Abstract
Epidermal growth factor receptor (EGFR) signaling has been implicated in hypoxia-associated resistance to radiation or chemotherapy. Non-small cell lung carcinomas (NSCLC) with activating L858R or ΔE746–E750 EGFR mutations exhibit elevated EGFR activity and downstream signaling. Here, relative to wild type (WT) EGFR, mutant (MT) EGFR expression significantly increases radiosensitivity in hypoxic cells. Gene expression profiling in human bronchial epithelial cells (HBEC) revealed that MT-EGFR expression elevated transcripts related to cell cycle and replication in aerobic and hypoxic conditions and down-regulated RAD50, a critical component of non-homologous end-joining (NHEJ) and homologous recombination (HR) DNA repair pathways. NSCLCs and HBEC with MT-EGFR revealed elevated basal and hypoxia-induced γ-H2AX-associated DNA lesions that were coincident with replication protein A in S-phase nuclei. DNA fiber analysis showed that, relative to WT-EGFR, MT-EGFR NSCLCs harbored significantly higher levels of stalled replication forks and decreased fork velocities in aerobic and hypoxic conditions. EGFR blockade by cetuximab significantly increased radiosensitivity in hypoxic cells, recapitulating MT-EGFR expression and closely resembling synthetic lethality of PARP inhibition.
Keywords: EGFR mutations, Non-small cell lung carcinoma, hypoxia-associated radioresistance, DNA repair, replication stress
INTRODUCTION
Relative to well-oxygenated tumors, hypoxic tumors tend to be significantly more resistant to ionizing radiation (IR). Radiation-induced free radicals chemically react with DNA and cause DNA damage through the formation of a DNA radical. For DNA damage to persist, oxygen is required to oxidize the DNA radical and extend its half-life. Hypoxic cells are more radioresistant because, in conditions of low oxygen, oxidation of the DNA radical and radiation-induced DNA damage are limited (1). Oxygen enhances radiosensitivity by a radiation dose modifying factor called oxygen enhancement ratio (OER). OER is defined as the ratio of the radiation dose under anoxic or hypoxic conditions to the dose under conditions of a specific partial pressure of oxygen to produce the same biological effect. Alternatively, the hypoxia reduction factor (HRF) has been defined as the ratio of radiation dose at a specific partial pressure of oxygen to the dose under fully aerobic conditions (21% O2) for the same biological effect (2).
Prolonged exposure to hypoxia itself can be cytotoxic and tumor survival depends on an adaptive pro-survival response to hypoxia that could also augment radioresistance. Activation and downstream signaling of the epidermal growth factor receptor (EGFR) in many cancers, including non-small cell lung cancer (NSCLC), has been linked to survival responses to hypoxia. These include inhibition of hypoxia-induced cell death (3), stimulation of pro-proliferative pathways (4), induction of epithelial-mesenchymal transition (5) and establishment of a positive feedback loop through induction of the hypoxia inducible factor 1 (HIF-1α) (6).
EGFR also has roles in the repair of radiation-induced DNA double strand breaks (DSBs). DSB repair in tumors is catalyzed by two signaling pathways, non-homologous end joining (NHEJ), and homologous recombination (HR). Several studies have demonstrated a role for EGFR in NHEJ, which involves radiation-induced nuclear translocation of EGFR, interactions with the NHEJ enzyme, DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and modulation of DNA-PKcs phosphorylation (7–11). Other studies have shown that EGFR-mediated DSB repair may also involve HR (12,13). Recent studies have uncovered a less understood, kinase-independent role for EGFR in HR-mediated repair of cisplatin induced DNA inter-strand crosslinks, but not radiation induced DSBs (14).
We previously demonstrated that NHEJ-mediated DSB repair is defective in NSCLCs harboring somatic, activating mutations in EGFR that were clinically linked to sensitivity to EGFR tyrosine kinase inhibitors (TKI) (15–17). We showed that, despite elevated EGFR tyrosine phosphorylation and downstream signaling, NSCLCs harboring ΔE746–E750, L858R or the TKI-resistant T790M mutant (MT) EGFR are profoundly radiosensitive (18). MT-EGFR-associated radiosensitivity manifests as pronounced delays in repair of radiation-induced DSBs, failure of EGFR nuclear import, abrogation of EGFR-DNA-PKcs interactions (8) and inhibition of DNA-PKcs phosphorylation at Threonine 2609 (T2609) (11), a pattern which is remarkably similar to EGFR blockade by the monoclonal antibody, cetuximab (19). How expression of signaling-hyperactive, NHEJ-defective MT-EGFR affects hypoxia response is not known.
Evidence from a number of studies shows that hypoxia induces a unique DNA damage response (DDR), which is a cascade of signaling events that collectively orchestrate DNA damage recognition, DNA repair, cell cycle arrest or apoptosis (20). DNA DSBs do not usually occur in hypoxic cells (21). However, in response to hypoxia, decrease in ribonucleotide reductase (RNR) activity (22) causes nucleotide insufficiency resulting in a rapid halt of DNA synthesis and transient cell cycle arrest through replication fork-bound replication protein A (RPA) and activation of the ataxia telangiectasia related-checkpoint kinase (ATR) Cdc25 pathway (ATR/Chk1/Cdc25 pathway) (23). Stalled replication forks are at a high risk for DNA DSBs and repair of fork-associated DSBs is essential for survival. During conditions of transient hypoxia (< 12 hours), HR but not NHEJ appears to be critical for survival (24). By contrast, during prolonged hypoxia (>12 hours), many components of the HR pathway are down-regulated with a significant up-regulation of several NHEJ components (25).
This study investigates the effect of activating L858R and ΔE746–E750 mutations on radiation resistance in conditions of hypoxia. Our study has uncovered a novel relationship between MT-EGFR expression and replication stress in oxygen-replete and hypoxic conditions. We demonstrate that in the context of an altered DDR in hypoxic cells, expression of NHEJ-defective MT-EGFR, or an EGFR blockade by cetuximab, exerts a synthetic lethality effect and sensitizes hypoxic cells to radiation. This study has important clinical implications in the treatment of NSCLC patients, especially those at greatest risk of therapy failure due to tumor hypoxia.
MATERIAL AND METHODS
Cell Culture
NSCLC cell lines with either WT-EGFR (NCI-A549), or ΔE746-E750-T790M (NCI-H820), E746–E750 (NCI-HCC827) or L858R-T790M (NCI-H1975) mutated forms of EGFR, used in this study were from the American Type Culture Collection (ATCC) and maintained as previously described (8). CDK4/hTERT immortalized Human Bronchial Epithelial (HBEC) cells stably expressing the V5-tagged wild-type EGFR (WT), L858R, and ΔE746–E750 forms of EGFR were maintained as described in (11). For induction of hypoxia, cells were cultured in poly L-Lysine (Sigma-Aldrich) or Attachment factor (AF) (ThermoFisher Scientific) coated glass petri dishes to avoid the problem of oxygen (O2) dissolved in plastic. Similarly, for imaging studies, cells were first seeded on glass coverslips and the coverslips were submerged in glass petri dishes containing medium. Petri dishes containing cells were incubated in an open Ziploc bag at 5% CO2, 0.1% O2, and 37 °C in a ProOx 110-controlled N2/CO2 gas environment chamber, i-Glove (Biospherix, NY). Dissolved oxygen in medium, measured using a ruthenium-based oxygen probe, reached desired levels (0.1%) within 30–45 minutes of incubation in hypoxia chamber. For irradiation under hypoxia, Ziploc bags were hermetically sealed while still under hypoxic conditions before being exposed to radiation in either XRAD320 (dose rate 117 cGy/minute, Precision X-ray,) or Cs137 irradiator (dose rate 347 cGy/minute, J.L. Shephard and associates).
All cell lines were tested weekly for mycoplasma contamination using the MycoAlert kit (Lonza) and authenticated twice annually using professional authentication services (Genetica-LabCorp Cell line testing).
Clonogenic survival assay
Clonogenic survival was measured as described before (8,11,18). For survival response in hypoxic state, cells were exposed to 0.1% O2 for 24 h, irradiated as described above, allowed to recover for 8 hours under hypoxic conditions and allowed to form colonies over 7–10 days in aerobic environment (21% O2). Where inhibitors were used, cells were incubated in aerobic or hypoxic environments as above and then received vehicle, PARP1/2 inhibitor, ABT-888, (Cayman Chemical) for 2 h, or 50 μg/mL (~345 nmoles/L) humanized monoclonal antibody anti-EGFR Cetuximab (Imclone/Bristol Myers Squibb/Eli Lilly) for 6 h prior to irradiation. Plating efficiency (PE) values were used to determine effective concentration for 50% survival (EC50) for ABT-888. Surviving fraction (SF) values were normalized to PE and plotted as a function of radiation dose. The data were fit with the linear quadratic equation using SigmaPlot version 12.5, (Systat Software). www.systatsoftware.com).
Microarray Gene expression analysis
HBEC cells stably expressing WT, L858R or ΔE746–E750 forms of EGFR were left under aerobic conditions (21% O2) or exposed for 24 hours to a hypoxic environment containing 0.1% O2. Illumina Whole Genome HumanHT12 v4 Expression BeadChip was used in this study. Total RNA was extracted, amplified, transcribed into biotin-labeled cRNA and hybridized with streptavidin-Cy3 (GE Healthcare) using standard Illumina protocols as described previously (26). Slides were scanned on an Illumina Beadstation. Summarized expression values for each probe sets were generated using BeadStudio 3.1 (Illumina Inc.). The data were background subtracted and quantile-quantile normalized across samples using MBCB algorithm (27). A two-sample t-test was performed between WT EGFR and MT-EGFR (L858R and ΔE746–E750 combined) samples or, separately, between 21% O2 and 0.1% O2 samples within WT, L858R and ΔE746–E750 cohorts. Genes with p < 0.01 and fold change greater than 2 were considered as changed with statistical significance. The microarray data are deposited in NCBI’s Gene Expression Omnibus (GEO) (28) and are accessible through GEO accession number GSE95564.
