Abstract
Ewing sarcoma is a bone malignancy driven by a translocation event resulting in the fusion protein EWS/FLI1 (EF). EF functions as an aberrant and oncogenic transcription factor that misregulates the expression of thousands of genes. Previous work has focused principally on determining important transcriptional targets of EF, as well as characterizing important regulatory partnerships in EF-dependent transcriptional programs. Less is known, however, about EF-dependent metabolic changes or their role in Ewing sarcoma biology. Therefore, the metabolic effects of silencing EF in Ewing sarcoma cells were determined. Metabolomic analyses revealed distinct separation of metabolic profiles in EF-knockdown vs. control-knockdown cells. Mitochondrial stress tests demonstrated that knockdown of EF increased respiratory as well as glycolytic functions. Enzymes and metabolites in several metabolic pathways were altered, including de novo serine synthesis and elements of one-carbon metabolism. Furthermore, phosphoglycerate dehydrogenase (PHGDH) was found to be highly expressed in Ewing sarcoma and correlated with worse patient survival. PHGDH knockdown or pharmacological inhibition in vitro caused impaired proliferation and cell death. Interestingly, PHGDH modulation also led to elevated histone expression and methylation. These studies demonstrate that the translocation-derived fusion protein EF is a master regulator of metabolic reprogramming in Ewing sarcoma, diverting metabolites toward biosynthesis. As such, these data suggest that the metabolic aberrations induced by EF are important contributors to the oncogenic biology of these tumors.
Keywords: sarcoma, metabolomics, EWS/FLI
Introduction
Ewing sarcoma is an aggressive malignancy that most often emerges from bone and bone-associated tissues of children and adolescents. It is the second most common pediatric bone tumor; second only to osteosarcoma. Surgical strategies have achieved very good outcomes for many of these patients. But truly effective molecular therapies continue to evade discovery, translating to especially poor prognosis for patients who fall victim to metastasis or relapse. Indeed, the rate of relapse after surgery is ~35%, and the probability of 5-year survival after relapse is ~20% (1, 2). Continued investigation of the molecular underpinnings of this disease are thus crucial, and have the promise of providing additional candidate therapies to these patients.
Ewing sarcoma tumors are uniquely devoid of recurrent mutations in classical oncogenes or tumor suppressors (3). Instead, oncogenesis of Ewing sarcoma is almost entirely due to the far-reaching consequences of a single genetic lesion: a translocation event between chromosomes 11 and 22. This chromosomal rearrangement generates an in-frame fusion product merging the EWSR1 and FLI1 genes, giving rise to the oncogenic fusion protein known as EWS/FLI (EF). EF contains the N-terminal transcriptional regulatory domain of EWS and the DNA-binding domain of FLI1. Interestingly, while EF can affect known targets of FLI1 by binding to canonical FLI1 binding sites, it also influences expression of genes that are not targets of FLI1 or EWSR1 (4). Thus, EF is an aberrant transcription factor with neomorphic functions and tumorigenic consequences.
The transcriptional reprogramming driven by EF has been well profiled (5). In-depth study of EF targets has revealed that this master regulator exerts its oncogenic effects on numerous processes, contributing to many of the hallmark capabilities of cancer cells. These include: proliferation and growth (6), resistance to cell death (7), immortality (8), angiogenesis (9), invasion and metastasis (7), and evading immune destruction (10). However, very little is known about how EF affects another characteristic feature of cancer cells – the pro-oncogenic reprogramming of cellular metabolism.
Cancer cells characteristically adopt programs that modify substrate utilization and alter metabolite abundance in ways that support the biosynthetic requirements of these highly proliferative and resilient cells (11). Evidence increasingly indicates that metabolic alterations observed in cancer are not mere spectator phenomena, but are active contributors to oncogenesis (11). Significant interrelationships exist linking cellular metabolic processes to nuclear control, and vice versa (11). As a result, defining the metabolic processes undergirding cancer cells provides promising new opportunities for discovery of potentially actionable therapeutic targets.
Currently there is a paucity of data concerning the role of EF in altering metabolic processes in Ewing sarcoma. Given its relatively low mutation burden compared to other cancers (3), Ewing sarcoma provides a unique model to study metabolic alterations that are important to oncogenesis. Changes in cellular metabolism that result from EF transcriptional misregulation likely contribute directly to oncogenesis. Thus, studying cellular metabolism in Ewing sarcoma promises to increase our understanding of metabolic reprogramming in general, and inform development of novel therapeutic strategies for Ewing sarcoma specifically. With these goals in mind, we set out to investigate EF-driven metabolic reprogramming in Ewing sarcoma.
Materials & Methods
Cell culture
Cell lines expressing EWS/FLI (A673, UTES-14-01872; see Fig. S2B & C) or EWS/ERG (TTC466) were grown in 10%FBS/DMEM or 10% FBS/RPMI (RPMI 1640 Glutamax; ThermoFisher), respectively. DMEM (Corning) contained 25 mM glucose, 4 mM L-glutamine and 1 mM sodium pyruvate. Media were supplemented with 3 µg/ml puromycin for selection of shRNA-expressing cells. 293T cells were grown in 10% FBS/DMEM. Mycoplasma testing was performed as previously described (12). Population doublings were assessed using a 3T5 growth assay as previously described (13). A673 and TTC466 lines were validated prior to experimentation by STR analysis (Genetica DNA Labs). The patient-derived cell lines A673 and TTC466 were acquired from stocks maintained S. Lessnick. UTES-14-01872 is a recently-derived cell line of Ewing sarcoma established in 2014 by S. Schiffman.
shRNA constructs, virus production and infections
EWS/FLI- and Luciferase-siRNA sequences were used previously for transcriptional profiling of Ewing sarcoma (14–16). siRNA sequences for PHGDH and PSPH were designed using the Whitehead Institute siRNA Selection Program (17). shRNA-encoding inserts containing these sequences were ligated into the pMKO.1p retroviral vector. siRNA sequences were: shEF, 5’-ATAGAGGTGGGAAGCTTAT-3’; shPHGDH, 5’-CCCTGTAGTACAGCAATAA-3’; shPSPH, 5’-GGCTGAAATTCTACTTTAA-3’. Retrovirus was produced by transfecting 293T cells with Optimem and Lipofectamine 2000, and viral supernatant was collected at 48h and 72h post-infection. Viral supernatant was filtered at 0.45 µm, aliquoted and stored at −80° C. Infections were performed by adding viral supernatant (plus 8 µg/ml polybrene) to sub-confluent cultures. Selection in puromycin-containing media was continued for 3 days, and puromycin efficacy was confirmed by observing cell death in uninfected cells. As EF is the primary oncogenic driver of Ewing sarcoma, most Ewing sarcoma cell lines do not tolerate its silencing well. A673 cells are known to best tolerate EF knockdown. Accordingly, we have had the most success using A673 cells, and limited success performing such experiments in other lines.
Mitotracker staining
A673 cells expressing shLUC or shEF were seeded onto 1.5 borosilicate chambered coverglass slides (Thermo Fisher) and incubated overnight. Media was replaced with phenol red-free DMEM containing 25 mM glucose, 1% Glutamax, 1% sodium pyruvate, 100 nM Mitotracker Red FM, 200 nM Mitotracker Green FM, and 5 µg/ml Hoechst stain (Thermo Fisher). Cells were incubated in staining medium for 10 minutes, after which staining medium was removed and cells were washed twice with phenol red-free medium. Cells were immediately imaged using a Zeiss Axio Observer Z1 microscope.
Images were quantified using ImageJ software. Three cells were selected in each of 5 high-power field images for a total of 15 quantified cells. Integrated density measurements were taken for Mitotracker Red and Mitotracker Green channels for each cell with identical areas, and corrected total cell fluorescence (CTCF) was calculated for each channel as described previously (18). CTCF calculations were done as: CTCF = integrated density − (area of selected cell × mean fluorescence of background readings).
Western blotting
Whole cell extract (WCE) was prepared using RIPA buffer containing protease inhibitor cocktail (Sigma). Total protein content was determined via BCA assay, and WCE was prepared for SDS-PAGE by adding Laemmli buffer. Immunoblotting was done using the following antibodies: Fli1 (Abcam, 1:1,000), PHGDH (Cell Signaling, 1:1,000), PSAT (Abcam, 1:1,000), PSPH (Abcam, 1:1,000), SHMT1 (Cell Signaling, 1:1,000), SHMT2 (Cell Signaling, 1:1000), TYMS (Cell Signaling, 1:1,000), DHFR (Abcam, 1:1,000), HK1 (Abcam, 1:1000), H3K9me3 (Abcam, 1:1,000), H3K27me3 (Cell Signaling, 1:1,000), Histone H3 (Abcam, 1:5,000), α-tubulin (Cell Signaling, 1:5,000), β-actin (Sigma, 1:5,000). Densitometry was performed using Image J software as previously described (19).
