Abstract
The zygomycete fungus Lichtheimia ramosa H71D, isolated from sugarcane bagasse compost, was identified by applying phylogenetic analysis based on the DNA sequence of the Internal Transcribed Spacer (ITS), and subsequent secondary structure analysis of ITS2. L. ramosa H71D was able to grow over a wide range of temperatures (25–45 °C), manifesting optimal growth at 37 °C. A 64 kDa xylanase (named LrXynA) was purified from the culture supernatant of L. ramosa H71D grown on 2% carboxymethylcellulose (CMC), as the only carbon source. LrXynA displayed optimal activity at pH 6 and temperature of 65 °C. The enzyme retained more than 50% of its maximal activity over a broad range of pH values (4.5–7.5). Enzyme half-life (t½) times at 55, 65 and 75 °C were 80, 25, and 8 min, respectively. LrXynA showed higher affinity (k M of 2.87 mg/mL) and catalytic efficiency (k cat/k M of 0.651 mg s/mL) towards Beechwood xylan in comparison to other substrates such as Birchwood xylan, Oat-spelt xylan, CMC, Avicel and Solka floc. The predominant final products from LrXynA-mediated hydrolysis of Beechwood xylan were xylobiose and xylotriose, suggesting that the enzyme is an endo-β-1,4 xylanase. Scanning electron microscopy (SEM) imaging of sugar cane bagasse (SCB) treated with LrXynA, alone or in combination with commercial cellulases, showed a positive effect on the hydrolysis of SCB. To our knowledge, this is the first report focusing on the biochemical and functional characterization of an endo-β-1,4 xylanase from the thermotolerant and fast-growing fungus Lichtheimia ramosa.
Keywords: Internal transcribed spacer (ITS), Zygomycete fungus, Lichtheimia ramosa, Xylanase, Sugarcane hydrolysis
Introduction
Xylan is next in order to cellulose, in terms of the major structural components of plant cell walls, and is the second most abundant renewable polysaccharide in nature (Collins et al. 2002). Xylan is a complex, highly branched heteropolysaccharide and its structure varies between different plant species. The homopolymeric backbone chain of xylan consists of 1,4-linked β-d-xylopyranosyl units, that to a varied extent can be substituted with glucuronopyranosyl, 4-O-methyl-d-glucuronopyranosyl, α-l-arabinofuranosyl, acetyl, feruloyl or p-coumaroyl side-chain groups (Kulkarni et al. 1999). The complete hydrolysis of xylan requires the action of several enzymes, including endo-1,4-β-d-xylanase (EC3.2.1.8), which is crucial for xylan depolymerization (Polizeli et al. 2005). Xylanases, as glycoside hydrolase members, are able to catalyze the hydrolysis of the glycosidic linkage (β-1,4) of xylosides, leading to the formation of a sugar hemiacetal and the corresponding free aglycone (Hatanaka 2012). Xylanases and the microorganisms that produce them are interesting because they have extensive biotechnological applications. These enzymes are currently used in waste management in order to degrade xylan for the production of renewable fuels and chemicals. Likewise, they are used in food, agro-fiber, and paper and pulp industries, where xylanases help to reduce environmental impact (Collins et al. 2002). Oligosaccharides produced from the action of xylanases are further used as functional food additives or alternative sweeteners with beneficial properties (Pellerin et al. 1991). In terms of biotechnological application, thermostable enzymes have several generic advantages as the high specific activity, that is often associated with this kind of enzymes, reduces the required amount of enzyme and prolongs hydrolysis time due to greater stability than that exhibited by mesophilic enzymes (Viikari et al. 2007).
Many reports exist on thermophilic and mesophilic microorganisms signifying that bacteria and fungi are major producers of thermostable xylanases (Sunna and Antranikian 1997; Collins et al. 2002). In terms of fungi, most xylanases characterized to date are derived from Ascomycetes and Basidiomycetes; in particular, mesophilic fungi belonging to the genera Aspergillus and Trichoderma are preeminent in xylanase production (Polizeli et al. 2005). Production of thermostable xylanases has been reported for several filamentous fungi including Laetiporus sulphureus (Lee et al. 2009), Talaromyces thermophiles (Maalej et al. 2009), Thermomyces lanuginosus (Singh et al. 2003), Nonomuraea flexuosa, Thermoascus aurantiacus (Zhang et al. 2011), and the Zygomycete fungus Rhizomucor miehei (Fawzi 2011).
Zygomycetes are able to grow on a wide variety of carbon sources at different temperatures, oxygenation rates, and pH values (Ferreira et al. 2013). Recently, Zygomycetes are receiving increased attention in the biotechnological context for the production of a wide range of metabolic products e.g., organic acids, enzymes, and biofuels such as bioethanol and biodiesel (Ferreira et al. 2013). The genus Lichtheimia (syn. Mycocladus, Absidia) belongs to the Zygomycete class and includes saprotrophic microorganisms that can be isolated from decomposing soil and plant material (Alastruey-Izquierdo et al. 2010). Members of this genus are considered to constitute thermotolerant fungi, as they can grow at a wide range of temperatures, from 20 to 53 °C, with 37 °C presenting the best temperature for growth, where it occurs most rapidly (Voigt et al. 1999; André et al. 2014). This rapid growth rate of filamentous fungi belonging to the Zygomycete genus Lichtheimia makes them pertinent to the study of enzymes involved in the breakdown of plant material and offers possible advantages for a biotechnological application.
There are few studies on carbohydrate-active enzymes in the Zygomycetes fungi belonging to the Lichtheimia genus. Lichtheimia blakesleeana was described as a producer of phytase and xylanase (Neves et al. 2011). Additionally, Lichtheimia ramosa has been reported as a producer of xylanase, carboxymethylcellulase (CMCase), an ample producer of β-glucosidase in wheat bran-based medium (Gonçalves et al. 2013), and also a producer of amylases, β-glucosidases, CMCase, and xylanases, via solid state bioprocess, utilizing fruit waste from the Brazilian savannah (de Silva et al. 2013). Nevertheless, to our knowledge, there are no studies on the biochemical and catalytical properties of xylanases from a filamentous fungus belonging to the Zygomycete genus Lichtheimia.
Taxonomy of Mucorales has traditionally been based on microscopic morphology and mating experiments; however, molecular phylogeny has revealed that diversity within and between species is much greater than anticipated, also leading to a proliferation of the number of taxa recognized (Walther et al. 2013). The internal transcribed spacer region (ITS) consists of three parts: ITS1, ITS2 and the highly conserved 5.8S rDNA exon located between them. ITS2 usually has a conserved secondary structure with four helices which appear to be essential for successful excision of ITS2 from the precursor rRNA (Caisová et al. 2011). The ITS2 has been viewed as a possible useful marker for taxonomic classification, at a wide range of levels (Coleman 2003), because of its high divergence in sequence and assumed conservation in structure (Schultz et al. 2005). Additionally, it has been suggested modeling this cloverleaf-like structure as a novel tool for phylogenetics (Wolf et al. 2005). Furthermore, the ITS2 has been proposed as a candidate for the DNA fungi barcodes because it possesses a number of valuable characteristics (Yao et al. 2010). In Mucorales, the ITS region turned out to be an appropriate barcoding marker (Walther et al. 2013).
Hence, the aim of this work was to use phylogenetic analysis to identify the H71D strain, isolated from sugarcane bagasse compost, and undertake the purification and biochemical characterization of a secreted xylanase from this thermotolerant and fast-growing fungus.
Materials and methods
Microorganisms and growth conditions
The strain H71D was isolated from composting soils and kindly donated by Dr. Sergio Trejo-Estrada research group (CIBA-IPN, Tlaxcala. México).
For spore production, the fungus was grown on potato dextrose agar (PDA) medium plates and incubated at 37 °C for 72 h. Spore collection was performed and a suspension thereof, which was read at λ = 650 nm and adjusted to achieve an Absorbance of 0.5, which is equivalent to 5 × 106 spores/mL (Tien and Kirk 1988).
