Abstract
The transsulfuration enzyme cystathionine-β-synthase (CBS) and its product hydrogen sulfide (H2S) are aberrantly upregulated in colorectal cancers, where they contribute to tumor growth and progression by both autocrine and paracrine mechanisms. However, it is unknown whether the CBS/H2S axis plays a role in colorectal carcinogenesis. Here, we report upregulation of CBS in human biopsies of precancerous adenomatous polyps and show that forced upregulation of CBS in an adenoma-like colonic epithelial cell line is sufficient to induce metabolic and gene expression profiles characteristic of colorectal cancer cells. Differentially expressed metabolites (65 increased and 20 decreased) clustered into the glycolytic pathway, nucleotide sugars, intermediates of the pentose phosphate pathway, and lipogenesis, including primarily phospholipids, sphingolipids, and bile acids. CBS upregulation induced broad changes in the NCM356 cell transcriptome with over 350 differentially expressed genes. These genes overlapped significantly with gene sets related to glycolysis, hypoxia, and a colon cancer cell phenotype, including genes regulated by NF-κB, KRAS, p53, and Wnt signaling, genes downregulated after E-cadherin knockdown, and genes related to increased extracellular matrix, cell adhesion, and epithelial-to-mesenchymal transition. The CBS-induced switch to an anabolic metabolism was associated with increased NCM356 cell bioenergetics, proliferation, invasion through Matrigel, resistance to anoikis, and CBS-dependent tumorigenesis in immune compromised mice. Genetic ablation of CBS in CBS heterozygous mice (CBS+/−) reduced the number of mutagen-induced aberrant colonic crypt foci. Taken together, these results establish that activation of the CBS/H2S axis promotes colon carcinogenesis.
Keywords: cancer metabolism, hydrogen sulfide, transsulfuration pathway, adenoma to carcinoma sequence, cystathionine-βsynthase
INTRODUCTION
Previously, we have reported the aberrant overexpression of cystathionine-β-synthase (CBS; EC 4.2.1.22) in colorectal tumor tissue and colon cancer-derived cell lines (1). CBS is the first and rate-limiting enzyme of the reverse transsulfuration pathway, the primary metabolic pathway for the synthesis of cysteine, and a key enzyme in regulating methionine metabolism. CBS synthesizes cystathionine from the condensation of homocysteine and serine. Cystathionine in turn is hydrolyzed by cystathionine-γ-lyase (CSE) to produce the amino acid cysteine. Cysteine is used for the biosynthesis of proteins, glutathione, and the gaseous transmitter hydrogen sulfide (H2S), which is produced by both CBS and CSE (2). In established colorectal cancers and cancer-derived cell lines, CBS-dependent production of H2S, and not CSE produced H2S, has been shown to regulate cellular bioenergetics in a context-specific and concentration-dependent manner (3,4). The CBS-H2S signaling axis promotes colorectal cancer growth and metastasis through cancer cell autonomous regulation of cellular proliferation, migration, and bioenergetics, as well as paracrine regulation of tumor angiogenesis (1). Subsequently, others have reported in various cancer types that CBS promotes cancer growth, metastasis and chemoresistance, confirming the importance of this metabolic enzyme in established cancers (5–9). However, it is unknown whether CBS plays a role in carcinogenesis.
We report that CBS expression is increased in human adenomatous polyps, and that forced expression of CBS in a premalignant colonic epithelial cell line causes metabolic reprograming and induction of an invasive tumorigenic phenotype. Furthermore, we demonstrate a direct correlation between CBS gene dosage and mutagen-induced aberrant crypt foci formation supporting the conclusion that enhanced CBS activity can promote colon carcinogenesis.
MATERIALS AND METHODS
Experimental Procedures
Cell lines
HCT116 colon cancer cell line (ATCC; Manassas, VA, purchased in 2012) and NCM356 cells (Incell Corporation, LLC; San Antonio, TX, purchased in 2010) were cultured in McCoy’s 5A or high glucose DMEM (Sigma-Aldrich) supplemented with 10% FBS, respectively. All cells were frozen at passages 2–3 since purchase. Experiments were performed using passages 3–15 since removal from liquid N2. The NCM356 parental cells were transduced with lentiviral particles (cat # LPP-Z0797-Lv103-050, GeneCopoeia; Rockville MD) containing a N-eGFP-tagged CBS cDNA (NM_000071.2) or the empty vector (Cat #LPP-EGFP-Lv103-050), respectively, according to the manufacturer’s protocol, and cultured in high glucose DMEM with 10% FBS and puromycin (2 µg/ml) to maintain selection. We confirmed cell line authentication using Short Tandem Repeat (STR) analysis. All cells used were mycoplasma free using MycoAlert™ Kit (Lonza); they are tested when removed from liquid-N2 storage and monitored monthly to insure sterility.
Western blots
Equal amounts of protein were separated on 10% Tris Gylcine gels (Invitrogen) and transferred to PVDF membranes using a Pierce Power Blotter. Membranes were blocked with 5% low fat milk in Tris buffered saline and 0.02% Tween 20 (TBST) and then probed overnight at 4°C with 1° antibodies to anti-CBS (1:1000 dilution) or anti- CSE (1:1000) (Proteintech). Membranes were washed three times with TBST and incubated in anti-rabbit horseradish peroxidase conjugated 2° antibody at 1:10,000 dilution (Santa Cruz). Signal was detected using chemiluminescent substrate. Protein content was normalized with anti-β actin antibody (Sigma) (1:5000); densitometry was performed using Image J Software.
Immunohistochemical staining
Normal colonic mucosa, colonic polyps, and colorectal cancer specimens were obtained using an approved institutional review board (IRB) protocol. Formalin-fixed paraffin-embedded tissues sections (4 µm) were deparaffinized in xylene and rehydrated through a graded series of ethanol solutions. Antigen retrieval was performed in 0.01 mol/L citrate buffer (pH 6.0) by microwave oven for 10 min at 98°C to 100°C. Endogenous peroxidase activity was inhibited with 3% H2O2 (in methanol) for 30 min at room temperature (RT). Tissue sections were blocked for 30 min with 5% goat serum (Vector Laboratories) and incubated with a CBS antibody (1:150 dilution, Proteintech, #14787-1-AP) for 1 h RT, then a goat anti-rabbit 2° antibody (Vector Laboratories) for 1 h RT. Positive staining was visualized with DAB (3,3’-diaminobenzidine; Vector Laboratories) and counterstained with hematoxylin. Slides were dehydrated by immersing in three alcohol baths for 5 min, cleared in two xylene baths, and then application of coverslip. Images were captured at 200X and 400X magnification using an Olympus BX51 microscope.
Metabolomic profiling
Global metabolic profiles were performed by Metabolon, Inc. (Durham, NC) (10,11). The samples were extracted and prepared per Metabolon’s standard solvent extraction method. Technical replicates (n=4) were normalized by protein content (Bradford assay). The extract was divided into four fractions: two for analysis by two separate reverse phase (RP)/UPLC-MS/MS methods with positive ion mode electrospray ionization (ESI), one for analysis by RP/UPLC-MS/MS with negative ion mode ESI, and one for analysis by HILIC/UPLC-MS/MS with negative ion mode ESI. Following log transformation and imputation with minimum observed values for each compound, Welch’s two-sample t-tests identified biochemicals that differed significantly between the two groups. A total of 85 named biochemicals were identified at a cutoff of p≤0.05 (Table 1).
Table 1.
Metabolites that are over- or under-expressed in NCM356 cells overexpressing CBS (CBS2) compared to vector-transduced control NCM356 cells.
