Abstract
We describe a novel B cell–associated cytokine, encoded by an uncharacterized gene (C17orf99; chromosome 17 open reading frame 99), that is expressed in bone marrow and fetal liver and whose expression is also induced in peripheral B cells upon activation. C17orf99 is only present in mammalian genomes, and it encodes a small (~27-kDa) secreted protein unrelated to other cytokine families, suggesting a function in mammalian immune responses. Accordingly, C17orf99 expression is induced in the mammary gland upon the onset of lactation, and a C17orf99−/− mouse exhibits reduced levels of IgA in the serum, gut, feces, and lactating mammary gland. C17orf99−/− mice have smaller and fewer Peyer’s patches and lower numbers of IgA-secreting cells. The microbiome of C17orf99−/− mice exhibits altered composition, likely a consequence of the reduced levels of IgA in the gut. Although naive B cells can express C17orf99 upon activation, their production increases following culture with various cytokines, including IL-4 and TGF-β1, suggesting that differentiation can result in the expansion of C17orf99-producing B cells during some immune responses. Taken together, these observations indicate that C17orf99 encodes a novel B cell–associated cytokine, which we have called IL-40, that plays an important role in humoral immune responses and may also play a role in B cell development. Importantly, IL-40 is also expressed by human activated B cells and by several human B cell lymphomas. The latter observations suggest that it may play a role in the pathogenesis of certain human diseases.
Cytokines are small secreted proteins that play an essential role in host defense, inflammation, the development of the immune system, and in immune responses. Cytokines exert their effects by binding specific receptors on the membrane of target cells. The elucidation of cytokine receptor/ligand pairs has furthered our understanding of the mechanisms through which cytokines regulate the development of immune responses (1). Cytokine genes likely arose through gene duplication from ancient precursors (2) and, therefore, exhibit common structural features in their sequences that reflect their common evolutionary origins. This characteristic has facilitated the identification of all of the members that belong to a particular cytokine superfamily (3). It follows that, if any cytokines remain to be discovered, they are not likely to be members of any known cytokine family.
We sought to identify novel immune system–associated genes encoding secreted or transmembrane proteins. To this end, we analyzed a comprehensive database of human gene expression (Body Index of Gene Expression [BIGE]) that includes 105 human tissues or cells (4, 5). These analyses led to the identification of 35 poorly characterized genes predicted to encode transmembrane or secreted proteins expressed by leukocytes or immune system–associated organs. We have reported three of these novel genes, including Isthmin 1, Tetraspanin 33 (TSPAN33), and Meteorin-like (6–8). In this article, we report an uncharacterized gene (C17orf99; chromosome 17 open reading frame 99) that encodes a small secreted protein (~27 kDa) produced by B cells upon activation. These characteristics strongly suggest that C17orf99 encodes a novel B cell–expressed cytokine. Therefore, we predicted that C17orf99 would have effects in the immune system. To test this hypothesis, we obtained and analyzed a mouse with a targeted deletion of C17orf99. Phenotypic analysis of this mouse indicates that it exhibits altered B cell numbers; low levels of IgA in the serum, mammary gland, and feces; and an altered microbiome. We conclude that C17orf99 encodes a novel B cell–derived cytokine. Recently, a new member of the IL-12 family has been identified and named IL-39 (9, 10). Therefore, we have named the novel cytokine encoded by C17orf99 IL-40 (11); in this report, we demonstrate that IL-40 is a novel cytokine involved in the regulation of humoral immunity.
Materials and Methods
Cells
The human B cell line 2E2 (derived from Burkitt lymphoma) has been described (12). The human T cell line Jurkat was obtained from the American Type Culture Collection (Manassas, VA). The human B cell lymphoma cell lines have been described previously (13) and were a generous gift from Dr. D. Fruman (University of California, Irvine). The murine cell line A20-2J has been described (14) and was a kind gift of Dr. P. Marrack (National Jewish Health, Denver, CO). Human peripheral blood B cells were purified by flow cytometry (>95%).
Quantitative PCR
Human cDNAs were obtained from Clontech (Mountain View, CA), and PBMCs were from Sanguine BioSciences (Sherman Oaks, CA). RNA was isolated from human cell lines/cells or tissues using the QIAGEN RNeasy Kit, according to the manufacturer’s instructions (QIAGEN, Valencia, CA). cDNA reactions were performed using QuantiTect Reverse Transcription (QIAGEN). Quantitative PCR (qPCR) was performed using the Roche LightCycler 480 Real-Time PCR system with probes designed to detect CD19 (B cell marker), C17orf99, and GAPDH (housekeeping gene) (Roche, Pleasanton, CA) (Supplemental Table I). The mouse gene homolog of human C17orf99 is 6030468B19Rik.
Mice
All animal protocols were approved by the Institutional Animal Care and Use Committee of the University of California, Irvine. MRL/MpJ-Faslpr/J mice (stock number 000485) and C57BL/6J mice (stock number 000664) were obtained from The Jackson Laboratory (Bar Harbor, ME). Il40−/− mice (B6/129S5-6030468B19Rik−/−) were obtained from the KOMP Repository (University of California, Davis, Davis, CA). B6/129S5-6030468B19Rik−/− mice obtained were backcrossed to C57BL/6J mice at least five generations. Wild-type (WT) mice used as controls were littermates from het/het pairings that also yielded homozygous Il40−/− mice. Serum samples were obtained through submandibular cheek bleed, as described (15). Fecal samples were collected, as described (16). Breast milk samples were obtained by collecting the stomach contents of neonate mice. Briefly, stomach contents were centrifuged at maximum speed for 15 min, and supernatants were collected.
ELISA
Serum Ig ELISAs were performed according to the manufacturer’s protocols (eBioscience, San Diego, CA). Plates were read at 450 nm. For fecal IgA ELISA, plates (Sigma-Aldrich, St. Louis, MO) were coated with anti-IgA mAb (clone RMA-1; BioLegend, San Diego, CA) at 2 μg/ml. Five-percentage milk in PBS-Tween (PBS-T) was used as blocking buffer. Plates were washed three times with PBS-T after each step. Alkaline phosphatase– conjugated anti-mouse IgA (SouthernBiotech, Birmingham, AL) was used as secondary Ab for 1 h at room temperature and washed three times with PBS-T. SureBlue substrate (KPL, Gaithersburg, MD) was added, and the reaction was stopped with 2 N H2SO4. Plates were read at 630 nm.
Western blot
To verify that IL-40 is secreted, human IL40 cDNA was cloned from human 2E2 B cells, a Burkitt lymphoma model of B cell activation and differentiation (12), and inserted into pTT5 vector (13), resulting in a recombinant gene encoding a fusion protein with a C-terminal 8× histidine (His) tag. HEK293 cells were transiently transfected with the pTT5-IL40 construct (or empty vector used as a control), and day-1 and day-3 supernatants were collected, concentrated in an anti-His column (GenScript), and analyzed for the presence of rIL-40 protein by Western blot (using anti-His Ab) (Bio-Rad).
Flow cytometry
Peyer’s patches (PPs) were isolated from the small intestines of WT or Il40−/− mice. PP B cells were stained with PE-labeled anti-mouse CD45R (B220), rat mAb (clone RA3-6B2; eBioscience), 7-aminoactinomycin D (BioLegend), and allophycocyanin-labeled IgM (BioLegend) and analyzed by flow cytometry using a BD FACSCalibur (BD Biosciences, San Jose, CA) and FlowJo data analysis software (TreeStar, Ashland, OR).
Bone marrow (BM) cells were isolated from femurs of C57BL/6J or Il40−/− mice by flushing them out with a 25G needle using FACS buffer (PBS 5% heat-inactivated FBS). The cells were stained with the following monoclonal conjugates: PE anti-mouse CD45 conjugate (clone 30-F11), FITC anti-mouse CD11b (clone M1/70), BV421 anti-mouse B220 (clone RA3-6B2), anti-mouse CD51 (clone RMV-7; all from BioLegend), and Fixable Viability Dye eFluor 760 (eBioscience) in FACS buffer for 20 min and sorted using a FACSAria Fusion sorter (BD Biosciences). The data were analyzed with FlowJo v10.0.8 software (TreeStar).
