Abstract
The physiological relevance of Matrigel as a cell-culture substrate and in angiogenesis assays is often called into question. Here, we describe an array-based method for the identification of synthetic hydrogels that promote the formation of robust in vitro vascular networks for the detection of putative vascular disruptors, and that support human embryonic stem cell expansion and pluripotency. We identified hydrogel substrates that promoted endothelial-network formation by primary human umbilical vein endothelial cells and by endothelial cells derived from human induced pluripotent stem cells, and used the hydrogels with endothelial networks to identify angiogenesis inhibitors. The synthetic hydrogels show superior sensitivity and reproducibility over Matrigel when evaluating known inhibitors, as well as in a blinded screen of a subset of 38 chemicals, selected according to predicted vascular disruption potential, from the Toxicity ForeCaster library of the US Environmental Protection Agency. The identified synthetic hydrogels should be suitable alternatives to Matrigel for common cell-culture applications.
Because of the increasing number of diseases associated with vascular disorders, the ability to detect compounds that affect the human vasculature is becoming more important. Vascular disorders include various forms of cancer, atherosclerosis, stroke, diabetic retinopathy and developmental complications, all of which can result from exposure to some chemicals present in the environment1,2. In this regard, early studies led to the development of an in vitro endothelial-network-formation assay3, in which cloned capillary endothelial cells formed interconnected endothelial networks after approximately 10 days in culture. A more rapid assay (<24 h) described in 1988 has become a gold-standard method for the identification of inhibitors and stimulators of angiogenesis in drug discovery4 and toxicological screens2,5–9.
The assay developed in 1988 has a number of inherent complexities, due in part to the use of a natural extracellular matrix derived from Englebreth-Holm-Swarm (EHS) tumors produced in mice, referred to as Matrigel™, EHS matrix™, or Geltrex™ (and hitherto referred to as Matrigel)10. Matrigel is used in multiple applications as substrates in human cell culture and organoid assembly, with two of the most common uses being angiogenesis assays and the expansion of undifferentiated human embryonic stem cells (hESCs)11,12. However, Matrigel is inherently limited by its compositional complexity and lack of lot-to-lot reproducibility. Recent proteomic analysis of normal and “growth-factor reduced” Matrigel identified a total of 1851 unique proteins, and individual tests from two manufacturers showed only 53% batch-to-batch similarity in proteins identified13. Matrigel’s low and variable elastic modulus, ranging from of 0.12 to 0.45 kPa14, results in poor handling characteristics, a need for precise temperature control, and user-to-user variability. Numerous confounding factors such as locally sequestered and matrix-bound growth factors15. as well as physiologically irrelevant mechanisms of inhibition such as bulk matrix dissolution by Suramin treatment16,17, have previously resulted in the identification of false positives and false negatives in Matrigel-based chemical compound screens. Moreover, the introduction of xenogenic components by Matrigel interferes with mechanistic studies of cell behavior and limits therapeutic applications of stem cells expanded in culture12.
Synthetic and natural extracellular matrices (ECMs; for example, collagen, fibrin and vitronectin) are suitable alternatives to Matrigel for assembling endothelial networks and for expanding stem cell populations. Natural ECMs, however, are often animal-derived (bringing xenogenic material into the culture environment) and often are presented as coatings that fail to mimic the mechanical properties of the native ECM. In endothelial-network-formation assays, other natural ECMs fail to form endothelial networks similar to those of Matrigel without the addition of a supporting cell type. Chemically defined synthetic hydrogels have received increased attention as suitable alternatives to Matrigel and to other natural ECMs due to their minimal batch-to-batch variation, increased reproducibility, defined material properties, compositions and controllable degradation properties18–20. Isolated components of Matrigel (such as laminin and collagen type IV) and synthetic poly(ethylene glycol) (PEG) hydrogels in the form of enzymatically crosslinking PEG-vinyl sulfone (PEG-VS) with varying stiffness were systematically screened to control early events in neurogenesis. Human embryonic stem cells were encapsulated in a broad range of hydrogel conditions and material properties were shown to impact the differentiation of ESCs towards an ectodermal fate and their later dorsal ventral patterning to model that of the developing hindbrain and spinal cord21. Another study demonstrated the ability of synthetic PEG hydrogels to optimize reprogramming efficiency of mouse and human fibroblasts into induced pluripotent stem cells and later maintain their phenotype in 3D environments22. Chemically defined synthetic hydrogels have additionally demonstrated the ability to support the expansion of intestinal stem cells and the formation of intestinal organoids23, control the formation of epithelial Madin-Darby Kidney cysts24, alter stem cell fate 25–29, generate tumorigenesis models30–33 that are similar or superior to Matrigel and their natural ECM counterparts.
In this study, we applied an array based method of optimizing synthetic hydrogels for use in vascular toxicity assays and hESC expansion. We used a synthetic hydrogel composed of a photo-crosslinked, cell-degradable polyethylene glycol (PEG) hydrogels formed via a step-growth reaction to generate hydrogels with controlled mechanical properties and presentation of bioactive peptides that mimic functional groups of larger ECM molecules. The chemistry used in this paper, comparisons to other materials, and array based methods are discussed in the Supplemental Information. We evaluated over 1200 distinct synthetic hydrogels and identified chemically defined, synthetic hydrogels that replaced the role of Matrigel in vascular screening assays and hESC expansion. The resulting poly (ethylene glycol) (PEG) hydrogels had defined characteristics, including cell-adhesion properties, shear modulus (stiffness), and non-covalent binding affinity for vascular endothelial growth factor (VEGF). These hydrogels supported robust formation of endothelial networks from endothelial cells derived from multiple primary human sources, unprecedented batch-to-batch reproducibility, and increased sensitivity to known angiogenesis inhibitors. The synthetic hydrogel-based assay was superior to the commonly used Matrigel-based assay in its ability to qualify the relative anti-angiogenic activities across a 38-chemical subset from the EPA’s ToxCast™ chemical library that was selected for evaluation based on in vitro cellular and molecular bioactivity profiles that predict their potential for vascular disruption, ranging from inactivity to strong inhibition. Similarly, the array-based hydrogel identification methods applied in these studies enabled discovery of suitable substrates for hESC adhesion and maintenance of pluripotency. These results indicate that an array-based discovery approach can be used to identify relatively simple, synthetic materials that provide superior utility when compared to more complex naturally-derived materials.
Results
Approach for Identification of Synthetic Hydrogel Formulations
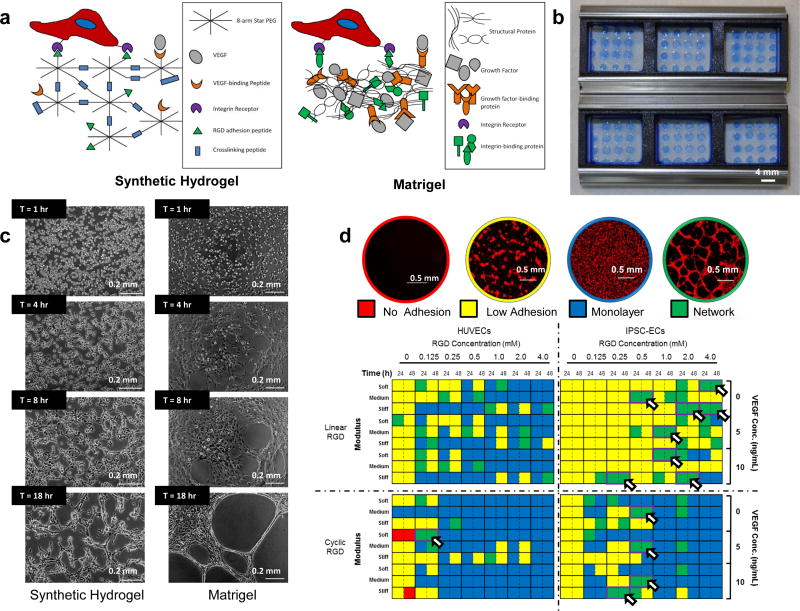
The approach to identify synthetic hydrogels that supported specific intended cell behaviors (e.g. tubulogenesis, hESC culture) used arrays of 120 µm-thickness synthetic hydrogels (thin hydrogel arrays) that mimicked properties of the native extracellular matrix (ECM). The inclusion of pendant linear H-Cys-Arg-Gly-Asp-Ser-NH2 (linear RGD) or head-to-tail cyclized Arg-Gly-Asp-[d-Phe]-Cys (cyclic RGD) peptides mediated cell adhesion to the hydrogels through the RGD motif commonly present in integrin-binding ECM proteins, such as laminin, vitronectin, and fibronectin34 (Figure 1A, D). The mechanical properties of the hydrogels were set at 0.45 ± 0.04 kPa (soft), 1.16 ± 0.08 kPa (medium) or 4.72 ± 0.17 kPa (stiff) by tuning concentrations of 20 kDa, 8-arm PEG-norbornene (PEGNB) and dithiol-terminated crosslinking molecules used to form the hydrogels (Figure 1D, Supp. Fig. 1A). All hydrogel solutions were polymerized using a crosslinking peptide H-Lys-Cys-Gly-Gly-Pro-Gln-Gly-Ile-Trp-Gly-Gln-Gly-Cys-Lys-NH2 that was degradable by matrix metalloproteinases (MMPs)35, thereby enabling cell-mediated remodeling of the hydrogel substrates36. All peptides included in the hydrogels were covalently attached to PEG via thiol-ene “click” chemistry, which couples thiols to norbornene groups on the PEG molecules37. Finally, the ability to bind and sequester vascular endothelial growth factor (VEGF) was provided by a receptor-mimicking VEGF binding peptide (VBP) included in the hydrogels38 (Figure 1A, Supp. Fig. 2). Further methods and results for evaluating hydrogel crosslinking efficiency and long-term swelling behavior may be found in the Supplemental Results and Discussion and the Supplemental Methods. Taken together, these hydrogels presented a simple, chemically defined set of cell-signaling cues relative to the complex ECM presented by Matrigel.
Figure 1.
An endothelial cell culture system identifies environments that enable endothelial network formation on synthetic PEG-based hydrogels. (A) Schematic of endothelial cell seeded on synthetic hydrogels and Matrigel. (B) Thin hydrogel arrays assembled with 3-well ProPlate® Cell Culture Isolators. Hydrogel spots were stained using Brilliant Blue for visualization. (C) Left: Endothelial network formation on synthetic hydrogels and Matrigel over time. Right: Typical endothelial networks formed on synthetic hydrogels and Matrigel between 1 and 18 hours after seeding. (D) Qualitative scoring system for HUVEC network formation and heat map showing initial screening results for HUVEC and iPSC-EC network formation. Arrowheads denote environments that enabled HUVEC network formation within 24 hours after seeding and maintained structural integrity over the 48-hour culture period (n = 3). The initial screen was performed once over the course of these studies. Red: Cell Tracker Red.
Identifying Materials to Promote Endothelial Network Formation
Thin hydrogel arrays (Figure 1B) identified cell culture environments that supported robust endothelial network formation by human umbilical vein endothelial cells (HUVECs) as well as endothelial cells derived from human induced pluripotent stem cells (iPSC-ECs). Successful culture environments promoted endothelial network assembly by endothelial cells (ECs) within 24 hours of seeding (Figure 1C), and maintained network stability 48 hours after seeding (Figure 1D). Multiple culture environments successfully supported HUVEC network formation after 24 hours, but HUVEC networks typically disassembled during the subsequent 24-hour period. A single condition, characterized as 2 mM PEG, 0.125 mM cyclic RGD, and 4 mM crosslinking peptide (50% cross-linked) in the substrate, with 5 ng/ml soluble VEGF present in media, supported prolonged endothelial network stability for 24 hours after initial endothelial network formation (Figure 1D). Interestingly, the 0.45 kPa stiffness of the identified hydrogel fell approximately within the average stiffness range of Matrigel14. As demonstrated during time-lapse microscopy, the synthetic hydrogel formulation identified here reproducibly formed endothelial networks within six to eight hours of seeding (Figure 1C, Supp. Movie 1). Additionally, the identified hydrogel formulation was suitable as embedding substrates in aortic ring sprouting assays (Supp. Fig. 5). Further discussion of this experiment may be found in the Supplemental Results and Discussion and the Supplemental Methods.