| Pathway | Biochemical name |
NCM356+CBS NCM356 |
p-value | q-value |
|---|---|---|---|---|
| Alanine and Aspartate Metabolism | N-acetylasparagine | 0.7568 | 0.0023 | 0.0554 |
| Glutamate Metabolism | N-acetylglutamine | 0.8459 | 0.0315 | 0.1203 |
| gamma-aminobutyrate (GABA) | 0.6944 | 0.0035 | 0.0577 | |
| glutamate, gamma-methyl ester | 1.3026 | 0.0231 | 0.1047 | |
| Phenylalanine and Tyrosine Metabolism | N-acetylphenylalanine | 0.7508 | 0.0073 | 0.0742 |
| N-acetyltyrosine | 0.6497 | 0.0295 | 0.1157 | |
| Tryptophan Metabolism | Indolelactate | 0.8672 | 0.0437 | 0.1496 |
| serotonin | 2.4659 | 4.49E-06 | 0.0006 | |
| Leucine, Isoleucine and Valine Metabolism | Tiglylcarnitine | 1.2628 | 0.0333 | 0.1235 |
| 2-hydroxy-3-methylvalerate | 0.6267 | 0.0461 | 0.1504 | |
| alpha-hydroxyisocaproate | 0.5781 | 0.0372 | 0.1343 | |
| Methionine, Cysteine, SAM and Taurine Metabolism | S-adenosylhomocysteine (SAH) | 0.6148 | 0.0024 | 0.0554 |
| Homocysteine | 0.5229 | 0.0028 | 0.0554 | |
| Cystathionine | 1.4358 | 0.0164 | 0.1 | |
| Cysteine sulfinic acid | 1.3029 | 0.0462 | 0.1504 | |
| Polyamine Metabolism | N-acetylputrescine | 0.6283 | 0.0063 | 0.0726 |
| Glutathione Metabolism | ophthalmate | 1.4876 | 0.0093 | 0.0829 |
| Gamma-glutamyl Amino Acid | gamma-glutamylalanine | 0.7866 | 0.0021 | 0.0554 |
| gamma-glutamyl-epsilon-lysine | 1.2492 | 0.0146 | 0.0972 | |
| gamma-glutamylvaline | 1.1864 | 0.0096 | 0.0829 | |
| Dipeptide Derivative | anserine | 1.2466 | 0.0175 | 0.1002 |
| Dipeptide | phenylalanylglycine | 1.9195 | 0.0033 | 0.0577 |
| tryptophylglycine | 1.5487 | 0.0274 | 0.1138 | |
| tyrosylglycine | 2.0093 | 0.0045 | 0.0598 | |
| valylglutamine | 1.4499 | 0.0218 | 0.1047 | |
| leucylglutamine | 1.4636 | 0.0029 | 0.0554 | |
| Glycolysis, Gluconeogenesis, Pyruvate Metabolism | Lactate | 1.3496 | 0.0029 | 0.0554 |
| glycerate | 1.7169 | 0.0105 | 0.0842 | |
| Pentose Phosphate Pathway | 6-phosphogluconate | 1.4264 | 0.0025 | 0.0554 |
| Glycogen Metabolism | maltotriose | 1.2892 | 0.0422 | 0.147 |
| Fructose, Mannose and Galactose Metabolism | mannitol/sorbitol | 1.3805 | 0.0019 | 0.0554 |
| Nucleotide Sugar | UDP-glucose | 1.532 | 0.0205 | 0.103 |
| UDP-galactose | 1.5104 | 0.0026 | 0.0554 | |
| UDP-acetylglucosamine/galactosamine | 0.4683 | 0.0106 | 0.0842 | |
| Aminosugar Metabolism | N-acetyl-glucosamine 1-phosphate | 1.4594 | 0.007 | 0.0742 |
| TCA Cycle | succinate | 0.8096 | 0.0184 | 0.1002 |
| Long Chain Fatty Acid | myristoleate (14:1n5) | 1.3327 | 0.0287 | 0.1144 |
| 10-nonadecenoate (19:1n9) | 1.5602 | 0.0489 | 0.1537 | |
| Fatty Acid, Branched | 15-methylpalmitate | 1.5051 | 0.0178 | 0.1002 |
| Fatty Acid Metabolism (Acyl Carnitine) | octanoylcarnitine | 1.64 | 0.0041 | 0.0586 |
| Phospholipid Metabolism | 1,2-dipalmitoyl-GPC (16:0/16:0) | 1.2133 | 0.026 | 0.1129 |
| 1,2-dioleoyl-GPC (18:1/18:1)* | 1.2038 | 0.0401 | 0.1429 | |
| 1-palmitoleoyl-2-oleoyl-GPC (16:1/18:1)* | 1.2242 | 0.047 | 0.1511 | |
| 1-palmitoyl-2-oleoyl-GPE (16:0/18:1) | 1.2722 | 0.0174 | 0.1002 | |
| 1-stearoyl-2-oleoyl-GPE (18:0/18:1) | 1.2325 | 0.0359 | 0.1312 | |
| 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | 1.7167 | 0.0127 | 0.0972 | |
| 1-palmitoyl-2-stearoyl-GPC (16:0/18:0) | 1.2002 | 0.0227 | 0.1047 | |
| 1,2-dioleoyl-GPE (18:1/18:1) | 1.3008 | 0.0079 | 0.0742 | |
| 1,2-dilinoleoyl-GPC (18:2/18:2) | 1.2698 | 0.0424 | 0.147 | |
| 1-oleoyl-2-linoleoyl-GPE (18:1/18:2)* | 1.266 | 0.0133 | 0.0972 | |
| 1,2-distearoyl-GPC (18:0/18:0) | 2.0252 | 0.0153 | 0.0995 | |
| Lysolipid | 1-lignoceroyl-GPC (24:0) | 1.2294 | 0.0194 | 0.1014 |
| Plamalogen | 1-(1-enyl-palmitoyl)-2-linoleoyl-GPE (P-16:0/18:2)* | 0.6839 | 0.019 | 0.1014 |
| Glycerolipid Metabolism | glycerophosphoglycerol | 1.2397 | 0.0331 | 0.1235 |
| Sphingolipid Metabolism | sphingomyelin (d18:1/22:1, d18:2/22:0, d16:1/24:1)* | 1.2077 | 0.028 | 0.1138 |
| sphingomyelin (d18:1/21:0, d17:1/22:0, d16:1/23:0)* | 1.4556 | 0.0068 | 0.0742 | |
| sphingomyelin (d18:2/23:0, d18:1/23:1, d17:1/24:1)* | 1.3062 | 0.0281 | 0.1138 | |
| tricosanoyl sphingomyelin (d18:1/23:0)* | 1.3659 | 0.0238 | 0.1059 | |
| glycosyl-N-stearoyl-sphingosine | 1.5483 | 0.0181 | 0.1002 | |
| glycosyl-N-palmitoyl-sphingosine | 1.3352 | 0.022 | 0.1047 | |
| Mevalonate Metabolism | 3-hydroxy-3-methylglutarate | 1.1996 | 0.0448 | 0.1504 |
| Primary Bile Acid Metabolism | glycocholate | 1.5488 | 0.0028 | 0.0554 |
| taurocholate | 1.577 | 0.0107 | 0.0842 | |
| glycochenodeoxycholate | 1.4125 | 0.0081 | 0.0742 | |
| Purine Metabolism | inosine 5’-monophosphate (IMP) | 0.3173 | 0.0015 | 0.0554 |
| 2’-deoxyinosine | 1.3469 | 0.0135 | 0.0972 | |
| N6-succinyladenosine | 1.1581 | 0.0479 | 0.1524 | |
| guanosine 5’- diphosphate (GDP) | 0.6358 | 0.0165 | 0.1 | |
| guanine | 2.0175 | 0.0143 | 0.0972 | |
| Pyrimidine Metabolism | uridine 5’-monophosphate (UMP) | 1.3071 | 0.0459 | 0.1504 |
| beta-alanine | 0.77 | 0.0061 | 0.0726 | |
| ribose 1-phosphate | 1.2259 | 0.0266 | 0.1129 | |
| Nicotinate and Nicotinamide Metabolism | nicotinamide ribonucleotide (NMN) | 1.6278 | 0.0075 | 0.0742 |
| nicotinamide adenine dinucleotide (NAD+) | 1.2129 | 0.0205 | 0.103 | |
| 1-methylnicotinamide | 0.652 | 0.0041 | 0.0586 | |
| adenosine 5’-diphosphoribose (ADP-ribose) | 2.6864 | 1.86E-05 | 0.0017 | |
| Riboflavin Metabolism | flavin adenine dinucleotide (FAD) | 1.3294 | 0.0208 | 0.103 |
| flavin mononucleotide (FMN) | 1.4536 | 0.0042 | 0.0586 | |
| Tocopherol Metabolism | alpha-tocopherol | 1.4689 | 0.0049 | 0.0618 |
| Folate Metabolism | 5-methyltetrahydrofolate (5MeTHF) | 1.3164 | 0.0157 | 0.0996 |
| Thiamine Metabolism | thiamine diphosphate | 0.7367 | 0.0299 | 0.1157 |
| Vitamin B6 Metabolism | pyridoxamine | 1.2353 | 0.0139 | 0.0972 |
| pyridoxal phosphate | 1.3339 | 0.0224 | 0.1047 | |
| Benzoate Metabolism | O-methylcatechol sulfate | 1.4732 | 0.0264 | 0.1129 |
| Chemical | lanthionine | 12.2969 | 1.54E-06 | 0.0004 |
H2S Measurements using AzMC Fluorescent Probe
H2S levels were determined using a 7-azido-4-methylcoumarin (AzMC) fluorescent probe (Sigma-Aldrich, # L511455) (12). Cells were lysed using non-denaturing lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40, 1% Triton X-100). The protein concentration was determined with DC Protein Assay (BioRad) with bovine serum albumin (BSA, Thermo Scientific) as a standard. Cell extracts, each containing 150 µg of total protein, reacted with D,L-homocysteine (2 mM), L-cysteine (2 mM), PLP (0.005 mM), and AzMC (0.01 mM). Fluorescence of the AzMC was measured using SpectraMax M2 (Molecular Devices) microplate reader (excitation λ=365, emission λ=450 nm). Inhibition of CBS-produced H2S was achieved by aminooxyacetic acid (AOAA, 1 mM).
Proliferation assay
NCM356 cells were seeded in triplicate in 6 well plates (1×106 cells/well) in DMEM media supplemented with 5% FBS and incubated at 37°C. Proliferation rates were detected using Coulter Cell Counter (Z-Series, Beckman-Coulter Life Sciences). At harvest, cell culture media was removed and centrifuged to collect floating cells; adherent cells were collected using trypsin. Cell suspensions (floating and attached cells pooled, 100 µL) were pipetted into vials containing 9.9 mL of Isotone and were counted. Oxythiamine and FX-11 (10 µM each, Selleck Chemicals) were added to the culture media every other day. Control cultures were treated with an equal volume of phosphate buffered saline (PBS).
Cell respiration analysis in NCM356 cell line
Cells were trypsinized, resuspended in 2 ml DMEM (2×106 cells/ml), and transferred to an Oxygraph-O2K respirometer chamber (Oroboros Instruments, Innsbruck, Austria). The temperature was maintained at 37°C and O2 set at a range of 100–200µM. Baseline respiratory flux was used to determine routine respiration; oligomycin (20 µM; Santa Cruz, # sc-201551A) was used to inhibit ATP synthase and measure leak rate. Carbonyl cyanide m-chlorophenylhydrazone (CCCP; 5 µM, Sigma #C-2759) was infused into the oxygraph chamber to determine uncoupled state. Cell O2 consumption rates were calculated using DatLab software version 6 (Oroboros Instruments). Reserve respiratory capacity was calculated as the difference between uncoupled and routine respiration. Citrate synthase activity was determined spectrophotometrically (at 412 nm) in lysed cells using Ellman’s regent to determine the linear rate of thiol group (coenzyme A) produced by the condensation of oxaloacetate and acetyl-CoA (13).
Migration/Invasion assay
NCM356 parental, Vector, and CBS2 cells were detached with 0.25% trypsin-EDTA and 105 cells resuspended in serum-free DMEM/0.1% BSA were seeded onto the upper chambers inserts with 8µm pores (Corning, Tewksbury, MA). The inserts were uncoated (migration assay) or coated with 70 µL of diluted Matrigel Basement Membrane Matrix (50 µg/ml; BD Biosciences) (invasion assay). The bottom chamber contained 600 µL of NIH3T3 fibroblast conditioned media. Cells were incubated at 37°C for 6 or 24 h for migration and invasion, respectively. After incubation, the migrated cells were fixed with methanol and then stained with 0.1% crystal violet. Each experiment was performed in triplicate.