BM B cell subsets
Eight-week-old mice were used for FACS analyses of the BM. Single BM cell suspensions were stained using anti-mouse Abs against B220 (clone RA3-6B2; eBioscience), CD19 (clone 1D3; eBioscience), CD43 (clone S7; BD Biosciences, Franklin Lakes, NJ), IgM (clone RMM-1; BioLegend), IgD (clone IA6-2; BioLegend), c-Kit (clone 2B8; eBioscience), Flk2 (clone A2F10; eBioscience) and Ter119 (clone Ter119; eBioscience). The gating was done as described (17, 18).
Splenic B cell subsets analyses
Spleens from 8-wk-old mice were used for analysis. Single-splenocyte suspensions were stained with anti-mouse Abs anti-CD19 (clone HIB19), anti-CD93 (clone AA4.1), anti-IgD (clone IA6-2), anti-IgM (clone RMM-1), anti-CD21/35 (clone CR2/CR1), and anti-CD23 (clone B3B4) (all from Bio-Legend). The gating strategy was done as described by Allman and Pillai (19).
Class switch recombination assay
This assay was performed as described (20, 21). Briefly, B cells were purified from splenocyte suspensions using a MojoSort Pan B Cell Isolation Kit (BioLegend), following the manufacturer’s protocol. B cells were cultured at 50 × 103 per milliliter in RPMI 1640 supplemented with 10% and 50 μM 2-ME and stimulated with LPS (10 μg/ml) for class switch recombination (CSR) to IgG3. Additionally, B cells were stimulated with LPS (10 μg/ml) and with the following cytokines and reagents: IL-4 (5 ng/ml) for CSR to IgG1; TGF-β (5 ng/ml) for CSR to IgG2b; and IL-4 (5 ng/ml), IL-5 (5 ng/ml), anti-IgD/dextran (10 ng/ml), and TGF-β for CSR to IgA. Cells were harvested by centrifugation at 500 × g, stained with Abs to B220, IgM, IgG1, IgG2b, IgG3, IgA, and CD138, and analyzed by flow cytometry, as described above.
Statistical analyses
We used the unpaired Student t test for all experiments. Differences with p < 0.05 were considered statistically significant and are labeled as follows: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Error bars represent mean ± SEM. Some experiments were also analyzed using ANOVA or an F test (variance).
Microbiome analyses
DNA extracted from fecal samples was amplified by PCR of 16S rDNA (V4 region) with primers 515F and 806R modified by addition of barcodes for multiplexing and then sequenced on an Illumina MiSeq system (University of California, Davis, Host Microbe Systems Biology Core Facility). Sequences were processed and analyzed using QIIME (22) pipeline v1.9.1 with default settings, except as noted. In brief, paired-end sequences were joined, quality filtered, and chimera filtered (usearch61 option, RDP gold database); operational taxonomic units (OTUs) were picked de novo (“pick_otus” options: enable_rev_strand_match True, otu_picking_method usearch61) at 97% similarity, using the SILVA rRNA gene database v123 (23, 24) (align_seqs:template_fp core_alignment_SILVA123.fasta; “filter_alignment” options: allowed_gap_frac 0.80, entropy_threshold 0.10, suppress_lane_mask_filter True); and taxonomy was assigned with the RDP classifier (“assign_taxonomy” options: assignment_method rdp, confidence 0.8, rdp_max_memory 24, 000, reference_seqs_fp 97_otus_16S.fasta, id_to_taxonomy_fp consensus_taxonomy_7_levels.txt). Samples were rarefied to 10,000 reads and then α (Shannon index) and β (unweighted and weighted UniFrac) diversity was assessed via QIIME. Prism 7 software (GraphPad) was used for statistical analyses (Mann–Whitney U test).
Results
C17orf99 encodes a novel cytokine
Analysis of the BIGE database led to the identification of several poorly characterized genes encoding transmembrane or secreted proteins and expressed by cells or organs of the immune system. Further bioinformatics analyses led to the identification of an uncharacterized gene annotated as C17orf99 that encodes a small secreted protein (~27 KDa). Other than its identification as one of 472 genes reported in a large study aimed at the identification of poorly characterized or unknown human genes encoding trans-membrane or secreted proteins (25), as well as a previous gene survey predicting that C17orf99 encodes a small secreted protein (26), there is no information on C17orf99 in the literature. The BIGE database, which consists of microarray data, indicates that C17orf99 is expressed in fetal liver and BM, as well as activated B cells (Fig. 1A). C17orf99 encodes a small secreted protein of 265 aa (including a 20-aa signal peptide) predicting a mature protein ~ 27 kDa (Fig. 1B). We confirmed these expression data in human tissues by end-point PCR (Fig. 1C) and in mouse and human tissues by qPCR (Supplemental Fig. 1). Because C17orf99 is an unannotated gene of unknown function, we performed further bioinformatics analyses and Basic Local Alignment Search Tool searches, specifically looking for shared synteny among different genomes. These analyses revealed that C17orf99 is only present in mammalian genomes (Fig. 1D). To confirm that it encodes a secreted protein, we transfected the predicted human C17orf99 open reading frame with a His tag into HEK293 cells. Supernatants were concentrated in an anti-His column and then analyzed by Western blot for the presence of recombinant C17orf99 protein using an anti-His Ab. A band between 23 and 30 kDa (consistent with an ~27-kDa protein) was detected in cell cultures transfected with the pTT5V5H8-C17orf99 construct (Fig. 1E), confirming that C17orf99 encodes a secreted protein.
FIGURE 1.
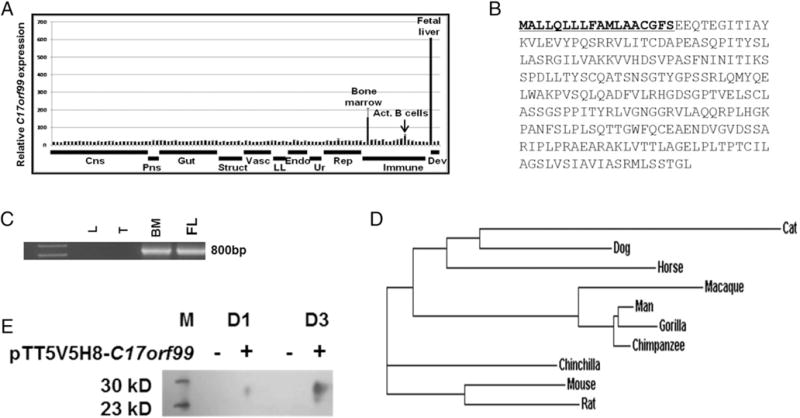
C17orf99 encodes a novel cytokine (IL-40). (A) C17orf99 expression in normal human tissues and immune cells from the BIGE (4, 5) database. IL-40 is expressed in fetal liver, BM, and peripheral blood B cells activated with anti-CD40 and IL-4. (B) C17orf99 amino acid sequence showing the predicted signal peptide (underlined). (C) qPCR of C17orf99 showing a positive band in human fetal liver (FL) and BM but not in thymus (T) or lung (L) (which were used as controls); one experiment out of three independent experiments shown. (D) Clustal Omega analyses of the C17orf99 amino acid sequence in 10 selected mammalian species. C17orf99 homologs exhibit a minimum sequence homology with human C17orf99 (at the amino acid level) of 55% and a maximum E value of 3 × e−70. (E) Western blot of supernatant from day 1 (D1) or day 3 (D3) of cultures of HEK293 cells transfected with IL-40pTT5V5H8-C17orf99-His vector. Supernatants were concentrated in an anti-His column before being run in Western blots with anti-His Ab. Representative of two independent experiments. M, m.w. ladder.
C17orf99 encodes a secreted protein produced by activated B cells
The expression of C17orf99 in BM and fetal liver suggested a hematopoietic function. We initially hypothesized that it would be expressed by stromal cells, because these cells express many hematopoietic cytokines (27). We analyzed various freshly harvested mouse BM cell populations for expression of the mouse homolog of human C17orf99 (6030468B19Rik) and observed that its expression decreased rapidly following culture in vitro. By 48 h following culture, the expression of C17orf99 was undetectable (data not shown). We then FACS-sorted various BM cell populations and performed qPCR for C17orf99. We found that the main source of C17orf99 expression in the BM is CD45− cells (representing mostly stromal cells), whereas very low expression was detected in the CD45+B220+ compartment, which is primarily made up of precursor and mature B cells, and no expression in CD45+CD11b+B220− or CD45+CD11b−B220− cells, which represent myeloid, T, and dendritic cells (data not shown).