Thin hydrogel arrays also identified culture conditions that promoted endothelial network assembly by iPSC-ECs39. Multiple culture environments supported iPSC-EC network formation within 24 hours after seeding and maintained network stability for 24 hours after initial endothelial network formation (Figure 1D). Successful culture environments detected by the thin-hydrogel arrays were adapted for use in 96-well plates for toxicity screening experiments. The amount of photoinitiator and volume of hydrogel used were optimized to 0.2% w/v Irgacure 2959 (I2959) and 9 µL solution per well, respectively, to produce hydrogels with the flattest and most uniform surfaces for seeding of HUVECs or iPSC-ECs (Supp. Fig. 6) without changing the fundamental swelling properties of the hydrogels (Supp. Fig. 4C). This reduced hydrogel buckling and out-of-plane imaging of the 96 well plate, resulting in reduced imaging time and non-uniformity between samples.
Confocal images showed the organization of HUVECs and iPSC-ECs into clearly branching, interconnected networks on both synthetic hydrogels and Matrigel (Figure 1D, Figure 2A). All ECs forming networks on synthetic hydrogels displayed an ability to migrate and develop cell-cell contacts consistent with the process of capillary morphogenesis in vivo (Figure 1C, Supp. Movies 1–3). The ability to form endothelial networks on the synthetic hydrogels was not limited to only ECs expanded in culture, but also demonstrated the ability to form similar endothelial networks directly from cryopreservation (Supp. Fig. 1B–C).
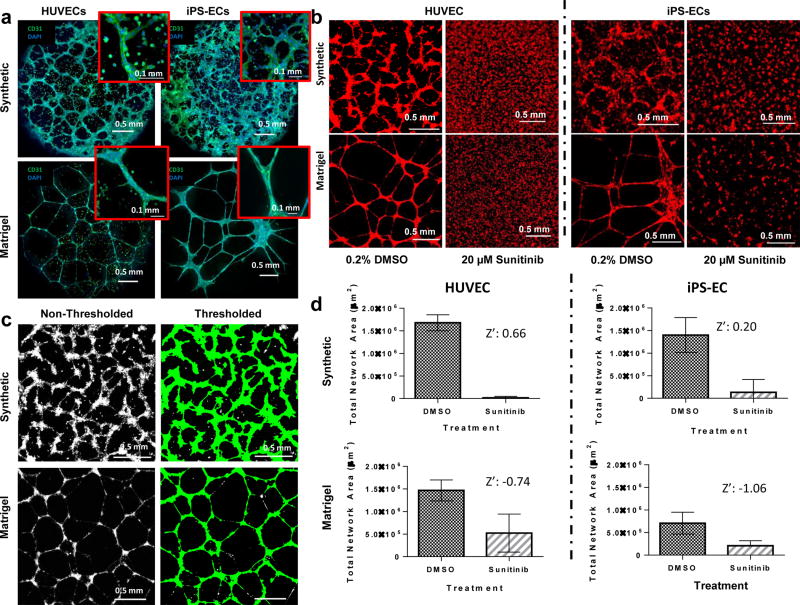
Figure 2.
Visualization and quantification of endothelial networks on synthetic hydrogels and Matrigel. (A) Confocal microscopy images of HUVECs and iPSC-ECs forming networks on synthetic versus Matrigel substrates. Insets: Enhanced magnification of multicellular structures. Scale bar: 0.1 mm (B) HUVECs and iPSC-ECs seeded on synthetic and Matrigel substrates form endothelial networks in DMSO-treated conditions. Network formation is disrupted by Sunitinib treatment. (C) HUVEC networks on synthetic and Matrigel substrates are identified by thresholding fluorescence intensity and object size. Network formation is quantified as total object area per substrate. (D) Endothelial network formation in synthetic hydrogel and Matrigel systems is quantified and subjected to the Z’ test to verify their efficacy of as toxicity screening systems (n = 48). Data represents means and error bars represent standard deviations. The Z’ test was performed twice over the course of these studies. Green: CD31. Red: Cell Tracker Red. Blue: DAPI nuclear stain.
Utility of Synthetic Hydrogels for Vascular Inhibition Assays
Synthetic hydrogel-based endothelial network formation assays accurately and reproducibly distinguished endothelial networks disrupted using the known angiogenesis inhibitor Sunitinib from non-inhibited networks. Quantification of total endothelial network area in the inhibited and non-inhibited conditions (Figure 2B–C) resulted in a Z’ score of 0.66 for the synthetic hydrogel-based assay (considered an “excellent” assay40), while HUVEC networks formed on Matrigel resulted in a Z’ score of −0.74, indicating that inconsistencies in the Matrigel-based assay interfere with the identification of inhibiting versus non-inhibiting conditions (Figure 2D). Similarly, screening for Sunitinib activity on iPSC-EC networks formed on synthetic hydrogels produced a favorable Z’ score of 0.20, while iPSC-EC networks formed on Matrigel resulted in a poor Z’ score of −1.06 (Figure 2D).
HUVEC Network Sensitivity to Known VEGF Signaling Inhibitors
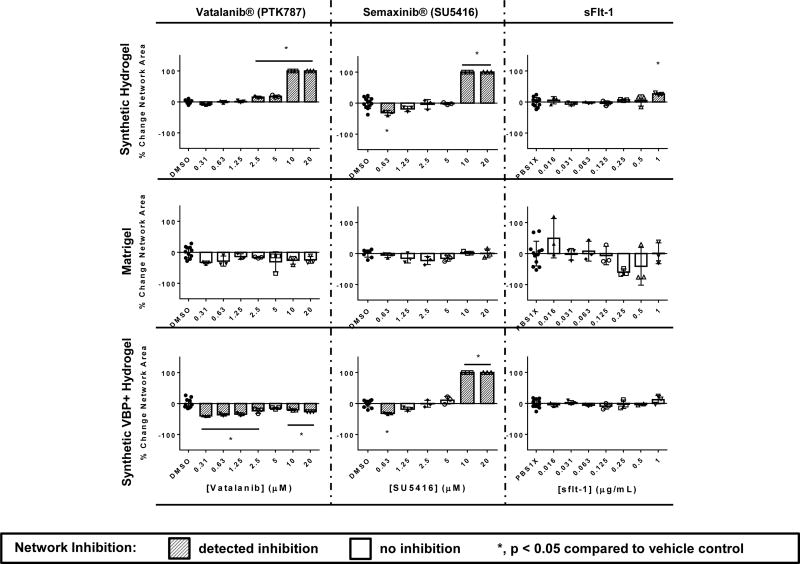
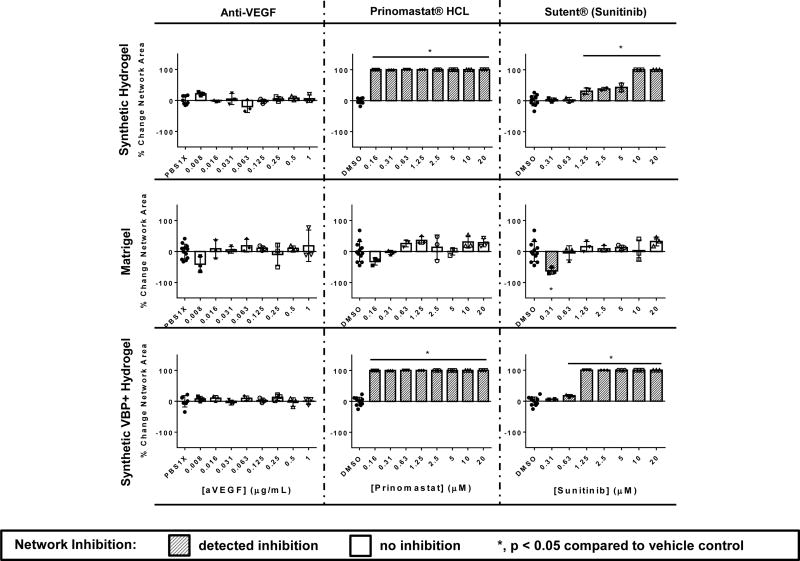
HUVEC networks on synthetic hydrogels demonstrated increased sensitivity to a number of known VEGF inhibitors in comparison to HUVEC networks on Matrigel, including Vatalanib®, Semaxanib®, Sunitinib®, and a soluble flt-1 (sFlt-1) receptor (Figure 3, Supp. Fig. 7A). In all of the above cases, the VEGF inhibitors demonstrated significant inhibition of network formation in a wider range of concentrations in the synthetic hydrogel-based assay compared to the Matrigel-based assay. Vatalanib® demonstrated inhibition at 2.5 – 20 µM concentrations on synthetic hydrogels, while inhibition was not detected Matrigel. Semaxanib® demonstrated inhibition at 0.63 µM and 10–20 µM on synthetic hydrogels, while inhibition was not detected on Matrigel. Anti-VEGF did not demonstrate inhibition on synthetic hydrogels or Matrigel. Sunitinib® demonstrated inhibition between concentrations 1.25 – 20 µM on the synthetic hydrogels, while inhibition was detected at 0.31 µM on Matrigel. On Matrigel this effective concentration was subject to change significantly across technical replicates. Comparisons of these results to those demonstrated in prior literature may be found in the Supplemental Results and Discussion.
Figure 3.
Concentration-dependent inhibition of endothelial network formation by known vascular inhibitors, measured as percent change in network area, compared to vehicle controls. Sensitivity of HUVECs to inhibitors is compared between the synthetic, VBP+ and Matrigel systems (n = 3, n = 8–12 for DMSO and PBS controls. Negative changes in area represent disconnected networks, while positive changes in area represent thickened network structures. A 100% change in area represents complete network disruption or monolayer formation.). Data represents means and error bars represent standard deviations. The test was performed twice over the course of these studies. Inhibited samples (*, and dark-gray bars) have a p-value < 0.05 by One-way ANOVA followed by Dunnett’s multiple comparisons analysis compared to the DMSO/1× PBS control conditions depending on the vehicle of the inhibitor.
To gain insight into the influence of VEGF sequestering on performance of the tubulogenesis assays, we generated a separate set of synthetic hydrogels that supported endothelial network formation (Supp. Fig. 2), but also included a VEGF-binding peptide. Interestingly, the presence of VBP in synthetic hydrogels increased the effectiveness of VEGF receptor tyrosine kinase (RTK) inhibitors Vatalanib® and Sunitinib® in disrupting HUVEC network formation. In contrast sFlt-1, which binds soluble VEGF in media, did not result in significant changes to network area at any tested concentration when the synthetic hydrogel contained VBP. The effective concentration ranges of Semaxanib® and Prinomastat HCl were not changed between hydrogels that contained or lacked VBP. These data indicate that VEGF sequestration changes sensitivity of HUVEC networks to VEGF inhibitors, depending on whether they affect receptor tyrosine kinase activity or the activity of soluble VEGF as possible mechanisms of inhibition (Figure 3, Supp. Fig. 8).
A Differential Response to MMP Inhibition and Known Matrigel Dissolution Agents
HUVEC networks on synthetic hydrogels were more sensitive to an MMP inhibitor than HUVEC networks on Matrigel. HUVEC network formation on each of the synthetic hydrogels – with or without VBP – was inhibited by Prinomastat HCl, a broad-based inhibitor of MMPs-2, −3, −9, −13, and −14, and resulted in confluent cell sheet formation rather than network formation. In contrast, HUVEC network formation on Matrigel was unaffected by this MMP inhibitor (Figure 3, Supp. Fig. 7B, Supp. Fig. 9). Interestingly, the presence of VBP in synthetic hydrogels did not interfere with the inhibitory activity of Prinomastat HCl, and treatment also resulted in cell sheet formation rather than network formation. This is consistent with the expectation that Prinomastat HCl acts independently of VEGF signaling (Figure 3).
HUVEC networks on synthetic hydrogels were unaffected by a putative matrix dissolution agent, which dramatically disrupted network formation on Matrigel. Suramin Hydrochloride, an inhibitor to EGF, PDGF and TGFβ signaling16, inhibited network formation on Matrigel in a dose-dependent manner, and resulted in cell clustering indicative of apparent Matrigel dissolution17 and cell settling on an underlying polystyrene cell culture substrate. On synthetic hydrogels, Suramin Hydrochloride treatment minimally affected HUVEC network formation and did not dissolve the underlying material, suggesting the inhibitory effect on Matrigel was attributable to substrate dissolution rather than vascular inhibition (Supp. Fig. 10), as demonstrated previously17. This result illustrates a concern in interpretation of results from Matrigel endothelial network formation assays, as Matrigel dissolution is not a process relevant to disorders of in vivo angiogenesis.