Soft agar assay
Low melting point agarose (0.8%) (Thermo Fisher Scientific, # 16520-050) in growth medium (high glucose DMEM supplemented with 10% FBS) was applied to 12 well plates (1 ml/well) to form the base layer. Cells (5 × 103 cells/well) were suspended in 0.4% agar in growth medium and applied to the top of the base layer. Cell cultures were fed every 4 days either with growth medium (1 ml) with PBS, or the CBS inhibitor AOAA (1 mM) or with 600 µM of the H2S donor, p-(4-methoxyphenyl)-P-4-morpholinyl-phosphinodithioic acid (GYY4137, Sigma-Aldrich, #C13408, #SML0100), respectively. Plates were incubated for 3 weeks. Cell colonies stained with 0.005% crystal violet and counted at 400X magnification.
NCM356 cell subcutaneous xenograft studies
All animal studies were approved by the Institutional Animal Care and Use Committee at the University of Texas Medical Branch. Athymic nude mice (Invigo Indianapolis, IN) were used for the in vivo studies (5 males and 5 females per group). Mice (6–10 wks) were injected s.c. in the dorsum with NCM356 vector or CBS overexpressing cells (2×106). Mice were monitored daily and body weight measured once/week. Tumor diameters were measured transcutaneously using a caliper 2–3 times per week for the duration of the experiment. Tumor volumes were calculated using the formula: V = (π/6)LxWxD. Graphs and statistical analyzes (2-way ANOVA) were preformed using GraphPad Prism version 7 (GraphPad Software, La Jolla, CA).
RNA extraction and quality control
Total cellular RNA was extracted from cells (N=3 each from independently cultured cells) using RNAqueous™ (Life Technologies). RNA was quantified using a NanoDrop ND-1000. Quality was assessed by visualization of 18S and 28S RNA bands using an Agilent BioAnalyzer 2100; electropherograms were used to calculate the 28S/18S ratio and the RNA integrity number.
Library construction and sequencing
Poly-A+ RNA was enriched from total RNA (1 µg) using oligo dT-attached magnetic beads. Bound RNA was fragmented by incubation at 94°C for 8 min in 19.5 µL of fragmentation buffer. First and second strand synthesis, adapter ligation and amplification of the library were performed using the Illumina TruSeq RNA Sample Preparation kit. “Index tags” incorporated into the adapters were used to track samples. Library quality was evaluated using an Agilent DNA-1000 chip on an Agilent 2100 Bioanalyzer. Quantification of library DNA templates was performed using qPCR and a known-size reference standard. Cluster formation of the library DNA templates was performed using the TruSeq PE Cluster Kit v3 and the Illumina cBot workstation. Template input was adjusted to obtain a cluster density of 700–900 K/mm2. Paired end 50 base sequencing by synthesis was performed using TruSeq SBS kit v3 on an Illumina HiSeq 1000. All protocols were followed per manufacturer and reagents were purchased from Illumina.
NGS run quality assessment
Quality is evaluated by Illumina’s internal assessment of each run. Overall run quality per lane, per cycle were reported. After demultiplexing the samples, FASTQC (http://www.bioinformatics.bbsrc.ac.UK/projects/Fastqc) is used to provide per sample information on base percentage distribution per cycle, GC content across the run, percents uncalled bases and duplication of sequences.
RNA-seq analysis
The alignment of NGS sequence reads was performed using the Spliced Transcript Alignment to a Reference (STAR) program version 2.4.2a, using default parameters (14). We used the human hg19 assembly as a reference; differential expression between the two cell lines was evaluated using the Cufflinks/Cuffdiff software, v2.2.1. Gene ontology, pathway analyses, and enrichment of transcription factors within the gene lists of interest were performed using the Gene Set Enrichment Analysis (GSEA; http://www.broadinstitute.org/gsea/index.jsp) using gene sets MSigDBv5.1 and Enrichr (15,16). Full gene list was deposited at GEOarchive, GSE91000.
Single nucleotide polymorphism (SNP) analysis and mutation detection
The Broad Institute GATK Best Practices workflow for SNP and indel calling on RNAseq data was used to identify variants in the NCM356 cell line (https://software.broadinstitute.org/gatk/best-practices). Genes known to be involved in carcinogenesis in the NCM cells lines were compared to those described in the COSMIC database (http://cancer.sanger.ac.uk/cosmic). SNP analysis identified mutations known in the adenoma-to-carcinoma sequence such as KRAS mutation (G12V), the tumor suppressor gene p53 (P72R) and the APC gene (Q1338*). However, other SNPs identified in colorectal cancer related genes are of unknown significance (Supplemental Table S1).
RESULTS
CBS expression is upregulated in colonic polyps
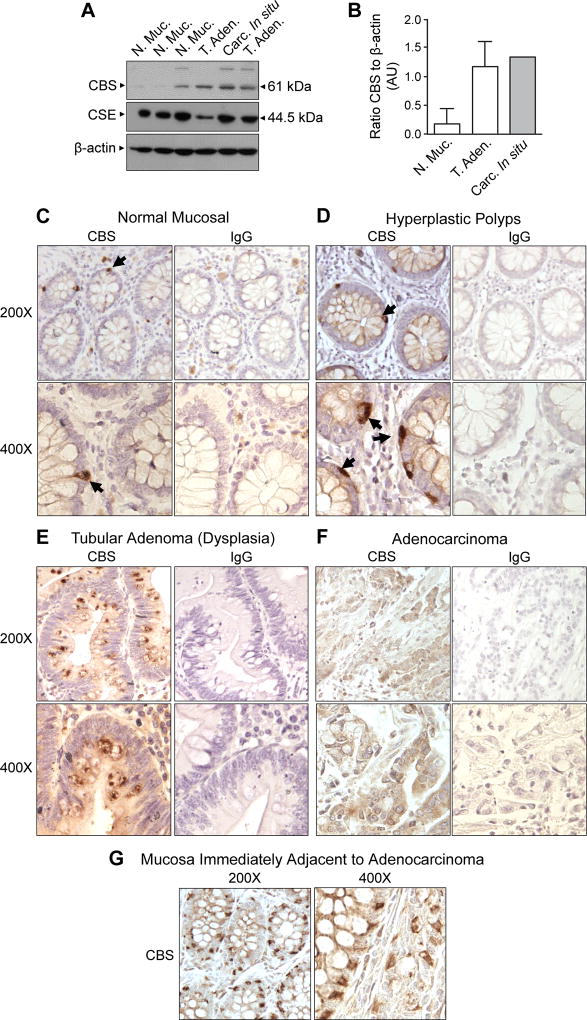
CBS expression was assessed in protein extracts prepared from biopsies of normal mucosa, tubular adenoma (low-grade dysplasia) and carcinoma in situ (high-grade dysplasia). CBS levels were relatively low in two of three biopsies of normal mucosa and elevated in polyps exhibiting both tubular adenoma and carcinoma in situ (Fig 1A, B). The levels of CSE showed little variation between specimens (Fig 1A). Immunohistochemical analyses of formalin-fixed/paraffin-embedded tissue sections of normal mucosa and hyperplastic polyps revealed CBS immunoreactivity in a small number of cells located along the basal laminar aspect of the colonic crypts in both normal and hyperplastic polyps (Fig 1C, D, arrows). A slight increase in cytoplasmic CBS staining also was noted in the epithelial cells of hyperplastic polyps when compared to normal crypt cells. In contrast, the epithelial cells of tubular adenoma specimens exhibited higher levels of diffuse cytoplasmic CBS staining with frequent focal areas of intense immunostaining adjacent to mucin-containing vesicles (Fig 1E, dark brown). We also observed increased CBS staining in cells of the lamina propria mucosa. Sections of adenocarcinoma exhibited diffuse CBS staining throughout the cytoplasm of cancer cells (Fig 1F). Additionally, in mucosal crypts immediately adjacent to the adenocarcinoma cells, CBS staining was generally increased in the cytoplasm of the epithelial cells and also expressed at high levels in the basal laminar aspect of a subset of mucin-producing goblet cells (Fig 1G). The increase in CBS expression with progression from benign hyperplastic polyps to premalignant adenomas and invasive adenocarcinoma suggests that the enzyme may play a functional role in colorectal carcinogenesis.
Figure 1. Cystathionine-β-synthase (CBS) expression is increased in premalignant polyps.
A) Western blot of protein extracts from freshly collected biopsy specimens probed with antibodies to CBS and cystathionine-γ-lyase (CSE). Under an IRB approved protocol, three polyps were biopsied and diagnosed to be dysplastic polyps by a pathologist [two tubular adenomas (T. Aden.) and one carcinoma in situ (Carc. In situ)]. Three biopsies of “normal” mucosa (N. Muc.) were also obtained from the same patients. B) Densitometry analysis of the Western blot shown in panel A. The bar graph shows mean expression level (with range bars) of CBS [expressed as a ratio of density (AU = arbitrary units) of CBS to β-actin]. Formalin-fixed paraffin-embedded sections of C) normal colonic mucosa, D) hyperplastic polyp, E) tubular adenoma, F) adenocarcinoma and G) normal mucosa immediately adjacent to the adenocarcinoma were stained with a primary antibody to CBS (1:150 dilution). Tissue architecture was visualized by counterstaining with hematoxylin. To assess non-specific staining, serially cut sections were stained with a rabbit isotype-matched (IgG) antibody (dilution 1:150, Santa Cruz Biotechnology, Cat# sc-2763).
