (B) Western blot analysis of SUZ12 and EZH2 protein levels in SF8628 stably expressing a SUZ12 specific shRNA compared to control. β-TUBULIN served as loading control. Image representative of at least 2 independent experiments. Uncropped images in Supplementary Figure 18. (C) Top 1000 transcription start sites (TSS) occupied by PRC2 and H3K27me3 and relative heat map representing the transcript levels (normalized RNA-seq intensities) of TSS associated genes in SF8628 and DIPG IV upon SUZ12 knockdown compared to control (CTRL). Occupancy is ranked and centered accordingly to SUZ12 1000 most bound TSS (extended 5kb downstream and upstream). RNA-seq data shown are from 2 independent experiments. Row z-score scale legend is shown. Scale bar represents relative intensities. (D) Venn diagram representing TSS bound by PRC2 (SUZ12 and EZH2) and H3K27me3 in SF8628 and DIPG IV. (E) UCSC genome browser snapshot of the CCND2 locus showing PRC2 and H3K27me3 enrichment in both SF8628 and DIPG IV. (F) Transcriptional levels of CCND2 in both SF8628 and DIPG IV upon SUZ12 knockdown compared to control. Normalized counts from RNA-seq (mean values from independent experiments, n=2). Error bars indicate SD. (G) Boxplot analyses representing the transcriptional levels of all the direct EZH2, SUZ12 and H3K27me3 targets in both SF8628 and DIPG IV upon SUZ12 knockdown compared to control. The Combined boxplot refers to the 923 common PRC2 and H3K27me3 target genes between the two DIPG cells. The Non Target boxplot represents all the genes non-PRC2 and non-H3K27me3 targets. P-values from a two-tailed unpaired t-test is shown. (H) Gene ontology (GO) analyses of common genes in both SF8628 and DIPG IV. Adjust p-value and relative enrichment (color coded) for each class is shown.
To identify potential targets of PRC2 driving DIPG cell proliferation, we focused our attention on CDKN2A/p16, a negative cell-cycle regulator and well-established target of PRC2 transcriptional repression12. SUZ12 or EED knockdown resulted in de-repression of p16 expression in SF8628 cells, consistent with CDKN2A being a PRC2 target in these cells. In contrast, DIPG IV cells do not upregulate p16 expression upon PRC2 depletion (Supplementary Fig. 10A–B and Supplementary Fig. 9D), despite the anti-proliferative effect of SUZ12 and EED knockdown in these cells (Fig. 2A–B, Supplementary Fig. 9A–D and Supplementary Fig. 10A–B).
To further explore the role of PRC2 in DIPG proliferation we employed a small molecule catalytic inhibitor of EZH2 methyltransferase activity, EPZ-6438. Consistent with the SUZ12 and EED knockdown results, treatment of H3K27M cells with EPZ-6438 diminished DIPG proliferation (Supplementary Fig. 10C). DIPG IV cells tolerate higher doses of EPZ-6438 than SF8628 (Supplementary Fig. 10C) and this likely reflects lower effectiveness of the chemical inhibitor in DIPG IV, because a common PRC2 target gene, Cyclin D2 (CCND2) (Fig 2C–F), simply requires increased doses to achieve de-repression similar to SF8628 (Supplementary Fig. 10D) Notably, target de-repression correlates with decreased proliferation in these cells (Supplementary Fig. 10C–D). As for EED and SUZ12 knockdown, EZH2 catalytic inhibition does not elicit p16 upregulation in DIPG IV (Supplementary Fig. 10C). In combination, our results indicate that PRC2 pro-proliferative activity in H3K27M tumor cells can act through p16-independent mechanisms as already demonstrated in other models 13.
To further investigate global PRC2 transcriptional regulation, we performed transcriptome analyses (RNA-sequencing) of SUZ12 shRNA and control vector transduced SF8628 and DIPG IV cells. Our analysis revealed that the several genes highly occupied by PRC2 in SF8628 and DIPG IV show a trend of de-repression upon SUZ12 depletion (Fig. 2C). As expected from our previous results, genomic regions bearing PRC2 at their TSS are characterized by the presence of H3K27me3 (87% in SF8628 and 92% in DIPG IV). Many PRC2 targets are shared between SF8628 and DIPG IV (70% and 40% respectively: Fig. 2D–E), and are significantly de-repressed upon SUZ12 knockdown as compared to non-PRC2 target genes (p<1.5E-22 in SF8628 and p<1.17E-22 in DIPG IV: Fig. 2F–G). Thus, the activation of these genes suggests they could functionally inhibit DIPG proliferation. Importantly, gene ontology (GO) analysis of common genes supports their importance in mediating neuronal lineage differentiation and establishing mature neuronal functions (Fig. 2H).