Western blot analysis
Approximately106 NSCLC or HBEC3KT cells were exposed to an aerobic (21% O2) or hypoxic (0.1% O2) environment for 24 hours. Whole cell lysates were prepared as previously described (8,11) Antibodies specific to RAD50 (# sc20155), RAD51 (# sc-8349), MRE11 (# sc5859), β-actin (# sc-69879), Santa Cruz Biotechnology, NBS1/Nibrin (# 05-616), RNR/RRM2 (# ABC106), Ku80 (# 05-393), EMD-Millipore (Upstate), and HIF1α (# GTX628480), GeneTex were used. Blots were imaged using the Chemidoc MP imaging system (Biorad) and band densitometric analysis was performed using the ImageLab software version 4.1 (Biorad). After background subtraction, density of a given band on a blot was normalized to density of β actin band from the same sample on that particular blot to obtain relative density.
Immunocytochemistry for γH2AX foci
NSCLC cells were seeded in duplicate on glass bottom 96-well plates and then exposed for 24 hours to aerobic or hypoxic environments. For re-oxygenation experiments, cells were returned to an aerobic environment and harvested at various time-points for immune-detection of 53BP1 and γH2AX foci as described previously (8,11,18).
For cell cycle distribution of γH2AX foci, while still under aerobic or hypoxic states, cells were pulsed for 10 minutes with 10 μM of the nucleoside analog, 5-ethynyl-2′-deoxyuridine (EDU). before being processed for immunocytochemistry using the Alexa Fluor-488 Click-iT EdU Imaging Kit (Invitrogen), mouse anti-γH2AX (pSer 139; # 05-636), rabbit anti-phospho Histone H3 (pSer 10; # 04-817) from EMD-Millipore (Upstate), and rabbit anti-cyclin B1 antibody (# 4138), Cell Signaling Technology. Fluorescent images were acquired under the IN Cell Analyzer 2000 high content automated imaging system (GE Healthcare) using a 40x objective and analyzed using the IN Cell Investigator software (GE Health care). γH2AX foci in EDU-positive S phase, Cyclin B1 positive G2 phase, and Histone H3-positive M-phase cells were enumerated. The G1 sub-population was estimated by subtracting the sum of S, G2, and M phase cells from the total number of nuclei. In each phase, the fraction of nuclei harboring > 5 foci was calculated.
In a separate experiment, to evaluate extent of radiation-induced DNA damage, NSCLC cells were processed for immunocytochemistry at 45 minutes following 0 Gy or 1 Gy radiation and 15 minutes after an EDU pulse. Images were acquired in multiple focal z-planes using a Nikon A1rsi scanning confocal microscope with an oil-immersion 100x (NA1.49) objective. Images within a Z-stack were collapsed to generate a single composite image and total number of nuclei, number of γH2AX foci per nucleus, and EDU+ S-phase nuclei were enumerated using the Cell Profiler image analysis software.
Immunocytochemistry for RPA-γH2AX foci in S-phase cells
NSCLC and HBEC3KT cells were seeded in 96 well plates and pulse-labeled as described above. To obtain clear RPA foci, cells were subjected to in situ fractionation as described in (29), labeled with Alexa 488 Click-it reagent and immuno-stained with rabbit anti-γH2AX and mouse anti-replication protein A (RPA) (# MAB286, EMD Millipore) followed by incubation with secondary antibodies and DAPI. Fluorescent images were acquired under the IN Cell Analyzer 2000 system using a 40x objective. γH2AX foci, RPA foci and EDU-positive (S phase) nuclei were enumerated and number of S-phase nuclei with merged RPA and γH2AX foci was obtained.
DNA Fiber analysis
Replication fork dynamics were evaluated by DNA fiber assay as described in (30). Briefly, NSCLC cells were seeded in AF-coated glass 60 mm dishes and exposed for 24 hours to aerobic or hypoxic environments. Cells were pulse-labeled with 25 μM 5′ iodo 2′ deoxyuridine (IdU) for 20 minutes followed by 10-fold excess (250 μM) of 5′ Chloro 2′ deoxy uridine (CldU) for a further 20 minutes. Cells were detached and diluted to a density of 7 × 105 cells/mL. Nuclear suspension (2 μL) was spotted on glass slides, dried, and fixed. Samples were blocked with 5% BSA in PBS and incubated with a 1:25 dilution of IdU-specific mouse anti-BrdU (BD Biosciences) and 1: 400 dilution of CldU-specific rat anti-BrdU (Abcam). Samples were incubated with a 1:500 dilution of Cy3-conjugated sheep anti-mouse and 1:400 dilution of Alexa-488 conjugated goat anti-rat secondary antibodies and mounted. Images were acquired with the Nikon A1rsi scanning confocal microscope. A minimum of 200 well-separated fibers were counted per sample. The number and juxtaposition of IdU- and CldU-stained fibers were manually measured using NIS Elements software (v 4.0). Replication structures such as stalled forks (IdU+/CldU−), active forks (IdU+/CldU+), new origins (IdU−/CldU+), elongating forks (CldU-IdU-CldU) and terminating forks (IdU-CldU-IdU) were enumerated. Length of DNA synthesized in 20 minute nucleotide pulses was measured using a DNA extension factor of 2.59 kbp/μm and relative fork velocity in aerobic and hypoxic conditions was calculated as a ratio of CldU/IdU lengths. A CldU/IdU ratio ~ 1.0 indicated unperturbed replication and CldU/IdU ratio <1 was considered slower replication.
RESULTS
Mutations in EGFR compromise hypoxia associated radiation resistance
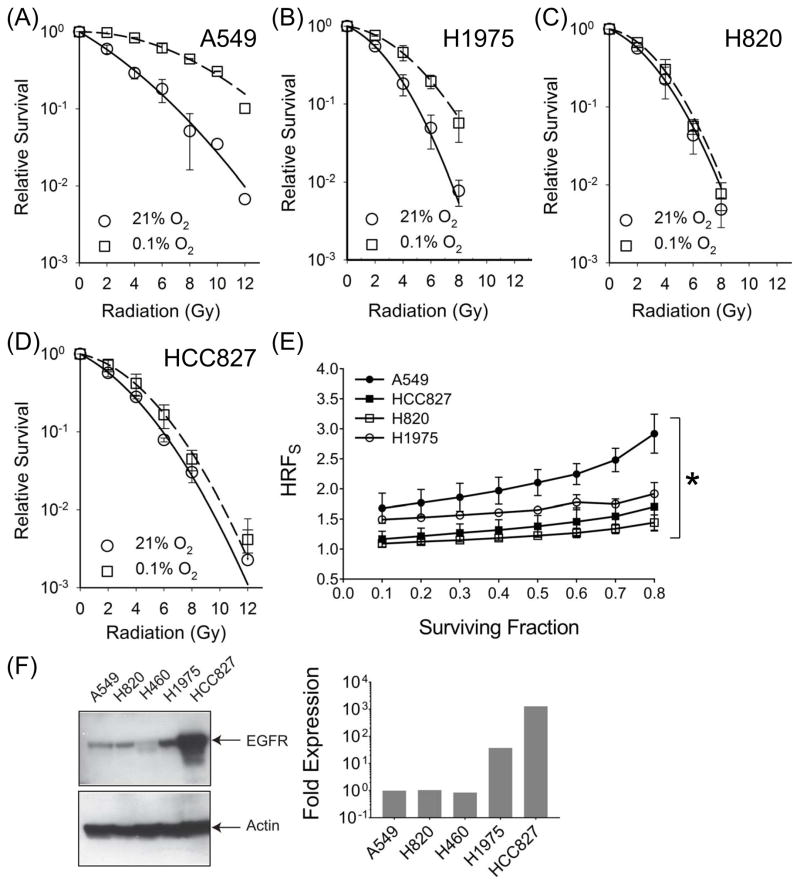
We compared radiation response in four different NSCLC cell lines following a 24 exposure to either an aerobic (21% oxygen) or a hypoxic (0.1% oxygen) environment. In all three MT-EGFR NSCLC cell lines, exposure to hypoxia reduced plating efficiency (PE) across a wide range of oxygen concentrations while WT-EGFR expressing A549 cells were relatively unaffected (Figures S1A–S1C). When normalized for PE, hypoxic WT-EGFR expressing A549 cells were significantly more radioresistant relative to aerobic A549 cells (Figure 1A). By contrast, hypoxia-associated radioresistance in MT-EGFR expressing NSCLCs was marked reduced (Figures 1B–1D).
Figure 1.
Hypoxia-associated radiation resistance is compromised in NSCLCs with activating mutations in EGFR. Clonogenic survival assay in NSCLC cell lines, A549 (A), H1975 (B), H820 (C) and HCC827 (D) following a 24 hour exposure to 21% O2 (circles, solid line) or 0.1% O2 (squares, broken lines). Symbols, representing mean SF normalized to PE and error bars representing standard deviation (SD) were derived from at least 3 independent experiments, each with samples in triplicate. (E) Hypoxia reduction factor (signaling) (HRFS) in NSCLCs. HRFS values at SF from 0.1 to 0.8 are shown. Symbols (mean HRFS) and error bars (SD). Asterisk represents summary of an ordinary one way ANOVA test performed between A549 and MT-EGFR NSCLCs where p < 0.001. (F) Western blot analysis (left panel) of whole cell lysates from indicated NSCLCs with WT (A549, H460) and MT-EGFR (H820, H1975, and HCC827). Densitometric analysis (right panel) of EGFR band intensities normalized to β-actin band intensities in each lane. Relative to A549, levels of MT forms of EGFR were at least 1.5 – 3 orders of magnitude higher.