CBS overexpression NCM356 cells
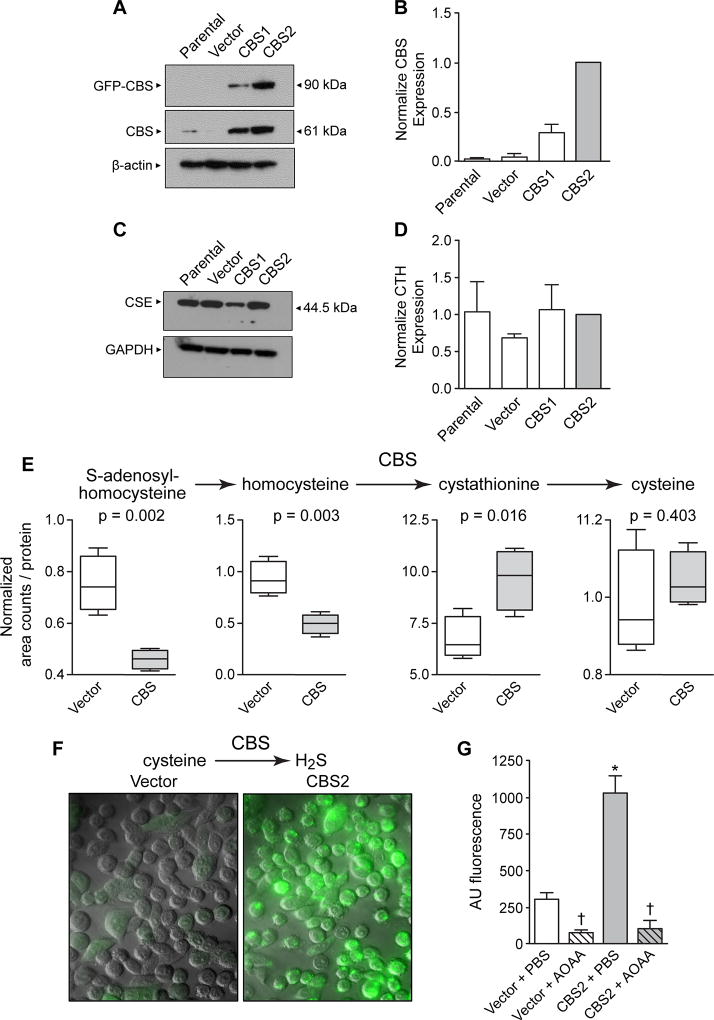
To assess the potential functional role of CBS in colon carcinogenesis, we used the NCM356 cell line as a model. This cell line was derived from the wide-margin normal mucosa of a colorectum resected to remove a rectal adenocarcinoma (17). Importantly for our studies, NCM356 cells express a very low level of CBS protein when compared to either colon cancer specimens or colon cancer-derived cell lines (18). Also, NCM356 cells do not grow in a soft agar assay, a surrogate for anchorage-independent growth, and are non-tumorigenic when xenografted into immunocompromised mice (17,19). Using Next Generation sequencing, we confirmed the existence of a mutant TP53 gene reported by Stauffer, et al. (17), and identified additional functional mutations in the KRAS and APC genes (Supplemental Table S1). These data suggest that the NCM356 cell line exhibit the characteristics of a late adenoma. We have, therefore selected NCM356 cells as a model to determine whether CBS upregulation can drive progression to an invasive carcinoma phenotype. To force overexpression of CBS, we used a lentiviral expression vector (pReceiver-Lv103, GeneCopoeia™) containing a full-length CBS cDNA (NM_000071.2) with an in-frame N-terminal green fluorescent protein (GFP) tag under control of a CMV promoter. We used a tagged fusion protein to differentiate recombinant protein from endogenous enzyme. Previously, we have shown that addition of an N-terminal protein tag does not affect the enzymatic activity of CBS (20). Non-transduced NCM356 cells (i.e., parental) and cells transduced with virus particles containing the empty pReceiver-Lv103 GFP expression vector (i.e., Vector) were used as controls throughout our studies. As expected, immunoblotting with a CBS antibody identified a protein at ~90 kDa representing the GFP-CBS fusion protein in extracts from cells transduced with the recombinant CBS expression vector (Fig 2A). This band was absent in both the parental NCM356 cell line and control vector transduced cells (Fig 2A). We also observed increased expression of endogenous CBS (~61 kDa) in cells expressing the recombinant enzyme (Fig 2A), suggesting that steady-state levels of cellular CBS may be primarily controlled by protein degradation. We also noted an approximately 3-fold difference in total CBS expression between CBS1 and CBS2 cells (Fig 2A, B), which were transduced at a multiplicity of infection (MOI) of 5 and 15, respectively. Similar to the human colonic biopsy specimens (Fig 1A), CSE protein was readily detectable in the NCM356 cells and its expression levels were not significantly affected by overexpression of CBS (Fig 2C, D).
Figure 2. Characterization of CBS forced overexpression in NCM356 cell proliferation and bioenergetics.
A) Representative Western blot comparing the expression of recombinant (GFP-CBS) and endogenous CBS levels in non-transduced NCM356 cells (Parental), NCM356 cells transduced with a lentivirus containing the empty pReceiver-Lv103 GFP expression vector (Vector), or NCM356 cells transduced with the pReceiver-Lv103 GFP vector containing full-length CBS cDNA at a multiplicity of infection (MOI) of 5 (CBS1) and 15 (CBS2). The level of β-actin in each sample was assessed to determine variation in protein loading and transfer. B) Densitometry analyses of Western blots. The bar graph shows a comparison of the mean ± SEM (n=4 blots) of total CBS expressed [i.e., sum of recombinant (90 kDa band) + endogenous (61 kDa band)] in each cell line. The ratio of total CBS to β-actin was determined for each cell line for each blot and, then the ratio for each cell line was normalized to the total CBS in the CBS2 cells across blots. C) Representative Western blot and D) densitometry analyses comparing expression of CSE in the different NCM356 cell lines. The level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used to control for protein loading and transfer. E) CBS overexpression increased flux through the reverse transsulfuration pathway and H2S production. F) 7-Azido-4-Methylcoumarin fluorescent images show increase levels of CBS-dependent H2S. G) Quantification of 7-Azido-4-Methylcoumarin fluorescents data (error bars are mean ± S.E.M, N=3) Cell were treated with aminooxyacetic acid (AOAA) or phosphate buffered saline (PBS).
CBS overexpression alters NCM356 cell metabolism including increased flux through the reverse transsulfuration pathway and production of H2S
Global metabolic profiling (Metabolon, Durham, NC) (21) was performed on cellular extracts from NCM365 overexpressing cells (CBS2) and vector-transduced controls. A total of 540 metabolites were identified in the cellular extracts. A comparison of CBS overexpressing NCM356 cells to vector controls identified 85 metabolites that were differentially expressed (65 increased and 20 decreased) at a significance of p≤0.05 (Welch’s Two-Sample t-Test) (Table 1). Metabolites clustered into carbohydrate metabolism, specifically glycolysis and pyruvate metabolism, nucleotide sugars and the pentose phosphate pathway; lipid biosynthesis, primarily phospholipids, sphingolipids, and bile acids; cofactors and vitamins including nicotinate and nicotinamide metabolism, riboflavin and vitamin B6 metabolism; and dipeptide production (Table 1). These data suggest a reprograming into an anabolic metabolism. However, there was not a significant change in cellular glucose levels between vector and CBS2 cells (Supplemental Fig S1A). CBS overexpression also increased flux through the reverse transsulfuration pathway (Fig 2E). Significant decreases in the levels of S-adenosyl homocysteine and homocysteine, and an increase in cystathionine levels were detected in CBS2 cells when compared to the vector-transduced cells (Fig 2E). Although cysteine or gamma-glutamylcysteine (Supplemental Fig S1B) levels were not significantly different in CBS2, several downstream metabolites were elevated. There was a small but significant increase in cysteine sulfinate levels (1.3-fold increase, p=0.0460) and a robust increase in lanthionine levels (12.3-fold increase, p=1.5E-06) (Table 1). CBS can catalyze the production of H2S from cysteine through three reactions: 1) the condensation of cysteine and homocysteine to produce cystathionine and H2S, 2) the hydration of cysteine to produce serine and H2S, and 3) the condensation of two molecules of cysteine to produce lanthionine and H2S (3,22). To assess whether the increase in lanthionine was associated with increased H2S production, we used the fluorescent probe AzMC to quantify H2S levels. Basal H2S production was increased approximately four-fold in CBS2 cells, when compared to vector controls (Fig 2F, G). Treatment with AOAA reduced the level of H2S approximately 10-fold in CBS2 cells and 4-fold in vector controls (Fig 2G).
Although the GSH levels were not different between vector control and CBS overexpressing cells (Supplemental Fig S1C, D; Supplemental Fig S2A), there was evidence of increased glutathione metabolism (i.e., increased levels of ophthalmate) and oxidative stress in the CBS2 cell when compared to vector controls. Ophthalmate is an indicator of glutathione depletion and it has been proposed as a biomarker for oxidative stress (23). Metabolite analyses showed the levels of ophthalmate increased in CBS2 cells 1.44-fold (p<0.01) (Table 1). Using the methyl ester form of the fluorescent probe for Reactive Oxygen Species (ROS), 2’,7’-dichlorofluorescin diacetate (DCFDA), we determined that the CBS2 cell also produced approximately a 25% increase in ROS (Supplemental Fig S2B). Although there was no change in Nrf2 transcriptional activity (Supplemental Fig S2C), the Nrf2 target heme oxygenase-1 (HO-1) was increased (Supplemental Fig S2D). However, the proliferation of CBS-overexpressing cells was not dependent on the synthesis of glutathione, because treating the cells with DL-buthionine-(S,R)-sulfoximine (BSO) had no effect on the growth rate of CBS2 cells, but did significantly inhibit vector control cultures (Supplemental Fig S3A, B).