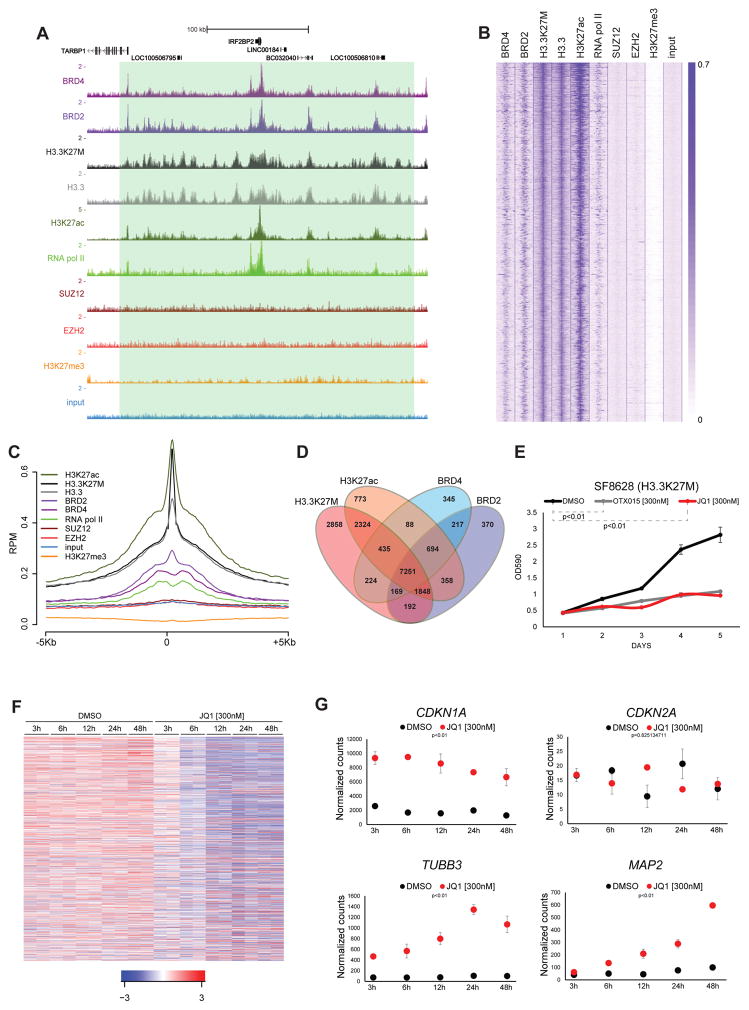
Although still functionally active, PRC2 catalytic property toward H3K27M containing nucleosomes is strongly inhibited4. Interestingly upon loss of H3K27me3 these nucleosomes acquire H3K27ac resulting in the formation of H3K27M-K27ac heterotypic nucleosomes4 (Figure 1D and Supplementary Fig. 5B–C). These results support the increased association of acetyl-binding bromodomain proteins to H3K27M containing nucleosomes5, and prompted us to investigate the role of these proteins in DIPG cells. Strikingly bromodomain-containing protein 2 (BRD2) and 4 (BRD4) show highly significant overlap with H3K27M occupied sites (Fig. 3A–D, Supplementary Fig. 2 and Supplementary Fig. 3A–D). BRD2/BRD4 and H3K27M co-occupied loci represent 85% of all the BRD2 or BRD4 positive regions in SF8628 (Fig. 3D). Histone H3K27M localizes to promoters, intragenic regions and at H3K4me1 positive intergenic regions (Supplementary Fig. 11A–B). We also found that H3K27M occupancy at active enhancers (H3K4me1 and H3K27ac positive regions) is asymmetric and characterized by the presence of high density occupancy over a fraction of active enhancers that also show increased presence of BRD2/4 and active transcription marks (Supplementary Fig. 11C–D), suggesting that H3K27M participates in the formation of super enhancer-like elements purported to play a key role in cancer14.
Figure 3. Bromodomain proteins inhibition impairs proliferation and triggers differentiation of H3K27M positive DIPG cells.
(A) Same as in Fig. 1A. (B–C) Same as in Fig. 1B–C. (D) Venn diagrams with numbers of individual and overlapping target genes in the indicated ChIP-seq samples in SF8628. (E) 5-days growth curve of SF8628 treated with JQ1 [300nM] and OTX015 [300nM] compared to DMSO. The plot represents a crystal violet absorbance quantification (λ=590 nm) measured at each day (one representative of at least 3 independent experiments is presented, each point is an average of cell culture replicates, n=3). Error bars indicate SD. (F) Heat map of relative transcriptional levels (RNA-seq) changes during JQ1 [300nM] compared to DMSO time course in SF8628 (independent experiments, n=2). Genes represented (n=415, 369 downregulated, 46 upregulated. Adjust pval<0.01). Row z-score scale legend is shown. (G) Transcriptional levels of CDKN1A, CDKN2A, TUBB3 and MAP2 in SF8628 during JQ1 [300nM] and DMSO time course treatment. Normalized counts from RNA-seq (each point is the mean of values from independent experiments, n=2). P-values from a two-tailed unpaired t-test. Error bars indicate SD.
The strong co-occupancy between BRD2/4 proteins and H3K27M/K27ac heterotypic nucleosomes combined to support a potential role of BRD proteins in DIPG pathogenesis. To address this point, we treated SF8628 and DIPG IV cells with JQ1, a well-known BET (Bromodomain and Extra Terminal domain) inhibitor15. Dose-response analysis revealed sub-micromolar IC50 values in the H3K27M mutant cell lines compared to glioblastoma cells not harboring that mutation (Supplementary Fig. 12A), with the growth inhibitory effect of single JQ1 administrations extending to 5 days post-treatment (Fig. 3E and Supplementary Fig. 12B–C). This effect was fully recapitulated by the treatment with another BET inhibitor (Fig. 3E and Supplementary Fig. 12D). Interestingly, the anti-proliferative effect of JQ1 was accompanied by a neuron-like morphological change (Supplementary Fig. 12E).
In accordance with the crucial function of bromodomain proteins in transcriptional elongation16 we performed RNA-seq analysis during a time-course treatment with JQ1. Although at any given time point we observed both downregulated and upregulated genes, data interpolation on the overall time course shows a prevalent shutdown of many actively transcribed genes upon JQ1 treatment (Fig. 3F and Supplementary Fig. 13A). Importantly, the genes transcriptionally modulated by JQ1 show H3K27M, active transcription marks and BRD2/4 occupancy around their promoters indicating they are direct targets affected by JQ1 treatment (Supplementary Fig. 13B–C). Among the genes transcriptionally upregulated we noticed CDKN1A (p21), marker of cell cycle arrest17, TUBB3 (Tuj1) and MAP2, markers of differentiation in mature neurons18, further supporting the morphology changed observed upon JQ1 treatment (Fig. 3G and Supplementary Fig. 13D–E). CDKN2A is unaffected by JQ1 suggesting also in this case that the phenotype is p16 independent (Fig. 3G and Supplementary Fig. 13D). FACS analyses also confirmed that the majority of JQ1-treated cells demonstrated upregulation of the mature neuron marker TUBB3 (Supplementary Fig. 13F). Interestingly, JQ1 treatment elicited a time-dependent decrease in the activating H3K27ac mark19, consistent with the observed trend in transcriptional downregulation (Fig. 3F and Supplementary Fig. 13A–E).
