Abstract
Detection of methylated genes in exfoliated cells from the lungs of smokers provides an assessment of the extent of field cancerization, is a validated biomarker for predicting lung cancer, and provides some discrimination when interrogated in blood. The potential utility of this 8-gene methylation panel for predicting tumor recurrence has not been assessed. The Eastern Cooperative Oncology Group initiated a prevention trial (ECOG-ACRIN5597) that enrolled resected Stage I non-small cell lung cancer patients who were randomized 2:1 to receive selenized yeast versus placebo for four years. We conducted a correlative biomarker study to assess prevalence for methylation of the 8-gene panel in longitudinally collected sputum and blood after tumor resection to determine if selenium alters their methylation profile and whether this panel predicts local and/or distant recurrence. Patients (n=1561) were enrolled into the prevention trial, 565 participated in the biomarker study with 122 recurrences among that group. Assessing the association between recurrence and risk of gene methylation longitudinally for up to 48 months showed a 1.4-fold increase in odds ratio for methylation in sputum in the placebo group independent of location (local or distant). Kaplan Meier curves evaluating the association between number of methylated genes and time to recurrence showed no increased risk in sputum, while a significant hazard ratio of 1.5 was seen in plasma. Methylation detection in sputum and blood is associated with risk for recurrence.
Keywords: gene methylation, lung cancer recurrence, biomarker
Introduction
Lung cancer remains the leading cause of cancer-related death for men and women in the US due largely to detection of most cancers at advanced stage and modest effectiveness of chemotherapy (1). Even for patients whose early stage I non-small cell lung cancer (NSCLC) is detected and surgically removed, rate of recurrence within 5 years is ~25% (2). A pooled analysis of five large trials with cisplatin-based chemotherapy revealed a five-year benefit of only 5.4% from adjuvant therapy (3). The ineffectiveness of adjuvant therapy in general may stem in part from the short treatment period (~three to six months) dictated by the systemic toxicity of the therapy and the likelihood that the occult local or regional lesion is quiescent over this time period. Developing lung cancer preventive drugs that can be given chronically has been challenging because of the extensive heterogeneity of this disease with respect to genetic and epigenetic alterations and the necessity of a non-genotoxic agent.
The addition of molecular biomarkers interrogated in accessible biologic fluids such as sputum and blood have shown promise for predicting lung cancer risk. In addition, biomarkers detected in sputum or blood could be used to monitor chemopreventive agents for affecting local and distant disease recurrence (4, 5). Gene silencing through methylation of cytosine in CpG islands in conjunction with chromatin remodeling leads to the development of heterochromatin of the gene promoter region, which denies access to regulatory proteins needed for transcription (6). This epigenetically driven process is a major and causal event silencing hundreds of genes involved in all aspects of normal cellular function during lung cancer initiation and progression (6). Our group showed that detecting gene methylation in exfoliated cells from the lungs of smokers provides an assessment of the extent of field cancerization, could predict cancer up to 18 months prior to clinical diagnosis, and was independently validated through case-control studies for predicting LC risk (7, 8). However, the potential utility of this validated gene methylation panel for predicting tumor recurrence has not been assessed.
One chemopreventive agent extensively studied is selenium. In vitro and in vivo studies showed that selenium supplementation reduced the development of several types of cancer by inhibiting chemical carcinogenesis at the initiation and post-initiation stages (9–12). Clark et al. (13) conducted the Nutritional Prevention of Cancer Trial in patients with a history of basal cell or SCC of the skin. All subjects received either daily L-selenomethionine or placebo for 4.5 years with up to 6.4 years of follow-up. Although the primary endpoint of skin cancer prevention was negative, secondary endpoints revealed significant protection for cancers of the lung, prostate, colon, and esophagus, albeit in a selenium deficient population.
Based on the protective effect of selenium against lung cancer, the Eastern Cooperative Oncology Group initiated a trial (ECOG-ACRIN5597) that enrolled completely resected Stage I NSCLC patients who were randomized 2:1 based on smoking status, sex and stage to receive selenized yeast versus placebo daily for four years. The primary endpoint for this tertiary chemoprevention trial was the effectiveness of selenium for reducing the incidence of second primary tumors (SPT). The incidence of local and metastatic tumor recurrence was also assessed. An interim analysis of the trial in October 2009 showed a trend in favor of the placebo group with a low likelihood that the trial would become positive; thus the study was stopped. Trial results with regard to SPTs and recurrence concluded that selenium was safe, but conferred no benefit over placebo in the prevention of SPT in patients with resected NSCLC (14).