To quantify hypoxia-associated radiation resistance, radiation dose modifying factors such as OER or HRF are frequently used. Here we use the term hypoxia reduction factor (signaling) (HRFS), which encompasses not only physiochemical modification of DNA but also the biological or enzymatic processes that can influence survival under hypoxic conditions. We define HRFS as the ratio of the radiation dose at a specific oxygen concentration (0.1%) to the radiation dose under fully aerobic conditions (21% oxygen) for the same reduction in clonogenic survival. Data in Figure 1E reveals that HRFS across a range of surviving fractions (SF) was consistently and significantly higher in WT-EGFR expressing A549 cells (Mean HRFS, 2.28) compared to mutant-EGFR expressing cells (Mean HRFS, 1.12 – 1.58). Our panel consisted of NSCLCs with low (H820), medium (H1975) and high (HCC827) expression of MT-EGFR (Figure 1F). Despite these wide differences in expression levels, all three cell lines exhibited marked reduction in HRFS relative to wild EGFR expressing NSCLC, A549, indicating that expression level was not a factor in HRFS reduction.
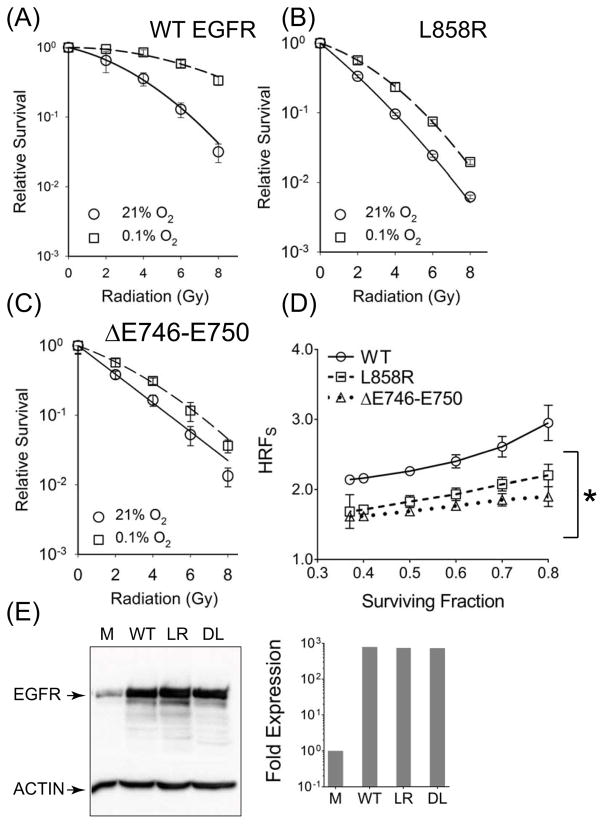
To rule out effects from genetic background, we stably over-expressed WT- and MT-EGFR forms in HBEC cells. Again, hypoxia alone significantly inhibited survival in HBEC cells over-expressing L858R or ΔE746–E750 EGFR but had no effect on cells over-expressing the WT-EGFR (Figure S1B). Cells expressing L858R or ΔE746–E750 EGFR expressing cells (Figure 2B and 2C) exhibited significantly reduced HRFS (Means, 1.66 and 1.76) compared to WT-EGFR (Mean 2.23) (Figure 2A). Figure 2E shows that WT-EGFR and MT-EGFR forms were over-expressed to similar levels in HBEC cells and were nearly 3 orders of magnitude higher relative to mock-transfected controls. These levels were comparable to levels of expression found in most MT-EGFR expressing NSCLCs such as H1975 and HCC827 (Figure 1E). Again, over-expression alone was not a factor in HRFS reduction because similar results were obtained when L858R or ΔE746–E750 EGFR were ectopically expressed at lower levels in A549 NSCLCs (FigureS5). Even though MT-EGFR expression was only 5.5- to 6.7-fold over endogenous WT EGFR (Figure S5A), this level of MT-EGFR expression was sufficient to exert a strong dominant negative effect in A549 cells by significantly reducing PE (Figure S5B, left panel), radioresistance (Figures S5B and S5C) and HRFS (Figure S5D).
Figure 2.
Ectopic expression of L858R and ΔE746–E750 EGFR significantly reduces hypoxia associated radiation resistance. Clonogenic survival assay in HBEC cells stably expressing WT (A), L858R (B) and ΔE746–E750 EGFR (C), following a 24 hour exposure to 21% O2 (circles, solid line) or 0.1% O2 (squares, broken lines). Symbols, representing mean SF normalized to PE and error bars representing SD were derived from at least 3 independent experiments, each conducted with samples in triplicate. (D) HRFS values at different SF. Symbols (mean HRFS) and error bars (SD). Asterisk represents summary of an one way ANOVA test performed between WT- and MT-EGFR expressing HBEC cells where p < 0.001. (E) Western blot analysis: Lysates from HBEC cells that were mock transfected (M) or stably transfected with WT-EGFR (WT), L858R (LR) and ΔE746–E750 (DL) forms of EGFR (left panel). Densitometric analysis (right panel) of EGFR band intensities normalized to β-actin band intensities in each lane. Relative to vector alone, levels of WT or MT forms of EGFR were at least 2.5 – 3 orders of magnitude higher
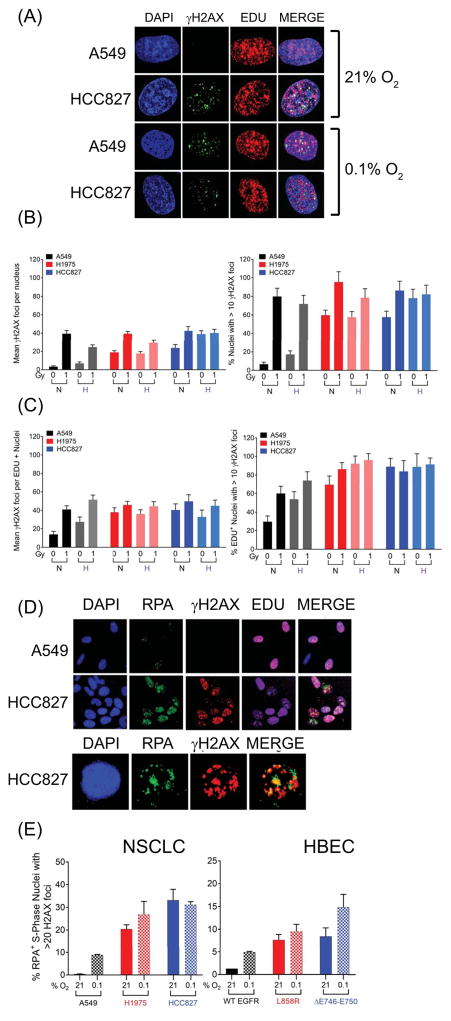
Figure 5.
Elevated MT-EGFR associated γH2AX foci. (A)–(C) NSCLC cells exposed for 24 h to an aerobic (21% O2) or hypoxic (0.1% O2) environment were either mock irradiated (0 Gy) or exposed to radiation (1 Gy), pulsed with EDU and allowed to recover in their respective environments for 45 minutes before being stained for γH2AX and EDU. (A) Representative images of un-irradiated cells in aerobic conditions from a typical experiment showing composite images generated by collapsing image stack of multiple confocal images acquired at 100x magnification. Mean γH2AX foci per nucleus (left panel) and fraction of γH2AX foci bearing nuclei (>10 foci, right panel) across (B) the entire population or (C) the EDU+, S-phase sub-population. Bars represent mean values and error bars are SEM from 2 independent experiments, each with N= ~100 nuclei per sample. NSCLCs and HBECs exposed for 24 h to an aerobic (21% O2) or hypoxic (0.1% O2) environment stained for total nuclei (DAPI), S phase nuclei (EDU, Alexa-647), Replication protein A (Alexa-488) and γH2AX (Alexa-547). (D) Representative images, acquired at 40 x magnification from a typical experiment with NSCLCs are shown. (E) S-phase nuclei with γH2AX and RPA foci were enumerated in NSCLC and HBEC cells by image analysis software. Left panel: NSCLCs, right panel: HBEC cells. Bars represent mean fraction of nuclei that are in the S-phase (EDU+) and contain >20 RPA foci, >20 γH2AX foci. Mean of means and SEM were derived from at least 2 independent experiments, each with ~1500 nuclei (N ≥ 1500) per sample.
