Abstract
We examined the effects of osteocyte secreted factors on myogenesis and muscle function. MLO‐Y4 osteocyte‐like cell conditioned media (CM) (10%) increased ex vivo soleus muscle contractile force by ∼25%. MLO‐Y4 and primary osteocyte CM (1% to 10%) stimulated myogenic differentiation of C2C12 myoblasts, but 10% osteoblast CMs did not enhance C2C12 cell differentiation. Because WNT3a and WNT1 are secreted by osteocytes, and the expression level of Wnt3a is increased in MLO‐Y4 cells by fluid flow shear stress, both were compared, showing WNT3a more potent than WNT1 in inducing myogenesis. Treatment of C2C12 myoblasts with WNT3a at concentrations as low as 0.5 ng/mL mirrored the effects of both primary osteocyte and MLO‐Y4 CM by inducing nuclear translocation of β‐catenin with myogenic differentiation, suggesting that Wnts might be potential factors secreted by osteocytes that signal to muscle cells. Knocking down Wnt3a in MLO‐Y4 osteocytes inhibited the effect of CM on C2C12 myogenic differentiation. Sclerostin (100 ng/mL) inhibited both the effects of MLO‐Y4 CM and WNT3a on C2C12 cell differentiation. RT‐PCR array results supported the activation of the Wnt/β‐catenin pathway by MLO‐Y4 CM and WNT3a. These results were confirmed by qPCR, showing upregulation of myogenic markers and two Wnt/β‐catenin downstream genes, Numb and Flh1. We postulated that MLO‐Y4 CM/WNT3a could modulate intracellular calcium homeostasis as the trigger mechanism for the enhanced myogenesis and contractile force. MLO‐Y4 CM and WNT3a increased caffeine‐induced Ca2+ release from the sarcoplasmic reticulum (SR) of C2C12 myotubes and the expression of genes directly associated with intracellular Ca2+ signaling and homeostasis. Together, these data show that in vitro and ex vivo, osteocytes can stimulate myogenesis and enhance muscle contractile function and suggest that Wnts could be mediators of bone to muscle signaling, likely via modulation of intracellular Ca2+ signaling and the Wnt/ β‐Catenin pathway. © 2017 American Society for Bone and Mineral Research
Keywords: WNT/β‐CATENIN/LRPS, BONE‐MUSCLE INTERACTIONS, MOLECULAR PATHWAYS, OSTEOCYTES, GENERAL MUSCLE PHYSIOLOGY
Introduction
Muscle and bone are connected both anatomically and functionally and interact as a functional unit.1 Musculoskeletal diseases are the most common cause of chronic disabilities worldwide. Osteoporosis/osteopenia (loss of bone tissue and function) and sarcopenia (progressive muscle loss with a greater and disproportional loss of muscle force/strength) are inevitable consequences of the aging process, prevalent in the frail elderly, and have become a growing public health concern.2
The emerging field of bone‐muscle crosstalk is helping to clarify the role of bones and muscles as endocrine organs because they produce and secrete “hormone‐like factors” that can mutually influence each other and other tissues.3, 4 Recently, evidence has emerged that osteocytes secrete factors that have profound effects on C2C12 muscle cells in culture, and vice versa,3, 5 suggesting that muscle and bone communicate at a molecular level via biochemical factors that are reciprocally important for optimal function.6
Bones secrete a host of factors that have clear paracrine and endocrine effects, such as FGF23, Sclerostin, Osteocalcin, WNTs, Prostaglandin E2 (PGE2), etc. (reviewed in Dallas and colleagues7). Sclerostin is one of these factors that has gained much attention because of recent clinical applications and trials.8 Sclerostin is one of the osteocyte‐specific proteins, and is product of the SOST gene,9 which is expressed by mature osteocytes.10, 11 Sclerostin is a negative regulator of the Wnt/β‐catenin signaling pathway by binding to the Wnt co‐receptors, low‐density lipoprotein receptor‐related proteins 5 and 6 (LRP5 and LRP6).12 In the presence of Sclerostin, Wnt‐receptor interaction is inhibited, and β‐catenin is phosphorylated by glycogen synthase kinase 3 and targeted for ubiquitination and degradation via the proteasome pathway.13 Studies using loss of function and gain of function mouse models of Sost have demonstrated increased and decreased bone mass, respectively.10 Our earlier studies demonstrated that many of the effects of bone cell conditioned medium in triggering acceleration of myogenesis could be partially mimicked by low nanomolar range concentrations of PGE2.3
The Wnt/β‐catenin signaling pathway is important for cell and tissue homeostasis because secreted WNTs act through specific receptors to control and modulate cell proliferation, differentiation, apoptosis, survival, migration, and polarity (reviewed in Clevers and Nusse14). They play critical roles during embryonic development (including muscle and skeletal patterning) as well as in postnatal health and diseases, including cancer and degenerative disorders. The Wnt/β‐catenin signal pathway has been shown to be an important component of bone mass accrual, regulation, and maintenance,15 and accumulating data show that the Wnt/β‐catenin signal pathway is strongly implicated in skeletal muscle development, growth, and regeneration.16
To determine whether osteocytes can potentially regulate muscle function, we tested the effects of MLO‐Y4 conditioned medium (MLO‐Y4 CM) on muscle contractility in soleus (SOL) muscles using a murine ex vivo muscle contractility assay and found out that MLO‐Y4 CM increased the contractile force of SOL muscles. To gain new insight into the mechanisms of bone to muscle signaling we have used MLO‐Y4 osteocyte‐like cells, osteoblast cells, primary osteocytes, and C2C12 myoblasts as in vitro models. We report that MLO‐Y4 cells and primary osteocytes secrete factors that potently stimulate myogenesis, accompanied by enhanced β‐catenin translocation, suggesting that the effect may be mediated via Wnt/β‐catenin signaling. However, 10% osteoblast CMs did not enhance C2C12 cell differentiation. We therefore investigated the expression of Wnts in osteocytes and showed that WNT3a, which is expressed in osteocytes, mirrored the effects of osteocyte conditioned medium on myogenesis. Knocking down Wnt3a in MLO‐Y4 osteocytes inhibited the effect of CM on C2C12 myogenic differentiation. Sclerostin inhibited the effects of CM or WNT3a on C2C12 cell differentiation. To determine potential mechanisms of contractile force enhancement, we examined the effects of osteocyte conditioned medium on calcium release from the sarcoplasmic reticulum (SR). Our in vitro and ex vivo data show that osteocytes secrete soluble factors that enhance myogenic differentiation, enhance both contractile force and calcium release from the SR, and provide evidence that WNT3a is a potential factor from osteocytes with the intrinsic potential to modulate these effects of bone‐muscle crosstalk.
Materials and Methods
Materials
DMEM high‐glucose media, α‐MEM media, penicillin‐streptomycin (P/S) 10,000 U/mL each and trypsin‐EDTA 1× solution were obtained from Mediatech Inc. (Manassas, VA, USA); calf serum (CS), fetal bovine serum (FBS), horse serum (HS), and caffeine were obtained from Thermo Fischer Scientific Inc. (Waltham, MA, USA); bovine serum albumin (BSA) and diamidino‐2‐phenylindole (DAPI) were from Sigma‐Aldrich (St. Louis, MO, USA); rat tail collagen type I was purchased from BD Biosciences (San Jose, CA, USA); 16% paraformaldehyde was from Alfa Aesar (Ward Hill, MA, USA); Recombinant Mouse Wnt‐3a and Recombinant Mouse SOST/Sclerostin protein was purchased from R&D Systems Inc. (Minneapolis, MN, USA); WNT1 Recombinant Human Protein was obtained from Life Technologies (Grand Island, NY, USA). Cy‐3 donkey anti‐mouse was purchased from Invitrogen (Carlsbad, CA, USA); this antibody has been previously validated.5 Mouse anti‐active β‐catenin antibody was from Millipore (Billerica, MA, USA); this antibody has been previously validated.5 Lipofectamine RNAiMAX Transfection Reagent was from ThermoFisher Scientific (Waltham, MA, USA); Wnt3a siRNA (antisense strand: 5′‐GCAUCCGCUCUGACACUUAAUACTC‐3′), negative control siRNA and TYE 563 DS Transfection Control were from Integrated DNA Technologies (Coralville, Iowa, USA); Tri reagent was obtained from Molecular Research Center, Inc. (Cincinnati, OH, USA); high‐capacity cDNA reverse transcription kit was from Applied Biosystems (Foster City, CA, USA); and RT2 Real‐Time SYBR green/Rox PCR master mix was from SABiosciences (Valencia, CA, USA). Anti‐human Myosin Heavy Chain‐CFS, Carboxyfluorescein (CFS)‐conjugated mouse monoclonal anti‐human Myosin Heavy Chain antibody was from R&D Systems Inc. (Minneapolis, MN, USA); this antibody has been previously validated.6 Fura‐2/AM was obtained from Life Technologies (Grand Island, NY, USA). C2C12 cells were obtained from American Type Culture Collection (ATCC) (Manassas, VA, USA). Gentamycin and 2‐APB; Wingless Type MMTV Integration Site Family, Member 3A (WNT3A) ELISA Kit was purchased from MyBioSource (San Diego, CA, USA).
Methods
No in vivo experiments were conducted in this study.
Animals
Five‐month‐old male C57BL/6 mice from The Jackson Laboratory (Bar Harbor, ME, USA) were used for isolation of bone cells and isolation of intact muscles for ex vivo contractility studies, following humane euthanasia cervical dislocation. All animal procedures were performed according to an approved IACUC protocol at the University of Missouri–Kansas City (UMKC) and conformed to relevant federal guidelines. The UMKC animal facility is operated as a specific pathogen‐free, AAALAC approved facility. Animal care and husbandry at UMKC meet the requirements in the Guide for the Care and use of Laboratory Animals (8th edition), National Research Council. Animals were group housed and maintained on a 12‐hour light/dark cycle with ad libitum food and water at a constant temperature of 72°F and humidity of 45% to 55%. Daily health check inspections were performed by qualified veterinary staff and/or animal care technicians.
Ex vivo muscle contractility
Dissection and setup
Mice were humanely euthanized and soleus muscles were carefully removed for contractility analysis, as reported17 and following the Protocol of the Treat‐NMD Network (DMD_M.1.2.002). Dissected soleus muscles were immediately placed in a dish containing a HEPES Ringer solution (143 mM NaCl; 5 mM KCl; 1.8mM MgCl2; 10mM HEPES; 2.5 mM CaCl2; pH 7.40) with 10 mM glucose. This solution was continuously aerated with 100% O2. Muscles were mounted vertically between two stimulating platinum electrodes and immersed in a 25‐mL bathing chamber containing the Ringer solution. Via the tendons, the muscles were suspended from adjustable isometric force transducers above the chambers and secured to the base of the tissue support within the chambers. The analog output of the force transducer was digitized, stored and analyzed with PowerLab Software (ADInstruments Inc., Colorado Springs, CO, USA). For each muscle the stimulatory voltage was provided by a S8800 dual pulse digital stimulator (Grass Products, West Warwick, RI, USA) (pulse duration, 1 ms; train duration, 500 ms). Optimal muscle length (L0) was determined by tetanic stimulations of 100 Hz at an interval of 1 min.
