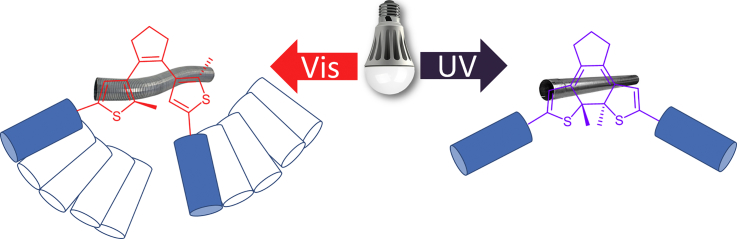
Abstract
For the first time, the photoisomerization of a diarylethene moiety (DAET) in peptide conjugates was used to probe the effects of molecular rigidity/flexibility on the structure and behavior of model peptides bound to lipid membranes. The DAET unit was incorporated into the backbones of linear peptide-based constructs, connecting two amphipathic sequences (derived from the β-stranded peptide (KIGAKI)3 and/or the α-helical peptide BP100). A β-strand-DAET-α-helix and an α-helix-DAET-α-helix models were synthesized and studied in phospholipid membranes. Light-induced photoisomerization of the linker allowed the generation of two forms of each conjugate, which differed in the conformational mobility of the junction between the α-helical and/or the β-stranded part of these peptidomimetic molecules. A detailed study of their structural, orientational and conformational behavior, both in isotropic solution and in phospholipid model membranes, was carried out using circular dichroism and solid-state 19F-NMR spectroscopy. The study showed that the rigid and flexible forms of the two conjugates had appreciably different structures only when embedded in an anisotropic lipid environment and only in the gel phase. The influence of the rigidity/flexibility of the studied conjugates on the lipid thermotropic phase transition was also investigated by differential scanning calorimetry. Both models were found to destabilize the lamellar gel phases.
Keywords: Molecular flexibility/rigidity, Membrane-active peptides, Molecular photoswitches, Diarylethene, Differential scanning calorimetry, Solid-state 19F-NMR spectroscopy
Graphical abstract
Highlights
-
•
DAET building blocks can be used to study rigidity/flexibility effects in supramolecular model systems.
-
•
Photoswitchable DAET linkers perturb only up to 3–4 adjacent amino acid residues.
-
•
Membrane-bound amphiphilic secondary structure elements exert a negligible influence on each other when linked by DAET.
-
•
The rigidity of peptide conjugates affected their structural behavior only in the lipid gel phase.
1. Introduction
A subtle balance between conformational flexibility and rigidity is crucial in proteins to exert their specific functions with optimal efficiency, e.g. in molecular recognition and/or enzymatic catalysis [1], [2], [3], [4]. This balance varies significantly across any large structural unit and may correlate with the particular functions of different molecular domains [5], [6]. The role of molecular rigidity/flexibility in biochemical processes should not be underestimated, as it affects the kinetic and thermodynamic parameters of chemical transformations and the supramolecular interactions in a complex manner. For example, it has long been believed that the entropy of rigid ligand binding to receptors should always be favorable compared to the binding of conformationally flexible analogs [7], [8]. While this might be true for simple supramolecular host-guest complexes [9], [10], quantitative investigations of proteins interacting with flexible and rigid ligands in water showed that this concept should be revised. In fact, the entropy penalty for the binding of rigid ligands to protein targets can be substantially higher than for their closest flexible analogs [11], [12], [13], [14], [15], [16], [17], [18]. This difference was attributed to the stronger freezing of protein molecular motion that occurs upon binding of a rigid ligand [19], [20] compared to the case of binding a flexible ligand. It was also noted that changes in non-bonding interactions throughout entire protein-ligand complexes, including interactions with water molecules and counter-ions, should be considered [21]. Only recently have powerful experimental techniques, in particular NMR and sensitive calorimetry, started to reveal the structural and energetic details of protein-ligand binding [22], [23]. The elucidation of such details is important for practical applications, especially in medicinal chemistry. The restriction of conformational mobility is one of the most general principles of drug design; there are numerous examples where rigidified molecules bind to biological targets more tightly and display higher efficacy and selectivity than their flexible analogs [24]. However, there are also many examples where this principle fails [25], [26], [27], [28], hence a deeper understanding of the role of conformational flexibility/rigidity in the interaction of drug candidates with their biological targets is of great value.
Even less studied than for enzymes and receptors is the role of conformational flexibility/rigidity in the case of polypeptides interacting with lipid bilayers. There are several reports in the literature indicating that protein flexibility is a key determining factor for important functions of membrane-active peptides. Bertocco et al. compared a fusion peptide GLFGAIAGFIEG-NHEt derived from the influenza virus hemagglutinin protein [29] with its conformationally restricted analog (containing three α-Me-valines at positions 2, 6 and 10 in place of the natural residues) and found that the restricted analog was less potent in promoting lipid mixing [30]. The authors concluded that the molecular flexibility of the fusion peptide and the resulting conformational plasticity were essential for the fusogenicity – the ability to destabilize the host membrane and facilitate transfection. An analogous study was carried out with the 22-mer antimicrobial peptide (AMP) piscidin 1; again, the rigid analog was shown to be less active [31]. In this case, a single replacement of a conformationally restricted proline by a flexible peptoid residue at the junction of two α-helical fragments resulted in a clear-cut enhancement of membranolytic activity. Other studies, however, demonstrated that more rigid AMPs might have stronger antimicrobial activities compared to their flexible counterparts [32]; and again there are also papers arguing that AMP activity may not change at all upon the purposeful rigidification of peptides [33]. Liu et al. systematically studied the “mechanical determinant” underlying the activity of amphipathic cationic AMPs, and established a “flexibility index” [34] that seems to be applicable to those AMPs that involve direct membrane damage. Furthermore, rigid and flexible molecules, when embedded in membranes, may affect the physical properties of lipid bilayers in a differential manner. A well-recognized example is the rigid molecule cholesterol, which causes a reduction in lipid chain conformational dynamics in the fluid phase, but leads to increased fluidity of the gel phase [35]. The influence of rigid/flexible peptides on membrane properties has also been studied, using various biophysical methods [36], [37], [38].
The studies cited above have prompted us to further address the role of conformational rigidity/flexibility in the membrane interactions of amphipathic peptides. We addressed these properties by designing linear model compounds, based on the two most common secondary structure elements (α-helix, β-strand), in which the junction between two formally independent peptide fragments would be either flexible or rigid. By using a molecular photoswitch as a cross-linking building block, a change in rigidity/flexibility can be achieved with a minimal difference in the number of atoms, the overall chemical bonding pattern and the chemical nature of the functional groups. In this paper, we describe the design of such model rigid/flexible peptide conjugates, and report the use of circular dichroism (CD) and solid-state 19F-NMR to study their structural differences in model membranes. Their influence on the membrane properties was also addressed, using differential scanning calorimetry (DSC). We have not focused on any particular type of biologically active peptide, nor have we tried to generate a useful photoswitchable AMP. Nonetheless, our model molecules are of biological relevance in as far as they will help to elucidate the role of conformational rigidity/flexibility in real biological systems, like peptides interacting with biomembranes.
2. Materials and methods
2.1. Materials
All Fmoc-protected amino acids and reagents for peptide synthesis (DIPEA, diisopropylethylamine; HOBt, N-hydroxybenzotriazole; HBTU, 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate; HOBt, 1-hydroxybenzotriazol; piperidine) were purchased from Iris Biotech (Marktredwitz, Germany) or Novabiochem (Nottingham, UK). The Fmoc-protected (L)-3-(trifluoromethyl)-bicyclo[1.1.1]pent-1-ylglycine (Bpg) was obtained from Enamine (Kyiv, Ukraine). Solvents for synthesis and purification were purchased from Biosolve (Valkenswaard, Netherlands) or Acros Organics (Geel, Belgium). Ultraviolet-grade chloroform and methanol for the sample preparation in biological and biophysical assays were obtained from VWR International (Bruchsal, Germany). Ultrapure laboratory grade Milli-Q water was used in all cases (prepared with an EMD Millipore system for water purification). The lipids were purchased either from Sigma-Aldrich (sodium dodecylsulfate, SDS) or from Avanti Polar Lipids (1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine, DMPC) and used without further purification. All other materials were of the highest purity available.