The BIGE database indicates that, in addition to BM and fetal liver, activated human peripheral blood B cells express C17orf99 (with anti-CD40 and IL-4) (Fig. 1A). Therefore, we studied the expression of C17orf99 by B cells. As shown in Fig. 2A, activation of the mouse B cell lymphoma A20-2J (14) with CD40L and IL-4 induced strong expression of C17orf99. Spleen B cells from WT C57BL/6 mice activated with anti-CD40 and IL-4 also express C17orf99 (Fig. 2B). We next tested several activating stimuli on these cells, including various cytokines, and observed that B cells require several stimuli for optimal expression of C17orf99. These include a BCR stimulus (anti-IgM), anti-CD40, and several cytokines. Interestingly, TGF-β1 strongly potentiates the expression of C17orf99 by activated B cells, and the strongest expression was observed when B cells were activated with anti-CD40+anti-IgM+IL-4+TGF-β1 (Fig. 2B). Taken together, the characteristics of C17orf99 (encoding a small (~27-kDa) secreted protein, expressed upon activation by B cells) strongly suggest that C17orf99 encodes a novel B cell–derived cytokine that we have called IL-40 (11), a nomenclature that we will use in this article.
FIGURE 2.
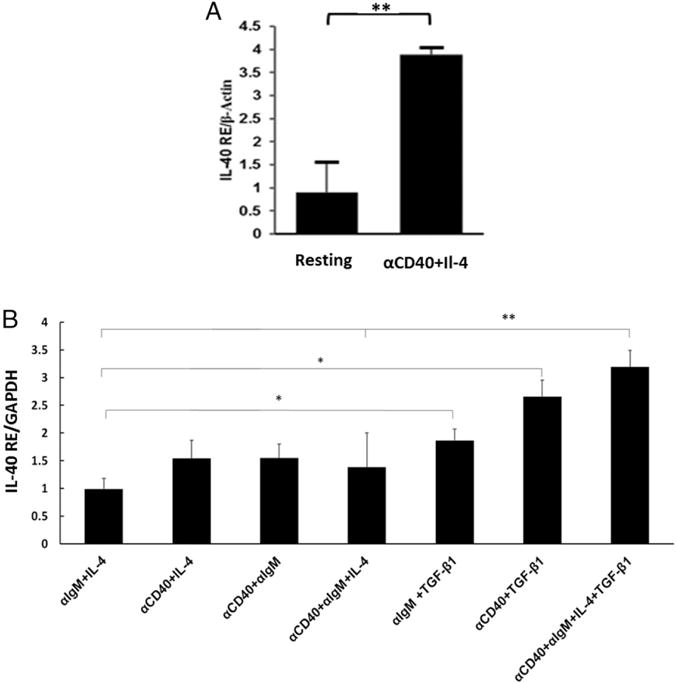
IL-40 production is induced upon B cell activation. (A) IL-40 production by the mouse A20-2J cell line stimulated with anti-CD40 mAb and IL-4 for 24 h; data are representative of two independent experiments (mean ± SEM) (qRT-PCR). (B) qRT-PCR analysis of Il-40 expression in mouse spleen B cells stimulated with anti-CD40 Ab and anti-IgM (5 μg/ml) alone or in presence of IL-4 or TGF-β1 (all at 10 ng/ml) for 2 d. Data are fold expression compared with resting cells and are representative of three independent experiments (mean ± SEM). *p < 0.05, **p < 0.01.
IL-40 expression is induced upon the onset of lactation in the mammary gland
IL-40 is 72% conserved at the amino acid level between mouse and human (Supplemental Fig. 1A) and exhibits a similar expression profile between the two species (human, Supplemental Fig. 1B; mouse, Supplemental Fig. 1C). As shown in Fig. 1D, C17orf99 is present only in mammalian genomes and is expressed by activated B cells (Fig. 2). Therefore, we hypothesized that it may be involved in mammalian-specific functions, such as lactation. To test this hypothesis, we measured the expression of Il40 in virgin, pregnant, 1-wk postnatal (PN), and 3-wk PN mammary glands of WT C57BL/6 mice. As shown in Fig. 3A, Il40 expression is induced upon the onset of lactation in the mammary glands. Interestingly, the expression of IL-40 correlates with the onset of IgA production in the mammary gland (Fig. 3B).
FIGURE 3.
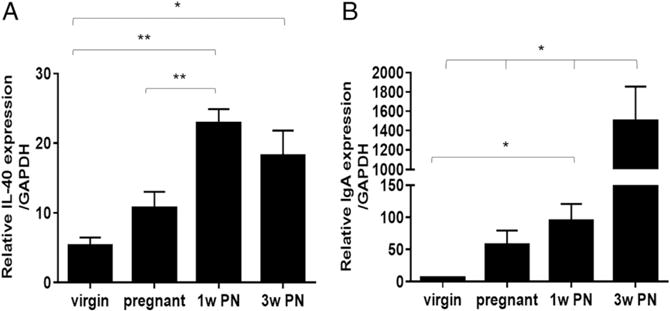
IL-40 expression is induced in the mammary gland upon the onset of lactation, and the Il40−/− mouse exhibits decreased IgA levels in milk. (A) qRT-PCR analysis of IL-40 expression in WT virgin, pregnant, 1 wk PN lactating, and 3 wk PN lactating mouse mammary glands (mean ± SEM). *p < 0.02, **p < 0.001. (B) qRT-PCR analysis of IgA expression in WT virgin, pregnant, 1 wk PN, and 3 wk PN lactating mouse mammary glands (mean ± SEM). *p < 0.029.
An Il40−/− mouse exhibits B cell abnormalities
To further explore the role of IL-40 in the immune response, we obtained a mouse with a targeted deletion of the mouse homolog of C17orf99 (6030468B19Rik), which has been described as part of a large mouse knockout database (25), and that results in the complete absence of IL-40 expression. The Il40−/− mouse is viable and reproduces normally. We hypothesized that this mouse would exhibit B cell abnormalities. The Il40−/− mouse has fewer B cells in the spleen compared with WT mice (Fig. 4A), indicating that the absence of IL-40 affects B cell homeostasis. To obtain more information about the effects of IL-40 in splenic B cells, we analyzed transitional and marginal zone B cells from Il40−/−mice by flow cytometry and, as shown in Fig. 4B, we observed a significant reduction in the number of transitional, follicular, and total marginal zone B cells. These observations suggest that IL-40 may be involved in peripheral B cell homeostasis; however, they could also be a result of altered B cell development. Therefore, we sought to analyze the latter possibility in more detail.
FIGURE 4.