For genomic DNA preparation and xylanase production, the strain H71D was grown in the liquid culture media described by Mandels and Sternburg (1967), with some modifications as follows. The culture basal medium contained in g/L: yeast extract 1; (NH4)2SO4 1.4; KH2PO4 2.0; Urea 0.3; CaCl2 0.3; MgSO4 7H2O 0.3; and 1 mL/L of trace element solution containing (g/L): FeSO4 7H2O 0.05; MnSO4 H2O 0.016; ZnSO4 7H2O 0.014; CoCl2 0.02. The culture basal medium was supplemented with 2% (w/v) CMC as the only carbon source, unless otherwise stated. The medium was sterilized for 15 min at 121 °C and 15 psi. Flasks of 250 mL containing 50 mL of medium were inoculated with 500 µL of spore solution of the desired concentration, and flasks were incubated at 37 °C for 9–12 days on an orbital shaker at 160 rpm.
Identification of the H71D strain
The identification of the strain H71D was made using morphological characteristics and the DNA sequence of the ribosomal DNA ITS2 region as a molecular marker. Morphological characterization followed the key published elsewhere (Hoffmann 2010). For molecular analysis, the genomic DNA was extracted from mycelia of H71D strain grown in liquid medium after 3 days of incubation at 37 °C and orbital agitation at 160 rpm, as described above. The mycelium was obtained by centrifugation (7000 rpm at 4 °C for 20 min); then, it was ground with liquid nitrogen, and this material was used for genomic DNA extraction, by using the DNeasy Blood & Tissue kit (QIAGEN, Valencia, CA). The ITS2 region was amplified from genomic DNA by PCR using the HotStar HiFidelity Polymerase Kit (Qiagen, Valencia, CA), and the barcoding primer pair ITS4 and ITS5 previously reported (White et al. 1990). The DNA sequence of the ITS2 region from H71D was compared with those ITS2 sequences from strains deposited at NCBI-GenBank, by using BLASTn available at the NCBI server (https://www.ncbi.nlm.nih.gov/). The phylogenetic analysis was carried out using ITS2 sequences from Lichtheimia species (Table 1), employing as outgroup the ITS2 sequence from Dichotomocladium elegans CBS 695.76 (GenBank Accession: HM999950), which has previously been used for the same purpose (O’Donnell et al. 2001). The multiple alignments were conducted by using the Clustal X program (version 2.0) (Larkin et al. 2007), and the FindModel program (https://www.hiv.lanl.gov/content/sequence/findmodel/findmodel.html) was used to select the model that best describes the data to generate a better tree. The phylogenetic analysis was conducted by the method of maximum likelihood using PhyML with 100 bootstrap replicates. The secondary structure of the ITS2 from the strain H71D was predicted by using the RNAfold WebServer (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi), with the following setup: (i) minimum free energy (MFE), (ii) partition function and avoid isolated base pairs for fold algorithms, (iii) with no dangling end energies, and (iv) RNA parameters: Turner model (Mathews et al. 2004) at 37 °C. For comparison purpose, the ITS2 sequence from L. ramosa GQ342874 strain was selected and analyzed under the same conditions. Hence, the H71D strain was identified as L. ramosa H71D strain and deposited on “Colección de Cultivos Microbianos” (CDBB CINVESTAV-IPN, México) with access number CDBB–H–1939. The ITS2 DNA sequence from L. ramosa H71D strain was deposited in the GenBank database with Access Number KY311837.
Table 1.
GenBank accession numbers of the DNA sequences from members of the genus Lichteimia used in this study
| Strain | Specie | ITS GenBank Access Number |
|---|---|---|
| H71D | L. ramosa | KY311837 |
| CBS 100.17 | L. corymbifera | GQ342885 |
| CBS 100.31 | L. corymbifera | GQ342879 |
| CBS 100.51 | L. corymbifera | GQ342886 |
| CBS 429.75 | L. corymbifera | GQ342878 |
| CBS 519.71 | L. corymbifera | GQ342889 |
| CBS 109940 | L. corymbifera | GQ342881 |
| CBS 100.28 | L. hyalospora | GQ342896 |
| CBS 100.36 | L. hyalospora | GQ342898 |
| CBS 102.36 | L. hyalospora | GQ342895 |
| CBS 173.67 | L. hyalospora | GQ342893 |
| CBS 518.71 | L. hyalospora | GQ342894 |
| CBS 291.66 | L. ornata | GQ342891 |
| CBS 958.68 | L. ornata | GQ342890 |
| CNM-CM 4978 | L. ornata | GQ342892 |
| AS 3.4808 | L. ramosa | GQ342867 |
| CBS100.24 | L. ramosa | GQ342876 |
| CBS100.49 | L. ramosa | GQ342858 |
| CBS 223.78 | L. ramosa | GQ342877 |
| CBS 271.65 | L. ramosa | GQ342875 |
| CBS 582.65 | L. ramosa | GQ342874 |
| CBS 649.78 | L. ramosa | GQ342849 |
| CBS 124197 | L. ramosa | GQ342870 |
| CBS 124198 | L. ramosa | GQ342848 |
| CNM-CM 3148 | L. ramosa | GQ342872 |
| CNM-CM 3590 | L. ramosa | GQ342869 |
| CNM-CM 4261 | L. ramosa | GQ342854 |
| CNM-CM 4337 | L. ramosa | GQ342852 |
| CNM-CM 4427 | L. ramosa | GQ342853 |
| CNM-CM 4537 | L. ramosa | GQ342873 |
| CNM-CM 4849 | L. ramosa | GQ342855 |
| CNM-CM 5111 | L. ramosa | GQ342871 |
| CNM-CM 5171 | L. ramosa | GQ342864 |
| CBS 420.70 | L. sphaerocystis | GQ342900 |
| CBS 647.78 | L. sphaerocystis | GQ342899 |
| CBS 648.78 | L. sphaerocystis | GQ342901 |
| CBS695.76 | D. elegans | HM99995 |
Determination of optimum growth temperature
To determine the optimum growth temperature, L. ramosa H71D strain was analyzed based on its radial growth (cm) on Petri dishes with PDA at different temperatures (25, 30, 35, 37, 40 and 45 °C) during 6 days. PDA plates were inoculated with small circles taken from the edge of a 2 days old colony. The plates were incubated at the different temperatures indicated above, and the diameter was measured every 8 h for 48 h. The growth rate, measured in centimeters per hour, was calculated for each plate and temperature.
Enzyme and protein assays
Xylanase and cellulase activities were determined by measuring the amount of reducing sugars released, quantified by the DNS method at 540 nm (Miller 1959), using xylose or glucose as a standard. For xylanolytic activity, the assay mixture contained 25 µL of enzyme preparation, 975 µL of 0.2% Beechwood xylan (Sigma-Aldrich, St. Louis, MO, USA) in 50 mM citrate buffer, pH 6; subsequently, the mixture was stirred and incubated at 65 °C for 5 min. For cellulolytic activity, the assay mixture contained 100 µL of enzyme preparation, 900 µL of 0.3% CMC (Sigma-Aldrich, St. Louis, MO, USA) in 50 mM acetate buffer, pH 5.6; then, the mixture was stirred and incubated at 50 °C for 10 min. Enzyme activity was expressed as U/mL, where U corresponds to the µmoles of xylose/glucose released per minute, under assay conditions. All tests were carried out in triplicate and error bars represent the standard deviation. Protein concentration was measured using Bradford reagent (Sigma-Aldrich, St. Louis, MO, USA), and bovine serum albumin (Life Technologies, Grand Island, NY, USA) as the standard.
Xylanase and cellulase production
For enzyme production, the H71D strain was grown in the liquid culture media as described by Mandels and Sternberg (1967), with some modifications as described above. Culture basal medium was supplemented with 2% (w/v) Beechwood xylan or 2% (w/v) CMC. The culture medium was sterilized for 15 min at 121 °C and 15 psi. Flasks of 2.8 L with 500 mL of medium were inoculated with 5 mL of spore solution at the desired concentration. Then, cultures were incubated at 37 °C for 9 days and orbital agitation at 160 rpm. Every 12 or 24 h, aliquot samples of 5 mL were taken from each flask. The pellet was obtained by centrifugation at 7000 rpm at 4 °C for 20 min, and was used to determine fungal biomass by the dry weight method; whereas the culture supernatant was used for extracellular enzyme (xylanase and cellulase) assays. The results presented are expressed as the mean ± standard deviation of three replicates.