2.2. Synthesis of β/α-model peptides and their 19F-labeled analogs
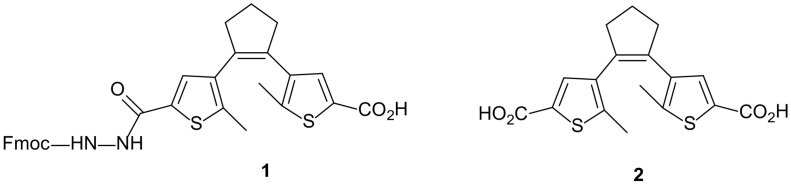
Standard Fmoc-based solid-phase peptide synthesis (SPPS) and commercially available reagents were used for the peptide synthesis. Leucine-preloaded Rink amide 4-methylbenzhydrylamine resin with a loading of 0.67 mmol/g (150 mg, 1 equiv) was used. Coupling of the amino acids was performed using the following molar ratios of the reagents: an Fmoc-amino acid (4 equiv), HOBt (4 equiv), HBTU (3.9 equiv), and DIPEA (8 equiv). A diarylethene-derived N-Fmoc-protected amino acid (Fig. 1, compound 1) was prepared as described [39] and used as an individual SPPS building block to incorporate the photoswitching linker at an appropriate stage in the linear peptide sequence. The photoswitch was incorporated by coupling with 1 (1.5 equiv), HOBt (1.5 equiv), HBTU (1.45 equiv), and DIPEA (3 equiv). The coupling time in all cases was 40 min. N-Fmoc deprotection was carried out by treating the resin with 20% piperidine in dimethylformamide for 20 min. After completing the synthesis, the resin was washed with dichloromethane and dried under vacuum for 24 h. The peptides (Table 1) were cleaved from the resin with a cleavage cocktail (trifluoroacetic acid, triisopropylsilane and water, 92.5:2.5:5 v/v, 10 ml, 30 min). The volatile products were blown off from the filtered solutions by an argon stream. The residual materials were dissolved in an acetonitrile-water (1:1) mixture and lyophilized. The crude peptides were purified on a preparative (22 × 250 mm) Vydac C18 column with a linear A:B gradient of 5% B/min slope (A:10% acetonitrile, 5 mM HCl; B:90% acetonitrile, 5 mM HCl). The purity of the peptides was determined to be in each case > 95% on an analytical (4.6 × 250 mm) Vydac C18 column with a linear A:B gradient of 1% B/min slope. The identity of each compound was confirmed by conventional MALDI-TOF mass spectrometry.
Fig. 1.
Diarylethene (DAET) building blocks for SPPS: Fmoc-protected amino acid (1), and a dicarboxylic acid (2) used to prepare the model compounds studied in this work.
Table 1.
Composition of the DAET-linked peptide conjugate, representing the β/α-model, and list of its 19F-labeled analogs.
| Name | Sequence |
|---|---|
| β/α-model (unlabeled) | KIKIGAKI-1-KKLFKKILKYL-NH2 |
| β2I/α | K-Bpg-KIGAKI-1-KKLFKKILKYL-NH2 |
| β4I/α | KIK-Bpg-GAKI-1-KKLFKKILKYL-NH2 |
| β6A/α | KIKIG-Bpg-KI-1-KKLFKKILKYL-NH2 |
| β8I/α | KIKIGAK-Bpg-1-KKLFKKILKYL-NH2 |
| β/α3L | KIKIGAKI-1-KK-Bpg-FKKILKYL-NH2 |
| β/α4F | KIKIGAKI-1-KKL-Bpg-KKILKYL-NH2 |
| β/α7I | KIKIGAKI-1-KKLFKK-Bpg-LKYL-NH2 |
| β/α8L | KIKIGAKI-1-KKLFKKI-Bpg-KYL-NH2 |
| β/α10Y | KIKIGAKI-1-KKLFKKILK-Bpg-L-NH2 |
2.3. Synthesis of α/α-model peptides and their 19F-labeled analogs
19F-Labeled fragments of peptide BP100 were synthesized as described in Section 2.2 to the point of Fmoc deprotection after the coupling of the 11th amino acid. Then, the resin was treated for 2 h with a mixture of the following reagents: 0.5 equiv. of the diarylethene-derived, commercially available (Enamine, Ukraine) dicarboxylic acid (Fig. 1, structure 2), HOBt (0.5 equiv), HBTU (0.5 equiv), and DIPEA (1 equiv). After the reaction was completed, the resin was washed with dichloromethane and dried under vacuum for 24 h. The peptides (Table 2) were cleaved, purified and analyzed as described in Section 2.2.
Table 2.
Composition of the DAET-linked peptide conjugate, representing the α/α-model, and list of its 19F-labeled analogs.
| Name | Sequence |
|---|---|
| α/α-model (unlabeled) | 2-(KKLFKKILKYL-NH2)2 |
| α/α3L | 2-(KK-Bpg-FKKILKYL-NH2)2 |
| α/α4F | 2-(KKL-Bpg-KKILKYL-NH2)2 |
| α/α7I | 2-(KKLFKK-Bpg-LKYL-NH2)2 |
| α/α8L | 2-(KKLFKKI-Bpg-KYL-NH2)2 |
| α/α10Y | 2-(KKLFKKILK-Bpg-L-NH2)2 |
2.4. Preparation of the compounds in the defined photoforms
The peptides were synthesized with the diarylethene linkers maintained in their ring-open photoforms (stable under visible light) and converted to their ring-closed photoforms by irradiation of their water/acetonitrile (3:1) solutions (100 μg/ml) with an ultraviolet (UV) lamp (XX-15F/F Spectroline, 365 nm, peak UV intensity of 1.1 mW/cm2 at 25 cm) for 75 min. The conversion progress was monitored by HPLC. All of the peptidomimetics were converted to their corresponding ring-closed form almost quantitatively. The ring-closed photoforms of the compounds were used immediately after their generation whenever possible. All the operations with the ring-closed forms of the conjugates were performed under dim light in order to avoid uncontrolled back-isomerization. Afterwards, we checked again by HPLC that no uncontrolled back-isomerization had taken place during sample preparation and experiments (see Supplementary Data).
2.5. Samples for solid-state NMR
The 19F-labeled analogs were reconstituted in oriented DMPC bilayers following the solid-state 19F-NMR strategy described elsewhere [40], [41]. All oriented samples were prepared at a molar peptide-to-lipid ratio (P/L) of 1:100. The peptides (0.206 μmol, 0.54–0.68 mg) and the lipid (20.6 μmol, 14 mg) were dissolved in methanol and spread over 14 glass plates (18 × 7.5 mm, Marienfeld Laboratory Glassware). For the sample preparation of the compounds in the ring-closed forms, the method was slightly modified: first, the peptides were dissolved in methanol and the solutions were additionally exposed to UV light for 15 min in order to ensure the stable photostate of the compounds in their ring-closed forms; then the required amount of the lipid solution was added and the mixtures were spread over the glass plates (this step and all the following operations with the ring-closed forms were performed under dim light). The solvent was allowed to evaporate for 40 min, and the glass plates were further dried in vacuum for 8 h in a dark vessel. The plates were stacked onto each other and hydrated in a dark chamber with saturated potassium sulfate (96% relative humidity) for 12 h, then wrapped in a Nescofilm and Sarogold foils to prevent their dehydration and light exposure during solid-state NMR.
2.6. Solid-state NMR measurements
Solid-state 31P-NMR measurements were done at 202.5 MHz on an Avance III Bruker NMR spectrometer with a wide bore 500 MHz magnet (Bruker Biospin, Karlsruhe, Germany). A standard flat-coil double-resonance 1H/X probe from Bruker was used. The Hahn-echo pulse sequence [42] with a 90° pulse of 5 μs and a 30 μs echo time and 1H SPINAL64 [43] decoupling during acquisition were used. The acquisition time was 10 ms, and the recycle time was set to 1 s. Typically, 128 scans were acquired. Observation of the 31P-NMR signals from the phospholipids allowed the quantitative assessment of the degree of lipid orientation. > 70% of the lipids were found to be well oriented in all of the prepared samples (see the Supplementary Data); therefore, an assumption was made that the homogenously incorporated peptides should also possess a similar high degree of alignment.
Solid-state 19F-NMR experiments were performed using double-tuned goniometer-equipped 19F/1H probes from Doty Scientific (Columbia, USA) at a frequency of 470.6 MHz using an anti-ringing pulse sequence [44] with a 90° pulse of 3.25 μs, a sweep width of 500 kHz, 4096 data points, and proton decoupling by a TPPM scheme [45]. The 19F chemical shifts were referenced using the 19F-NMR signal of a 100-mM solution of NaF, the chemical shift of which was set to − 119.5 ppm. Typically, 8000 scans were accumulated. The solid-state 19F-NMR measurements were done below and above the main lipid phase transition temperature (at 15 and 40 °C, respectively; the DMPC phase transition occurs at ~ 23 °C) for each prepared oriented sample.
In all solid-state NMR experiments, the samples were aligned with the bilayer normal parallel to the magnetic field.
2.7. Solid-state 19F-NMR analysis
Anisotropic dipolar 19F-NMR splittings that were measured in the spectra of Bpg-labeled peptide conjugates were used to assess the secondary structure and orientation in the lipid bilayers, in terms of the tilt angle (ρ) and the rotation angle (τ), as described in the literature [40], [41]. In short, for each peptide fragment the underlying structure was assumed (i.e., regular β-strand or an α-helix). The 19F-labels (Bpg) [46] were incorporated into the model in silico, using their known geometric positions relative to the peptide backbone (i.e., the orientation of the Cα—Cβ bond vector of the side chain (collinear to the CF3-axis director as given by the Bpg side chain geometry) was assumed to be described by the angles α = 46°, and β = 113°, ω = 100°) [47]. Next, a systematic grid search over all orientations of the 19F-label in space [0°, 180°; 1°-step] was performed, and the dipolar splittings for each labeled position were calculated and compared with the corresponding experimental values. The calculation of an individual Cα—Cβ vector position was performed in accordance to the known relationship between an observed dipolar splitting (ΔCF3obs) and the time-averaged value of the angle θ (the angle formed by the rotating CF3-group symmetry axis and the membrane normal): ΔCF3obs = ½(3cos2θ-1) × ΔCF3max. The CF3-group was assumed to freely rotate (i.e. ΔCF3max = 16 kHz) [48]. For every structural assumption, the comparison at each ρ/τ combination was implemented by calculating the root-mean-square deviation (rmsd) between the predicted and observed dipolar splittings. In addition, any averaging by segmental and/or global molecular motions (fast on the NMR-timescale) was taken into account by scaling with an order parameter Smol, which was systematically varied in the grid search between 1 and 0 (assuming 1 = completely rigid; 0 = isotropically mobile molecule) in steps of 0.01. A self-consistent solution with minimal rmsd thus confirmed the assumed structural model and yielded the corresponding orientational angles ρ and τ, and the mobility factor Smol.