Immunohistochemistry
Procedures for immunohistochemistry (IHC) are previously described (20–22). Briefly, paraffin was removed and tissue was subjected to high pressure antigen retrieval (Vector Unmasking Solution; Vector Laboratories, Burlingame, CA). Samples were incubated in primary antibody (PHGDH, Cell Signaling) overnight at 4°C. Secondary antibodies, raised in donkey (Jackson Immunoresearch, West Grove, PA), were used at a dilution of 1:250 at room temperature for 1-hour. Vectastain reagents and diaminobenzidine (DAB) (Vector Laboratories) were used to develop IHC. Images were subsequently taken on a Zeiss Axio Observer Z1 microscope. Tissue samples from human patients were obtained with patient consent and approval from the University of Utah Institutional Review Board.
PHGDH inhibition, 2-HG & %5-mC
The PHGDH inhibitors NCT502 and NCT503 (Cayman) and the inactive control molecule called PHGDH-inactive (Cayman) were reconstituted in DMSO. Final concentrations of inhibitors were 3.7 uM, 18.5 uM and 37 uM for PHGDH-inactive and NCT502; and 2.5 uM, 12.5 uM and 25 uM for NCT503. These concentrations were chosen to achieve maximal levels of inhibition at the highest doses based on previously published IC50 data (23).
Global 5-methylcytosine levels were assessed using the Global DNA Methylation Assay – LINE1 (Active Motif). D-2-hydroxyglutarate was quantified using a colorimetric D2HG assay kit (BioVision). All procedures were carried out as indicated by the manufacturer.
Seahorse
Cells were seeded into 96-well format Seahorse analyzer plates 24 hours prior to performing Seahorse experiments. We performed side-by-side Mito Stress Tests and a Glycolysis Stress Tests in 96-well format using the XF96e analyzer. The Mito Stress Test was performed in standard assay media (DMEM, 25 mM Glucose, 1 mM pyruvate, 2 mM Glutamine, pH 7.4) with the final concentrations of drugs as follows [Oligomycin: 2 µM; carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP): 2 µM; Rotenone: 0.5 µM; Antimycin A: 0.5 µM]. Assay protocol was standard (3 measurements per phase, acute injection followed by 3 min mixing, 0 min waiting, and 3 min measuring). Data were normalized to cell number because morphological changes that result from EF silencing potentially confound normalization to total cellular protein (Fig. S1D) (24). The Glycolysis Stress test was performed in standard assay media (DMEM, 2 mM Glutamine, pH 7.4) and the following final concentrations (glucose: 10 mM; Oligomycin: 2µM; 2-deoxy-glucose: 50 mM). Results were normalized as above and analyzed in WAVE software and processed through the XF Mito Stress Test Report and Glycolysis Stress Test Generators. All statistics were generated through GraphPad Prism 7 using unpaired two-tailed student t-tests.
Gene Ontology, Gene Set Enrichment Analysis, and Survival-Expression Analyses
Gene ontology analysis was performed on lists of differentially expressed genes from RNA-seq data (Lg2Rto: 0.585; AdjP≥10). Gene lists and data are available in Supplemental Data. Methodology of the PANTHER gene ontology platform and gene set enrichment analysis (GSEA) are described elsewhere (25,26). Ewing sarcoma patient survival data were compared with gene expression by analyzing a publicly available dataset (GSE17679) with the SurvExpress online tool (27,28). Risk groups were defined by prognostic index, with low-risk being defined as less than the median prognostic index, and high-risk being defined as greater than the median prognostic index.
RNA-Seq dataset
RNA-Seq of A673 cells treated with shLUC or shEF was previously performed (16). The datasets we acquired are publicly available in the NCBI BioProject database, accession # PRJNA176544, and the Sequence Read Archive (SRA059329). We re-analyzed these data to determine differential expression analysis using USeq (useq.sourceforge.net) version 8.8.3. Lists of differentially expressed genes are provided in Supplementary Data.
Metabolomics
All GC-MS analysis was performed with a Waters GCT Premier mass spectrometer fitted with an Agilent 6890 gas chromatograph and a Gerstel MPS2 autosampler. Dried samples were suspended in 40 µL of a 40 mg/mL O-methoxylamine hydrochloride (MOX) in pyridine and incubated for one hour at 30°C. To autosampler vials was added 25 uL of this solution. 40 µL of N-methyl-N-trimethylsilyltrifluoracetamide (MSTFA) was added automatically via the autosampler and incubated for 60 minutes at 37°C with shaking. After incubation 3 µL of a fatty acid methyl ester standard (FAMES) solution was added via the autosampler then 1 µL of the prepared sample was injected to the gas chromatograph inlet in the split mode with the inlet temperature held at 250°C. A 10:1 split ratio was used for analysis. The gas chromatograph had an initial temperature of 95°C for one minute followed by a 40°C/min ramp to 110°C and a hold time of 2 minutes. This was followed by a second 5°C/min ramp to 250°C, a third ramp to 350°C, then a final hold time of 3 minutes. A 30 m Phenomex ZB5-5 MSi column with a 5 m long guard column was employed for chromatographic separation. Helium was used as the carrier gas at 1 mL/min. Due to the high amounts of several metabolites the samples were analyzed once more at a 10-fold dilution. Data were collected using MassLynx 4.1 software (Waters). Metabolites were identified and their peak area was recorded using QuanLynx. This data was transferred to an Excel spread sheet (Microsoft, Redmond WA). Metabolite identity was established using a combination of an in-house metabolite library developed using pure purchased standards and the commercially available NIST library. Metabolomic data were further analyzed using MetaboAnalyst 3.0 (29).
Results
EWS/FLI misregulates numerous genes involved in metabolism
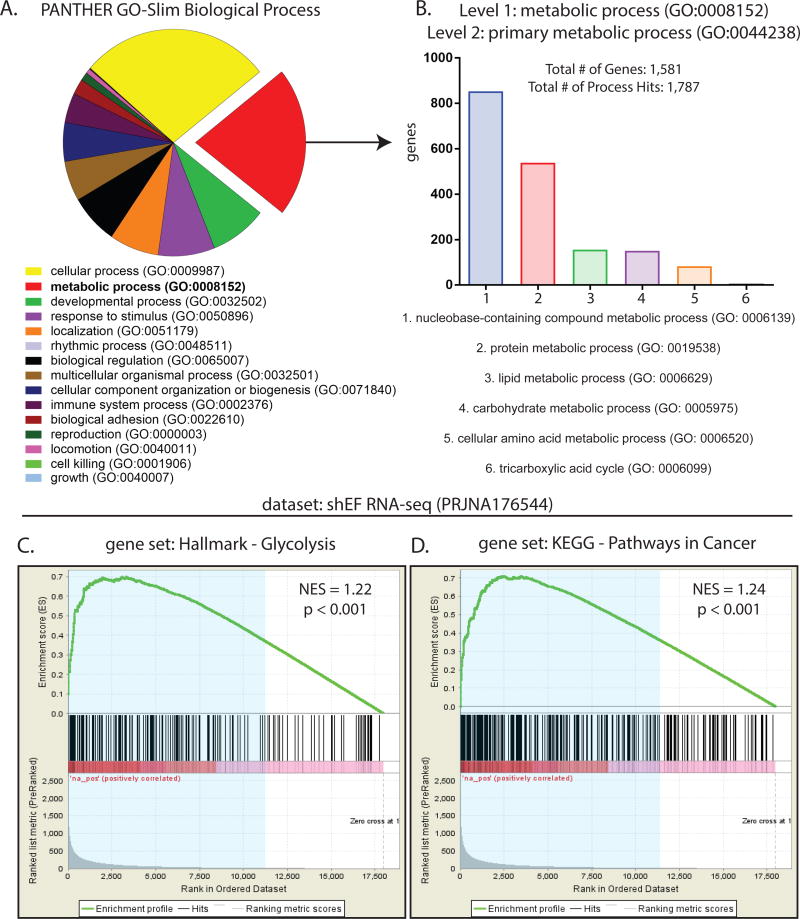
Transcription profiling of Ewing sarcoma cells has been previously performed by microarray and RNA-seq experiments after silencing EWS/FLI (EF) in A673 cells (14,16,30). We analyzed data from a publicly available RNA-seq dataset wherein EF was silenced by shRNA (shEF) and compared to control knockdown (shLUC). Our analysis identified 7,094 genes that are differentially expressed in shEF vs. control shLUC cells (gene lists found as Excel files in Supplemental Data). We defined significant changes in expression as a fold change of ≥ 1.5-fold and a p-value of < 0.05. After EF knockdown, 4,140 genes exhibited increased expression, while 2,954 genes were less expressed. Gene ontology (GO) analysis was performed on the list of all EF-misregulated genes, revealing metabolic processes (GO: 0008152) as the second most enriched GO term (2,058 genes; 33.7%) (Fig. 1A). These genes were further subdivided into primary metabolic processes (GO: 0044238), revealing that the majority of these genes were involved in nucleobase-containing compound metabolism (GO: 0006139) and protein metabolism (GO: 0019538) (Fig. 1B).