Enzyme purification
The culture supernatant (800 mL) was treated with ammonium sulfate (70% saturation). The precipitate was collected by centrifugation (8500 rpm, 4 °C for 15 min), then the pellet was resuspended and dialyzed against buffer A (50 mM Tris- HCl buffer pH 8, 0.1 mM PMSF, and 5% (v/v) glycerol). After dialysis, the protein preparation was loaded onto anion exchange UNOsphere Q (Bio-Rad), and cation exchange UNOsphere S (Bio-Rad) columns (column volume, 15 mL). Absorbed proteins were eluted from the column with a linear gradient of KCl (0.025–1 M) in buffer A, at a constant flow rate of 2 mL/min, and 2 mL fractions were collected. Fractions with xylanase activity were pooled and analyzed by 10% SDS-PAGE.
Electrophoretic analysis
SDS–Polyacrylamide gel electrophoresis (SDS–PAGE) was performed using a polyacrylamide gel 10% according to the method described by Laemmli (1970). The gel was stained with Coomassie Brilliant Blue R-250 (Bio-Rad). Molecular weight (MW) was estimated by linear regression with reference to a broad range molecular weight protein standard (Bio-Rad).
Zymogram analysis
The zymogram of xylanolytic activity was performed according to the methodology described by Royer and Nakas (1990), with some modifications as follows. Briefly, protein samples were separated on 10% polyacrylamide gels co-polymerized with 1% Remazol Brilliant Blue linked to xylan (RBB-X), under denaturing conditions. Protein samples were resuspended in SDS sample buffer with 5% (v/v) 2-β-mercaptoethanol, then samples were boiled in a water bath for 5 min. After electrophoresis, the gel was rinsed with distilled water and incubated in 50 mM citrate buffer, pH 6 at 50 °C for 2.5 h.
Carbohydrate content
The amount of carbohydrates was determined by the Anthrone method (Leyva et al. 2008). A 0.2% of cold Anthrone solution was prepared in sulfuric acid. One milliliter of solution was slowly mixed with 500 µL of a sample preparation. This mixture was incubated at room temperature for 5 min, boiled for 10 min in a water bath, and then, tubes were placed on ice for 5 min. The samples were read at 640 nm, and the percentage of glycosylation was calculated according to the total amount of protein present in the sample. The standard curve was made with mannose.
Biochemical properties
Optimal pH and pH stability
The effect of pH on the xylanolytic activity of LrXynA was determined by varying the pH of the reaction mixtures using 50 mM citrate–phosphate buffer (pH 3–7), and 50 mM phosphate buffer (pH 6 to 8). Reaction mixtures were incubated at 50 °C for 10 min. For pH stability assay, the enzyme was preincubated in the above mentioned buffers, without substrate, at 50 °C for 3 h. Subsequently, the remaining xylanolytic activity was measured under standard conditions (65 °C and pH 6.0 for 5 min), and compared to the activity displayed by the untreated enzyme.
Effect of temperature on LrXynA activity and stability
The effect of temperature on the enzymatic activity of LrXynA was estimated by conducting the activity assay at different temperatures ranging from 30 to 80 °C in 50 mM citrate–phosphate buffer, pH 6. Reaction mixtures were incubated for 5 min under standard conditions. The thermostability of the enzyme was investigated after preincubation at 55, 65 and 75 °C without substrate. Residual enzyme activities at specific time points were determined under standard conditions, (65 °C, pH 6.0, for 5 min). To determine half-life (t½) of the enzyme, aliquot samples were withdrawn at different time intervals and residual enzymatic activity was measured under standard conditions.
Substrate specificity of LrXynA and kinetic parameters
The xylanolytic activity of LrXynA was determined under optimal assay conditions using 1% (w/v): Beechwood xylan, Birchwood xylan, Oat-spelt xylan, CMC, Avicel or Solka floc as the substrate. The kinetic parameters k M and V max of LrXynA were determined under optimal conditions for enzyme activity using Beechwood xylan as substrate, at a concentration ranging from 0.05 to 1%. The kinetic parameters k M and V max were determined and calculated from the Nonlinear least squares regression applied to the Michaelis and Menten (http://statpages.org/nonlin.html).
Effect of metal ions and EDTA
To study the effect of various metal ions (Ca2+, Cu2+, Fe2+, Hg2+, Li, Mg2+, Mn2+, Na+, Ni2+ and Zn2+), and the chelating agent EDTA on the activity of LrXynA, the enzyme was independently incubated with metal ions or EDTA, at final concentrations of 1 and 5 mM under optimal assay conditions (65 °C, pH 6 for 5 min). The activity was expressed as the percentage of the activity observed in the absence of any compound.
Analysis of LrXynA hydrolysis products
Thin-layer chromatography (TLC) of LrXynA hydrolysis product was carried out as follows. Purified enzyme (50 µL, 3 U/mL) was mixed with 50 µL of 1% (w/v) Beechwood xylan in 50 mM citrate buffer, pH 6. The reaction proceeded at 40 °C, and aliquot samples were taken after 0, 12, 24, 36, 48 h of incubation. TLC was carried out at room temperature using as mobile phase a mix of butanol:ethanol:water 5:3:2 (v/v). The plate was air dried, sprayed with H2SO4 15% and bake at 100 °C for 2 h after color development.
Enzymatic hydrolysis of sugar cane bagasse (SCB)
SCB used in this work was previously characterized by Pavón-Orozco et al. (2012). The hydrolysis experiments were carried out to a final concentration of 15 mg/mL of SCB in a 50 mM citrate buffer pH 5, at 37 °C for 88 h and orbital shaking at 120 rpm. Enzymes used were, LrXynA (X) from the L. ramosa H71D strain (this work), a commercial cellulase from Aspergillus niger (A) (Sigma-Aldrich, USA) and a commercial cellulase from Trichoderma viridae (T) (Calbiochem, USA), kindly donated by Dr. Plinio Guzmán Villate (CINVESTAV-Irapuato). In addition, reaction mixtures with different molar ratios from 0 to 100% of X combined with A or T were used. In all assays, the final molar concentration of the enzymes was kept constant at 34 mM. All preparations were supplemented with 0.01 mM β-glucosidase (Sigma-Aldrich, USA), to prevent potential inhibition by product, and with 0.02% sodium azide, to avoid contamination during kinetics. Aliquot samples (400 µL) were taken every 8 h during 88 h, time points were analyzed for reducing sugars (glucose as standard) by the DNS method (Miller 1959). The hydrolysis reaction was stopped by boiling the samples 5 min, followed by centrifugation at 10,000 rpm for 5 min. Hydrolysis experiments were performed in triplicate. A total amount of 15 mg/mL of SCB in 50 mM citrate buffer, pH 5.0, 0.01 mM β-glucosidase and sodium azide 0.02%, was used as a blank. For control one, the blank was supplemented with the specified proportion of X. For control two, the blank was supplemented with the specified proportion of A or T. Therefore, a blank and two controls were used for each reaction mixture and data obtained were used to calculate the amount of reducing sugars released from each reaction.
Scanning electron microscopy (SEM) analysis
The effect of xylanase/cellulase activity on the surface of SCB, before and after enzymatic treatments of SCB with xylanase (X) from the L. ramosa H71D strain (this work), and a commercial cellulase from Aspergillus niger (A) or from Trichoderma viridae (T), were analyzed by SEM imaging. Sample preparations for SEM were dried in a muffle at 50 °C, and then placed directly on a graphite layer, coated with gold and finally observed at 10 kV with an amplification of ×50 and ×3,000 on a JEOL (JSM 6510 LB) at the Electronic Microscopy Laboratory (CGSE, CINVESTAV-IPN, México).
Results
Identification of H71D strain
The H71D strain was identified based on morphological characteristics and molecular markers. Morphological characteristics of H71D strain were comparable to those described for the type species of L. ramosa (Hoffmann 2010), e.g., sporangia were light gray colored, subsporangial septum was absent, and sporangiospores were ellipsoidal (data not shown).