Equilibration
The intact muscles were allowed a 30‐min equilibration period during which time they were stimulated with pairs of alternating high (100 Hz) and low (20 Hz) frequency pulse‐trains administered with a periodicity of 1 min. Utilization of the proposed paradigm of stimulation helps with the study of the relative contributions of the contractile proteins (100 Hz) and the SR (20 Hz) to contractile function.17 After equilibration and stabilization of submaximal and maximal forces, blank medium or MLO‐Y4 CM was added to the control and test muscle, respectively, and force was followed for 30 min after the addition of the media.
Force normalization
Muscle force was reported as relative force (mN) and force normalized to muscle physiological cross sectional area (N/cm2) as reported by our group.17, 18, 19 Five animals and 10 muscles were used for final analyses.
Cell cultures and generation of CM from primary osteocytes (PO CM), MLO‐Y4 cells (MLO‐Y4 CM), 2T3 cells (2T3 CM), and MC3T3‐E1 (MC3T3‐E1 CM)
Cell culture
C2C12 myoblasts were cultured following our own previously published protocols.3, 5, 6 Briefly, cells were grown at 37°C in a controlled humidified 5% CO2 atmosphere in growth medium (GM), DMEM/high glucose +10% FBS (100 U/mL P/S), and maintained at 40% to 70% cell density. Under these conditions, myoblasts proliferate, but do not differentiate into myotubes. During myoblast proliferation, medium was changed every 48 hours. To induce differentiation of myoblasts into myotubes, when cells reached 75% confluence, the culture media were switched from GM to differentiation medium (DM), DMEM/high glucose + 2.5% horse serum (HS) (100 U/mL P/S), and DM was changed every 48 hours. Fully differentiated, functional, contracting myotubes were formed within 5 to 7 days.
MLO‐Y4, a murine long bone‐derived osteocyte‐like cell line (referred to hereafter as MLO‐Y4 cells), were cultured as described.20 Briefly, cells were seeded onto type I collagen‐coated plates and cultured in α‐MEM + 2.5% FBS + 2.5% CS (100 U/mL P/S). Cells were maintained at ∼60% confluence throughout the culture period.
2T3 osteoblast‐like cells (referred to hereafter as 2T3 cells) were cultured as described,21 with a few modifications. Briefly, cells were cultured in α‐MEM + 10% FBS (100 U/mL P/S). Cells were maintained at ∼60% confluence throughout the culture period.
MC3T3‐E1 preosteoblast cells (referred to hereafter as MC3T3‐E1 cells) were cultured as described.22 Briefly, cells were cultured in α‐MEM + 10% FBS (100 U/mL P/S). Cells were maintained at ∼60% confluence throughout the culture period.
Generation of CMs from primary osteocytes (PO CM), MLO‐Y4 cells (MLO‐Y4 CM), 2T3 cells (2T3 CM), and MC3T3‐E1 cells (MC3T3‐E1 CM)
The method of isolation of primary osteocytes was described by Stern and colleagues23 and Stern and Bonewald,24 with minor modifications. Briefly, long bones were dissected and muscles were removed taking care not to remove the periosteum. Epiphyses were cut and bone marrow was then flushed out. The bones were cut into small pieces (1 to 2 mm2) and incubated for 25 min in collagenase solution (300 units/mL) with constant shaking in an incubator at 37°C and 5% CO2. The solution was then removed and centrifuged. Cells were then resuspended in 10% FBS containing α‐MEM and plated in collagen‐coated plates (fraction 1: fibroblast‐enriched). Bone chips were then subsequently incubated with fresh collagenase solution to obtain fractions 2 to 4 (osteoblast‐enriched). Then bone chips were digested in alternating EDTA (5mM) and collagenase solutions to obtain fractions 5 to 9 (osteocyte‐enriched). The remaining bone pieces were transferred into collagen‐coated dishes and cells migrated from the bone pieces after 3 to 5 days of culture (fraction 10: bone particles). All of the cells obtained from fractions 5 to 9 that displayed an osteocyte‐like morphology stained positive for the osteocyte marker E11/GP38. The osteocyte phenotype was further confirmed by a lack of staining for alkaline phosphatase and the absence of collagen1a1 expression. The osteocytes also expressed additional osteocyte‐specific genes such as Sost and Mepe. To generate PO CM, cells of fractions 5 to 9 were cultured in 60 cm2 collagen‐coated plates with α‐MEM + 5% FBS + 5% CS (1000 U/mL P/S) culture media, 3600 cells/cm2 and allowed to attach and grow for 48 hours. The medium was collected and centrifuged at 500g for 10 min. CM was aliquotted and flash frozen in liquid nitrogen then stored at −80°C.
To prepare MLO‐Y4 CM, 2T3 CM, and MC3T3‐E1 CM, MLO‐Y4, 2T3, and MC3T3‐E1 cells were plated in 10‐cm dishes, 5000 cells/cm2 and allowed to attach and grow for 48 hours. The medium was collected and centrifuged at 500g for 10 min. CM was aliquotted and flash frozen in liquid nitrogen then stored at −80°C.
Fluid flow shear stress (FFSS) treatment of MLO‐Y4 and 2T3 cells
At day 1, MLO‐Y4 and 2T3 cells were seeded onto glass slides coated with type I collagen at a density of 5600 cells/cm2 in α‐MEM supplemented with either 2.5% CS, 2.5% FBS, and 1% P/S (for MLO‐Y4 cells) or 10% FBS and 1% P/S (for 2T3 cells) for 24 hours. At day 2, shear stress was applied to the slides at 2 dynes/cm2 for 2 hours in α‐MEM supplemented with 0.1% CS, 0.1% FBS, and 1% P/S. For fluid flow shear stress, 350 mL media was used to run the system (six slides per run). For static controls, three slides were placed in a 150 mm round tissue culture dish with 175 mL of α‐MEM media (1% P/S) containing 0.1% CS and 0.1% FBS. Media was collected after 2 hours of shear stress. Three slides of seeded cells were used for RNA isolation at the end of the 2 hours of shear stress (or static culture). The other three slides were incubated in regular media (α‐MEM supplemented with 2.5% CS, 2.5% FBS, and 1% P/S) for 24 hours. RNA was extracted from both static and shear stressed slides, 2 hours and 24 hours after FFSS for RT‐qPCR detection of gene expression.
ELISA for WNT3a in PO CM and MLO‐Y4 CM
WNT3a in PO CM and MLO‐Y4 CM was detected by Wingless Type MMTV Integration Site Family, Member 3A (WNT3A) ELISA Kit according to the manufacturer's protocol. Serum‐free CM were collected from the cells that were cultured for 24 hours, and 100 µL CM was added into the ELISA plate wells coated with a biotin‐conjugated WNT3a antibody. Next, Avidin conjugated to horseradish peroxidase is added and incubated for 1 hour. After TMB (3, 3′, 5, 5′‐Tetramethylbenzidine) substrate solution is added, the color change is measured spectrophotometrically at 450 nm. The concentration of WNT3a is then determined by comparing the optical density (O.D.) of the samples to the standard curve. Three independent experiments with two replicates were performed for these experiments.
Immunolabeling of active β‐catenin for nuclear translocation
These studies followed our previous protocols25 with minor modifications for C2C12 cells. Briefly, C2C12 cells were seeded onto positively‐charged glass slides and cultured overnight. CMs or blank media was added to produce 10% final concentration. Cells were incubated for 30 min, 60 min, and 90 min, respectively, rinsed two times in cold PBS, fixed in 2% paraformaldehyde containing 0.2% Triton X‐100 for 10 min and then washed three times in PBS at ambient temperature for 10 min each. Slides were blocked with 2.5 % BSA and 1% normal donkey serum in PBS overnight at 4°C. Primary antibody against the active form of β‐catenin was used at 1:100 dilution in blocking buffer for 4 hours at ambient temperature. Slides were washed three times with PBS and incubated for 1 hour with Cy3‐conjugated donkey anti‐mouse antibody. The quantification of the intensity of β‐catenin immunofluorescence was performed as described using a Leica TCS SP5 II confocal microscopy system (Leica, Wetzlar, Germany) on an inverted microscope platform.26 Sixteen‐bit images were obtained and were pseudocolored in green and analyzed using ImageJ software (NIH, Bethesda, MD, USA; https://imagej.nih.gov/ij/). The nuclear and cytoplasmic histograms from each image were used to quantify staining intensity. Three independent experiments with five to 10 cells monitored on each individual experiments.
C2C12 cell morphometry and immunostaining
Cell morphology
Phase contrast images were taken with a LEICA DMI‐4000B inverted microscope equipped with a 14‐BIT CoolSNAP CCD camera (Photometrics), using the LEICA LAS imaging software for calibration (Leica Microsystems).
Immunostaining
Experiments were performed following our published protocols.5, 6, 27 Briefly, C2C12 cells were fixed with neutral buffered formalin and permeabilized with 0.1% Triton X‐100. Myosin heavy chain (MHC) was detected with Anti‐Human Myosin Heavy Chain‐CFS (1:50) at room temperature for 30 min and counterstained with DAPI. Images were taken using a 20× LEICA FLUO objective with the LEICA system described above.
Fusion index
To quantify myogenic differentiation of C2C12 cells after treatments, the Fusion Index (FI) was calculated, where FI is defined as: (nuclei within myosin heavy chain‐expressing myotubes/total number of myogenic nuclei) × 100.28 We conducted three to five independent experiments, two replicates, and three to five areas per replicate were randomly selected for the measurements.
Treatment of C2C12 cells with PO CM, MLO‐Y4 CM, 2T3 CM, MC3T3‐E1 CM, and WNT3a
C2C12 cells were plated in six‐well plates, 10 × 104/well, and allowed to attach and grow overnight. Medium of C2C12 myoblasts were changed from GM to DM with CMs and WNT3a, respectively. Forty eight hours later, medium was changed with fresh DM. CMs and WNT3a were removed from the medium. At day 3 of differentiation, C2C12 cells were analyzed according to “C2C12 Cell Morphometry and Immunostaining” described above.
