Summary
Factors impacting the developing neonatal gut microbiome and immune networks may increase risk for developing complex immune disorders such as inflammatory bowel diseases (IBD). In particular, peripartum antibiotics have been suggested as risk factors for human IBD, although direct evidence are lacking. We therefore examined the temporal impact of the commonly used antibiotic, cefoperazone, on both maternal and offspring microbiota when administered to dams during the peripartum period in the IL-10 deficient murine colitis model. By rigorously controlling for cage, gender, generational, and murine pathobiont confounders, we observed that offspring from cefoperazone-exposed dams develop a persistent gut dysbiosis into adulthood associated with skewing of the host immune system and increased susceptibility to spontaneous and chemically (DSS)-induced colitis. Thus, early life exposure to antibiotic-induced maternal dysbiosis during a critical developmental window for gut microbial assemblage and immune programming elicits a lasting impact in genetically susceptible offspring to increase IBD risk.
Keywords: Microbiome, host-microbe interactions, microbiome intervention, immune development, microbial assemblage, dysbiosis, inflammatory bowel diseases, complex immune disorders, vertical transmission
Graphical abstract
Introduction
Inflammatory bowel diseases (IBD) are chronic disorders that include two main clinical phenotypes, ulcerative colitis (UC) and Crohn’s disease (CD). Diseases like IBD are often referred to as “new age” disorders because of the alarming increase in their incidence and prevalence over the past century, particularly in populations that have undergone rapid changes in industrialization, hygiene, and diet (Bager et al., 2012; Benchimol et al., 2015; Devkota et al., 2012; Pugazhendhi et al., 2011). While there is a genetic basis (Anderson et al., 2011; Franke et al., 2010; Jostins et al., 2012; Liu et al., 2015; Liu and Stappenbeck, 2016; Van Limbergen et al., 2014), it is unlikely that genetic drift alone over this short period of time accounts for these diseases, raising the more likely role of environmental risk factors in triggering disease in genetically-predisposed individuals. These factors can affect individuals in many ways, but their impact on the gut microbiome, resulting in disturbances in host-microbe interactions, may be of relevance to the development and pathogenesis of IBD (Frank et al., 2007; Gophna et al., 2006; Manichanh et al., 2006; Ott et al., 2004; Seksik et al., 2003). This may be particularly relevant during the early stages of life when critical events in the development of the gut microbiome and immune system are taking place. The identification and avoidance of tipping factors in early life therefore represents a logical and important strategy for lowering risk of disease. In this regard, the promiscuous use of antibiotics during the preterm and post-natal (peripartum) periods that affect both maternal and neonatal microbiota has been suggested as a risk factor for human IBD, although compelling scientific evidence is lacking.
In the United States, it is estimated that antibiotics are prescribed with unclear indications at ~21% of pediatric ambulatory visits, where half of the prescriptions are broad-spectrum (Hersh et al., 2011). Approximately 40% of pregnant women at term and greater than 30% of neonates are exposed to antibiotics (Broe et al., 2014; Stokholm et al., 2013). In developed countries, broad-spectrum antibiotics, such as cephalosporins, are prescribed more frequently during pregnancy (Petersen et al., 2010). Furthermore, several studies have shown an association between early life exposure to antibiotics with increased risk IBD development, especially treatment-naïve pediatric CD (Gevers et al., 2014; Hviid et al., 2011; Kronman et al., 2012; Ungaro et al., 2014; Virta et al., 2012). However, these studies were limited in their ability to establish a causal link due to large differences in inter-individual gut microbiomes, challenges in controlling retrospectively for confounding variables, and constraints of observational clinical design. To address this issue, we took an alternate approach, employing the well-accepted IL-10 knock-out (KO) mouse model, where the immunomodulatory cytokine IL-10 was genetically deleted (Kuhn et al., 1993). Genetic risk in IL-10 KO mice is not sufficient to cause disease, because these animals rarely develop disease if they are raised germ-free (GF) or housed in a Helicobacter hepaticus-free environment (Keubler et al., 2015; Sellon et al., 1998). H. hepaticus rarely causes colitis in wild-type mice, but can cause nearly a 100% disease penetrance in IL-10 KO mice (Kullberg et al., 1998). We examined the temporal impact of the broad-spectrum antibiotic, cefoperazone (CPZ), on both the maternal and offspring microbiota when administered to dams during the peripartum period. Here, we reasoned that vertical transmission of maternal microbiota is the major source of microbes for the development of the infant gut microbiome (Adlerberth and Wold, 2009; Asnicar et al., 2017; Caufield et al., 1993; Caufield et al., 2007; Funkhouser and Bordenstein, 2013; Jimenez et al., 2008; Nayfach et al., 2016; Pantoja-Feliciano et al., 2013). Under experimental conditions that controlled for cage, gender, and age effects as well as the confounding presence of common murine pathobionts, we observed that offspring from CPZ-exposed dams develop gut dysbiosis that persists into adulthood. This effect is associated with a skewing of the host immune system and increased susceptibility to the development of spontaneous and chemically (dextran sodium sulfate; DSS)-induced colitis in IL-10 KO mice. Fecal microbiota transplantation (FMT) of CPZ-exposed dam’s microbiota into GF IL-10 KO mice resulted in a similar skewing of the host immune response in the offspring of FMT-CPZ recipients to that observed in offspring of donor animals. Together, our findings support the notion that early life exposure to antibiotic-induced maternal dysbiosis during a critical developmental window for gut microbial assemblage and immune programming can have a lasting impact in genetically susceptible offspring, increasing risk for complex immune-related disorders such as IBD.
Results
Antibiotic treatment in early life increases the risk for colitis in offspring
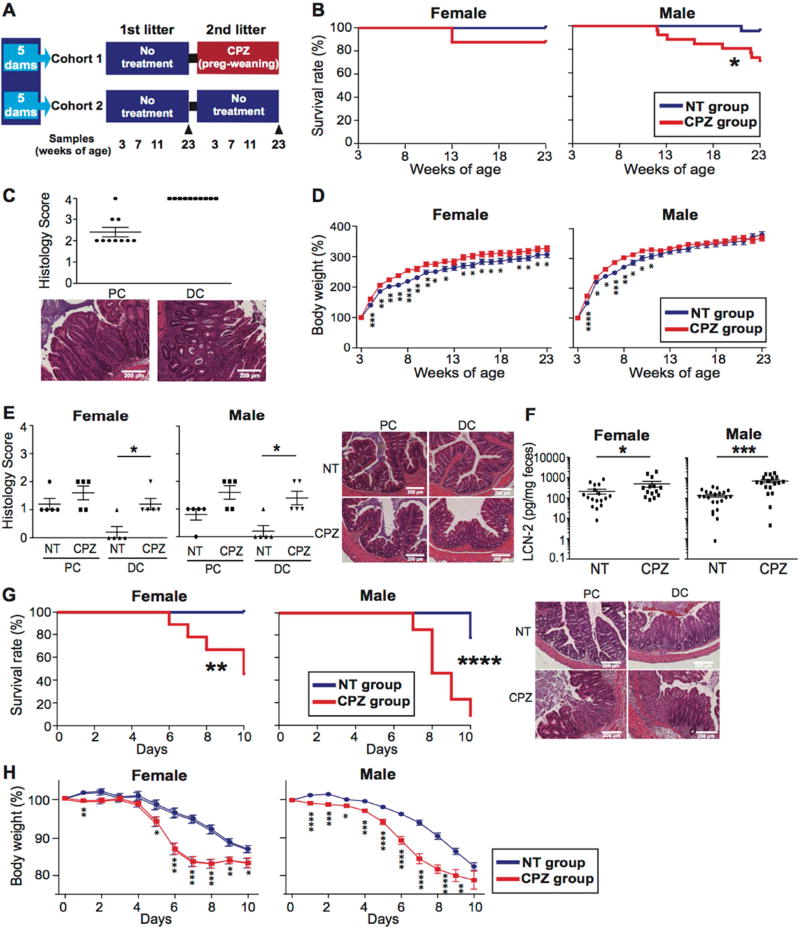