Figure 1. EWS/FLI exerts significant effects on expression of genes important for metabolism.
A. Gene ontology analysis of 5,698 EWS/FLI-regulated genes from publicly available RNA-seq data (27,37) using the PANTHER GO-Slim Biological Process annotation dataset. Threshold of significance for differential expression was log2(ratio) ≥ 0.585 and AdjP ≥ 10. Of these, 1,920 (33.7%) were annotated as metabolic processes (GO: 0008152). B. Among these metabolic processes (GO: 0008152), genes annotated as primary metabolic processes (GO: 0044238) were mostly involved in nucleobase-containing compound metabolism (GO: 0006139) and protein metabolism (GO: 0019538). C & D. Gene set enrichment analysis comparing a rank-ordered dataset (ranked by AdjP) of EWS/FLI knockdown RNA-seq data to the gene sets Hallmark-Glycolysis and KEGG-Pathways In Cancer, available from the Molecular Signatures Database (MSigDB) version 5.2. Differentially expressed genes (blue shaded region) in shEF vs. shLUC correlate significantly with genes important for glycolysis and pathways commonly altered in cancer. NES = normalized enrichment score.
To further assess whether EF significantly alters genes of metabolic importance, gene set enrichment analysis (GSEA) was performed comparing our dataset of differentially expressed genes in shEF cells to gene sets from the Molecular Signatures Database (MSigDB) (25). Significant correlations were observed between differentially expressed genes and genes in the Hallmark-Glycolysis gene set and the KEGG-Pathways in Cancer gene set. (Fig. 1C & 1D). Significant correlations were also observed with gene sets of Hallmark-Reactive Oxygen Species, Hallmark-Epithelial-Mesenchymal Transition, but not in the Hallmark-Oxidative Phosphorylation gene set (Fig. S1A).
Knockdown of EWS/FLI causes changes in cellular metabolism
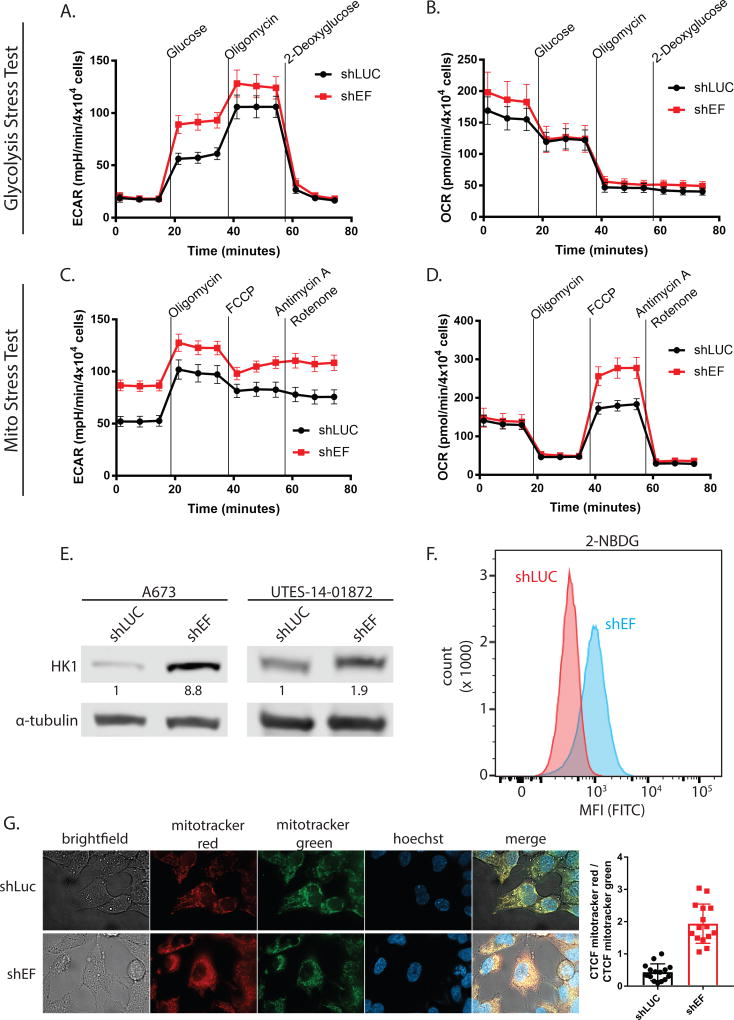
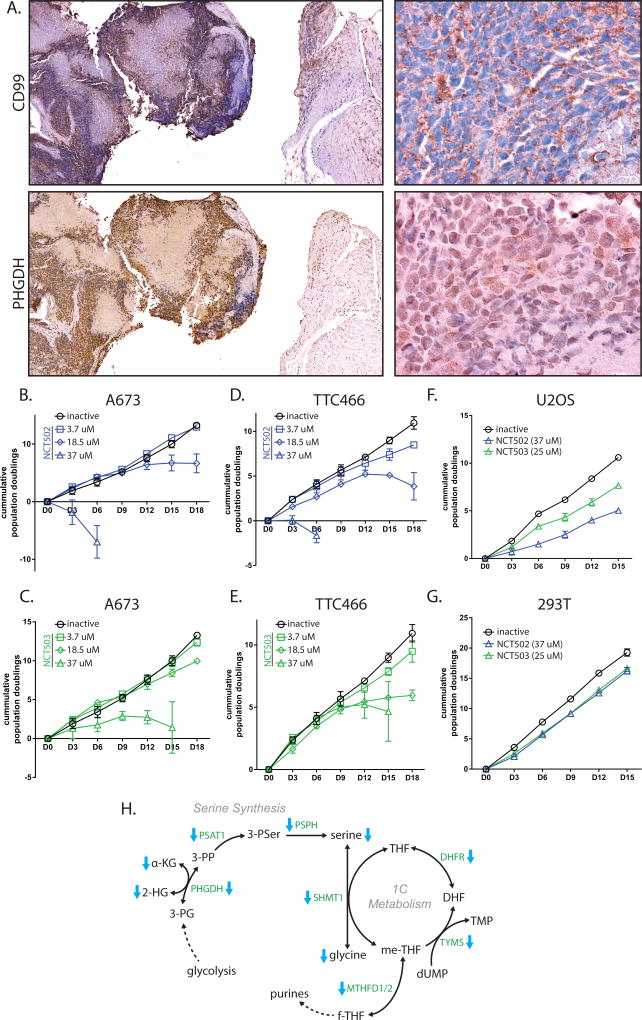
To more directly study whether EF alters cellular metabolism, we performed stress tests on glycolysis and mitochondrial function using the Seahorse XF96 analysis system. During the glycolysis stress test, addition of glucose and oligomycin caused greater increases of extracellular acidification rate (ECAR) in shEF vs. shLUC cells (Fig. 2A). Oxygen consumption rate was not different between shEF and shLUC cells throughout the glycolysis stress test (Fig. 2B). These data suggest that silencing of EF results in increased utilization of glucose by glycolysis (Table 1 & Fig. S1B). Basal ECAR was not different between shEF and shLUC cells at the beginning of the glycolysis stress test, reflecting the lower glucose (10 mM) conditions required for a glycolysis stress test (Fig. 2B). However, in the mitochondrial stress test glucose is present at typical cell culture levels (25 mM), allowing us to observe that basal ECAR is higher in shEF vs. shLUC cells (Fig. 2C). The apparent increase of glycolytic rate caused by EF knockdown is surprising, and is possibly explained by de-repression of hexokinase 1 (HK1) by shEF (Fig. 2E). Indeed, uptake of 2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-D-glucose (2-NBDG; a fluorescently-labeled analog of deoxyglucose) is elevated in shEF vs. shLUC A673 cells (Fig. 2F).
Figure 2. Silencing EWS/FLI results in functional metabolic changes.