Treatment of MLO‐Y4 cells with Wnt3a siRNA and generation of CM from Wnt3a siRNA treated MLO‐Y4 cells (MLO‐Y4 siRNA‐treated CM)
MLO‐Y4 cells were plated in six‐well plates, 5 × 104 cells/well, and allowed to attach and grow overnight. Cells were transfected with Lipofectamine RNAiMAX Transfection Reagent following the manufacturer's instructions. First, TYE 563 DS Transfection Control, which is tagged with a red fluorescent probe was used to detect the transfection efficiency. To knock down Wnt3a, MLO‐Y4 cells were treated with 10 nM Wnt3a siRNA+ 9 μL Lipofectamine RNAiMAX reagent (MLO‐Y4‐siRNA–treated cells), while negative control cells were treated with 10nM negative control siRNA + 9 μL Lipofectamine RNAiMAX reagent. We further employed a vehicle control by treating the cells with only 9 μL Lipofectamine RNAiMAX reagent without siRNA. After 24 hours of transfection, total RNA was isolated from the cells with Tri reagent and Wnt3a expression was detected by RT‐qPCR. To prepare the CM of MLO‐Y4 cells transfected with Wnt3a siRNA, 24 hours after siRNA transfection, medium was changed with fresh medium and the cells were cultured for 48 hours, then CM was collected and centrifuged at 500g for 10 min. CM was aliquotted and flash frozen in liquid nitrogen then stored at −80°C. The CMs of negative control and Lipofectamine RNAiMAX reagent‐only vehicle treated cells (vehicle control) were also collected. Three independent experiments were performed.
Treatment of C2C12 cells with MLO‐Y4 siRNA‐treated CM
C2C12 cells were plated in six‐well plates, 10 × 104/well, and allowed to attach and grow overnight. Medium of C2C12 myoblasts were changed from GM to DM with 10% CMs, respectively. Forty eight hours later, medium was changed with fresh DM. CMs were removed from the medium. At day 3 of differentiation, C2C12 cells were analyzed according to “C2C12 Cell Morphometry and Immunostaining” described above.
Treatment of C2C12 cells with Sclerostin
C2C12 cells were plated in six‐well plates, 10 × 104/well, and allowed to attach and grow overnight. C2C12 myoblasts in DM without Sclerostin treatment were used as a negative control. C2C12 myoblasts were treated with 100 ng/mL Sclerostin in DM for 6 hours. C2C12 myoblasts were treated in DM with 10% MLO‐Y4 CM for 48 hours as one of our positive controls. C2C12 myoblasts were treated in DM with 10 ng/mL WNT3a for 48 hours as our second positive control. C2C12 myoblasts were treated with 100 ng/mL Sclerostin in DM for 6 hours, followed by 10% MLO‐Y4 CM for 48 hours; C2C12 myoblasts were treated with 100 ng/mL Sclerostin in DM for 6 hours, followed by 10 ng/mLWNT3a for 48 hours; C2C12 myoblasts were treated with 100 ng/mL Sclerostin in DM for 6 hours, followed by the combination of 10% MLO‐Y4 CM + 10 ng/mLWNT3a for 48 hours. Under any experimental conditions where Sclerostin, CM, and WNT3a were used, they were maintained during treatments for 48 hours. After 48 hours, medium was changed with fresh DM. Sclerostin, CM, and WNT3a were removed from the media. At day 3 of differentiation, C2C12 cells were analyzed according to “C2C12 Cell Morphometry and Immunostaining” described above.
RNA isolation and real‐time quantitative PCR (RT‐qPCR)
Total RNA was isolated from the cells with Tri reagent according to the manufacturer's protocol and cDNA was synthesized using a high‐capacity cDNA reverse transcription kit. RT‐qPCR was performed using RT2 Real‐Time SYBR green/Rox PCR master mix. RT‐qPCR primers used in this study are summarized in Table 1. RT‐qPCR was run in a 25‐µL reaction volume on 96‐well plates using a StepOnePlus instrument (Applied Biosystems, Foster city, CA, USA). Data were analyzed using RT2 Profiler PCR Array Data Analysis (SABiosciences); Ct values were normalized to Glyceraldehyde‐3‐phosphate dehydrogenase (Gadph) as a reference gene. Gene expression was determined as fold upregulation or downregulation of the gene of interest compared to the controls. All reactions were done in duplicate and all experiments were repeated at least three times.
Table 1.
RT‐qPCR Primers Used in This Study
| Primer | Primer sequence (5′ to 3′) |
|---|---|
| GAPDHF | TGCGATGGGTGTGAACCACGAGAA |
| GAPDHR | GAGCCCTTCCACAATGCCAAAGTT |
| NumbF | AACCAGCCTTTGTCCCTACC |
| NumbR | GGCGACTGATGTGGATGAG |
| Fhl1F | TTCTGGATCCCATCCTGTGTGAG |
| Fhl1R | ACACGGTACCTGGTTCCAATGCC |
| MYHCF | CAAGTCATCGGTGTTTGTGG |
| MYHCR | TGTCGTACTTGGGCGGGTTC |
| MyoDF | CCCCGGCGGCAGAATGGCTACG |
| MyoDR | GGTCTGGGTTCCCTGTTCTGTGT |
| MyoGF | TGAGCATTGTCCAGGCCAG |
| MyoGR | GCTTCTCCCTCAGTGTGGCT |
| Wnt1F | ATGAACCTTCACAACAACGAG |
| Wnt1R | GGTTGCTGCCTCGGTTG |
| Wnt3aF | TAGATGGGTGCGACCTGTTG |
| Wnt3aR | GAACCCTGCTCCCGTGTTAG |
| Wnt4F | CGAGGAGTGCCAATACCAGT |
| Wnt4R | GCCACACCTGCTGAAGAGAT |
| Wnt5aF | GGCGAGCTGTCTACCTGTGG |
| Wnt5aR | GGCGAACGGGTGACCATAGT |
| Wnt7aF | CGACTGTGGCTGCGACAAG |
| Wnt7aR | CTTCATGTTCTCCTCCAGGATCTTC |
RT‐PCR gene arrays
The Mouse Signal Transduction PathwayFinder PCR Array from SABiosciences was used to simultaneously detect gene expression changes of 10 signaling pathways (see details in Table 2). cDNA was synthesized using the RT2 First Strand Kit (this kit contains genomic DNA elimination process) and the PCR Array was run according to the manufacturer's protocol including a threshold of 0.25 and validation of each gene tested by the identification of single peaks in melting curves. Data were analyzed using the RT2 Profiler PCR Array Data Analysis Software; Ct values were normalized to six built‐in reference housekeeping genes, genomic DNA control, reverse transcription control, and positive PCR control. We used this analytical software (http://www.qiagen.com/us/shop/genes-and-pathways/data-analysis-center-overview-page/) to set the statistical significance of upregulation/downregulation of all tested genes at twofold difference. Three independent experiments were performed.
Table 2.
Pathways and Genes in Mouse Signal Transduction PathwayFinder PCR Array
| Signaling pathways | Genes |
|---|---|
| TGFβ pathway | Atf4, Cdkn1b (p27Kip1), Emp1, Gadd45b, Herpud1, Ifrd1, Myc, Tnfsf10 |
| WNT pathway | Axin2, Ccnd1, Ccnd2, Dab2, Fosl1 (Fra‐1), Mmp7 (Matrilysin), Myc, Ppard, Wisp1 |
| NFκB pathway | Bcl2a1a (Bfl‐1/A1), Bcl2l1 (Bcl‐x), Birc3 (c‐IAP1), Ccl5 (Rantes), Csf1 (Mcsf), Icam1, Ifng, Stat1,Tnf |
| JAK/STAT pathway | |
| JAK1, 2/STAT1 | Irf1 |
| STAT3 | Bcl2l1 (Bcl‐x), Ccnd1, Cebpd, Lrg1, Mcl1, Socs3 |
| STAT5 | Bcl2l1 (Bcl‐x), Ccnd1, Socs3 |
| JAK1, 3/STAT6 | Fcer2a, Gata3 |
| p53 pathway | Bax, Bbc3, Btg2, Cdkn1a (p21Cip1/Waf1), Egfr, Fas (Tnfrsf6), Gadd45a, Pcna, Rb1 |
| Notch pathway | Hes1, Hes5, Hey1, Hey2, Heyl, Id1, Jag1, Lfng, Notch1 |
| Hedgehog pathway | Bcl2, Bmp2, Bmp4, Ptch1, Wnt1, Wnt2b, Wnt3a, Wnt5a, Wnt6 |
| PPAR pathway | Acsl3, Acsl4, Acsl5, Cpt2, Fabp1, Olr1, Slc27a4, Sorbs1 |
| Oxidative stress | Fth1, Gclc, Gclm, Gsr, Hmox1, Nqo1, Sqstm1, Txn1, Txnrd1 |
| Hypoxia | Adm, Arnt, Car9, Epo, Hmox1, Ldha, Serpine1 (PAI‐1), Slc2a1, Vegfa |
Intracellular Ca2+ measurements
A Photon Technology International (PTI) imaging system was used to measure intracellular calcium transients. The intracellular calcium transients produced by the stimulation of calcium release from sarcoplasmic reticulum (SR) with 20mM caffeine were measured. Each myotube imaged was loaded with Fura‐2, a ratiometric calcium dye and imaged in real time with the 14‐BIT CoolSNAP CCD camera. All calcium imaging was analyzed with PTI EasyRatioPro fluorescence imaging software. We conduced three to four independent experiments, resulting in 12 to 18 cells analyzed per group.6, 27
Statistical analysis
Data are presented as mean ± SD. Comparisons between more than two groups were made using one‐way ANOVA followed by Tukey's post hoc test for multiple comparisons. For comparisons of differences between two groups the t test was used. A value of p < 0.05 was considered as being significantly different.
Statistical analyses of muscle contractility
Data points that were three SDs away from the mean were considered outliers and removed from the data set. Kolmogorov‐Smirnov test was conducted to test for the normality assumption of the outcomes variables. All variables passed the normality test under the significance level of 0.05. A mixed model approach was conducted to test for the treatment effect while accounting for the repeated measures on the same individual. Pairwise contrasts were estimated and tested for significance between different treatments. Because of the multiple comparisons of the contrasts, a more strict alpha level of 0.025 was used to establish the statistical significance.
Results
MLO‐Y4 CM increased the contractile force of soleus muscle
SOL muscle was treated with blank media as control and with 10% MLO‐Y4 CM. The muscles were stimulated with two alternating frequencies of 20 Hz and 100 Hz. MLO‐Y4 CM increased the contractile force of SOL muscles at the 20 Hz frequency by ∼25% (Fig. 1 A) compared to blank media.
Figure 1.