Taxonomic identification was carried out based on the DNA sequence of the ITS2, as a molecular marker. Thirty-six ITS2 DNA sequences from members of the genus Lichtheimia were selected from the GenBank for phylogenetic analysis and listed in Table 1. The phylogenetic tree was created by the method of maximum likelihood using the PhyML (HYK85 model) with 100 bootstrap replicates. Findings here indicate that the H71D strain belongs to L. ramosa clade, which was further corroborated by a high bootstrap value (Fig. 1). This molecular analysis also revealed that the ITS2 DNA sequence from H71D strain is very similar (98% of identity) to those sequences from L. ramosa GQ342876, GQ342875, and GQ342874.
Fig. 1.
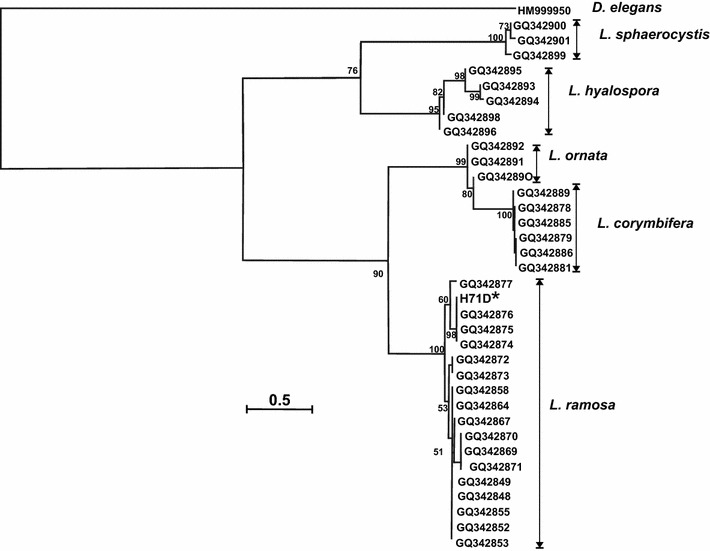
Phylogenetic tree inferred from ITS DNA sequences from Lichtheimia species. The tree was inferred under PhyML algorithm using an HKY85 model. The numbers on the branches correspond to the robustness (bootstrap values) obtained from 100 replicates
To confirm the identity of L. ramosa H71D, the secondary structure of the ITS2 region from strain H71D was compared to that from L. ramosa GQ342874, as both ITS2 regions are very similar (99% identity, 100% cover), and analyzed by RNAfold WebServer using the RNA Turner model, (Mathews et al. 2004) at 37 °C. Notably, the ITS2 DNA sequences from L. ramosa H71D and L. ramosa GQ342874 differ in only three nucleotides, corresponding to changes U→C, A→G and C→A, indicated by arrows (Fig. 2). These nucleotide changes did not result in different specific structures; however, the secondary structures showed differences in the MFE values of − 242.60 and − 245.30 kcal/mol for L. ramosa H71D and L. ramosa GQ342874, respectively.
Fig. 2.
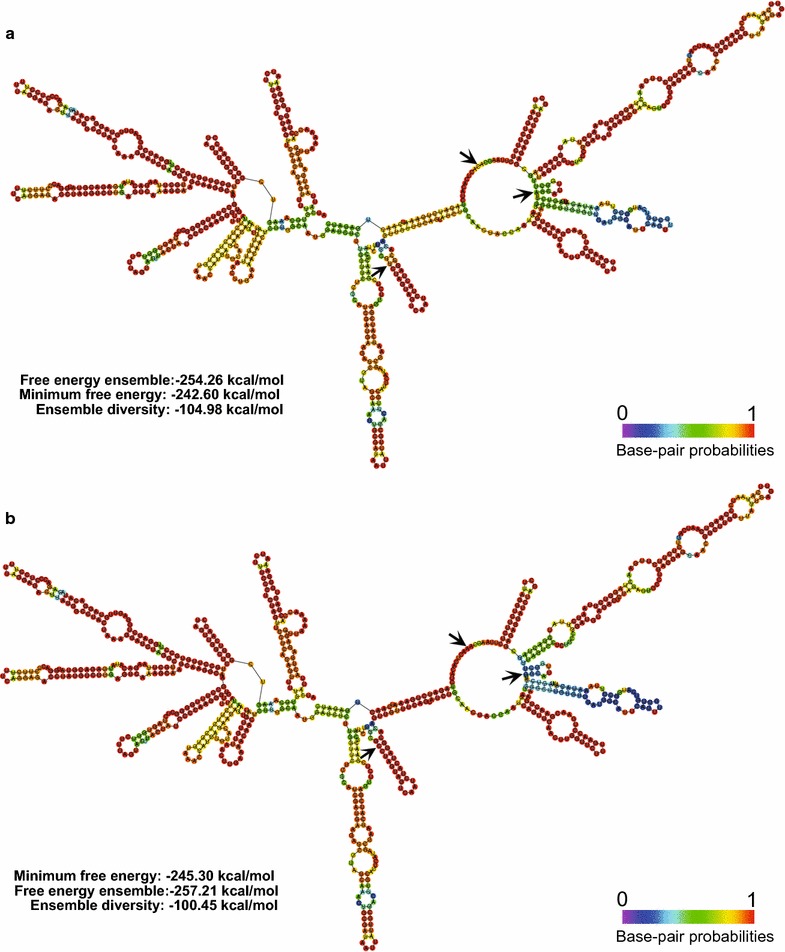
ITS2 secondary structures of strains a H71D and b GQ342874, determined with the RNA model Turner, 2004 (RNAfold Web Server); the most significant differences in the sequences that determined the secondary structure are indicated with black arrows
Determination of optimum growth temperature
To determine the optimum growth temperature of L. ramosa H71D, the fungus was cultured on Petri dishes with PDA medium at 25, 30, 35, 37, 40 and 45 °C for 6 days. The optimum growth temperature of L. ramosa H71D was 37 °C, with a radial growth of 3.6 cm after 48 h of incubation (Fig. 3a).
Fig. 3.
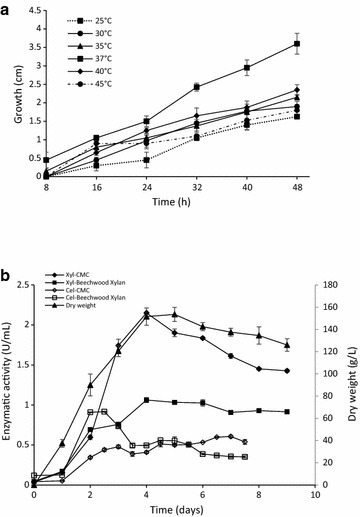
Xylanase and cellulose activities during growth of L. ramosa H71D. a Radial growth kinetic of L. ramosa H71D at different growth temperatures on PDA agar, the diameter was measured every 8 h for 48 h. b Growth and enzyme activities (xylanase and cellulose) of L. ramosa H71D at 37 °C. Dry weight in Mandels and Sternberg culture medium (Black up-pointing triangle). Xylanolytic activity produced on CMC (Black diamond suit) or Beechwood xylan (Black square), as a carbon source. Cellulolytic activity produced on CMC (Lozenge) or Beechwood xylan (Square), as a carbon source
Xylanase and cellulase production
To evaluate the production of xylanase and cellulase activities, L. ramosa H71D was cultured at 37 °C in modified Mandels and Sternberg liquid media, using 2% (w/v) CMC or 2% (w/v) Beechwood xylan as carbon source. Xylanase and cellulase synthesis was induced with both carbon sources. Greatest xylanase activity was produced on CMC (2.1 U/mL) after 4 days of incubation, whereas the greatest cellulase activity was observed in the presence of Beechwood xylan (0.091 U/mL), after two and a half days of incubation (Fig. 3b). The xylanolytic activity onset occurred on the first day of culture, whereas highest activity was reached after 4 days of incubation; subsequently, xylanase activity remained constant for the next 4 days. Due to the xylanolytic activity was higher on CMC than on Beechwood xylan, the growth kinetic of L. ramosa H71D was investigated using CMC as the only carbon source (Fig. 3b). Zymogram analysis of the culture supernatant from L. ramosa H71D grown on CMC, after 4 days of incubation at 37 °C, revealed at least five bands with xylanolytic activity (Fig. 4a).