Identification of Putative Vascular Disrupting Chemicals from the ToxCast Compound Library
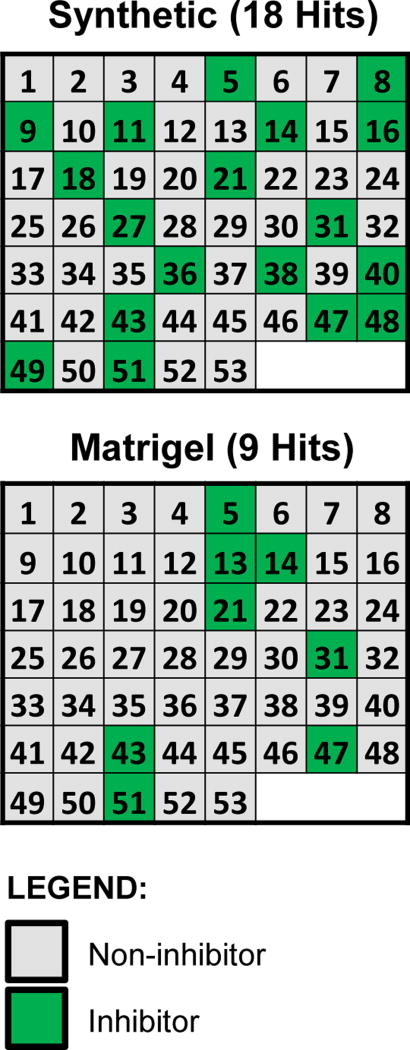
As a further investigation of the enhanced utility of the synthetic hydrogel system, we ran a comparison utilizing a subset of candidate chemical compounds from the ToxCast library. This test set included 38 unique chemicals ranging in their predicted potential as vascular disruptors (pVDCs) based on a signature derived from the ToxCast Phase I screen of 309 chemicals screened across 500 high-throughput screening (HTS) assays41,42. The ToxCast signature is translated by a pVDC cutoff provisionally set to 0.100, wherein 26 chemicals were predicted to have some anti-angiogenic activity and 12 chemicals were predicted inactive1,41,43–47. To best qualify this evaluation, the test panel was firstly blinded to the experimenters and secondly included 38 unique chemicals and an assortment of replicates to yield 53 samples. Each of these 53 samples were ran in triplicate. Results for the positive and negative calls after 24 hours in culture are shown for the blinded sample matrix (Figure 4A) and for the decoded list (Figure 4B). The synthetic hydrogel assay confirmed 11 out of the 26 provisional positives, while the Matrigel assay detected only five of them (overlap of four detected by both platforms) (Figure 4, Supp. Fig. 11, Table 1). In both cases, all positive calls fell into the higher pVDC scores (> 0.1) predicting greater disruptive activity (Table 1). Both the synthetic hydrogel and Matrigel assays identified an increasing number of inhibitors as pVDC scores increased. Inhibitory ‘hit’ conditions and non-inhibitory ‘miss’ conditions were compared to predicted inhibitory or non-inhibitory calls in a confusion matrix anchored to the pVDC domain (Table 1, Table 2)48. The test yielded an accuracy value of 60.5% in the synthetic hydrogel assay and 44.7% in the Matrigel assay. Sensitivity was 42.3% and 19.2% for the synthetic hydrogel and Matrigel assays, respectively, and the specificity in both systems was 100%. An F1 score, which measures the harmonic mean of precision and recall was 0.59 in the synthetic and 0.32 in the Matrigel assay (scored from 0 to 1). Matthew’s Correlation Coefficient, which measures the quality of binary classification, was 0.43 in the synthetic hydrogel versus 0.26 in Matrigel assay (scored from 0 to 1). The balanced accuracy, which takes into account both the sensitivity and specificity of both systems, and removes potential bias from unbalanced datasets49, gave a value of 72.2% in the synthetic versus 68.1% in the Matrigel assay.
Figure 4.
Identification of vascular inhibitors from a subset of candidate chemical compounds from the ToxCast library through the use of synthetic hydrogel versus Matrigel systems. The criterion for a positive call was total area of inhibitor-treated endothelial networks at two standard deviations above or below the mean of vehicle (DMSO)-treated network areas. The test panel was blinded to the experimenters and included 38 unique candidate chemical compounds and an assortment of replicates to yield 53 total samples. Positive and negative calls are shown in the blinded sample matrix.
Table 1.
Results gathered from the 53 blinded samples were compiled into a decoded list of the 38 unique chemicals and sorted according to the previously-derived pVDC scores from ToxCast utilizing a provisional score of 0.1 to discriminate between pVDCs and non-pVDCs. Blue: provisional negative. Red: provisional positive.
| Chemical Name: | Matrigel | Synthetic | pVDC score |
|---|---|---|---|
| 1,2,4-Trichlorobenzene | 0 | 0 | 0.000 |
| Decane | 0 | 0 | 0.000 |
| Tris(2-chloroethyl) phosphate | 0 | 0 | 0.000 |
| 1,2,3-Trichloropropane | 0 | 0 | 0.002 |
| Pymetrozine | 0 | 0 | 0.002 |
| Methimazole | 0 | 0 | 0.002 |
| Diethanolamine | 0 | 0 | 0.002 |
| Imazamox | 0 | 0 | 0.007 |
| D-Mannitol | 0 | 0 | 0.007 |
| Methylparaben | 0 | 0 | 0.010 |
| Valproic acid | 0 | 0 | 0.016 |
| 2,4-Diaminotoluene | 0 | 0 | 0.069 |
| Bisphenol A | 0 | 0 | 0.146 |
| Haloperidol | 0 | 0 | 0.177 |
| Tris(2-ethylhexyl) phosphate | 0 | 0 | 0.182 |
| Tris(1,3-dichloro-2-propyl)phosphate | 0 | 0 | 0.188 |
| Cladribine | 0 | 0 | 0.196 |
| TNP-470 | 0 | 0 | 0.238 |
| Oxytetracycline dihydrate | 0 | 0 | 0.260 |
| Celecoxib | 0 | 1 | 0.269 |
| Docusate sodium | 0 | 0 | 0.304 |
| C.I. Solvent Yellow 14 | 0 | 1 | 0.306 |
| Reserpine | 0 | 0 | 0.307 |
| Quercetin | 0 | 1 | 0.309 |
| Phenolphthalein | 0 | 0 | 0.327 |
| 5HPP-33 | 1 | 0 | 0.327 |
| tert-Butylhydroquinone | 0 | 0 | 0.336 |
| Triclocarban | 1 | 1 | 0.362 |
| Triclosan | 0 | 1 | 0.372 |
| Pyridaben | 0 | 1 | 0.379 |
| 1-Hydroxypyrene | 1 | 1 | 0.386 |
| Sodium dodecylbenzenesulfonate | 0 | 0 | 0.429 |
| Disulfiram | 1 | 1 | 0.432 |
| Fluazinam | 1 | 1 | 0.434 |
| Octyl gallate | 0 | 1 | 0.450 |
| Bisphenol AF | 0 | 1 | 0.457 |
| PFOS | 0 | 0 | 0.460 |
| 4-Nonylphenol, branched | 0 | 0 | 0.461 |
pVDC Score: Non-Inhibitory
 Inhibitory
Inhibitory
Table 2.
Performance of the synthetic hydrogel and Matrigel systems evaluated by a confusion matrix anchored to the provisional ToxCast pVDC scoring system.
| Assay Outcome | Matrigel (n=3) | Synthetic Hydrogel (n=3) |
|---|---|---|
| Number of Compounds Screened (n) | 38 | 38 |
| Predicted positive (P’) | 26 | 26 |
| Identified Positive Conditions (P) | 5 | 11 |
| True positive (TP) | 5 | 11 |
| False positive (FP) | 0 | 0 |
| Predicted negative (N’) | 12 | 12 |
| Identified Negative Conditions (N) | 33 | 27 |
| True negative (TN) | 12 | 12 |
| False negative (FN) | 21 | 15 |
| False positive rate (FPR) | 0% | 0 % |
| False negative rate (FNR) | 80.8% | 57.7% |
| False Discovery Rate (FDR) | 0 % | 0 % |
| True positive rate (TPR/ Specificity) | 100% | 100 % |
| True negative rate (TNR/Sensitivity) | 19.2% | 42.3% |
| Prevalence (PRV) | 0.68 | 0.68 |
| Positive Predictive Value (TP/TP+FP = PPV) | 1 | 1 |
| Accuracy (ACC) | 44.7 % | 60.5 % |
| Balanced Accuracy (BACC) | 68.2 % | 72.2% |
| F1 Score (F1) | 0.32 | 0.59 |
| Matthew’s Correlation Coefficient (MCC) | 0.26 | 0.43 |
Identifying Hydrogels to Maintain Human Embryonic Stem Cell Pluripotency during Initial Expansion
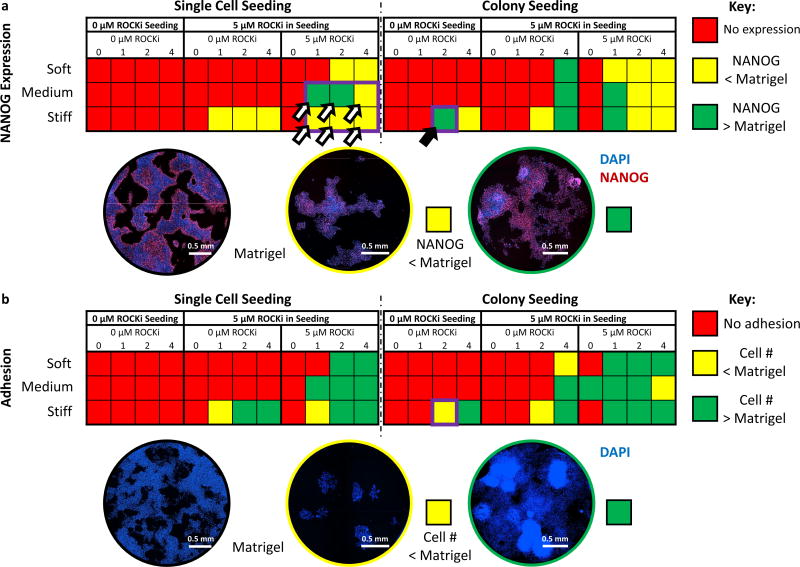
Hydrogel arrays (Figure 1B) identified multiple hydrogel formulations that maintained short-term NANOG expression by hESCs at greater or equal levels compared to hESCs cultured on Matrigel, 4 days after seeding (Figure 5A). Defined environmental properties included three varying levels of hydrogel stiffness, i.e. 1.5 kPa (soft), 3 kPa (medium) and 10 kPa (stiff) (Figure 5, Supp. Fig. 12), a range of cyclic RGD cell adhesion peptide concentrations in the hydrogels, and ROCK inhibitor (Y-27632) concentrations in the medium during seeding and during the 4-day maintenance period. Out of a total of 64 possible conditions (Figure 5), a number of hydrogel formulations successfully maintained NANOG expression to levels greater than or equal to Matrigel controls (Figure 5A). Particularly, one hydrogel formulation containing 4 mM PEG, 2 mM cyclic RGD, and 12 mM 3.4 kDa dithiolated PEG crosslinking molecule (10 kPa), increased NANOG expression without the need to include ROCK Inhibitor in either the cell seeding or during maintenance on the hydrogel substrate. Other conditions, which contained 4 mM PEG, 4 mM cyclic RGD, and 12 mM 3.4 kDa dithiolated PEG crosslinking molecule (10 kPa), enabled increased NANOG expression along with increased cell adhesion relative to Matrigel (Figure 5A, B). These conditions encompassed situations in which hESCs were seeded as single cells or colonies, with or without ROCK Inhibitor during culture maintenance.
Figure 5.
Material-dependent maintenance of hESC pluripotency. (A) Quantitative heat map of hESC NANOG expression relative to Matrigel in varying synthetic hydrogel-based culture conditions (n=3, n=5 in colony seeding conditions where ROCK inhibitor was removed, n=10 in single-cell seeding conditions where ROCK inhibitor was removed). Cells were cultured with either 0 or 5 µM ROCK inhibitor in maintenance culture, and hydrogels contained 0, 1, 2 or 4 mM cyclic RGD. The screen was performed once over the course of these studies. Conditions highlighted with a black arrowhead denote environments that maintained both hESC pluripotency and cell attachment over a 96-hour culture period without the use of ROCK Inhibitor. Conditions highlighted with a white arrowhead denote conditions that were further investigated for OCT3/4 expression in addition to NANOG expression. (B) Quantitative heat map of hESC attachment in varying culture conditions (n=3, n=5 in colony seeding conditions where ROCK inhibitor was removed, n=10 in single-cell seeding conditions where ROCK inhibitor was removed). The screen was performed once over the course of these studies.