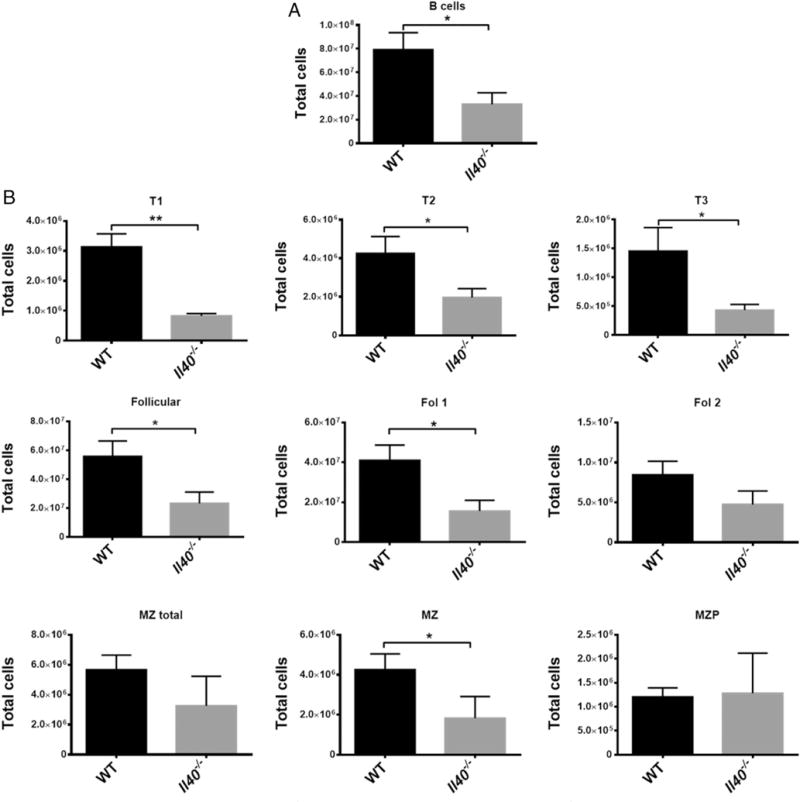
B cell populations are altered in the Il40−/− mouse. (A) Total B cells (CD19+) in WT or Il40−/− mice. (B) B cell subpopulation analysis in the spleen. Transitional B cell subpopulations 1, 2, and 3 (T1: CD23−CD93+IgMhighIgD−/lowCD21/35−/low; T2: CD23+CD93+IgMhighIgDhighCD21/35low; T3: CD23+CD93+IgMlowIgDhighCD21/35low respectively), total follicular B cells (Follicular: CD23+CD93−CD21/35int), follicular 1 cells (Fol 1: CD23+ CD93−IgMlowIgDhighCD21/35int), follicular 2 cells (Fol 2: CD23+CD93−/lowsIgMhighsIgDhighCD21/35int), total marginal zone B cells (MZ total: CD93−/low IgMhighCD21/35high), marginal zone cells (MZ: CD23−CD93−IgMhighIgDlowCD21/35high), and marginal zone precursors (MZP: CD23+CD93−/lowIgMhigh IgDhighCD21/35high). All of the cells were CD19+. Bars represent mean ± SEM; n = 4 per group. Data are representative of two independent experiments. *p # 0.05, **p # 0.002.
B cell populations are altered in the BM of Il40−/− mice
An Il40−/− mouse exhibits a deficiency of B220+ cells in the BM. B220 is a B cell–lineage marker that is expressed by B cell progenitors (pre-B cells) during development in the mouse BM (28). B220+ cells account for ~20% of the cells in the BM of normal adult mice (29). We stained for stromal and progenitor cells (CD45−) and leukocytes (CD45+), which, in turn, were divided into the B cell compartment (CD45+B220+), the myeloid compartment (CD45+CD11b+), and T cell progenitors (CD45+CD11b−) (30, 31). We observed a significant reduction in the number of B220+ cells in the BM of Il40−/− mice (Fig. 5A, 5B), which include precursor and mature B cells in the BM. These data suggested that IL-40 is involved in the development of B cells. Because BM expresses IL-40 (Fig. 1A), we tried to further identify the cells that produce IL-40. We initially observed that CD45− (but not CD45+) BM cells express IL-40 (data not shown). The BM CD45+ population contains B cell precursors (pro-B, pre-B cells) and macrophage and dendritic cell precursors (CD45+CD11b+ or CD45+ CD11b−), whereas the CD45−lineage− (CD3, Ly-6G/Ly-6C, CD11b, B220, and Ter-119) population represents mostly stromal cells. We sought to further identify the IL-40–producing cells by subdividing the CD45− population based on the expression of lineage markers and CD51 (αV integrin) (32). As shown in Fig. 5C, IL-40 is strongly expressed by the CD45−lineage−CD51− population, which primarily contains mature stromal cells (32).
FIGURE 5.
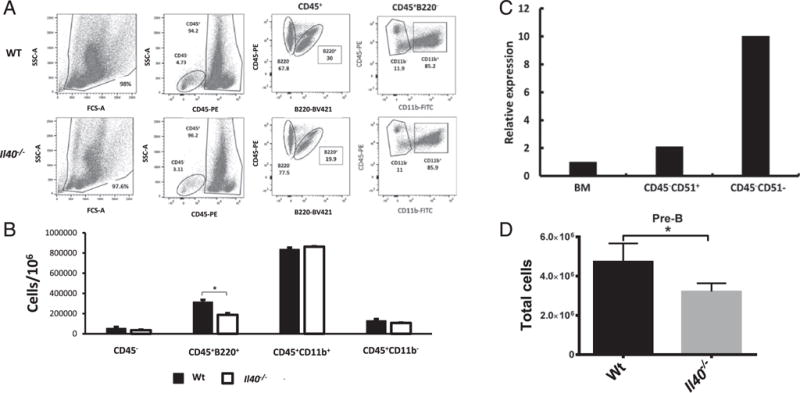
IL-40 is highly expressed in BM stroma and is required for normal B cell homeostasis. (A) BM staining of different cell compartments: stromal cells (CD45−), leukocytes (CD45+), the B cell compartment (CD45+B220+), the myeloid compartment (CD45+CD11b+), and T cell progenitors (CD45+CD11b−). Shown is a representative plot of n = 5 per group, which is representative of two independent experiments. (B) Total cell count for each compartment in (C). Results shown are representative of two independent experiments. (C) qPCR of sorted stromal populations in the BM. Whole BM expression, stromal precursors (Lin−CD45−CD51+), and mature stroma (Lin−CD45−CD51−). Relative expression compared with whole BM. Data are representative of two independent experiments. (D) Number of pre-B cells (CD19+, B220+, IgM−, IgD−, CD43−) in BM of WT and Il40−/− mice. *p < 0.05.
We also analyzed the B cell precursor populations in the BM and found reduced numbers of pre-B cells (Fig. 5D, Supplemental Fig. 2). Taken together, the fact that some BM stromal cells produce IL-40, whereas there are alterations in the B cell precursor compartment in the BM, strongly suggests that IL-40 plays a role in B cell development.
We have also analyzed hematopoietic stem cell populations in the BM (short-term hematopoietic stem cells, long-term hematopoietic stem cells, common myeloid progenitors, granulocyte-macrophage progenitors, and megakaryocyte and erythrocyte progenitors) and found no significant alterations in Il40−/− mice BM compared with WT mice (data not shown). These data suggest that IL-40 does not affect hematopoiesis.
Il40−/− mice exhibit reduced levels of IgA in mucosal secretions and exhibit GALT abnormalities
Lactation triggers the onset of IgA production in the mammary gland. Several genes have been implicated in triggering IgA production in the mammary gland. For example, a CCR10−/− mouse has no IgA in the milk, because the CCL28/CCR10 chemokine axis mediates the recruitment of IgA plasmablasts to the lactating mammary gland (33). Therefore, we next explored whether IgA production is affected by the absence of IL-40. To this end, we analyzed GALT, which contains a large number of immune cells and is a preferred site of IgA responses (34), in Il40−/− or WT mice. PPs are GALTs that participate in immune surveillance of the digestive tract and contain germinal center B cells and IgA-secreting plasma cells (34). The Il40−/− mouse has fewer and smaller PPs than WT mice (Fig. 6A) (35, 36). Because IgA is the main Ig produced at this site (35–37), we measured total IgA switched cells (IgA+) and found a 5-fold reduction in total IgA+ cells (Fig. 6B–D). Taken together, these data indicate that IL-40 participates in the development of IgA responses in GALT, especially in the PP. Given these results, we next measured IgA levels in the serum, feces, or milk of WT or Il40−/− mice and observed a significant reduction in IgA levels in the Il40−/− mouse (Fig. 6E–G). The lower levels of IgA in feces of the Il40−/− mouse correlate with defects observed in the PPs (Fig. 6A–D). Serum levels of IgM and total IgG were unaffected (data not shown). We then hypothesized that the defects in IgA production may be due to defects in CSR (20). To test this, we performed a CSR assay but did not detect differences when using cells from Il40−/− or WT mice, suggesting that this mechanism is not affected by lack of IL-40 (Supplemental Fig. 3). Based on these observations, we conclude that IL-40 is involved in the development/control of humoral immune responses, especially those involving IgA production.
FIGURE 6.