Fig. 4.
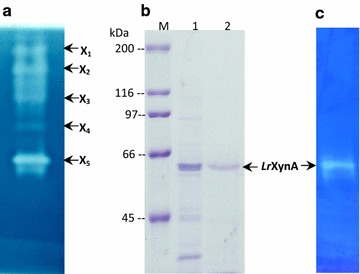
Protein and zymogram analysis of LrXynA from L. ramosa H71D on 10% SDS-PAGE. a Zymogram analysis of crude extract from L. ramosa H71D, using 1% RBB-X as the substrate. b 10% SDS-PAGE analysis of purified LrXynA. Lanes: M, molecular weight standard; 1, crude extract; 2, purified LrXynA. c Zymogram analysis of purified LrXynA, using 1% RBB-X as substrate
Purification of LrXynA
An extracellular xylanase was purified from the culture supernatant of L. ramosa H71D grown on CMC as the only carbon source. All purification steps are summarized in Table 2. The xylanase was purified 6.4-fold to homogeneity with a recovery yield of 38.5% and a specific activity of 126.43 U/mg of protein. The purified xylanase was separated on 10% SDS-PAGE, and its molecular weight was estimated to be 64 kDa (Fig. 4b), and named LrXynA. Zymogram analysis of the purified LrXynA, using 1% RBB-X as the substrate, showed a single clear band (Fig. 4c), thus confirming the xylanase activity of LrXynA.
Table 2.
Purification steps of xylanase LrXynA from L. ramosa H71D
| Sample | Total activity (U) | Total protein (mg) | Specific activity (U/mg) | Yield (%) | Purificación (n) |
|---|---|---|---|---|---|
| Crude extract | 1878.01 | 95.12 | 19.75 | 100 | 1 |
| 70% ammonium sulfate precipitation | 991.69 | 26.24 | 37.80 | 52.81 | 1.9 |
| Ion exchange chromatography | 723.12 | 5.52 | 131 | 38.5 | 6.6 |
Carbohydrate content
Most xylanases produced by bacteria and fungi reported so far are glycosylated. The carbohydrate content of the enzyme was determined by the Antrone-sulfuric acid method, in order to reveal whether LrXynA is a glycoprotein. However, no carbohydrate was detected.
Biochemical properties
The purified xylanase LrXynA from L. ramosa H71D was biochemically characterized and the results are described below.
Effect of pH on LrXynA activity and stability
The influence of pH on the xylan hydrolysis of LrXynA was determined at pH values ranging from 3 to 8 at 50 °C. LrXynA showed maximum activity at pH 6 and exhibited about 50% of its maximal activity at different pH values ranging from 4 to 7.5 (Fig. 5a). The pH stability of LrXynA at different pH values in the range from 3 to 8, after 3 h of incubation at 50 °C was evaluated. LrXynA was stable at a broad range of pH (4.5–7), retaining more than 50% of its original activity (Fig. 5a).
Fig. 5.
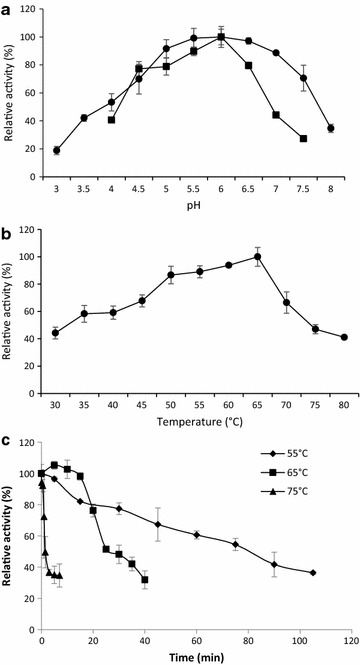
Effect of pH and temperature on LrXynA activity and stability. a Effect of pH on xylanolytic activity (Black circle) and stability (Black square) of LrXynA. LrXynA was incubated in 50 mM citrate–phosphate (3–7) or phosphates (6–8) buffer and incubated at 50 °C for 10 min; for pH stability, LrXynA was preincubated at 50 °C for 3 h in the same buffers. b Effect of temperature on the xylanolytic activity of LrXynA. The enzyme was incubated in 0.2% (w/v) Beechwood xylan in 50 mM citrate–phosphate buffer, pH 6.0 at different temperatures (30–80 °C). c Thermostability of LrXynA at 75 °C (Black up-pointing triangle), 65 °C (Black square) and 55 °C (Black diamond suit)
Effect of temperature on LrXynA activity and stability
The effect of temperature on the xylan hydrolysis of LrXynA was determined at different temperatures ranging from 30 to 80 °C, at pH 6. LrXynA showed optimal activity at 65 °C, and displayed 50% of its maximal activity over a wide temperature range; from 35 to 70 °C (Fig. 5b). In order to evaluate enzyme thermostability, the purified LrXynA was incubated at 55, 65 and 75 °C in 50 mM citrate–phosphate buffer, pH 6. The half-lifes of LrXynA at 55 and 65 °C were 80 and 25 min, respectively (Fig. 5c).
Substrate specificity of LrXynA and kinetic parameters
The substrate specificity of LrXynA was determined under optimal assay conditions, using 1% (w/v) Beechwood xylan, Birchwood xylan, Oat-spelt xylan, CMC, Avicel and Solka floc. LrXynA showed high specificity for all of the xylans assayed, manifesting highest affinity to Beechwood xylan, whereas no activity was detected on CMC, Avicel and Solka floc. To determine the kinetic parameters k M and V max, the initial reaction rates for LrXynA were studied under optimal conditions for enzyme activity using Beechwood xylan as the substrate, at a concentration ranging from 0.05 to 1%. LrXynA exhibited typical kinetics of Michaelis–Menten, with k M and V max values of 2.421 mg/mL and 6.325 U/mg, respectively (Table 3).
Table 3.
Kinetic properties of LrXynA
| k M (mg/mL) | V max (U/mg) | kcat (s−1) | Catalytic efficiency (mL/mg∙s) | |
|---|---|---|---|---|
| Beechwood xylan | 2.421 | 6.325 | 0.403 | 0.166 |
| Birchwood xylan | 5.155 | 8.726 | 0.553 | 0.107 |
| Oat-spelt xylan | 11.66 | 11.924 | 0.756 | 0.065 |
| CMC | 0 | 0 | 0 | 0 |
| Solka floc | 0 | 0 | 0 | 0 |
| Avicel | 0 | 0 | 0 | 0 |
Effect of metal ions on enzyme activity
The effect of several metal ions and EDTA on the enzymatic activity of LrXynA was determined at a final concentration of 1 and 5 mM each (Table 4). Xylanase activity of LrXynA increased 170, 217 and 298% in the presence of the metal ions Ca2+, Mn2+ and Fe2+ (5 mM), respectively. The Mn2+ ion increased the activity of LrXynA to 137 and 217% at a concentration of 1 and 5 mM, respectively; in contrast, the quelant agent EDTA decreased the enzymatic activity of LrXynA by 3 and 16%, at concentrations of 1 and 5 mM, respectively. The activity of LrXynA was almost completely inhibited by the Hg2+ ion at 1 and 5 mM (Table 4).
Table 4.
Influence of metal ions and EDTA on the xylanolytic of LrXynA
| Metal ions and EDTA | Relative xylanase activity (%) | |
|---|---|---|
| 1 mM | 5 mM | |
| Control | 100 | 100 |
| Ca2+ | 95.2 ± 2.6 | 170.5 ± 9.7 |
| Cu2+ | 96.7 ± 8.2 | 72.9 ± 2.4 |
| Fe2+ | 157.7 ± 0.8 | 298.3 ± 4.2 |
| Hg2+ | 7.5 | 7.9 ± 6.0 |
| Li+ | 98.4 ± 6.2 | 121.8 ± 4.4 |
| Mg2+ | 119.4 ± 0.5 | 123.1 ± 11 |
| Mn2+ | 137.3 ± 4.6 | 217.7 ± 4.1 |
| Na+ | 105.9 ± 5.15 | 119.6 ± 6.3 |
| Ni2+ | 103.7 ± 7.9 | 122.5 ± 13 |
| Zn2+ | 132.2 ± 2.5 | 143.1 ± 11.4 |
| EDTA | 97.3 ± 4.9 | 84.6 ± 9.9 |
Analysis of LrXynA hydrolysis products
The mode of action of LrXynA towards Beechwood xylan was examined by analyzing the production of reducing-sugar at different times, and the hydrolysis products by silica gel thin-layer chromatography (TLC) (Fig. 6). The mobility of hydrolysis products after 48 h of incubation showed that the main products were xylotriose and xylobiose (Fig. 6).