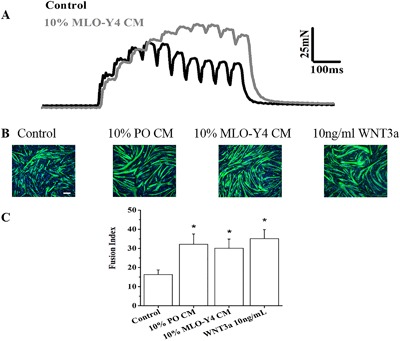
Osteocyte CM and WNT3a modulate the function of skeletal muscle. (A) MLO‐Y4 CM increased the contractile force of ex vivo intact SOL muscles. Original record shows the increase in 20‐Hz force produced by 10% MLO‐Y4 CM, on average the force increased by 25% (5 mice, 10 muscles, p < 0.05). (B) MLO‐Y4 CM, PO CM, and WNT3a enhance myogenic differentiation. Representative fluorescence images of DAPI‐stained nuclei (blue) and myosin heavy chain antibody (MHC, green)‐stained myocytes/myotubes of C2C12 myoblasts after 3 days in 100% DM media (control), 10% PO CM, 10% MLO‐Y4 CM, and 10 ng/mL WNT3a. (C) Summary data for FI of control, PO CM, MLO‐Y4 CM, and WNT3a treatment. The treatment of 10% PO CM, 10% MLO‐Y4 CM, and10 ng/mL WNT3a promoted myogenic differentiation compared to the control (n = 3–5, compared to control, 10% PO CM, p = 0.001; 10% MLO‐Y4 CM, p = 0.004; 10 ng/mL WNT3a, p < 0.001). Scale bar = 100 μm.
MLO‐Y4 CM, PO CM, and WNT3a enhance myogenic differentiation of C2C12 cells, while osteoblast CMs do not enhance myogenesis
C2C12 cells were treated with MLO‐Y4 CM, PO CM, and WNT3a, respectively. At day 3 of differentiation, control and treatment groups cells were stained with Anti‐human Myosin Heavy Chain‐CFS antibody, which only stains maturing myocytes/myotubes not myoblasts, and DAPI, which stains only the nuclei (Fig. 1 B). We also detected the specific FI of each experimental group. These data are summarized in Fig. 1 C, showing that FI is significantly increased in the treatment groups compared with control. When C2C12 cells were treated with the 10% CM deriving from two different osteoblast cell lines, 2T3 (an established osteoblast cell line) and MC3T3‐E1 (a preosteoblast cell line), respectively, there is no significant effects on the myogenic differentiation (Fig. 2 A, B).
Figure 2.
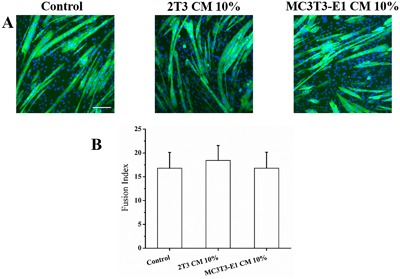
Osteoblast CM does not increase myogenic differentiation. (A) Representative fluorescence images of DAPI‐stained and MHC‐stained C2C12 myoblasts after 3 days in DM media (control), 10% 2T3 CM, and 10% MC3T3‐E1 CM. (B) Summary data for FI of control, 2T3 CM, and MC3T3‐E1 CM treatment. The treatment of 10% 2T3 CM and 10% MC3T3‐E1 CM had no significant effect on myogenic differentiation compared to the control (n = 3–5, compared to control, 10% 2T3 CM, p = 0.670; 10% MC3T3‐E1 CM, p = 1.000). Scale bar = 100 μm.
Wnt3a mRNA is expressed in MLO‐Y4 cell and 2T3 osteoblast cells and the expression increased after FFSS
Wnt1, Wnt3a, Wnt4, Wnt5a, and Wnt7a are involved in myogenesis (reviewed in von Maltzahn and colleagues16). To detect the gene expression of Wnt1, Wnt3a, Wnt4, Wnt5a, and Wnt7a, we performed RT‐qPCR in MLO‐Y4 and 2T3 cells. Our results show that Wnt3a is expressed in bone cells and is modulated by FFSS. These results are summarized and explained in detail in the legend of Fig. 3 A–F.
Figure 3.
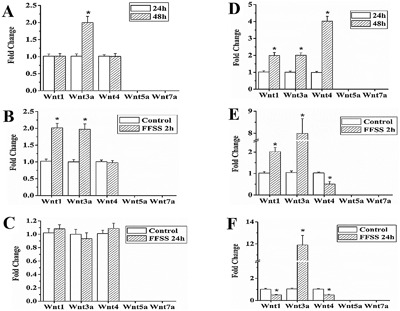
Expression of Wnt3a and other Wnt genes in MLO‐Y4 cells and 2T3 cells. Summary data of RT‐qPCR results at 24 and 48 hours of culture and 2 hours and 24 hours after FFSS in MLO‐Y4 and 2T3 cells (A–F). (A) Wnt3a, Wnt1, and Wnt4 gene expression was detected in MLO‐Y4 cells cultured for 24 and 48 hours. At 48 hours, compared to 24 hours, the expression of Wnt3a increased significantly by 1.995‐fold, while the expression of Wnt1 and Wnt4 was not changed. (B) In MLO‐Y4 cells after FFSS treatment, compared to control, the expression of Wnt1 and Wnt3a increased significantly by 1.95‐fold and 2.0‐fold, respectively. (C) No significant difference in expression of Wnt3a, Wnt1, and Wnt4 was detected at 24 hours after FFSS treatment, compared to control. (D) In 2T3 cells, at 48 hours, compared to 24 hours, the expression of Wnt3a, Wnt1, and Wnt4 increased significantly by 2.01‐fold, 1.99‐fold, and 4.02‐fold, respectively. (E) In 2T3 cells after FFSS treatment, compared to control, the expression of WNT3a and Wnt1 increased significantly by 2.02‐fold and 7.98‐fold, respectively, and expression of Wnt4 decreased by 50% (p = 0.004). (F) Compared to control, 24 hours after FFSS, the expression of Wnt3a increased by 11.9‐fold and Wnt1 and Wnt4 decreased by 50.1% (p = 0.001) and 50.7% (p = 0.001), respectively (n = 3). The expression of Wnt5a and Wnt7a was undetectable under all conditions.
ELISA for WNT3a in PO CM and MLO‐Y4 CM
The only commercially available ELISA kit was used to detect WNT3a in MLO‐Y4 CM and PO CM and the factor was detected in the 100 to 200 pg/mL range. Although this level might be considered low, we do see effects in vitro with WNT3a concentrations as low as 500 pg/mL. Furthermore, there are many technical reasons that might explain these results, including the fact that it is technically challenging to isolate and purify WNT3A (http://web.stanford.edu/group/nusselab/cgi-bin/wnt/purification).
Physiological concentrations of WNT3a and low concentrations of MLO‐Y4 CM stimulate myogenesis of C2C12 cells
Studies were performed with concentrations of WNT3a ranging from 0.1 to 10 ng/mL as well as 1% MLO‐Y4 CM. A concentration as low as 0.5 ng/mL to 10 ng/mL WNT3a consistently enhanced the C2C12 cell differentiation (Fig. 4 A, C). In agreement with lower concentrations of WNT3a accelerating myogenesis, we also found that the myogenic differentiation of C2C12 cells was enhanced with as low as 1% MLO‐Y4 CM. After 3 days of differentiation, a significant increase of the FI of MLO‐Y4 CM‐treated cells was found as compared with the control cells (Fig. 4 B, D).
Figure 4.
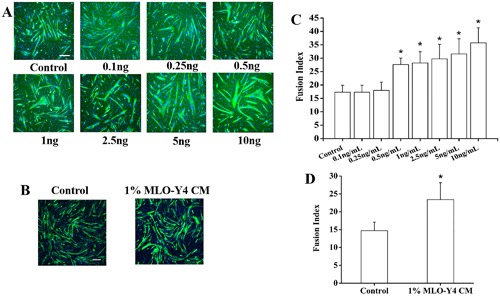
Lower concentrations, more physiological levels of WNT3a and 1% MLO‐Y4 CM enhance C2C12 myoblast differentiation. Fluorescence images of DAPI‐stained and MHC‐stained myocytes/myotubes of C2C12 cells at differentiation day 3 after WNT3a treatment (A) and 1% MLO‐Y4 CM (B). (C) Summary data for FI for treatments ranging from 0.1 ng/mL to 10 ng/mL WNT3a. FI increased significantly for concentration ranging from 0.5 to 10 ng/mL WNT3a compared to control (n = 3–5; compared to control, 0.5 ng/mL WNT3a, p = 0.028; 1 ng/mL WNT3a, p = 0.016; 2.5 ng/mL WNT3a, p = 0.004; 5 ng/mL WNT3a, p = 0.001; 10 ng/mL WNT3a, p < 0.001). (D) Summary data for 1% MLO‐Y4 CM‐treated C2C12 cells at differentiation day 3. FI increased significantly for 1% MLO‐Y4 CM compared to control (n = 3–5, p = 0.014). Scale bar = 100 μm.
MLO‐Y4 CM, PO CM, and WNT3a induced translocation of β‐catenin to the nucleus of C2C12 cells
To explore the possible roles of Wnt/β‐catenin pathway in C2C12 cells, confocal microscopy was used to study the time course of the subcellular localization of β‐catenin after C2C12 cells were treated with 10% MLO‐Y4 CM, which demonstrated that peak translocation occurred at 60 min (Fig. 5 A, B). Next, we tested the effects of C2C12 GM, C2C12 DM, MLO‐Y4 CM, PO CM, WNT3a, and LiCl as a positive control. The results demonstrated that β‐catenin translocation to the nucleus was enhanced significantly by the different treatments when compared to GM. Furthermore, MLO‐Y4 CM, PO CM, WNT3a, and LiCl treatments were more potent than DM in inducing nuclear translocation of β‐catenin (Fig. 5 C, D).
Figure 5.