In order to understand the direct molecular effectors of the phenotype induced by JQ1, we assessed MYC transcription, a well-known downstream target of JQ1 in many tumors20. Surprisingly, we found a modest but consistent increase of MYC transcripts in both the SF8628 and DIPG IV cell lines upon JQ1 treatment, whereas its closest homolog, MYCN, is silenced by PRC2 in those cells (Supplementary Fig. 14A–B). These results exclude that JQ1 induces growth arrest and upregulation of differentiation markers through the canonical reduction in MYC transcription. Focusing on genes downregulated by JQ1 at an early time point (6h), we performed gene ontology (GO) analysis. Interestingly both SF8628 and DIPG IV share the top downregulated molecular functions rather than the upregulated ones. Among them emerged the GO term “transcription factor activity”, suggesting that JQ1 may affect an important transcriptional network in DIPG (Supplementary Fig. 15A–B). Interestingly one of the main downregulated transcripts is GLI2, a crucial downstream effector of the Sonic Hedgehog (SHH) pathway21, that is a BRD2/4 and H3K27M/K27ac heterotypic nucleosomes target (Supplementary Fig. 16A–B). The Hedgehog pathway is implicated in DIPG pathogenesis as the candidate DIPG cell-of-origin is proposed to be a SHH-responsive cell9. Moreover, inhibition or activation of Hedgehog signaling negatively or positively affects DIPG neurosphere formation ability, respectively9. Interestingly, a fraction of genes previously reported to be preferentially expressed in H3.3K27M DIPG compared to H3 wild type gliomas6 is target of heterotypic nucleosomes and BRD2/4 as shown by their decreased expression induced by JQ1 treatment (Supplementary Fig. 16C–E).
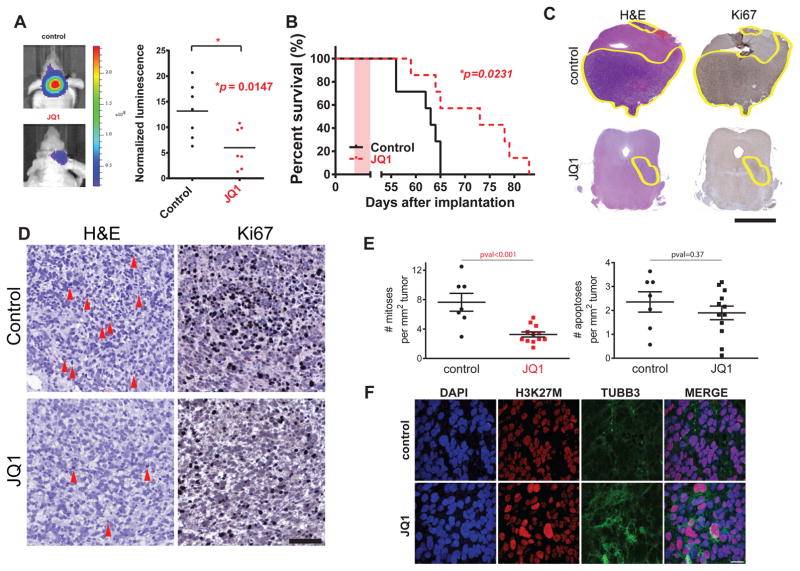
To investigate JQ1 activity against K27M DIPG in vivo, we employed a xenograft mouse model of DIPG based on injection of SF8628 cells into the brainstem of athymic mice8. Remarkably, mice treated with JQ1 for 10 days exhibited significantly reduced tumor size, as indicated by bioluminescence imaging, and prolonged animal survival (Fig. 4A–B). Haematoxylin and eosin (H&E) and KI67, a marker of cell proliferation22, staining of brainstem tumors from mice euthanized at the completion of 10 days treatment confirmed a dramatic reduction of tumor burden compared to controls (Fig. 4C). As JQ1 treatment causes marked reduction of global H3K27ac levels in DIPG cells (Supplementary Fig. 13E) we performed an immunohistochemistry analysis for H3K27ac on tumors coming from JQ1 treated and control mice as a surrogate marker for JQ1 target inhibition. This analysis revealed significantly lower H3K27ac levels in tumors coming from JQ1 treated mice compared to control, thus reflecting direct target inhibition in vivo (Supplementary Fig. 17A). To further support our in vivo data we used another BET inhibitor (I-BET151) to treat our xenograft DIPG model (Supplementary Fig. 17B) and also tested JQ1 against H3.1K27M DIPG IV xenografts (Supplementary Fig. 17C) with both approaches showing significant impairment of DIPG growth in vivo. Importantly, treatment with JQ1 strongly inhibits proliferation of DIPG cells in vivo as evidenced by reduced mitotic events in JQ1 treated tumors compared to control, without significantly altering apoptosis (Fig. 4D–E). Furthermore, we were also able to demonstrate induction of TUBB3 levels in JQ1 treated tumors compared to control (Fig. 4F) thus providing in vivo support for our in vitro observation that JQ1 tumor growth inhibition is accompanied by an upregulation of differentiation markers in DIPG cells. Finally JQ1 treated DIPG xenograft mice show significantly improved survival even compared to treatment with GSK-J4, a compound previously proposed as a candidate drug for DIPG treatment (Supplementary Fig. 17D). Altogether our data strongly support BET inhibitors as a promising therapeutic strategy in DIPG.
Figure 4. Bromodomain proteins inhibition significantly extends survival of DIPG xenograft model.
(A) Tumor volume was measured by bioluminescence imaging after 10 days treatment with JQ1 [50 mg/Kg] or vehicle (control). (left) tumor bioluminescence from representative JQ1 and control treated mice, (right) normalized bioluminescence values control and JQ1 treated mice. Bars indicate the mean of each group. A two-tailed unpaired t-test was used to compare control and JQ1 treated groups (n=7, each group). (B) DIPG xenografts mice were generated by SF8628 injection in the brainstem. Survival curves for JQ1 and control treated mice. Pink shaded areas indicate the duration of treatment. P-value was obtained using a long-rank test. (C) Digital scan of representative sections of xenograft DIPG showing haematoxylin and eosin (H&E) and Ki67 immunohistochemical (IHC) stainings in control and JQ1 treated mice. Scale bar = 2.5mm. (D) High magnification of representative photomicrograph of H&E and Ki67 IHC in tumor sections from control and JQ1 treated mice. Red arrows indicate mitotic events. Scale bar = 100μm. (E) Quantification of mitotic and apoptotic events in JQ1 and control treated mice, (n=7 for control group, n=12 JQ1 treated group). Each dot represents an average of 10 independent power fields for each tumor. P-value from a two-tailed unpaired t-test is shown. (F) Representative confocal images showing double immunostaining for H3K27M and TUBB3 in DIPG xenograft sections from control and JQ1 treated mice. Nuclei are identified by DAPI staining. Scale bar = 20μm.