Fig. 6.
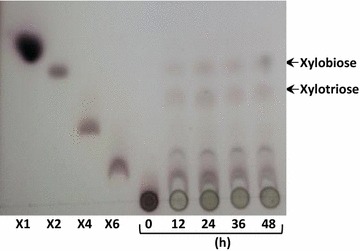
TLC analysis of Beechwood xylan by LrXynA trough kinetic time of 0, 12, 24, 36 and 48 h. Standards: X1 (xylose), X2 (xylobiose), X4 (xylotetrahose) and X6 (xylohexosa)
Enzymatic hydrolysis of sugarcane bagasse (SCB)
The xylanase LrXynA from L. ramosa H71D (named as X) was functionally characterized by its ability to released reduced sugars from SCB alone, or in combination with the commercial cellulase from Aspergillus niger (named as A) or the commercial cellulase from Trichoderma viridae (named as T). The enzymatic hydrolysis of SCB (15 mg/mL) was evaluated by either X, A or T (100%), and with mixtures of X-A or X-T at different molar ratios (25:75, 50:50 and 75:25). In all cases, the reaction mixtures were supplemented with β-glucosidase to avoid inhibition by product, maintaining a final enzyme concentration at 34 mM. In control experiments with all substrates used, β-glucosidase alone did not produce any detectable reducing sugars. The tests were carried out for up to 88 h, however, 64 h was considered the final point because, after this time lapse, the release of reducing sugars ceases to be linear. After 64 h of hydrolysis, the xylanase X released 1.54 ± 0.07 µmol/mL, the cellulase A released 4.98 ± 0.28 µmol/mL and the cellulase T released 4.16 ± 0.99 µmol/mL of reducing sugars. The hydrolysis at different molar ratios (100, 25:75, 50:50 and 75:25) was evaluated for the mixtures of X-A or X-T. The maximum degradation of SCB was detected in the molar ratio of 25X: 75A/T at 64 h. The mix X-T released 4.77 ± 0.38 µmol/mL, whereas X-A released 5.66 ± 0.37 µmol/mL of reducing sugars after 64 h.
Scanning electron microscopy (SEM) analysis
To evidence the impact of the purified xylanase LrXynA (X) on the surface of SCB, as well as the effect of this enzyme in combination with commercial cellulases (A, T), SEM imaging of saccharified SCB was analyzed after 64 h of incubation at 37 °C (Fig. 7). First, to determine the effect of xylanase X on the surface of SCB, the reaction mix 100% X was assayed. Then, this methodology was used to evaluate a putative cooperative effect between xylanase X and a commercial cellulase A or T. For these experiments, reaction mixtures with different molar ratios (from 0 to 100%) were prepared. For all treatments involving a single enzyme, the biomass surface and fibrils became rough and disordered, possibly due to the removal of a polysaccharide (Fig. 7, panels C to H), compared to that observed for SCB without enzymatic treatment (blank), where the biomass surface appears integrated and the fibers look plane, smooth and continuous (Fig. 7, panels a, b). When LrXynA was used in isolation, the fiber showed greater porosity and separation of microfibrils (Fig. 7, panel d). Interestingly, when the SCB was treated with a mix of xylanase/cellulase (25X:75 A or T), the surface was disrupted and looked scaly or cracked (Fig. 7, panels i–l).
Fig. 7.
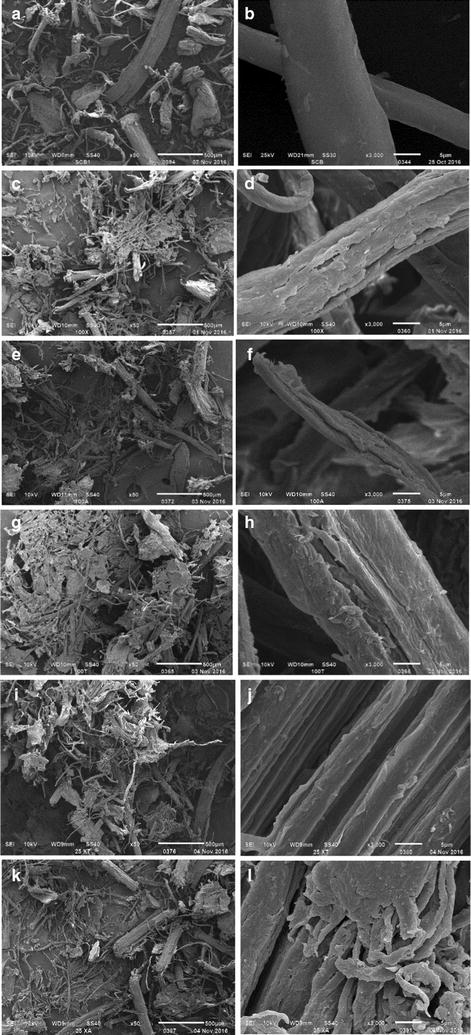
SEM analysis of untreated SCB samples (a, b) and with enzymatic treatment at 37 °C during 64 h of incubation. Using 100% of LrXynA from L. ramosa H71D (c, d); 100% of cellulase from A. niger (e, f); 100% of the cellulase from T. viridae (g, h); a mixture (25:75) of LrXynA/cellulase from A. niger (i, j), and a mixture (25:75) of LrXynA/cellulase from T. viridae (k, l). On the left side the micrographs are shown with an amplification of ×50, and on the right side are shown with an amplification of ×3000
Discussion
Several lignocellulolytic enzymes produced by different microorganisms have been studied. In the context of fungi, Ascomycetes and Basidiomycetes have been widely studied; however, very few studies focus on Zygomycetes and their enzymes. This is why our study focused on the biochemical and functional characterization of one of the enzymes involved in the xylanolytic activity produced by the thermotolerant Zygomycete, L. ramosa H71D.
Firstly, we identified the H71D strain by its morphological characteristics and the molecular marker ITS2 as L. ramosa H71D. Phylogenetically, Mucorales constitute a very old group with considerable molecular distances between species. The weighted intraspecific ITS variability for Zygomycetes is 3.2% (Pawłowska et al. 2013); whereas for Ascomycetes vary by 1.96% (Nilsson et al. 2008). Furthermore, Walther et al. (2013) emphasize the fact that the intraspecific variability in Mucorales differs among species but can reach more than 5%, as occurs in Mucor circinelloides (5.3%) or L. ramosa (7.6%). It has been reported that the ITS2 secondary structure analysis can improve the phylogenetic resolution obtained from the primary sequence (Keller et al. 2008), and the combination and simultaneous analysis of sequence and structural ITS2 RNA data supplemented with indel coding binaries yielded robust phylogenetic hypotheses as measured by bootstrap values for ancestral haplotypes (Poczai et al. 2015). Moreover, Alastruey-Izquierdo et al. (2010) studied species boundaries in Lichtheimia using genealogical concordance phylogenetic species recognition and established that the ITS region is the marker of choice for molecular identification of species in Lichtheimia because of its high degree of variability and the possibility of direct sequencing in most cases. Therefore, due to the intraspecific ITS variability observed for L. ramosa, and with the aim to give more robustness to the identification of the H71D strain, the secondary structure of the ITS2 region from the H71D strain was analyzed and compared to that from L. ramosa GQ342874, because it is one of the sequences with which the H71D strain showed greater identity (99% identity, 100% cover). The ITS2 secondary structures obtained are the same; however, they differ in terms of MFE, − 242.60 kcal/mol (H71D strain) and − 245.30 kcal/mol (L. ramosa GQ342874), due to the three differences in the nucleotide sequence. Hence, phylogenetic analysis based on the ITS2 DNA sequence, and subsequent ITS2 secondary structure analysis allow us to identify the H71D strain as L. ramosa H71D, with a high degree of certainty.