The major objectives for the current study was to conduct a correlative biomarker study to determine the prevalence for methylation of a gene panel in longitudinally collected sputum and blood after tumor resection of ECOG-ACRIN5597 patients and to follow trial participants beyond four years to determine if selenium alters their methylation profile and whether this panel can predict local and/or distant recurrence.
Materials and Methods
Experimental design
All biospecimens were collected and processed using standardized and validated protocols. The assay for gene methylation described briefly below is also standardized. Personnel conducting the methylation assays were blinded to study group. All data obtained are shown in the manuscript, and none were excluded, even if they were outliers.
ECOG-ACRIN5597 recruitment and biospecimen collection
Eligibility criteria for participation in the prevention trial included the following: age ≥ 18 years; 6 to 36 months from complete resection of histologically proven stage IA (pT1N0) or stage IB (pT2N0) NSCLC (carcinoid tumors were excluded [14]). The institutional review board for human studies approved the protocols and written consent was obtained from subjects. Following consent onto the correlative study, the Lovelace study coordinator sent a collection kit to the study site. Blood and sputum were collected at time of entry onto study, and at 6, 12, 24, and 48 months during the study. Patients were followed for ten-years to collect tumor recurrence data.
Blood (32 cc) was drawn into Citrate Vacutainer CPT tubes (Bectin Dickinson), which are designed to separate plasma and mononuclear cells from whole blood. Blood was processed within 2 hours of collection, and approximately 10 ml of plasma was recovered. Plasma and mononuclear cell pellets were shipped frozen overnight to the study coordinator within 5 days of processing.
Each participant was asked to provide two consecutive spontaneous sputum samples collected at home at each time point as described previously (7). Study participants placed the sputum cups in the postage-paid mailer addressed to the study coordinator at Lovelace. Material from the second 3-day pooled sputum was used for this study. The collection of two sputum samples at each time point was based on the finding by Kennedy et al. (15) that the second sample has a higher success rate (80%) in producing an adequate sputum sample based on established cytologic standards, attributed to a “learning effect” in adequate sputum collection. Following receipt, the sputum samples were pelleted and washed in Saccomanno’s fixative. A small portion was smeared onto two or three slides and stained with Papanicoleau prior to cytologic diagnosis with the remaining sample stored at −80°C until time for DNA isolation. Sputum containing epithelial cells from the upper or lower airways has proven satisfactory for methylation assays and using these criteria, virtually 100% of samples were adequate for study (7).
DNA isolation and methylation specific PCR
Sputum and plasma DNA was isolated using methods previously described with yields of DNA that ranged from 5–100 μg and 0.3–6 μg (7, 16). The eight genes selected were based on positive performance in our initial nested, case-control study in a Colorado cohort (7). These genes included CDKN2, MGMT, DAPK1, RASSF1, GATA4, GATA5, PAX5α and PAX5β. These genes are cancer specific genes methylated solely in epithelial cells. DNA was bisulfite modified and two-stage, nested methylation specific PCR (MSP) assays were used for increased sensitivity for detection of promoter methylation in sputum and plasma as described (7, 16). The immense cellular heterogeneity in sputum where the epithelial fraction is typically <3% of the specimen limits the ability to quantitate methylation, thus methylation was scored as positive or negative. Methylation data was obtained for 8 genes in sputum. Methylation data for GATA4 and DAPK1 in plasma were missing for some ECOG-ACRIN5597 participants at baseline, 6- and 12-months because these genes were added to the methylation panel two years after study initiation and DNA was exhausted. Thus, primary analyses described below evaluated methylation of 8 and 6 genes interrogated in sputum and plasma, respectively.
Statistical analysis
Demographic variables for ECOG-ACRIN5597 participants were summarized by recurrence status with percentages for sex, tumor stage, and smoking status and mean and SD for age at randomization. Differences in demographic variables between recurrence and non-recurrence groups were assessed with Pearson’s χ2 and Student’s t-tests when appropriate.