Mutations in EGFR alter DNA damage response patterns in hypoxic cells
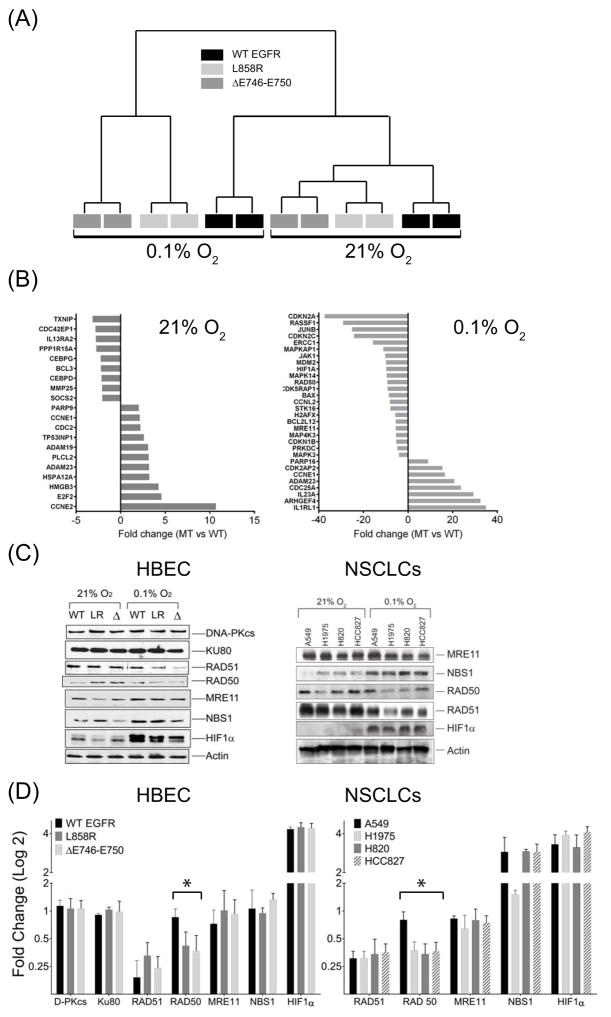
To examine whether mutation status of EGFR influenced hypoxia-induced changes in gene expression and whether these effects could account for reduced HRF, we compared gene expression patterns associated with WT, L858R or ΔE746–E750 EGFR stably expressed in the isogenic background of HBEC cells. In aerobic conditions, relative to WT-EGFR expressing cells, 226 genes were differentially expressed 2-fold or more (p < 0.01) in MT-EGFR HBECs. By contrast, in hypoxic conditions, the number was nearly 8 times larger, with 1793 genes differentially expressed between the two cohorts. Unsupervised clustering revealed significant differences between WT- and MT-EGFR in both aerobic and hypoxic conditions (Figure 3A) and gene expression changes in L858R and ΔE746–E750 expressing cells clustered together. In aerobic conditions (Figure 3B), relative to WT-EGFR, MT-EGFR expression was associated with up-regulation of cell cycle activators, including CCNE1 and CCNE2, and down-regulation of cell cycle inhibitors such as CDKN2A, CDKN2D, and CDKN1A. This is consistent with the elevated pro-proliferative signaling associated with the MT-EGFR. In hypoxic conditions, a very different pattern emerged. While expression of NHEJ genes, PRKDC (DNA-PKcs), XRCC6 (KU70), XRCC5 (KU80) or LIG4 (Ligase IV) was unchanged, the hypoxic state was associated with down-regulation of many HR DNA repair components including, RAD50, RAD51, MRE11 and NBN (NBS1). Of these, mRNA levels of RAD50 and MRE11 appeared significantly lower in MT-EGFR expressing cells relative to WT-EGFR cells. Data from western blot and densitometric analysis (Figures 3C, 3D) largely validated the microarray profile. For example, at both mRNA and protein levels, RAD51 levels reduced to the same extent in hypoxic WT- and MT-EGFR expressing cells. Likewise, at both transcript and protein levels, RAD50 remained largely unaffected by hypoxia in WT-EGFR cells but was significantly reduced in L858R or ΔE746–E750 EGFR expressing HBEC cells. However, MRE11 and NBS1 appeared to be differentially down-regulated at the mRNA level but not at the protein level. Remarkably similar trends were observed in WT- and MT-EGFR expressing NSCLC cells. The data in Figure 3 not only confirm hypoxia-associated HR down-regulation but also suggest that MT-EGFR expression may exert an inhibitory effect on additional HR components such as RAD50.
Figure 3.
Microarray gene expression analysis of mRNA isolated from HBEC cells stably expressing wild type (WT, black), L858R (dark gray) or ΔE746–E750 (light gray) forms of EGFR in aerobic (21% O2) conditions or after 24 hours exposure to hypoxic (0.1% O2) environments (A) Unsupervised clustering of genes from different samples. Gene expression patterns of L858R or ΔE746–E750 clustered as one group (MT) which was distinct from WT in both aerobic and hypoxic conditions (B) Fold differences in gene expression in MT-EGFR relative to WT-EGFR cells grown in aerobic (left panel) or hypoxic conditions (right panel). Positive values indicate fold increase; negative values represent fold decrease in expression in MT-EGFR expressing cells, relative to WT-EGFR expressing cells. (C) Western blot analysis of lysates from HBEC cells stably transfected with WT-EGFR (WT), L858R (LR) and ΔE746–E750 (Δ) forms of EGFR (left panel) or indicated NSCLCs (right panel) isolated from cells in aerobic or hypoxic conditions. Representative blots from multiple gels from at least 3 independently performed experiments are shown. (D) Densitometry analysis: Band intensities of individual proteins in 3C were normalized to intensities of the corresponding β-actin band from the same gel as the target protein. Bars represent mean fold change in hypoxic conditions relative to aerobic state and error bars represent SEM (N=3 independent experiments). Asterisk summarizes an ordinary one way ANOVA test performed between WT- and MT-EGFR expressing HBEC or NSCLC cells where p < 0.001.
MT-EGFR expressing NSCLCs and HBEC exhibit unique patterns of hypoxia associated DNA lesions
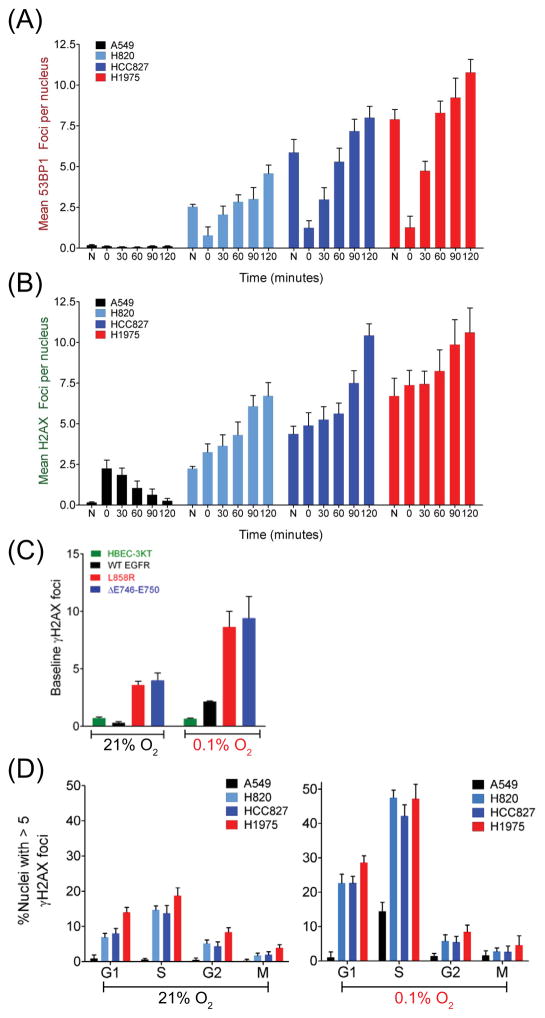
We next examined the effect of hypoxia and re-oxygenation on two well-established surrogates of DNA damage, 53BP1 and γH2AX, which localize to sites proximal to DSBs and form distinct foci (Figure 4). Relative to WT-EGFR expressing A549 cells (0.5 foci per nucleus), MT-EGFR expressing H820, HCC827 and H1975 cells harbored dramatically elevated levels of basal 53BP1 (Figure 4A) and γH2AX foci (2.5 to 7.5 foci per nucleus) (Figure 4B). A 24-hour exposure to 0.1% O2 resulted in a modest 5-fold increase in γH2AX foci in A549 cells, which returned to near baseline levels, 2 hours following re-oxygenation. Interestingly, 53BP1 foci were undetectable in hypoxic A549 cells, even after re-exposure to 21% O2. In striking contrast, all three MT-EGFR expressing NSCLCs tested showed high basal levels of 53BP1 and γH2AX foci. Exposure to a 0.1% O2 environment resulted in 2.5- to 7-fold decrease in 53BP1. However, re-exposure to an aerobic environment not only led to a further increase in γH2AX foci but also a dramatic resurgence of 53BP1 foci to levels that were nearly 2-fold higher compared to aerobic conditions. Highly similar results were observed when WT- and MT- EGFR were ectopically expressed in HBEC cells (Figure 4C). Relative to WT-EGFR expressing cells, cells expressing ΔE746–E750 and L858R exhibited significantly higher levels of γH2AX foci at 21% O2 that further increased upon exposure to a hypoxic environment.
Figure 4.
MT-EGFR expression is associated with elevated levels of 53BP1 and γH2AX foci in aerobic and hypoxic state. NSCLC cells exposed to an aerobic environment (N) or exposed for 24 h to 0.1% O2 (0) and then re-exposed to the 21% O2 for the indicated time-points. (A) 53BP1 foci or (B) γH2AX foci enumerated by analysis of images acquired by automated fluorescence microscopy. Bars represent mean foci per nucleus and error bars represent standard error of mean (SEM) from 2 independent experiments with N = ~ 2500 nuclei per sample in each experiment (C) HBEC cells expressing WT- and MT-EGFR at 24 h after exposure to 21% or 0.1% O2 were processed by as above to detect nuclear γH2AX foci. Bars represent mean foci per nucleus and error bars are SEM from 2 independent experiments with N = ~ 1500 nuclei per sample in each experiment. (D) Cell cycle distribution of basal γH2AX foci in nuclei stained with cyclin B (G2), p-histone H3 (M), Alexa-488 linked 5-ethynyl-2′-deoxyuridine (EdU) (incorporated only in S-phase) in aerobic (left panel) and hypoxic (right panel) conditions. Number of G1 cells was determined by subtracting the sum of S, G2, M phase cells from total number of nuclei. Bars represent the mean fraction of nuclei with > 5 foci and error bars represent SEM from two independent experiments, each with N = ~ 5000 nuclei per sample.