Here we provide the first characterization of the genome-wide deposition of H3K27M mutant histone in human DIPG cells. Strikingly, H3K27M co-localizes with transcriptionally active chromatin regions, and is largely excluded from regions occupied by PRC2 and H3K27me3. Moreover, H3K27M containing nucleosomes are enriched for acetylated H3K274 (Fig. 1D) and, in agreement with our previous biochemical data5, heterotypic nucleosomes demonstrate high co-occupancy with bromodomain proteins (Fig. 3A–D). In our DIPG cells, PRC2 is almost entirely confined in chromatin regions marked with H3K27me3 and this mark is mutually exclusive with H3K27ac23. Given the high correlation between H3K27M and H3K27ac the possibility that H3K27M somehow co-occupies many chromatin regions with EZH2 or PRC2 is unlikely. Collectively, these results strongly indicate that H3K27M in DIPG cells does not appear to recruit or sequester EZH2 (or in general PRC2), as suggested by other studies4,6,7. Rather, our data support a model where H3K27M excludes PRC2 binding, thus inducing aberrant accumulation of H3K27ac at H3K27M binding sites consistent with PRC2 loss of function in other systems24,25. PRC2 activity is redirected during the neuronal differentiation of mouse embryonic stem cells, suggesting that external signals shape PRC2 recruitment, resulting in the repression of distinct gene sets at specific developmental stages26. Heterotypic H3K27M/K27ac nucleosomes may on one hand create an impediment for PRC2 repressive activity at a critical subset of genes involved in the maintenance of an undifferentiated state while on the other hand confining PRC2 to genes that should be activated during the differentiation process. This, in cooperation with other genetic alterations characterizing DIPG (e.g. TP53, PDGFRA, ACVR1, etc.), would increase the proliferation potential and decrease the differentiation ability, two events widely known to be hallmarks of cancer27,28. Therefore a logical conclusion from our data supports the possibility that while a locus specific reduction of PRC2 activity/binding by H3K27M could promote DIPG formation the overall PRC2 function is essential for the maintenance of their oncogenic status. Abrogating the anti-differentiation effect of H3K27M/K27ac heterotypic nucleosomes through use of BET inhibitors such as JQ1 could conceivably provide the dual benefit of decreasing DIPG cell proliferation while promoting terminal neuronal differentiation of the tumor cells. Recently, a comprehensive drug screening for promising agents for DIPG therapy identify HDAC inhibitors as potentially suitable for DIPG treatment29. Interestingly HDAC inhibitors can synergize with JQ1 in pancreatic cancer mouse model30 and in a murine lymphoma model where they were suggested to induce similar transcriptional outcome31, thus paving the ground for a combinatorial therapy in DIPG. In conclusion our work provides new important molecular insights about H3K27M function in DIPG oncogenesis and identified PRC2 and BET bromodomain proteins as potential therapeutic targets for this disease.
ONLINE METHODS
Cell sources and propagation
Primary pediatric human glioma cell lines, SF8628, SF7761 and SU-DIPG-IV, pcGBM2, SF9402 and SF9427 were obtained from UCSF medical center and Stanford University, and in accord with institutionally approved protocol at each institution. Establishment of SF8628 and SF7761 cell cultures, from surgical specimens, and tumor cell modification for expression of firefly luciferase, for in vivo bioluminescence imaging, have been described 8,32,33. The SU-DIPG-IV were described earlier 9 and were adapted to the following growth conditions: DMEM 1X (Thermo Fisher), 10% serum (Corning), 1X glutamine (Life Technologies), 1X penicillin/streptomycin (Life Technologies), cells were grown in a CO2 incubator (5% CO2) at 37°C. 293T and HCT116 cells were grown using the same conditions. Cell images were taken using a Leica inverted microscope (DM IL) 40 X objective and a QICAM fast 1394 camera (QImaging).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed following previously described protocols34–36. ~5e7 cells for each immunoprecipitation were crosslinked using freshly prepared 1% formaldehyde in complete cell medium (described in Cell sources and propagation) for 10 min at RT and subsequently quenched with [0.125M] glycine for at least 5 min and then rinsed twice in PBS. Cells were gently scraped from the plates and centrifuged in a 15mL tube (Falcon) at 1350 g for 5 min at 4°C. Cells were resuspended in 10 mL Buffer 1 ([50 mM] HEPES at pH=7.5, [140 mM] NaCl, [1 mM] EDTA, 10% Glycerol, 0.5% Igepal, 0.25% Triton X-100) for 10 min at 4°C. Then centrifuged at 1350 g for 5 min at 4°C. Pellet was resuspended in 10 mL Buffer 2 ([10 mM] Tris-HCl at pH=8.0, [200 mM] NaCl, [1 mM] EDTA, [0.5 mM] EGTA) at RT for 10 min. Then centrifuged at 1350 g for 5 min at 4°C. Pellet was resuspended in 1 mL Buffer 3 ([10 mM Tris-HCl] at pH=8.0, [1 mM] EDTA, [0.1%] SDS) and transferred to a milliTUBE (1mL, Covaris) and sonicated using E220 focused-ultrasonicator (Covaris) using following parameters: 20% duty cycle, 175 PIP, 200 cycles/burst, 1500 seconds. 50 μL of chromatin were de-crosslinked by adding 200 μL of Elution Buffer ([50 mM] Tris-HCl at pH=8.0, [10 mM] EDTA, 1.0% SDS) for 1.5–2 hrs at 65°C and the DNA was extracted using a PRC purification kit (QIAGEN). DNA fragments were loaded in a 1% agarose gel and checked for their size (average range 200–600 bp). The remaining sheared chromatin (~950μl) was centrifuge at 20,000 g for 15 min at 4°C to remove cellular debris. 100μL of 10 X ChIP dilution buffer (10% Triton x-100, [1 M] NaCl, 1% Na-Deoxycholate, 5% N-Lauroylsarcosine) was added to sheared chromatin. 10 μL was saved as input, the remaining was subsequently incubated with a specific antibody overnight at 4°C on a nutator (for antibody list refer to table 2). The following day, 50μL of protein A/G agarose beads (Santa Cruz biotech) were added and incubated for 3hrs at 4°C on a nutator. Beads were then pelleted by centrifugation at 2500g for 1 min and washed with 1mL of RIPA buffer ([50 mM] HEPES at pH=7.5, [500 mM] LiCl, [1 mM] EDTA, 1.0% Igepal, 0.7% Na-Deoxycholate). The washing procedure was repeated 4 times. An extra wash was finally performed with 1mL of [50mM] NaCl in TE 1X. Inputs and beads were then resuspended in 200 μL of Elution Buffer for 30 min at 65°C then centrifuged for 2 min at 15000 g. The supernatant was transferred in new tubes and de-crosslinked overnight at 65°C. The following day DNA was extracted using a PCR purification kit (QIAGEN) in 70 μl final volume.