When seeded on synthetic hydrogels, hESCs that expressed elevated levels of NANOG also expressed elevated levels of OCT3/4 and SOX-2 compared to hESCs on Matrigel. Following the completion of the initial screen, hESCs were seeded as single cells onto a subset of hydrogels containing 1 to 4 mM cyclic RGD and having stiffnesses of either 3 or 10 kPa. The 3 kPa hydrogels containing 1 and 2 mM cyclic RGD increased NANOG expression by hESCs compared to hESCs on Matrigel (Figure 5A,C, Supp. Fig. 13), while all other hydrogels in the tested range of conditions decreased NANOG expression. On most hydrogels in this range of conditions, hESCs expressed elevated OCT3/4 and SOX-2 relative to Matrigel (Supp. Fig. 13). This change in marker expression was detectable using NANOG as a marker of pluripotency, but was not detectable using OCT3/4 or SOX-2.
Discussion
Through the use of hydrogel arrays, we identified alternative synthetic substrates to Matrigel for use in angiogenesis assays (Figure 1–4) and short-term hESC expansion (Figure 5). In angiogenesis assays, synthetic hydrogels mediated endothelial network formation by HUVECs and iPSC-ECs, enabled evaluation of known pharmacological inhibitors of angiogenesis at concentrations previously demonstrated as effective in vitro50–53, and enabled accurate detection of blinded vascular disruptive chemicals. We demonstrated distinct advantages of synthetic hydrogel-based assays over Matrigel-based assays in terms of reproducibility, as suggested by Z prime scores, and in accuracy, as suggested by the blinded 38 compound screen. The rationale behind the use of thiol-ene photocrosslinking chemistry for hydrogel crosslinking and the optimization of hydrogel properties may be found in the Supplemental Introduction and Supplemental Results and Discussion. The screening techniques described here are expected to enable the discovery of hydrogels that mediate network formation by a variety of EC types beyond those utilized in these studies, including tissue-specific human ECs and ECs from non-human species. In hESC expansion, cells seeded on synthetic hydrogels expressed greater or equal levels of the pluripotency markers NANOG, OCT3/4 and SOX-2 compared to cells seeded on Matrigel. These outcomes can be attributed to the fact that synthetic hydrogels provided defined levels of adhesion molecule concentration, substrate stiffness, and growth factor sequestration to control cell behavior, while minimizing variable and unwanted signaling that is normally present in poorly-defined materials such as Matrigel. This supports previous studies where encapsulation of hESCs in a synthetic PEG hydrogel demonstrated enhanced reprogramming efficiency and maintenance of pluripotency in three dimensions22, andhow controlling material parameters, using array-based methods, could affect stem cell pluripotency and colony formation from individual mouse embryonic stem cells (mESCs)29.
The physiological relevance of Matrigel as a cell culture substrate as an angiogenesis assay is often called into question as multiple non-endothelial cell types, including melanoma, glioblastoma, numerous breast cancer cell lines, retinal epithelial cells, lens cells, murine Leydeg cells and human fibroblasts54–57 have formed cellular networks resembling vasculature while on Matrigel. It has been suggested that these networks form via cell traction and malleability of the matrix57,58 rather than relevant mechanisms of vascular morphogenesis, such as single-cell migration demonstrated by HUVECs on synthetic hydrogels (Figure 1C, Supp. Movies 1–2)59. The process by which cancer cells form vascular networks has been viewed as similar to an in-vivo process dubbed vasculogenic mimicry60, and processes such as these likely generate misleading insights into vascular network formation and false negatives when screening for vascular disrupting chemicals. For example, we suggest that the differential processes of morphogenesis on synthetic hydrogels and Matrigel impact endothelial cell morphologies which may distinguish between various mechanisms of vascular inhibition. We demonstrated that on synthetic hydrogels, inhibitors to VEGF-receptor binding (e.g. sflt-1) disrupted HUVEC networks into isolated cell clusters; inhibitors to receptor-tyrosine-kinase activity (e.g. Sunitinib®) disrupted cell-cell contacts and induced the onset of rounded HUVEC morphologies; finally, inhibitors to MMP activity (e.g. Prinomastat HCl) induced the formation of confluent HUVEC monolayers rather than networks (Supp. Fig. 7, 9). However, while these morphological outcomes were identifiable on a defined synthetic substrate, they were masked if similar assays were performed on Matrigel (Figure 3, Supp. Fig. 7, 9–11). This can be attributed to interfering, poorly defined signals provided by Matrigel, as well as the aforementioned contraction mechanism utilized by cells on Matrigel. One such source of interference is binding and sequestration of growth factors, including VEGF, by Matrigel. A discussion of this mechanism may be found in the Supplemental Results and Discussion.
The goal of the synthetic hydrogel and Matrigel-based angiogenesis assays was to leverage enough EC functionality, including adhesion, migration, and establishment of cell-cell contacts, to generate endothelial networks as a functional readout of pVDC activity. We did not expect to recapitulate other aspects of EC functionality, such as MMP-mediated angiogenic sprouting46, in these assays due the use of 2D surfaces as culture environments. Interestingly, our studies suggest that network formation on synthetic hydrogels still requires MMP activity, as MMP inhibition by Prinomastat HCl resulted in monolayer formation rather than network formation (Figure 3, Supp. Fig. 7, 9). Though the synthetic hydrogels used in these studies were MMP labile, endothelial network assembly has been documented on substrates that are not MMP labile61. Therefore, it is unlikely that MMP-driven substrate degradation was the sole process driving network formation on the synthetic substrates here. We expect that MMP activity mediates other cellular functions during network formation on synthetic hydrogels, including the functions of integrins, growth factor receptors, and VE-Cadherin62. While future studies are necessary to characterize the roles of MMP in the context of network formation, the synthetic hydrogels are well-suited for these studies as MMP inhibition was difficult to detect on Matrigel (Figure 3, Supp. Fig. 7, 9).
Synthetic hydrogel and Matrigel-based angiogenesis assays identified putative vascular disrupting chemicals in a blinded subset of 38 chemical compounds taken from the EPA’s ToxCast library. Results from this experiment contributed to an ongoing effort by the EPA to identify potential developmental neurotoxins and the significance of this work is highlighted in an OECD adverse outcome pathway ((i.e. OECD AOP43 - https://aopwiki.org/aops/43). Both assays were more likely to positively identify candidate positives if their pVDC scores were high, and neither assay generated false positives (Figure 4). While the synthetic hydrogel-based angiogenesis assay displayed superior sensitivity to pVDCs than the Matrigel-based assay, neither assay identified all high-scoring compounds as disrupting chemicals (Figure 4). Since only a single concentration was tested here (10 µM), versus a range of test concentrations comparable to the high levels in ToxCast that generated the pVDC list, we hypothesize that the sensitivity of both assays might be enhanced by increasing the test concentrations. For example, 5HPP-33, which is known to inhibit cell proliferation and migration63, was not detected by the synthetic hydrogel assay here. In cases like these, some inhibitors were clearly beginning to show effects of destabilizing endothelial networks (Supp. Fig. 11B), and a higher concentration of inhibitor may have sufficiently changed network area to enable detection of inhibition. Alternatively, the angiogenesis assays here focused only on network formation and did not encompass the full range of cell functions exhibited in angiogenesis whereas the ToxCast predictions are based on knowledge about the full range of the angiogenic cycle, including vasculogenesis, angiogenesis and angio-adaptation. For example, TNP-470, a clinical anti-angiogenesis compound which reduces permeability and dilation of mature blood vessels while not necessarily affecting developing vessels64,65, was not detected by either assay here. We expect that mechanisms such as these would not have a detectable impact on cell activity in a single functional angiogenesis assay. The pVDC scores of the ToxCast compounds were originally derived from numerous in vitro technology platforms, literature searches and computational methods to measure direct chemical interactions with cell receptors and resulting changes in gene expression levels1,41,43–47. However, caution should be exercised when using a high pVDC score as an absolute assurance of disruptive activity, and emerging functional angiogenesis assays may identify high-scoring chemicals as false positive inhibitors. As our assay and other functional angiogenesis assays46 emerge and highlight varying endothelial cell functionalities in angiogenesis, pVDC scores should be revised over time.
To explore the versatility of the screening methods described here, synthetic hydrogels were customized to replace Matrigel’s ability to maintain short-term hESC pluripotency. Hydrogel arrays identified multiple culture conditions that maintained hESC NANOG expression to greater or equal levels measured on equivalent Matrigel substrates, and these conditions spanned a wide parameter field (Figure 5A). It should be noted that NANOG was initially used as the marker for pluripotency as, compared to OCT3/4, SOX2 and C-myc, it is the first pluripotency marker lost upon differentiation66–68. Over the course of pluripotency evaluation here, NANOG, OCT3/4 and SOX-2 were highly co-expressed in pluripotent hESCs, but in multiple environments hESCs expressed elevated OCT3/4 and SOX-2 but not elevated NANOG compared to Matrigel, and we expect this to signify differentiation (Supp. Fig. 13). These initial discoveries will enable future explorations of how multiple cell culture variables, such as single-cell seeding and colony seeding techniques, affect long-term hESC expansion. Additionally, as the well-defined nature of the synthetic hydrogels enabled control of cell morphology during expansion, hESC expansions may be performed in environments that promote either low or high cell adhesion (Figure 5B). Notably, the hydrogel arrays detected a synthetic hydrogel substrate that increased NANOG expression without the inclusion of ROCK Inhibitor Y-27632 in media during cell seeding or maintenance. Previous studies have utilized synthetic cell culture substrates for long-term expansion of pluripotent stem cells, but these procedures required the use of ROCK Inhibitor69,70 which has poorly understood effects on long-term hESC expansion71. Additionally, previous studies were not always done using human ESCs. Instead mouse ESCs which may demonstrate different responses to underlying material properties have been used29. The hydrogels discovered here are synthetic, chemically defined, and enable human ESC expansion without the use of ROCK inhibitor.
Outlook
This manuscript demonstrates that simple synthetic hydrogels with 3 controllable parameters (adhesion peptides, degradability, stiffness) can outperform a natural biomaterial with over 1500 unique proteins. The versatility of the synthetic hydrogels, as demonstrated in vascular biology and stem cell biology applications here, can be applied to a number of emerging biomanufacturing applications. In one area, the hydrogels can be used to construct increasingly complex multicellular tissue models for drug and toxicity screening. Examples include vascularized tissues46,72, hierarchically assembled organoid models73, and models of developing embryonic tissue74. In this context, the synthetic materials can offer improved reproducibility, and superior screening performance. In another area, synthetic hydrogels can provide a reproducible, chemically defined, xeno-free environment for production of stem cells and their derivatives. Examples include pluripotent stem cells (described here), mesenchymal stem cells, and other emerging therapeutic candidates. In this context, the synthetic hydrogels can control the phenotype of the manufactured cells and decrease cost. Taken together, we predict that the use of synthetic hydrogel-based culture systems will ultimately lead to a significant reduction in false positives and false negatives in drug and toxicity screening, improved performance of compounds taken to preliminary animal testing, and an improvement in the safety and efficacy of cell therapies.
Methods
Endothelial Cell Culture and Maintenance
Human umbilical vein endothelial cells (HUVECs) were purchased from Lonza (Walkersville, MD) and cultured in growth medium consisting of medium 199 (M199) (Mediatech Inc, Manassas, VA) supplemented with EGM-2 Bulletkit™ (Lonza). The medium supplement contained 2% fetal bovine serum as well as hydrocortisone, hFGF-B, VEGF, R3-IGF-1, Ascorbic Acid, Heparin, FBS, hEGF, and GA-1000. Growth medium was changed every other day and cells were passaged every 5–7 days. Cell passages were performed using 0.05% trypsin solution (Thermo Fisher Scientific, Inc., Carlsbad, CA) and detached cells were recovered in M199 supplemented with 10% cosmic calf serum (Thermo Fisher Scientific, Inc., Carlsbad, CA). All medium was supplemented with 100 U/mL Penicillin/100 µg/mL Streptomycin (Thermo Fisher Scientific, Inc., Carlsbad, CA). The cells were maintained in a humidified 37 °C incubator with 5% CO2 and used between 5 and 16 population doublings in all experiments.