To determine whether hypoxia-associated increase in γH2AX foci affected all cells or only cells in specific phases of cell cycle progression, cell cycle distribution of basal and hypoxia-induced γH2AX foci in WT- and MT-EGFR NSCLCs was assessed (Figure 4D). In aerobic A549 cells, the fraction of foci-bearing cells (with >5 γH2AX) was extremely low (0.2 to 0.8%) in all phases of the cell cycle. but increased ~23-fold (up to 14.5%) following a 24-hour exposure to 0.1% O2. The increase in foci-bearing fractions was observed almost exclusively in the S-phase sub-population, while those in G1, G2, and M sub-populations remained essentially unchanged. In striking contrast, foci-bearing fractions in aerobic MT-EGFR expressing NSCLCs were significantly elevated in all phases of the cell cycle, although the most dramatic difference was in the S-phase sub-population (24- to 31-fold higher relative to A549). At 24 hours of exposure to 0.1% O2, foci-bearing fractions in all three MT-EGFR NSCLC cell lines were significantly higher (2- to 3.5-fold) compared to aerobic state but, again, these increases were most pronounced in the G1 and S sub-populations. The data suggest that NSCLCs with activating ΔE746–E750 and L858R mutations differ significantly from WT-EGFR cells in basal and hypoxia-associated DNA damage response.
We next used confocal microscopy to examine the extent and magnitude of radiation induced DNA damage in aerobic and hypoxic WT- and MT-EGFR expressing NSCLC cells 45 minutes after exposure to 1 Gy radiation. Representative images of confocal scans are shown in Figure 5A. Regardless of EGFR status, in aerobic conditions, all three NSCLCs, regardless of EGFR status, registered essentially similar levels of γH2AX foci per nucleus (39.5 to 42.5 ± 3.5) in response to 1 Gy (Figure 5B). Moreover, the fraction of foci bearing nuclei (with >10 foci) in aerobic state after 1 Gy was also essentially similar; 80 to 85% (± 10.11) of nuclei had >10 γH2AX foci in all three cell lines. As expected, exposure to 1 Gy in an oxygen-deficient environment reduced the number of γH2AX foci but to differing extents depending on EGFR status. A549 cells irradiated under hypoxic conditions showed a ~40% decrease in foci relative to aerobic irradiated cells but only 25% and 2% reductions in H1975 and HCC827, respectively (Figure 5B). When foci bearing EDU+, S-phase sub-population was considered, a different pattern emerged (Figure 5C). First, in all three NSCLCs, relative to aerobic cells, the mean number of radiation-induced γH2AX foci in EDU+ nuclei remained unchanged with hypoxia. Moreover, S-phase foci-bearing fractions (>10 foci) in all three cell lines was either the same (HCC827) or slightly higher (A549 and H1975) with hypoxia. The data in Figures 4D and 5C suggested that hypoxia-associated increases in S-phase γH2AX foci likely involved replication events in the S-phase.
To verify whether this is indeed the case, we enumerated fractions of S-phase nuclei harboring >20 γH2AX foci that also contained >20 foci of replication protein A (RPA). In NSCLCs and HBEC cells, WT-EGFR expression was associated with a 5-fold (HBEC) to 9-fold (A549) hypoxia-associated increase in the RPA+/H2AX+ S-phase fraction (Figures 5D, 5E). In aerobic and hypoxic cells expressing MT-EGFRs, this fraction was significantly higher compared to WT-EGFR cells. Camptothecin, a selective inhibitor of topoisomerase I (Top1), which stabilizes Top1-linked single stranded nicks, also caused a dramatic increase in the RPA+/H2AX+ S-phase fraction (Figure S3A) but this increase was evident in both WT- and MT-EGFR cells. By contrast, the effects of hydroxyurea (Figure S3B), which blocks initiation and elongation phases of replication, resembled those of hypoxia, with modest HU-induced increase in RPA+/H2AX+ fractions in WT-EGFR NSCLCs and HBECs but dramatically high basal and HU-induced fractions of these events in MT-EGFR cells. The data indicate that in both aerobic and hypoxic states, replicating MT-EGFR expressing cells harbor a significantly higher burden of γH2AX foci compared to WT-EGFR cells.
MT-EGFR expression is associated with replication stress
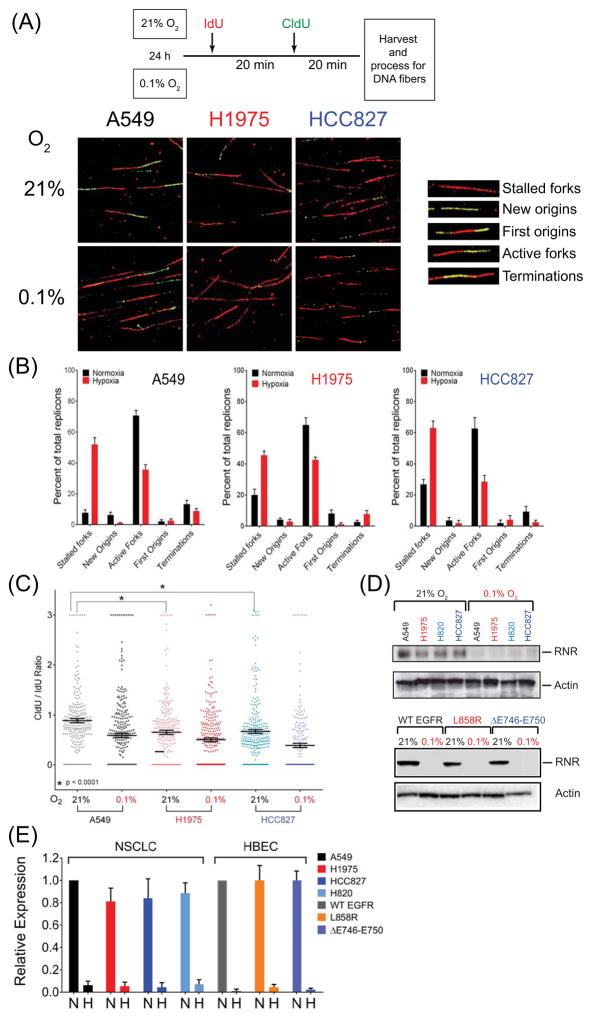
To conclusively determine whether MT-EGFR expression has any effect on kinetics of replication fork progression, we examined kinetics of fork initiation, fork extension and fork progression in NSCLCs A549, H1975, HCC827 after a 24 h exposure to 21% or 0.1% O2 using the DNA fiber assay (30). This assay involves sequential 20-minute pulses of two nucleotide analogs, IdU and CldU, and detection of their incorporation in replicating DNA by differently labeled fluorescent antibodies. Stalled forks (red only fibers) active forks (red-green fibers), terminating forks (red-green-red) and new origins (green only) fibers (Figure 6A and 6B) were enumerated. Ratio of CldU:IdU lengths was used to determine relative rates of fork progression with lower ratios representing replication stalling or slowing and higher ratios representing unperturbed replication (Figure 6C). Data in Figure 6B reveal that, in aerobic WT-EGFR expressing A549 NSCLCs, a small fraction (7.5%) of all replicons occurred as stalled replication forks, while the majority of structures were active forks (70%) or successful terminating replicons (13.5%). Exposure to 0.1% for 24 hours resulted in a 7-fold increase in the fraction of stalled replication forks (52%), with a 50% reduction in the fraction of active forks (35%) and terminating replicons (6%). In striking contrast, even in the aerobic state, almost 20–26% of replicons in MT-EGFR expressing H1975 and HCC827 NSCLCs appeared as stalled forks, a ~3–4 higher baseline relative to WT-EGFR expressing A549 cells. In hypoxic conditions, this fraction of stalled forks formed the majority of replicons (~ 63%) in HCC827 and equaled active forks (45%) in H1975. Active and terminating forks constituted a combined 83% of all replicons in aerobic A549 cells, but formed a significantly lesser proportion (65–70%) of all replicons in H1975 and HCC827. In hypoxia, there was a further reduction in the combined fraction of active and terminating forks (31% and 45% in HCC827 and H1975, respectively). The high proportion of stalled replication forks clearly seems to have affected fork velocity. Even in aerobic conditions, relative fork velocity measured as CldU/IdU ratio was significantly reduced in H1975 (0.65 ± 0.04) and HCC827 (0.66 ± 0.03), compared to A549 cells (0.89 ± 0.04) (Figure 5C). In hypoxic conditions, whereas WT-EGFR NSCLC registered a ratio of 0.60 ± 0.035, H1975 and HCC827 exhibited ratios of 0.49 and 0.39, respectively.
Figure 6.