A & B. Glycolysis stress tests demonstrate an increased extracellular acidification rate (ECAR) in response to a glucose bolus in shEF vs. shLUC A673 cells. No difference was seen in oxygen consumption rate in the glycolysis stress test. C & D. Basal ECAR is higher in shEF vs. shLUC cells, a reflection of different basal glucose concentrations in different test media. Mito stress tests reveal increased oxygen consumption rate (OCR) after treatment with the FCCP mitochondrial uncoupler, indicating increased maximal respiration in shEF vs. shLUC A673 cells. Basal respiration was similar between groups. E. Hexokinase 1 (HK1) was increased in A673 and UTES-14-01872 cells after EF knockdown. F. Uptake of the fluorescent glucose molecule 2-NBDG was increased in shEF vs. shLUC cells, as measured by flow cytometry. G. Mitotracker red signal was elevated in shEF vs. shLUC cells, suggesting increased mitochondrial membrane potential. A representative image from A673 cells is shown, and quantification was done on 3 cells from each of 5 high-power field images, shown as the corrected total cell fluorescence (CTCF) ratio of mitotracker red / mitotracker green (mean ± standard deviation).
Table 1.
Glycolysis and Mitochondrial Stress Tests
| shLuc | shEF | p-value | |
|---|---|---|---|
| Mito Stress Test: OCR (pmol/min/4×104 cells) | |||
| Maximal Respiration | 154.87 ± 13.60 | 243.46 ± 26.39 | 0.0046 |
| Spare Respiratory Capacity | 53.93 ± 7.64 | 140.05 ± 13.19 | <0.0001 |
| Spare Respiratory Capacity (%) | 153.75 ± 8.28 | 237.99 ± 20.39 | 0.0004 |
| Basal Respiration | 100.95 ± 9.54 | 103.41 ± 17.0 | 0.9002 |
| Proton Leak | 17.58 ± 3.42 | 14.47 ± 4.07 | 0.5615 |
| Non-Mitochondrial Respiration | 28.59 ± 3.86 | 34.41 ± 4.20 | 0.3132 |
| ATP Production | 83.37 ± 8.53 | 88.94 ± 13.93 | 0.7347 |
| Coupling Efficiency (%) | 82.58 ± 3.03 | 86.15 ± 2.51 | 0.3692 |
| Glycolysis Stress Test: ECAR (mpH/min/4×104 cells) | |||
| Glycolysis | 43.64 ± 4.71 | 74.95 ± 5.22 | <0.0001 |
| Non-Glycolytic Acidification | 17.57 ± 2.72 | 18.15 ± 2.83 | 0.8832 |
| Glycolytic Capacity | 88.36 ± 8.59 | 110.00 ± 10.30 | 0.1138 |
| Glycolytic Reserve | 44.72 ± 5.31 | 35.05 ± 6.45 | 0.2533 |
Data are mean ± SEM; OCR = oxygen consumption rate; ECAR = extracellular acidification rate
Tests of mitochondrial respiration showed enhanced response to mitochondrial uncoupling by carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP) in shEF vs. shLUC cells (Fig. 2D). These data indicate that silencing EF results in increased maximal respiration and spare respiratory capacity, with no change in basal respiration, ATP production, proton leak, non-mitochondrial respiration, and coupling efficiency (Table 1 & Fig. S1B). To additionally assess mitochondrial respiratory function, cells were treated with the membrane potential-sensitive Mitotracker Red FM dye. Comparisons were made relative to a membrane potential-independent mitochondrial dye (Mitotracker Green). Consistent with increased maximal respiration and respiratory capacity, shEF cells also had elevated mitochondrial membrane potential compared to shLUC cells (Fig. 2G). Together, these data suggest that EF increases the proportion of glucose-derived carbon that is shunted away from oxidative metabolism consistent with a biosynthetic, pro-oncogenic metabolic program.
EWS/FLI is required for global changes in metabolite abundance
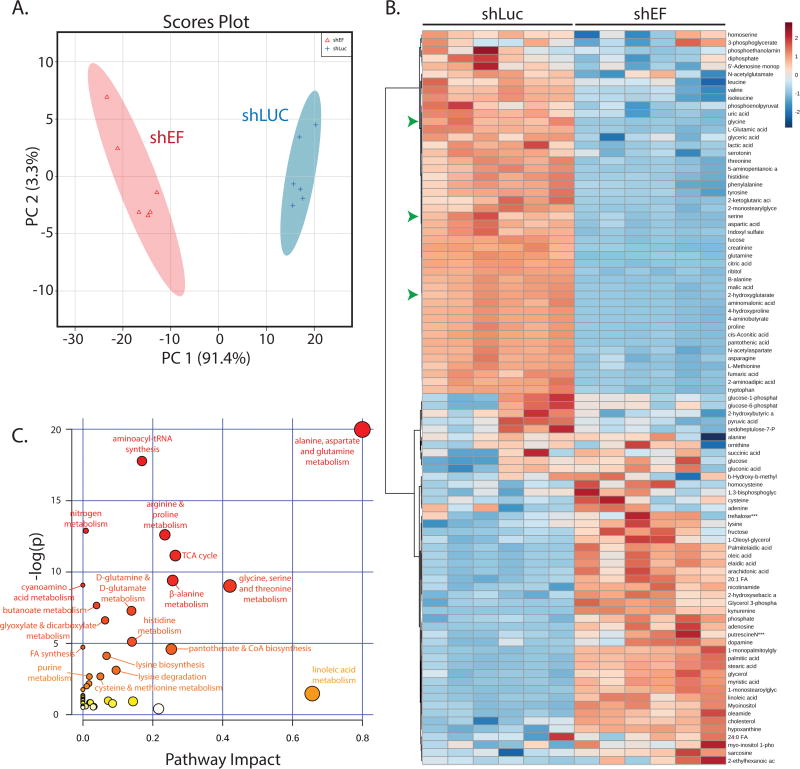
Global changes in metabolite abundance were assessed by performing steady-state metabolomics analyses comparing shEF with shLUC A673 cells. Examining metabolomic data by principal components analysis revealed that EF knockdown results in a distinct metabolic profile compared to control cells (Fig. 3A & 3B). Topological analysis of metabolites within metabolic pathways highlighted several metabolic pathways that were more significantly impacted by silencing of EF (Fig. 3C) (31). Pathways significantly affected by EF silencing included alanine, aspartate and glutamine metabolism, and glycine, serine and threonine metabolism.
Figure 3. The global profile of cellular metabolite abundance is altered by EWS/FLI.
A. Principal components analysis of metabolite abundance data (assessed by GC-MS) demonstrates distinct metabolic profiles in shEF vs. shLUC A673 cells. B. Hierarchichal clustering of differential metabolite abundance after EF knockdown was done by Euclidean distances and a Ward statistic. Metabolite abundances cluster into two distinct groups corresponding to shEF and shLUC conditions. Serine, glycine, and 2-hydroxyglutarate are indicated by green arrowheads. C. Topological analysis of metabolite data was performed by MetaboAnalyst 3.0. Circle color represents p value and node size represents pathway impact value. Pathway impact is calculated as the cumulative percentage of centrality measures of metabolites notes within pathways.
Additionally, metabolomic profiling data were combined with transcription profiling data to perform analyses of pathways in which both metabolite abundance and gene expression were significantly altered in shEF vs. shLUC cells. This allowed us to determine which pathways were impacted the most by shEF, potentially by altering expression of enzymes and metabolites in that pathway. Intriguingly, the enzymes of serine and glycine synthesis were found to be significantly downregulated by EF silencing in the RNA-seq dataset, and corresponding decreases in serine and glycine were observed in our metabolomic dataset (Fig. 3C & Fig. S2A).