Induced pluripotent stem cell derived endothelial cells (iPSC-ECs) were kindly provided by Cellular Dynamics International (Madison, WI) and cultured in complete Vasculife® medium (Lifeline® Cell Technology, Frederick, MD) supplemented with 10% FBS and iCell Endothelial Cells Medium Supplement (Cellular Dynamics International). Culture flasks were coated with 30 µg/ml human plasma fibronectin (Corning) for 30 minutes prior to use. Growth medium was changed every other day and passaged every 3–4 days. Cells were detached using 0.05 % trypsin and recovered in basal medium supplemented with 10 % cosmic calf serum. The cells were maintained in a humidified 37 °C incubator with 5% CO2 and used between 5 and 16 population doublings in all experiments.
Human Embryonic Stem Cell Culture and Maintenance
WA09 H1 hESCs (WiCell Research Institute, Madison, WI) at passage 34 were thawed in E8 medium (Life Technologies, Madison, WI) containing 5 µM ROCK Inhibitor Y-27632 (EMD Millipore, Billerica, MA). Tissue-culture polystyrene plates were coated using Matrigel (Corning, NY) at a density of 0.0087 µg/cm2. The hESCs were seeded onto the plates and cultured for 24 hours, then maintained in E8 medium at 37 °C in a 5% CO2 atmosphere. During routine maintenance, media were changed every day and colonies showing morphological differentiation were manually removed prior to media change.
For colony passaging, the cells were incubated in Versene (Life Technologies, Madison, WI) for 2–3 minutes at 37 °C until edges of colonies were weakly detached. Weakly adherent colonies were collected and centrifuged at 200 × g for 5 minutes, and colonies were seeded between a 1:10 to 1:6 split ratio onto fresh Matrigel-coated plates.
For single-cell passaging, the cells were incubated in TrypLE (Life Technologies, Madison, WI) for 3–4 minutes at 37 °C until the majority of colonies were lifted. To dilute the TrypLE, 3 mL of E8 medium with 5 µM Y-27632 per 1 mL of TrypLE were added to TrypLE-treated cells. Afterward, the cells were collected, centrifuged at 200 × g for 5 minutes, resuspended in E8 and 5 µM Y-27632, and mixed to singularize. Cells were seeded at 5,000 cells/cm2 onto fresh Matrigel-coated plates.
Labeling Cells with Cell Tracker Red
One day prior to seeding the HUVECs/iPSC-ECs onto the hydrogel spots, the cells were stained with cell tracker red to aid in automated tubulogenesis quantification. Briefly, the HUVECs were rinsed in basal M199 for five minutes and stained with 1.3 µM Cell Tracker Red (Invitrogen) in M199 for 45 minutes. Afterward, the cells were rinsed again in basal M199 for five minutes before incubating in growth medium overnight to allow the cells to sufficiently recover.
Poly (ethylene glycol) (PEG) Functionalization with Norbornene
Modification of PEG-OH molecules with terminal norbornene groups was performed using methods similar to previous studies 72,75. Briefly, PEG-OH (20 kDa molecular weight, 8-arm, tripentaerythritol core, Jenkem USA, Allen TX), dimethylaminopyridine and pyridine (Sigma Aldrich, St. Louis, MO) were dissolved in anhydrous dichloromethane (Fisher Scientific, Waltham, MA). In a separate reaction vessel, N,N’-dicyclohexylcarbodiimide (Thermo Scientific, Waltham, MA) and norbornene carboxylic acid (Sigma Aldrich) were dissolved in anhydrous dichloromethane and reacted for 30 minutes to activate the norbornene. Norbornene carboxylic acid was covalently coupled to the PEG-OH through the carboxyl group by combining the PEG solution and norbornene solutions and stirring the reaction mixture overnight under anhydrous conditions. Urea byproduct was removed from the reaction mixture using a glass fritted funnel and the filtrate was precipitated in cold diethyl ether (Thermo Fisher Scientific, Inc., Carlsbad, CA) to extract the norbornene-functionalized PEG (PEGNB). The precipitated PEGNB was collected and dried overnight in a ceramic fritted filter. To remove impurities, the PEGNB was dissolved in chloroform (Sigma Aldrich), precipitated in diethyl ether and dried a second time in a buchner funnel. To remove excess norbornene carboxylic acid, PEGNB was dissolved in de-ionized H2O, dialyzed in de-ionized H2O for one week and filtered through a Millex® 0.45 µm pore-size PVDF syringe filter (Millipore). The aqueous PEGNB solution was frozen using liquid nitrogen and lyophilized. Functionalization of PEG was quantified using proton nuclear magnetic resonance spectroscopy (NMR) to detect protons of the norbornene-associated alkene groups located at 6.8–7.2 PPM. Functionalization efficiency for norbornene coupling to PEG-OH arms was above 90% for all PEGNB used in these experiments.
Preconjugation of Adhesion Peptides to PEG
Lyophilized PEGNB was dissolved in de-ionized H2O at a 0.5 mM concentration and combined with 0.05% w/v Irgacure 2959 photoinitiator (I2959) (Ciba Specialty Chemicals, Tarrytown, NY) as well as a 2× molar excess of either head-to-tail cyclized Arg-Gly-Asp-[d-Phe]-Cys (cyclic RGD, Genscript, Piscataway, NJ) adhesion peptide; 2× molar excess of linear H-Cys-Arg-Gly-Asp-Ser-NH2 (linear RGD, Genscript, Piscataway, NJ); 2× molar excess of non-functional H-Cys-Arg-Asp-Gly-Ser-NH2 scrambled adhesion peptide (CRDGS, Genscript, Piscataway, NJ); 3× molar excess of H-Cys-Glu-[d-Phe]-[d-Ala]-[d-Tyr]-[d-Leu]-Iso-Asp-Phe-Asn-Trp-Glu-Tyr-Pro-Ala-Ser-Lys-NH2 (VBP), or 3× molar excess of the non-functional scrambled VBP peptide H-Cys-Asp-[d-Ala]-Pro-Tyr-Asn-[d-Phe]-Glu-Phe-Ala-Trp-Lys-[d-Tyr]-Iso-Ser-[d-Leu]-Glu-NH2. The mixtures were reacted under 365 nm ultraviolet (UV) light for 5 min at a dose rate of 4.5 mW/cm2 to covalently attach the peptides to norbornene groups via the thiol-ene reaction76. To remove buffer salts and unreacted peptides from the decorated PEGNB, the reaction mixtures were dialyzed in de-ionized H2O for two days. The dialyzed solutions were frozen in liquid nitrogen and lyophilized. The coupling efficiency of peptides to the PEGNB was quantified using proton NMR to detect disappearances of alkene protons at 6.8–7.2 PPM caused by covalent bonding of the peptides to the norbornene group (Supp. Fig. 14).
Preparation of Patterned Gold Slides with Hydrophobic and Hydrophilic Regions
Gold coated glass slides (EMF, Ithaca, NY) were sonicated in 100% ethanol for 5 minutes and immersed in a 0.1 mM FlouroSAM solution (HS-C11-O-C2-(CF2)5-CF3, Pro Chimia, Sopot, Poland) prepared in 100% ethanol for two hours protected from light at room temperature. This created a hydrophobic region on the surface of the gold. A PDMS mask with the pattern of choice was aligned with the gold slide and adhered to the surface. The exposed hydrophobic regions of the mask were etched by surface plasma treatment using a Diener Plasma Treatment Chamber (Diener Electronic, Ebhausen, Germany) at 40 sccm and 50 W for approximately one minute. After etching, the PDMS mask was removed and slides were rinsed in 100% ethanol. The etched gold slide was placed into a 0.25 mM solution of [HS–C11–(O–CH2–CH2)3–OH] (EG3-OH) in 100% ethanol for two hours at room temperature to create hydrophilic regions on the surface of the gold slide.
Silanization of Glass Slides for Screening of Substrate Conditions
Glass slides were sonicated for 45 minutes in 100% acetone to remove any surface impurities. Slides were rinsed three times in 100% ethanol to remove excess acetone from the surface. Cleaned glass slides were surface plasma treated using the Diener Plasma Treatment Chamber on both sides of slides for five minutes at 40 sccm and 50 W. The activated slides were transferred to 2.5 % 3-mercatopropyl trimethoxylsilane (3-MPTS) (Sigma Aldrich) in toluene overnight. After retrieval, samples were cleaned by subsequent rinses of toluene, 1:1 toluene: ethanol and three rinses of 100 % ethanol respectively. Slides were cured in a nitrogen-purged pressure chamber at 100 °C for one hour. After curing, silanized glass slides were placed in an airtight container and protected from light until use.
Preparation of PEG solutions for Array Formation
Hydrogels for assessing endothelial cell network formation consisted of PEGNB molecules, PEG molecules preconjugated to RGD adhesion peptides and VBP, MMP-degradable H-Lys-Cys-Gly-Gly-Pro-Gln-Gly-Ile-Trp-Gly-Gln-Gly-Cys-Lys-NH2 crosslinking peptide (Genscript) and 0.05% w/v I2959 photoinitiator dissolved in phosphate buffered saline (1× PBS). PEGNB, adhesion peptide and VBP concentrations were adjusted to varying levels to achieve different levels of stiffness, cell adhesion and VEGF-binding capabilities in the hydrogels. Prior to inclusion in precursor solutions, the purity of the crosslinking peptide was verified using the Ellman’s Assay (Thermo Fisher). To vary endothelial cell adhesion to the hydrogels, precoupled-PEGNB-linear RGD and PEGNB-cyclic RGD molecules were added to the solutions to achieve desired adhesion peptide concentrations between 0–4 mM. PEGNB-CRDGS molecules were added along with the functional adhesion molecules to provide a total of 4 mM pendant adhesion peptide included in every solution. To vary VEGF binding to the hydrogels, precoupled-PEGNB-VBP molecules were added to the solutions to achieve desired peptide concentrations of 0.3 mM VBP. To vary the modulus of the hydrogels PEGNB was added at 2, 2.5 or 3.5 mM concentrations and crosslinking molecules were added to the solution to achieve crosslinking of 50% total norbornene functional groups present in solution. Once completed, all hydrogel solutions were immediately stored at −80°C, thawed immediately before use, and subjected to only one freeze-thaw cycle prior to use.
Hydrogels for assessing embryonic stem cell pluripotency consisted of PEGNB molecules, PEG molecules preconjugated to cyclic RGD adhesion peptides, and 3.4 kDa dithiolated PEG crosslinking molecule (Laysan Bio) and I2959 photoinitiator dissolved in 1× PBS. PEGNB and adhesion peptide concentrations were adjusted to varying levels to achieve different levels of stiffness and cell adhesion capabilities in the hydrogels. To vary hESC adhesion to the hydrogels, precoupled-PEGNB-RGDFC molecules were added to the solutions to achieve desired adhesion peptide concentrations between 0–4 mM. PEGNB-CRDGS molecules were added along with the functional adhesion molecules to provide a total of 4 mM pendant adhesion peptide included in every solution. To vary the modulus of the hydrogels PEGNB was added at 2, 2.5 or 4 mM concentrations and crosslinking molecules were added to the solution to achieve crosslinking of 50 or 75% total norbornene functional groups present in solution. Once completed, all hydrogel solutions were immediately stored at −80°C, thawed immediately before use, and subjected to only one freeze-thaw cycle prior to use.
Mechanical Properties of PEG Hydrogels
Shear modulus was measured in bulk samples of hydrogel spot formulations. To measure shear modulus, 72 µL of hydrogel solutions, each containing 0.125 mM RGDFC, were pipetted into 8.0 mm diameter, 1.2 mm depth Teflon wells and cured for eight seconds using 365 nm UV light at a 90 mW/cm2 dose rate. The resulting hydrogels were swollen in 1× PBS for 24 hours and cut, if necessary, to a final diameter of 8 mm using a hole punch. The samples were tested using an Ares-LS2 rheometer (TA Instruments, New Castle, DE). A 20 g force was applied to the samples via parallel plate crossheads and a strain sweep test at 1Hz fixed frequency was performed from 0.1 to 20% strain. If the sample was not robust enough to withstand a 20 g force the gap between the parallel plates of the rheometer was set to 1.0 mm distance instead. Complex shear modulus of each sample was as the average of measurements taken at 10 Hz, 2 to 10% strain.