Generalized estimating equation (GEE) was used to assess the association between selenium treatment, tumor recurrence, and risk for gene methylation (17). A vector of the methylation status of the 8 genes in sputum or 6 genes in plasma (1 for methylated and 0 for unmethylated) for each individual at each time point was used as the outcome in the models. Age, sex, time between tumor resection and patient randomization, and time between patient randomization and sputum or plasma collection were selected a priori as covariates for adjustment. In addition, because methylation outcome of each individual gene was binary, an alternating logistic regression (ALR) algorithm was applied to modify the GEE, which modeled the association between pairs of responses with log odds ratio, instead of with correlations. Specifically, each individual was considered as a cluster, and each gene within an individual was measured repeatedly as a sub-cluster. ALR was applied by using the PROC GENMOD procedure in SAS software with NEST1 regression structure and SUBCLUST options specified on the REPEATED statement. PROC GENMOD in SAS ignores data with missing gene methylation values for each individual, and the ALR assumes the data missing are completely at random, which is the case with our dataset. This model benefits the most from the repeated measurements of multiple genes in each sputum or plasma sample that were collected for up to 4 years from participants. Tumor recurrence was treated as a variable reflecting a latent biological process that leads to recurrence in the models. Similar results were observed for analyses repeated for subjects with the 8-gene panel in plasma and thus are not shown.
The association between the number of genes methylated in sputum or plasma and time from sputum or plasma collection to recurrence was assessed using the Kaplan-Meier estimator and Cox proportional hazard model. Patients without tumor recurrence were censored at last follow-up. Because our previous study suggested that gene methylation measured in sputum samples collected within 18 months prior to cancer diagnosis was predictive for lung cancer, the methylation data from sputum samples proximal to tumor recurrence or last follow-up were assessed in the models (7). The Kaplan-Meier estimator was used to estimate the time to recurrence for tumor patients with low (0–1), medium (2–3), or high (≥ 4) methylation index (sum of methylated genes from the panel) in sputum. Because methylation prevalence was much lower in plasma, comparison was restricted to having zero versus one or more methylated genes. The Cox proportional hazard model was also used to assess the association between methylation index and the time to recurrence with adjustment for age, sex, stage, and selenium treatment. All statistical analyses used two-sided tests and were conducted using SAS 9.3 and R 3.1.
Results
Demographics, clinical variables, outcomes and biomarker collection for study cohort
The ECOG-ACRIN5597 trial randomized 1561 patients 1:2 to placebo or L-selenomethione and 565 patients were enrolled into the correlative biomarker study. Of the 565 patients enrolled onto the biomarker study, 30 were excluded based on lack of sample submission (n=12) or not randomized (n=18). Four hundred and eight subjects provided a plasma sample, 515 provided sputum, and 388 provided both specimens. More than 50% of participants provided a 6- and 12-month biospecimen, while sample collection diminished as expected due to drop off from the trial over the 24- and 48-month collection periods (14).
Recurrence in the form of second primary, recurrent local or metastatic disease was observed in 33, 44, and 45 patients, respectively for a total of 122 events among biomarker study participants. Demographics with respect to age, sex, and tumor stage at resection or taking selenium did not significantly differ between disease free and recurrent patients (Table 1). Patients that recurred were more often current smokers. Demographics and rate of recurrence within the biomarker cohort reflected that of the composite trial participants (14).
Table 1.
Baseline demographics and clinical characteristics of ECOG-ACRIN5597 comparing lung cancer recurrence to disease free patient population
| Variables1 | Cancer recurrence4 | Cancer free | P value5 |
|---|---|---|---|
| N | 122 | 413 | |
| Age (Mean ± SD) | 67.4 + 8.6 | 66.3 + 6.1 | 0.17 |
| Sex (Male) | 61 (50%) | 217 (53%) | 0.62 |
| Stage6 | |||
| 1a | 82 (67%) | 302 (75%) | 0.07 |
| 1b with chemo | 3 (2%) | 16 (4%) | |
| 1b without chemo | 37 (30%) | 83 (21%) | |
| Smoking status | |||
| Current | 44 (36%) | 116 (28%) | 0.08 |
| Former2 | 64 (52%) | 263 (64%) | |
| Never3 | 14 (12%) | 34 (8%) | |
| Selenium use | 90 (74%) | 268 (65%) | 0.07 |
| Time to event7 | 35 (20, 56) | ||
| Time to censor7 | 69 (50, 94) | ||
All demographics represent baseline information.
Quit ≥ 1 y before study
Less than 100 cigarettes smoked
Includes second primary, local and distant recurrence.