EWS/FLI up-regulates expression of serine and glycine synthesis enzymes
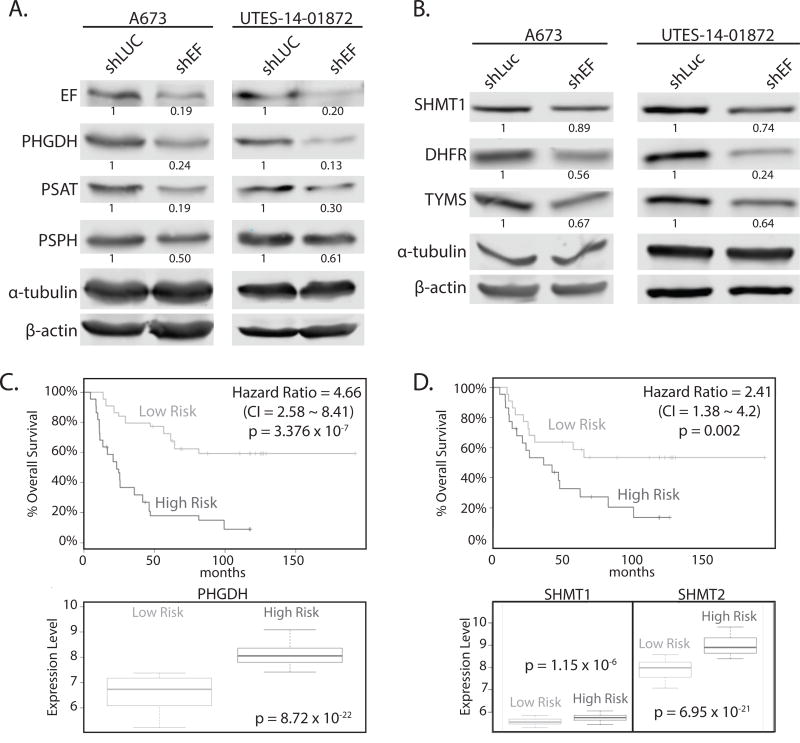
The de novo serine synthesis pathway is one way by which glycolytic intermediates can be shunted toward biosynthetic processes, and this has been implicated in many cancers (32,33). Specifically, phosphoglycerate dehydrogenase (PHGDH) commits the glycolytic intermediate 3-phosphoglycerate to serine and glycine synthesis, providing important precursors for protein, lipid and nucleic acid synthesis. Increased PHGDH has been linked to the oncogenic biology of other tumors (32,34). In fact, PHGDH expression is higher in Ewing sarcoma than all other cancer cell lines in the Cancer Cell Line Encyclopedia (Fig. S3A). In concordance with RNA-seq data, protein levels of PHGDH were decreased in shEF vs. shLUC cells (Fig. 4A). Protein levels of phosphoserine aminotransferase (PSAT) and phosphoserine phosphatase (PSPH) – the two enzymes downstream of PHGDH in the serine synthesis pathway – were also lower upon silencing of EF (Fig. 4A). Similar patterns of expression were observed for serine hyrdroxymethyltransferase (SHMT1 and SHMT2), which converts serine to glycine and feeds into 1-carbon folate metabolism cycles, as well as dihydrofolate reductase (DHFR) and thymidylate synthetase (TYMS) – important enzymes in 1-carbon folate metabolism cycles (Fig. 4B and Fig. S2D & S2E).
Figure 4. Key enzymes of serine & glycine synthesis, and 1-carbon metabolism, are altered by silencing EWS/FLI.
A. Western blots of enzymes of serine synthesis in A673 and UTES-14-01872 cells treated by shEF and shLUC. PHGDH, PSAT, and PSPH are lower in shEF vs. shLUC. B. SHMT1, DHFR and TYMS are also decreased in A673 and UTES-14-01872 cells after EF knockdown. C & D. SurvExpress analysis of overall survival of patients correlated with expression of PHGDH, SHMT1 and SHMT2, demonstrating higher expression correlates with poorer overall survival in Ewing sarcoma patients. CI = confidence interval.
PHGDH is elevated in high-risk Ewing sarcoma patients
To explore correlations between PHGDH expression and clinical outcome, we used the SurvExpress tool to perform Cox survival analysis of a Ewing sarcoma dataset (27,28). Patients who were categorized as high-risk had significantly higher expression of PHGDH and SHMT1/2 (Fig. 4C & D). Similarly, higher expression of PSPH, DHFR, TYMS, MTHFD2 and SHMT2 was correlated with poorer survival in Ewing sarcoma patients (Fig. S3B & S3C). Similar correlations were also observed between expression levels and event-free survival (data not shown).
Furthermore, PHGDH expression was assessed in histological samples from Ewing sarcoma patients. PHGDH staining was much higher in tumor (defined by IHC for CD99) than in adjacent normal tissue (Fig. 5A). CD99 exhibited staining along cell surface boundaries, while PHGDH staining appeared diffusely cytoplasmic.
Figure 5. PHGDH is high in Ewing sarcoma tumors, and is important for cell viability.
A. Immunohistochemistry of CD99 and PHGDH in Ewing sarcoma tumors from a Ewing sarcoma patient. Cell surface staining of CD99 is diagnostic for Ewing sarcoma. Cytosolic staining of PHGDH closely correlates with regions of tumor, indicating elevated PHGDH levels in tumor vs. adjacent tissue. B & C. PHGDH inibition in A673 cells by NCT502 and NCT503 causes decreased cell growth and eventual cell death. D & E. PHGDH inhibition also impaired cell growth of TTC466 cells. F & G. High-dose treatment of osteosarcoma cells (U2OS) and 293T cells show some growth defect, but not as much as in Ewing sarcoma cell lines. H. Diagramatic representation of serine/glycine synthesis and 1-carbon metabolism pathways that are decreased by EF knockdown. Enzymes are green text and metabolites are black text. Blue arrows indicate enzymes or metabolites that were observed as decreased in shEF vs. shLUC cells. Nucleotide and folate analogs were not measured. Co-factors and some intermediates are ommitted for clarity.
PHGDH is required for proliferation and normal chromatin modification
Inhibition of PHGDH function by NCT502 and NCT503 caused growth defects and cell death in Ewing sarcoma cell lines, including the EWS/ERG-harboring TTC466 line (Fig. 5B–5E). This effect was less prominent in 293T or U2OS cells (Fig. 5F & G). This and previous data indicate that upregulated expression of serine synthesis and 1-carbon metabolism contributes importantly to the oncogenic phenotype in Ewing sarcoma (Fig. 5H).
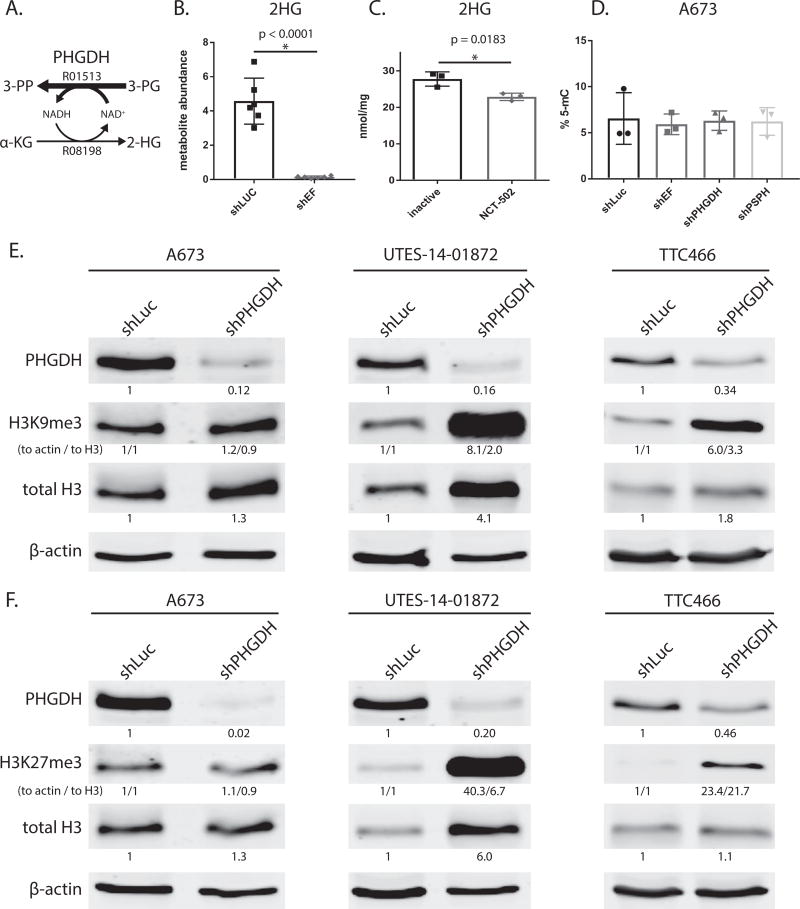
While the major product of PHGDH is 3 phosphohydroxypyruvate (3-PP) from 3-phosphoglycerate (3-PG), the enzyme also produces relatively small amounts of 2-hydroxyglutarate (2-HG) from α-ketoglutarate (Fig. 6A)(35). 2-HG has been shown to be oncogenic in high amounts, typically resulting from neomorphic mutations of isocitrate dehydrogenase enzymes (36). Interestingly, we observed that 2-HG was lower in shEF vs. shLUC cells (Fig. 6B), suggesting that EF might be driving elevated production of 2-HG via increased expression of PHGDH.
Figure 6. Modulation PHGDH causes decreased 2-hydroxyglutarate, and altered expression and methylation of histones.