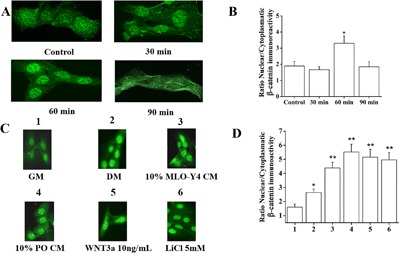
MLO‐Y4 CM, PO CM, and WNT3a induced nuclear translocation of β‐catenin in C2C12 myoblasts. (A) Immunostaining for β‐catenin of C2C12 myoblasts at 30 min, 60 min, and 90 min after treatment with 10% MLO‐Y4 CM. (B) Quantification of the nuclear β‐catenin staining showed that the maximum induction of nuclear translocation of β‐catenin in C2C12 myoblasts occurred at 60 min (n = 3, 5–10 cells per experiment, p < 0.001). (C) Confocal images of immunostaining for β‐catenin of C2C12 cells cultured in GM, DM, treated 60 min in DM with 10% PO CM, 10% MLO‐Y4 CM, 10 ng/mL WNT3a, and LiCl was used as positive control. (D) Summary data show that compared to GM, DM promoted significantly higher nuclear translocation of β‐catenin (n = 3, 5–10 cells per experiment, p = 0.023) and compared to DM, PO CM, MLO‐Y4 CM, WNT3a, and LiCl induced significantly higher nuclear translocation of β‐catenin (compared to DM, PO CM, p < 0.001; MLO‐Y4 CM, p = 0.003; WNT3a, p < 0.001; LiCl, p < 0.001).
Treatment of C2C12 cells with MLO‐Y4 siRNA‐treated CM
To explore the roles of WNT3a from MLO‐Y4 CM on C2C12 cells, MLO‐Y4 osteocytes were transfected with Wnt3a siRNA. To detect siRNA transfection efficiency, MLO‐Y4 osteocytes were transfected with 10 nM TYE 563 DS Transfection Control siRNA. Twenty‐four hours after transfection, images were taken, and the percentage of fluorescent positive cells was determined. The result revealed that the siRNA transfection efficiency of MLO‐Y4 osteocytes was 80.02% ± 7.18% (Fig. 6 A). RT‐qPCR was conducted to examine Wnt3a expression after knockdown by Wnt3a siRNA. The results indicated that the relative Wnt3a mRNA expression in the siRNA‐treated cells was 20.47% ± 4.72% of the negative control cells when normalized to expression of Gapdh, which was not altered by the negative control, vehicle control, or siRNA treatments (Fig. 6 A); therefore, ∼80% of Wnt3a expression was knocked down. MLO‐Y4 cells were treated with Wnt3a siRNA, and the CM (MLO‐Y4 siRNA‐treated CM) was collected. C2C12 cells were treated with MLO‐Y4 siRNA‐treated CM. At day 3 of differentiation, we measured the specific FI of each experimental group. These data are summarized in Fig. 6 B, C, showing that in the cells treated with MLO‐Y4 CM or the MLO‐Y4 CM siRNA negative control, FI significantly increases, while in the cells treated with the MLO‐Y4 siRNA‐treated CM, FI did not increase and it is very similar to the DM control. (There was no difference between negative control and vehicle control).
Figure 6.
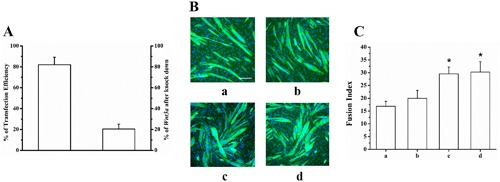
Wnt3a knockdown in MLO‐Y4 cells impairs CM capacity to enhance myogenic differentiation. MLO‐Y4 cells were transfected with Wnt3a siRNA. (A) Twenty hours after transfection, the transfection efficiency equaled 80.02% ± 7.18%, leading Wnt3a expression decreased to 20.47% ± 4.72% compared to negative control (n = 3). (B) Fluorescence images of DAPI‐stained and MHC‐stained C2C12 cells at differentiation day 3 after treatment of MLO‐Y4 siRNA‐treated CM. (C) Summary data for FI for treatments ranging from DM control (a), 10% MLO‐Y4 siRNA‐treated CM (b), 10% CM from MLO‐Y4 cells treated with negative control siRNA (c) and 10% MLO‐Y4 CM as positive control (d). FI significantly increased in 10% CM from MLO‐Y4 cells treated with negative control siRNA and positive control compared to DM control, while no significant difference was found in 10% MLO‐Y4 siRNA‐treated CM treatment (n = 3; compared to DM control, b, p = 0.318; c, p < 0.001; d, p < 0.001). Scale bar = 100 μm.
Treatment of C2C12 cells with Sclerostin inhibits the effects of MLO‐Y4 CM and WNT3a on myogenic differentiation
Sclerostin is a Wnt signaling inhibitor.(12) To study the effect of Sclerostin on C2C12 myoblast differentiation stimulation by MLO‐Y4 CM and WNT3a, C2C12 myoblasts were pretreated with 100 ng/mL Sclerostin for 6 hours prior to the addition of MLO‐Y4 CM and WNT3a. At day 3 of differentiation, we measured the specific FI of each experimental group which was represented in Fig. 7 A. FI data of each group are summarized in Fig. 7 B, showing that compared to negative control (a) no effect was detected in groups b, e, and f, while a significant effect was found for groups c, d, and g; FI of treatment group (b): C2C12 myoblasts were treated with 100 ng/mL Sclerostin in DM for 6 hours; treatment group (e): C2C12 myoblasts were treated with 100 ng/mL Sclerostin in DM for 6 hours, followed by 10% MLO‐Y4 CM for 48 hours; treatment group (f): C2C12 myoblasts were treated with 100 ng/mL Sclerostin in DM for 6 hours, followed by 10 ng/mLWNT3a for 48 hours was not significantly changed, and significantly reduced when compared to the positive controls (c, d), 10% MLO‐Y4 CM and 10 ng/mL WNT3a, respectively. The FI of treatment group (g): C2C12 myoblasts were treated with 100 ng/mL Sclerostin in DM for 6 hours, followed by the combination of 10% MLO‐Y4 CM + 10 ng/mLWNT3a for 48 hours; increased significantly compared with negative control (a), suggesting that only the MLO‐Y4 CM + WNT3a together can overcome the inhibitory effect of Sclerostin.
Figure 7.
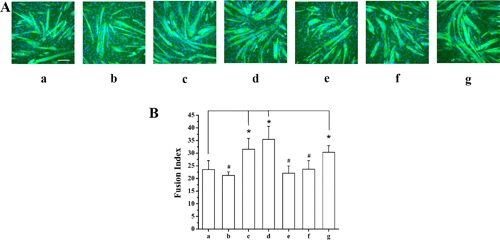
Sclerostin inhibits the effects of MLO‐Y4 CM and WNT3a on myogenic differentiation. (A) Fluorescence images of DAPI‐stained and MHC‐stained C2C12 cells at differentiation day 3. (B) Summary data for FI for treatment groups: (a) DM without Sclerostin treatment as a negative control, (b) DM with 100 ng/mL Sclerostin for 6 hours, (c) DM with 10% MLO‐Y4 CM for 48 hours as one of our positive controls, (d) DM with 10 ng/mL WNT3a for 48 hours as our second positive control, (e) DM with 100 ng/mL Sclerostin for 6 hours, followed by 10% MLO‐Y4 CM for 48 hours, (f) DM with 100 ng/mL Sclerostin for 6 hours, followed by 10 ng/mL WNT3a for 48 hours, and (g) DM with 100 ng/mL Sclerostin for 6 hours, followed by the combination of 10% MLO‐Y4 CM + 10 ng/mL WNT3a for 48 hours. #No significant difference of FI was found for groups (b), (e), and (f), compared to negative control (a), respectively. (n = 3–6; compared to negative control, b, p = 0.925; e, p = 0.971; f: p = 1.000). *FI increased significantly for group (g) and positive controls (c and d), compared to negative control. (n = 3–6; compared to negative control, g, p = 0.030; c, p = 0.007; d, p < 0.001). Scale bar = 100 μm.
Modulation of gene expression by MLO‐Y4 CM and WNT3a
We employed the Mouse Signal Transduction PathwayFinder PCR Array to detect differences in expression of signaling molecules between control versus MLO‐Y4 CM (10%) and WNT3a (10 ng/mL) treated C2C12 cells at 24 hours after treatment. A major advantage of this technology is that it monitors 10 different pathways simultaneously and is validated at the protein level. We found a clear common shared gene regulation change between 10% MLO‐Y4 CM (Fig. 8 A) and 10 ng/mL WNT3a (Fig. 8 B), which was the upregulation of the Wnt pathway detected by the overexpression of Wnt1 in the cells treated with CM and Wnt5a and Axin2 in the cells treated with WNT3a.
Figure 8.
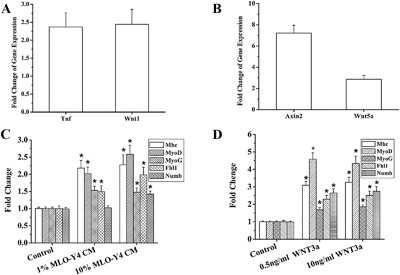
RT‐PCR Gene Array and RT‐qPCR detection of gene alterations in C2C12 cells after MLO‐Y4 CM and WNT3a treatment. Summary data from 3 independent experiments for the Mouse Signal Transduction Pathway Finder PCR Array. (A) In 10% MLO‐Y4 CM, the expression of Tnf and Wnt1 were significantly increased (p = 0.031, p = 0.022, respectively). (B) When cells treated with 10 ng/mL WNT3a, the expression of Axin2 and Wnt5a increased significantly (p < 0.001, p = 0.011, respectively). Summary data of RT‐qPCR detection of gene expression changes for Mhc, MyoD, MyoG, Fhl1, and Numb at differentiation day3 after 1% MLO‐Y4 CM and 10% MLO‐Y4 CM (C) and 0.5 ng/mL and 10 ng/mL WNT3a treatment (D). The results demonstrated that both 1% and 10% MLO‐Y4 CM, as well as 0.5 ng and 10 ng/mL WNT3a (except Numb in 1% MLO‐Y4 CM, p = 0.084) significantly increased the expression of these five specific genes as compared to control (n = 3, 2 replicates). Mhc, MyoD, MyoG, and Fhl1 increased significantly after 1% MLO‐Y4 CM treatment (p = 0.002, p = 0.003, p = 0.004, p < 0.043, respectively). Mhc, MyoD, MyoG, Fhl1, and Numb increased significantly after 10% MLO‐Y4 CM treatment (p = 0.001, p < 0.001, p = 0.006, p = 0.002, p < 0.001, respectively). After 0.5 ng/mL WNT3a treatment, Mhc, MyoD, MyoG, Fhl1, and Numb increased significantly (p = 0.004, p = 0.002, p = 0.001, p = 0.001, p < 0.001, respectively). After 10 ng/mL WNT3a treatment, Mhc, MyoD, MyoG, Fhl1, and Numb increased significantly (p = 0.006, p = 0.001, p < 0.001, p < 0.001, p < 0.001, respectively).