The mice used for this study were derived from GF IL-10 KO mice on a C57Bl/6 background conventionalized with H. hepaticus-free donor gut microbiota and subsequently housed in a H. hepaticus-free room under continuous monitoring. To control for genetics and maternal contribution, the 10 breeding pairs used for two sequential parturitions underwent a vetted normalization protocol involving mixed bedding transfers between cages of the same generation before pairs were set up for breeding (Figure 1A). In cohort 1, the first litters from each dam were used as the non-treatment (NT) group. For their second pregnancy, dams were treated with CPZ from day 14 of gestation and throughout the pre-weaning period (Figure S1A). The rationale for this protocol was to expose pups to maternal microbiota that had been conditioned with CPZ at the time of the pup’s birth, analogous to common practice scenarios in human populations. Cohort 2 served as additional controls with dams untreated throughout both sequential parturitions to control for the potential impact of sequential litters on colitis development. Pups were tracked from 3 weeks (wks) of age through 23 wks of age. Both genders were studied with 17 females and 23 males used for the NT tracking group and 16 females and 26 males used for the CPZ tracking group. No differences in the incidence of colitis (0%) were observed between the first and second litters of cohort 2 NT dams (Figure S1B). In contrast, peripartum CPZ exposure led to a significantly decreased survival rate in IL-10 KO pups, particularly in males (p=0.018) (Figure 1B). This included 12.5% of female (2/16) and 30.8% of male (8/26) mice that were sacrificed because of severe weight loss (a euthanasia criteria). Histological examination of their intestines confirmed the presence of severe colonic inflammation (Figure 1C). As noted by others, both female and male mice exposed to peripartum CPZ treatment gained more weight initially (Figure 1D), which can be attributed to the obesogenic effects of antibiotic-induced mucosal inflammation and gut dysbiosis (Cho et al., 2012). Females from CPZ-exposed dams continued to exhibit heavier body weights throughout the 23 wks relative to NT controls, however, 8 out of 26 males exposed to CPZ began to lose weight at as early as 11 wks of age due to spontaneous colitis development. Despite this, no significant differences were observed in histopathology scores for colitis at 3 and 7 wks of age between NT and CPZ treatment groups (Figure S1C) prior to decreases in body weight. Thus, peripartum CPZ exposure in the absence of H. hepaticus, a known pathobiont, appears to promote increased risk for spontaneous colitis in genetically susceptible offspring later in life.
Figure 1. Maternal peripartum exposure to cefoperazone increases the risk for colitis in offspring.
(A) Study design using IL-10 knock-out (KO) mouse model. All mice (cohort 1 and 2) were obtained from 10 breeding pairs subjected to mixed bedding protocol to normalize microbiota among parent cages. Cohort 1 (1st row) included two sequential litters from 5 breeders divided into non-treatment (NT) tracking group (litter 1) and cefoperazone (CPZ) tracking group (litter 2). Cohort 2 (2nd row) included two sequential litters from 5 breeders that were used as additional NT controls to assess/control for generational drift in gut microbiota across litters 1 and 2. (B) Survival rates of NT and CPZ offspring during the observation period (3 to 23 wks of age) (NT group n=17 females; 23 males. CPZ group n=16 females, 26 males). (C) Histology of proximal (PC) and distal colon (DC) for CPZ mice euthanized due to colitis onset (n=2 females, 8 males). Representative H&E histological sections of PC and DC for NT and CPZ. (D) Percent weight change (expressed as % of starting weight) of tracked pups beginning at 3 wks of age (weaning). (E) Histology of PC and DC in mice that did not develop colitis at 23 wks of age (n=5/gender/treatment). (F) Fecal lipocalin-2 (LCN-2) levels were measured in mice that did not develop colitis at 23 wks of age (NT group n=17 females; 22 males. CPZ group n=14 females, 18 males). (G) Survival curves for NT and CPZ mice without frank colitis treated with 2.5% dextran sulfate sodium (DSS) at 23 wks of age. Representative H&E histology of PC and DC are presented for NT and CPZ, respectively following DSS. (H) Weight change following onset of DSS treatment. Mice showing more than 20% body weight loss were euthanized. *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001. Data are represented as mean ± SEM for (C)-(F) and (H). See also Figure S1.
We next examined the mice that remained grossly healthy and never developed overt signs of spontaneous colitis (NT group: 17 females, 22 males; CPZ group: 14 females, 18 males). At 23 wks of age, histologic grading of colons revealed higher inflammatory scores, particularly in the distal colons of the CPZ group compared to NT (n=5 mice/gender, p=0.021 in females and p=0.019 in males) (Figure 1E). Fecal lipocalin-2 (LCN-2), a marker for colitis (Chassaing et al., 2012), showed that both females and males from CPZ-exposed dams had significantly higher LCN-2 levels as compared to the NT group (p=0.037 in females and p<0.001 in males) (Figure 1F). Thus, even in IL-10 KO mice that did not develop frank disease, peripartum exposure to CPZ is associated with subclinical histologic evidence of colonic inflammation.
To examine how CPZ-exposed IL-10 KO mice that did not develop frank colitis would respond to a colitogenic challenge, mice were administered 2.5% DSS in drinking water at 23 wks of age. We observed that both female and male mice from CPZ-exposed dams had significantly lower survival rates, lost significantly more weight, and exhibited more severe histologic injury from the DSS challenge compared to their 23-wk-old NT counterparts (p=0.003 in females and p<0.0001 in males) (Figure 1G and H). Moreover, as expected, pups from wild-type C57Bl/6 (WT) dams that underwent an identical CPZ exposure protocol did not develop colitis. Furthermore, at 23 wks of age there were no differences in LCN-2 levels or DSS-induced colitis susceptibility regardless of gender (Figure S1D and S1E), highlighting the importance of genetic susceptibility for disease development in the case of maternal antibiotic-induced dysbiosis. Together, these data revealed that maternal CPZ exposure during the peripartum period in IL-10 KO mice leads to an increased susceptibility to developing both spontaneous and chemically-induced colitis later in life.