2.8. Sample preparation for CD
Stock solutions (1 mg/ml) of all measured conjugates were prepared in 50% ethanol, and their exact concentrations were determined with analytical HPLC, as detailed above. The aliquots containing the amount of the peptides required for the experiments were placed in 2-ml glass vials, and the solvent was evaporated in vacuum. The dry material in the vials was then used for the CD sample preparation, by either dissolving them in a phosphate buffer (PB, 20 mM, pH 7.4) or mixing them with detergent micelles or vesicle suspensions prepared as described below. The stock solutions and all samples of conjugates in the ring-closed forms were kept in darkness.
Stock solutions of the detergent SDS or the lipid DMPC, (10 mg/ml) were prepared in a 1:1 mixture of methanol/chloroform and aliquoted into 2-ml glass vials to supply 0.4 mg of lipid per vial. The solvents were removed by a flow of argon, and the vials were dried under vacuum for 8 h. In total, 300 μl of PB was added to each vial, and the suspensions were vortexed for 10 min (r.t.) and further sonicated for 30 min at 40 °C. To prepare the peptide/lipid mixtures, PB (100 μl) was added to the glass vials containing the dry peptides (to obtain a particular peptide-to-lipid ratio, see above), and the mixtures were sonicated for 5 min, followed by the addition of the suspension of vesicles (300 μl) or detergent micelles to yield 400 μl volumes in each case. The resulting samples had lipid concentrations of 1 mg/ml, and the pure conjugate photoforms were present in peptide/lipid ratios of 1:100, 1:20, or 1:5.
2.9. CD measurements
CD spectra were recorded on a Jasco J-815 spectrophotometer between 400 and 180 nm at 0.1 nm intervals, using 1 mm Suprasil® (Hellma) quartz cuvettes at 25 °C. Three consecutive scans at a scan-rate of 10 nm/min, 8 s response time and 1 nm bandwidth were averaged for each sample and for the background spectra. After subtracting the appropriate background spectra, the corrected CD data were processed using Jasco Spectra Analysis software.
2.10. DSC measurements
DSC analysis was performed using a MicroCal VP-DSC microcalorimeter (Malvern, UK). Stock suspensions of DMPC multilamellar vesicles (2 mg/ml) were prepared in PB by homogenization of the lipid at 40 °C (incubation for 1 h with repeated vortexing). The stocks were combined with PB solutions of the peptide material (prepared as above) or pure buffer to achieve the final concentration of the lipid (1 mg/ml) and appropriate peptide/lipid ratios. The samples were prepared fresh, and as a common procedure, both the reference (pure PB) and the samples were degassed for 10 min before being introduced into calorimeter cells. Thermograms were collected over the range 5–45 °C in the following order: 1st (heating) scan at 1.5°/min, 2nd (cooling) scan at 1 °C/min, 3rd (heating) scan at 0.5°/min, and 4th (cooling) scan at 1 °C/min. In all the experiments, only the heating scans were analyzed. The experiments were carried out in triplicate to check the reproducibility of the results. Buffer subtraction and baseline correction were performed using MicroCal software. The enthalpy values were obtained from the 3rd (heating) scans by integrating the area under the phase transition signals, as recommended by the vendor.
3. Results and discussion
3.1. Design and synthesis of model compounds
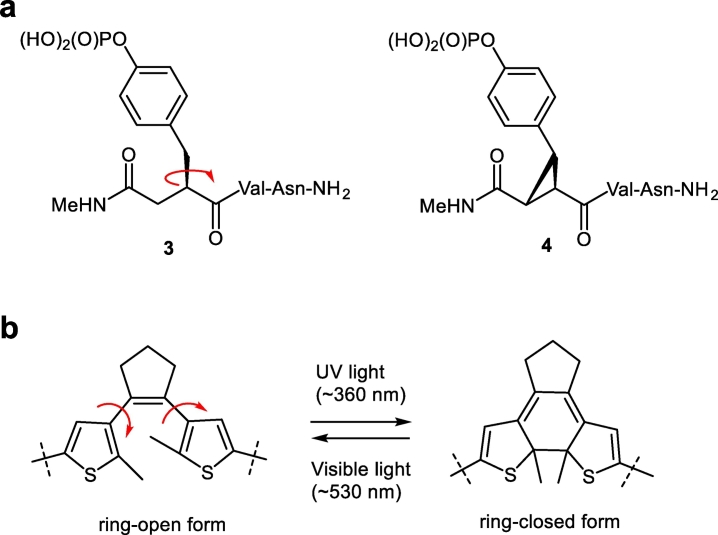
The principles of our design of flexible and rigid peptidomimetic models are inspired by and therefore similar to those described in [21], in which the flexible and rigidified pseudopeptide models 3 and 4 differed by only one C—C bond (Fig. 2a). The cyclopropane ring in 4 severely restricts intra-molecular motions compared with the relatively flexible 3. At the same time, the overall molecular constitution (atom connectivities, the number and the kind of atoms in the backbone and side chains) is similar for 3 and 4. Here, we propose the use of a molecular photoswitchable linker in the same fashion, allowing to control the rigidity/flexibility of the model compounds by a benign external stimulus – light. Recently, we reported on photoswitchable peptidomimetics containing a photocontrollable diarylethene fragment (DAET, Fig. 2b) that was directly incorporated into the polypeptide backbone [39]. Reversible photoisomerization of the DAET occurs through the formation or cleavage of a single C—C bond. The ring-closed photoform (obtained upon irradiation with UV light) and the ring-open photoform (produced by visible light) differ only slightly in their constitution but have substantially different conformational mobility: due to its polycyclic planar structure the ring-closed form is obviously more rigid than the ring-open form.
Fig. 2.
Design principles of the flexible/rigid module in peptidomimetic molecules. (a) Flexible (3) and rigidified (4) pseudopeptides studied in [21]; (b) a diarylethene moiety (within the dashed lines) in the flexible ring-open and the rigidified ring-closed forms. The additional conformational freedom of the flexible forms in both molecular fragments is indicated by the red arrows.
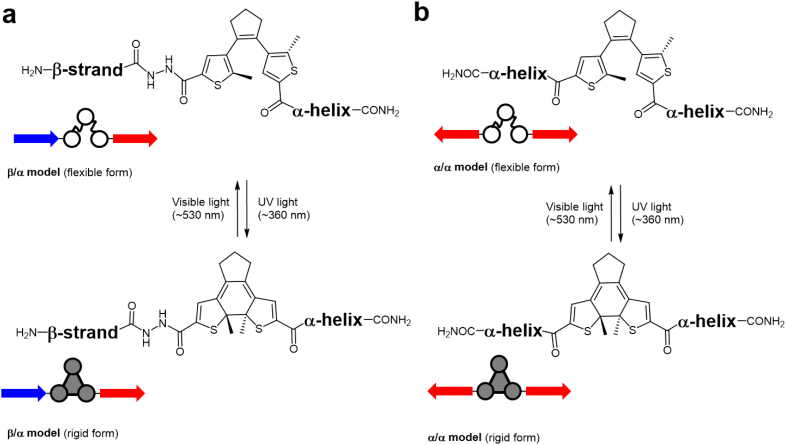
In this work, we placed the photoswitchable DAET module as a linker between two short polypeptide sequences derived from membrane-active peptides with defined conformational preferences. The choice of the two flanking peptide fragments was made by considering (i) their differential secondary structures, (ii) their amphipathic nature that determines their interaction with lipid bilayers, and (iii) our previous knowledge of their orientational preferences in lipid membranes. In particular, we used a truncated form of the (KIGAKI)3 peptide, which forms extended β-strands, and the well-studied 11-mer BP100 (KKLFKKILKYL-NH2), which is structured as an α-helix. Both are known to fold into ideal amphipathic secondary structure elements in the presence of lipid bilayers [49], [50]. Two types of model DAET-crosslinked conjugates were thus designed: an asymmetric “β/α-model”, and a symmetric “α/α-model” (Fig. 3). The first one comprised of an octapeptide “β-strand” KIKIGAKI that was connected to the DAET linker (using 1), which was then connected to the short “amphipathic α-helix” of BP100. The second conjugate had two identical “α-helices” (BP100) connected via their N-termini to the DAET-derived linker (in its dicarboxylic acid form 2). The ring-closed photoforms of DAET in both molecules render the linker rigid, while the ring-open photoforms provide a flexible connection for the flanking peptide fragments, as illustrated in Fig. 3. It should be noted at this point that there was no intention to insert a photoswitch within a continuous beta-strand or a helix, but we rather used the DAET linker to mimic a loop connecting two different domains. This way, it would be possible to compare and contrast the flexible forms of our hybrid-compounds with their corresponding rigidified forms.
Fig. 3.
Model compounds designed in this study: (a) β/α-model and (b) α/α-model with the DAET photoswitchable residue in both photoforms.