Chip-seq library preparation
ChIP-sequencing libraries were prepared using the KAPA HTP Library Preparation Kit complemented with NEXTflex DNA Barcodes from Bioo Scientific. 10 ng of DNA was used as starting material for input and ip samples. Libraries were amplified using 13 cycles on the thermocycler. Post amplification libraries were size selected at 250–450bp in length using Agencourt AMPure XP beads from Beckman Coulter. Libraries were validated using the Agilent High Sensitivity DNA Kit.
RNA extraction and RNA-seq library preparation
Cell pellets were generated after 6–7 days from antibiotic selection after shSUZ12, and 2 days shEED and relatively control vectors infection. For JQ1 treated cells the pellets were generated at specific time points indicated in the figures. DMSO treated cell pellets were collected at 48 hrs. Pellets were usually stored at −80°C or processed immediately for RNA extraction. RNA was extracted using RNeasy Plus micro kit (QIAGEN). RNA-sequencing libraries were prepared using the Illumina TruSeq Stranded Total RNA Preparation Kit with Ribo-Depletion. Input RNA quality was validated using the Agilent RNA 6000 Nano Kit. 1 μg of total RNA was used as starting material. Libraries were validated using the Agilent DNA 1000 Kit.
Next Generation Sequencing
ChIP-seq and RNA-seq libraries were single read (SR) sequenced on the Illumina NextSeq 500 Sequencing System.
ChIP-seq analysis
ChIP-seq reads were trimmed from the 3′ end until the final base had a quality score > 30, using Trimmomatic (10.1093/bioinformatics/btu170), and were aligned to the human genome (UCSC hg19, using Bowtie version 1.1.2) 37. Only uniquely mapping reads with at most two mismatches over the entire length of the read were considered for further analysis. To generate the UCSC genome browser tracks, locations of the mapped ChIP-seq reads were extended to 150 bases to represent sequenced fragments, were normalized to total reads aligned (reads per million, rpm), and were formatted as bigWig coverage plots. Peaks were called from the aligned reads using SICER version 1.1 (10.1093/bioinformatics/btp340). SICER was run with a window size of 200, a fragment size of 150, an effective genome fraction of 0.8, a gap size of 600 and an FDR threshold of 1e-8. In each case, peaks were determined relative to an input control. Genes and transcription start sites nearest to peaks were identified from Ensembl release 72. For large-scale analyses, rpm numbers were summed for each 25 bp window tiling the 5 kb immediately upstream and downstream of the highest rpm signal in a peak (or upstream and downstream of transcription start sites, as in Figure 2C). The rpms in each window were either plotted separately for each locus to generate heatmaps (exported from R in a “cdt” table, which was plotted within Java TreeView; 10.1093/bioinformatics/bth349) or averaged and plotted in R with the gplots package to create metaplots.
RNA-Seq analysis
Total RNA-seq reads were trimmed from the 3′ end until the final base had a quality score > 30, using Trimmomatic (10.1093/bioinformatics/btu170), and then aligned to the human genome (UCSC hg19, using Tophat version 2.1.0) (doi:10.1186/gb-2013-14-4-r36) with gene annotations from Ensembl release 72, allowing only uniquely mapped reads. Raw read counts were normalized to reads per million (rpm) per sample and then displayed in the UCSC genome browser as bigWig-formatted coverage tracks. Exonic reads were assigned to specific genes from Ensembl release 72 using htseq-count from the python package HTSeq-0.6.1 (http://bioinformatics.oxfordjournals.org/content/31/2/166). We used the R Bioconductor package edgeR, version 3.12.0 38, to fit the data to a negative binomial generalized log-linear model and estimate a dispersion parameter. We used an adjusted p-value threshold of 0.01 to identify genes significantly differentially expressed in one experimental condition relative to another. In addition to finding differentially expressed genes for each time point individually (two replicates of JQ1 treated cells vs two replicates of DMSO treated cells, repeated for each cell line), we also used edgeR to fit splines with different degrees of freedom to the time courses. We also identified those genes that fit a single degree spline in the JQ1 but not DMSO time course, treating each cell line independently. All groups of differentially expressed genes were analyzed for functional enrichment for specific biological processes or molecular functions using the R Bioconductor package TopGO (Alexa A and Rahnenfuhrer J (2010)) using a background gene set from hg19 in Ensembl release 75.