Peripartum antibiotic exposure leads to aberrant development of the offspring immune system
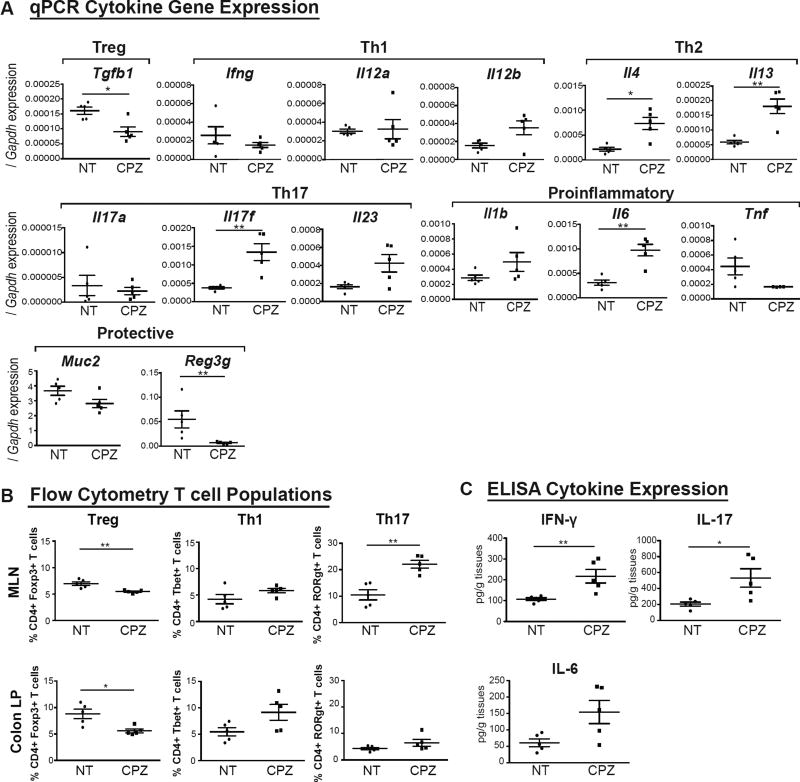
To determine if and how maternal CPZ exposure during early development alters immune profiles, we first compared mRNA levels of cytokine genes indicative of T-cell subtypes (Treg, Th1, Th2 and Th17 cells), pro-inflammatory cytokine genes, and mucosal protective factors relevant to IBD in colons of 3-wk-old pups from CPZ-exposed dams versus NT controls. Male data are shown in Figure 2 and female data in Figure S2. Exposure to maternal CPZ-induced dysbiosis elevated mRNA levels of key immune mediators such as Il17f (p<0.01 in males), Il4 (p<0.05 in males), Il13 (p<0.01 in males), Il1b (p<0.05 in females), and Il6 (p<0.01 in males). Conversely, anti-inflammatory and trophic mediators including Tgfb1 (p<0.05 in males), Muc2 (p<0.05 in females) and Reg3g (p<0.01 in females and males) were significantly decreased (Figure 2A and Figure S2A). In spite of the increased mRNA expression of Il4 and Il13, there were no significant differences in plasma IgE level in CPZ pups compared to NT controls (Figure S2D). Next, we examined markers of several T-cell populations prior to the onset of colitis in NT and CPZ 3-wk-old IL-10 KO pups using flow cytometry. We determined the proportion of T-cells expressing transcription factors indicative of both regulatory and inflammatory T-cell populations, including Forkhead box P3 (Foxp3), T-box transcription factor (T-bet) and RAR-related orphan receptor gamma t (RORγt). We harvested cells from the mesenteric lymph nodes (MLN) and colonic lamina propria (LP) of 3-wk-old NT and CPZ pups. Here, we focused on CD4+ T cell populations, as shown in the gating strategies used for flow cytometry in Figure S2F. Specifically, we examined the percentage of live CD45+TCRb+CD4+ cells (CD4+ T cells) that expressed Foxp3+ (regarded as regulatory T cells; Treg), T-bet+ (Th1 cells) or RORγt+ (Th17 cells). We observed that male CPZ IL-10 KO pups exhibited a significant reduction in Tregs in both the MLN (p<0.01) and colonic LP (p<0.05) with significant increases in Th17 cells in the MLN (p<0.01) as compared to NT counterparts (Figure 2B). Similarly, CPZ IL-10 KO female pups showed significant decreases in MLN Tregs (p<0.01) and significant increases in LP Th17 cells as compared to NT (p<0.05) (Figure S2B). These immune changes were not observed in 3-wk-old NT versus CPZ group WT mice with the exception for significant differences detected in MLN Th1 cells from WT females (Figure S2E). As shown in Figure 2C, 23-wk-old male CPZ IL-10 KO mice that survived and did not develop spontaneous colitis still showed significantly higher levels of IFN-γ (p<0.01) and IL-17 (p<0.05) compared to their NT counterparts. In contrast, females at 23 wks of age did not show such drastic differences (Figure S2C). These findings suggest that an early immune skewing induced by maternal peripartum CPZ exposure in IL-10 KO mice contributes to a proinflammatory milieu that lasts into adulthood.
Figure 2. Maternal peripartum exposure to cefoperazone leads to aberrant development of the host immune system in IL-10 KO mice.
(A) Real-Time qPCR mRNA levels of cytokine and pro- vs. anti-inflammatory genes involved in colonic inflammation from colonic mucosal scrapings in non-treated (NT) vs. cefoperazone (CPZ) male IL-10 KO mice at 3 wks of age. mRNA levels expressed as ΔΔCT relative to housekeeper gene Gapdh. (B) Flow cytometric analyses of live CD45+TCRβ+CD4+ T cells expressing Foxp3+ (Treg), T-bet+ (Th1) or RORγt+ (Th17) in MLNs and colonic LPs of NT vs. CPZ males at 3 wks of age. Data represent percentage of live CD4+ cells. (C) Protein levels of inflammatory cytokines in MLNs of NT vs. CPZ-exposed males at 23 wks of age determined via ELISA. N=4–5/group. NT (black circles), CPZ (black squares). *p<0.05, **p<0.01 via Mann-Whitney U-test. Data are represented as mean±SEM. Female data shown in supplemental data. See also Figure S2.
Maternal peripartum antibiotic exposure induces a persistent maternal and neonatal gut dysbiosis
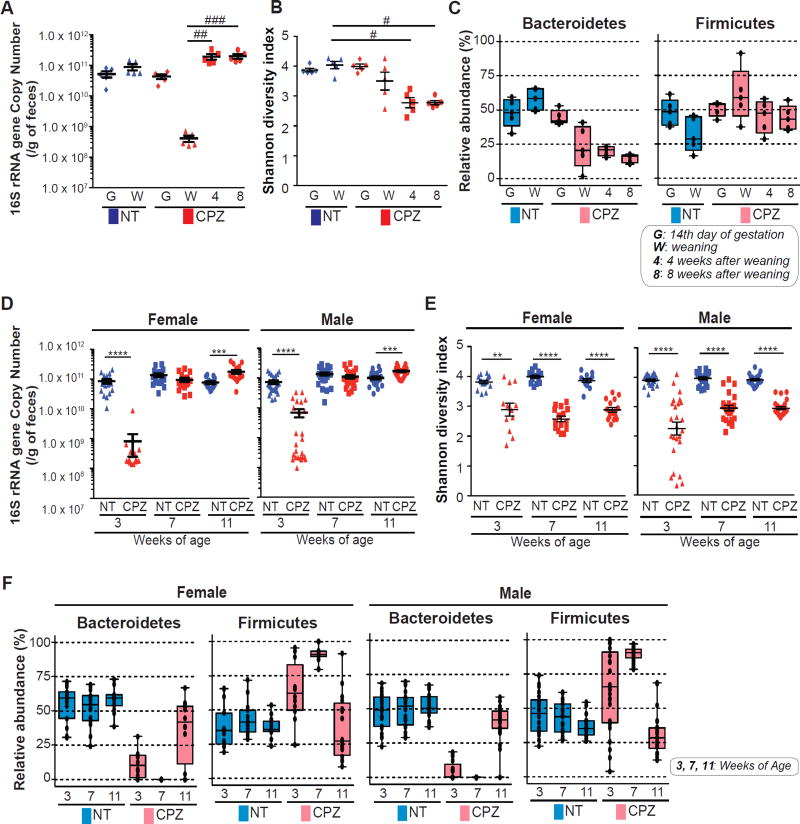
Examination of 16S ribosomal RNA (rRNA) gene copy number, as determined by qPCR using universal 16S primers (shown in Table S2), showed a significant decrease in dams following CPZ exposure as compared to NT, which recovered by 4 and 8 wks following CPZ cessation (Figure 3B). Despite recovery of 16S gene copy number, the microbial community structure of dams based on 16S rRNA gene amplicon sequencing revealed peripartum administration of CPZ caused persistent shifts in the microbiota (Figure 3B and Figure 3C). Shannon diversity index was decreased (Figure 3B) and microbial β-diversity (Figure 3C) did not recover, even after 8 wks of CPZ cessation. At the taxonomic level of Phyla shown in Figure S3A, CPZ dramatically reduced the relative abundance of Bacteroidetes and increased the relative abundance of Firmicutes and Verrucomicrobia. These changes, particularly in Bacteroidetes and Verrucomicrobia, were also observed 4 and 8 wks following cessation of CPZ exposure (Figure S3A). Interestingly, despite the persistent changes in the maternal gut microbiota, none of the dams exposed to CPZ developed colitis. This observation suggested that disruption of the microbial community due to CPZ exposure in adult IL-10 KO with already maturated gut microbiota may not predispose these animals to develop colitis. To test this, we treated adult female and male IL-10 KO mice with CPZ for 4 wks (identical to IL-10 KO dams) and tracked their microbial community structure as well as body weight. Here, we observed no significant differences in survival rate between NT and CPZ-treated IL-10 KO adults, and both females and males exhibited a significant increase in body weight following CPZ exposure (Figure S3B). Contrary to the 16S rRNA changes observed in IL-10 KO dams, no appreciable differences were observed in regards to relative abundance of Bacteroidetes in adult IL-10 KO of either gender following the cessation of CPZ, however, both Firmicutes and Verrucomicrobia appeared to be elevated (Figure S3C). Together, this data suggests that exposure to CPZ following microbial maturation may be beyond the window of immune maturation and may not significantly impact disease prognosis, even in genetically susceptible mice.