Preparation of Thin Hydrogel Arrays for Identifying Hydrogels that Promote Endothelial Network-Formation
To prepare the silanized glass slides for formation of the hydrogel arrays, samples were treated in 10 mM dithiothreitol (Sigma) in 1× PBS at 37°C for one hour to increase the number of free thiols on the surface of the slide. After incubation, slides were sequentially rinsed in 1× PBS, 100% ethanol and dried with Nitrogen gas. Patterned gold slides were rinsed with ethanol and dried using Nitrogen gas. A 120 µm-thickness PDMS spacer was applied to the surface of the gold slide to control the height of the hydrogels. 0.8 µL of the prepared PEG solutions were pipetted onto the hydrophilic spots of the glass slide in a humidity chamber 75,77. A silanized glass slide was slowly placed onto the surface of the gold slide and transferred under a UV lamp. The hydrogel spots were exposed to 365 nm ultraviolet light at 4.5 mW/cm2 for eight minutes. Following polymerization, the glass slide was removed from the underlying gold slide. This resulted in a patterned hydrogel array on the surface of the glass slide. Samples were stored in 1× PBS overnight at 4°C prior to sterilization and seeding (Supp. Movie 4).
Assembling and Seeding of Hydrogel Arrays with Endothelial Cells
On the day of culture, the arrays were assembled within a three chamber Proplate Isolator assemblies (Grace Bio-labs, Bend, OR). Arrays were allowed to warm to room temperature and removed from the 1× PBS rinse. Arrays were subsequently sterilized by immersion in 70% ethanol for 30 minutes, followed by two rinses in 1× PBS. Residual PBS was carefully aspirated from the regions surrounding the hydrogel spots to guarantee adherence to the grace bio isolators. Using sterile technique, the hydrogel arrays were subsequently assembled within the Grace Bio Isolator system and the individual wells were bathed into basal M199 until use. Cell Tracker Red stained cells were removed from the incubator and subsequently rinsed with 1× PBS. Cells were passaged by incubating the cells in 0.05% trypsin (Hyclone) for 5 minutes, quenching the enzyme through addition of basal M199 with 10% cosmic calf serum, subsequently counted and resuspended to give a cell count of approximately 85,000 cells/cm2. Residual medium was removed from the assembled arrays and replaced with the resuspended cell solution. When seeding HUVECs the cells were resuspended in M199 containing either 0, 5 or 10 ng/ml VEGF and EGM2 (without VEGF) as per Figure 1D. When seeding iPSC-ECs the cells were resuspended in full Vasculife® growth medium supplemented with 0, 5 or 10 ng/mL additional VEGF. After seeding, the assembled constructs were transferred to a 37° C incubator for 72 hours. During this period, hydrogel spots were imaged using a Nikon TI Eclipse inverted florescent microscope and individual spots were subsequently qualitatively scored on their ability to promote endothelial network formation (Figure 1D). Briefly, spots were qualitatively assigned a score from 0 – 3 where: 0 = no adhesion; 1 = low cell adhesion; 2 – monolayer formation; and 3 = network formation. Each condition was tested using at least n = 3 spots per condition, and a final qualitative score was assigned based on the behavior observed in the majority of replicates.
Assembling and Seeding of Hydrogel Arrays with Embryonic Stem Cells
hESCs cultured on hydrogel arrays were seeded as colonies (1:10 split ratio) or single cells (5,000 cells/cm2) in either E8 media (with or without 5 µM Y-27632) and maintained E8 media (with or without 5 µM Y-27632). 24 hours after seeding, seeding media were replaced with the designated maintenance media. Media were changed on a daily basis for four days prior to fluorescent imaging.
Quantification of Human Embryonic Stem Cell Behavior on Hydrogel Arrays
Hydrogel arrays were incubated on an environmentally controlled stage on the Nikon TI Eclipse microscope to match atmospheric conditions of a humidified 37°C incubator and photographed at 24, 48, 72, and 96 hours after cell seeding. At 96 hours, cells were fixed via 15-minute incubation in formalin. Immunofluorescence staining for NANOG was conducted using rabbit monoclonal antibody to NANOG (1:400 dilution) (Abcam, Cambridge, MA, Cat# ab109250) and Alexa Fluor 488 goat anti rabbit secondary antibody (1:200 dilution) (Life Technologies, Carlsbad, CA, Cat# A11008). Immunofluorescence staining for OCT3/4 was conducted using mouse monoclonal antibody to OCT3/4 (1:100 dilution) (Santa Cruz Biotechnology, Dallas, TX, Cat# SC-5279) and Alexa Fluor 594 goat anti mouse secondary antibody (1:200 dilution) (Life Technologies, Carlsbad, CA, Cat# A11005). Immunofluorescence staining for SOX-2 was conducted using mouse monoclonal antibody to SOX-2 (1:200 dilution) (Abcam, Cambridge, MA, Cat# ab75485) and Alexa Fluor 594 goat anti mouse secondary antibody (1:200 dilution) (Life Technologies, Carlsbad, DA, Cat# A11005). Nuclei were stained using a 1:5000 dilution of DAPI (MP Biomedicals, Santa Ana, CA, Cat# 157574).
Cell number was determined by thresholding area of nuclei stained using DAPI, NANOG, OCT3/4 and SOX-2 using the Nikon NIS Elements software. Percent marker expression was determined using automated measurements on the stained cells and normalized against DAPI-stained cells.
Adapting Network-Forming Hydrogels for Use in 96-Well Plates
To ensure substrate stability and repeatable network area measurements in 96-well Angiogenesis plates (Ibidi, USA, Madison WI) photoinitiator concentration, solution volume and cell seeding density were optimized. For all hydrogel formation and cell seeding operations screening arrays were constructed as follows: the 96-well angiogenesis plates were coated using 150 – 300 kDa poly(L-lysine) (PLL, Sigma Aldrich). A 0.01% v/v solution of PLL in de-ionized H2O was pipetted into the wells at 8 µL volume to evenly coat the bottoms of the wells. After 5 minutes, incubation at room temperature, the solution was aspirated from all wells. Each well was then washed with de-ionized H2O three times before drying. Hydrogel solutions identified as enabling endothelial network formation were pipetted into the wells, cured for 8 minutes under 365 nm, 4.5 mW/cm2 UV light and swollen in 70 µL 1× PBS overnight. For HUVECs, hydrogel solutions consisted of 2 mM PEG, 0.125 mM cyclic RGD, and 4 mM crosslinking peptide, For iPSC-ECs, hydrogel solutions consisted of 2 mM PEG, 1 mM linear RGD, and 4 mM crosslinking peptide. Afterward the PBS was aspirated and replaced with 35 µL medium (network-forming medium was identified in the thin-hydrogel screening experiments) by itself, or containing vehicle or inhibitors at desired concentrations. For HUVECs, media consisted of M199 containing 5 ng/ml VEGF and EGM2 (without VEGF). For iPSC-ECs, media consisted of full Vasculife growth medium supplemented 10 ng/mL additional VEGF. Cell suspensions were added as 35 µL volumes on top of the other 35 µL medium. Cells were left undisturbed for 24 hours when endothelial networks were photographed by epifluorescence and phase-contrast microscopy using a Nikon Eclipse microscope. After photography, cells were fixed via 30-minute incubation in formalin and prepared for confocal microscopy on a Nikon A1RS Confocal Microscope. Immunofluorescence staining for CD31 was conducted using mouse monoclonal antibody to CD31 (1:200 dilution) (Dako, Carpinteria, CA, Cat# M082301-2) and Alexa Fluor 488 Donkey anti mouse secondary antibody (1:200 dilution) (Life Technologies, Carlsbad, CA, Cat# A21202). Nuclei were stained using 1:5000 dilution of DAPI (MP Biomedicals, Santa Ana, CA, Cat# 157574).
To enhance substrate stability in the 96 well plates photoinitiator concentration was increased from the original 0.05% w/v concentration used in the thin hydrogel arrays to 0.1, 0.2 and 0.4% w/v I2959. Hydrogel stability was measured as swollen diameters of hydrogels retrieved from the 96 well plates. Test hydrogels were cured as 8 µL volumes per well using the aforementioned array assembly procedure. A 3.5 mm diameter hole punch was used to retrieve hydrogel samples from the wells before the diameter of the samples was measured using a micrometer. Unstable hydrogels were evaluated as having the largest diameters after removal from the wells, indicating high swelling at equilibrium (Supp. Fig. 6A).
To reduce the impact of meniscus formation on image of endothelial networks 7, 8, 9 and 10 µL volumes of hydrogel were cured in the well plates using the aforementioned array assembly procedure. The hydrogels contained 0.2% w/v photoinitiator as was found to be the optimal concentration in the swelling experiments. HUVECs were stained with cell tracker red and seeded at a density of 1.2 × 105 cells/cm2 onto the hydrogels. Meniscus formation was evaluated by observing in-focus and out-of-focus areas in a typical image of HUVEC network formation 24 hours after seeding (Supp. Fig. 6B).
To ensure the consistent formation of endothelial networks in the arrays HUVEC seeding density was explored at 1.2 × 105 cells/cm2, 1.8 × 105 cells/cm2, and 2.4 × 105 cells/cm2. iPSC-EC seeding density was also explored at 1.8 × 105 cells/cm2, and 2.4 × 105 cells/cm2. Here hydrogels were pipetted into the wells as 9 µL volumes as was determined to be optimal for meniscus reduction. Network formation was evaluated 24 hours after seeding (Supp. Fig. 6C).
Initial Inhibitor Screen for Assessing Optimized Synthetic Hydrogel Assay in Comparison to Matrigel
Synthetic hydrogel and Matrigel conditions were included in the screening arrays as follows: synthetic hydrogels were added to the 96 well plates in 9 µL volumes with HUVECs seeded at a density of 2.4 × 105 cells/cm2 and iPSC-ECs seeded at a density of 1.8 × 105 cells/cm2. Matrigel was added to the 96 well plates in 10 µL volumes and incubated in a humidified 37°C incubator for 30 minutes. Afterward either HUVECs or IPS-ECs were seeded at a density of 1.2 × 105 cells/cm2 using the aforementioned seeding procedure in 96 well plates. All HUVEC experiments were conducted in M199 containing 5 ng/ml VEGF and EGM2 (without VEGF). All iPSC-EC experiments were conducted in full Vasculife growth medium supplemented 10 ng/mL additional VEGF. Network formation was evaluated 24 hours after seeding in the 96 well plates. To conduct time-lapse microscopy of HUVEC network formation on synthetic hydrogels or Matrigel HUVECs were seeded onto substrates as dictated for the initial inhibitor screens, treated with 0.2% DMSO in medium, incubated on an environmentally controlled stage on the Nikon TI Eclipse microscope to match atmospheric conditions of a humidified 37°C incubator and photographed every 15 minutes using phase contrast microscopy.
The Z-prime statistic (Equation 1) assessed the accuracy of the synthetic hydrogel and Matrigel screening systems assays in distinguishing endothelial networks disrupted using the known angiogenesis inhibitor Sunitinib from non-inhibited networks. The statistic accounts for differences in positive and negative controls, as well as standard deviations across the assay40. PEG and Matrigel substrates were formed in separate 96 well plates. HUVEC plates were divided into four 24 well corners with 0.1% DMSO controls occupying two opposite corners and 20 µM Sunitinib® controls occupying the other two opposite corners. iPSC-EC plates were divided into four 24 well corners with 0.2% DMSO controls occupying two opposite corners and 40 µM Sunitinib® controls occupying the other two opposite corners. Network formation was quantified and compared using the total endothelial network area identified by NIS Elements software, with a maximum size cutoff applied to exclude the area of networks completely disrupted by Sunitinib®.
| Equation 1|Z’ calculation |
The optimized PEG formulations with or without a VEGF binding component were compared directly to growth factor reduced Matrigel™ using an initial panel of five inhibitors i.e. 1) Vatalanib® – a clinically tested inhibitor to vascular endothelial growth factor receptor activity78 2) SU5416 – a clinically approved inhibitor of vascular endothelial growth factor receptor activity 79; 3) soluble fms-like tyrosine kinase 1 (sFlt-1) – an antagonist of VEGF and placental growth factor 80–82; 4) Anti-VEGF – a human monoclonal antibody to bind soluble VEGF83; 5) Sunitinib, a clinically approved inhibitor of vascular endothelial growth factor receptor activity84,85. Endothelial cells were additionally treated on PEG and Matrigel substrates with inhibitors acting independently of VEGF signaling, including 1) Prinomastat HCl – an antagonist to the activity of MMPs 2, 3, 9, 13, and 14; and 2) Suramin HCL, an inhibitor to EGF, PDGF and TGFβ signaling16. Network formation was quantified and compared using the total endothelial network area identified by NIS Elements software, with a maximum size cutoff applied to exclude the area of networks completely disrupted by Vatalanib® or Sunitinib®.