The unlabeled conjugates for the DSC and CD studies were prepared from the respective peptide fragments consisting of their usual amino acid residues. In addition, a series of singly or doubly 19F-labeled analogs were prepared for the solid-state 19F-NMR measurements (see Table 1, Table 2 in the methods section). These analogs contained the artificial fluorine-labeled amino acid Bpg [46] in place of a non-polar residue in one (β/α-model) or two (α/α-model) positions of the peptide conjugate. Bpg had been originally designed as a 19F-NMR label for the substitution of hydrophobic (Ala, Val, Leu, Ile, and Met) residues. Moreover, our previous NMR data on the full-length [KIGAKI]3 and the free BP100 peptides had been obtained using the Bpg labeled analogs [51], [52], hence they can now serve as direct references for the present study. Notably, as was shown in [46], [51], [52] 19F-labelling with a single Bpg substituent is essentially non-perturbing. The structure and membrane interactions of the parent peptides were not affected by labelling.
3.2. Lipid polymorphism of DMPC vesicles in the presence of conjugates
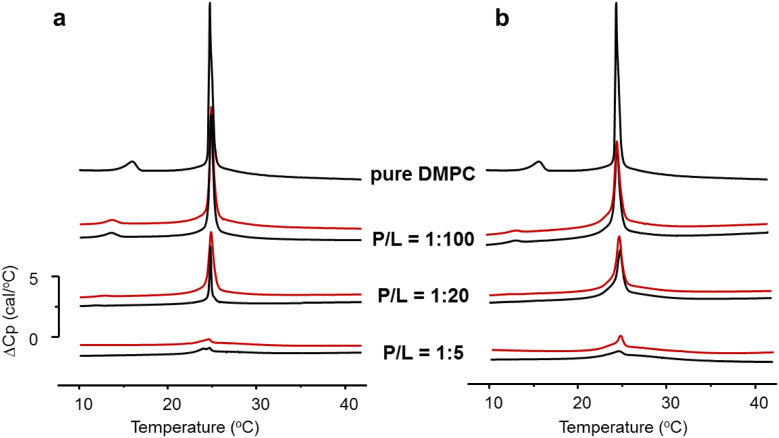
First, we examined whether the two model peptides are still able to interact with lipid membranes. DSC was performed with DMPC vesicles in the presence of increasing amounts of the rigid/flexible forms of both models. In the absence of the peptides, as expected, the multilamellar vesicles of the saturated lipid DMPC exhibited two endothermic events (Fig. 4a). A less enthalpic pre-transition (Tp) arising from a conversion of the lamellar gel phase to the rippled gel phase occurred at ~ 16 °C [53], [54]. A second, sharp main transition (Tm), arising from a conversion of the rippled gel phase to the lamellar liquid-crystalline fluid phase, was observed at 23.4 °C, in accordance with previous data (e.g., [53], [54]). The enthalpies for the pre-transition (∼ 1 kcal/mol) and the main transition (∼ 6.8 kcal/mol) are also close to the reported values (e.g., [54]). The addition of the peptide conjugates to the vesicle suspensions systematically influenced the Tp and Tm transitions in both cases, i.e. for both the β/α- and α/α-models (Fig. 4). Hence, both amphiphilic conjugates interact with the zwitterionic DMPC bilayers, as anticipated. For all four molecules, an increase in the amount of peptide leads to a reduction in the Tm and Tp values. At the highest P/L ratio used (1:5), no pre-transition peaks were observed. In addition, both models, irrespective of their rigidity/flexibility, caused a concentration-dependent broadening of the transitions, which may reflect a reduction in the cooperativity and possibly a disintegration of the multilamellar vesicles. Interestingly, all thermograms collected after the addition of peptides were skewed towards lower temperatures, indicating that both compounds preferentially distribute into fluid regions of the membrane, i.e., they stabilize the fluid lamellar phase. The rigid forms of both models behave like the flexible forms, but higher concentrations were required to elicit the same qualitative effects, suggesting a decreased binding affinity of the rigid forms to the gel states of the membranes.
Fig. 4.
DSC traces (heating scans) illustrating the thermotropic phase behavior of DMPC vesicles without (a, b topmost trace) or in the presence of different amounts of the (a) β/α-model and (b) α/α-model. Concentrations are reported as molar peptide-to-lipid ratios (P/L), the black lines indicate the flexible ring-open forms of the conjugates, and the red lines indicate the rigid ring-closed forms.
3.3. Conformational differences between rigid and flexible conjugates in solution
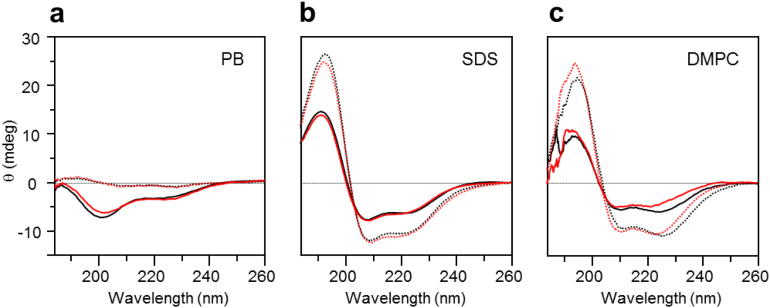
Next, we studied the conformational preferences of the two models by means of CD, monitoring the overall structure of the peptidic parts. The CD spectra (Fig. 5) of both β/α- and α/α-models were recorded in order to assess the impact of the flexibility/rigidity of the DAET linkers in isotropic and membrane-mimicking environments. No significant differences were observed between the two forms of either model, when dissolved in phosphate buffer alone or in the presence of SDS micelles. The CD spectra of the rigidified and the flexible forms were essentially the same. Spectra of the flexible forms in the presence of DMPC revealed a systematically increased negative intensity at 222 nm and a red shift of the 208 nm band. This result may correspond to either a slight decrease in the helicity of the rigid forms, or to differential binding affinities of the two forms to DMPC. The latter suggestion is corroborated by the results of the DSC experiments described above. Since there was no correlated increase in the intensity of the 196 nm CD band, which actually decreased, we conclude that a conformational perturbation by the rigid DAET form is unlikely. These measurements suggest that the CD spectral range should be generally useful for the structural analysis of DAET-containing peptides, at it is free of any spectral contribution from the photoswitching linker (in contrast to some cyclic peptides containing aromatic residues [39]).
Fig. 5.
CD spectra of the peptide conjugates in different environments at a concentration of 100 μg/ml. β/α-Model (solid lines) and α/α-model (dotted lines) in PB solutions (a); in the presence of negatively charged SDS micelles (b, 25 mM SDS); and in the presence of sonicated DMPC vesicles (c). The peptides are compared here in their flexible form (black lines) and in the rigid form (red lines).
In addition, the CD data provide useful insights into the conformational properties of the individual peptidic units of the two models. According to previous studies, the parent peptides are unstructured in aqueous buffers [51], [52], [55], [56]. In the presence of lipid bilayers, both peptides bind to the membranes and undergo structural transitions to either a β-strand or an α-helix. In full accordance with this observation, our β/α-model in both forms was unstructured in buffer solution, as its CD spectra resembled that of a random coil. The α/α-model gave only a weak signal in aqueous buffer, independent of the state of the DAET linker, which may be due to aggregation. As expected, both models produced CD lineshapes with a dominant α-helical signature in SDS micelles and in DMPC vesicles, indicating the binding to membranes and structuring of the amphiphilic peptides under these conditions.
3.4. Structural studies in oriented lipid bilayers by solid-state 19F-NMR
3.4.1. Rigid and flexible forms of the β/α-model in DMPC bilayers
3.4.1.1. β-Fragment
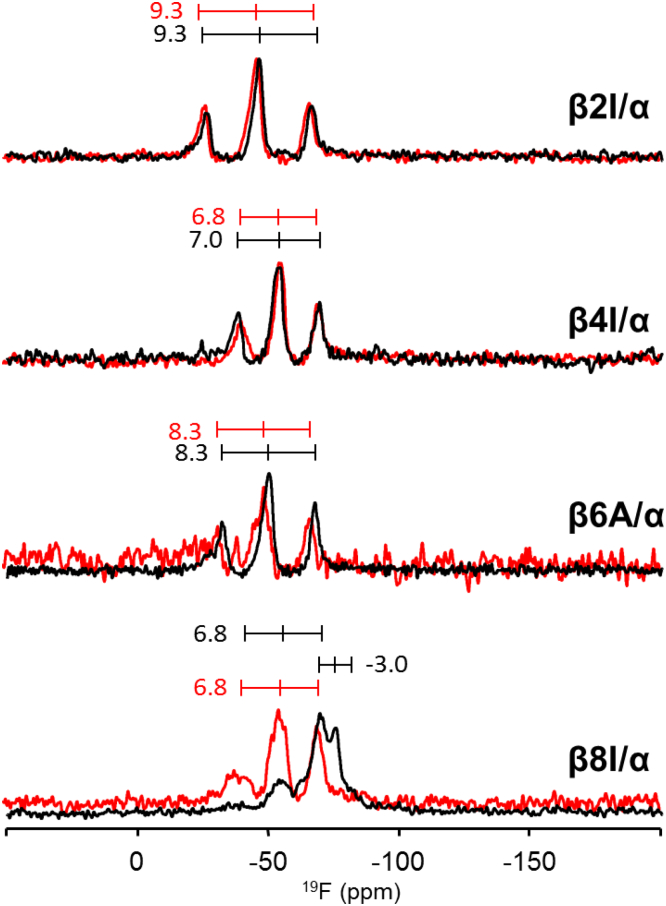
The parent peptide [KIGAKI]3 was originally designed as an ideal amphipathic β-strand, composed of alternating hydrophobic and polar/cationic residues. It was shown to engage in a concentration-dependent equilibrium between a flexible monomeric form and aggregated β-sheets in the membrane-bound state [51], [56]. Both forms could be distinguished by solid-state 19F-NMR of selectively 19F-labeled peptides in macroscopically oriented lipid samples. In the case of the monomeric form, a triplet splitting of about + 8 kHz is observed for the Bpg reporter group independently of the label position, and the peptide is rather dynamic (given a molecular oder parameter of Smol ≈ 0.5). The aggregated β-sheets are characterized by a dipolar coupling of about + 15 kHz (because the Smol values are in this case close to the maximum of 1.0). Fig. 6 illustrates the solid-state 19F-NMR spectra of the β/α-model containing a single 19F-reporter group at different positions along the β-stranded fragment. As can be seen, each of the labeled peptides shows a single triplet, with positive coupling constants in the range of 6.8–9.3 kHz, when the conjugate was in the rigid form (see Table 3 for the coupling values). In general, the 19F-NMR spectra of the rigidified β/α-model closely resembled the monomeric state of the parent full-length [KIGAKI]3 peptide, which was earlier shown to align parallel to the membrane surface [51], [56]. The slightly lower coupling constant values obtained for our analogs labeled at positions 4 and 8 can be explained by their proximity to the two perturbing sites in the molecule (Gly5 and the DAET residues, respectively), assuming that the C-terminally placed photoswitching unit behaves as a β-strand breaker.