NCM356 cells overexpressing CBS exhibit an increased rate of proliferation and enhanced cellular bioenergetics
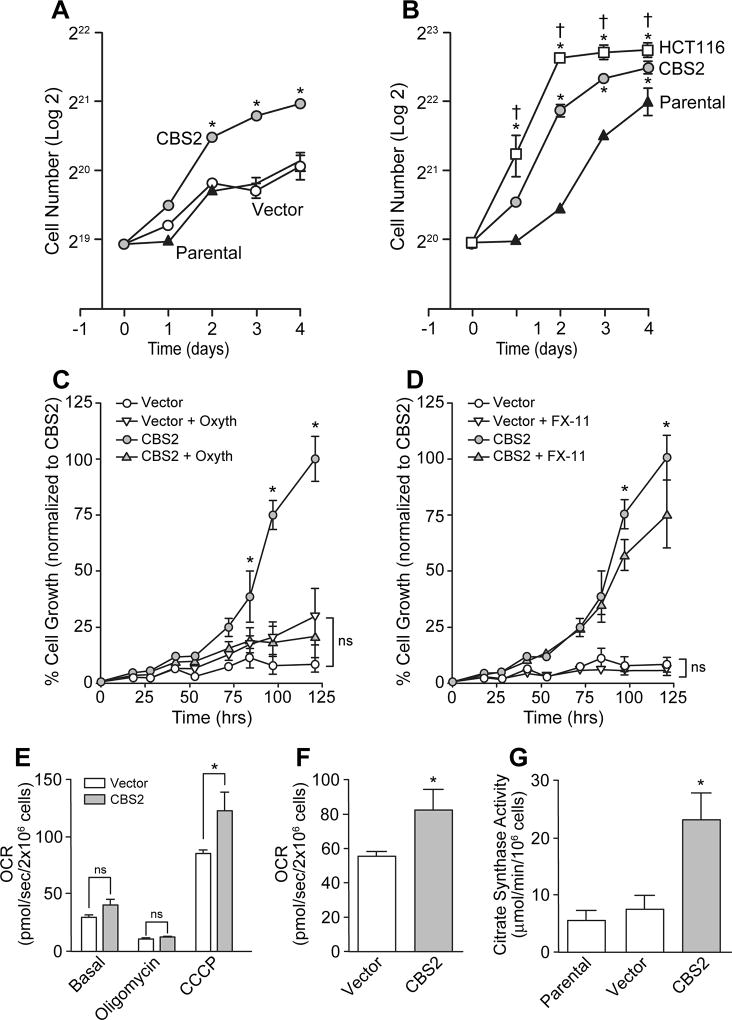
The log-phase doubling-time of CBS overexpressing NCM356 cells was approximately 1.3 times faster than control cultures, resulting in a significant increase in cell number at days 2, 3 and 4 (Fig 3A, B). By comparison, the doubling-time of the colon cancer cell line HCT116 was 1.4 times faster than the doubling-time of CBS overexpressing cells and 1.9 times faster than parental NCM365 cells (Fig 3B). Metabolomic profiling (Table 1) also suggested that overexpression of CBS increased flux through both the pentose phosphate and glycolytic pathways. Therefore, we tested the sensitivity of NCM356 cell proliferation to a transketolase inhibitor, oxythiamine, and the lactate dehydrogenase inhibitor FX-11. Inhibition of the pentose phosphate pathway with oxythiamine (10 µM) significantly inhibited the growth of CBS2 cells (reducing growth by 90% at day 5), but had no effect on the growth rate of vector-transduced control cultures (Fig 3C). Similarly, treatment with the 6-Aminonicotinamide (6-AN) decreased CBS2 cell numbers by 74% at 120 h vs. a 45% reduction of vector control cells at the same time point (Supplemental Fig S3A, B). Inhibition of lactate dehydrogenase with FX-11 (10 µM) reduced the growth rate of CBS2 cells (~25% at day 5), but had no significant effect on the slower growing vector cells (Fig 3D). Using high-resolution respirometry (Oroboros Oxygraph-2k), we assessed the effects of CBS overexpression on NCM356 cell bioenergetics. When compared to vector control cells, CBS2 cells exhibited a significant increase in maximum uncoupled respiration rate (Fig 3E) and reserve respiratory capacity (Fig 3F). Citrate synthase activity was also increased (Fig 3G), suggesting that CBS overexpression can increase diversion of TCA cycle intermediates to provide cytoplasmic acetyl-CoA for lipogenesis (Table 1).
Figure 3. Effects of CBS overexpression on NCM356 cell growth and bioenergetics.
A) A comparison of the growth rates of non-transduced NCM356 cells (Parental), vector-transduced NCM356 cells (Vector), and NCM356 cell overexpressing CBS (CBS2) (* p ≤ 0.05, CBS2 vs. Parental or Vector). B) A comparison of the growth rates of Parental, CBS2 and HCT116 colon cancer cells (* p ≤ 0.05 HCT116 or CBS2 v. Parental; † p ≤ 0.05 HCT116 vs. CBS2). Data show the change in mean cell number (Log2) ± SEM per well over time. C) Effects of the transketolase inhibitor, oxythiamine, and D) the lactate dehydrogenase inhibitor, FX-11 on CBS-dependent NCM356 cell growth (* p<0.05, CBS2 vs. Vector, Vector + oxythiamine (Oxyth) or XF-11, or CBS2 + Oxyth or FX-11; two-way ANOVA with Tukey’s multiple comparison test, ns = not significant). E) Effects of CBS overexpression on NCM356 cell respiration. Baseline respiratory flux was used to determine routine respiration (Basal), oligomycin (20 µM) was used to inhibit ATP synthase and so that leak respiration could be recorded, and the proton ionophore carbonyl cyanide m-chlorophenylhydrazone (CCCP, 5 µM) was used to determine maximal uncoupled respiration. F) Reserve Respiratory Capacity was calculated as the difference between uncoupled and routine respiration (* p = 0.0049, Student t-test). G) Citrate synthase assay comparing CBS overexpressing NCM356 cells (CBS2) to vector and parental NCM356 controls (*p<0.01). Statistical analyses (one- or two-way ANOVA with Tukey’s multiple comparison test) were performed using Prism 7 (GraphPad Software, Inc. La Jolla, CA).
To better define the capacity of Complex I and Complex II to support respiration in vector and CBS2 cells, we performed additional respirometry experiments, where we selectively inhibited Complex I and Complex II with rotenone and malonic acid, respectively. In agreement with our previous results, total routine respiration was significantly greater in CBS2 cells vs. vector (p<0.001). Further, absolute rates of respiration supported by Complex I (p<0.01) and Complex II (p<0.05) were greater in CBS2 cells compared to vector. To account for the greater respiratory capacity of CBS2 cells, we calculated the respiratory response to both rotenone and malonic acid in both cell lines, and normalized these values to total routine respiration. While Complex I supported respiration normalized to total routine respiration was significantly greater in CBS2 cells vs. vector (p=0.01), this was not the case for Complex II supported respiration (p=0.1 vs. vector). Collectively, these data suggest that greater respiratory capacity in CBS2 cells is the result of a greater capacity for both Complex I and Complex II to support electron transfer. Whether there are qualitative differences between Complex I and II function in the mitochondria in CBS2 cells remains to be seen. While we saw a marginally greater response to rotenone in CBS2 cells vs. vector, suggesting that perhaps there are differences in Complex I function in CBS2 cells, we should note that our experiments in intact cells were not designed to assay mitochondrial quality in terms of Complex I and II function.
The contribution of various complexes to electron transfer is influenced by substrate availability. Since glucose is the principal substrate in our growth media (in addition to pyruvate and glutamine), respiration was likely predominately supported by Complex I and mitochondrial glycerol-3-phosphate dehydrogenase in our experiments. While endogenous succinate will be produced by the TCA to support Complex II electron transfer, no exogenous succinate was provided. In fact, metabolite analyses indicate that succinate levels are lower in CBS2 cells (0.809, p<0.02) compared to vector, suggesting that H2S is providing the electrons to maintain Complex II activity.
Overexpression of CBS enhances NCM356 cell migration, invasion and anchorage-independent growth
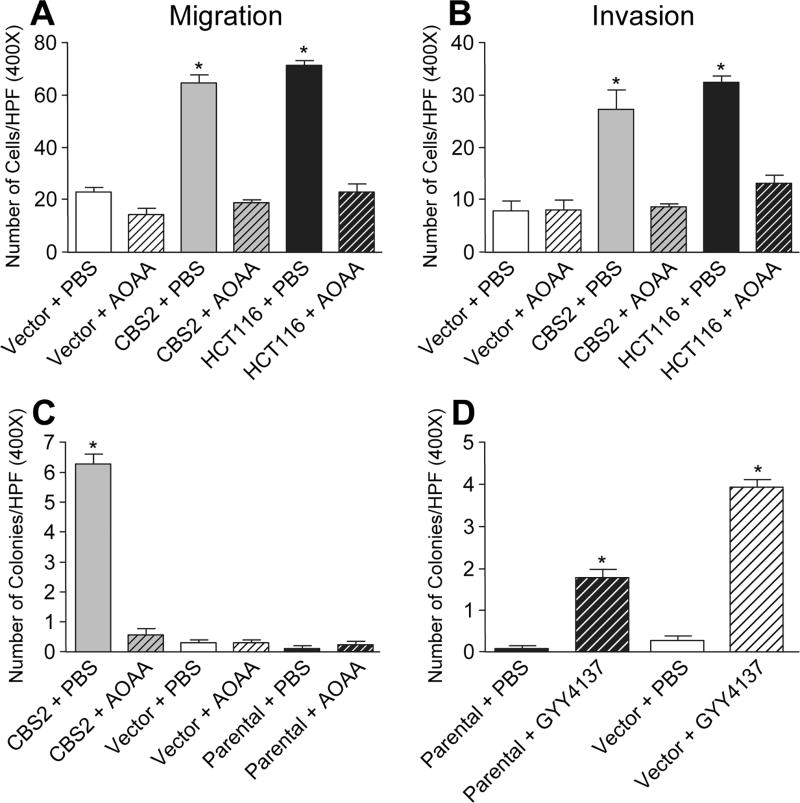
To evaluate the effects of CBS overexpression on cell migration and invasion, we used a transwell culture chamber assay as previously described (24). Compared to vector NCM356 cells, CBS2 cells showed an approximately 3-fold increase in cell migration toward NIH3T3 fibroblast-conditioned medium after 6 h (Fig 4A), and a similar 3-fold increase in cell invasion through Matrigel after 24 h (Fig 4B). The fold-increase in CBS2 cell migration was comparable to that of HCT116 colon cancer cell (Fig 4A, B), which expressed higher levels of endogenous CBS (1). Treatment of the cells with the CBS inhibitor AOAA completely blocked the stimulatory effects of CBS overexpression on NCM365 cell migration and invasion, but did not significantly affect the invasion of control NCM356 cells (Fig 4A, B, respectively).
Figure 4. Effects of CBS upregulation on NCM356 cell migration, invasion and anchorage-independent growth.