Lichtheimia ramosa H71D was able to grow over a wide range of temperatures (25-45 °C), manifesting optimal growth at 37 °C. A 64 kDa xylanase (named LrXynA) was purified from the culture supernatant of L. ramosa H71D grown on 2% carboxymethylcellulose (CMC), as the only carbon source.
The optimum growth temperature of L. ramosa H71D was determined as 37 °C and the colony reached 3.6 cm after 48 h of incubation. In agreement to our data, an optimum temperature for L. ramosa growth of 35 °C, based on its extensive radial growth (5 cm) after 40 h of incubation, was reported (Gonçalves et al. (2013). According to the optimum growth temperature of L. ramosa H71D (37 °C), this fungus is mesophilic in nature; however, L. ramosa H71D can be considered a thermotolerant fungus because it is able to grow over a wide range of temperature (25–45 °C). Findings here concur with previous reports, e.g., in a study on Mucorales it was observed that thermotolerant species had optimum growth temperatures above 37 °C, between 37 and 45 °C (Hoffmann et al. 2007); in particular, it was reported that L. ramosa grew at a temperature that ranged from 24 to 49 °C (Alastruey-Izquierdo et al. 2010).
The xylanolytic activity produced by L. ramosa H71D in the presence of Beechwood xylan or CMC was assessed, indicating that xylanase activity was greater when the fungus was cultured on CMC compared to than that observed for Beechwood xylan as carbon source; thus, indicating that CMC is more effective for the production of xylanase activity by L. ramosa H71D, under the culture conditions tested. Similarly, it was reported that cellulose, cellobiose, and even heterodisaccharide, composed of glucose and xylose, induce the production of xylanolytic enzymes in Aspergillus terreus (Hrmová et al. 1991). The fungus T. reesei also exhibits cellulolytic and xylanolytic activity in the presence of cellulose, xylan, or mixtures of plant polymers (Amore et al. 2013). Interestingly, and in agreement to findings here, when Neurospora crassa was cultured on Avicel as the sole carbon source, both cellulases and hemicellulases encoding genes were induced, and the expression levels of some hemicellulases genes were much higher than those observed when N. crassa was cultured on xylan (Sun et al. 2012; Amore et al. 2013). The production of xylanases by fungi grown on cellulose as the only carbon source has been reported in fungi as Hypocrea jecorina (Stricker et al. 2008) and Trichoderma harzianum (Hrmová et al. 1989). Xylanase production by L. ramosa H71D (2.1 U/mL) is comparable to that reported for L. ramosa (1.80 U/mL) via solid state bioprocess, utilizing waste from Brazilian savannah fruit (de Silva et al. 2013); and for L. ramosa (2.54 U/mL) grown in wheat bran-based medium (Gonçalves et al. 2013).
In this study, we have purified, characterized, and quantitatively evaluated the activity of a xylanase from L. ramosa H71D. Zymogram analysis of LrXynA using 10% SDS-PAGE revealed a band with an estimated MW of approximately 64 kDa, with xylanolytic activity. Most xylanases produced by bacteria and fungi are proteins pertaining to a subunit with a wide molecular weight range of 8–145 kDa (Beg et al. 2001). LrXynA is not a glycoprotein; however, it has been estimated that over half the proteins in nature are glycosylated (Apweiler et al. 1999). Reports indicate that in the case of xylanases, carbohydrate decoration on β-xylosidases contributes 10–30% of their molecular weight. Exceptionally, fungal β-xylosidases from Humicola grisea var. thermoidea and Paecilomyces thermophila are not glycosylated (Knob and Carmona 2010) and four xylanases (xyn10A, xyn10B, xyn11A, xy11B) from Penicillium oxalicum GZ-2 are not glycosylated (Liao et al. 2015).
We compared certain biochemical characteristics of LrXynA with those of other fungal xylanases. The optimal pH assay showed that the enzyme had maximal activity at 6, a value which falls within the range (2–8) of optimal pH values for several fungal xylanases (Beg et al. 2001). Xylanases obtained from different microorganisms with optimal function at pH 6 have been reported, such as those from P. oxalicum GZ-2 (Liao et al. 2015), Humicola insolens Y1 (Shi et al. 2015), Remersonia thermophila CBS 540.69 (McPhillips et al. 2014). The pH stability data of LrXynA (4.5–7) was similar to other isolated xylanases, e.g., the xylanase from R. miehei retained more than 90% of its activity at pH values of 5 and 6.5 after 60 min at 50 °C (Fawzi 2011); whereas, the xylanase from Thielaviopsis basicola exhibited alkaline stabilities ranging from pH 3–11 (Goluguri et al. 2012), and the xylanase from Chaetomium sp. retained more than 80% of its activity after 30 min at 50 °C, when tested at the pH range from 4.5 to 11 (Jiang et al. 2010). The optimal temperature for LrXynA was 65 °C, xylanases from other microorganisms exhibited an optimal activity at 65 °C, for example, that from Paenibacillus sp. DG-22 (Lee and Lee 2014) and Remersonia thermophila CBS 540.69 (McPhillips et al. 2014). This value falls within the range of optimal temperature values of several fungal xylanases. For example, 40 °C is the optimal temperature for the xylanase from Leucoagaricus gongylophorus (Moreira et al. 2014). Recently, our research group reported an optimal temperature of 85 °C for the xylanase TtXynA from the thermophilic fungus Thielavia terrestris Co3Bag1, which at that moment was the highest optimal temperature for fungal xylanases (García-Huante et al. 2017). Nevertheless, there are reports of non-fungal xylanases with optimal temperatures higher than 85 °C, such as SSO1354 from Sulfolobus solfataricus at 95 °C (Maurelli et al. 2008) and XYNB from Dictyoglomus thermophilum at 100 °C (Li et al. 2013). Thermostability assays indicated that LrXynA displays low thermostability at 75 °C; however, the enzyme exhibited half-lifes of 25 and 80 min at 65 and 55 °C, respectively. At 65 °C, the thermostability of LrXynA (t½ = 25 min) is lower than that displayed by the xylanase TtXynA (t½ = 23 days) from T. terrestris Co3Bag1 (García-Huante et al. 2017) but higher than that reported for the xylanase XynAS9 (t½ = 16 min) from Streptomyces (Wang et al. 2011).
It has been reported that over 90% of Beechwood and Birchwood xylan are composed of xylose. In Beechwood xylan, xyloses are mainly linked by 2,4 and 1,4-linkages; in Birchwood xylan, xyloses are mainly linked by 1,4-linkages, whereas most of the Oat-spelt xylan contains xylose and arabinose with minor amounts of glucose and galactose (Liab et al. 2000). Hence, our results suggest that LrXynA has higher affinity towards 1,4-linkages between xyloses, present in Beechwood and Birchwood xylans, but when the amount of xylose decreases, as in Oat-spelt xylan, its affinity also decreases. Other reported xylanases show higher affinity for Beechwood xylan than for Birchwood xylan, e.g., XynGR40 (k M = 1.8 mg/mL) from the environmental DNA of goat rumen (Wang et al. 2011) and TtXynA (k M = 0.41 mg/mL) from T. terrestris Co3Bag1 (García-Huante et al. 2017). However, there are reports of other xylanases that show greater affinity for Oat-spelt xylan than for Beechwood xylan (Liao et al. 2015). LrXynA did not show activity on CMC, Solka floc, and Avicel, and reports describe some other xylanases such as xyn10A, xyn10B, xyn11A, xy11B) from P. oxalicum GZ-2 to be active towards polymeric xylans; although not on other substrates (Liao et al. 2015). The kinetic parameters of LrXynA and the k M and V max values, determined for each of the xylan substrates used in this study, are similar to those obtained for other fungal xylanases. The LrXynA k M value of 2.42 mg/mL is similar to those reported for rXynSW3 xylanase from Streptomyces sp. SWU10 of k M = 2.3 mg/mL (Sukhumsirichart et al. 2014) and xylanase XYN2 from T. reesei of k M = 2.1 mg/mL (He et al. 2009). Likewise, xylanases with k M values higher than that observed for LrXynA have been reported, e.g., endo-1,4-beta xylanase B (k M = 8.9 mg/mL) from Aspergillus niger BCC14405 (Krisana et al. 2005) and Xyn II (k M = 5.56 mg/mL) from Aspergillus usamii (Zhou et al. 2008). The V max value (6.325 U/mg) of LrXynA is higher than the V max value reported for rXynSW3 (0.35 U/mg) of Streptomyces sp. (Sukhumsirichart et al. 2014), but it is lower than those reported for other xylanases, e.g., xylanase (V max = 1235 U/mg) of Talaromyces thermophilus (Maalej et al. 2009) and xylanase (V max = 113.5 U/mg) of R. miehei (Fawzi 2011).