Replication stress in MT-EGFR expressing cells. (A) Schematic representation of DNA fiber assay (top panel). Representative images acquired by confocal microscopy at 100x magnification (bottom panel) from NSCLC cells exposed to aerobic or hypoxic conditions showing of IdU stained (red) and CldU stained (green) fibers. Inset: replication structures identified (B) Bars, representing mean percentage of specific replication structures relative to total number of replicons, and error bars, representing SD from two independent experiments, each with at least N = 200 replicons per sample are shown. (C) Dot density plot showing frequency distribution of relative fork velocity derived from CldU/IdU ratios. Horizontal bars represent mean ratio. Asterisk represents results of a t-test analysis of between A549 and H1975 or A549 and HCC827 cells. (D) Western blot analysis: Representative blots probed for ribonucleotide reductase (RNR) and β-actin in lysates of NSCLCs (top) and HBEC (bottom) cells expressing WT, L858R or ΔE746–E750 forms of EGFR (E) Densitometric analysis of EGFR band intensities normalized to β-actin band intensities in aerobic conditions (N) and hypoxic (H) conditions. Bars represent mean RNR expression relative to A549 RNR levels for NSCLCs, or WT-EGFR expressing cells in HBEC. Error bars are SEM from 2 independent experiments.
Many studies have shown that, during prolonged hypoxia, levels of a key enzyme in nucleotide metabolism, ribonucleotide reductase (RNR) are drastically reduced (22) and nucleotide depletion inhibits or delays replication initiation, leading to stalled replication forks and reduced fork velocities (31,32). We tested whether replication stress encountered in aerobic MT-EGFR NSCLCs was due to RNR deficiency. Consistent with other reports, (22,33), prolonged 24 hours exposure to 0.1% O2 almost completely abrogated RNR levels in all cell lines, regardless of EGFR status (Figure 6D, 6E). The data indicate that, despite adequate levels of RNR, replication fork progression in MT-EGFR NSCLCs is significantly compromised in aerobic conditions and RNR–depleted hypoxic conditions may exacerbate replication stress.
EGFR blockade by Cetuximab significantly reduces radioresistance of hypoxic cells
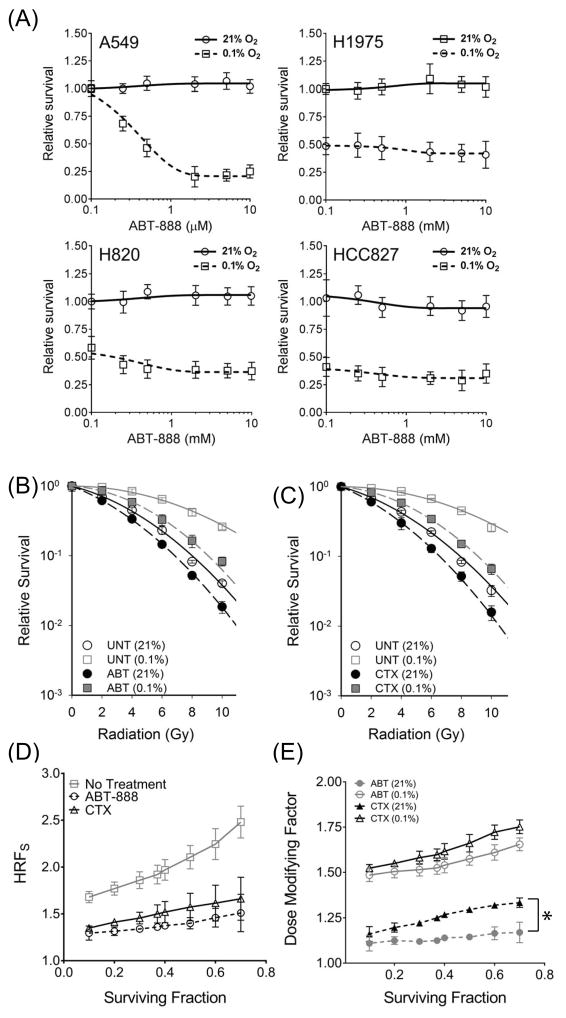
Synthetic lethality occurs when simultaneous inactivation of two complementary genes or pathways causes loss of cell viability or reproductive capacity, whereas inactivation of only one gene/pathway does not. Synthetic lethality has been demonstrated by inhibition of the Poly ADP-Ribose Polymerase (PARP), an abundant nuclear protein with an important function in an alternate non-homologous end-joining DSB repair pathway involving XRCC1 and ligase III (34). Multiple studies have shown that inhibition of PARP-mediated alternate NHEJ has a profound synthetic lethality effect in the context of HR-deficiency in BRCA-deficient breast cancers (35) as well as HR-down-regulated hypoxic tumors (36–39). Since MT-EGFR expression (8,11,18) or EGFR blockade by the anti EGFR monoclonal antibody, Cetuximab (CTX) (7,19) also compromises NHEJ, we reasoned that CTX might have a similar synthetic lethality effect on HR-down-regulated hypoxic tumors. As a positive control for synthetic lethality, we first confirmed the effects of the PARP inhibitor, ABT-888, on PE in NSCLCs in aerobic and hypoxic conditions (Figure 7A). Treatment with ABT-888 had no effect on survival in aerobic A549 cells but significantly reduced of A549 cells in the hypoxic state with an effective concentration (EC50) of 0.5 μM. Consistent with results in Figure 1E), hypoxia alone was sufficient to reduced PE in MT-EGFR expressing NSCLCs. Surprisingly, ABT-888 had no effect on PE of any of the MT-EGFR NSCLCs cells even in hypoxic conditions. ABT-888 at a concentration of 1μM had no effect on radiosensitivity of aerobic A549 NSCLC cells but significantly increased A549 radiosensitivity in hypoxic conditions (Figure7B). D37 is the radiation dose required to reduce the SF down to 37% (SF = 0.37). In aerobic conditions, the difference in D37 values between untreated and ABT-888-treated A549 cells was only 0.88 Gy but it was ~ 3.5-fold higher in hypoxic conditions. EGFR blockade by CTX had surprisingly similar effect (Figure 7C). The difference in D37 values between untreated and CTX-treated was 1.02 Gy in aerobic and 3.05 Gy in hypoxic conditions, again 3-fold higher. Both ABT-888 and CTX reduced HRF to similar extents (Figure 7D). Moreover, both ABT-888 and CTX were significantly more effective in hypoxic conditions (Figure 7E). The dose-modifying factor (DMF) is defined as the ratio of the radiation dose without agent to the radiation dose with agent to decrease SF to the same extents. In aerobic conditions, compared to ABT-888, CTX was slightly more effective in augmenting radiation in aerobic A549 cells (Figure 7E). However, mean DMF for both ABT-888 and CTX were significantly higher in hypoxic conditions (2.0 Gy and 1.78 Gy respectively) compared to aerobic conditions (1.12 Gy and 1.29 Gy respectively). The data indicate that, like ABT-888, EGFR blockade by CTX was significantly more effective in radiosensitizing A549 cells in the hypoxic state compared to cells in the aerobic state.
Figure 7.
EGFR blockade sensitizes hypoxic A549 to radiation. (A) Clonogenic assay in indicated NSCLCs following treatment with indicated concentrations of ABT-888 in aerobic or hypoxic conditions. (B) Clonogenic survival assay in A549 NSCLC cells, pre-exposed for 24 hours to aerobic or hypoxic environment, treated for 2 hours with vehicle (UNT), 1 μM ABT-888 (B), or 50 μg/mL cetuximab (C) and exposed to indicated doses of radiation. Symbols represent mean SF relative to vehicle treated samples (A) or SF normalized to PE (B and C). Mean of means and mean SD from at least 3 independent experiments, each with samples in triplicate are shown. (D) HRFS values determined across a range of SFs in A549 NSCLCs that were left untreated (square symbol), ABT-888-treated (circle) and CTX-treated (triangle). (E) Dose modifying factor for ABT-888 (circles) and CTX (triangles) in aerobic (filled symbols) and hypoxic conditions (open symbols). Symbols are mean HRFS (D) or mean DMF (E) and error bars are SEM from 3 independent experiments, each with samples in triplicate. Asterisk summarizes a simple, paired, two-tailed t-test comparing DMF trends between CTX and ABT treatments in aerobic conditions where p < 0.001.
DISCUSSION
EGFR activation and downstream signaling has well-documented roles in pro-survival mechanisms that contribute to radiation resistance not only in aerobic but also in hypoxic environments. This study proffers evidence that activating ΔE746–E750 and L858R mutations in EGFR have the opposite effect on hypoxia associated radiation-resistance and survival. Compared to WT-EGFR, MT-EGFR expression in NSCLCs or HBEC significantly compromised survival (Figure S1A, S1B) and diminished HRF (Figures 1, 2). Deficiency of the NHEJ enzyme, DNA-PKcs, similarly reduced survival in hypoxic but not aerobic conditions (Figure S1C). Hypoxia-induced PARP cleavage, a reliable indicator of apoptosis, was observed in all NSCLCs, regardless of EGFR status, but not in HBEC cells, (Figure S1D). Thus, the difference in survival between WT- and MT-EGFR expressing cells is unlikely due to apoptosis.
We used HRFS as a measure of hypoxia-associated radiation resistance from physicochemical modification of DNA as well as biochemical signaling. While oxygen concentration can significantly influence HRFS, variations in cellular reducing environment can also impact HRFS. Reduction of the DNA radical by thiol (–SH) containing compounds restores DNA to its original (reduced) form, thereby limiting radiation-induced DNA damage. Thus, cells with high thiol content are generally less sensitive to radiation and the effect of oxygen (or hypoxia) on HRFS in these cases is less apparent (40). Thiol content was not a factor in the decreased HRFS because thiol levels were similar between WT- and MT-EGFR NSCLCs and ectopic MT-EGFR expression in HBEC cells had no impact on thiol levels (Figure, S2). Moreover, microarray analysis of WT- and MT-EGFR expressing HBEC cells (Figure 3, GEO accession number GSE95564) or Oncomine database analysis of a large data set with 99 WT- and127 MT-EGFR tumor samples from NSCLC patients (GEO accession number GSE31210) showed no significant differences in antioxidant metabolic pathway between WT- and MT-EGFR NSCLCs. It appears unlikely that the reduced HRFS in MT-EGFR expressing cells was due to physicochemical modulation of the DNA radical by oxygen or thiols.