Figure 3. Maternal peripartum exposure to cefoperazone induces a persistent gut dysbiosis in dams and pups.
(A) Fecal 16s rRNA gene copy number in non-treated (NT) versus cefoperazone (CPZ)-exposed dams at 14th day of gestation (start of CPZ treatment), at weaning (end of CPZ treatment), 4 wks and 8 wks after CPZ cessation. (B) Shannon diversity index in dams at 14th day of gestation (G), at weaning (W), 4 wks and 8 wks after CPZ cessation. (C) Bacterial community composition in NT and CPZ-exposed dams tracked until 8 wks after CPZ cessation. (D) Fecal 16s rRNA gene copy number in NT and CPZ IL-10 KO offspring at 3, 7 and 11 wks of age. (E) Shannon diversity index in NT vs. CPZ IL-10 KO offspring at 3, 7 and 11 wks of age. (F) Bacterial community composition was assessed at 3, 7 and 11 wks of age in female and male offspring. Two dominant phyla, Bacteroidetes and Firmicutes are presented for (C) and (F). Additional phyla shown in Supplemental Figure 3. #p<0.05, ##p<0.01 and ###p<0.001 via Dunn’s test. **p<0.01, ***p<0.001 and ****p<0.0001 via Mann-Whitney U-test. Oligotype phyla taxa presented as relative abundance and represented as box plots. Dots represent individual samples. See also Figure S3.
We next examined the microbial community composition of pups from NT and CPZ dams at 3, 7, and 11 wks of age. Analysis of 16S rRNA gene copy number revealed significant reduction at 3 wks of age in both male and female CPZ pups (p<0.0001) with a significant compensatory increase at 7 and 11 wks of age relative to NT pups (Figure 3D). The pups from CPZ-exposed dams showed a significantly lower Shannon diversity index compared to those in the NT group (p<0.01 at 3 wks of age and p<0.0001 at 7 and 11 wks in females; p<0.0001 at 3, 7 and 11 wks of age in males) (Figure 3E). A persistently low α-diversity was observed as late as 11 wks of age in CPZ pups. At 3 wks of age, the microbial community composition differed dramatically between NT and CPZ groups. However, each group exhibited community profiles similar to their respective dams. The alterations to the microbiota in CPZ pups were persistent, showing different patterns of maturation through adulthood (11 wks of age), 8 wks post-exposure to maternal CPZ-induced dysbiois and at a time point prior to the onset of inflammation in either group (Figure 3F and Figure S3D).
Exposure to antibiotics in the peripartum period induces persistent and significant changes in gut microbiota in IL-10 KO dams and offspring
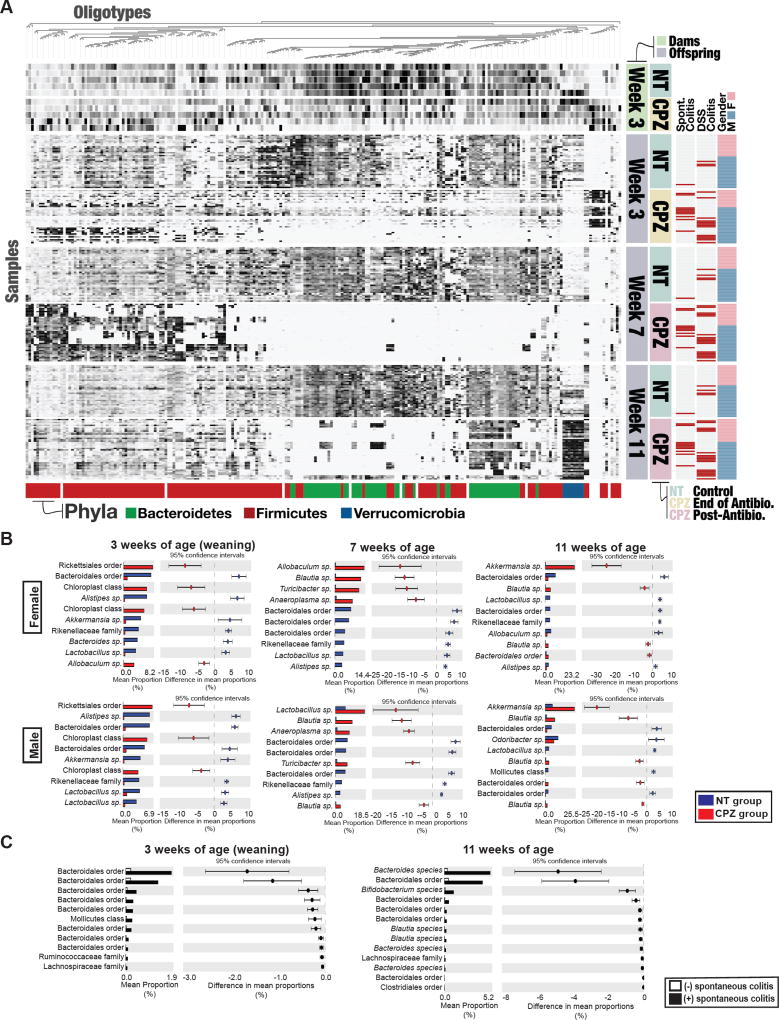
A high-resolution analysis of 16S rRNA gene amplicons using Minimum Entropy Decomposition (MED) revealed that CPZ exposure of dams resulted in significant changes in bacterial oligotypes as compared to NT dams at the time of weaning (Figure 4A, top heatmap). Interestingly, the samples from dams suggest that CPZ exposure leads to persistent changes in specific oligotypes, even following CPZ cessation, in that little to no recovery of oligoypes was observed at 4 or 8 wks post-treatment (Figure S4A). We observed specific alterations in oligotype abundance in maternal microbiota of NT versus CPZ-exposed dams at weaning (3 wks post birth; approximately 4 wks after the start of CPZ treatment) that were consistent with changes in the microbiota of the offspring at weaning (3 wks of age) (Figure 4A, top and bottom heatmap). As described, nearly 100% of the mice that developed either spontaneous colitis or severe DSS-induced colitis were from CPZ-exposed dams, where dysbiosis was clearly evident, suggesting that maternal vertical transmission of specific oligotypes, and not vertical transmission of CPZ itself, may play a role in colitis development. To rule out a significant contribution of direct exposure of CPZ to the pups’ microbiota via maternal transmission, we performed antimicrobial assays using cecal contents from NT versus CPZ-exposed dams and their respective offspring at weaning. We observed that only cecal contents from CPZ-exposed dams elicited antimicrobial effects against E. coli, while no appreciable killing effect was observed in NT or CPZ pups, suggesting that maternal transmission of CPZ was minimal (Figure S4B). Together, this data provides evidence that maternal CPZ-induced gut microbiota is vertically transferred to the offspring and does not fully recover despite cessation of antibiotic intake.
Figure 4. Maternal peripartum exposure to cefoperazone induces persistent and significant changes in specific microbial oligotypes in IL-10 KO dams and offspring.