Screening of Putative Vascular Inhibitors from the ToxCast Library
To demonstrate the capability of the screening system to identify vascular inhibitors from a library of unknown chemical compounds samples from the ToxCast library of chemicals were applied to HUVECs similarly to the known vascular inhibitors screened on the 96 well plate. Of the 1066 compounds in the ToxCast library, 38 chemicals (36 environmental chemicals and 2 reference compounds) were provided by the EPA to encompass a range of predicted activity levels, from “inactive” to “strongly active” inhibitory compounds based on a predicted vascular disrupting chemical (pVDC) score from the ToxCast in vitro bioactivity profile1,46. The pVDC score/rank was previously generated from a filtered list of 23 assays and 309 chemicals (initially >500 in vitro assays and 1066 chemicals) based on six biological assay targets related to embryonic blood vessel development in the ToxCast Library using the iCSS dashboard (http://actor.epa.gov/dashboard)41. The pVDC rank of the 38-compound test set is described elsewhere46,47. The compounds were presented as 53 samples, including replicates, the identities of which were not made known until after the data were analyzed. Compounds were assigned ID numbers (coded by EPA/NCCT in a database, and only decoded after data was delivered and analyzed) along with stock concentrations (25 mM or 100 mM respectively), and while the samples were delivered as compounds in transparent 96 well plates and the colors of stock solutions were visible to experimenters, no other information was provided to reveal the identities of unknown compounds. Additional details on chemicals, QA/QC and assays performed can be found in ref. 86. The DMSO stock solutions from the library were diluted using tubulogenic medium at a 1:1000 dilution prior to their additions into the screening system. Endothelial network inhibition was evaluated by quantifying the total area of endothelial networks treated with unknown compounds, with a maximum size cutoff applied to exclude the area of networks completely disrupted by Sunitinib®, and comparing them to mean total areas of non-inhibited control networks treated with vehicle only (0.2% DMSO). Networks with areas greater than 2 standard deviations away from the mean were identified as being inhibited by a candidate compound. All tests were performed in triplicate and compounds that resulted in inhibition for a majority of replicates were counted as inhibitors. These were given a binary output of either 1 for inhibited conditions or 0 for uninhibited conditions. Compounds identified as inhibitors in a majority of the triplicate plates were counted as inhibitors in the synthetic hydrogel and Matrigel systems.
Statistics
All data analyzed were unpaired (samples independent from each other). Prior to conducting multiple comparisons tests, the Brown-Forsythe test was performed to determine homogeneity of variance between data sets. Normal distributions of n = 48 HUVEC and iPSC-EC network areas were determined using the Shapiro-Wilk normality test prior to inhibitor treatment. To compare multiple data sets, Tukey’s multiple comparisons test was used as a single-step multiple comparison procedure to find means significantly different from each other. To compare data sets to a DMSO or 1×PBS vehicle control, Dunnett’s test was used to find means significantly different from the control. All statistical tests were two-tailed (two-sided test). All statistical analyses were performed with Graphpad software. P-values were as follows: P-value > 0.05 (nonsignificant), *P < 0.05. Variances between each group of data were represented by the s.d. Sample sizes to ensure adequate power were as follows: initial network formation screen, n = 3 sample replicates; z-prime tests, n = 48 sample replicates; known inhibitor panel, n = 3 sample replicates, for DMSO and PBS controls n = 8–12 sample replicates; blinded ToxCast screen, 3 repeated experiments, with chemical replicates blinded to experimenters during each experiment; embryonic stem cell NANOG, OCT3/4, SOX-2 adhesion and confluence screen, n = 3 sample replicates, and in conditions where cell viability was expected to be compromised, such as in the cases of ROCK inhibitor removal, sample sizes were increased to 5 and 10; mechanical, swelling, cell density and hydrogel volume optimization, n = 3 sample replicates; Ellmans assays to determine free thiols in precursor solutions after freeze-thaw cycles, as well as before and after UV curing, n = 3 sample replicates; volumetric swelling assays, n = 3sample replicates; detection of non-crosslinked PEG molecules after UV curing, n = 3 sample replicates. Samples were excluded from analysis if they were damaged during the testing procedure, or were determined to be outliers through the Grubbs Outlier Test.
Supplementary Material
Acknowledgments
The authors would like to acknowledge funding from the National Institutes of Health (NIH R01HL093282-01A1, R21EB016381-01, 1UH2TR000506-01, T32HL007889, T32HL07936, R01EB10039, R24 EY022883, R01 EY026078, P30 EY016665, P30 CA 014520, 5 P30 CA 014520-01, and the Biotechnology Training Program NIGMS5T32GM08349), the National Science Foundation (GE-0718123), the UW-Madison Graduate Engineering Research Scholars program, the Environmental Protection Agency (STAR grant no. 83573701, the Chemical Safety for Sustainability Research Program, the Office of Research and Development, the Virtual Tissue Models Project and the National Center for Computational Biology), the UW-Madison Molecular and Environmental Toxicity Center Training Program (NIH T32 ES007015), the Gates Millennium Scholars Program, the Retina Research Foundation, and an unrestricted departmental award from Research to Prevent Blindness. Mechanical testing data was obtained using the Ares LS2 rheometer at the UW-Madison Soft Materials Laboratory. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grant P41GM103399 (NIGMS), old number: P41RR002301. Equipment was purchased with funds from the University of Wisconsin-Madison, the NIH P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, S10RR029220), the NSF (DMB-8415048, OIA-9977486, BIR-9214394), and the USDA. The authors would like to acknowledge Mai Lien Dombroe for her assistance in HUVEC culture. The authors would like to acknowledge the laboratory of Olachi Mezu-Ndubuisi for providing mice for use in aortic ring sprouting assays. The US Environmental Protection Agency (EPA), through its Office of Research and Development, funded and managed part of the research described here. The views expressed in this paper are those of the authors and do not necessarily reflect the views or policies of the EPA.
Footnotes
Data availability
The authors declare that all data supporting the findings of this study are available within the article and its Supplementary Information files. Source data for the figures in this study are available in Figshare with the identifying doi: 10.6084/m9.figshare.c.3791386 87.
Competing Financial Interests
C.S.L. is an employee and stockholder of StemPharm, Inc. W.L.M. is a founder and stockholder for StemPharm, Inc.
Author Contributions
EHN, WTD, CSL, MF, DGB, TBK and GEA contributed to conception and design of endothelial network experiments.
EHN, WTD, MF, MPS, CSL and MAS contributed to execution of endothelial network experiments.
EHN, WTD, MF, MPS, CSL, DGB, MAS and TBK contributed to analysis and figure preparation of endothelial network experiments.
NNTL contributed to conception, design and execution of hESC experiments.
EHN and NNTL contributed to analysis and figure preparation of hESC experiments.
EHN, CSL contributed to the conception, design and execution of hydrogel characterization experiments.
EHN, WTD, TBK and WLM drafted the manuscript.
NS and WLM supervised work throughout data collection and manuscript preparation.
WLM approved the final version of the manuscript.
References
- 1.Knudsen TB, Kleinstreuer NC. Disruption of embryonic vascular development in predictive toxicology. Birth Defects Res C Embryo Today. 2011;93:312–323. doi: 10.1002/bdrc.20223. [DOI] [PubMed] [Google Scholar]
- 2.De Falco S. Antiangiogenesis therapy: an update after the first decade. Korean J Intern Med. 2014;29:1–11. doi: 10.3904/kjim.2014.29.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Folkman J, Haudenschild C. Angiogenesis in vitro. Nature. 1980;288:551–556. doi: 10.1038/288551a0. [DOI] [PubMed] [Google Scholar]
- 4.Kubota Y, Kleinman HK, Martin GR, Lawley TJ. Role of laminin and basement membrane in the morphological differentiation of human endothelial cells into capillary-like structures. The Journal of cell biology. 1988;107:1589–1598. doi: 10.1083/jcb.107.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faulkner A, et al. A thin layer angiogenesis assay: a modified basement matrix assay for assessment of endothelial cell differentiation. BMC Cell Biol. 2014;15:41. doi: 10.1186/s12860-014-0041-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnaoutova I, Kleinman HK. In vitro angiogenesis: endothelial cell tube formation on gelled basement membrane extract. Nat Protoc. 2010;5:628–635. doi: 10.1038/nprot.2010.6. [DOI] [PubMed] [Google Scholar]
- 7.Crawford Y, Ferrara N. VEGF inhibition: insights from preclinical and clinical studies. Cell and Tissue Research. 2009;335:261–269. doi: 10.1007/s00441-008-0675-8. [DOI] [PubMed] [Google Scholar]
- 8.Kerbel R, Folkman J. Clinical translation of angiogenesis inhibitors. Nature Reviews Cancer. 2002;2:727–739. doi: 10.1038/nrc905. [DOI] [PubMed] [Google Scholar]
- 9.Sennino B, McDonald DM. Controlling escape from angiogenesis inhibitors. Nature Reviews Cancer. 2012;12:699–709. doi: 10.1038/nrc3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleinman HK, Martin GR. Matrigel: basement membrane matrix with biological activity. Matrigel: basement membrane matrix with biological activity. 2005;15:378–386. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Xu C, et al. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 12.Villa-Diaz LG, Ross AM, Lahann J, Krebsbach PH. Concise review: The evolution of human pluripotent stem cell culture: from feeder cells to synthetic coatings. Stem Cells. 2013;31:1–7. doi: 10.1002/stem.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes C, Postovit L, Lajoie G. Matrigel: A complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 14.Wood JA, Liliensiek SJ, Russell P, Nealey PF, Murphy CJ. Biophysical Cueing and Vascular Endothelial Cell Behavior. Materials. 2010;3:1620–1639. doi: 10.3390/ma3031620. [DOI] [Google Scholar]
- 15.Vukicevic S, et al. Identification of multiple active growth factors in basement membrane Matrigel suggests caution in interpretation of cellular activity related to extracellular matrix components. Exp Cell Res. 1992;202:1–8. doi: 10.1016/0014-4827(92)90397-q. [DOI] [PubMed] [Google Scholar]
- 16.Stein CA, Larocca RV, Thomas R, Mcatee N, Myers CE. Suramin - an Anticancer Drug with a Unique Mechanism of Action. Journal of Clinical Oncology. 1989;7:499–508. doi: 10.1200/JCO.1989.7.4.499. [DOI] [PubMed] [Google Scholar]
- 17.Prigozhina NL, Heisel AJ, Seldeen JR, Cosford ND, Price JH. Amphiphilic suramin dissolves Matrigel, causing an 'inhibition' artefact within in vitro angiogenesis assays. Int J Exp Pathol. 2013;94:412–417. doi: 10.1111/iep.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy WL, McDevitt TC, Engler AJ. Materials as stem cell regulators. Nat Mater. 2014;13:547–557. doi: 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murrow LM, Weber RJ, Gartner ZJ. Dissecting the stem cell niche with organoid models: an engineering-based approach. Development. 2017;144:998–1007. doi: 10.1242/dev.140905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cruz-Acuna R, Garcia AJ. Synthetic hydrogels mimicking basement membrane matrices to promote cell-matrix interactions. Matrix Biol. 2017;57–58:324–333. doi: 10.1016/j.matbio.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranga A, et al. Neural tube morphogenesis in synthetic 3D microenvironments. Proc Natl Acad Sci U S A. 2016;113:E6831–E6839. doi: 10.1073/pnas.1603529113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caiazzo M, et al. Defined three-dimensional microenvironments boost induction of pluripotency. Nat Mater. 2016;15:344–352. doi: 10.1038/nmat4536. [DOI] [PubMed] [Google Scholar]