Mono-nucleosome purification and immunoprecipitation
H3.3 wild-type and H3.3K27M containing mono-nucleosomes were purified from HEK293 TRex FLP-in cells as previously described 5. Briefly, cells were induced with 1.0μg/ml Doxycycline to express histone transgenes. Cells were harvested and lysed in Buffer A (10mM HEPES, pH7.9, 10mM KCl, 1.5mM MgCl2, 340mM sucrose, 10% glycerol, 0.1% Triton X-100, 0.5mM DTT, 0.5x Protease inhibitor, 10mM Sodium Butyrate) and nuclei were isolated by centrifugation through a sucrose cushion (30% sucrose, 10mM HEPES, pH7.9, 1.5mM MgCl2, 10mM Sodium Butryate). Nuclei were digested using micrococcal nuclease (New England Biolabs, M0247S) in Buffer A containing 1mM CaCl2 for 15 minutes at 37°C. MNase digestion was inhibited by addition of 2mM EGTA, 1mM EDTA, 100mM KCl and 0.05% Triton X-100. Cleared lysates were incubated with 100μl Anti-FLAG M2 agarose (Sigma, #A2220) for 2 hours at 4°C. Beads were washed with washing buffer (50mM HEPES, pH 7.9, 100mM KCl, 10% glycerol, 1mM EDTA, 2mM EGTA, 10mM Sodium Butyrate, 0.05% Triton X-100) and proteins were eluted in 20mM HEPES, pH7.9, 100mM NaCl, 10mM Sodium Butyrate, 0.1mg/ml 3x FLAG peptide (Sigma, #F4799).
Plasmid vectors and lentiviral infections
H3.3 wild-type and H3.3K27M cDNA full length fused to a short sequence codifying 6 histidine residues were cloned into the pINDUCER lentiviral vector (Addgene plasmid # 44012, 39) using gateway technology (Thermo Fisher). Lentiviral particle assembly and infection procedure were performed as previously described. HCT116 cells were transduced with H3.3 wild-type and H3.3K27M expressing lentiviral vectors. Neomycin [1mg/mL] was added to select infected cells for 5 days. Transgene expression was induced for 4 days by adding doxycyline [200ng/mL] every two days. For the experiment in Fig. S5 doxycycline was initially added for 7 days and then withdrawn for 21 days. SF8628 and SU-DIPG-IV were transduced with SUZ12 shRNA, EED shRNA and control vectors as previously described. Briefly, 1e6 cells were plated the day before the infection. The following day 0.45μL filtered medium enriched with viral particles was applied to the cells and 10μg/ml polybrene (Sigma) was added. After 6–8 hrs, cells were washed in PBS 1X and fresh medium was applied overnight. The entire procedure was repeated the following day. After that the cells were selected with 150μg/mL hygromicin B (Invitrogen) for 3–4 days (SUZ12 shRNA and control) or with Puromycin 2μg/mL (EED shRNA and control).
Western Blot
Cell pellets were generated after 6–7 days from antibiotic selection after shSUZ12, shEED and control vectors infections or after 7–8 days treatment with EPZM-6438 or DMSO. For JQ1 treated cells the pellets were generated at specific time points indicated in the figures. DMSO treated cell pellets were collected at 48 hrs. Pellets were usually stored at −80°C or processed immediately for protein extraction. Cell pellets were lysed in an appropriate volume (typically 5–7 times the pellet volume) of S300 buffer ([20 mM] Tris-HCl at pH=7.6, [300 mM] NaCl, 5% glycerol, 0.2% Igepal) supplemented with protease cocktail inhibitor (Roche) at 4°C. Subsequently the protein lysate was transferred to one or more milliTUBE (1mL, Covaris) or microTUBE (100 μL, Covaris), accordingly to the volume, and sonicated using E220 focused-ultrasonicator (Covaris) using following parameters: 20% duty cycle, 175 PIP, 200 cycles/burst, 300 seconds. The sonicated protein extracts were centrifuge at 20,000 g for 15 min at 4°C to remove cellular debris. Bradford total protein quantification was performed using Coomassie Plus (Thermo Fisher) and all the proteins extract volumes were adjusted to the same concentration. 40–60 μg of total protein extracts were resolved in SDS-PAGE. Proteins were then transferred to a nitrocellulose membrane (Thermo Fisher) and blocked with Tween 0.1% TBS (T-TBS), 5% milk powder overnight at 4 °C. Blocked membranes were then incubated in primary antibody diluted in blocking solution for 2 hrs at room temperature (for antibodies, see Table 2), washed 3 × with T-TBS, and incubated for 1 h in secondary antibody diluted in blocking solution. Membranes were again washed 3 × with T-TBS, ECL (Millipore) was added to the membrane and the signal was detected on film (Central Infusion Alliance Inc.) or digitally acquired using ChemiDoc Touch imaging system (Biorad).
Growth curves
Growth curves were performed as previously described 13. Briefly, 5e4 cells/well were plated in a 6 well plate (corning) for each day in triplicate. Every day a single plate of cells was fixed using 4% formaldehyde diluted in PBS (Life Technologies) for 10 min at RT, washed with water twice and let air dry overnight. At the end of the curve (5 days) all the cells were stained using a 0.1% solution of crystal violet (Sigma) for 30 min at RT, washed four times in water and air dried overnight. Crystal violet was solubilized in 10% acetic acid (Fisher), and absorbance measured at λ=590 nm. For shRNA SUZ12 and EED experiments, cells were seeded after 5–6 days upon antibiotic selection after infection. JQ1 (Selleckchem), OTX015 (Selleckchem) and EPZM-6438 was resuspended in DMSO at [10mM], stock aliquots were kept at −20°C. For growth curves experiments with JQ1 and OTX015 treatment, the drug and the vehicle control (DMSO) were added once at the beginning of the experiment (day 1) to all the plates. For EPZM-6438 treatment the cells were pretreated for 7 days (fresh inhibitor was added every other day), the treatment continued (same schedule as pretreatment) for the entire growth curve experiment.