(A) Comprehensive comparison of gut microbes in fecal samples collected from non-treatment (NT) and cefoperazone (CPZ)-treated IL-10 KO dams at weaning (wk 3) and their offspring at 3, 7 and 11 wks. Oligotype abundance was determined in samples from dams and offspring that were tracked for spontaneous colitis or DSS-treated. Sixteen samples were excluded due to low number of sequences. Columns represent an individual oligotype and rows represent individual samples. Samples with red horizontal bars represent mice that developed spontaneous colitis (Spont. Colitis) or DSS-colitis following exposure at 23 wks of age and euthanized due to frank colitis. Colored bars at the bottom of the figure represent dominant phyla. (B) Significant changes in mean proportions of oligotypes between NT and CPZ offspring at 3, 7, and 11 wks of age determined by STAMP. The top 10 oligotypes with the greatest changes in relative abundance are presented as extended error bar plots (bars represent 95% confidence intervals). All oligotypes significantly different between groups are presented in the supplementary table. See Table S1. (C) Oligotypes with significant differences in relative abundance were identified at 3 and 11 wks of age between males from CPZ-exposed dams that did or did not develop spontaneous colitis. All oligotypes identified as significantly altered between groups are sorted by changes in relative abundance. See also Figure S4.
Further analysis of gut microbes of the offspring from NT or CPZ-exposed dams at 3, 7, and 11 wks of age showed persistent dysbiosis over time in CPZ pups (Figure 4A, bottom). The changes in oligotypes in pups from CPZ-exposed dams persisted well beyond maternal influence, in that CPZ pups do not exhibit recovery at 7 or 11 wks of age, while oligotypes in NT pups appear to retain some stability over time (Figure 4A, bottom heatmap). Pups from CPZ-exposed dams showed significant alterations at 3, 7, and 11 wks of age in oligotypes belonging to the Phyla Bacteroidetes, Firmicutes and Verrucomicrobia relative to age-matched NT controls. Overall, offspring from CPZ-exposed dams exhibited a decrease in a large portion of oligotypes assigned to Bacteroidetes across all time points, despite some evidence for a reemergence of several of these oligotypes, along with several others belonging to both Firmicutes and Verrucomicrobia at 11 wks of age. Oligotypes across all samples visualized in the heatmap can be further explored in depth at https://anvi-server.org/merenlab/il10ko_peripartum_cpz.
At each time point, significant differences were observed in relative abundance of particular oligotypes between offspring from NT versus CPZ-exposed dams. To determine which oligotypes were the most significant contributors to the overall differences between NT and CPZ microbial communities, we utilized Statistical Analysis of Metagenomic Profiles (STAMP) to identify significantly enriched oligotypes. The top ten oligotypes exhibiting the greatest differences in mean relative abundance among females and males and across all 3 time points are shown in Figure 4B. Specifically, at 3 wks of age, CPZ males and females exhibited a larger proportion of oligotypes belonging to Rickettsiales order from the phyla Proteobacteria, as compared to NT controls. At 7 wks of age both male and females from CPZ-exposed dams exhibited a significant increase in specific Allobaculum sp., Blautia sp., Turicibacter sp., Lactobacillus sp., and Anaeroplasma sp. oligotypes, respectively. By 11 wks of age, Akkermansia sp. and Blautia sp. were significantly elevated in female and male CPZ offspring as compared to NT. Furthermore, analysis of the male offspring from CPZ-exposed dams at 3 and 11 wks of age showed unique oligotype signatures between those mice that eventually developed spontaneous colitis as compared to mice that remained healthy. These oligotypes that are significantly different are presented in Figure 4C and include bacteria belonging to the order Bacteroidales at both 3 and 11 wks of age.
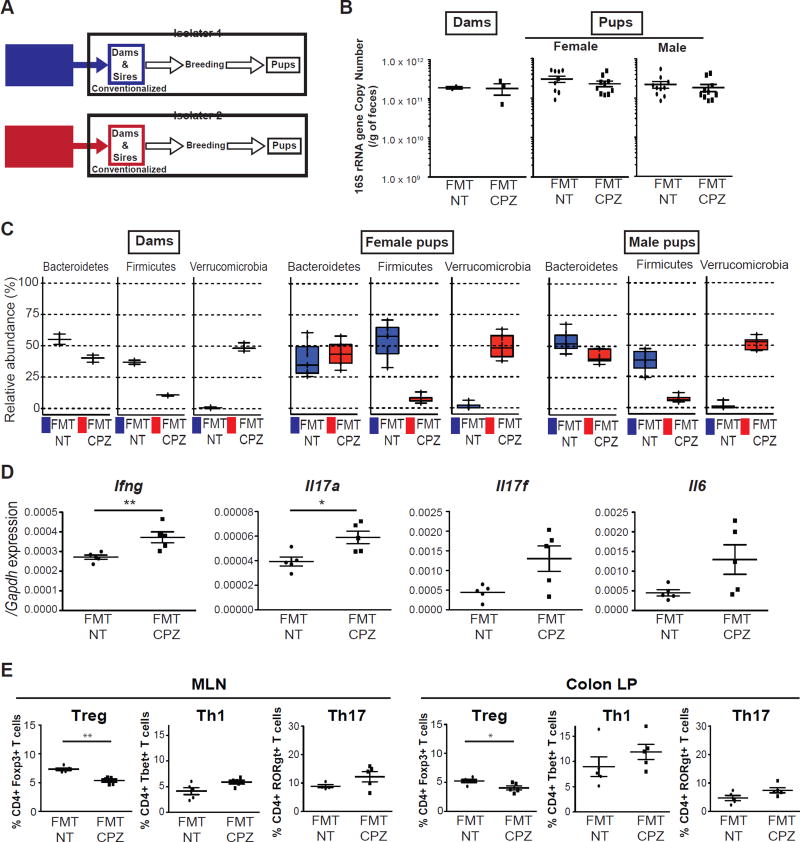
Fecal microbiota transplant of maternal CPZ-induced dysbiosis into germ-free IL-10 KO dams elicits a pro-inflammatory milieu in offspring
We next wanted to examine whether CPZ-induced dysbiosis observed in dams would transfer an inflammatory phenotype in the absence of CPZ exposure. Here, we performed fecal microbiota transplantation (FMT) using stool obtained from NT or CPZ-exposed dams at weaning into GF IL-10 KO females maintained in separate flexible film isolators (Figure 5A). Following FMT, these mice were set-up into breeding pairs. Examination of dams and pups at the time of weaning revealed identical 16S rRNA gene copy number as measured by qPCR in recipients regardless of donor treatment (Figure 5B). Analysis of 16S rRNA amplicons via MED showed that dams, female and male pups within the FMT-CPZ group exhibited significant decreases in Firmicutes with concomitant increases in Verrucomicrobia relative to FMT-NT (Figure 5C). Slight but insignificant changes in Bacteroidetes were observed only in dams and male FMT-CPZ pups (Figure 5C). Significant changes were not observed in other Phyla between FMT-NT and FMT-CPZ dams and pups (Figure S5A and S5B). At 3 wks of age, we examined similar immune parameters in FMT pups as shown in SPF IL-10 pups from NT or CPZ-exposed dams in Figure 2. Male FMT-CPZ pups exhibited significantly increased expression of Ifng and Il17a in MLNs relative to FMT-NT, and increased but not significant trends of Il17f and Il6 (Figure 5D). Similar observations were also seen in female FMT-CPZ 3-wk-old pups (Figure S5C). We next examined markers of T-cell populations in FMT-NT or FMT-CPZ 3-wk-old pups via flow cytometry. Again, we determined the proportion of T-cells expressing transcription factors, including Foxp3, T-bet, and RORγt in the MLN and colonic LP, focusing on CD4+ T cell populations (Figure S2E). We observed that FMT-CPZ male IL-10 KO pups exhibited a significant reduction in Tregs in both the MLN (p<0.01) and colonic LP (p<0.05) with increases, although not significant, in Th1 and Th17 cells in the MLN and LP as compared to FMT-NT counterparts (Figure 5B). Interestingly, FMT-CPZ female IL-10 KO pups exhibited no significant differences in MLN or LP proportions of Tregs, however, both Th1 and Th17 cells were significantly increased in the MLN (p<0.05) (Figure S5D). To rule out the direct impact of CPZ on these immune parameters, we analyzed 3-wk-old pups from GF IL-10 dams that had or had not received peripartum CPZ in an identical manner presented in Figure 1A. Analysis of the proportion of T-cells expressing Foxp3, T-bet, or RORγt were not significantly altered by maternal exposure to CPZ under germ-free conditions (Figure S5E), suggesting that exposure to maternal dysbiosis induced by CPZ under SPF conditions in early life served as the main driver of changes.