Χ2 was used for all variables except age that was evaluated by Student’s t test.
Stage was missing from 12 cancer-free participants.
Median months (Q1, Q3).
Age and Temporal Effects on Prevalence for Sputum and Plasma Gene Methylation
The prevalence for methylation of each gene within the 8-gene panel ranged from 10 to 71% in baseline sputum samples (Table 2). Compared to sputum, plasma has considerably lower methylation prevalence ranging from 4 to 38%, corroborating our initial studies in this population (Tables 2 & 3 [16]). GEE models showed that every ten-year increase in age for trial participants was associated with increased risk for methylation in sputum (OR = 1.10; 95% CI = 1.01–1.19, p = 0.026) and plasma (OR = 1.21, 95% CI = 1.08–1.36, p = 0.0012). Every 6-month increase in time between baseline and longitudinal sputum collection was associated with an increased risk for methylation (OR = 1.04, 95% CI = 1.03–1.07, p = 0.033). The risk for gene methylation in plasma did not change over time.
Table 2.
Gene Methylation Prevalence Over Time in Sputum
| Gene | Baseline | 6-Month | 12-Month | 24-Month | 48-Month | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | N | % | |
| CDKN2 | 507 | 35 | 313 | 42 | 220 | 39 | 154 | 45 | 75 | 49 |
| MGMT | 504 | 43 | 313 | 42 | 223 | 47 | 154 | 49 | 75 | 53 |
| RASSF1 | 509 | 10 | 313 | 4 | 223 | 4 | 154 | 0 | 75 | 3 |
| DAPK1 | 502 | 43 | 307 | 42 | 223 | 46 | 154 | 36 | 75 | 31 |
| GATA4 | 505 | 71 | 309 | 75 | 222 | 84 | 153 | 78 | 75 | 85 |
| GATA5 | 505 | 45 | 311 | 51 | 222 | 52 | 153 | 51 | 75 | 45 |
| PAX5α | 504 | 31 | 312 | 35 | 223 | 32 | 154 | 29 | 75 | 45 |
| PAX5β | 507 | 31 | 313 | 41 | 222 | 39 | 154 | 44 | 75 | 53 |
Minor sample size differences across genes reflect the lack of detection of an unmethylated or methylated gene product, thus leading to unknown status.
Table 3.
Gene Methylation Prevalence Over Time in Plasma
| Gene | Baseline | 6-Month | 12-Month | 24-Month | 48-Month | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | N | % | |
| CDKN2 | 404 | 25 | 343 | 19 | 267 | 28 | 207 | 21 | 111 | 15 |
| MGMT | 402 | 10 | 342 | 12 | 266 | 8 | 206 | 12 | 110 | 7 |
| RASSF1 | 398 | 4 | 342 | 7 | 265 | 8 | 206 | 11 | 110 | 5 |
| DAPK11 | 252 | 17 | 294 | 16 | 246 | 13 | 205 | 10 | 111 | 11 |
| GATA41 | 222 | 38 | 292 | 30 | 243 | 27 | 197 | 36 | 111 | 27 |
| GATA5 | 393 | 18 | 338 | 18 | 249 | 14 | 193 | 14 | 107 | 7 |
| PAX5α | 394 | 8 | 333 | 3 | 261 | 3 | 187 | 3 | 93 | 3 |
| PAX5β | 396 | 5 | 338 | 5 | 262 | 3 | 195 | 4 | 105 | 6 |
Sample sizes for GATA4 and DAPK1 are reduced at baseline, 6- and 12-months because these genes were added to the methylation panel two years after study initiation and DNA was exhausted from some plasma samples.
Minor sample size differences across other genes reflect the lack of detection of an unmethylated or methylated gene product, thus leading to unknown status.