Xenograft model
Details about mice and procedures used in this study were presented in previously 8. All protocols, described below, were approved by the Northwestern University Institutional Animal Care and Use Committee. For the in vivo therapy-response analysis, ten mice were randomly assigned to vehicle (DMSO, n = 9) and JQ1 (n = 9) treatment groups, with JQ1 administration by ip injection at 50 mg−1 kg−1, with once daily administration for ten days. Additional two mice, not included in the survival analysis, were sacrificed 6 h after administration of final treatment, with the brains of these mice resected and placed in 4% paraformaldehyde (PFA). To investigate the efficacy of JQ1 treatment against H3K27M mutant DIPG, 4 × 106 SU-DIPG IV cells, in 0.4 ml of cell culture media with matrigel (BD Bioscience) were injected in the right flank of mice. Mice were randomly assigned to vehicle (DMSO, n = 4) and JQ1 (n = 5) treatment groups, with JQ1 administration by ip injection at 50 mg/kg, with once daily administration for ten days. All of these treatments were initiated on day 7 following tumor cell implantation. Tumor growth and response to therapy were determined twice weekly by bioluminescence imaging. Bioluminescence measurements for each animal at each time point were normalized against corresponding readings obtained at the beginning of therapy. To investigate the efficacy of I-BET (I-BET-762, ApexBio) treatment against H3K27M mutant DIPG, 1 × 105 SF8628 cells in 1 μl Hanks’ Balanced Salt Solution, without Ca2+ and Mg2+, were slowly injected into the pontine tegmentum as described above. Mice were randomly assigned to vehicle (OraPlus, n = 9) and I-BET (n = 5) treatment groups, with I-BET administration by oral gavage at 50 mg/kg, with once daily administration for ten days. All of these treatments were initiated on day 48 following tumor cell implantation. GSKJ4 treatment protocol was described before 8. Mice monitoring, tumor growth imaging and quantification, survival plots and relative statistical analyses were performed as described in previous studies8. Two mice from the control group in Figure 4A died immediately after the end of the vehicle administration therefore they were excluded from the survival study. Four mice from the treatment group and one mouse from the vehicle group in the experiments presented respectively in Supplementary Figure 17B and C were sacrificed for tumor unrelated problems and therefore excluded from the study.
Histological analyses
Histologic scoring of apoptotic bodies and mitoses was performed on 4 μm-thick sections of PDX DIPG at 200x, cut from formalin-fixed, paraffin embedded blocks and stained with hematoxylin and eosin. Anti-KI67 and anti-H3K27ac were diluted in antibody diluent and applied to slides at room temperature for 1 hour. The Dako EnVision+ System-HRP (DAB) for use with rabbit antibody was used as a visualization system (K4011, DAKO). TUNEL staining was performed using the ApopTag Peroxidase In Situ Apoptosis Detection Kit (S7100, Millipore), according to the manufacturer’s protocol. The intensity of the H3K27ac in each cell from JQ1 and control treated PDX tumor slides (n=50 each group) was measured using imageJ.
Slide scan
Coronal sections of mouse brainstems were stained with H&E and KI67 were digitized via a Hamamatsu Nanozoomer 2.0 HT Digital Pathology Slide Scanner (Hamamatsu, Skokie, IL). Images were processed with Adobe Photoshop CC (San Jose, CA).
Double immunofluorescence on tissue sections
Serial sections of 4 μm were cut from paraffin blocks of PDX tissue samples, and used for immunohistochemical and double immunofluorescence assays. Slides were placed at 60°C for 1 hour; deparaffinized through 3 changes of xylene; hydrated to water by two changes of 100% ethanol, two changes of 95% ethanol, one of 80% ethanol, one of 70% ethanol, and two changes of distilled water. Antigen retrieval was performed using 1X Dako Target Retrieval Solution pH6 (cat #S1699, Carpinteria, CA). The slides were incubated in Biocare Medical Decloaking Chamber at 110C for five minutes; rinsed in distilled water two times and in PBS for five minutes. If the staining was performed on mouse tissue, an additional mouse IgG blocking (cat #MKB-2213, Vector. Burlingame, CA) step was added, the excess was tapped off and the tissue was incubated with 10% NGS (cat #S1000, Vector) blocking serum for twenty minutes. Following the removal of NGS, primary antibodies were applied as follows: (1) Anti-Histone H3K27M (cat #ABE419, Millipore, Billerica, MA), 1:1000; (2) TUBB3 β3 Tubulin (TU-20) Mouse mAb (cat #4466, Cell Signaling, Danvers, MA), 1:100; (3) Ki67 mouse monoclonal, clone MIB-1 (cat #M7240, Dako), 1:150. H3K27 Acetyl Histone 3 (Lys27) (D5E4) XP rabbit monoclonal antibody (cat #8173, Cell Signaling), 1:200. All primary antibodies were diluted in antibody diluent with background reducing components (cat #S3022, Dako) and were incubated overnight at 4°C. Slides were then washed for 15 minutes in PBS; 3 minutes in TBST (cat #S3306, Dako) and transferred to PBS. Secondary antibodies were diluted in PBS. Goat anti-rabbit IgG, DyLight 488 conjugated Highly Cross-adsorbed (Invitrogen by Thermo Fisher Scientific) was diluted 1:200; 568 Alexa Fluor goat anti-mouse IgG (H+L) (A11004) was diluted 1:1000. Slides were incubated in the dark for 30 minutes with both secondary antibodies at the same time. Secondary antibodies were rinsed off with PBS, washed in PBS and transferred to TBST for 3 minutes. Tissue was then incubated in 0.1% solution Sudan Black B for 30 minutes at room temperature in the dark. Slides were washed with PBS and mounted in fluorescent mounting medium with DAPI (Prolong Antifade reagent with DAPI, cat #P36935, Molecular Probes, Eugene, OR). Images were acquired using a Nikon C2 confocal microscope (Melville, NY) and processed with Nikon NIS Elements software. All images within each multi-panel figure were taken with the same confocal settings, and were processed in NIS Elements the exact same way.
Statististical methods
For statistical analyses, GraphPad Prism 6 and Microsoft Excel were used.
All the data where a statistical analysis was reported meet the criteria to use the appropriate statistical test, (for the normal distribution of data, the empirical rule was used to infer the distribution). F-test was performed to determine if different groups have the same variance or not. For growth curves and time-course RNA-seq t-tests were calculated between the area under the curve (AUC) values, similarly to what described in previous studies13. Statistical tests used are reported in the figure legends. P values <0.05 were considered to be statistically significant. All the experiments were not conducted in blind conditions.