Fig. 6.
Solid-state 19F-NMR spectra of the β/α-model labeled with Bpg in the β-strand. Conjugates were reconstituted in oriented DMPC bilayers (P/L = 1:100) and measured at 40 °C in the rigid (red lines) and flexible (black lines) forms. Triplet splitting values are indicated for clarity and as an example of the spectra readout (see Table 3).
Table 3.
19F-19F dipolar couplings of the β/α-model labeled in the β-strand, as determined in DMPC at 40 °C. Values for the full-length (KIGAKI)3 parent peptide labeled at the same positions are also given as a reference for its monomeric and aggregated states (taken from [51], [56], respectively).
| DAET state | Dipolar coupling (kHz ± 0.5) |
|||
|---|---|---|---|---|
| β2I/α | β4I/α | β6A/α | β8I/α | |
| Flexible | + 9.3 | + 7.0 | + 8.3 | + 6.8/− 3.0a |
| Rigid | + 9.3 | + 6.8 | + 8.3 | + 6.8 |
| Reference, monomeric | + 8.8 | + 8.2 | + 8.0 | + 8.2 |
| Reference, aggregated | + 15.3 | + 14.4 | + 15.4 | + 16.4 |
Two sets were observed, see text and Fig. 6.
The 19F-NMR spectra of the corresponding flexible forms of the β/α-model are identical to those of the rigid forms, suggesting that the orientational state of the β-stranded fragment is stable, and that the β-fragment folds and behaves essentially independently from the α-helix and the DAET modules. A single exception to this behavior was observed for the label at position Ile8, which is directly adjacent to the photoswitchable linker. As can be seen in Fig. 6, the + 6.8 kHz splitting persisted, but an additional triplet signal with − 3.0 kHz splitting emerged. In the membrane-bound state, photoconversion back to the closed-ring form led to the disappearance of this new signal, confirming that it was caused by photoswitching. The signal could be assigned to one of the two possible conformations of the open-ring state of the linker (i.e., with parallel or antiparallel aromatic rings), which is in line with the above-suggested β-breaking ability of the DAET unit.
Measurements at the temperatures below the lipid chain melting temperature Tm did not reveal any difference from the spectra of the observations described above, except that the 19F-NMR signals were broadened, as expected for surface-bound peptides in the more viscous gel phase lipids. Again, the orientation of the β-stranded fragment in both, the rigidified and the flexible forms of the β/α-model, is independent of the membrane fluidity.
3.4.1.2. α-Fragment
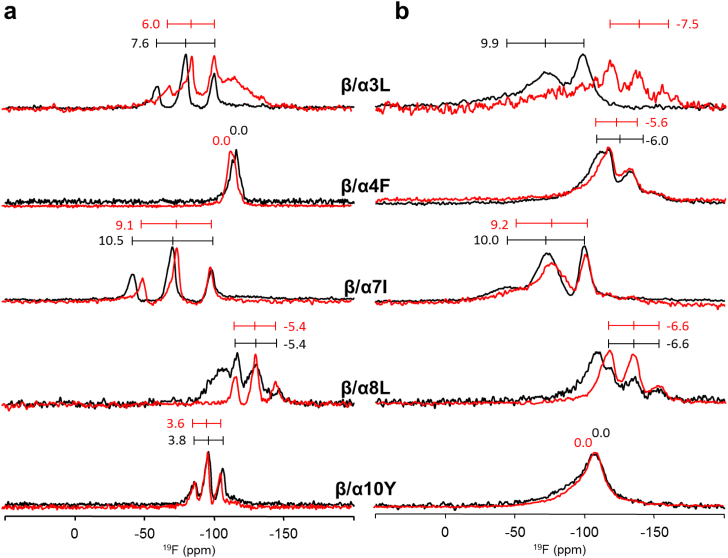
The 19F-NMR spectra of the α-helical part of the β/α-model (labeled with a single Bpg reporter group in different positions along the α-helix) are shown in Fig. 7, and the values of the triplet splittings are summarized in Table 4.
Fig. 7.
Solid-state 19F-NMR spectra of the β/α-model labeled with Bpg in the α-helix. Conjugates were reconstituted in oriented DMPC bilayers DMPC (P/L = 1:100) and measured at 40 °C (a) 15 °C (b). The compounds were in their rigid (red lines) and flexible (black lines) forms.
Table 4.
19F-19F dipolar couplings of the β/α-model labeled in the α-helix, as determined in DMPC bilayers in the fluid (40 °C) and gel (15 °C) states. Values for the non-conjugated BP100 parent peptide labeled at the same positions are also given as a reference for fluid and gel bilayers (taken from [49], [52] and [57], respectively. Results of the 19F solid-state NMR analysis of the alignment of the α-helix fragment of the β/α-model are shown. The same analysis of the non-conjugated BP100 parent peptide (labeled at the same positions) is also given as a reference for fluid and gel bilayers.
| DAET state | Lipid phase | Dipolar coupling (kHz ± 0.5) |
Helix orientation |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β/α3L | β/α4F | β/α7I | β/α8L | β/α10Y | τ [°] | ρ [°] | Smol | rmsd [kHz] | ||
| Flexible | fluid | + 7.6 | 0.0 | + 10.5 | − 5.4 | + 3.8 | 53 | 13 | 0.75 | 0.9 |
| Rigid | fluid | + 6.0 | 0.0 | + 9.1 | − 5.4 | + 3.6 | 48 | 12 | 0.70 | 0.8 |
| Reference | fluid | + 5.1 | + 1.0 | + 8.2 | − 5.2 | + 3.2 | 46 | 10 | 0.66 | 1.0 |
| Flexible | gel | + 9.9 | − 6.0 | + 10.0 | − 6.6 | 0.0 | 89 | 16 | 1.00 | 1.4 |
| Rigid | gel | − 7.5 | − 5.6 | + 9.2 | − 6.6 | 0.0 | n.aa | − | − | − |
| Reference | gel | + 4.0 | − 3.5 | + 8.7 | − 6.5 | 0.0 | 90 | 9 | 0.77 | 0.8 |
Was not analyzed, best-fit for an α-helix has rmsd > 4.1 kHz.
It is known from our earlier 19F-NMR structure analysis of BP100 that this α-helical parent peptide binds to fluid bilayers independently of the membrane composition [58], with a tilt angle (τ) of around 50° [52]. Note that the accuracy of the τ determination is limited, because the hydrophobic Bpg labels cannot be optimally distributed around the helix, but it is already highly informative to simply compare the underlying dipolar splittings.
The 19F-NMR splittings observed in the α-helical fragment of the β/α-model are very similar to those of the stand-alone parent peptide BP100, which had been labeled at the same positions (Table 4). From these splitting values above the Tm we deduced a tilted alignment of the BP100 fragment, with a best-fit solution of τ = 53° and ρ = 13° (Smol = 0.75, rmsd = 0.9 kHz), in line with the earlier data from MD, 2H-NMR, 15N-NMR and 19F-NMR see [52], [58], [59], [60]). In fluid bilayers, this alignment is found to remain unaffected by rigidifying the molecule, as the splittings are essentially the same or only slightly reduced in their absolute values. The appearance of broad featureless signals in the case of β/α3L and β/α8L, which could stem from aggregated peptides, seems to depend on the flexibility/rigidity of the DAET linker, but this interpretation cannot be generalized.
It is known that the membrane alignment of the parent BP100 peptide changes significantly below the lipid phase transition temperature (Table 4, “Reference, gel”). The corresponding NMR data reveal a re-orientation of the helix into a surface-bound state with a best-fit of τ = 90° and ρ = 9° (Smol = 0.77, rmsd = 0.8 kHz). The observed spectral changes for the BP100-derived α-fragment in our β/α-model are largely coherent, with a slight difference for only the N-terminal labels that are most closely adjacent to the DAET linker. Having shown above that the β-strand fragment is not affected by the presence of the DAET-linked α-helical fragment, we can now conclude that both peptide fragments of our β/α-model behave essentially independently from one another in DMPC membranes. They fold and re-orient in the same way as their respective β-stranded and α-helical parent peptides, regardless of the state of the photoswitchable junction. Thus, the rigidity/flexibility of the DAET linker has no significant influence on the structure and alignment of the β/α-model in the lipid bilayers, and this holds true both for fluid and gel phase lipids. This conclusion is in agreement with the CD and DSC measurements presented above.