A) Summary of migration data (n=4). B) Summary of invasion data (* p<0.001 vs. Vector + PBS, Vector + AOAA, CBS2 + AOAA or HCT116 + AOAA). C) Quantification of soft agar colony-form assay data comparing CBS overexpressing NCM356 cells to the vector and parental controls with and without 1mM AOAA treatment. Data represent the mean number of colonies per high power field (HPF) ± SEM from 2 independent experiments (*p<0.001). D) Quantification of the effects of GYY4137 treatment on anchorage-independent growth (*p<0.05). For all soft agar experiments, 8 non-overlapping HPFs were counted per well at 400X magnification. Statistical analyses (one-way ANOVA with Tukey’s multiple comparison test) were performed using Prism 7 software.
A hallmark of the malignant transformation of epithelial cells is acquired resistance to anoikis or apoptosis in response to detachment from the extracellular matrix (25). The soft agar colony formation assay, which assesses the ability of a cell to proliferate in an anchorage-independent manner, is a surrogate for resistance to anoikis (26). Only CBS overexpressing cells formed colonies in soft agar (Fig 4C, Supplemental Fig S4A). After 3 weeks in culture, an average of 6 colonies (20–50 µm in diameter) were observed per high-powered field (HPF) in CBS2 cell cultures (Fig 4C). By comparison, we observed only occasional small cell clusters (< 1 per HPF), containing only a few cells in cultures of parental NCM356, vector-transduced, and AOAA-treated cells (Fig 4C). To determine if H2S could substitute for CBS overexpression, we treated cultures of parental and vector-transduced cells with the slow-release H2S donor GYY4137. After 3 weeks, colonies of cells 20–25 µm in diameter were observed in both the parental and vector cell cultures treated with GYY4137 (Fig 4D, Supplemental Fig S4B).
Overexpression of CBS enhances NCM356 cell tumorigenicity
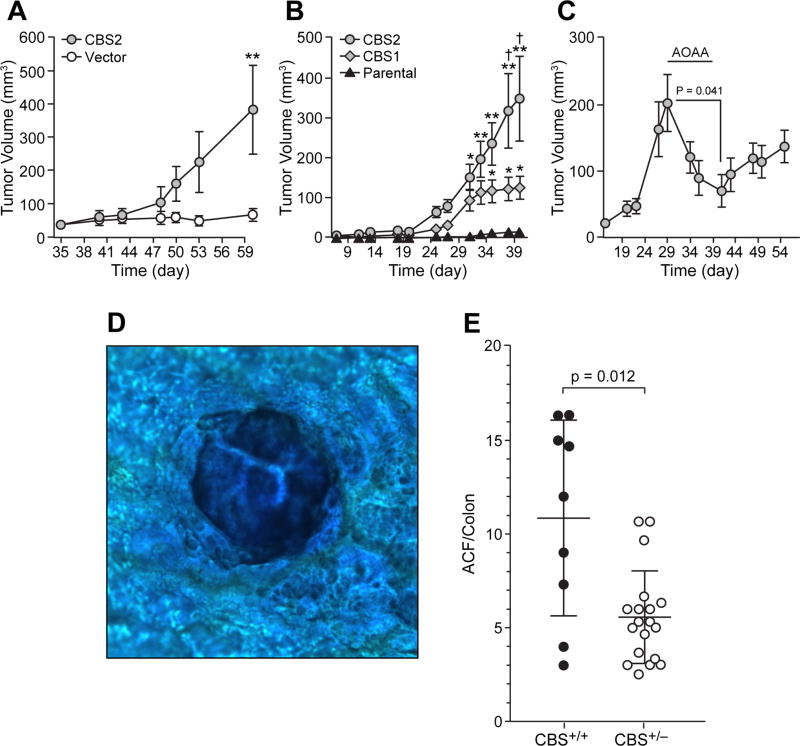
To evaluate the effects of CBS upregulation on NCM356 cell tumorigenicity, we injected 106 CBS2 or vector control cells subcutaneously into the flanks of athymic nude mice, 10 animals per group. Palpable nodules were detected at the injection site of all mice at day 35 (Fig 5A). Tumor growth was detected by post-injection day 48 in mice injected with CBS2 cells (Fig 5A). By day 60, the tumors in the CBS2 group increased in size to a mean volume of approximately 400 mm3. By comparison, the palpable nodules measured in the vector control mice maintained a size of approximately 50 mm3 for the duration of the experiment (Fig 5A). Next, we assessed the effects of different levels of CBS expression on in vivo tumorigenicity by comparing CBS2 cells to CBS1 cells, which express about one-third less CBS protein than CBS2 cells (Fig 2B). The parental NCM356 cells were used as a control. Ten mice per group were injected subcutaneously with 2×106 cells each. Tumor growth was detected in both CBS overexpressing groups by day 25 (Fig 5B). By day 35, tumors in mice injected with CBS1 or CBS2 cells were significantly larger than the small palpable nodules at the injection site of the parental NCM356 cell group (Fig 5B). By days 37 and 40, the tumors in the CBS2 group where significant larger than those in the CBS1 group (Fig 5B), demonstrating that in vivo NCM356 tumorigenicity and growth rate is proportional to the level of CBS expression and presumed CBS activity. To address the question of CBS activity, we injected 2×106 CBS2 cells into 10 mice and allowed tumors to grow to a mean size of approximately 200 mm3 (Fig 5C, day 29). We then treated mice with AOAA (9 mg/kg) daily for a total of 10 days. The CBS inhibitor caused a significant reduction in tumor volume (student-t test, day 29 vs. day 40, p = 0.041), confirming that CBS activity is critical for NCM356 cell tumorigenicity. When AOAA treatment was stopped tumor growth resumed but at a slightly slower growth rate (Fig 5C).
Figure 5. Effects of CBS overexpression on NCM356 tumorigenicity in vivo.
A) Comparison of the tumorigenicity of CBS2 cells to vector-transduce control cells (**p<0.01, CBS vs. Vector). 106 cells were injected subcutaneously into athymic nude mice. Tumor size was measured with a caliper and volume calculated using the equation V = (π/6) LxWxD. B) Comparison of different levels of CBS expression of NCM356 tumor growth rates (*p<0.01 CBS1 vs. Parental; **p<0.001 CBS2 vs. Parental; †p<0.05 CBS2 vs. CBS1). Two-way ANOVA with Tukey’s multiple comparisons test were performed using Prism 7 (GraphPad Software, Inc. La Jolla, CA). C) Effect of AOAA on CBS tumor growth. Effect of CBS gene dosage on azoxymethane (AOM)-induce aberrant crypt foci (ACF) formation in CBS heterozygous (CBS+/−) and wild-type (CBS+/+) control mice. D) Methylene blue stained ACF. E) Quantification of ACF per colon [Mann-Whitney test (unpaired, nonparametric, two-tailed) was performed using Prism 7 software].
Overexpression of CBS has a broad impact on the gene expression profile of NCM356 cells
Whole transcriptome RNA sequencing (RNA-seq) was used to compare the mRNA expression profiles of CBS over-expressing NCM356 cells to vector-transduced controls. At a q-value (false discovery rate-adjusted p-value) of < 0.01, 351 differentially expressed genes (243 genes upregulated and 108 downregulated) were identified in the CBS2 cells. Gene Set Enrichment Analysis (GSEA) (27) gene family categorization shows a prevalence of transcriptions factors (20 upregulated and 10 downregulated genes), cytokine and growth factors (18 up and 3 down), protein kinases (7 up and 3 down), and oncogenes (5 up and 2 down) (Supplemental Table S2). GSEA gene set overlap analyses (Table 2) compares our genes of interest to 8 major Molecular Signature Database gene sets (MSigDB v 5.1) which include Hallmark gene sets, Curated gene sets, and Oncogenic signatures (28). Upregulated genes from CBS overexpressing cells overlapped significantly with gene sets related to glycolysis, hypoxia and a colon cancer cell phenotype including: NF-κB, KRAS, and p53 signaling, apical junction gene set, genes downregulated after E-cadherin knockdown by RNAi, and genes regulated by the Wnt pathway. Finally, there was significant overlap with genes related to increased extracellular matrix (ECM) and ECM-related proteins, cell adhesion molecules, and epithelial to mesenchymal transition, consistent with a migratory and invasive phenotype (Table 2). Together, these changes in gene expression support the more aggressive phenotype of CBS2 cells when compared either to the vector or parental NCM356 cells.
Table 2.
GSEA: Gene Families for 351 genes (q<0.01).
| Gene Families | Genes Up (243) | Genes Down (108) |
|---|---|---|
| Tumor Suppressors | 1 (CDH1) | 1 (PIK3R1) |
| Oncogenes | 5 (ASPSCR1, PER1, PRDM16, HOXD11, CCND2) | 2 (LCP1, DDX10) |
| Translocated Cancer Genes | 5 (ASPSCR1, PER1, PRDM16, HOXD11, CCND2) | 3 (LCP1, DDX10) |
| Protein Kinases | 7 (DDR1, MAP3K5, CDC42BPG, SBK1, RPS6KA2, PTK7, TRIB3) | 3 (PLK2, AXL, DCLK1) |
| Cell Differentiation Markers | 4 (CXCR1, DDR1, CDH1, CD8B) | 2 (NGFR, NRP1) |
| Homeodomain Proteins | 2 (IRX3, HOXD11) | 2 (ISL1, ZEB1) |
| Transcription Factors | 20 (E2F2, IRX3, CEBPB, ELF3, FAM20C, HES6, PRDM16, JUNB, HOXD11, HES1, ASCL2, FOS, EBF4, DBP, IDI, HES4, ETS2, HES2, PER1, ID3) | 10 (CRIP1, ZNF45, ZFP30, ZNF107, ZFPMZ, DAH1, ZEB1, ISL1, ZNF20, AHR) |
| Cytokines and Growth Factors | 18 (FGF19, S100A6, TNC, FGF11, TNSF15, KITLG, IGF2, TNFSF9, MDK, INHBB, SEMA6B, ADM, INHBE, VEGFA, ADM2, GDF15, APLN, FGF3) | 3 (CXCL1, SAA2, SAA1) |
Loss of a CBS allele reduces the number of mutagen-induced aberrant crypt foci (ACF)
To further test the hypothesis that CBS plays a role in promoting colon carcinogenesis, we assessed the effect of CBS gene-dosage on azoxymethane (AOM)-induced aberrant crypt foci (ACF) formation in mice. CBS heterozygous mice (CBS+/−) and wild-type (CBS+/+) controls were treated with the mutagen AOM at a dose of 10 mg/kg via intraperitoneal injection once per week for 5 weeks. At the end of 18 weeks, the colon from each animal was harvested, transected longitudinally, flattened on a glass microscope slide, fixed in 10% formalin for 24 h and then transferred to 70% ethanol. To aid visualization of ACF, the colons were stained with 0.05% methylene blue (Fig 5D). The number of ACF per colon were determined by 3 different observers counting independently and blinded to the identity of the individual slides. AOM treatment induced significantly less ACF in the colons of CBS+/− mice compared to wild-type controls (Fig 5E). The loss of one allele of CBS reduced the number of AOM-induced ACF by half. The mean number of ACF ± SD for CBS+/+ and CBS+/− were 10.85 ± 5.2 and 5.57 ± 2.5, respectively (Fig 5E).