Figure 5. Conventionalization of germ-free IL-10 KO mice with cefoperazone-induced dysbiosis skews host immune status in the offspring.
(A) Fecal microbiota transplant (FMT) study design. GF IL-10 KO mice were gavaged with fecal slurries prepared from non-treated (NT) or cefoperazone (CPZ)-treated dams at weaning from cohort 1 in separate flexible film isolators. Breeding pairs were then set up (isolator 1: 2 FMT-NT dams, isolator 2: 3 FMT-CPZ dams). Offspring from FMT dams in each isolator were analyzed at weaning (n=10/gender/group). (B) Fecal 16s rRNA gene copy number in dams and offspring at weaning. (C) Bacterial community composition in dams and offspring. Three dominant phyla, Bacteroidetes, Firmicutes, and Verrucomicrobia are presented. Additional phyla are shown in supplemental Figure 5. (D) Real-Time qPCR mRNA levels of inflammatory cytokines from colonic mucosal scrapings in FMT-NT (black circles) versus FMT-CPZ (black squares) male IL-10 KO offspring at 3 wks of age. mRNA levels expressed as ΔΔCT relative to housekeeper gene Gapdh. (E) Flow cytometric analyses of live CD45+TCRβ+CD4+ T cells expressing Foxp3+ (Treg), T-bet+ (Th1) or RORγt+ (Th17) in MLNs and colonic LPs of 3-wk-old FMT-NT versus FMT-CPZ male offspring (n=4–5/group). Data represent percentage of live CD4+ cells. *p<0.05, **p<0.01 via Mann-Whitney U-test. See also Figure S5.
Discussion
The findings of this study support a potential role of broad-spectrum antibiotics as a risk factor for IBD when used during the peripartum period. While this possibility has been entertained by several retrospective human studies, a clear causal link between peripartum antibiotics and IBD risk has never been proven (Gevers et al., 2014; Hviid et al., 2011; Kronman et al., 2012; Ungaro et al., 2014; Virta et al., 2012). Using a genetic risk model, the IL-10 KO mouse, and employing a prospective study design that rigorously controls for starting gut microbiomes and generational drift, we were able to observe the impact of cefoperozone (CPZ), a broad-spectrum antibiotic, on maternal and offspring gut microbiota during and after antibiotic exposure. Our data indicates that peripartum CPZ exposure of dams causes gut microbial dysbiosis in dams, which is vertically transmitted to the offspring and is still evident following cessation of CPZ exposure. These changes were associated with a skewing of the immune profile in IL-10 KO pups from dams exposed to peripartum CPZ, rendering them at higher risk for the development of spontaneous and DSS-induced colitis later in life.
We designed our study to follow a common practice scenario in humans where antibiotics are used widely among late term pregnancy and during the neonatal period. By using a carefully designed bedding transfer protocol, we ensured that the various treatment groups had similar starting microbiomes and, by using a model of sequential parturitions, we controlled for generational drift in microbiota. Finally, we used MED to characterize the microbial community structure at high-resolution. The impact of CPZ-induced perturbations in maternal microbiota on the development of offspring gut dysbiosis is particularly notable, as the changes in respective taxa over time were similar, except more pronounced in the offspring. We speculate that the severe gut dysbiosis observed in the offspring of CPZ-exposed dams stems from the antibiotic-induced selective pressure on maternal microbiota that reduces the abundance, diversity, and likelihood of susceptible organisms to establish in the neonate. The CPZ-induced changes in maternal gut microbiota (Figure 3A) show that Bacteroidetes are significantly reduced at the time of CPZ cessation, which persists in the dams up to 8 wks after cessation of the antibiotic (Figure S3B). However, these changes were not to the same extreme as to those observed in offspring, perhaps indicative of a greater resilience of a mature adult gut microbiota to broad-spectrum antibiotics. Consistent with this notion, dams were only exposed to CPZ during adulthood and none developed colitis. We further showed that exposure of both male and female IL-10 KO mice in adulthood failed to produce the same degree of dysbiosis and also did not result in colitis. In contrast, neonates that acquired microbes from maternal antibiotic-induced gut dysbiosis during this critical window of microbial assemblage displayed a dysbiotic gut microbiome from the very start, which was associated with immediate and long-term skewing of their immune system.
As shown in Figure 4, it is striking to see the near absence of most members of the Bacteroidetes phylum and the elevated abundance of members of the Firmicutes phylum at 3 (weaning) and 7 wks of age (4 wks post-exposure to maternal CPZ-induced dysbiosis). We attribute the nadir of Bacteroidetes at 7 wks of age to the possibility that other more fit groups (Firmicutes, Verrucomicrobia) developed stable networks that limited the ability of Bacteroidetes to establish (Figure S3D). By 11 wks of age, we observe some appearance of Bacteroidetes taxa, but their diversity remained limited. This gut dysbiosis persisted to adulthood (11 wks of age), even before frank colitis was observed in any of the groups. Additionally, we identified oligotypes that appeared to be significant contributors to the overall differences observed between NT and CPZ groups as well as among CPZ male pups that did or did not develop colitis (Figure 4). Furthermore, FMT of maternal CPZ-induced communities into GF IL-10 KO mice resulted in immune skewing in these offspring that was consistent with that of observed in SPF IL-10 KO pups exposed to maternal CPZ-induced dysbiosis. This reveals that maternal CPZ-induced dysbiosis alone in the absence of direct exposure to CPZ can elicit a negative impact on immunological outcomes and possibly further predispose genetically-susceptible offspring to develop colitis. Future studies involving shotgun metagenomics should provide additional insights into maternal-offspring influences, host disease development, and potential predictive markers of risk.
The maternal CPZ-induced gut dysbiosis transmitted to the offspring is associated with disruptions in immune development and protective processes that would normally occur at the time of weaning to adulthood (Figure 2). The increased mRNA expression of Il4 and Il13 implied the possibility that Th2-mediated mechanisms, including invariant natural killer T (iNKT) cells may play a role in development of colitis as previously reported (Olszak et al., 2012). However, in our model, plasma IgE was not significantly altered by maternal CPZ exposure (Figure S2D), hence we focused on Th1 and Th17 cells, which have been shown to be relevant in the IL-10 KO mouse (Keubler et al., 2015). While we observed significant increases specifically in Th17 RORyt cells, previous work in WT C57Bl/6 mice from dams experiencing repeat exposure to subtherapeutic doses of penicillin exhibited decreases in IL-17 and IFNg T cell populations. However, in our model, genetically susceptible hosts such as the IL-10 KO mouse respond in a different manner relative to WT mice, resulting in a relative increased risk for “spontaneous” colitis, as was observed (Figure 1). It is notable that this risk continues to exist for mice that do not develop frank spontaneous colitis, as these mice exhibited increased histological and subclinical mucosal inflammation and were highly susceptible to the colitogenic effects of DSS compared to NT controls. These findings are likely relevant to human IBD where genetic risk, while necessary, is by itself insufficient to cause frank disease. It is also possible that a significant number of these at-risk subjects have subclinical disease that would be detectable histologically or through examination of immune/inflammatory markers, which has been previously suggested (Howarth et al., 2002; Sakata et al., 2001). Moreover, these individuals are likely to be more sensitive to risk factors that set the stage for events that trigger the immune response to cause frank clinical disease. We speculate that this may involve improper imprinting of immune networks of genetically susceptible individuals created by the gut dysbiosis caused through peripartum antibiotic exposure.
In summary, our studies provide compelling evidence that peripartum maternal antibiotic exposure skews maternal gut microbiota and that of the subsequent offspring, and increases IBD risk in genetically susceptible offspring by affecting a critical stage of their microbial and immune development. These effects can be persistent into adulthood and promote the risk of complex immune disorders such as IBD in genetically susceptible hosts. Identification of potential risk factors and a better understanding of the underlying mechanisms that lead to increased IBD risk is paramount for developing strategies for the prevention and/or treatment of human IBD. In this case, we may have to rethink common practices of indiscriminate and empirical use of antibiotics during pregnancy and infancy and perhaps give thought to the development of microbial and host metrics that can assess states of host-microbe interactions so that course corrections can be made to promote good health.