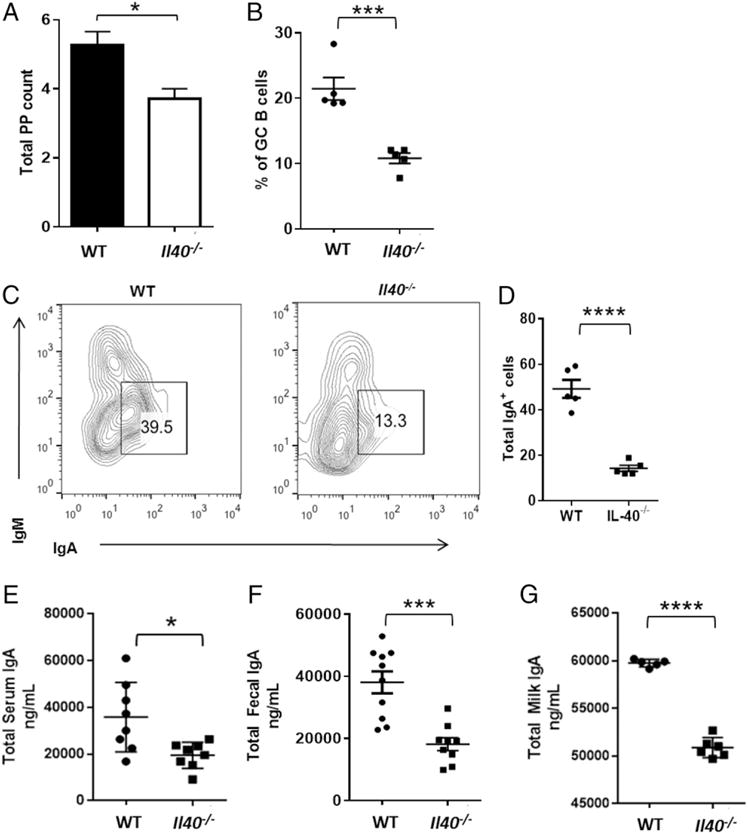
Il40−/− mice exhibit defects in GALT B cell populations. (A) Total counts of PPs from WT or Il40−/− mice (mean ± SEM; n = 7). *p < 0.0102. (B) Percentage of cells representing germinal center B cells in PPs of (A). (C) Flow cytometry analysis of germinal center B cells from PPs showing lower numbers of IgA+ B cells. (D) Total number of IgA+ cells from (B) (× 10−5). ****p < 0.01. (E) Serum IgA levels in Il40−/− mice compared with WT controls, as measured by sandwich ELISA (mean ± SEM; n = 8). *p < 0.0114. (F) Measurements of total IgA in fecal pellets by sandwich ELISA of WT versus Il40−/− mice (mean ± SEM; n = 10). ***p < 0.006. (G) Measurement of total IgA in breast milk by sandwich ELISA of WT (n = 5) versus Il40−/− mice (n = 6) (mean ± SEM). ****p < 0.0001. Results shown are representative of three independent experiments, each performed with five mice per group.
An Il40−/− mouse exhibits microbiome abnormalities
The gut is one of the major producers of IgA in the body (38, 39). IgA is an important regulator of commensal bacteria in the gut and controls infections in mucosal secretions (39). We hypothesized that the low levels of IgA measured in Il40−/− mice could be affecting the composition of the intestinal microbiome. To test this hypothesis, we collected fecal samples from WT and Il40−/− mice to perform total DNA extraction and 16S sequencing. Eubacterial diversity was reduced in the Il40−/− mouse, and the community composition and structure were substantially different by the absence of IL-40 expression (Fig. 7A, 7B). At the phylum level (Fig. 7C), we observed a significant reduction in Firmicutes in Il40−/− mice relative to WT, with a concomitant increase in Bacteriodetes. Deeper analysis of the affected phyla revealed significantly altered families and genera in the Il40−/− mouse. Among the Bacteroidetes, the uncultured Bacteroidales family S24-7 (40) was significantly increased (mean = WT 36.99%, Il40−/− 70.02%, p < 0.001). For the Firmicutes, the family Lachnospiraceae was the most prominent among those that were significantly reduced (WT 45.54%, Il40−/− 11.93%, p < 0.001), reflecting decreases in genera, such as Blautia (WT 0.48%, Il40−/− 0.02%, p < 0.001), Coprococcus 1 (WT 0.58%, Il40−/− 0.07%, p < 0.001), Roseburia (WT 2.37%, Il40−/− 0.13%, p < 0.001), and UCG-001 (WT 11.08%, Il40−/− 0.22%, p < 0.001). There were also significant changes in the phylum Proteobacteria; the genus Desulfovibrio was not detected in three Il40−/− mice (WT 0.62%, Il40−/− 0.01%, p < 0.001), whereas the genus Bilophila was detected in all Il40−/− mice but not in WT mice (WT 0%, Il40−/− 0.06%, p < 0.001). These data strongly suggest that the lower levels of IgA in the gut of Il40−/− mice affect the diversity of their microbiome.
FIGURE 7.
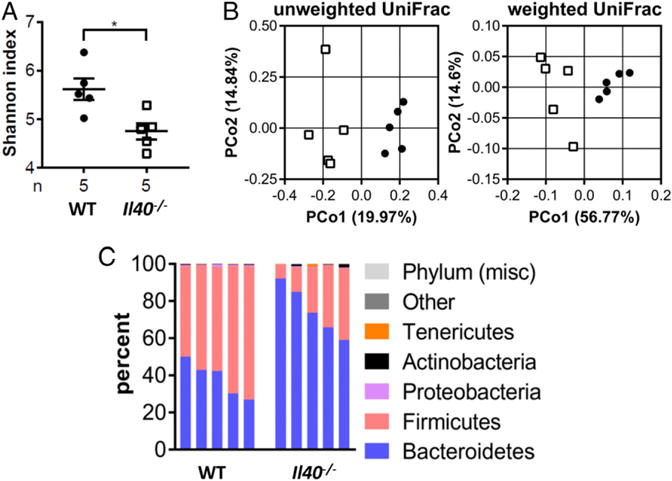
The microbiome of Il40−/− mice is altered. (A) The α diversity of fecal samples from WT or IL40−/− mice. (B) The β diversity of WT or IL40−/− mice. (C) Relative abundances (%) of different microbial phyla represented in WT or IL40−/− mice. *p < 0.05. n = 5 per group.
Human IL-40 is expressed by activated human B cells and human B cell lymphomas
Human peripheral blood B cells activated with anti-CD40 Ab plus IL-4 for 24 h showed strongly upregulated expression of human C17orf99/Il40 (Fig. 8A). We also tested various human non-Hodgkin B cell lymphoma lines (41) for IL-40 expression by qPCR and observed that several of them (OCI-Ly1, HL-2, and Val) produce IL-40 constitutively (Fig. 8B). The latter observation suggests that IL-40 may play a role in the pathogenesis of human lymphomas (42). Interestingly, the Il40+ lymphomas also express the B cell activation Ag TSPAN33 (8), indicating that they exhibit an “activated” B cell phenotype (42). Taken together, we conclude that C17orf99 encodes a novel B cell–derived cytokine (IL-40) that has significant effects in IgA production, B cell homeostasis, and, likely, B cell development.
FIGURE 8.
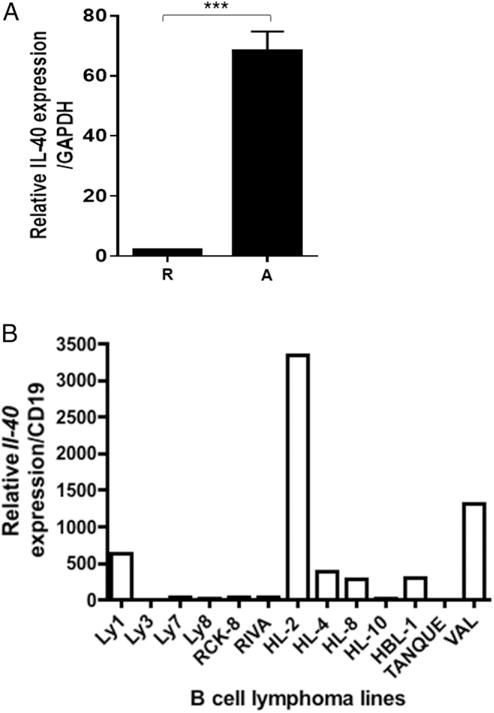
Human B cells produce IL-40. (A) qRT-PCR analysis of IL-40 expression in human resting (R) and activated (A) B cells. Cells were activated for 24 h with anti-CD40 Ab + IL-4. Data are representative of three independent experiments (mean ± SEM). (B) IL-40 expression pattern in several human diffuse large B cell lymphoma cell lines (qPCR). A representative experiment out of three is shown. ***p < 0.0005.