A. Diagramatic representation of PHGDH enzyme function. PHGDH primarily catalyzes converion of 3-phospho-D-glycerate (3-PG) to 3-phosphoonoxypyruvate (3-PP), but also produces a minor product via conversion α-ketoglutarate (α-KG) to 2-hydroxyglutarate (2-HG). KEGG reaction numbers are indicated. B & C. 2-hydroxyglutarate abundance as measured by GC-MS is lower in shEF vs. shLUC A673 cells. Inhibition of PHGDH by NCT-502 also decreases levels of 2-HG in A673 cells. D. LINE-1 methylation, a surrogate of bulk DNA methylation, is not different in shEF, shPHGDH or shPSPH vs. shLUC cells. E & F. In UTES-14-01872 and TTC466 cells, knockdown of PHGDH results in increased levels of histone H3 and increased methylation of H3 histones at lysine 9 and 27. This was not seen in A673 cells. A673 and UTES-14-01872 cell lines express EWS/FLI; TTC466 cells express the EWS/ERG fusion protein. For H3K9me3 and H3K27me3, both normalization to actin and normalization to total H3 are shown (indicated as “to actin / to total H3”).
To test whether elevated levels of 2-HG in Ewing sarcoma cells was due to PHGDH, we assessed 2-HG levels after inhibition of PHGDH function with NCT502. 2-HG was lower in cells treated by NCT502 vs. its inactive analog (Fig. 6C). Importantly, the levels of 2-HG in Ewing sarcoma cells was lower than previously observed in cells harboring mutations in isocitrate dehydrogenase (IDH) enzymes (37). To test whether 2-HG generated from PHGDH had effects on DNA methylation, we silenced PHGDH expression and determined 5-methylcytosine (5-mC) content of LINE-1 elements (a surrogate for bulk DNA methylation). However, knockdown of EF or PHGDH had no effect on 5-mC levels (Fig. 6D). PSPH was also silenced to distinguish between serine synthesis and 2-HG producing functions of PHGDH, and disruption of serine synthesis downstream of PHGDH also had no effect on 5-mC levels.
To further assess whether modulation of PHGDH affects chromatin, we immunoblotted histone proteins. This was performed with A673 cells, as well as another more recently established patient-derived EF-harboring Ewing sarcoma cell line (UTES-14-01872) (Fig. S2C). Interestingly, methylation of histone H3 at lysine (K) 9 and K27 was increased by PHGDH knockdown in UTES-14-01872 and TTC466 cells, but not in A673 cells (Fig. 6E & F). Additionally, total histone protein was increased by PHGDH knockdown in two of the three cell lines. These data raise the possibility of a role for PHGDH in chromatin regulation, possibly by affecting abundance of metabolites used as cofactors for histone modifying enzymes.
Discussion
EF has long been recognized as the primary oncogenic driver of Ewing sarcoma. Overexpression of EF in NIH3T3 murine fibroblasts results in oncogenic transformation, and silencing EF expression in patient-derived cell lines results in a loss of tumorigenesis (5,38,39). However, effective therapeutic strategies directly targeting transcription factors have been notoriously elusive. For this reason, studies of the biology of Ewing sarcoma have primarily focused on identifying important EF target genes, as well as the interacting partners EF requires to elicit its effects. Indeed, numerous EF targets have been shown to be necessary, but not sufficient, for oncogenesis in these tumors (6,14,40,41). Unfortunately, many of these critical EF targets and partners lack enzymatic function and are themselves not susceptible to pharmacological modulation. With this in mind, we sought to determine whether EF generates an oncogenic program of cellular metabolism by misregulating the transcription of genes encoding key metabolic enzymes.
By analyzing existing EF transcriptional datasets, we were able to identify significant patterns of altered metabolic gene expression (Fig. 1). Specifically, gene ontology analysis revealed significant alterations in the expression of genes involved in glycolysis, nucleobase-containing metabolic processes, and protein metabolism. Consistent with a cancerous phenotype, Ewing sarcoma cells are hyperproliferative and require increased macromolecule synthesis to sustain accelerated growth and proliferation. Additionally, EF-driven alterations in nucleobase-containing metabolic processes such as (NAD salvage) could be sensitive to inhibition and therapeutic exploitation (42). The increased metabolic demands of rapid proliferation and growth could be exploited in similar ways.
We also observed that EF knockdown resulted in increased maximal respiration and mitochondrial membrane potential (Fig. 2D & 2G). This is in line with studies of other malignancies, wherein various oncogenic processes lead to a metabolic program of aerobic glycolysis – diverting metabolites toward pathways that are important for various kinds of macromolecule synthesis (11). As more carbon is diverted to biosynthetic pathways from glycolysis, flux through the TCA cycle may decrease, resulting in oncogene-induced reduction in respiratory capacity, as suggested by our data. These findings reinforce the concept that such a shift away from oxidative metabolism is a characteristic feature of oncogenesis, important in all cancers, and not a spectator phenomenon merely correlating with oncogenic processes. Furthermore, they demonstrate that EF is a driver of oncogenic metabolic reprogramming, in addition to and as a consequence of transcriptional reprogramming.
Surprisingly, we also observed an increase of glycolysis in shEF vs. shLUC cells (Fig. 2A & C). Such observations appear to run contrary to the known glycolytic metabolism that predominates in the vast majority of cancer cells. These observations could be explained by transcriptional downregulation of genes important for glucose import or utilization. Indeed, we observed that hexokinase 1 (HK1) is increased in shEF vs. shLUC cells (Fig. 2E). In such a scenario, EF may restrict glucose entry into glycolysis, effectively reducing the cell’s ability to metabolize additional glucose (as seen in our Seahorse analysis data). However, if this restriction is not so severe to cause cell death by starvation, glucose uptake may remain sufficient to fulfill cellular energy requirements. Simultaneously, EF upregulates genes of biosynthetic pathways such as de novo serine synthesis (Fig. 4A), resulting in an increased proportion of glycolytic intermediates being shunted into biosynthetic pathways. Thus it may be possible to generate the net effect of oncogenic and biosynthetic reprogramming of metabolism by increasing the proportion of glucose-derived carbon shunted towards biosynthetic pathways, even in the face of EF-induced downregulation of HK1. Additionally, considering that the putative cell of origin for Ewing sarcoma is a mesenchymal stem cell, and that EF is known to repress mesenchymal features, the baseline glycolytic metabolism of these stem cells may provide ample flexibility to be permissive to decreased HK1 within the context of oncogenic metabolic reprogramming induced by EF (24,43). Thus, any decrease in glycolysis caused by EF is not likely to be pro-oncogenic or pro-proliferative. Instead, we believe it is merely part of a constellation of changes, not all being programatically consistent, that together contribute to transformation. Rather than increasing glucose utilization in order to provide metabolites for biosynthetic pathways, EF could increase the proportion of glycolytic intermediates being diverted to pathways such as de novo serine synthesis. These and other hypotheses are a subject of current and future study.
Our findings also suggest that EF drives global shifts in metabolite abundance in Ewing sarcoma cells (Fig. 3). Integrating metabolomic and transcriptomic data enabled us to identify EF-driven upregulation of de novo serine synthesis via PHGDH as an important candidate for further study. PHGDH is amplified in various tumors, and has been implicated previously in tumor biology (32,35). PHGDH commits the glycolytic intermediate 3-PG to de novo serine synthesis, and EF-driven upregulation of PHGDH expression appears to be an important factor in diverting metabolites toward macromolecule synthesis (Fig. 4A). Importantly, elevated expression of PHGDH also correlates with a poor prognosis in Ewing sarcoma patients, suggesting that these phenomena have important clinical implications (Fig. 4C). Our findings are similar to previous findings demonstrating upregulation of serine synthesis by other oncogenes. PHGDH, PSPH and PSAT are upregulated in some Myc-driven lymphomas (44). Additionally, oncogene-driven transcriptional misregulation in other malignancies (e.g., HER2, SP1, NFY, NRF2, ATF4) has been shown to increase expression of enzymes of de novo serine synthesis (45–47). EF thus appears to generate a metabolic program of upregulated serine synthesis similar to other malignancies. Existing transcription profiling data suggest that SP1 is an upregulated target of EF, while NFY, NRF2 and ATF4 are not differentially expressed after EF knockdown; HER2 may be slightly downregulated by EF (data not shown). Further study is required to determine whether EF exerts its effects by coopting any of the aforementioned regulatory factors. Moreover, existing ChIP-seq data are inconsistent on whether EF binds directly at the promoter of PHGDH (48,49). However, the fact that EF binding does appear to occur in some datasets suggests that it could be directly regulating expression of PHGDH, and further assessment of EF-directed regulation of PHGDH deserves more in-depth study.