Experimental Procedures
Animals
Germ-free IL-10 KO mice on a C57Bl/6J genetic background were bred in the University of Chicago Gnotobiotic Research Animal Facility (GRAF) and fecal microbiota transplanted (conventionalized) with Helicobacter hepaticus-free microbiota (kindly provided by Dr. Cathryn Nagler, University of Chicago) as previously described (Gillilland et al., 2012). Following conventionalization, these SPF IL-10 KO breeding pairs were bred in-house under H. hepaticus-free conditions (Institutional Animal Care and Use Committee (IACUC) protocol 71084). Five initial breeding pairs were prepared and their progeny were transferred to bedding to normalize gut microbes. Bedding was mixed at 4 days and 9 days following cage changes every 2 wks until breeding was initiated. Following this normalization procedure, 10 breeding pairs were prepared to obtain two parturitions for no treatment (NT) and CPZ treatment groups (Figure 1A). In cohort 1, the 1st and 2nd litters were used as NT controls or the CPZ group, respectively. In cohort 2, the 1st and 2nd litters were used as NT controls. Wild-type C57Bl/6J mice underwent the same breeding, microbial normalization, and CPZ treatment protocol for comparison.
Antibiotic treatment
CPZ sodium salt was purchased from Sigma-Aldrich (St. Louis, MO). CPZ (0.5 mg/ml) was administered in drinking water beginning at the 3rd wk of gestation until weaning of pups (Figure S1A). Mice had free access to water throughout the treatment period (IACUC protocol 72101). The same dose of CPZ was administered to adult IL-10 KO mice between 12 to 20 (18.9 ±2.9) wks of age for 4 wks.
Clinical evaluation of spontaneous colitis
Body weights of mice in cohort 1 and 2 were measured weekly beginning at weaning. The euthanasia criteria for spontaneous colitis included rectal prolapse, more than 15% body weight loss, or signs of pain/distress including poor grooming, decreased activity, and hunched posture (Hale and Greer, 2012).
DSS-induced colitis
Mice were given 2.5% DSS (36–50 kDa) (MP Biomedicals, Santa Ana, CA) in drinking water for 10 days (IACUC protocol 72101). All mice were weighed and monitored daily. Mice exhibiting more than 20% body weight loss were euthanized.
Fecal Microbiota Transplantation (FMT) in GF IL-10 KO mice
Fecal samples collected from NT and CPZ-exposed dams at weaning were used to gavage (FMT) 200uL of fecal supernatant (100mg feces per 1mL sterile PBS) into GF IL-10 KO female recipients maintained in separate flexible film isolators (See model, Figure 5A). NT and CPZ-FMT females were paired with GF IL-10 KO males for breeding. Pups were utilized for flow cytometry and qPCR analysis at 3 wks of age (weaning). Fecal pellets were collected from dams and offspring for microbial analysis.
Histological analysis
Colon samples were fixed in 4% formaldehyde and embedded in paraffin followed by H&E staining. Colitis histological score for colitis was employed as previously described (Erben et al., 2014).
Fecal DNA extraction and 16S rRNA gene amplicon analysis
Fecal samples of tracked offspring in cohort 1 were harvested and rapidly frozen at −80°C at 3 7, 11 and 23 wks of age. DNA was extracted as previously described (Wang et al., 2009), and the V4-V5 region of the 16S rRNA gene was amplified following EMP protocols (http://www.earthmicrobiome.org/emp-standard-protocols/16s/). Sequencing was performed on an Illumina MiSeq sequencer at the High-Throughput Genome Analysis Core, Argonne National Laboratory, Illinois. Raw sequencing data were de-multiplexed, and partially overlapping paired-end reads were merged using illumina-utils (Eren et al., 2013b). Mismatches at the overlapping regions of pairs were resolved using the base with the higher Q-score, and the merged sequences were kept for downstream analyses only (1) if they contained three or less mismatches at the overlapping region, and (2) 66% of the bases in the first half of each read had an average Q-score of 30. The quality filtered reads were partitioned into ecologically relevant units using Minimum Entropy Decomposition (MED) (Eren et al., 2015b) with default parameters. Using Shannon entropy, MED resolves a given amplicon dataset iteratively into high-resolution oligotypes (Eren et al., 2013a). Taxonomy was assigned to oligotypes using GAST (Huse et al., 2008), and Anvi’o v2.3.1 (Eren et al., 2015a) was used to visualize the relative abundance of each oligotype across samples in the context of metadata. Oligotype community data were normalized and taxonomic relative abundances in NT and CPZ groups were compared for female and male separately at each time point. Oligotypes with significant differences between NT and CPZ communities were filtered for oligotypes with greater than 10-fold changes in relative abundance, then further filtered for the top ten oligotypes exhibiting the greatest difference in mean relative abundance. Extended error plots were created using Statistical analysis of taxonomic and functional profiles (STAMP) v2.1.3 (Parks et al., 2014).
Statistical analysis
Spontaneous and DSS-induced colitis incidence by NT or CPZ treatment was estimated by “survival analysis”, performed using log-rank test followed by Kaplan-Meyer plot. Mann-Whitney U test was used to compare body weights, histological scores, T cell populations, mRNA expression levels, protein levels, or 16s rRNA gene copy number between NT and CPZ groups. Kruskal-Wallis test and Dunn’s test were also employed for 16s rRNA gene copy number. These tests were performed with GraphPad Prism (GraphPad Software, CA, USA). Statistical significance was assumed when p≤0.05. Raw counts of oligotypes were normalized using the “decostand” function implemented in R/CRAN package “vegan”. Alpha diversity via Shannon diversity index was computed using “diversity” function implemented in the “vegan” package. Student’s t-test was used to compare oligotype enrichment at 3, 7, and 11 wks of age between IL-10 KO pups from NT versus CPZ-exposed dams and between male pups from CPZ-exposed dams that eventually developed spontaneous colitis versus male pups that remained healthy. The p-values were adjusted for multiple-test using Benjamini-Hochberg method (Benjamini and Hochberg, 1995). The criterion of significance was set at false discovery rate (FDR)<0.05.
Supplemental Experimental Procedures
Methods for immunological analyses are included in the supplemental file.
Supplementary Material
Supplemental Table 1, Olygotype unit numbers and corresponding taxons with significant changes between no-treatment (NT) and cefoperazone (CPZ)-exposed offspring (sorted by mean population), Related to Figure 4.
Acknowledgments
The present research was supported by the NIDDK Digestive Disease Core Research Center (NIH P30 DK42086), NIDDK grants R37 DK47722 (EBC), T32 DK007074 (AMB), F31 DK107297 (AMB), and the GI Research Foundation of Chicago. We acknowledge the generous support of the David and Ellen Horing Research Fund and thank the Human Tissue Resource Center for histological processing as well as the Gnotobiotic Research Core Facility staff for GF animal husbandry. We also thank Karen Yang and Dr. Mark W. Musch for sample acquisition/analysis as well as Dr. Mrinalini Rao for proofreading the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.M., A.M.B., V.L. and E.B.C. conceived the study, designed experiments, and prepared the manuscript. J.M., A.M.B., and S.M. performed experiments and analyzed data. J.M., Y.H., N.H., T.O.D., and A.M.E. analyzed microbial data sets. E.B.C. oversaw the entire project.
Distribution of Data
Mouse sample information and microbial dataset can be found via BioProject ID PRJNA376026 and Sequence Read Archive accession SRP108147.