We also employed RT‐qPCR to detect the specific expression of myogenic differentiation markers (Mhc, MyoD, and MyoG) and two Wnt/β‐catenin downstream genes directly associated with C2C12 myogenesis (Fhl1,29 Numb 30). At day 3 of myotube differentiation, we found that both 1% and 10% MLO‐Y4 CM, as well as 0.5 ng and10 ng/mL WNT3a (except Numb in 1% MLO‐Y4 CM) significantly increased the expression of these five specific genes (Fig. 8 C, D).
MLO‐Y4 CM and WNT3a enhance caffeine‐induced SR calcium release
Calcium homeostasis is very important in myoblast differentiation,31 and is a major surrogate of skeletal muscle function.27, 31 Avila and colleagues32 have demonstrated that WNT3a modulates intracellular Ca2+ in hippocampal neurons. Furthermore, because our ex vivo muscle contractility experiments showed that the force produced by the lower frequency of stimulation was increased, to determine if SR calcium release was influenced by either MLO‐Y4 CM or WNT3a, we employed an effective approach by testing the direct effects of caffeine's ability to release calcium from the SR in Fura‐2 loaded myotubes. In day 5 of myotube differentiation, caffeine responses were significantly larger in myotubes treated with 10% MLO‐Y4 CM and 10 ng/mL WNT3a as indicated by multiple and oscillatory peaks and a prolongation of the relaxation phase of the calcium transient response induced by caffeine (Fig. 9 A).
Figure 9.
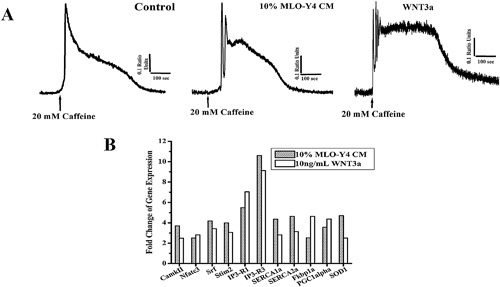
MLO‐Y4 CM and WNT3a enhance Ca2+ release from the SR and modulate expression of key Ca2+ signaling/homeostasis genes. (A) Representative Ca2+ transient of C2C12 myotubes loaded with Fura‐2/AM exposed to 20 mM caffeine (arrows). MLO‐Y4 CM and WNT3a treatment increased caffeine induced Ca2+ release in C2C12 myotubes. In 10% MLO‐Y4 CM and 10 ng/mL WNT3a‐treated C2C12 myotubes, multiple oscillatory Ca2+ peaks in response to caffeine were observed and the relaxation phase of the transient was significantly longer (p < 0.05). (B) Increased expression of key genes related to Ca2+ signaling/hypertrophy, Ca2+ homeostasis, mitochondrial biogenesis, and oxidative stress in C2C12 cells at 60 hours of differentiation after treatments was detected by a focused‐muscle custom‐built RT‐PCR gene array. Two PCR arrays provided similar results.
MLO‐Y4 CM and WNT3a induced common gene overexpression profile of Ca2+ signaling/hypertrophy, Ca2+ homeostasis, mitochondrial biogenesis, and oxidative stress genes
Expression of important genes related to Ca2+ signaling/hypertrophy, Ca2+ homeostasis, mitochondrial biogenesis, and oxidative stress in C2C12 cells at 60 hours of differentiation after treatments was detected by a focused custom‐built RT‐PCR gene array using the same technology that the PathwayFinder was used. In agreement with both the enhanced contractile force and SR calcium release by MLO‐Y4 CM and WNT3a, the expression of these key regulatory pathways was significantly upregulated (Fig. 9 B, Table 3).
Table 3.
Pathways and Genes in Focused Custom‐Built RT‐PCR Gene Array
Discussion
Recent studies have shown that bone (osteoblasts and osteocytes) acts as an endocrine organ by the production and secretion of at least two circulating factors, FGF23 and osteocalcin.7 Osteocytes also secreted large amounts of PGE2, which we previously demonstrated to be a promoter of C2C12 myoblasts myogenesis3 and proliferation.33
Our concept is that bone‐muscle crosstalk enhances myogenic differentiation but may also have effects on muscle function. In this study, we showed that 10% MLOY4 CM and 10% PO CM enhanced C2C12 cell differentiation in vitro. In contrast, the differentiation was not significantly affected when C2C12 cells were treated with 10% 2T3 CM and 10% MC3T3‐E1 CM, respectively. In support of this we found that 10% MLOY4 CM increased the force production of SOL muscles stimulated ex vivo at lower frequencies of stimulation, suggesting that osteocytes release soluble factors that can either promote increased calcium release from the SR or enhance the sensitivity of the contractile machinery to calcium. Consistent with this, we showed that the caffeine‐induced calcium release from the SR was significantly increased in C2C12 myotubes treated with MLO‐Y4 CM and WNT3a. Another major motivation for the current study was the observation that WNT3a can activate MyoD and Myogenin,34 and these two genes are important myogenic regulatory factors. WNT3a, which is found in the extracellular matrix of different cell types,35 is expressed in C2C12 cells and 2T3 osteoblasts,36, 37, 38 and is detected at relatively high levels in serum of both humans (16.9 ± 2.4 ng/mL) and mice (0.225 to 3.74 ng/mL),39 suggesting a potential role of WNT3a in bone‐muscle crosstalk. In this study, we used RT‐qRCR to detect Wnt3a expression after C2C12 myoblasts were treated with MLO‐Y4 CM, and found that Wnt3a expression was not altered (data not shown). We detected WNT3a in the ∼200 pg/mL range in MLO‐Y4 CM and PO CM CMs by using the only available commercial ELISA kit, and observed stimulation of myogenesis with as little as 500 pg/mL. Although this difference seems large, when considering the log scale for the effects of drugs (ie, dose‐response relationship) perhaps this difference is not as profound as it appears to be. In addition, there are many technical reasons that might explain these results, including the fact that it is technically challenging to isolate and purify WNT3a (see http://web.stanford.edu/group/nusselab/cgi-bin/wnt/purification). Furthermore, it is possible that the continuous and chronic exposure of cells to picogram levels of WNT3a might lead to a host of cell biological effects.
To determine Wnt3a expression in MLO‐Y4 and 2T3 cells, we investigated it at different time points and under static and FFSS conditions; and found that Wnt3a expression was modulated by FFSS. This finding is consistent with the results of Santos and colleagues,40 who reported that MLO‐Y4 cells subjected to Pulsating Fluid Flow (PFF) showed increased expression of Wnt3a. Jia and colleagues41 found similar results in osteoblast cells stimulated with PFF. These results might suggest that with increased levels of activity, bone‐secreted WNT3a could increase, leading to signaling to muscle cells that modulate muscle function.
When C2C12 cells were treated with either 10% MLO‐Y4 CM or PO CM, and 10 ng/mL WNT3a, there was an enhancement of β‐catenin translocation into the nucleus, providing evidence that the Wnt/β‐catenin pathway is activated by CM and also by WNT3a. In contrast, when C2C12 cells were treated with either 10% 2T3 CM or 10% MC3T3‐E1 CM (osteoblast CMs), we did not observe any enhancement of myogenic differentiation, suggesting that factors secreted in the osteoblast CM are either different in nature or in concentration. This concept is further supported by the data from osteocytes showing that even 1% CM can enhance myogenic differentiation (Fig. 4 B, D).
To directly test the involvement of WNT3a as a mediator of the effects of CM, we treated MLO‐Y4 cell with Wnt3a siRNA and collected the CM (MLO‐Y4 siRNA‐treated CM) (Fig. 6). When C2C12 cells were treated with 10% MLO‐Y4 siRNA‐treated CM, there was no significant change of C2C12 cell differentiation. This indicated that WNT3a is a key factor in MLO‐Y4 CM responsible for the enhancement effect of C2C12 cell differentiation.
Next, to further explore if Wnt/β‐catenin pathway was involved in the effect of MLO‐Y4 CM and WNT3a on C2C12 cell differentiation enhancement, we pretreated C2C12 cells with Sclerostin, which is a negative regulator of the Wnt/β‐catenin signaling pathway. Sclerostin inhibited the capacity of 10% MLO‐Y4 CM and 10 ng/mL WNT3a to enhance C2C12 cell differentiation, respectively, but failed to inhibit myogenic differentiation acceleration when CM and WNT3a were combined in the same treatment. These results support that the Wnt/β‐catenin signaling pathway is involved in MLO‐Y4 CM and WNT3a enhancement of C2C12 cell differentiation. The reason that the combined treatment with MLO‐Y4 CM and WNT3a together enhanced C2C12 cell differentiation may be because factors in MLO‐Y4 CM (such as WNT3a, WNT1, etc.) and WNT3a together reached a certain threshold that they competed with Sclerostin to bind to Lrp5 and Lrp6, then decreased the inhibition of Sclerostin on the Wnt/β‐catenin signaling pathway. Furthermore, other factors in CM such as PGE2 could be contributing to the combined effects of CM and WNT3a.
To search for molecular insights into these effects of CM and WNT3a, we used the Mouse Signal Transduction PathwayFinder PCR Array and found that in 10% MLO‐Y4 CM treatment, the expression of two protein coding genes Tnf and Wnt1 were significantly increased (Fig. 8 A). Tnf has been found to exert many of its effects on skeletal muscle through NFκB signaling.42 It has been reported that mechanical stimulation of myoblasts leads to release of TNF, which is necessary for myogenic differentiation.43 TNF is known to be one of the key regulators of skeletal muscle cell responses to injury. Whereas physiological levels of Tnf are transiently upregulated during myoblast regenerative responses to injury and stimulate differentiation, sustained high levels of TNF are associated with chronic inflammatory diseases and especially with muscle pathology associated with impairment of differentiation and muscle wasting. Wnt1 is directly involved in myogenesis.44 Based on this, we propose that the myogenic effect of MLO‐Y4 CM may be predominantly due to Tnf and Wnt1 activation. Because our data also show that WNT1 can enhance myogenesis (Supporting Fig. 1A–C), the Wnt1 induced by MLOY4 CM might contribute to autocrine stimulation of the Wnt/β‐catenin signaling pathway. An important question raised by our experiments with CMs and WNT3a was whether other Wnts might exert similar effects. We tested this possibility by treating C2C12 myoblasts with WNT1 ranging from 0.5 to 100 ng/mL. Interestingly, we found that the lower concentrations of WNT1 did not increase differentiation, but 10 ng/mL, 50 ng/mL, and 100 ng/mL of WNT1 significantly enhanced myogenic differentiation in C2C12 cells, although even at these higher concentrations WNT1 was less potent than WNT3a.