A slight structural difference is observed between α-helical fragment and the parent B100 peptide only in close vicinity to the photoswitchable linker at position Leu3 (and less so for Phe4) in the lipid gel phase. Considering also the data for the β-fragment (see above), we may thus conclude that the local conformational perturbation of the DAET linker extends over approximately 3–4 adjacent residues.
3.4.2. Rigid and flexible forms of the α/α-model in DMPC bilayers
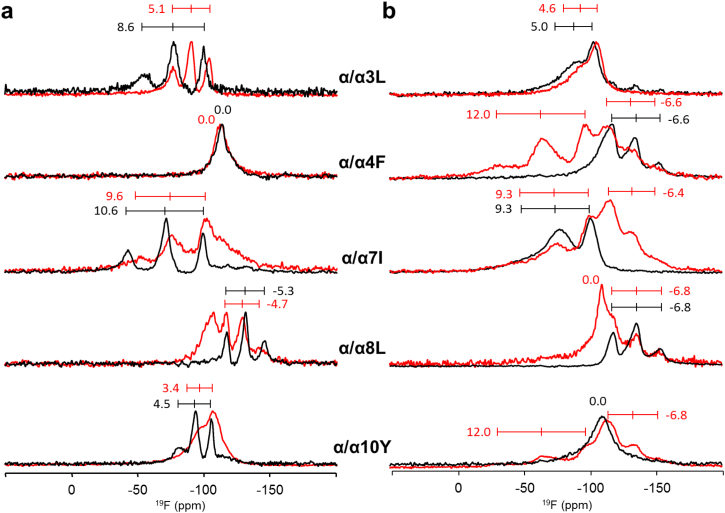
The spectra of the doubly labeled α/α-model with the photoswitching linker in the flexible form in fluid bilayers (Fig. 8a) showed only a single 19F-NMR signal for each analog, suggesting that the two identical and symmetrically attached α-helical fragments are structured and aligned in the same way. This result is in line with our underlying assumption that the conformationally flexible ring-open form of the DAET linker in these constructs allows the two peptidic fragments to behave fully autonomously in the lipid membranes. Also in the gel phase lipids, the ring-open linker was found to permit independent re-alignment of the two BP100-derived helices (Fig. 8b, Table 5). However, this was not the case for the ring-closed form.
Fig. 8.
Solid-state 19F-NMR spectra of the α/α-model labeled with Bpg. The constructs were reconstituted in DMPC and measured in lipids in the (a) fluid (40 °C) and (b) gel phase (15 °C). The compounds are in the rigid (red lines) and flexible (black lines) forms. The intensities of the red lines were scaled up in order to align the unchanged components in the spectra with the spectra of the flexible forms.
Table 5.
19F-19F dipolar couplings of the doubly-labeled α/α-model, as determined in oriented DMPC bilayers in the fluid (40 °C) and gel (15 °C) states. Values for the parent peptide BP100, labeled at the same positions are also included as a reference from [49], [52] and [57]. Results of the 19F solid-state NMR analysis of the alignment of the α-helix fragment of the α/α-model are shown. The same analysis of the non-conjugated BP100 parent peptide (labeled at the same positions) is also given as a reference for fluid and gel bilayers.
| DAET state | Dipolar coupling (kHz ± 0.5) |
Helix orientation |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lipid phase | α/α3L | α/α4F | α/α7I | α/α8L | α/α10Y | τ [°] | ρ [°] | Smol | rmsd [kHz] | |
| Flexible | fluid | + 8.6 | 0.0 | + 10.6 | − 5.3 | + 4.5 | 54 | 15 | 0.76 | 1.1 |
| Rigid | fluid | + 5.1 | 0.0 | + 9.6 | − 4.7 | + 3.4 | 52 | 11 | 0.65 | 0.9 |
| Reference | fluid | + 5.1 | + 1.0 | + 8.2 | − 5.2 | + 3.2 | 46 | 10 | 0.66 | 1.0 |
| Flexible | gel | + 5.0 | − 6.6 | + 9.3 | − 6.8 | 0.0 | 96 | 13 | 1.00 | 1.0 |
| Rigid | gel gela |
+ 4.6 + 5.0 |
− 6.6 + 12 |
+ 9.3 − 6.4 |
− 6.8 0.0 |
− 6.8 + 12.0 |
99 n.ab |
17 n.a |
1.00 n.a |
1.8 n.a |
| Reference | gel | + 4.0 | − 3.5 | + 8.7 | − 6.5 | 0.0 | 90 | 9 | 0.77 | 0.8 |
Two sets of signals were observed, see text and Fig. 8.
Was not analyzed, best-fit for an α-helix has rmsd > 4.1 kHz.
Interestingly, the rigidified DAET linker was found to perturb the helical fragments in the α/α-model to a much greater extent than in the β/α-model. As can be seen in Fig. 8 (red traces), the doubly labeled rigid α/α-model analogs gave complex 19F-NMR spectra, particularly in gel phase bilayers. Closer inspection of the spectra run under these conditions shows that about a half of the peaks in most cases are at positions very close to and with similar splittings as in the spectra of the β/α-model peptides and of the wild-type BP100. Therefore, we can conclude that one of the two BP100-derived units in the α/α-model aligns in essentially the same way as the free parent peptide BP100. However, the second BP100-derived unit in the α/α-model experiences a significant influence from its neighbor via the rigid ring-closed DAET linker, when embedded in the highly viscous environment of the gel state lipids. Under these conditions it has no chance to adopt its preferred orientation in the membrane.
As outlined for the β/α-model above (Table 4), we also formally evaluated the structural parameters τ and ρ for the α/α-model from the measured 19F-NMR dipolar splittings (Table 5). In fluid lipid bilayers, the helical parts of the flexible and rigidified conjugates are aligned very similarly in both models with τ around 50°, though the deviation is somewhat more pronounced for the symmetrical α/α-model than for the β/α-model. In these dynamical fluid bilayers, this situation did not result in two distinct sets of signals (i.e. from the two 19F–labels in each α-helical fragment). On the other hand, the spectra acquired below Tm clearly revealed two sets of signals (Fig. 8b). One set in the flexible form agrees well with the formal alignment of the parent BP100 peptide (τ = 90°, ρ = 9°) and corresponds to the situation seen in the BP100-derived fragment of the β/α-model (τ = 89°, ρ = 16°).
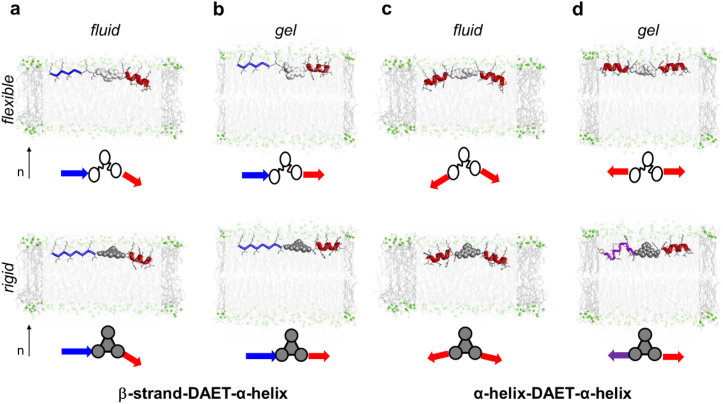
Overall, we can conclude that the flexible ring-open DAET linker allows both α-helical units to align almost parallel to the bilayer surface, similar to the free parent peptide BP100 and to the α-helical fragment of the β/α-conjugate. The rigidified junction, however, prevents the two fragments from taking on their favorable alignment at the same time; one set of signals remains intact (Fig. 8b) and can be attributed to an α-helix aligned “normally” (i.e., with the same orientation and structure to that observed in the parent BP100 and in the helical part of the β/α-model). The second set of signals, which in few cases clearly corresponds to about a half of the spectral intensity, represents an entirely different membrane-bound structure. It was not possible to deduce any well-defined conformation or alignment from this second set of signals, suggesting a partial unfolding in the gel phase lipids. Fig. 9 summarizes all observations.
Fig. 9.
Schematic representation of the structure and alignment of the model conjugates with a conformationally flexible (top) and rigidified (bottom) DAET linker. β/α-Model in lipid bilayers in the fluid (a) and gel phase (b). α/α-Model in lipid bilayers in the fluid (c) and gel phase (d). Conjugates are shown with space-filling for the DAET linkers (flexible – white, rigid - gray); peptide fragments are without hydrogens as stick-models with the secondary structures of the backbones highlighted as blue β-strands and red α-helices. Each observed situation is illustrated by a molecular model and a corresponding schematic summary; flexible DAET – white; rigid DAET – gray. All molecules are drawn to scale, but the depth of insertion is not known. The distortion of one α-helix in the symmetric model (d, bottom) is indicated by the purple color.