Discussion
We initially sought to identify novel immune system–associated genes. To this end, we analyzed a comprehensive database of human gene expression (BIGE) for genes expressed by immune system–associated organs, including tonsil, lymph nodes, spleen, thymus, BM, and fetal liver. This analysis yielded 35 poorly characterized genes predicted to encode transmembrane or secreted proteins, many of which are completely uncharacterized and were only annotated in the genome through their chromosomal location. For most of these, there is no information linking them with the immune system. We have reported three of these genes: Isthmin 1, a large secreted protein originally described in the brain of Xenopus, but which is expressed by NK, NKT, and Th17 cells in mammals (7); TSPAN33, a membrane protein that represents a novel biomarker of activated B cells (8); and Meteorin-like (6), a secreted small protein related to a known neurotrophic growth factor (meteorin) that is expressed by activated macrophages. In the current study, we focused on C17orf99, because it is predicted to encode a small secreted protein (~27 kDa) and is expressed by activated B cells, fetal liver, and BM. Further analyses indicated that the cellular source of mouse C17orf99 in the BM is CD45− cells, which are enriched in stromal cells. This is not surprising because these cells are known to produce many cytokines (27). Mouse C17orf99-producing cells in the BM are further enriched in the lineage−CD45−CD51− BM population (Fig. 5C). Interestingly, CD45−CD51+ cells include stromal cell precursors that support the development of bone and cartilage, whereas CD45−CD51− cells include mature stromal cells that support hematopoiesis and lymphopoiesis (32). These observations strongly suggest that mouse C17orf99 is produced by stromal cells that participate in lymphopoiesis. Along with this observation, we observed reduced numbers of B220+ cells and pre-B cells in the BM of C17orf99−/− mice (Fig. 5B, 5D). Importantly, lymphopoiesis occurs in the fetal liver and BM, and both sites express C17orf99 (Fig. 1A). Taken together, these observations strongly suggest that C17orf99 plays a role in B cell development in the fetal liver and BM. However, further studies are necessary to confirm this hypothesis and to identify its potential role in lymphopoiesis. Conversely, we have not detected abnormalities in hematologic precursors in C17orf99−/− mice (data not shown), suggesting that C17orf99 is mainly linked to B cell development and function.
Data from the BIGE database indicate that C17orf99 is expressed by human peripheral blood B cells activated with anti-CD40 and IL-4 (Fig. 1A). We confirmed the latter observation with the murine B cell lymphoma A20-2J (Fig. 2A) and mouse spleen B cells (Fig. 2B). The fact that C17orf99 encodes a small secreted protein (~27 kDa), and that its expression is inducible in B cells upon activation, strongly suggests that C17orf99 encodes a novel cytokine. Recently, a new member of the IL-12 family has been identified and named IL-39 (6, 43). Therefore, we have named the small secreted protein encoded by C17orf99 IL-40, because that is the next number in the IL sequence (11). In the periphery, the main source of IL-40 is likely to be activated B cells, suggesting that it plays a role in B cell homeostasis or humoral responses.
Cytokines are very important molecules in the immune system. Therefore, it is relevant to ask why C17orf99/Il40 has not yet been identified as a cytokine. Several factors may account for this, including that it is expressed by a limited number of tissues (BM and fetal liver); it does not belong to any recognized cytokine family (structural features would have facilitated its identification); and its main producers in the periphery (B cells) are not known to be prolific cytokine producers (44). Regarding this last point, we should note that some B cell subsets have recently achieved notoriety precisely through their ability to differentiate into cytokine producers. An example is the B10 regulatory cells, which produce IL-10 and are believed to be involved in autoimmunity (45). In contrast, C17orf99 has been recognized to encode a small secreted protein (26), and Tang et al. (25) included it in a library of knockout mice that encode transmembrane or secreted proteins.
B cells require several signals for optimal expression of IL-40. One of these involves the BCR (represented by the anti-IgM stimulus), and anti-CD40 Ab, or CD40L. The second signal is represented by cytokines like IL-4, which induces B cell activation and proliferation. Interestingly, among several cytokines tested, we found that TGF-β1 potentiates the expression of Il40 by activated B cells (Fig. 2B). The ability of cytokines like IL-4 and TGF-β1 to potentiate IL-40 production by B cells suggests that optimal IL-40 expression by B cells may require a specific differentiation mechanism to achieve optimal production (44). We predict that these “B40” cells could develop in vivo under certain conditions. In support of this, other B cell subsets have been described that require a specific differentiation mechanism to become strong cytokine producers. For example, IL-21 is required for B10 cells to develop in vitro (46). Similarly, an IL-40–producing subset of cells (B40 cells) may be associated with specific immune responses or human diseases.
As represented in a phylogram (Fig. 1D), C17orf99 is only present in mammalian genomes. This observation led us to hypothesize that IL-40 may have a mammalian-specific function. Lactation is such a function, and its onset triggers the production of IgA in the mammary gland. Importantly, IL-40 expression is induced upon the onset of lactation, and it correlates with the expression of IgA in the milk (Fig. 3). Further support for a role for IL-40 in IgA responses comes from the phenotype of an Il40−/− mouse, which shows a reduction in the size and numbers of PPs, IgA+ B cells, and in IgA levels (Fig. 6), as well as significant differences in its microbiota (Fig. 7). The reduction in serum Igs is reminiscent of the phenotype of the B cell–activating factor BAFF−/− mouse (47), which exhibits a severe reduction in serum Igs; however, unlike the Il40−/− mouse, the BAFF−/− mouse only exhibits a moderate reduction in IgA levels. Ablation of another TNF family member, APRIL, also resulted in reduction of IgA serum levels (48). The inability of the Il40−/− mouse to produce normal levels of IgA suggests that the ablation of Il40 may be altering a mechanism that specifically affects this B cell subpopulation. Furthermore, the changes observed in the microbiome of the Il40−/− mouse are likely a consequence of the altered IgA production in mucosal tissues (49). Conversely, the number of lymphocytes in PPs and other GALT is influenced by microbial colonization (50), reflecting common evolutionary mechanisms. Taken together, these observations suggest an important role for IL-40 in the proliferation or differentiation of IgA-committed B cells.
Another interesting IL-40–linked cytokine is TGF-β1, which is also involved in IgA responses (51) and, as we show in Fig. 2B, potentiates IL-40 production by B cells. This raised the possibility that abnormal production of TGF-β1 in the Il40−/− mouse could account for the lower IgA levels observed in this mouse. However, we have tested the capacity of activated T cells from Il40−/− mice to produce TGF-β1, and it appears normal (data not shown).
Activated human peripheral blood B cells also produce IL-40 (Fig. 8A). Importantly, some human B cell lymphomas can produce IL-40 constitutively (Fig. 8B). B cell–ablation therapies (i.e., anti-CD20 mAbs) have shown significant therapeutic benefit in several autoimmune diseases, including multiple sclerosis (52) and rheumatoid arthritis (53). Yet, the mechanism(s) through which B cells participate in the pathology of these diseases remains undefined (44), although it is unlikely to be due to auto-antibodies, because the ablation of B cells [e.g., using rituximab (54)] does not affect plasma cells or autoantibody titers. An emerging concept is that B cell–derived cytokines (45, 46) may affect the development of autoimmunity. The latter observations suggest that IL-40, as a B cell–associated novel cytokine, is an excellent candidate to be involved in the pathology of human diseases, including autoimmune diseases (55) and lymphomas (56), for which activated B40 cells may play an important role in their pathogenesis (44). Future experiments will aim to address some of the questions raised by the results described in this article.
Supplementary Material
Acknowledgments
We thank Emmanuel Dotsey, Phil Felgner, Jose Luis Maravillas, and Mathew Inlay (University of California, Irvine) for helpful discussions; David Fruman for the human B cell lymphoma lines and Philippa Marrack for the A20-2J mouse B cell line; and Clayton White for help with CSR assays.