Downstream of de novo serine synthesis, conversion of serine to glycine is carried out by serine hydroxymethyltransferase (SHMT). This reaction is coupled to the conversion of tetrahydrofolate to 5,10-methylenetetrahydrofolate, and is a major provider of 1-carbon units needed for multiple cellular functions (50). Our data suggest that EF is also driving upregulation of these processes, further establishing a pro-oncogenic, biosynthetic metabolic program in these cells (Fig. 5H). It is likely that this is also clinically relevant, as patients with high levels of SHMT also have worse overall survival (Fig. 4D). Together these data also raise the interesting possibility of utilizing expression levels of metabolic enzymes such as PHGDH and SHMT to inform predictions of patient prognosis, or even using metabolic indices to define different patient populations.
The nature of the relationship between patient survival and expression levels of these genes deserves further consideration. Several hypotheses exist for why increased expression of these enzymes correlate with poorer prognosis. For instance, upregulation of de novo serine synthesis may contribute importantly to increased synthesis of glycine and cysteine. Such a process would contribute more amino acids to meet the biosynthetic requirements of proliferative cancer cells. Additionally, since glycine and cysteine both contribute to production of glutathione, increased serine synthesis also potentially contributes to greater redox buffering. Indeed, serine metabolism has been shown to be activated under conditions of increased ROS production, perhaps contributing to increased capacity to confront redox stress (51). Furthermore, the conversion of serine to glycine contributes importantly to one-carbon metabolism and the folate cycle. The de novo synthesis of adenosine, guanosine and thymidylate require one-carbon units derived from serine. Thus, increased serine synthesis could contribute to greater nucleotide synthesis in general. Upregulation of SHMT1/2 and TYMS is consistent with these hypotheses. Upregulation of such processes could permit cancer cells to become more proliferative or aggressive, thus contributing to poorer prognosis in patients. However, as these hypotheses have yet to be tested, more in-depth investigation is clearly needed.
Upregulation of de novo serine synthesis and 1-carbon metabolism has far-reaching consequences for cancer cells, as flux through these pathways is important for regulation of methyl donor (methionine) recycling and epigenetic states. In addition, previous work has suggested that amplified PHGDH may play a role in cancer through production of 2-HG (52). 2-HG is a known oncometabolite when overproduced by neomorphic mutations in IDH enzymes (36). Intriguingly, our data also suggest that important links exist between de novo serine synthesis (specifically PHGDH) and nuclear processes. We observed decreased amounts of 2-HG in shEF vs. shLUC cells (Fig. 6B & C). However, the amounts of 2-HG generated by EF-induced upregulation of PHGDH are lower than levels seen in other malignancies with IDH mutations, and 2-HG likely has less of a role in Ewing sarcoma biology. Additionally, IDH mutations are rare in Ewing sarcoma (53). Some work has shown that the small amounts of 2-HG produced by PHGDH may be sufficient to promote histone methylation, contributing to an oncogenic hypermethylation of histones and, possibly, DNA (52). However, our results suggest that this is not the case in Ewing sarcoma. Rather, knockdown of PHGDH did not affect DNA methylation (Fig. 6D), but did cause significant increases in histone methylation (Fig. 6E & F). Interestingly, these histone alterations were observed in EF-expressing cells (UTES-14-01872) as well as in TTC466 cells, which harbor the alternate translocation EWS/ERG (found in ~10% of Ewing sarcomas). While the mechanisms responsible for these chromatin changes are unclear, we hypothesize that abundance of metabolites important for the function of other enzymes (either as substrates or cofactors) might play a role. Future efforts are aimed at testing these hypotheses and further elucidating the mechanisms linking metabolism and nuclear control.
Taken together our findings demonstrate that EF drives metabolic misregulation resulting in diversion of metabolites toward biosynthetic pathways in order to meet the increased demands of oncogenesis. EF induces increased expression of PHGDH, PSAT and PSPH, shunting glycolytic intermediates into serine and glycine synthesis, and contributing to 1-carbon metabolism through upregulated SHMT. While most research of Ewing sarcoma has previously focused on transcriptional regulation, epigenetic modification and other nuclear processes, little attention has been focused on metabolic pathways. Indeed, metabolic systems that are co-opted or altered by the presence of EWS/FLI might include targetable enzymes that could be modulated by new therapies. An EWS/FLI-specific metabolic program could provide a targetable surrogate for EWS/FLI itself, providing a way to specifically destroy EWS/FLI-harboring cells through pharmacological modulation of metabolic or other enzymes. Such efforts aspire to inform the development of hitherto elusive molecular targeted therapies for Ewing sarcoma.
Supplementary Material
Implications.
This previously unexplored role of EWS/FLI1-driven metabolic changes expands the understanding of Ewing sarcoma biology, and has potential to significantly inform development of therapeutic strategies.
Acknowledgments
The authors would like to thank Dr. Kevin Jones, Dr. Emily Theisen, Dr. Ranajeet Saund, and Dr. Kathleen Pishas, and all members of the Lessnick and Rutter labs for many useful and thought-provoking discussions throughout the course of these studies.
Financial Support: J. Rutter was supported by HHMI and NIH grant GM 110755. J. Schiffman was supported by St. Baldrick’s Foundation, Sarcoma Alliance for Research through Collaboration (SARC) Career Development Award, Damon Runyon Clinical Investigator Award, and the Eunice Kennedy Shriver Children’s Health Research Career Development Award NICHD 5K12HD001410. Metabolomics analysis was performed at the Metabolomics Core Facility at the University of Utah which is supported by 1 S10 OD016232-01, 1 S10 OD021505-01 and 1 U54 DK110858-01. The authors also acknowledge the support of the Huntsman Cancer Institute (P30CA042014) & the Huntsman Cancer Foundation.
References
- 1.Linabery AM, Ross JA. Childhood and adolescent cancer survival in the US by race and ethnicity for the diagnostic period 1975–1999. Cancer. 2008;113:2575–96. doi: 10.1002/cncr.23866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lahl M, Fisher VL, Laschinger K. Ewing’s sarcoma family of tumors: an overview from diagnosis to survivorship. Clin J Oncol Nurs. 2008;12:89–97. doi: 10.1188/08.CJON.89-97. [DOI] [PubMed] [Google Scholar]
- 3.Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, et al. The Genomic Landscape of the Ewing Sarcoma Family of Tumors Reveals Recurrent STAG2 Mutation. PLOS Genet. 2014;10:e1004475. doi: 10.1371/journal.pgen.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gangwal K, Sankar S, Hollenhorst PC, Kinsey M, Haroldsen SC, Shah AA, et al. Microsatellites as EWS/FLI response elements in Ewing’s sarcoma. Proc Natl Acad Sci. 2008;105:10149–54. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hancock JD, Lessnick SL. A transcriptional profiling meta-analysis reveals a core EWS-FLI gene expression signature. Cell Cycle Georget Tex. 2008;7:250–6. doi: 10.4161/cc.7.2.5229. [DOI] [PubMed] [Google Scholar]
- 6.Cironi L, Riggi N, Provero P, Wolf N, Suvà M-L, Suvà D, et al. IGF1 is a common target gene of Ewing’s sarcoma fusion proteins in mesenchymal progenitor cells. PloS One. 2008;3:e2634. doi: 10.1371/journal.pone.0002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lissat A, Joerschke M, Shinde DA, Braunschweig T, Meier A, Makowska A, et al. IL6 secreted by Ewing sarcoma tumor microenvironment confers anti-apoptotic and cell-disseminating paracrine responses in Ewing sarcoma cells. [cited 2017 Mar 29];BMC Cancer [Internet] 2015 :15. doi: 10.1186/s12885-015-1564-7. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4517368/ [DOI] [PMC free article] [PubMed]
- 8.Douglas D, Hsu JH-R, Hung L, Cooper A, Abdueva D, van Doorninck J, et al. BMI-1 promotes Ewing sarcoma tumorigenicity independent of CDKN2A-repression. Cancer Res. 2008;68:6507–15. doi: 10.1158/0008-5472.CAN-07-6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katuri V, Gerber S, Qiu X, McCarty G, Goldstein SD, Hammers H, et al. WT1 regulates angiogenesis in Ewing Sarcoma. Oncotarget. 2014;5:2436–49. doi: 10.18632/oncotarget.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters HL, Yan Y, Nordgren TM, Cutucache CE, Joshi SS, Solheim JC. Amyloid precursor-like protein 2 suppresses irradiation-induced apoptosis in Ewing sarcoma cells and is elevated in immune-evasive Ewing sarcoma cells. Cancer Biol Ther. 2013;14:752–60. doi: 10.4161/cbt.25183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uphoff CC, Drexler HG. Detecting mycoplasma contamination in cell cultures by polymerase chain reaction. Methods Mol Biol Clifton NJ. 2011;731:93–103. doi: 10.1007/978-1-61779-080-5_8. [DOI] [PubMed] [Google Scholar]