Conflict of Interests
The authors declare no conflict of interests
References
- Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98:229–238. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asnicar F, Manara S, Zolfo M, Truong DT, Scholz M, Armanini F, Ferretti P, Gorfer V, Pedrotti A, Tett A, et al. Studying Vertical Microbiome Transmission from Mothers to Infants by Strain-Level Metagenomic Profiling. mSystems. 2017:2. doi: 10.1128/mSystems.00164-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bager P, Simonsen J, Nielsen NM, Frisch M. Cesarean section and offspring’s risk of inflammatory bowel disease: a national cohort study. Inflamm Bowel Dis. 2012;18:857–862. doi: 10.1002/ibd.21805. [DOI] [PubMed] [Google Scholar]
- Benchimol EI, Mack DR, Guttmann A, Nguyen GC, To T, Mojaverian N, Quach P, Manuel DG. Inflammatory bowel disease in immigrants to Canada and their children: a population-based cohort study. Am J Gastroenterol. 2015;110:553–563. doi: 10.1038/ajg.2015.52. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Broe A, Pottegard A, Lamont RF, Jorgensen JS, Damkier P. Increasing use of antibiotics in pregnancy during the period 2000–2010: prevalence, timing, category, and demographics. BJOG. 2014;121:988–996. doi: 10.1111/1471-0528.12806. [DOI] [PubMed] [Google Scholar]
- Caufield PW, Cutter GR, Dasanayake AP. Initial acquisition of mutans streptococci by infants: evidence for a discrete window of infectivity. J Dent Res. 1993;72:37–45. doi: 10.1177/00220345930720010501. [DOI] [PubMed] [Google Scholar]
- Caufield PW, Saxena D, Fitch D, Li Y. Population structure of plasmid-containing strains of Streptococcus mutans, a member of the human indigenous biota. J Bacteriol. 2007;189:1238–1243. doi: 10.1128/JB.01183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B, Srinivasan G, Delgado MA, Young AN, Gewirtz AT, Vijay-Kumar M. Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS One. 2012;7:e44328. doi: 10.1371/journal.pone.0044328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erben U, Loddenkemper C, Doerfel K, Spieckermann S, Haller D, Heimesaat MM, Zeitz M, Siegmund B, Kuhl AA. A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int J Clin Exp Pathol. 2014;7:4557–4576. [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Esen OC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015a;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML. Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol. 2013a:4. doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML. Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J. 2015b;9:968–979. doi: 10.1038/ismej.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Vineis JH, Morrison HG, Sogin ML. A filtering method to generate high quality short reads using illumina paired-end technology. PLoS One. 2013b;8:e66643. doi: 10.1371/journal.pone.0066643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funkhouser LJ, Bordenstein SR. Mom knows best: the universality of maternal microbial transmission. PLoS Biol. 2013;11:e1001631. doi: 10.1371/journal.pbio.1001631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillilland MG, 3rd, Erb-Downward JR, Bassis CM, Shen MC, Toews GB, Young VB, Huffnagle GB. Ecological succession of bacterial communities during conventionalization of germ-free mice. Appl Environ Microbiol. 2012;78:2359–2366. doi: 10.1128/AEM.05239-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale LP, Greer PK. A novel murine model of inflammatory bowel disease and inflammation-associated colon cancer with ulcerative colitis-like features. PLoS One. 2012;7:e41797. doi: 10.1371/journal.pone.0041797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh AL, Shapiro DJ, Pavia AT, Shah SS. Antibiotic prescribing in ambulatory pediatrics in the United States. Pediatrics. 2011;128:1053–1061. doi: 10.1542/peds.2011-1337. [DOI] [PubMed] [Google Scholar]
- Howarth GF, Robinson MH, Jenkins D, Hardcastle JD, Logan RF. High prevalence of undetected ulcerative colitis: data from the Nottingham fecal occult blood screening trial. Am J Gastroenterol. 2002;97:690–694. doi: 10.1111/j.1572-0241.2002.05586.x. [DOI] [PubMed] [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Mark Welch D, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hviid A, Svanstrom H, Frisch M. Antibiotic use and inflammatory bowel diseases in childhood. Gut. 2011;60:49–54. doi: 10.1136/gut.2010.219683. [DOI] [PubMed] [Google Scholar]
- Jimenez E, Marin ML, Martin R, Odriozola JM, Olivares M, Xaus J, Fernandez L, Rodriguez JM. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008;159:187–193. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keubler LM, Buettner M, Hager C, Bleich A. A Multihit Model: Colitis Lessons from the Interleukin-10-deficient Mouse. Inflamm Bowel Dis. 2015;21:1967–1975. doi: 10.1097/MIB.0000000000000468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronman MP, Zaoutis TE, Haynes K, Feng R, Coffin SE. Antibiotic exposure and IBD development among children: a population-based cohort study. Pediatrics. 2012;130:e794–803. doi: 10.1542/peds.2011-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- Kullberg MC, Ward JM, Gorelick PL, Caspar P, Hieny S, Cheever A, Jankovic D, Sher A. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infection and immunity. 1998;66:5157–5166. doi: 10.1128/iai.66.11.5157-5166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TC, Stappenbeck TS. Genetics and Pathogenesis of Inflammatory Bowel Disease. Annu Rev Pathol. 2016 doi: 10.1146/annurev-pathol-012615-044152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayfach S, Rodriguez-Mueller B, Garud N, Pollard KS. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 2016;26:1612–1625. doi: 10.1101/gr.201863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, Timmis KN, Schreiber S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantoja-Feliciano IG, Clemente JC, Costello EK, Perez ME, Blaser MJ, Knight R, Dominguez-Bello MG. Biphasic assembly of the murine intestinal microbiota during early development. ISME J. 2013;7:1112–1115. doi: 10.1038/ismej.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen I, Gilbert R, Evans S, Ridolfi A, Nazareth I. Oral antibiotic prescribing during pregnancy in primary care: UK population-based study. J Antimicrob Chemother. 2010;65:2238–2246. doi: 10.1093/jac/dkq307. [DOI] [PubMed] [Google Scholar]
- Pugazhendhi S, Sahu MK, Subramanian V, Pulimood A, Ramakrishna BS. Environmental factors associated with Crohn’s disease in India. Indian J Gastroenterol. 2011;30:264–269. doi: 10.1007/s12664-011-0145-1. [DOI] [PubMed] [Google Scholar]
- Sakata T, Niwa Y, Goto H, Hirooka Y, Hayakawa T, Ohmiya N, Kobayashi S. Asymptomatic inflammatory bowel disease with special reference to ulcerative colitis in apparently healthy persons. Am J Gastroenterol. 2001;96:735–739. doi: 10.1111/j.1572-0241.2001.03614.x. [DOI] [PubMed] [Google Scholar]
- Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, Jian R, Dore J. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infection and immunity. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokholm J, Schjorring S, Pedersen L, Bischoff AL, Folsgaard N, Carson CG, Chawes BL, Bonnelykke K, Molgaard A, Krogfelt KA, et al. Prevalence and predictors of antibiotic administration during pregnancy and birth. PLoS One. 2013;8:e82932. doi: 10.1371/journal.pone.0082932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungaro R, Bernstein CN, Gearry R, Hviid A, Kolho KL, Kronman MP, Shaw S, Van Kruiningen H, Colombel JF, Atreja A. Antibiotics associated with increased risk of new-onset Crohn’s disease but not ulcerative colitis: a meta-analysis. Am J Gastroenterol. 2014;109:1728–1738. doi: 10.1038/ajg.2014.246. [DOI] [PubMed] [Google Scholar]
- Van Limbergen J, Radford-Smith G, Satsangi J. Advances in IBD genetics. Nat Rev Gastroenterol Hepatol. 2014;11:372–385. doi: 10.1038/nrgastro.2014.27. [DOI] [PubMed] [Google Scholar]
- Virta L, Auvinen A, Helenius H, Huovinen P, Kolho KL. Association of repeated exposure to antibiotics with the development of pediatric Crohn’s disease--a nationwide, register-based finnish case-control study. American journal of epidemiology. 2012;175:775–784. doi: 10.1093/aje/kwr400. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, Sun J, Antonopoulos DA, Chang EB, Claud EC. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3:944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1, Olygotype unit numbers and corresponding taxons with significant changes between no-treatment (NT) and cefoperazone (CPZ)-exposed offspring (sorted by mean population), Related to Figure 4.