This work was supported by National Institutes of Health Grants R21 AI096278-01 and R21 AI117556 (to A.Z.) and Grants AI 079705, AI 105813, and AI 060573 (to P.C.). M.I.V. was supported by NSF-GK-12 Grant DGE-0638751 and National Institutes of Health MBRS-IMSD Grant GM055246. V.P.L. and I.U. were supported by National Institute of Allergy and Infectious Diseases Training Grant T32 AI060573. M.H.-R. was a UCMEXUS-Consejo Nacional de Ciencia y Tecnología (CONACyT, Mexico) Post-Doctoral Scholar. J.C.-D. was supported by UCMEXUS-CONACyT/Secretaría de Educación Pública (Mexico) Grant 329416/BC-1455.
Abbreviations
- BIGE
Body Index of Gene Expression
- BM
bone marrow
- C17orf99
chromosome 17 open reading frame 99
- CSR
class switch recombination
- His
histidine
- OTU
operational taxonomic unit
- PBS-T
PBS-Tween
- PN
postnatal
- PP
Peyer’s patch
- qPCR
quantitative PCR
- TSPAN33
Tetraspanin 33
- WT
wild-type
Footnotes
ORCIDs: 0000-0002-9683-9278 (S.-P.N.); 0000-0002-8982-5177 (P.C.); 0000-0001-6487-4215 (M.R.); 0000-0002-1906-5846 (P.A.H.).
The online version of this article contains supplemental material.
Disclosures
V.P.L., P.A.H., A.M.B., I.U., and A.Z. are inventors named in a patent application (U.S. Provisional Patent Application number 61/906,855) describing uses of IL40 filed by the Regents of the University of California. The other authors have no financial conflicts of interest.
References
- 1.Lunney JK. Cytokines orchestrating the immune response. Rev - Off Int Epizoot. 1998;17:84–94. doi: 10.20506/rst.17.1.1094. [DOI] [PubMed] [Google Scholar]
- 2.Zlotnik A, Yoshie O. The chemokine superfamily revisited. Immunity. 2012;36:705–716. doi: 10.1016/j.immuni.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clavel G, Thiolat A, Boissier MC. Interleukin newcomers creating new numbers in rheumatology: IL-34 to IL-38. Joint Bone Spine. 2013;80:449–453. doi: 10.1016/j.jbspin.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Lee J, Hever A, Willhite D, Zlotnik A, Hevezi P. Effects of RNA degradation on gene expression analysis of human postmortem tissues. FASEB J. 2005;19:1356–1358. doi: 10.1096/fj.04-3552fje. [DOI] [PubMed] [Google Scholar]
- 5.Roth RB, Hevezi P, Lee J, Willhite D, Lechner SM, Foster AC, Zlotnik A. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7:67–80. doi: 10.1007/s10048-006-0032-6. [DOI] [PubMed] [Google Scholar]
- 6.Ushach I, Burkhardt AM, Martinez C, Hevezi PA, Gerber PA, Buhren BA, Schrumpf H, Valle-Rios R, Vazquez MI, Homey B, Zlotnik A. METEORIN-LIKE is a cytokine associated with barrier tissues and alternatively activated macrophages. Clin Immunol. 2015;156:119–127. doi: 10.1016/j.clim.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valle-Rios R, Maravillas-Montero JL, Burkhardt AM, Martinez C, Buhren BA, Homey B, Gerber PA, Robinson O, Hevezi P, Zlotnik A. Isthmin 1 is a secreted protein expressed in skin, mucosal tissues, and NK, NKT, and Th17 cells. J Interferon Cytokine Res. 2014;34:795–801. doi: 10.1089/jir.2013.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luu VP, Hevezi P, Vences-Catalan F, Maravillas-Montero JL, White CA, Casali P, Llorente L, Jakez-Ocampo J, Lima G, Vilches-Cisneros N, et al. TSPAN33 is a novel marker of activated and malignant B cells. Clin Immunol. 2013;149:388–399. doi: 10.1016/j.clim.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Liu X, Zhang Y, Wang Z, Zhu G, Han G, Chen G, Hou C, Wang T, Ma N, et al. Interleukin (IL)-39 [IL-23p19/Epstein-Barr virus-induced 3 (Ebi3)] induces differentiation/expansion of neutrophils in lupus-prone mice. Clin Exp Immunol. 2016;186:144–156. doi: 10.1111/cei.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Wei Y, Xiao H, Liu X, Zhang Y, Han G, Chen G, Hou C, Ma N, Shen B, et al. A novel IL-23p19/Ebi3 (IL-39) cytokine mediates inflammation in Lupus-like mice. Eur J Immunol. 2016;46:1343–1350. doi: 10.1002/eji.201546095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.IUIS/WHO Standing Committee on Interleukin Designation. Nomenclature for secreted regulatory proteins of the immune system (interleukins): update. Bull World Health Organ. 1997;75:175. [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z, Fulop Z, Wu G, Pone EJ, Zhang J, Mai T, Thomas LM, Al-Qahtani A, White CA, Park SR, et al. 14-3-3 adaptor proteins recruit AID to 59-AGCT-39-rich switch regions for class switch recombination. Nat Struct Mol Biol. 2010;17:1124–1135. doi: 10.1038/nsmb.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loignon M, Perret S, Kelly J, Boulais D, Cass B, Bisson L, Afkhamizarreh F, Durocher Y. Stable high volumetric production of glycosylated human recombinant IFNalpha2b in HEK293 cells. BMC Biotechnol. 2008;8:65. doi: 10.1186/1472-6750-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones B, Tite JP, Janeway CA., Jr Different phenotypic variants of the mouse B cell tumor A20/2J are selected by antigen- and mitogen-triggered cytotoxicity of L3T4-positive, I-A-restricted T cell clones. J Immunol. 1986;136:348–356. [PubMed] [Google Scholar]
- 15.Golde WT, Gollobin P, Rodriguez LL. A rapid, simple, and humane method for submandibular bleeding of mice using a lancet. Lab Anim (NY) 2005;34:39–43. doi: 10.1038/laban1005-39. [DOI] [PubMed] [Google Scholar]
- 16.Carvalho JJ, Walter MA, Baermann-Stapel Y, Weller MG, Panne U, Schenk JA, Schneider RJ. Non-invasive monitoring of immunization progress in mice via IgG from feces. In Vivo. 2012;26:63–69. [PubMed] [Google Scholar]
- 17.Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, Plevritis SK, Dill DL, Weissman IL. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inlay MA, Serwold T, Mosley A, Fathman JW, Dimov IK, Seita J, Weissman IL. Identification of multipotent progenitors that emerge prior to hematopoietic stem cells in embryonic development. Stem Cell Rep. 2014;2:457–472. doi: 10.1016/j.stemcr.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allman D, Pillai S. Peripheral B cell subsets. Curr Opin Immunol. 2008;20:149–157. doi: 10.1016/j.coi.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pone EJ, Xu Z, White CA, Zan H, Casali P. B cell TLRs and induction of immunoglobulin class-switch DNA recombination. Front Biosci (Landmark Ed) 2012;17:2594–2615. doi: 10.2741/4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Z, Zan H, Pone EJ, Mai T, Casali P. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat Rev Immunol. 2012;12:517–531. doi: 10.1038/nri3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO. The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014;42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang T, Li L, Tang J, Li Y, Lin WY, Martin F, Grant D, Solloway M, Parker L, Ye W, et al. A mouse knockout library for secreted and transmembrane proteins. Nat Biotechnol. 2010;28:749–755. doi: 10.1038/nbt.1644. [DOI] [PubMed] [Google Scholar]
- 26.Clark HF, Gurney AL, Abaya E, Baker K, Baldwin D, Brush J, Chen J, Chow B, Chui C, Crowley C, et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and trans-membrane proteins: a bioinformatics assessment. Genome Res. 2003;13:2265–2270. doi: 10.1101/gr.1293003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sison EA, Brown P. The bone marrow microenvironment and leukemia: biology and therapeutic targeting. Expert Rev Hematol. 2011;4:271–283. doi: 10.1586/ehm.11.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baumgarth N. B-cell immunophenotyping. Methods Cell Biol. 2004;75:643–662. doi: 10.1016/s0091-679x(04)75027-x. [DOI] [PubMed] [Google Scholar]