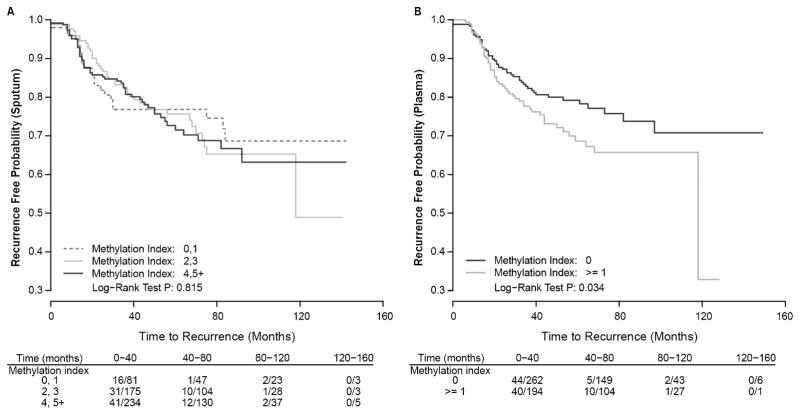
Association between gene methylation index and time to recurrence
Because individual gene methylation prevalence were high in sputum, we assessed associations with respect to three indexes: 0–1, 2–3, or ≥ 4 using the biospecimen collected most proximal to disease recurrence compared to the last sample collected from patients that remained disease free for up to 160 months. No significant association was seen (Figure 1A). In contrast, in plasma where methylation prevalence was much lower than sputum, subjects with one or more genes methylated were at increased risk for local or distant recurrence compared to those with zero genes methylated (HR = 1.51, 95% CI, 1.01–2.25, p = 0.045 [Figure 1B]). A random sampling approach was further applied to assess whether the methylation index in plasma measured at any single visit from patients with multiple visits can predict tumor recurrence (not shown). The median HR and p values were 1.21 and 0.35, respectively; suggesting only the methylation index in plasma samples proximal to recurrence is predictive. Kaplan Meier curves did not differ when stratified by treatment status (not shown).
Fig. 1.
Kaplan-Meier plots show recurrence free probability as a function of methylation index detected in sputum (A) and plasma (B) over 160 months. Methylation index reflecting the number of genes methylated in the panel was assessed with comparison to number of events/number of persons at risk over four time intervals.
Effect of selenium treatment and tumor recurrence on the risk for gene methylation
GEE was used to assess the association between selenium treatment and recurrence for risk of gene methylation in sputum and plasma evaluated longitudinally for up to 48 months. The risk for methylation in sputum or plasma was not increased among subjects taking selenium who had recurrent disease relative to subjects without recurrence (Table 4). In contrast, a significant 1.4-fold increase in odds ratio for methylation in sputum was seen in association with recurrence that was independent of location (local or distant) in subjects taking placebo (Table 4). No association was seen in plasma for the placebo group. Selenium treatment also had no significant effect on risk for methylation in sputum or plasma in subjects who remained cancer free (Table 4).
Table 4.
Effect of selenium treatment and tumor recurrence on risk for gene methylation in sputum and plasma
| Se Treatment1 | Recurrence | OR (95% CI)2 | P value | OR (95% CI)2 | P value |
|---|---|---|---|---|---|
| (Sputum) | (Plasma) | ||||
| Yes | Yes | 0.86 (0.73–1.03) | 0.11 | 1.03 (0.78–1.34) | 0.85 |
| Yes | No | ||||
| No | Yes | 1.43 (1.16–1.77) | 0.00086 | 1.15 (0.75–1.34) | 0.51 |
| No | No | ||||
| Yes | No | 1.12 (0.98–1.29) | 0.10 | 0.81 (0.65–1.01) | 0.06 |
| No | No | ||||
A highly significant interaction was observed between selenium treatment, and tumor recurrence when assessing risk for methylation (p < 0.001).
Adjusted for age, sex, time between tumor resection and patient randomization, and sputum collection up to 4 years.
Discussion
Our findings provide the first evidence that for patients taking placebo who relapse with lung cancer, their existing extensive field of injury as detected by methylation in sputum continues to increase significantly. In addition, detection of gene methylation in plasma was predictive for disease recurrence.
The GEE model that incorporates change in methylation prevalence over time showed that increased prevalence for methylation of the eight-gene panel detected in sputum was associated with local recurrence or a SPT in the placebo group, consistent with the hypothesis that for some patients, the field of injury continues to expand. Reduced DNA repair capacity is a validated determinant for increased gene methylation and risk factor for lung cancer (18–20). Spontaneous sputum production likely coincides with an active inflammatory process that continues to foster DNA damage. However, local tumor recurrence in patients taking selenium occurred in the absence of increased methylation prevalence. In vitro studies supporting an effect of selenium on cell cycle regulation, apoptosis, mitigating inflammatory mediated DNA damage and potential cytosine demethylation could blunt any progressive increase in the field of injury, thereby abrogating the increase in methylation seen in the placebo group (21–23). However, these effects if operative, were insufficient to prevent local cancer recurrence. A potential explanation for these findings is that recurrent and SPTs are already committed lesions that do not require additional genetic and epigenetic changes, rather host-environment interactions that stimulate clonal expansion. Based on these findings we speculate that selenium treatment masks the utility of change in sputum methylation seen in placebo that is likely an indicator for elevated inherent disease susceptibility, that in turn has prognostic value for predicting local recurrence and surprisingly distant metastasis.