A more likely mechanism could be defective DNA damage response (DDR). We previously demonstrated that NSCLCs and HBEC expressing MT-EGFR are defective in NHEJ-mediated repair of radiation induced DNA damage. Chronically hypoxic tumors exhibit a markedly altered DNA damage response (DDR) relative to aerobic tumors. A number of studies have demonstrated that, due to a selective down-regulation of HR, hypoxic cells are over-reliant on NHEJ as the sole DSB repair pathway (25,41). In agreement with these reports, our data confirm that key proteins in the HR DNA repair pathway, including RAD50 and RAD51 are down regulated during prolonged hypoxia. Interestingly, ectopic expression of ΔE746–E750 and L858R mutant EGFR not only reduced RAD51 levels, as did WT-EGFR, but also significantly down-regulated levels of RAD50, which were relatively unaffected by WT-EGFR expression. RAD51 exclusively participates in HR (42). By contrast, RAD50, a crucial component of the MRE11-RAD50-NBS1 (MRN) complex, is required for both NHEJ and HR (43). Thus, in addition to their reported defects in NHEJ (8,10,11,18), MT-EGFRs may exert an additional level of NHEJ and HR suppression in hypoxic conditions.
In hypoxic state, replication stress appears to be independent of EGFR mutation status and could be, in part, due to RNR down-regulation (Figure 5D) which is known to cause nucleotide depletion and replication arrest (33). However, in aerobic conditions, where RNR levels were similar, MT-EGFR NSCLCs showed elevated replication stress compared to WT-EGFR NSCLC. One possibility is that MT-EGFRs are defective in NHEJ (8,10,11,18) and an accumulation of unresolved DSBs in S-phase may arrest or slow down fork progression (Figure 4). An alternative mechanism may involve deregulation of S-phase checkpoint control resulting in fork associated DNA damage. A recent study found that, in addition to NHEJ-mediated DSB repair, DNA-PKcs facilitates S-phase check-point control (44) which involves Ataxia telangiectasia-related (ATR) protein, check point kinase 1, Chk1, and Cdc25a (45). DNA-PKcs is phosphorylated at T2609 by ATR in response to replication stress (46). We previously demonstrated that MT-EGFR expression in NSCLCs and HBEC cells completely abrogates DNA-PKcs T2609 phosphorylation (11). However, unlike MT-EGFR expression (Figure 5C), DNA-PKcs deficiency had no effect on CldU/IdU ratio in aerobic conditions (44). Thus, although S-phase checkpoint impairment may elevate DSBs, the replication stress in aerobic MT-EGFR NSCLCs may involve additional factors.
Parallels can be drawn between MT-EGFR and MYC oncogene associated replication stress. MYC influences replication at multiple levels including, direct interaction with pre-replication complex proteins, regulation of replication proteins, CDC6, CDK and mini chromosome maintenance (MCM) proteins such as MCM 2–7 and 10 at origin sites (47), and up-regulation of Cyclin E. Microarray gene expression analysis showed no differences in MYC mRNA between WT- and MT-EGFR expressing HBEC. However, many of MYC up-regulated proteins including, Cyclin E, MCM 2–7 and 10 were significantly over-expressed in MT-EGFR expressing cells relative to WT-EGFR cells (Figure S4). Like MT-EGFR expressing cells (Figure 4), MYC over-expression also results in fork associated DSBs (48) and slowing of replication due to premature origin firing and increased origin density (47). It is therefore conceivable that overabundance of replication factors, together with pre-existing DSBs due to NHEJ defects may, in part, be responsible for replication stress in aerobic MT-EGFR NSCLCs.
Our study finds interesting parallels between CTX-mediated EGFR blockade and PARP inhibition. Similar to PARP inhibition, expression of MT-EGFR exerted lethality, possibly through inhibition of NHEJ in an altered, HR-deficient DDR. First, MT-EGFR (Figures S1A, S1B), or DNA-PKcs deficiency (Figure S1C), similarly reduced survival of hypoxic cells but did not affect survival of aerobic cells. Second, MT-EGFR expression resulted in markedly reduced HRFS in both NSCLCs and HBEC (Figures 1 and 2) to the same extent as PARP inhibition (Figure 7). Third, like PARP inhibition (Figure 7) and (39), EGFR blockade by CTX was significantly more effective in increasing radiosensitivity of A549 cells in the hypoxic state (DMF: 1.78) compared to aerobic state (DMF: 1.29).
Interestingly, PARP inhibition did not affect survival of NHEJ-compromised MT-EGFR NSCLCs in either aerobic or hypoxic conditions (Figure 7A), indicating that EGFR and PARP may be epistatic, with inter-dependent activities and mutually supportive roles in NHEJ-mediated DSB repair (49). Compared to the transient 2 hour ABT-888 treatment in our study, a 72 hour PARP inhibition by olaparib did show MT-EGFR NSCLCs to be slightly more sensitive than WT-EGFR NSCLCs (14). Alternatively, EGFR and PARP may share a synthetic lethal interaction. EGFR blockade by lapatinib in triple negative breast cancers (50) and CTX in Head and Neck cancers (13) augmented ABT-888 cytotoxicity through possible suppression of NHEJ, HR, or both (12).
Our data support a model to explain MT-EGFR-associated radiosensitivity in aerobic and hypoxic conditions. In aerobic conditions, expression of NHEJ-defective MT-EGFR compromises repair of radiation-induced as well as fork-associated DSBs, resulting in increased radiosensitivity. In the hypoxic state, in the context of an altered HR down-regulated DDR, expression of NHEJ-defective MT-EGFR or blockade of EGFR-mediated NHEJ has a catastrophic effect on DSB repair and causes synthetic lethality.
Understanding how EGFR mediates repair of fork-associated DSBs could elucidate novel therapeutic targets. Our finding that MT-EGFR expression or EGFR blockade has a synthetic lethal effect in hypoxic conditions suggest that anti-EGFR therapy in combination with radiotherapy could potentially be effective in treating not only hypoxic NSCLCs but also NSCLCs with mutations in HR DSB repair genes.
Supplementary Material
Implications.
This study demonstrates that within an altered DNA damage response of hypoxic NSCLC cells, mutant EGFR expression, or EGFR blockade by cetuximab exerts a synthetic lethality effect and significantly compromises radiation resistance in hypoxic tumor cells.
Acknowledgments
FUNDING
This work was supported by the National Institutes of Health [R01CA129364 to C.S.N., RO1CA149461, RO1CA197796, R21CA202403 to S.B., R21CA175879 to D.S.]; the National Aeronautics and Space Administration [NNX16AD78G to S.B., NNJ05HD36G to M.D.S.]; and startup funds from the University of South Alabama Mitchell Cancer Institute.
The authors gratefully acknowledge Dr. David Boothman (University of Texas Southwestern Medical Center), Dr. Robert W. Sobol (University of South Alabama, Mitchell Cancer Institute) and Dr. Conchita Vens (Netherlands Cancer Institute) for their critical inputs during the course of this study.