- 29.Cimmino L, Dawlaty MM, Ndiaye-Lobry D, Yap YS, Bakogianni S, Yu Y, Bhattacharyya S, Shaknovich R, Geng H, Lobry C, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol. 2015;16:653–662. doi: 10.1038/ni.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagasawa T. Microenvironmental niches in the bone marrow required for B-cell development. Nat Rev Immunol. 2006;6:107–116. doi: 10.1038/nri1780. [DOI] [PubMed] [Google Scholar]
- 31.Kincade PW, Igarashi H, Medina KL, Kouro T, Yokota T, Rossi MI, Owen JJ, Garrett KP, Sun XH, Sakaguchi N. Lymphoid lineage cells in adult murine bone marrow diverge from those of other blood cells at an early, hormone-sensitive stage. Semin Immunol. 2002;14:385–394. doi: 10.1016/s1044532302000738. [DOI] [PubMed] [Google Scholar]
- 32.Chan CK, Seo EY, Chen JY, Lo D, McArdle A, Sinha R, Tevlin R, Seita J, Vincent-Tompkins J, Wearda T, et al. Identification and specification of the mouse skeletal stem cell. Cell. 2015;160:285–298. doi: 10.1016/j.cell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morteau O, Gerard C, Lu B, Ghiran S, Rits M, Fujiwara Y, Law Y, Distelhorst K, Nielsen EM, Hill ED, et al. An indispensable role for the chemokine receptor CCR10 in IgA antibody-secreting cell accumulation. J Immunol. 2008;181:6309–6315. doi: 10.4049/jimmunol.181.9.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerutti A. The regulation of IgA class switching. Nat Rev Immunol. 2008;8:421–434. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park SR, Zan H, Pal Z, Zhang J, Al-Qahtani A, Pone EJ, Xu Z, Mai T, Casali P. HoxC4 binds to the promoter of the cytidine deaminase AID gene to induce AID expression, class-switch DNA recombination and somatic hypermutation. Nat Immunol. 2009;10:540–550. doi: 10.1038/ni.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tezuka H, Abe Y, Iwata M, Takeuchi H, Ishikawa H, Matsushita M, Shiohara T, Akira S, Ohteki T. Regulation of IgA production by naturally occurring TNF/iNOS-producing dendritic cells. Nature. 2007;448:929–933. doi: 10.1038/nature06033. [DOI] [PubMed] [Google Scholar]
- 37.Pone EJ, Zhang J, Mai T, White CA, Li G, Sakakura JK, Patel PJ, Al-Qahtani A, Zan H, Xu Z, Casali P. BCR-signalling synergizes with TLR-signalling for induction of AID and immunoglobulin class-switching through the non-canonical NF-kB pathway. Nat Commun. 2012;3:767. doi: 10.1038/ncomms1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindner C, Wahl B, Föhse L, Suerbaum S, Macpherson AJ, Prinz I, Pabst O. Age, microbiota, and T cells shape diverse individual IgA repertoires in the intestine. J Exp Med. 2012;209:365–377. doi: 10.1084/jem.20111980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macpherson AJ, McCoy KD, Johansen FE, Brandtzaeg P. The immune geography of IgA induction and function. Mucosal Immunol. 2008;1:11–22. doi: 10.1038/mi.2007.6. [DOI] [PubMed] [Google Scholar]
- 40.Ormerod KL, Wood DL, Lachner N, Gellatly SL, Daly JN, Parsons JD, Dal’Molin CG, Palfreyman RW, Nielsen LK, Cooper MA, et al. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome. 2016;4:36. doi: 10.1186/s40168-016-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehra S, Messner H, Minden M, Chaganti RS. Molecular cyto-genetic characterization of non-Hodgkin lymphoma cell lines. Genes Chromosomes Cancer. 2002;33:225–234. doi: 10.1002/gcc.10025. [DOI] [PubMed] [Google Scholar]
- 42.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 43.Bensen JT, Dawson PA, Mychaleckyj JC, Bowden DW. Identification of a novel human cytokine gene in the interleukin gene cluster on chromosome 2q12-14. J Interferon Cytokine Res. 2001;21:899–904. doi: 10.1089/107999001753289505. [DOI] [PubMed] [Google Scholar]
- 44.Luu VP, Vazquez MI, Zlotnik A. B cells participate in tolerance and autoimmunity through cytokine production. Autoimmunity. 2014;47:1–12. doi: 10.3109/08916934.2013.856006. [DOI] [PubMed] [Google Scholar]
- 45.Bouaziz JD, Yanaba K, Tedder TF. Regulatory B cells as inhibitors of immune responses and inflammation. Immunol Rev. 2008;224:201–214. doi: 10.1111/j.1600-065X.2008.00661.x. [DOI] [PubMed] [Google Scholar]
- 46.Yoshizaki A, Miyagaki T, DiLillo DJ, Matsushita T, Horikawa M, Kountikov EI, Spolski R, Poe JC, Leonard WJ, Tedder TF. Regulatory B cells control T-cell autoimmunity through IL-21-dependent cognate interactions. Nature. 2012;491:264–268. doi: 10.1038/nature11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schiemann B, Gommerman JL, Vora K, Cachero TG, Shulga-Morskaya S, Dobles M, Frew E, Scott ML. An essential role for BAFF in the normal development of B cells through a BCMA-independent pathway. Science. 2001;293:2111–2114. doi: 10.1126/science.1061964. [DOI] [PubMed] [Google Scholar]
- 48.Castigli E, Scott S, Dedeoglu F, Bryce P, Jabara H, Bhan AK, Mizoguchi E, Geha RS. Impaired IgA class switching in APRIL-deficient mice. Proc Natl Acad Sci USA. 2004;101:3903–3908. doi: 10.1073/pnas.0307348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mueller C, Macpherson AJ. Layers of mutualism with commensal bacteria protect us from intestinal inflammation. Gut. 2006;55:276–284. doi: 10.1136/gut.2004.054098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamanaka T, Helgeland L, Farstad IN, Fukushima H, Midtvedt T, Brandtzaeg P. Microbial colonization drives lymphocyte accumulation and differentiation in the follicle-associated epithelium of Peyer’s patches. J Immunol. 2003;170:816–822. doi: 10.4049/jimmunol.170.2.816. [DOI] [PubMed] [Google Scholar]
- 51.Ingman WV, Robertson SA. Mammary gland development in transforming growth factor beta1 null mutant mice: systemic and epithelial effects. Biol Reprod. 2008;79:711–717. doi: 10.1095/biolreprod.107.067272. [DOI] [PubMed] [Google Scholar]
- 52.Bar-Or A, Calabresi PA, Arnold D, Markowitz C, Shafer S, Kasper LH, Waubant E, Gazda S, Fox RJ, Panzara M, et al. Rituximab in relapsing-remitting multiple sclerosis: a 72-week, open-label, phase I trial. Ann Neurol. 2008;63:395–400. doi: 10.1002/ana.21363. [Published erratum appears in 2008 Ann. Neurol. 63: 803] [DOI] [PubMed] [Google Scholar]
- 53.Wang D, Li Y, Liu Y, Shi G. The use of biologic therapies in the treatment of rheumatoid arthritis. Curr Pharm Biotechnol. 2014;15:542–548. doi: 10.2174/138920101506140910150612. [DOI] [PubMed] [Google Scholar]
- 54.Reddy V, Dahal LN, Cragg MS, Leandro M. Optimising B-cell depletion in autoimmune disease: is obinutuzumab the answer? Drug Discov Today. 2016;21:1330–1338. doi: 10.1016/j.drudis.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 55.Youinou P, Taher TE, Pers JO, Mageed RA, Renaudineau Y. B lymphocyte cytokines and rheumatic autoimmune disease. Arthritis Rheum. 2009;60:1873–1880. doi: 10.1002/art.24665. [DOI] [PubMed] [Google Scholar]
- 56.Mahadevan D, Fisher RI. Novel therapeutics for aggressive non-Hodgkin’s lymphoma. J Clin Oncol. 2011;29:1876–1884. doi: 10.1200/JCO.2010.32.7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