Supplementary Material
Acknowledgments
We thank all the members of the Shilatifard’s Lab for useful comments and suggestions. We thank Dr. M. Monje (Stanford University) for use of the SU-DIPG-IV cell line. We thank Dr. D. Pasini (European Institute of Oncology, IEO) for providing the SUZ12 and Control vectors and C. Rivetta for technical support. pInducer20 was a gift from Dr. S. Elledge (deposited in Addgene). A.P. is supported by a Long-Term EMBO fellowship (ALTF 372-2015) and his work in the Shilatifard’s Lab is supported by AIRC and Marie Curie Actions – People – COFUND. R.H. is supported by the US National Institutes of Health RO1NS093079, the Matthew Larson Foundation and the Bear Necessities Pediatric Cancer Foundation and Rally Foundation. Proteomics services were performed by the Northwestern Proteomics Core Facility, generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center and the National Resource for Translational and Developmental Proteomics supported by P41 GM108569. Studies in regards to the development of targeted therapeutics for DIPG within our groups are partially supported by the generous support from John McNicholas Pediatric Brain Tumor Foundation. Studies in Shilatifard’s laboratory are supported by NCI grant R35CA197569.
Footnotes
ACCESSION CODES
RNA-seq and ChIP-seq raw data are available in the GEO database: Series GSE78801
AUTHOR CONTRIBUTIONS
A.P. and A.S. designed the study. A.P. performed the majority of the experiments, part of the analyses and wrote the manuscript. R.H., C.D.J., A.P. and A.S. designed the in vivo studies. R.H. and Q.M. performed and analyzed the in vivo experiments. M.A.M. performed the MNase-IP experiment. A.R.W. performed the initial bioinformatics analyses on the studies related to the PRC2 role in DIPG. E.T.B. performed all the other bioinformatics analyses. C.M.H. performed and analyzed the immunohistochemistry studies. S.A.M and E.J.R generate and sequenced the NGS libraries. Y.T. provided technical help. A.V.M performed the FACS studies. N.A.A. and N.L.K. designed the mass spectrometry studies. N.A.A. performed the mass spectrometry experiments. R.R.L. and A.M.S. provided clinical supervision in the interpretation of data. A.P., M.A.M., C.D.J. and A.S. revised the manuscript. All the authors commented on the manuscript and approved data included in it.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Schroeder KM, Hoeman CM, Becher OJ. Children are not just little adults: recent advances in understanding of diffuse intrinsic pontine glioma biology. Pediatr Res. 2014;75:205–209. doi: 10.1038/pr.2013.194. [DOI] [PubMed] [Google Scholar]
- 2.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 3.Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nature genetics. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herz HM, et al. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science. 2014;345:1065–1070. doi: 10.1126/science.1255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender S, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Chan KM, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes & development. 2013;27:985–990. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashizume R, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20:1394–1396. doi: 10.1038/nm.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monje M, et al. Hedgehog-responsive candidate cell of origin for diffuse intrinsic pontine glioma. Proc Natl Acad Sci U S A. 2011;108:4453–4458. doi: 10.1073/pnas.1101657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Justin N, et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun. 2016;7:11316. doi: 10.1038/ncomms11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piunti A, Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016;352:aad9780. doi: 10.1126/science.aad9780. [DOI] [PubMed] [Google Scholar]
- 12.Bracken AP, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes & development. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piunti A, et al. Polycomb proteins control proliferation and transformation independently of cell cycle checkpoints by regulating DNA replication. Nat Commun. 2014;5:3649. doi: 10.1038/ncomms4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi J. Bromodomain and extraterminal domain inhibitors (BETi) for cancer therapy: chemical modulation of chromatin structure. Cold Spring Harb Perspect Biol. 2014;6:a018663. doi: 10.1101/cshperspect.a018663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nature reviews. Molecular cell biology. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galderisi U, Jori FP, Giordano A. Cell cycle regulation and neural differentiation. Oncogene. 2003;22:5208–5219. doi: 10.1038/sj.onc.1206558. [DOI] [PubMed] [Google Scholar]
- 18.Gordon J, Amini S, White MK. General overview of neuronal cell culture. Methods Mol Biol. 2013;1078:1–8. doi: 10.1007/978-1-62703-640-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.An W, Palhan VB, Karymov MA, Leuba SH, Roeder RG. Selective requirements for histone H3 and H4 N termini in p300-dependent transcriptional activation from chromatin. Molecular cell. 2002;9:811–821. doi: 10.1016/s1097-2765(02)00497-5. [DOI] [PubMed] [Google Scholar]
- 20.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nature reviews. Drug discovery. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 21.Gupta S, Takebe N, Lorusso P. Targeting the Hedgehog pathway in cancer. Ther Adv Med Oncol. 2010;2:237–250. doi: 10.1177/1758834010366430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nature reviews. Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 23.Weiner A, et al. Co-ChIP enables genome-wide mapping of histone mark co-occurrence at single-molecule resolution. Nat Biotechnol. 2016;34:953–961. doi: 10.1038/nbt.3652. [DOI] [PubMed] [Google Scholar]
- 24.Pasini D, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic acids research. 2010;38:4958–4969. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrari KJ, et al. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Molecular cell. 2014;53:49–62. doi: 10.1016/j.molcel.2013.10.030. [DOI] [PubMed] [Google Scholar]
- 26.Mohn F, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Molecular cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Jones C, Baker SJ. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nature reviews. Cancer. 2014;14 doi: 10.1038/nrc3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grasso CS, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med. 2015;21:827. doi: 10.1038/nm0715-827a. [DOI] [PubMed] [Google Scholar]
- 30.Mazur PK, et al. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat Med. 2015;21:1163–1171. doi: 10.1038/nm.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhadury J, et al. BET and HDAC inhibitors induce similar genes and biological effects and synergize to kill in Myc-induced murine lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2721–2730. doi: 10.1073/pnas.1406722111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashizume R, et al. Characterization of a diffuse intrinsic pontine glioma cell line: implications for future investigations and treatment. J Neurooncol. 2012;110:305–313. doi: 10.1007/s11060-012-0973-6. [DOI] [PubMed] [Google Scholar]
- 33.Mueller S, et al. Targeting Wee1 for the treatment of pediatric high-grade gliomas. Neuro Oncol. 2014;16:352–360. doi: 10.1093/neuonc/not220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen FX, et al. PAF1, a Molecular Regulator of Promoter-Proximal Pausing by RNA Polymerase II. Cell. 2015;162:1003–1015. doi: 10.1016/j.cell.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen F, Gao X, Shilatifard A. Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide. Genes & development. 2015;29:39–47. doi: 10.1101/gad.246173.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nature protocols. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meerbrey KL, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.