Footnotes
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
References
- 1.Gray LH, Conger AD, Ebert M, Hornsey S, Scott OC. The concentration of oxygen dissolved in tissues at the time of irradiation as a factor in radiotherapy. Br J Radiol. 1953;26:638–48. doi: 10.1259/0007-1285-26-312-638. [DOI] [PubMed] [Google Scholar]
- 2.Carlson DJ, Keall PJ, Loo BW, Jr, Chen ZJ, Brown JM. Hypofractionation results in reduced tumor cell kill compared to conventional fractionation for tumors with regions of hypoxia. Int J Radiat Oncol Biol Phys. 2011;79:1188–95. doi: 10.1016/j.ijrobp.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y, Henson ES, Xiao W, Huang D, McMillan-Ward EM, Israels SJ, et al. Tyrosine kinase receptor EGFR regulates the switch in cancer cells between cell survival and cell death induced by autophagy in hypoxia. Autophagy. 2016;12:1029–46. doi: 10.1080/15548627.2016.1164357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee SM, Lee CT, Kim YW, Han SK, Shim YS, Yoo CG. Hypoxia confers protection against apoptosis via PI3K/Akt and ERK pathways in lung cancer cells. Cancer Lett. 2006;242:231–8. doi: 10.1016/j.canlet.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Misra A, Pandey C, Sze SK, Thanabalu T. Hypoxia activated EGFR signaling induces epithelial to mesenchymal transition (EMT) PLoS One. 2012;7:e49766. doi: 10.1371/journal.pone.0049766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franovic A, Gunaratnam L, Smith K, Robert I, Patten D, Lee S. Translational up-regulation of the EGFR by tumor hypoxia provides a nonmutational explanation for its overexpression in human cancer. Proc Natl Acad Sci U S A. 2007;104:13092–7. doi: 10.1073/pnas.0702387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, et al. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–9. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- 8.Das AK, Chen BP, Story MD, Sato M, Minna JD, Chen DJ, et al. Somatic Mutations in the Tyrosine Kinase Domain of Epidermal Growth Factor Receptor (EGFR) Abrogate EGFR-Mediated Radioprotection in Non-Small Cell Lung Carcinoma. Cancer Res. 2007;67:5267–74. doi: 10.1158/0008-5472.CAN-07-0242. [DOI] [PubMed] [Google Scholar]
- 9.Kriegs M, Kasten-Pisula U, Rieckmann T, Holst K, Saker J, Dahm-Daphi J, et al. The epidermal growth factor receptor modulates DNA double-strand break repair by regulating non-homologous end-joining. DNA Repair (Amst) 2010;9:889–97. doi: 10.1016/j.dnarep.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Liccardi G, Hartley JA, Hochhauser D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 2011;71:1103–14. doi: 10.1158/0008-5472.CAN-10-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Javvadi P, Makino H, Das AK, Lin YF, Chen DJ, Chen BP, et al. Threonine 2609 phosphorylation of the DNA-dependent protein kinase is a critical prerequisite for epidermal growth factor receptor-mediated radiation resistance. Mol Cancer Res. 2012;10:1359–68. doi: 10.1158/1541-7786.MCR-12-0482-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myllynen L, Rieckmann T, Dahm-Daphi J, Kasten-Pisula U, Petersen C, Dikomey E, et al. In tumor cells regulation of DNA double strand break repair through EGF receptor involves both NHEJ and HR and is independent of p53 and K-Ras status. Radiother Oncol. 2011;101:147–51. doi: 10.1016/j.radonc.2011.05.046. [DOI] [PubMed] [Google Scholar]
- 13.Nowsheen S, Bonner JA, Lobuglio AF, Trummell H, Whitley AC, Dobelbower MC, et al. Cetuximab augments cytotoxicity with poly (adp-ribose) polymerase inhibition in head and neck cancer. PLoS One. 2011;6:e24148. doi: 10.1371/journal.pone.0024148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaffle HN, Wang M, Gheorghiu L, Ferraiolo N, Greninger P, Borgmann K, et al. EGFR-activating mutations correlate with a Fanconi anemia-like cellular phenotype that includes PARP inhibitor sensitivity. Cancer Res. 2013;73:6254–63. doi: 10.1158/0008-5472.CAN-13-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 16.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–7. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 18.Das AK, Sato M, Story MD, Peyton M, Graves R, Redpath S, et al. Non-Small Cell Lung Cancers with Kinase Domain Mutations in the Epidermal Growth Factor Receptor Are Sensitive to Ionizing Radiation. Cancer Res. 2006;66:9601–8. doi: 10.1158/0008-5472.CAN-06-2627. [DOI] [PubMed] [Google Scholar]
- 19.Dittmann K, Mayer C, Rodemann HP. Inhibition of radiation-induced EGFR nuclear import by C225 (Cetuximab) suppresses DNA-PK activity. Radiother Oncol. 2005;76:157–61. doi: 10.1016/j.radonc.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 20.Olcina M, Lecane PS, Hammond EM. Targeting hypoxic cells through the DNA damage response. Clin Cancer Res. 2010;16:5624–9. doi: 10.1158/1078-0432.CCR-10-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammond EM, Dorie MJ, Giaccia AJ. ATR/ATM targets are phosphorylated by ATR in response to hypoxia and ATM in response to reoxygenation. J Biol Chem. 2003;278:12207–13. doi: 10.1074/jbc.M212360200. [DOI] [PubMed] [Google Scholar]
- 22.Chimploy K, Tassotto ML, Mathews CK. Ribonucleotide reductase, a possible agent in deoxyribonucleotide pool asymmetries induced by hypoxia. J Biol Chem. 2000;275:39267–71. doi: 10.1074/jbc.M006233200. [DOI] [PubMed] [Google Scholar]
- 23.Hammond EM, Denko NC, Dorie MJ, Abraham RT, Giaccia AJ. Hypoxia links ATR and p53 through replication arrest. Mol Cell Biol. 2002;22:1834–43. doi: 10.1128/MCB.22.6.1834-1843.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sprong D, Janssen HL, Vens C, Begg AC. Resistance of hypoxic cells to ionizing radiation is influenced by homologous recombination status. Int J Radiat Oncol Biol Phys. 2006;64:562–72. doi: 10.1016/j.ijrobp.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 25.Chan N, Koritzinsky M, Zhao H, Bindra R, Glazer PM, Powell S, et al. Chronic hypoxia decreases synthesis of homologous recombination proteins to offset chemoresistance and radioresistance. Cancer Res. 2008;68:605–14. doi: 10.1158/0008-5472.CAN-07-5472. [DOI] [PubMed] [Google Scholar]
- 26.Ding LH, Park S, Peyton M, Girard L, Xie Y, Minna JD, et al. Distinct transcriptome profiles identified in normal human bronchial epithelial cells after exposure to gamma-rays and different elemental particles of high Z and energy. BMC Genomics. 2013;14:372. doi: 10.1186/1471-2164-14-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding LH, Xie Y, Park S, Xiao G, Story MD. Enhanced identification and biological validation of differential gene expression via Illumina whole-genome expression arrays through the use of the model-based background correction methodology. Nucleic Acids Res. 2008;36:e58. doi: 10.1093/nar/gkn234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirzoeva OK, Petrini JHJ. DNA Damage-Dependent Nuclear Dynamics of the Mre11 Complex. Molecular and Cellular Biology. 2001;21:281–8. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwab RA, Niedzwiedz W. Visualization of DNA replication in the vertebrate model system DT40 using the DNA fiber technique. Journal of visualized experiments: JoVE. 2011:e3255. doi: 10.3791/3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenberg GR, Hilfinger JM. Regulation of synthesis of ribonucleotide reductase and relationship to DNA replication in various systems. Prog Nucleic Acid Res Mol Biol. 1996;53:345–95. doi: 10.1016/s0079-6603(08)60150-6. [DOI] [PubMed] [Google Scholar]
- 32.Odsbu I, Morigen, Skarstad K. A reduction in ribonucleotide reductase activity slows down the chromosome replication fork but does not change its localization. PLoS One. 2009;4:e7617. doi: 10.1371/journal.pone.0007617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graff P, Amellem O, Andersson KK, Pettersen EO. Role of ribonucleotide reductase in regulation of cell cycle progression during and after exposure to moderate hypoxia. Anticancer Res. 2002;22:59–68. [PubMed] [Google Scholar]
- 34.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–26. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 35.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 36.Bindra RS, Schaffer PJ, Meng A, Woo J, Maseide K, Roth ME, et al. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol. 2004;24:8504–18. doi: 10.1128/MCB.24.19.8504-8518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan N, Bristow RG. “Contextual” synthetic lethality and/or loss of heterozygosity: tumor hypoxia and modification of DNA repair. Clin Cancer Res. 2010;16:4553–60. doi: 10.1158/1078-0432.CCR-10-0527. [DOI] [PubMed] [Google Scholar]
- 38.Kumareswaran R, Ludkovski O, Meng A, Sykes J, Pintilie M, Bristow RG. Chronic hypoxia compromises repair of DNA double-strand breaks to drive genetic instability. J Cell Sci. 2012;125:189–99. doi: 10.1242/jcs.092262. [DOI] [PubMed] [Google Scholar]
- 39.Liu SK, Coackley C, Krause M, Jalali F, Chan N, Bristow RG. A novel poly(ADP-ribose) polymerase inhibitor, ABT-888, radiosensitizes malignant human cell lines under hypoxia. Radiother Oncol. 2008;88:258–68. doi: 10.1016/j.radonc.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Astor MB. Oxygen concentration and the OER for acutely or chronically thiol deficient cells. Int J Radiat Oncol Biol Phys. 1986;12:1131–4. doi: 10.1016/0360-3016(86)90242-7. [DOI] [PubMed] [Google Scholar]
- 41.Bindra RS, Schaffer PJ, Meng A, Woo J, Maseide K, Roth ME, et al. Alterations in DNA repair gene expression under hypoxia: elucidating the mechanisms of hypoxia-induced genetic instability. Annals of the New York Academy of Sciences. 2005;1059:184–95. doi: 10.1196/annals.1339.049. [DOI] [PubMed] [Google Scholar]
- 42.Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23:247–51. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 43.Petrini JH. The mammalian Mre11-Rad50-nbs1 protein complex: integration of functions in the cellular DNA-damage response. Am J Hum Genet. 1999;64:1264–9. doi: 10.1086/302391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin YF, Shih HY, Shang Z, Matsunaga S, Chen BP. DNA-PKcs is required to maintain stability of Chk1 and Claspin for optimal replication stress response. Nucleic Acids Res. 2014;42:4463–73. doi: 10.1093/nar/gku116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shechter D, Costanzo V, Gautier J. Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair (Amst) 2004;3:901–8. doi: 10.1016/j.dnarep.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 46.Yajima H, Lee KJ, Chen BP. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol Cell Biol. 2006;26:7520–8. doi: 10.1128/MCB.00048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–51. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 48.Maya-Mendoza A, Ostrakova J, Kosar M, Hall A, Duskova P, Mistrik M, et al. Myc and Ras oncogenes engage different energy metabolism programs and evoke distinct patterns of oxidative and DNA replication stress. Molecular oncology. 2015;9:601–16. doi: 10.1016/j.molonc.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veuger SJ, Curtin NJ, Smith GC, Durkacz BW. Effects of novel inhibitors of poly(ADP-ribose) polymerase-1 and the DNA-dependent protein kinase on enzyme activities and DNA repair. Oncogene. 2004;23:7322–9. doi: 10.1038/sj.onc.1207984. [DOI] [PubMed] [Google Scholar]
- 50.Nowsheen S, Cooper T, Stanley JA, Yang ES. Synthetic lethal interactions between EGFR and PARP inhibition in human triple negative breast cancer cells. PLoS One. 2012;7:e46614. doi: 10.1371/journal.pone.0046614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.