DISCUSSION
Aberrant overexpression of the reverse transsulfuration pathway enzyme, cystathionine-β-synthase (CBS) has been reported in established cancers of the colon (1), ovary (5), breast (7), prostate (6), and lung (9), where it promotes cancer progression, metastasis and chemoresistance, in part, by H2S-dependent regulation of cancer cell proliferation, bioenergetics, migration and tumor angiogenesis. In this study, we demonstrate that CBS upregulation begins in premalignant polyps of the colon. CBS protein expression is relatively low in normal colon mucosa and hyperplastic colon polyps when compared to adenomatous polyps and colorectal carcinomas (Fig 1), suggesting that CBS may play a functional role in colon carcinogenesis. To begin testing this hypothesis, we forced overexpression of CBS in the immortalized, but non-invasive colonic epithelial cell line NCM356, and determined the effects of CBS gene-dosage in a mouse model of mutagen-induced carcinogenesis.
CBS upregulation in NCM356 cells induced an adenocarcinoma-like phenotype. With a background of mutations in the APC, KRAS and TP53 genes, the NCM356 parental and vector-transduced control cell lines did not migrate or invade through Matrigel in vitro, were poorly tumorigenic in athymic nude mice, and did not grow in soft agar. However, upregulation of CBS was sufficient to drive progression of the NCM356 cell line to an invasive carcinoma-like phenotype. Indeed, we show that overexpression of CBS was sufficient to induce an increased proliferation, migration and invasion, enhanced cellular bioenergetics, anchorage-independent growth in soft agar (i.e., resistance to anoikis), and tumor formation in immunocompromised mice. Additionally, ablation of one CBS allele reduced the number of mutagen-induced ACF formation, supporting the conclusion that the CBS/H2S axis plays a critical role in the multistep development of colorectal carcinogenesis.
CBS upregulation induced a shift to anabolic metabolism. Metabolic reprograming is a hallmark of cancer cells (29–31). In contrast to the catabolic metabolism of terminally differentiated cells, which under normal oxygen conditions maximizes ATP production through the oxidation of glucose to CO2, rapidly dividing cancer cells need to enhance the synthesis of glycerol and citrate for lipogenesis, ribose sugars for nucleotide production, and nonessential amino acids to support increased cell growth and proliferation. This metabolic reprograming is accomplished, in part, by enhancing the import of glucose and glutamine, increasing aerobic glycolysis (Warburg effect), and diverting glycolytic and tricarboxylic acid (TCA) cycle intermediates to the production of biosynthetic precursors. CBS upregulation stimulated NCM356 cell aerobic glycolysis as evidenced by increases in both lactate and nicotinamide adenine dinucleotide (NAD+) level in CBS2 cells relative to vector-transduced controls (Table 1). Glucose can be metabolized through the glycolytic pathway to produce pyruvate that can either enter the TCA cycle and oxidative phosphorylation in mitochondria to produce ATP, or be converted to lactate by the enzyme lactate dehydrogenase (LDH). LDH activity appears to play a partial role in CBS-dependent NCM356 cell growth because inhibition with FX-11 reduced the growth rate of CBS2 cells by about 25% (Fig 2). By comparison, CBS2 growth was more dependent on the diversion of glucose-6-phosphate from the glycolytic pathway to the pentose phosphate pathway (PPP). The PPP is an anabolic pathway that uses glucose-6-phosphate for the production of NADPH, and ribose-5-phosphate. CBS upregulation in NCM356 cells is associated with an increase in the PPP intermediate 6-phosphogluconate (Table 1), which is converted first to ribulose 5-phosphate, reducing NADP+ to NADPH, and then to ribose 5-phosphate, which is used for the synthesis of nucleotides, nucleic acids and coenzymes, such as the prosthetic group electron acceptors: flavin mononucleotide (FMH) for NADH-coenzyme Q oxidoreductase (complex 1), and flavin adenine dinucleotide (FAD) for Succinate-Q oxidoreductase (SQR; complex II), all of which were at higher levels in CBS overexpressing NCM356 cells (Table 1). The NADPH is used in the reductive biosynthesis of fatty acids and sterols (i.e., lipogenesis). CBS-stimulated NCM356 cell growth appears to depend on the NADPH produced by the PPP, because inhibition of transketolase, which with a transaldolase recycles ribulose 5-phosphate back to glucose 6-phosphate, significantly slowed the rate of CBS2 cell growth to that of the vector-transduced controls (Fig 3G). This observation is not surprising given the function of NADPH in lipogenesis.
Lipogenesis is required for membrane biosynthesis and essential for cell growth and proliferation. CBS upregulation caused enhanced lipid biosynthesis (Table 1). The essential building-block intermediate for lipogenesis is cytosolic acetyl-CoA. However, acetyl-CoA, which is produced from pyruvate or beta-oxidation of fatty acids in the mitochondrial matrix, cannot be directly exported to the cytoplasm. It must first be condensed with oxaloacetate to form citrate, which requires the enzyme citrate synthase. CBS upregulation increased citrate synthase activity approximately 5-fold (Fig. 3G), which with the NADPH produced by the PPP, would support the increased lipogenesis observed in CBS2 cells when compared to vector-transduced control NCM356 cells (Table 1).
CBS overexpressing NCM356 cells also exhibit increase flux through the hexosamine biosynthetic pathway (HBP). The HBP pathway uses fructose-6-phosphate from the glycolytic pathway to produce the end product UDP-N-acetylglucosamine, which is then used in the enzymatic post-translational modification of many cytosolic and nuclear proteins by O-linked β -N-acetylglucosamine (O-GlcNAc). Increased HBP flux and hyper-O-GlcNAcylation of proteins have been observed in the same cancers exhibiting increased levels of CBS protein and activity including, prostate (32), lung and colorectal cancers (33). Importantly, inhibition of hyper-O-GlcNAc in cell lines derived from these cancers decreased cell proliferation, anchorage-independent growth, orthotopic tumor growth, and triggered apoptosis. Here, we show that CBS2 cells exhibit a reduced level of O-GlcNAc when compared to vector control NCM356 cells (Table 1), suggesting increased utilization of O-GlcNAc for glycoprotein and glycolipid synthesis. O-linked β-N-acetylglucosamine addition to proteins is an important link between metabolism and transcriptional regulation via the modification of transcription factors (34), epigenetic modulators (35) and RNA polymerase II (36). CBS upregulation is associated with broad changes in the NCM356 cell transcriptome; over 350 genes were found to be differential expressed by overexpression of this single enzyme. GSEA pathway analyses of the transcriptome data are concordant with the metabolic data and show that CBS overexpression regulates defined gene sets that significantly overlapped with metabolic reprograming and an oncogenic phenotype (Table 2, Supplemental Table S2). Transcriptomic pathway analysis shows significant upregulation of genes associated with metabolism include insulin growth factor-2 (IGF-2) and ASPSCR1 (Alveolar Soft-part Sarcoma Chromosome Region, Candidate 1), both of which promote cellular glucose uptake via glucose transporter type 4 translocation to the plasma membrane. Other upregulated genes due to forced CBS overexpression have been shown to increase hypoxia responses (PER1, APLN), increased inflammation-mediated responses (ETS2, GDF15), cellular proliferation (CCND2, MAP3K5, PTK7, RPS6K2, E2F2, FGF19, MDK, KIT, JUNB, CXCR1, ELF3, FOS) cell migration (DDR1, PTK7, KIT, ELF3, S100A6, TNC), invasion (CDC42BPG, DDR1, HOXD11), and promotion of angiogenesis (ADM, ADM2, VEGFA, CXCR1, FOS).
How can upregulation of a single enzyme have such a broad impact on cellular phenotype? CBS plays an essential role in the synthesis of cysteine from homocysteine. In addition to being an essential amino acid constituent of cellular antioxidants such as glutathione, glutaredoxins, thioredoxins and peroxredoxin, cysteine is an important source of endogenous H2S. Our data show that despite evidence for increase flux through the reverse transsulfuration pathway, there is not a significant change between the levels of cysteine in CBS2 cells when compared to vector transduced controls (Fig 2C). This is likely due to the increased CBS-dependent synthesis of H2S by CBS2 cells (Fig 2D, E) and the accumulation of lanthionine (Table 1). H2S can directly stimulate cellular bioenergetics by contributing electrons to SQR in complex II to reduce coenzyme 10 to ubiquinol and, indirectly, by the post-translational persulfidation of F1F0 of ATP synthase/Complex V with the result of enhancing ATP production (37–39). Persulfidation, also known as sulfhydration is the modification of cysteine residues on the target protein by H2S to form a persulfide or–SSH group. H2S mediated persulfidation also has been shown to increase the activity of glyceraldehybe-3-phosphate dehydrogenase (GAPDH) increasing glycolytic flux, to enhance actin polymerization (40) and to stimulate NF-κB p65 activity inhibiting apoptosis (41). We show evidence for both increased glycolytic flux and resistance to apoptosis in NCM356 cell overexpressing CBS. H2S has also been shown to impact gene expression by altering histone H3 acetylation and DNA methylation (42). Thus, H2S-dependent and independent (i.e., increased O-GlcNAc of protein) processes could be responsible for the robust effects of CBS overexpression on NCM356 transcriptome.