4. Conclusions
The rigidified ring-closed form of the photoswitchable diarylethene linker and its flexible ring-open form do not differ in atomic composition, but they can influence the conformation of the adjacent residues when incorporated into the backbone of a linear peptide. This perturbation reaches up to 3–4 adjacent residues. This small range suggests that diarylethene building blocks are useful tools for studying the influence of peptide rigidity/flexibility in lipid membranes. Placing the photoswitchable linker into model peptide conjugates did not impair the autonomous alignment of the two linked fragments, when it was flexible in the ring-open state. The rigidified forms of our model peptides, on the other hand, could adopt different structures and orientations in lipid membranes, but these were only significant for the α/α-model in the viscous lipid gel phase.
Transparency document
Transparency document.
Acknowledgments
Acknowledgements
This project has received funding from the European Union's Horizon 2020 research and innovation programme (H2020-MSCA-RISE-Action) under grant agreement No 690973. This work was also partially supported by GRK 2039 from the German Research Society (DFG) and by the Helmholtz Association program “BIFTM”. We thank Dr. Wadhwani (KIT, Karlsruhe) for access to the peptide synthesis facility, and Dr. Mykhailiuk (Enamine, Kyiv) for discussing the study design; I.V.K. acknowledges the Alexander von Humboldt Foundation (Germany) for financial support as a recipient of the Georg Forster Research Prize in 2015.
Footnotes
The Transparency document associated with this article can be found, in the online version.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbamem.2017.09.021.
Contributor Information
Igor V. Komarov, Email: ik214@mail.univ.kiev.ua.
Anne S. Ulrich, Email: Anne.Ulrich@kit.edu.
Appendix A. Supplementary data
Supplementary material
References
- 1.Okamoto A., Ishii S., Hirotsu K., Kagamiyama H. The active site of Paracoccus denitrificans aromatic amino acid aminotransferase has contrary properties: flexibility and rigidity. Biochemistry. 1999;38:1176–1184. doi: 10.1021/bi981921d. [DOI] [PubMed] [Google Scholar]
- 2.Collins T., Meuwis M.-A., Gerday C., Feller G. Activity, stability and flexibility in glycosidases adapted to extreme thermal environments. J. Mol. Biol. 2003;328:419–428. doi: 10.1016/s0022-2836(03)00287-0. [DOI] [PubMed] [Google Scholar]
- 3.Liang X., Arunima A., Zhao Y., Bhaskaran R., Shende A., Byrne T.S., Fleeks J., Palmier M.O., Van Doren S.R. Apparent tradeoff of higher activity in MMP-12 for enhanced stability and flexibility in MMP-3. Biophys. J. 2010;99:273–283. doi: 10.1016/j.bpj.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinha N., Smith-Gill S.J. Protein structure to function via dynamics. Protein Pept. Lett. 2002;9:367–377. doi: 10.2174/0929866023408508. [DOI] [PubMed] [Google Scholar]
- 5.Bhalla J., Storchan G.B., MacCarthy C.M., Uversky V.N., Tcherkasskaya O. Local flexibility in molecular function paradigm. Mol. Cell. Proteomics. 2006;5:1212–1223. doi: 10.1074/mcp.M500315-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.N. Sinha, S.J. Smith-Gill, Molecular dynamics simulation of a high-affinity antibody–protein complex, Cell Biochem. Biophys. 43 (2005) 253–273. [DOI] [PubMed]
- 7.Smith W.W., Bartlett P.A. Macrocyclic inhibitors of penicillopepsin. 3. Design, synthesis, and evaluation of an inhibitor bridged between P2 and P1. J. Am. Chem. Soc. 1998;120:4622–4628. [Google Scholar]
- 8.Khan A.R., Parrish J.C., Fraser M.E., Smith W.W., Bartlett P.A., James M.N.G. Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes. Biochemistry. 1998;37:16839–16845. doi: 10.1021/bi9821364. [DOI] [PubMed] [Google Scholar]
- 9.Cram D.J. Preorganization – from solvents to spherands. Angew. Chem. Int. Ed. 1986;25:1039–1057. [Google Scholar]
- 10.Houk K.N., Leach A.G., Kim S.P., Zhang X. Binding affinities of host–guest, protein–ligand, and protein–transition-state complexes. Angew. Chem. Int. Ed. 2003;42:4872–4897. doi: 10.1002/anie.200200565. [DOI] [PubMed] [Google Scholar]
- 11.DeLorbe J.E., Clements J.H., Teresk M.G., Benfield A.P., Plake H.R., Millspaugh L.E., Martin S.F. Thermodynamic and structural effects of conformational constraints in protein-ligand interactions. Entropic paradoxy associated with ligand preorganization. J. Am. Chem. Soc. 2009;131:16758–16770. doi: 10.1021/ja904698q. [DOI] [PubMed] [Google Scholar]
- 12.Davidson J.P., Lubman O., Rose T., Waksman G., Martin S.F. Calorimetric and structural studies of 1,2,3-trisubstituted cyclopropanes as conformationally constrained peptide inhibitors of Src SH2 domain binding. J. Am. Chem. Soc. 2002;124:205–215. doi: 10.1021/ja011746f. [DOI] [PubMed] [Google Scholar]
- 13.Edwards A.A., Mason J.M., Clinch K., Tyler P.C., Evans G.B., Schramm V.L. Altered enthalpy-entropy compensation in picomolar transition state analogues of human purine nucleoside phosphorylase. Biochemistry. 2009;48:5226–5238. doi: 10.1021/bi9005896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin S.F. Preorganization in biological systems: are conformational constraints worth the energy? Pure Appl. Chem. 2007;79:193–200. [Google Scholar]
- 15.Wang Y., Kirschner A., Fabian A.-K., Gopalakrishnan R., Kress C., Hoogeland B., Koch U., Kozany C., Bracher A., Hausch F. Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control. J. Med. Chem. 2013;56:3922–3935. doi: 10.1021/jm400087k. [DOI] [PubMed] [Google Scholar]
- 16.DeLorbe J.E., Clements J.H., Whiddon B.B., Martin S.F. Thermodynamic and structural effects of macrocyclic constraints in protein-ligand interactions. ACS Med. Chem. Lett. 2010;1:448–452. doi: 10.1021/ml100142y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Udugamasooriya D.G., Spaller M.R. Conformational constraint in protein ligand design and the inconsistency of binding entropy. Biopolymers. 2008;89:653–667. doi: 10.1002/bip.20983. [DOI] [PubMed] [Google Scholar]
- 18.Martin S.F., Clements J.H. Correlating structure and energetics in protein-ligand interactions: paradigms and paradoxes. Annu. Rev. Biochem. 2013;82:267–293. doi: 10.1146/annurev-biochem-060410-105819. [DOI] [PubMed] [Google Scholar]
- 19.Edwards A.A., Tipton J.D., Brenowitz M.D., Emmett M.R., Marshall A.G., Evans G.B., Tyler P.C., Schramm V.L. Conformational states of human purine nucleoside phosphorylase at rest, at work, and with transition state analogues. Biochemistry. 2010;49:2058–2067. doi: 10.1021/bi902041j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Mol N.J., Catalina M.I., Dekker F.J., Fischer M.J.E., Heck A.J.R., Liskamp R.M.J. Protein flexibility and ligand rigidity: a thermodynamic and kinetic study of ITAM-based ligand binding to Syk tandem SH2. Chembiochem. 2005;6:2261–2270. doi: 10.1002/cbic.200500141. [DOI] [PubMed] [Google Scholar]
- 21.Benfield A.P., Teresk M.G., Plake H.R., DeLorbe J.E., Millspaugh L.E., Martin S.F. Ligand preorganization may be accompanied by entropic penalties in protein–ligand interactions. Angew. Chem. Int. Ed. 2006;45:6830–6835. doi: 10.1002/anie.200600844. [DOI] [PubMed] [Google Scholar]
- 22.Frederick K.K., Marlow M.S., Valentine K.G., Wand A.J. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325–330. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wieprecht T., Seelig J. Isothermal titration calorimetry for studying interactions between peptides and lipid membranes. Curr. Top. Membr. 2002;52:31–56. [Google Scholar]
- 24.Fang Z., Song Y., Zhan P., Zhang Q., Liu X. Conformational restriction: an effective tactic in ‘follow-on’-based drug discovery. Future Med. Chem. 2014;6:885–901. doi: 10.4155/fmc.14.50. [DOI] [PubMed] [Google Scholar]
- 25.Wamberg M., Pedersen E.B., Nielsen C. Synthesis of furoannelated analogues of emivirine (MKC-442) Arch. Pharm. Pharm. Med. Chem. 2004;337:148–151. doi: 10.1002/ardp.200300815. [DOI] [PubMed] [Google Scholar]
- 26.Busnel O., Carreaux F., Carboni B., Pethe S., Vadon-Le Goff S., Mansuy D., Boucher J.-L. Synthesis and evaluation of new ω-borono-α-amino acids as rat liver arginase inhibitors. Bioorg. Med. Chem. 2005;13:2373–2379. doi: 10.1016/j.bmc.2005.01.053. [DOI] [PubMed] [Google Scholar]