- 23.Gjorevski N, et al. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 24.Enemchukwu NO, et al. Synthetic matrices reveal contributions of ECM biophysical and biochemical properties to epithelial morphogenesis. J Cell Biol. 2016;212:113–124. doi: 10.1083/jcb.201506055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huebsch N, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nature Materials. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable Hydrogels for Dynamic Tuning of Physical and Chemical Properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khetan S, et al. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nature Materials. 2013;12:458–465. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher OZ, Khademhosseini A, Langer R, Peppas NA. Bioinspired materials for controlling stem cell fate. Acc Chem Res. 2010;43:419–428. doi: 10.1021/ar900226q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ranga A, et al. 3D niche microarrays for systems-level analyses of cell fate. Nat Commun. 2014;5:4324. doi: 10.1038/ncomms5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh SP, et al. A synthetic modular approach for modeling the role of the 3D microenvironment in tumor progression. Sci Rep. 2015;5:17814. doi: 10.1038/srep17814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gill BJ, et al. A synthetic matrix with independently tunable biochemistry and mechanical properties to study epithelial morphogenesis and EMT in a lung adenocarcinoma model. Cancer Res. 2012;72:6013–6023. doi: 10.1158/0008-5472.CAN-12-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck JN, Singh A, Rothenberg AR, Elisseeff JH, Ewald AJ. The independent roles of mechanical, structural and adhesion characteristics of 3D hydrogels on the regulation of cancer invasion and dissemination. Biomaterials. 2013;34:9486–9495. doi: 10.1016/j.biomaterials.2013.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raza A, Ki CS, Lin CC. The influence of matrix properties on growth and morphogenesis of human pancreatic ductal epithelial cells in 3D. Biomaterials. 2013;34:5117–5127. doi: 10.1016/j.biomaterials.2013.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 35.Nagase H, Fields GB. Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptides. Biopolymers. 1996;40:399–416. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C399::AID-BIP5%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 36.West JL, Hubbell JA. Polymeric biomaterials with degradation sites for proteases involved in cell migration. Macromolecules. 1999;32:241–244. [Google Scholar]
- 37.Morgan CR, Magnotta F, Ketley AD. Thiol/Ene Photocurable Polymers. Journal of Polymer Science. 1977;15:627–645. [Google Scholar]
- 38.Belair DG, Murphy WL. Specific VEGF sequestering to biomaterials: Influence of serum stability. Acta Biomaterialia. 2013 doi: 10.1016/j.actbio.2013.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belair DG, et al. Human vascular tissue models formed from human induced pluripotent stem cell derived endothelial cells. Stem Cell Rev. 2015;11:511–525. doi: 10.1007/s12015-014-9549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 41.Kleinstreuer NC, et al. Environmental impact on vascular development predicted by high-throughput screening. Environ Health Perspect. 2011;119:1596–1603. doi: 10.1289/ehp.1103412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Masri H, et al. Integration of Life-Stage Physiologically Based Pharmacokinetic Models with Adverse Outcome Pathways and Environmental Exposure Models to Screen for Environmental Hazards. Toxicol Sci. 2016;152:230–243. doi: 10.1093/toxsci/kfw082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knudsen TB, et al. Activity profiles of 309 ToxCast™ chemicals evaluated across 292 biochemical targets. Toxicology. 2011;282:1–15. doi: 10.1016/j.tox.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 44.Sipes NS, et al. Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013;26:878–895. doi: 10.1021/tx400021f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Houck KA, et al. Profiling bioactivity of the ToxCast chemical library using BioMAP primary human cell systems. J Biomol Screen. 2009;14:1054–1066. doi: 10.1177/1087057109345525. [DOI] [PubMed] [Google Scholar]
- 46.Belair DG, Schwartz MP, Knudsen T, Murphy WL. Human iPSC-derived endothelial cell sprouting assay in synthetic hydrogel arrays. Acta Biomater. 2016;39:12–24. doi: 10.1016/j.actbio.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tal T, et al. Screening for chemical vascular disruptors in zebrafish to evaluate a predictive model for developmental vascular toxicity. Reproductive Toxicology. 2016 doi: 10.1016/j.reprotox.2016.12.004. (Submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fawcett T. An introduction to ROC analysis. Pattern Recognition Letters. 2006;27:861–874. doi: 10.1016/j.patrec.2005.10.010. [DOI] [Google Scholar]
- 49.Brodersen KH, Cheng Soon O, Stephan KE, Buhmann JM. 20th International Conference on Pattern Recognition (ICPR); pp. 3121–3124. [Google Scholar]
- 50.Latham AM, et al. Indolinones and anilinophthalazines differentially target VEGF-A- and basic fibroblast growth factor-mediated responses in primary human endothelial cells. Br J Pharmacol. 2012;165:245–259. doi: 10.1111/j.1476-5381.2011.01545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brossa A, et al. Sunitinib but not VEGF blockade inhibits cancer stem cell endothelial differentiation. Oncotarget. 2015;6:11295–11309. doi: 10.18632/oncotarget.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friis T, Engel AM, Bendiksen CD, Larsen LS, Houen G. Influence of levamisole and other angiogenesis inhibitors on angiogenesis and endothelial cell morphology in vitro. Cancers (Basel) 2013;5:762–785. doi: 10.3390/cancers5030762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wood JM, et al. PTK787/ZK. a novel and potent inhibitor of vascular endothelial growth factor receptor tyrosine kinases, impairs vascular endothelial growth factor-induced responses and tumor growth after oral administration. Cancer Res. 2000;60:2178–2189. [PubMed] [Google Scholar]
- 54.Donovan D, Brown NJ, Bishop ET, Lewis CE. Comparison of three in vitro human 'angiogenesis' assays with capillaries formed in vivo. Angiogenesis. 2001;4:113–121. doi: 10.1023/a:1012218401036. [DOI] [PubMed] [Google Scholar]
- 55.Francescone RA, 3rd, Faibish M, Shao R. A Matrigel-based tube formation assay to assess the vasculogenic activity of tumor cells. J Vis Exp. 2011 doi: 10.3791/3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song J, Rolfe BE, Hayward IP, Campbell GR, Campbell JH. Reorganization of structural proteins in vascular smooth muscle cells grown in collagen gel and basement membrane matrices (Matrigel): a comparison with their in situ counterparts. J Struct Biol. 2001;133:43–54. doi: 10.1006/jsbi.2001.4327. [DOI] [PubMed] [Google Scholar]
- 57.Vernon R, Angello J, Iruelaarispe M, Lane T, Sage E. REORGANIZATION OF BASEMENT-MEMBRANE MATRICES BY CELLULAR TRACTION PROMOTES THE FORMATION OF CELLULAR NETWORKS INVITRO. Laboratory Investigation. 1992;66:536–547. [PubMed] [Google Scholar]
- 58.Vernon RB, Sage EH. Between molecules and morphology. Extracellular matrix and creation of vascular form. Am J Pathol. 1995;147:873–883. [PMC free article] [PubMed] [Google Scholar]
- 59.De Smet F, Segura I, De Bock K, Hohensinner P, Carmeliet P. Mechanisms of Vessel Branching Filopodia on Endothelial Tip Cells Lead the Way. Arteriosclerosis Thrombosis and Vascular Biology. 2009;29:639–649. doi: 10.1161/ATVBAHA.109.185165. [DOI] [PubMed] [Google Scholar]
- 60.Maniotis AJ, et al. Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Am J Pathol. 1999;155:739–752. doi: 10.1016/S0002-9440(10)65173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saunders R, Hammer D. Assembly of Human Umbilical Vein Endothelial Cells on Compliant Hydrogels. Cellular and Molecular Bioengineering. 2010;3:60–67. doi: 10.1007/s12195-010-0112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rundhaug JE. Matrix metalloproteinases and angiogenesis. J Cell Mol Med. 2005;9:267–285. doi: 10.1111/j.1582-4934.2005.tb00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rashid A, Kuppa A, Kunwar A, Panda D. Thalidomide (5HPP-33) suppresses microtubule dynamics and depolymerizes the microtubule network by binding at the vinblastine binding site on tubulin. Biochemistry. 2015;54:2149–2159. doi: 10.1021/bi501429j. [DOI] [PubMed] [Google Scholar]
- 64.Satchi-Fainaro R, et al. Inhibition of vessel permeability by TNP-470 and its polymer conjugate, caplostatin. Cancer Cell. 2005;7:251–261. doi: 10.1016/j.ccr.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 65.Niwano M, et al. Inhibition of tumor growth and microvascular angiogenesis by the potent angiogenesis inhibitor, TNP-470, in rats. Surg Today. 1998;28:915–922. doi: 10.1007/s005950050252. [DOI] [PubMed] [Google Scholar]
- 66.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 67.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 68.Radzisheuskaya A, Silva JC. Do all roads lead to Oct4? the emerging concepts of induced pluripotency. Trends Cell Biol. 2014;24:275–284. doi: 10.1016/j.tcb.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Musah S, et al. Glycosaminoglycan-binding hydrogels enable mechanical control of human pluripotent stem cell self-renewal. ACS Nano. 2012;6:10168–10177. doi: 10.1021/nn3039148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang R, et al. A thermoresponsive and chemically defined hydrogel for long-term culture of human embryonic stem cells. Nat Commun. 2013;4:1335. doi: 10.1038/ncomms2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gharechahi J, et al. The effect of Rho-associated kinase inhibition on the proteome pattern of dissociated human embryonic stem cells. Mol Biosyst. 2014;10:640–652. doi: 10.1039/c3mb70255c. [DOI] [PubMed] [Google Scholar]
- 72.Nguyen EH, Zanotelli MR, Schwartz MP, Murphy WL. Differential effects of cell adhesion, modulus and VEGFR-2 inhibition on capillary network formation in synthetic hydrogel arrays. Biomaterials. 2014;35:2149–2161. doi: 10.1016/j.biomaterials.2013.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwartz MP, et al. Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc Natl Acad Sci U S A. 2015;112:12516–12521. doi: 10.1073/pnas.1516645112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schukur L, Zorlutuna P, Cha JM, Bae H, Khademhosseini A. Directed differentiation of size-controlled embryoid bodies towards endothelial and cardiac lineages in RGD-modified poly(ethylene glycol) hydrogels. Adv Healthc Mater. 2013;2:195–205. doi: 10.1002/adhm.201200194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hansen TD, et al. Biomaterial arrays with defined adhesion ligand densities and matrix stiffness identify distinct phenotypes for tumorigenic and nontumorigenic human mesenchymal cell types. Biomater Sci. 2014;2:745–756. doi: 10.1039/C3BM60278H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fairbanks BD, et al. A Versatile Synthetic Extracellular Matrix Mimic via Thiol-Norbornene Photopolymerization. Advanced Materials. 2009;21:5005–5010. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Le NNT, Zorn S, Schmitt SK, Gopalan P, Murphy WL. Hydrogel arrays formed via differential wettability patterning enable combinatorial screening of stem cell behavior. Acta Biomaterialia. 2015;34:93–103. doi: 10.1016/j.actbio.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hojjat-Farsangi M. Small-molecule inhibitors of the receptor tyrosine kinases: promising tools for targeted cancer therapies. Int J Mol Sci. 2014;15:13768–13801. doi: 10.3390/ijms150813768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fong TAT, et al. SU5416 is a potent and selective inhibitor of the vascular endothelial growth factor receptor (Flk-1/KDR) that inhibits tyrosine kinase catalysis, tumor vascularization, and growth of multiple tumor types. Cancer Research. 1999;59:99–106. [PubMed] [Google Scholar]
- 80.Maynard SE, et al. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfimction, hypertension, and proteinuria in preeclampsia. Journal of Clinical Investigation. 2003;111:649–658. doi: 10.1172/Jci200317189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clark DE, et al. A vascular endothelial growth factor antagonist is produced by the human placenta and released into the maternal circulation. Biology of Reproduction. 1998;59:1540–1548. doi: 10.1095/biolreprod59.6.1540. [DOI] [PubMed] [Google Scholar]
- 82.Belgore FM, Blann AD, Lip GYH. sFlt-1, a potential antagonist for exogenous VEGF. Circulation. 2000;102:E108–E108. doi: 10.1161/01.cir.102.15.e108. [DOI] [PubMed] [Google Scholar]
- 83.Hashizume H, et al. Complementary actions of inhibitors of angiopoietin-2 and VEGF on tumor angiogenesis and growth. Cancer Res. 2010;70:2213–2223. doi: 10.1158/0008-5472.CAN-09-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rosenberg SA. Interleukin 2 for patients with renal cancer. Nat Clin Pract Oncol. 4:497. doi: 10.1038/ncponc0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao Y, Langer R. Optimizing the delivery of cancer drugs that block angiogenesis. Sci Transl Med. 2010;2:15ps13. doi: 10.1126/scitranslmed.3000399. [DOI] [PubMed] [Google Scholar]
- 86.Richard AM, et al. ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chem Res Toxicol. 2016;29:1225–1251. doi: 10.1021/acs.chemrestox.6b00135. [DOI] [PubMed] [Google Scholar]
- 87.Nguyen EH, et al. Dataset for Versatile Synthetic Alternatives to Matrigel for Vascular Toxicity Screening and Stem Cell Expansion. 2017 doi: 10.1038/s41551-017-0096. figshare http://dx.doi.org/10.6084/m9.figshare.c.3791386. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.