The elevated methylation burden present in lung cancer patients was likely a limiting factor for the lack of association between methylation in the proximal sputum samples and recurrence assessed by the Cox proportional hazards model. However, for plasma where baseline methylation was low, the cumulative change reflected by methylation in the proximal sample was associated with recurrence. The accumulation of circulating DNA in blood likely arises from a combination of cell lysis by necrosis, apoptosis or local angiogenesis from all tissue compartments. Our findings showed that increased methylation seen in proximal plasma samples was independent of location of tumor recurrence (lung versus distant) thereby reflecting the increased burden of field cancerization and the developing tumor from the lung or the recurrent distant tumor.
The clinical translation of this work should be to provide oncology physicians with the option of ordering a low cost (≤ $200) insurance reimbursable test for predicting risk for recurrence to guide treatment options. Our patented technology (24) is amenable to a CLIA setting through development of robotic/liquid handling for DNA isolation, bisulfite modification, and assembling of the Stage I and II MSP reactions in a 96-well format in conjunction with low cost SYBER-Green based detection of methylated products using real time PCR. Once established, this high throughput assay will be used in a validation study needed prior to its implementation in the clinic.
Acknowledgments
We would like to thank Dr. Christine Stidley for advice regarding data analysis strategy and Ms. Donna Klinge and Elia Casas with assistance in methylation assays and recruitment of patients.
Grant support
This work was primarily supported by National Cancer Institute grant R01 CA095568 (SAB). The State of New Mexico as a direct appropriation from the Tobacco Settlement Fund to SAB through collaboration with University of New Mexico provided initial support to establish the LSC. R01CA097356 (SAB) and NIH/NCI P30 CA118100 provided additional support. This study was also supported in part by the ECOG-ACRIN Cancer Research Group (Robert L. Comis, MD and Mitchell D. Schnall, MD, PhD, Group Chairs Co-Chairs) by Public Health Service Grants CA180794, CA180820, CA180864, CA180844, CA180858, CA189828 and the National Cancer Institute, National Institutes of Health and the Department of Health and Human Services. The National Cancer Institute had no role in the design and conduct of the study: collection, management analysis, and interpretation of the data, preparation, review, or approval of the manuscript, and the decision to submit the manuscript for publication.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors declare no potential conflict of interests.
Disclaimer
The contents of this study are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer Statistics. CA Cancer J Clinc. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Goodgame B, Viswanathan A, Zoole J, Gao F, Miller CR, Subramanian J, et al. Risk of recurrence of resected stage I non-small cell lung cancer in elderly patients as compared with young patients. J Thorac Oncol. 2009;4:1370–74. doi: 10.1097/JTO.0b013e3181b6bc1b. [DOI] [PubMed] [Google Scholar]
- 3.Pignon JP, Tribodet H, Scagliotti GV, Douillard JY, Shepherd FA, Spiro SG, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by LACE Collaborative Group. J Clin Oncol. 2008;26:3552–59. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis, and treatment selection. Nat Rev Cancer. 2005;5:845–56. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 5.Belinsky SA. Gene promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–17. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 6.Belinsky SA. Unmasking the Lung Cancer Epigenome. Ann Rev Physiol. 2015;77:453–74. doi: 10.1146/annurev-physiol-021014-072018. [DOI] [PubMed] [Google Scholar]