- 27.Hazeldine S.T., Polin L., Kushner J., White K., Corbett T.H., Horwitz J.P. Synthesis and biological evaluation of conformationally constrained analogs of the antitumor agents XK469 and SH80. Part 5. Bioorg. Med. Chem. 2006;14:2462–2467. doi: 10.1016/j.bmc.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 28.Park J.H., Lee S.U., Kim S.H., Shin S.Y., Lee J.Y., Shin C.-G., Yoo K.H., Lee Y.S. Chromone and chromanone derivatives as strand transfer inhibitors of HIV-1 integrase. Arch. Pharm. Res. 2008;31:1–5. doi: 10.1007/s12272-008-1111-z. [DOI] [PubMed] [Google Scholar]
- 29.Han X., Bushweller J.H., Cafiso D.S., Tamm L.K. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 30.Bertocco A., Formaggio F., Toniolo C., Broxterman Q.B., Epand R.F., Epand R.M. Design and function of a conformationally restricted analog of the influenza virus fusion peptide. J. Pept. Res. 2003;62:19–26. doi: 10.1034/j.1399-3011.2003.00063.x. [DOI] [PubMed] [Google Scholar]
- 31.Kim J.-K., Lee S.-A., Shin S., Lee J.-Y., Jeong K.-W., Nan Y.H., Park Y.S., Shin S.Y., Kim Y. Structural flexibility and the positive charges are the key factors in bacterial cell selectivity and membrane penetration of peptoid-substituted analog of Piscidin 1. Biochim. Biophys. Acta. 2010;1798:1913–1925. doi: 10.1016/j.bbamem.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 32.Liu L., Fang Y., Huang Q., Wu J. A rigidity-enhanced antimicrobial activity: a case for linear cationic α-helical peptide HP(2–20) and its four analogues. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radchenko D.S., Michurin O.M., Grygorenko O.O., Scheinpflug K., Dathe M., Komarov I.V. Confining the χ space of basic natural amino acids: cyclobutane-derived χ1,χ2-constrained analogues of arginine, lysine and ornithine. Tetrahedron. 2013;69:505–511. [Google Scholar]
- 34.Liu L., Fang Y., Wu J. Flexibility is a mechanical determinant of antimicrobial activity for amphipathic cationic α-helical antimicrobial peptides. Biochim. Biophys. Acta. 2013;1828:2479–2486. doi: 10.1016/j.bbamem.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 35.Kessel A., Ben-Tal N., May S. Interactions of cholesterol with lipid bilayers: the preferred configuration and fluctuations. Biophys. J. 2001;81:643–658. doi: 10.1016/S0006-3495(01)75729-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seelig J. Thermodynamics of lipid–peptide interactions. Biochim. Biophys. Acta. 2004;1666:40–50. doi: 10.1016/j.bbamem.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 37.May S. Membrane perturbations induced by integral proteins: role of conformational restrictions of the lipid chains. Langmuir. 2002;18:6356–6364. [Google Scholar]
- 38.Yano Y., Matsuzaki K. Membrane insertion and dissociation processes of a model transmembrane helix. Biochemistry. 2002;41:12407–12413. doi: 10.1021/bi026191v. [DOI] [PubMed] [Google Scholar]
- 39.Babii O., Afonin S., Berditsch M., Reiβer S., Mykhailiuk P.K., Kubyshkin V.S., Steinbrecher T., Ulrich A.S., Komarov I.V. Controlling biological activity with light: diarylethene-containing cyclic peptidomimetics. Angew. Chem. Int. Ed. 2014;53:3392–3395. doi: 10.1002/anie.201310019. [DOI] [PubMed] [Google Scholar]
- 40.Ulrich A.S., Wadhwani P., Dürr U.H.N., Afonin S., Glaser R.W., Strandberg E., Tremouilhac P., Sachse C., Berditchevskaia M., Grage S.L. Solid-state 19F-Nuclear magnetic resonance analysis of membrane-active peptides. In: Ramamoorthy A., editor. NMR Spectroscopy of Biological Solids. CRC Press; 2006. pp. 215–236. [Google Scholar]
- 41.Ulrich A.S. Solid state 19F NMR methods for studying biomembranes. Prog. Nucl. Magn. Reson. Spectrosc. 2005;46:1–21. [Google Scholar]
- 42.Rance M., Byrd R.A. Obtaining high-fidelity spin-1/2 powder spectra in anisotropic media - phase-cycled Hahn echo spectroscopy. J. Magn. Reson. 1983;52:221–240. [Google Scholar]
- 43.Fung B.M., Khitrin A.K., Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 44.Zhang S., Wu X.L., Mehring M. Elimination of ringing effects in multiple-pulse sequences. Chem. Phys. Lett. 1990;173:481–484. [Google Scholar]
- 45.Bennett A.E., Rienstra C.M., Auger M., Lakshmi K.V., Griffin R.G. Heteronuclear decoupling in rotating solids. J. Chem. Phys. 1995;103:6951–6958. [Google Scholar]
- 46.Mykhailiuk P.K., Afonin S., Chernega A.N., Rusanov E.B., Platonov M., Dubinina G., Berditsch M., Ulrich A.S., Komarov I.V. Conformationally rigid trifluoromethyl-substituted α-amino acid designed for peptide structure analysis by solid state 19F-NMR spectroscopy. Angew. Chem. Int. Ed. 2006;45:5659–5661. doi: 10.1002/anie.200600346. [DOI] [PubMed] [Google Scholar]
- 47.Radchenko D.S., Kattge S., Kara S., Ulrich A.S., Afonin S. Does a methionine-to-norleucine substitution in PGLa influence peptide-membrane interactions? Biochim. Biophys. Acta. 2016;1858:2019–2027. doi: 10.1016/j.bbamem.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 48.Glaser R.W., Sachse C., Dürr U.H.N., Wadhwani P., Ulrich A.S. Orientation of the antimicrobial peptide PGLa in lipid membranes determined from 19F-NMR dipolar couplings of 4-CF3-phenylglycine labels. J. Magn. Reson. 2004;168:153–163. doi: 10.1016/j.jmr.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 49.Blazyk J., Wiegand R., Klein J., Hammer J., Epand R.M., Epand R.F., Maloy W.L., Kari U.P. A novel linear amphipathic β-sheet cationic antimicrobial peptide with enhanced selectivity for bacterial lipids. J. Biol. Chem. 2001;276:27899–27906. doi: 10.1074/jbc.M102865200. [DOI] [PubMed] [Google Scholar]
- 50.Badosa E., Ferre R., Planas M., Feliu L., Besalú E., Cabrefiga J., Bardají E., Montesinos E. A library of linear undecapeptides with bactericidal activity against phytopathogenic bacteria. Peptides. 2007;28:2276–2285. doi: 10.1016/j.peptides.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 51.Wadhwani P., Reichert J., Strandberg E., Bürck J., Misiewicz J., Afonin S., Heidenreich N., Fanghänel S., Mykhailiuk P.K., Komarov I.V., Ulrich A.S. Stereochemical effects on the aggregation and biological properties of the fibril-forming peptide [KIGAKI]3 in membranes. Phys. Chem. Chem. Phys. 2013;15:8962–8971. doi: 10.1039/c3cp50896j. [DOI] [PubMed] [Google Scholar]
- 52.Wadhwani P., Strandberg E., van den Berg J., Mink C., Bürck J., Ciriello R.A.M., Ulrich A.S. Dynamical structure of the short multifunctional peptide BP100 in membranes. Biochim. Biophys. Acta. 2014;1838:940–949. doi: 10.1016/j.bbamem.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y.P., Lewis R.N., Hodges R.S., McElhaney R.N. Interaction of a peptide model of a hydrophobic transmembrane alpha-helical segment of a membrane protein with phosphatidylcholine bilayers: differential scanning calorimetric and FTIR spectroscopic studies. Biochemistry. 1992;31:11579–11588. doi: 10.1021/bi00161a042. [DOI] [PubMed] [Google Scholar]
- 54.Marsh D. CRC Press; Boca Raton, Florida, USA: 1990. CRC Handbook of Lipid Bilayers. [Google Scholar]
- 55.Eggenberger K., Mink C., Wadhwani P., Ulrich A.S., Nick P. Using the peptide Bp100 as a cell-penetrating tool for the chemical engineering of actin filaments within living plant cells. Chembiochem. 2011;12:132–137. doi: 10.1002/cbic.201000402. [DOI] [PubMed] [Google Scholar]
- 56.Wadhwani P., Strandberg E., Heidenreich N., Bürck J., Fanghänel S., Ulrich A.S. Self-assembly of flexible β-strands into immobile amyloid-like β-sheets in membranes as revealed by solid-state 19F NMR. J. Am. Chem. Soc. 2012;134:6512–6515. doi: 10.1021/ja301328f. [DOI] [PubMed] [Google Scholar]
- 57.Wadhwani et al., unpublished results.
- 58.Misiewicz J., Afonin S., Grage S.L., van den Berg J., Strandberg E., Wadhwani P., Ulrich A.S. Action of the multifunctional peptide BP100 on native biomembranes examined by solid-state NMR. J. Biomol. NMR. 2015;61:287–298. doi: 10.1007/s10858-015-9897-8. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y., Zhao T., Wie D., Strandberg E., Ulrich A.S., Ulmschneider J.P. How reliable are molecular dynamics simulations of membrane active antimicrobial peptides? Biochim. Biophys. Acta. 2014;1838:2280–2288. doi: 10.1016/j.bbamem.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 60.Zamora-Carreras H., Strandberg E., Mühlhäuser P., Bürck J., Wadhwani P., Jiménez M.Á., Bruix M., Ulrich A.S. Alanine scan and 2H NMR analysis of the membrane-active peptide BP100 point to a distinct carpet mechanism of action. Biochim. Biophys. Acta. 2016;1858:1328–1338. doi: 10.1016/j.bbamem.2016.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.
Supplementary material