When C2C12 cells were treated with 10 ng/mL WNT3a, the expression of two genes (Axin2, Wnt5a) increased significantly (Fig. 8 B). Axin2 associates directly with β‐catenin, GSK‐3β, and adenomatous polyposis coli (APC) and is induced by active Wnt signaling and acts in a negative feedback loop.16 Thus, upregulation of this gene indicates activation of the Wnt/β‐catenin pathway. Wnt5a is involved in modulation of the noncanonical Wnt pathway, which regulates calcium inside cells.45 Therefore providing strong evidence that WNT3a is acting via the Wnt/β‐catenin pathway and mobilizing intracellular calcium.
In order to further confirm the enhancement of myogenic differentiation by both CM and WNT3a, we monitored with RT‐qPCR the expression of two muscle regulatory factors (MRFs) (MyoD and Myogenin) and one marker of terminal differentiation (Mhc) (Fig. 8 C, D). Our results showed upregulation of these genes. To further dissect the mechanisms underlying these effects, we also measured the expression of two Wnt/β‐catenin pathway downstream genes (Fhl1, Numb), which were both upregulated. Fhl1 was recently shown to be a downstream gene of the Wnt signaling pathway that promotes differentiation of C2C12 cells,29 and it has been reported that the expression of Numb was increased during C2C12 cell differentiation and that treatment with WNT3a increased both Numb gene and protein expressions.30
Therefore, although not exactly the same genes, CM and WNT3a seem to be activating the Wnt/β‐catenin pathway and upregulating MRFs. In addition, striking similarities of overlapping gene regulation by MLO‐Y4 CM and WNT3a were revealed by our custom‐built muscle‐specific RT‐PCR gene array as summarized in Fig. 9 B. Our muscle‐specific RT‐PCR gene array results allowed us to show that both CM and WNT3a upregulated genes associated to pathways linked to Ca2+ signaling/hypertrophy, Ca2+ homeostasis, mitochondrial biogenesis, and oxidative stress (Table 3). In support of our observations of enhanced force generation by SOL muscles induced by MLOY4 CM and increased myogenic differentiation and enhanced calcium released induced by both CM and WNT3a, we found overexpression of specific genes associated with SR calcium release (IP3Rs 46, 47, 48 and Fkbp1a 49, 50), SR calcium uptake (SERCAs 51), and store‐operated calcium entry/myoblast differentiation (Stim 2).52 These studies also revealed that three key Ca2+ signaling genes (CamkII,53, 54 Nfatc3,52 and Srf 55) critical for myotube homeostasis and cellular hypertrophy were also upregulated as well as two genes associated with mitochondrial biogenesis and reduction–oxidation reaction (REDOX) regulation (PGC1α 56 and SOD1 57, 58). Our results are also in agreement with previous reports that suggested the maintenance of normal intracellular Ca2+ concentration is vital to myocyte gene expression and differentiation.31 It is also intriguing that Avila and colleagues32 reported that WNT3a modulates intracellular Ca2+ in hippocampal neurons, in fact, in a manner similar to that we have observed in muscle cells, suggesting that this might be a common mechanism in excitatory cells. We believe that the modulation in intracellular Ca2+ might work as the biophysical linker between the molecular‐genetic adaptations and the changes (phenotypes) induced by WNT3a in muscle cells.
A recurring question in this emerging field of bone‐muscle crosstalk is how secreted factors from bone and muscle can reach each other. Experiments performed by Beno and colleagues59 show that mouse tail vein injection of small dyes and molecules up to 70 kDa can permeate the osteocyte‐lacunar‐canalicular network in just a few minutes, which is bigger than the size of both WNT3a and WNT1(∼40 kDa), and in fact the majority of WNTs. This demonstrates that the canalicular fluid has ready access to the circulation and suggests that factors secreted by osteocytes could enter the blood and have effects on distant target cells, including muscle. Furthermore, osteocytes are known to be the main producers of FGF23, which is readily detected in the serum, supporting an endocrine role for the osteocyte (reviewed in Dallas and colleagues7). Another potential mechanism for the transport of factors between tissues is through microvesicles or extracellular vesicles (EVs), cell‐derived membrane vesicles secreted by all cell types. The release of EVs is now recognized as an important modulator of crosstalk between many cells types, and in disease states.60 Recently, McBride and colleagues61 demonstrated that bone marrow mesenchymal stem cell–derived exosomes transport WNT3a and enhance dermal fibroblast proliferation and angiogenesis in vitro.
Conclusion
Obviously, a limitation of the current study is that we are not studying bone‐muscle crosstalk in vivo because this was beyond the scope of the current work. Importantly, the cell models and the ex vivo work did support the concept of bone to muscle signaling. In conclusion, our data showed that MLO‐Y4 cells and primary osteocytes produce WNT3a. Bone‐secreted factors contained in CM and WNT3a enhanced C2C12 cell differentiation through mechanisms associated with stimulation of the Wnt/β‐catenin pathway and muscle regulatory factors. Osteocyte CM also increased the contractile function of ex vivo soleus muscles, which might be explained by the increased SR calcium release and the concurrent modulation of intracellular Ca2+ signaling/homeostasis. Furthermore, this modulation of Ca2+ signaling might serve another purpose, by linking the effects of WNT3a through the activation of the Wnt/β‐catenin pathway, leading to specific phenotypic changes. From our in vitro and ex vivo data, WNT3a could be one of the bone cell secreted factors that promote differentiation of C2C12 myoblast. Our results support a concept that muscle and bone are not only communicating via mechanical interaction, but also that bone can secrete factor(s) which can act in a paracrine or endocrine fashion to support muscle differentiation and function.
Disclosures
All authors state that they have no conflicts of interest.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Supporting Figure S1.
Acknowledgments
This study was directly funded by NIH‐NIA P01 AG039355 (to LFB, MB), the George W. and Hazel M. Jay Endowment (to MB), and the UT System Science and Technology Acquisition and Retention Program (STARS) (to MB). Dr. Yukiko Kitase prepared MLO‐Y4 CM and MC3T3E‐1 CM and Ms. Jennifer Rosser prepared 2T3 CM for this study.
Authors’ roles: Study design: SLD, MLJ, LFB, and MB. Study conduct: JH, SR, NL, CM, SK, and LB. Data collection: JH, SR, NL, CM, SK, and LB. Data analysis: JH, SR, NL, LFB, and MB. Data interpretation: JH, SLD, MLJ, LFB, and MB. Drafting manuscript: JH and MB. Revising manuscript: JH, NL, CM, SLD, MLJ, LFB, and MB. Approving final version of manuscript: MB. MB takes responsibility for the integrity of the data analysis.
Current address: Simon Kaja, Department of Ophthalmology, Stritch School of Medicine, Loyola University Chicago, 2160 S First Ave., Maywood, IL, 60153, USA.
Current address: Lynda F Bonewald, Indiana Center for Musculoskeletal Health, School of Medicine, Indiana University, Barnhill Drive, Indianapolis, IN 46202, USA.
Current address: Jian Huang, Chenglin Mo, Leticia Brotto, and Marco Brotto, College of Nursing and Health Innovation, University of Texas at Arlington, 411 S. Nedderman Dr, Arlington, TX 76019, USA.
Current address: Katharina Jähn, Emmy Noether Research Group for Bone Material Quality, Department of Osteology and Biomechanics (IOBM), University Medical Center Hamburg‐Eppendorf (UKE) Lottestr. 55a, 22529 Hamburg, Germany.
References
- 1. Kaji H. Linkage between muscle and bone: common catabolic signals resulting in osteoporosis and sarcopenia. Curr Opin Clin Nutr Metab Care. 2013; 16 (3):272–7. [DOI] [PubMed] [Google Scholar]
- 2. Burge R, Dawson‐Hughes B, Solomon DH, Wong JB, King A, Tosteson A. Incidence and economic burden of osteoporosis‐related fractures in the United States, 2005–2025. J Bone Miner Res. 2007; 22(3):465–75. [DOI] [PubMed] [Google Scholar]
- 3. Mo C, Romero‐Suarez S, Bonewald L, Johnson M, Brotto M. Prostaglandin E2: from clinical applications to its potential role in bone‐ muscle crosstalk and myogenic differentiation. Recent Pat Biotechnol. 2012; 6(3):223–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hamrick MW. A role for myokines in muscle‐bone interactions. Exerc Sport Sci Rev. 2011; 39 (1):43–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jahn K, Lara‐Castillo N, Brotto L, et al. Skeletal muscle secreted factors prevent glucocorticoid‐induced osteocyte apoptosis through activation of beta‐catenin. Eur Cell Mater. 2012; 24:197–209; discussion 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang J, Hsu YH, Mo C, et al. METTL21C is a potential pleiotropic gene for osteoporosis and sarcopenia acting through the modulation of the NF‐kappaB signaling pathway. J Bone Miner Res. 2014; 29(7):1531–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dallas SL, Prideaux M, Bonewald LF. The osteocyte: an endocrine cell… and more. Endocr Rev. 2013; 34 (5):658–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shah AD, Shoback D, Lewiecki EM. Sclerostin inhibition: a novel therapeutic approach in the treatment of osteoporosis. Int J Womens Health. 2015; 7:565–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brunkow ME, Gardner JC, Van Ness J, et al. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot‐containing protein. Am J Hum Genet. 2001; 68 (3):577–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Winkler DG, Sutherland MK, Geoghegan JC, et al. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 2003; 22 (23):6267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poole KE, van Bezooijen RL, Loveridge N, et al. Sclerostin is a delayed secreted product of osteocytes that inhibits bone formation. FASEB J. 2005; 19 (13):1842–4. [DOI] [PubMed] [Google Scholar]
- 12. Li X, Zhang Y, Kang H, et al. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem. 2005; 280 (20):19883–7. [DOI] [PubMed] [Google Scholar]
- 13. Semenov M, Tamai K, He X. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J Biol Chem. 2005; 280 (29):26770–5. [DOI] [PubMed] [Google Scholar]
- 14. Clevers H, Nusse R. Wnt/beta‐catenin signaling and disease. Cell. 2012; 149 (6):1192–205. [DOI] [PubMed] [Google Scholar]