In conclusion, the present study provides evidence for a functional role for the CBS/H2S axis in colon carcinogenesis. The studies in the NCM356 cell model demonstrated that upregulation of CBS in a premalignant cell was sufficient to induce the metabolic reprograming and cellular phenotype characteristic of invasive cancer. Gene dosage studies in genetically modified mice confirmed a role for CBS in colonic crypt transformation. Future studies will attempt to elucidate the pathophysiological context and molecular mechanisms underlying the carcinogenetic effects of the CBS/H2S axis on colonocytes.
Supplementary Material
Acknowledgments
The authors thank Jiyang Cai, PhD, for his assistance with the NRF2 transcriptional assay and Eileen Figueroa and Steve Schuenke for manuscript preparation.
Grant Support: This work was supported by grants from the National Institutes of Health, Cancer Prevention Research Institute of Texas, and Shriners Hospital for Children (R01 CA175803, T32 DK007639, UL1 TR001439, CPRIT DP150074, and SHC 84090).
Footnotes
Conflict of Interest: M.R.H., C.S., and C.C. are officers and/or shareholders of CBS Therapeutics Inc., an UTMB spin-off company involved in research and development of H2S biosynthesis inhibitors for the therapy of cancer. The other authors declare no potential conflicts of interest.
References
- 1.Szabo C, Coletta C, Chao C, Modis K, Szczesny B, Papapetropoulos A, et al. Tumor-derived hydrogen sulfide, produced by cystathionine-beta-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proc Natl Acad Sci U S A. 2013;110:12474–9. doi: 10.1073/pnas.1306241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang R. Physiological implications of hydrogen sulfide: a whiff exploration that blossomed. Physiol Rev. 2012;92:791–896. doi: 10.1152/physrev.00017.2011. [DOI] [PubMed] [Google Scholar]
- 3.Hellmich MR, Coletta C, Chao C, Szabo C. The therapeutic potential of cystathionine beta-synthetase/hydrogen sulfide inhibition in cancer. Antioxid Redox Signal. 2015;22:424–48. doi: 10.1089/ars.2014.5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellmich MR, Szabo C. Hydrogen sulfide and cancer. Handb Exp Pharmacol. 2015;230:233–41. doi: 10.1007/978-3-319-18144-8_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharyya S, Saha S, Giri K, Lanza IR, Nair KS, Jennings NB, et al. Cystathionine beta-synthase (CBS) contributes to advanced ovarian cancer progression and drug resistance. PLoS One. 2013;8:e79167. doi: 10.1371/journal.pone.0079167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu M, Wu L, Montaut S, Yang G. Hydrogen sulfide signaling axis as a target for prostate cancer therapeutics. Prostate Cancer. 2016;2016:8108549. doi: 10.1155/2016/8108549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sen S, Kawahara B, Gupta D, Tsai R, Khachatryan M, Roy-Chowdhuri S, et al. Role of cystathionine beta-synthase in human breast Cancer. Free Radic Biol Med. 2015;86:228–38. doi: 10.1016/j.freeradbiomed.2015.05.024. [DOI] [PubMed] [Google Scholar]
- 8.Szabo C. Gasotransmitters in cancer: from pathophysiology to experimental therapy. Nat Rev Drug Discov. 2016;15:185–203. doi: 10.1038/nrd.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szczesny B, Marcatti M, Zatarain JR, Druzhyna N, Wiktorowicz JE, Nagy P, et al. Inhibition of hydrogen sulfide biosynthesis sensitizes lung adenocarcinoma to chemotherapeutic drugs by inhibiting mitochondrial DNA repair and suppressing cellular bioenergetics. Sci Rep. 2016;6:36125. doi: 10.1038/srep36125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform. 2010;2:9. doi: 10.1186/1758-2946-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–67. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 12.Chao C, Zatarain JR, Ding Y, Coletta C, Mrazek AA, Druzhyna N, et al. Cystathionine-β-synthase inhibition for colon cancer: enhancement of the efficacy of aminooxyacetic acid via the prodrug approach. Mol Med. 2016;22:361–79. doi: 10.2119/molmed.2016.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srere PA. Citrate synthase. Methods Enzymol. 1969;13:3–11. [Google Scholar]
- 14.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stauffer JS, Manzano LA, Balch GC, Merriman RL, Tanzer LR, Moyer MP. Development and characterization of normal colonic epithelial cell lines derived from normal mucosa of patients with colon cancer. Am J Surg. 1995;169:190–6. doi: 10.1016/S0002-9610(99)80135-4. [DOI] [PubMed] [Google Scholar]
- 18.Modis K, Coletta C, Asimakopoulou A, Szczesny B, Chao C, Papapetropoulos A, et al. Effect of S-adenosyl-L-methionine (SAM), an allosteric activator of cystathionine-beta-synthase (CBS) on colorectal cancer cell proliferation and bioenergetics in vitro. Nitric Oxide. 2014;41:146–56. doi: 10.1016/j.niox.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chao C, Han X, Ives K, Park J, Kolokoltsov AA, Davey RA, et al. CCK2 receptor expression transforms non-tumorigenic human NCM356 colonic epithelial cells into tumor forming cells. Int J Cancer. 2010;126:864–75. doi: 10.1002/ijc.24845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asimakopoulou A, Panopoulos P, Chasapis CT, Coletta C, Zhou Z, Cirino G, et al. Selectivity of commonly used pharmacological inhibitors for cystathionine beta synthase (CBS) and cystathionine gamma lyase (CSE) Br J Pharmacol. 2013;169:922–32. doi: 10.1111/bph.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhri VK, Salzler GG, Dick SA, Buckman MS, Sordella R, Karoly ED, et al. Metabolic alterations in lung cancer-associated fibroblasts correlated with increased glycolytic metabolism of the tumor. Mol Cancer Res. 2013;11:579–92. doi: 10.1158/1541-7786.MCR-12-0437-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh S, Padovani D, Leslie RA, Chiku T, Banerjee R. Relative contributions of cystathionine beta-synthase and gamma-cystathionase to H2S biogenesis via alternative trans-sulfuration reactions. J Biol Chem. 2009;284:22457–66. doi: 10.1074/jbc.M109.010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soga T, Baran R, Suematsu M, Ueno Y, Ikeda S, Sakurakawa T, et al. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J Biol Chem. 2006;281:16768–76. doi: 10.1074/jbc.M601876200. [DOI] [PubMed] [Google Scholar]
- 24.Chao C, Lotz MM, Clarke AC, Mercurio AM. A function for the integrin alpha6beta4 in the invasive properties of colorectal carcinoma cells. Cancer Res. 1996;56:4811–9. [PubMed] [Google Scholar]
- 25.Buchheit CL, Weigel KJ, Schafer ZT. Cancer cell survival during detachment from the ECM: multiple barriers to tumour progression. Nat Rev Cancer. 2014;14:632–41. doi: 10.1038/nrc3789. [DOI] [PubMed] [Google Scholar]
- 26.Orford K, Orford CC, Byers SW. Exogenous expression of beta-catenin regulates contact inhibition, anchorage-independent growth, anoikis, and radiation-induced cell cycle arrest. J Cell Biol. 1999;146:855–68. doi: 10.1083/jcb.146.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–25. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 30.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Munoz-Pinedo C, El Mjiyad N, Ricci JE. Cancer metabolism: current perspectives and future directions. Cell Death Dis. 2012;3:e248. doi: 10.1038/cddis.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch TP, Ferrer CM, Jackson SR, Shahriari KS, Vosseller K, Reginato MJ. Critical role of O-Linked beta-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis. J Biol Chem. 2012;287:11070–81. doi: 10.1074/jbc.M111.302547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mi W, Gu Y, Han C, Liu H, Fan Q, Zhang X, et al. O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim Biophys Acta. 2011;1812:514–9. doi: 10.1016/j.bbadis.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 34.Ozcan S, Andrali SS, Cantrell JE. Modulation of transcription factor function by O-GlcNAc modification. Biochim Biophys Acta. 2010;1799:353–64. doi: 10.1016/j.bbagrm.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanover JA, Krause MW, Love DC. Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat Rev Mol Cell Biol. 2012;13:312–21. doi: 10.1038/nrm3334. [DOI] [PubMed] [Google Scholar]
- 36.Kelly WG, Dahmus ME, Hart GW. RNA polymerase II is a glycoproteinModification of the COOH-terminal domain by O-GlcNAc. J Biol Chem. 1993;268:10416–24. [PubMed] [Google Scholar]
- 37.Lagoutte E, Mimoun S, Andriamihaja M, Chaumontet C, Blachier F, Bouillaud F. Oxidation of hydrogen sulfide remains a priority in mammalian cells and causes reverse electron transfer in colonocytes. Biochim Biophys Acta. 2010;1797:1500–11. doi: 10.1016/j.bbabio.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 38.Modis K, Bos EM, Calzia E, van Goor H, Coletta C, Papapetropoulos A, et al. Regulation of mitochondrial bioenergetic function by hydrogen sulfide. Part II. Pathophysiological and therapeutic aspects. Br J Pharmacol. 2014;171:2123–46. doi: 10.1111/bph.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Modis K, Ju Y, Ahmad A, Untereiner AA, Altaany Z, Wu L, et al. S-Sulfhydration of ATP synthase by hydrogen sulfide stimulates mitochondrial bioenergetics. Pharmacol Res. 2016;113:116–24. doi: 10.1016/j.phrs.2016.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, et al. H2S signals through protein S-sulfhydration. Sci Signal. 2009;2:ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sen N, Paul BD, Gadalla MM, Mustafa AK, Sen T, Xu R, et al. Hydrogen sulfide-linked sulfhydration of NF-kappaB mediates its antiapoptotic actions. Mol Cell. 2012;45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rios EC, Szczesny B, Soriano FG, Olah G, Szabo C. Hydrogen sulfide attenuates cytokine production through the modulation of chromatin remodeling. Int J Mol Med. 2015;35:1741–6. doi: 10.3892/ijmm.2015.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.