- 7.Belinsky SA, Liechty KC, Gentry FD, Wolf HJ, Rogers J, Vu K, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338–44. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- 8.Leng S, Do K, Yingling CM, Picchi MA, Wolf HJ, Kennedy TC, et al. Defining a gene promoter methylation signature in sputum for lung cancer risk assessment. Clin Cancer Res. 2012;18:3387–95. doi: 10.1158/1078-0432.CCR-11-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy BS, Sugie S, Maruyama H, El-Bayoumy K, Mara P. Chemoprevention of colon carcinogenesis by dietary organoselenium, benzylselenocyanate, in F344 rats. Cancer Res. 1987;47:5901–04. [PubMed] [Google Scholar]
- 10.Reddy BS, Rivenson A, Kulkarni N, Upadhyaya P, El-Bayoumy K. Chemoprevention of colon carcinogenesis by the synthetic organoselenium compound 1,4-phenylenebis(methylene)-selenocyanate. Cancer Res. 1992;52:5635–40. [PubMed] [Google Scholar]
- 11.Tanaka T, Makita H, Kawabata K, Mori H, El-Bayoumy K. 1,4-Phenylenebis(methylene)-selenocyanate exerts exceptional chemopreventive activity in rat tongue carcinogenesis. Cancer Res. 1997;57:3644–48. [PubMed] [Google Scholar]
- 12.Richie JP, Kleinman W, Desai DH, Das A, Amin SG, Pinto JT, et al. The organoselenium compound 1,4-phenylenebis(methylene) selenocyanate inhibits 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced tumorigenesis and enhances glutathione-related antioxidant levels in A/J mouse lung. Chem Biol Interact. 2006;161:93–103. doi: 10.1016/j.cbi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Clark LC, Combs GF, Turnbull BW, Slate EH, Chalker DK, Chow J, et al. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. JAMA. 1996;276:1957–63. [PubMed] [Google Scholar]
- 14.Karp DD, Lee SJ, Keller SM, Wright GS, Aisner S, Belinsky SA, et al. A randomized, double blind, placebo-controlled, Phase III chemoprevention trial of selenium supplementation in persons with resected stage I non-small cell lung cancer (ECOG 5597) J Clin Oncol. 2013;31:4179–87. doi: 10.1200/JCO.2013.49.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy TC, Proudfoot SP, Piantadosi S, Wu L, Saccomanno G, Petty TL, et al. Efficacy of two sputum collection techniques in patients with airflow obstruction. Acta Cytol. 1999;434:630–36. doi: 10.1159/000331157. [DOI] [PubMed] [Google Scholar]
- 16.Belinsky SA, Klinge DM, Dekker JD, Smith MW, Bocklage TJ, Gilliland FD, et al. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin Cancer Res. 2005;11:6505–11. doi: 10.1158/1078-0432.CCR-05-0625. [DOI] [PubMed] [Google Scholar]
- 17.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–30. [PubMed] [Google Scholar]
- 18.Leng S, Liu Y, Weissfeld JL, Thomas CL, Han Y, Picchi MA, et al. 15q12 variants, gene promoter hypermethylation, and lung cancer risk: A GWAS in smokers. J Natl Cancer Inst. 2015;107:djv035. doi: 10.1093/jnci/djv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leng S, Stidley C, Willink R, Bernauer A, Do K, Picchi M, et al. Double-strand break damage and associated DNA repair genes predispose smokers to gene methylation. Cancer Res. 2008;68:3049–56. doi: 10.1158/0008-5472.CAN-07-6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sigurdson AJ, Jones IM, Wei Q, Wu X, Spitz MR, Stram DA, et al. Prospective analysis of DNA damage and repair markers of lung cancer risk from the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Carcinogenesis. 2011;32:69–73. doi: 10.1093/carcin/bgq204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Werz O, Steinhilber D. Selenium-dependent peroxidases suppress 5-lipoxygenase activity in B-lymphocytes and immature myeloid cells. The presence of peroxidase-insensitive 5-lipoxygenase activity in differentiated myeloid cells. Eur J Biochem. 1996;242:90–97. doi: 10.1111/j.1432-1033.1996.0090r.x. [DOI] [PubMed] [Google Scholar]
- 22.Fleming J, Ghose A, Harrison PR. Molecular mechanisms of cancer prevention by selenium compounds. Nutr Cancer. 2001;40:42–49. doi: 10.1207/S15327914NC401_9. [DOI] [PubMed] [Google Scholar]
- 23.Fiala ES, Staretz ME, Pandya GA, El-Bayoumy K, Hamilton SR. Inhibition of DNA cytosine methyltransferase by chemopreventive selenium compounds, determined by an improved assay for cytosine methyltransferase and DNA cytosine methylation. Carcinogenesis. 1998;19:597–604. doi: 10.1093/carcin/19.4.597. [DOI] [PubMed] [Google Scholar]
- 24.Belinsky SA. Gene methylation as a biomarker in sputum. 9,512,483. US Patent. 2016