- 15. Kato M, Patel MS, Levasseur R, et al. Cbfa1‐independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002; 157 (2):303–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. von Maltzahn J, Chang NC, Bentzinger CF, Rudnicki MA. Wnt signaling in myogenesis. Trends Cell Biol. 2012; 22 (11):602–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Park KH, Brotto L, Lehoang O, Brotto M, Ma J, Zhao X. Ex vivo assessment of contractility, fatigability and alternans in isolated skeletal muscles. J Vis Exp. 2012;(69):e4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brotto M, Brotto L, Jin JP, Nosek TM, Romani A. Temporal adaptive changes in contractility and fatigability of diaphragm muscles from streptozotocin‐diabetic rats. J Biomed Biotechnol. 2010; 2010:931903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Paula Brotto M, van Leyen SA, Brotto LS, Jin JP, Nosek CM, Nosek TM. Hypoxia/fatigue‐induced degradation of troponin I and troponin C: new insights into physiologic muscle fatigue. Pflugers Arch. 2001; 442(5):738–44. [DOI] [PubMed] [Google Scholar]
- 20. Kato Y, Windle JJ, Koop BA, Mundy GR, Bonewald LF. Establishment of an osteocyte‐like cell line, MLO‐Y4. J Bone Miner Res. 1997; 12(12):2014–23. [DOI] [PubMed] [Google Scholar]
- 21. Yang W, Lu Y, Kalajzic I, et al. Dentin matrix protein 1 gene cis‐regulation: use in osteocytes to characterize local responses to mechanical loading in vitro and in vivo. J Biol Chem. 2005; 280 (21):20680–90. [DOI] [PubMed] [Google Scholar]
- 22. Bonewald LF, Harris SE, Rosser J, et al. von Kossa staining alone is not sufficient to confirm that mineralization in vitro represents bone formation. Calcif Tissue Int. 2003; 72 (5):537–47. [DOI] [PubMed] [Google Scholar]
- 23. Stern AR, Stern MM, Van Dyke ME, Jahn K, Prideaux M, Bonewald LF. Isolation and culture of primary osteocytes from the long bones of skeletally mature and aged mice. Biotechniques. 2012; 52 (6):361–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stern AR, Bonewald LF. Isolation of osteocytes from mature and aged murine bone. Methods Mol Biol. 2015; 1226:3–10. [DOI] [PubMed] [Google Scholar]
- 25. Kitase Y, Barragan L, Qing H, et al. Mechanical induction of PGE2 in osteocytes blocks glucocorticoid‐induced apoptosis through both the beta‐catenin and PKA pathways. J Bone Miner Res. 2010; 25 (12):2657–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Noursadeghi M, Tsang J, Haustein T, Miller RF, Chain BM, Katz DR. Quantitative imaging assay for NF‐kappaB nuclear translocation in primary human macrophages. J Immunol Methods. 2008; 329 (1–2):194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shen J, Yu WM, Brotto M, et al. Deficiency of MIP/MTMR14 phosphatase induces a muscle disorder by disrupting Ca(2+) homeostasis. Nat Cell Biol. 2009; 11 (6):769–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Filigheddu N, Gnocchi VF, Coscia M, et al. Ghrelin and des‐acyl ghrelin promote differentiation and fusion of C2C12 skeletal muscle cells. Mol Biol Cell. 2007; 18 (3):986–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee JY, Chien IC, Lin WY, et al. Fhl1 as a downstream target of Wnt signaling to promote myogenesis of C2C12 cells. Mol Cell Biochem. 2012; 365 (1–2):251–62. [DOI] [PubMed] [Google Scholar]
- 30. Liu XH, Wu Y, Yao S, et al. Androgens up‐regulate transcription of the Notch inhibitor Numb in C2C12 myoblasts via Wnt/beta‐catenin signaling to T cell factor elements in the Numb promoter. J Biol Chem. 2013; 288 (25):17990–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Porter GA Jr, Makuck RF, Rivkees SA. Reduction in intracellular calcium levels inhibits myoblast differentiation. J Biol Chem. 2002; 277 (32):28942–7. [DOI] [PubMed] [Google Scholar]
- 32. Avila ME, Sepulveda FJ, Burgos CF, et al. Canonical Wnt3a modulates intracellular calcium and enhances excitatory neurotransmission in hippocampal neurons. J Biol Chem. 2010; 285 (24):18939–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mo C, Zhao R, Vallejo J, et al. Prostaglandin E2 promotes proliferation of skeletal muscle myoblasts via EP4 receptor activation. Cell Cycle. 2015; 14 (10):1507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ridgeway AG, Petropoulos H, Wilton S, Skerjanc IS. Wnt signaling regulates the function of MyoD and myogenin. J Biol Chem. 2000; 275 (42):32398–405. [DOI] [PubMed] [Google Scholar]
- 35. Willert K, Brown JD, Danenberg E, et al. Wnt proteins are lipid‐modified and can act as stem cell growth factors. Nature. 2003; 423 (6938):448–52. [DOI] [PubMed] [Google Scholar]
- 36. Vaes BL, Dechering KJ, van Someren EP, et al. Microarray analysis reveals expression regulation of Wnt antagonists in differentiating osteoblasts. Bone. 2005; 36 (5):803–11. [DOI] [PubMed] [Google Scholar]
- 37. Kalajzic I, Staal A, Yang WP, et al. Expression profile of osteoblast lineage at defined stages of differentiation. J Biol Chem. 2005; 280 (26):24618–26. [DOI] [PubMed] [Google Scholar]
- 38. Tanaka S, Terada K, Nohno T. Canonical Wnt signaling is involved in switching from cell proliferation to myogenic differentiation of mouse myoblast cells. J Mol Signal. 2011; 6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Korkosz M, Gasowski J, Leszczynski P, et al. High disease activity in ankylosing spondylitis is associated with increased serum sclerostin level and decreased wingless protein‐3a signaling but is not linked with greater structural damage. BMC Musculoskelet Disord. 2013; 14:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Santos A, Bakker AD, Zandieh‐Doulabi B, Semeins CM, Klein‐Nulend J. Pulsating fluid flow modulates gene expression of proteins involved in Wnt signaling pathways in osteocytes. J Orthop Res. 2009; 27 (10):1280–7. [DOI] [PubMed] [Google Scholar]
- 41. Jia YY, Li F, Geng N, et al. Fluid flow modulates the expression of genes involved in the Wnt signaling pathway in osteoblasts in 3D culture conditions. Int J Mol Med. 2014; 33 (5):1282–8. [DOI] [PubMed] [Google Scholar]
- 42. Li H, Malhotra S, Kumar A. Nuclear factor‐kappa B signaling in skeletal muscle atrophy. J Mol Med (Berl). 2008; 86 (10):1113–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhan M, Jin B, Chen SE, Reecy JM, Li YP. TACE release of TNF‐alpha mediates mechanotransduction‐induced activation of p38 MAPK and myogenesis. J Cell Sci. 2007; 120 (Pt 4):692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Porter JD, Baker RS. Absence of oculomotor and trochlear motoneurons leads to altered extraocular muscle development in the Wnt‐1 null mutant mouse. Brain Res Dev Brain Res. 1997; 100 (1):121–6. [DOI] [PubMed] [Google Scholar]
- 45. Miller JR, Hocking AM, Brown JD, Moon RT. Mechanism and function of signal transduction by the Wnt/beta‐catenin and Wnt/Ca2+ pathways. Oncogene. 1999; 18 (55):7860–72. [DOI] [PubMed] [Google Scholar]
- 46. Rizzuto R, Duchen MR, Pozzan T. Flirting in little space: the ER/mitochondria Ca2+ liaison. Sci STKE. 2004; 2004 (215):re1. [DOI] [PubMed] [Google Scholar]
- 47. Hayashi T, Su TP. Sigma‐1 receptor chaperones at the ER‐mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell. 2007; 131(3):596–610. [DOI] [PubMed] [Google Scholar]
- 48. Powell JA, Carrasco MA, Adams DS, et al. IP(3) receptor function and localization in myotubes: an unexplored Ca(2+) signaling pathway in skeletal muscle. J Cell Sci. 2001; 114 (Pt 20):3673–83. [DOI] [PubMed] [Google Scholar]
- 49. Gant JC, Blalock EM, Chen KC, et al. FK506‐binding protein 1b/12.6: a key to aging‐related hippocampal Ca2+ dysregulation? Eur J Pharmacol. 2014; 739:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zalk R, Lehnart SE, Marks AR. Modulation of the ryanodine receptor and intracellular calcium. Annu Rev Biochem. 2007; 76:367–85. [DOI] [PubMed] [Google Scholar]
- 51. Periasamy M, Kalyanasundaram A. SERCA pump isoforms: their role in calcium transport and disease. Muscle Nerve. 2007; 35 (4):430–42. [DOI] [PubMed] [Google Scholar]
- 52. Phuong TT, Yun YH, Kim SJ, Kang TM. Positive feedback control between STIM1 and NFATc3 is required for C2C12 myoblast differentiation. Biochem Biophys Res Commun. 2013; 430(2):722–8. [DOI] [PubMed] [Google Scholar]
- 53. Tavi P, Westerblad H. The role of in vivo Ca(2)(+) signals acting on Ca(2)(+)‐calmodulin‐dependent proteins for skeletal muscle plasticity. J Physiol. 2011; 589 (Pt 21):5021–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Witczak CA, Jessen N, Warro DM, et al. CaMKII regulates contraction‐ but not insulin‐induced glucose uptake in mouse skeletal muscle. Am J Physiol Endocrinol Metab. 2010; 298 (6):E1150–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guerci A, Lahoute C, Hebrard S, et al. Srf‐dependent paracrine signals produced by myofibers control satellite cell‐mediated skeletal muscle hypertrophy. Cell Metab. 2012; 15 (1):25–37. [DOI] [PubMed] [Google Scholar]
- 56. Vina J, Gomez‐Cabrera MC, Borras C, et al. Mitochondrial biogenesis in exercise and in ageing. Adv Drug Deliv Rev. 2009; 61(14):1369–74. [DOI] [PubMed] [Google Scholar]
- 57. Buettner GR. Superoxide dismutase in redox biology: the roles of superoxide and hydrogen peroxide. Anticancer Agents Med Chem. 2011; 11 (4):341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Buettner GR, Ng CF, Wang M, Rodgers VG, Schafer FQ. A new paradigm: manganese superoxide dismutase influences the production of H2O2 in cells and thereby their biological state. Free Radic Biol Med. 2006; 41(8):1338–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Beno T, Yoon YJ, Cowin SC, Fritton SP. Estimation of bone permeability using accurate microstructural measurements. J Biomech. 2006; 39 (13):2378–87. [DOI] [PubMed] [Google Scholar]
- 60. Rufino‐Ramos D, Albuquerque PR, Carmona V, Perfeito R, Nobre RJ, de Almeida LP. Extracellular vesicles: novel promising delivery systems for therapy of brain diseases. J Control Release. 2017; 262:247–58. [DOI] [PubMed] [Google Scholar]
- 61. McBride JD, Menocal‐Rodriguez L, Candanedo A, Guzman W, Garcia‐Contreras M, Badiavas EV. Bone marrow mesenchymal stem cell‐derived CD63+ exosomes transport Wnt3a exteriorly and enhance dermal fibroblast proliferation, migration and angiogenesis in vitro. Stem Cells Dev. Forthcoming. Epub 2017. Jul 5. DOI: 10.1089/scd.2017.0087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Supporting Figure S1.