- 13.Lessnick SL, Dacwag CS, Golub TR. The Ewing’s sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell. 2002;1:393–401. doi: 10.1016/s1535-6108(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 14.Smith R, Owen LA, Trem DJ, Wong JS, Whangbo JS, Golub TR, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing’s sarcoma. Cancer Cell. 2006;9:405–16. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Braunreiter CL, Hancock JD, Coffin CM, Boucher KM, Lessnick SL. Expression of EWS-ETS fusions in NIH3T3 cells reveals significant differences to Ewing’s sarcoma. Cell Cycle Georget Tex. 2006;5:2753–9. doi: 10.4161/cc.5.23.3505. [DOI] [PubMed] [Google Scholar]
- 16.Sankar S, Theisen ER, Bearss J, Mulvihill T, Hoffman LM, Sorna V, et al. Reversible LSD1 Inhibition Interferes with Global EWS/ETS Transcriptional Activity and Impedes Ewing Sarcoma Tumor Growth. Clin Cancer Res. 2014;20:4584–97. doi: 10.1158/1078-0432.CCR-14-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan B, Latek R, Hossbach M, Tuschl T, Lewitter F. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 2004;32:W130–4. doi: 10.1093/nar/gkh366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13:1400–12. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janes KA. An analysis of critical factors for quantitative immunoblotting. Sci Signal. 2015;8:rs2. doi: 10.1126/scisignal.2005966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O J-PDL, Emerson LL, Goodman JL, Froebe SC, Illum BE, Curtis AB, et al. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci. 2008;105:18907–12. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keefe MD, Wang H, O J-PDL, Khan A, Firpo MA, Murtaugh LC. β-catenin is selectively required for the expansion and regeneration of mature pancreatic acinar cells in mice. Dis Model Mech. 2012;5:503–14. doi: 10.1242/dmm.007799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopinke D, Brailsford M, Pan FC, Magnuson MA, Wright CVE, Murtaugh LC. Ongoing Notch signaling maintains phenotypic fidelity in the adult exocrine pancreas. Dev Biol. 2012;362:57–64. doi: 10.1016/j.ydbio.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pacold ME, Brimacombe KR, Chan SH, Rohde JM, Lewis CA, Swier LJYM, et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol. 2016;12:452–8. doi: 10.1038/nchembio.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaturvedi A, Hoffman LM, Welm AL, Lessnick SL, Beckerle MC. The EWS/FLI Oncogene Drives Changes in Cellular Morphology, Adhesion, and Migration in Ewing Sarcoma. Genes Cancer. 2012;3:102–16. doi: 10.1177/1947601912457024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–9. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos A, et al. SurvExpress: An Online Biomarker Validation Tool and Database for Cancer Gene Expression Data Using Survival Analysis. PLOS ONE. 2013;8:e74250. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savola S, Klami A, Myllykangas S, Manara C, Scotlandi K, Picci P, et al. High Expression of Complement Component 5 (C5) at Tumor Site Associates with Superior Survival in Ewing’s Sarcoma Family of Tumour Patients. ISRN Oncol. 2011;2011:168712. doi: 10.5402/2011/168712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia J, Wishart DS. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr Protoc Bioinforma. 2016;55:14.10.1–14.10.91. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- 30.Sankar S, Gomez NC, Bell R, Patel M, Davis IJ, Lessnick SL, et al. EWS and RE1-Silencing Transcription Factor Inhibit Neuronal Phenotype Development and Oncogenic Transformation in Ewing Sarcoma. Genes Cancer. 2013;4:213–23. doi: 10.1177/1947601913489569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6:743–60. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 32.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–74. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Locasale JW. Serine, glycine and the one-carbon cycle: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–83. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–50. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan J, Teng X, Liu L, Mattaini KR, Looper RE, Vander Heiden MG, et al. Human Phosphoglycerate Dehydrogenase Produces the Oncometabolite d-2-Hydroxyglutarate. ACS Chem Biol. 2015;10:510–6. doi: 10.1021/cb500683c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S-H, et al. Oncometabolite 2-Hydroxyglutarate Is a Competitive Inhibitor of α-Ketoglutarate-Dependent Dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.May WA, Lessnick SL, Braun BS, Klemsz M, Lewis BC, Lunsford LB, et al. The Ewing’s sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol. 1993;13:7393–8. doi: 10.1128/mcb.13.12.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chansky HA, Barahmand-pour F, Mei Q, Kahn-Farooqi W, Zielinska-Kwiatkowska A, Blackburn M, et al. Targeting of EWS/FLI-1 by RNA interference attenuates the tumor phenotype of Ewing’s sarcoma cells in vitro. J Orthop Res. 2004;22:910–7. doi: 10.1016/j.orthres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Kinsey M, Smith R, Lessnick SL. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing’s sarcoma. Mol Cancer Res MCR. 2006;4:851–9. doi: 10.1158/1541-7786.MCR-06-0090. [DOI] [PubMed] [Google Scholar]
- 41.Beauchamp E, Bulut G, Abaan O, Chen K, Merchant A, Matsui W, et al. GLI1 is a direct transcriptional target of EWS-FLI1 oncoprotein. J Biol Chem. 2009;284:9074–82. doi: 10.1074/jbc.M806233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mutz CN, Schwentner R, Aryee DNT, Bouchard EDJ, Mejia EM, Hatch GM, et al. EWS-FLI1 confers exquisite sensitivity to NAMPT inhibition in Ewing sarcoma cells. Oncotarget. 2017 doi: 10.18632/oncotarget.14976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riggi N, Cironi L, Provero P, Suvà M-L, Kaloulis K, Garcia-Echeverria C, et al. Development of Ewing’s sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res. 2005;65:11459–68. doi: 10.1158/0008-5472.CAN-05-1696. [DOI] [PubMed] [Google Scholar]
- 44.Sun L, Song L, Wan Q, Wu G, Li X, Wang Y, et al. cMyc-mediated activation of serine biosynthesis pathway is critical for cancer progression under nutrient deprivation conditions. Cell Res. 2015;25:429–44. doi: 10.1038/cr.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jun DY, Park HS, Lee JY, Baek JY, Park H-K, Fukui K, et al. Positive regulation of promoter activity of human 3-phosphoglycerate dehydrogenase (PHGDH) gene is mediated by transcription factors Sp1 and NF-Y. Gene. 2008;414:106–14. doi: 10.1016/j.gene.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 46.Bollig-Fischer A, Dewey TG, Ethier SP. Oncogene activation induces metabolic transformation resulting in insulin-independence in human breast cancer cells. PloS One. 2011;6:e17959. doi: 10.1371/journal.pone.0017959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeNicola GM, Chen P-H, Mullarky E, Sudderth JA, Hu Z, Wu D, et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet. 2015;47:1475–81. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bilke S, Schwentner R, Yang F, Kauer M, Jug G, Walker RL, et al. Oncogenic ETS fusions deregulate E2F3 target genes in Ewing sarcoma and prostate cancer. Genome Res. 2013;23:1797–809. doi: 10.1101/gr.151340.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suvà ML, et al. EWS-FLI1 Utilizes Divergent Chromatin Remodeling Mechanisms to Directly Activate or Repress Enhancer Elements in Ewing Sarcoma. Cancer Cell. 2014;26:668–81. doi: 10.1016/j.ccell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang M, Vousden KH. Serine and one-carbon metabolism in cancer. Nat Rev Cancer. 2016;16:650–62. doi: 10.1038/nrc.2016.81. [DOI] [PubMed] [Google Scholar]
- 51.Ye J, Fan J, Venneti S, Wan Y-W, Pawel BR, Zhang J, et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 2014;4:1406–17. doi: 10.1158/2159-8290.CD-14-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan J, Teng X, Liu L, Looper R, Rabinowitz J. Human phosphoglycerate dehydrogenase produces the oncometabolite D-2-hydroxyglutarate and promotes histone methylation. Cancer Metab. 2014;2:P75. doi: 10.1021/cb500683c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Na KY, Noh B-J, Sung J-Y, Kim YW, Santini Araujo E, Park Y-K. IDH Mutation Analysis in Ewing Sarcoma Family Tumors. J Pathol Transl Med. 2015;49:257–61. doi: 10.4132/jptm.2015.04.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.