The general consensus opines that some metal ions and reagents significantly affect xylanase activities (Juturu and Wu 2012). Therefore, we evaluated the effect of metal ions and EDTA on the xylanolytic activity of LrXynA. The Fe2+ ion 5 mM is presented as the major activator for increasing the activity of the xylanase LrXynA from L. ramosa by 298%. However, it has been reported that Fe2+ 1 mM inhibits the activity of the XYN11A from P. oxalicum by 68% (Liao et al. 2014). The Mn2+ ion increased the activity of LrXynA by 137 and 217% at a concentration of 1 and 5 mM, respectively. The activity of a xylanase from T. lanuginosus DSM 5826 was also stimulated by 137% (Lin et al. 1999), whereas a 40% decrease was observed for a xylanase from Streptomyces rameus (Li et al. 2010), in the presence of the metal ion Mn2+. The metal ion Hg2+ is known to be toxic to enzymes, as it binds to thiol groups present in the active sites of the enzyme, causing irreversible inactivation. This ion Hg2+ (1 and 5 mM) decreased the xylanolytic activity of LrXynA by 93%. It also inhibits 3 out of 4 xylanases (xyn10A, xyn10B, xyn11B) from P. oxalicum (Liao et al. 2015). Other reports state that the Hg2+ ion did not completely inhibit xylanase activity e.g., the xylanase xyn11A (24%, at 10 mM) from P. oxalicum (Liao et al. 2015), and the xylanase TtXynA (55%, at 1 mM) from T. terrestris (García-Huante et al. 2017). We also assessed EDTA, a metal chelator that may decrease xylanase activity. This would suggest that the enzyme needs a metal as a cofactor (Knob and Carmona 2010). The chelating agent EDTA, at final concentrations of 1 and 5 mM, decreased the activity of LrXynA by 3 and 16%, respectively. A decrease of 9% in the activity of a xylanase from L. sulphureus in the presence of EDTA 5 mM was reported by (Lee et al. 2009).
TLC analysis of final products from hydrolysis of Beechwood xylan by LrXynA were mainly xylotriose and xylobiose; therefore, LrXynA can be classified as an endo-xylanase without β-xylosidase activity, as xylose was not observed as a product even after 48 h of incubation. According to Knob and Carmona (2010) xylotriose is the smallest oligomer produced by most known xylanases. Nevertheless, other xylanases from fungi, such as L. sulphureus (Lee et al. 2009) and P. oxalicum (Liao et al. 2014) hydrolyze xylans to predominantly produce xylobiose and xylotriose.
The individual action of xylanase LrXynA (X), a commercial cellulase from Aspergillus niger (A), and a commercial cellulase from Trichoderma viridae (T) in the hydrolysis of SCB, as well as the effect of LrXynA in combination with a commercial cellulase (A or T), was quantified by the liberation of reducing sugars during the hydrolysis of SCB. Data obtained indicated that a positive effect in the hydrolysis of SCB occurred mainly in both of the mixtures studied; X-T and X-A. To reveal the impact of xylanase LrXynA, alone and in combination with commercial cellulases A or T, on the surface of SCB, samples of this substrate before and after enzymatic treatments were analyzed by SEM imaging. The micrographs showed the damage carried out by each of the enzymes separately; nevertheless, the damage was more noticeable when the enzymes were mixed (X-T and X-A). In this case, when xylanase LrXynA (X) was used in isolation, the fiber showed greater porosity and separation of microfibrils; a similar effect was observed with the xylanase QG-11-3 from Streptomyces sp. on eucalyptus kraft pulp (Beg et al. 2000). Furthermore, when SCB was treated with the cellulases A or T, the layers of fiber appeared to break, exposing the inner channels of the fiber. Our findings concur with those that describe the action of CELULASE CE 2 from Trichoderma longbrachiatum (Proenzimas, Cali, Colombia) on SCB (Quintero and Cardona 2009). Hence, a clear positive effect on the hydrolysis of SCB was observed when LrXynA was combined with any of the commercial cellulase preparations. This is probably due to the removal of hemicellulose by the action of LrXynA, which leads to an improved enzymatic hydrolysis of cellulose fibers by the action of the commercial cellulases. A synergic effect that occurs between two individual non-complexed pure enzymes of two aerobes: CflXyn11A xylanase from Cellulomomas flavigena and TrCel7B cellulase from Trichoderma reesei, during the hydrolysis of sugarcane bagasse has been previously reported by our research group (Pavón-Orozco et al. 2012). Overall, data obtained in this work suggest that LrXynA may represent an efficacious candidate for the degradation of plant cell biomass.
On the basis of morphological characteristics, the H71D strain was identified as L. ramosa (sporangia were light gray colored, subsporangial septum was absent, and sporangiospores were ellipsoidal). Phylogenetic analysis was based on the molecular marker ITS and its secondary structure. The study of new strains, especially those exhibiting fast-growing, as well as the biochemical characterization of its enzymes is important not only from an evolutionary point of view but also from an economic one, as demand has increased in the agricultural context with the production of biofuels from agricultural wastes.
There are few reports on glycosyl hydrolases produced by members of the Lichtheimia genus. To our knowledge, this study represents the first report on biochemical and functional characterization of a xylanase from a filamentous fungus belonging to the Zygomycete genus Lichtheimia. The xylanase LrXynA from L. ramosa H71D is considered heat-tolerant and heat stable; biochemical properties that make it appropriate for application in different biotechnological processes such as the biofuel industry, the manufacture of bread and animal feed and in the treatment of lignocellulosic residues. The bioconversion of hemicellulose to value-added products using xylanases thus offers many promising applications.
Authors’ contributions
MTAZ conducted the study and collected the data, created figures. MTAZ, JEC, and MEHL designed the study, analyzed and interpreted the data, and wrote the manuscript. Co-authors helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We kindly appreciate the service of Microscopía Electrónica, Laboratorio Nacional de Servicios Experimentales, CINVESTAV-IPN, and the DNA sequencing service at UBIPRO FES-Iztacala, UNAM. México.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All datasets were presented in the main paper.
Consent for participation
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
The project was funded by Departamento de Biotecnología y Bioingeniería, CINVESTAV-IPN. M.T. A-Z was the recipient of PhD. Scholarship (243329) from Consejo Nacional de Ciencia y Tecnología, México.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ITS
internal transcribed spacer
- LrXynA
xylanase from L. ramosa
- t½
enzyme half-life
- CMC
carboxymethylcellulose
- CMCase
carboxymethylcellulase
- SCB
sugarcane bagasse
- PDA
potato dextrose agar
- MFE
minimum free energy
- SDS–PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- MW
molecular weight
- RBB-X
Remazol Brilliant Blue linked to xylan
- TLC
thin layer chromatography
- X
xylanase LrXynA from L. ramosa
- A
cellulase from A. niger
- T
cellulase from T. viridae
- SEM
scanning electron microscopy
Contributor Information
María Teresa Alvarez-Zúñiga, Email: mtalvarez@cinvestav.mx.
Alejandro Santiago-Hernández, Email: jsantiago@cinvestav.mx.
Johan Rodríguez-Mendoza, Email: rodriguezmj@cinvestav.mx.
Jorge E. Campos, Email: jcampos@unam.mx
Patricia Pavón-Orozco, Email: optap@hotmail.com.
Sergio Trejo-Estrada, Email: sertre@hotmail.com.
María Eugenia Hidalgo-Lara, Email: ehidalgo@cinvestav.mx.
References
- Alastruey-Izquierdo A, Hoffmann K, De Hoog GS, Rodriguez-Tudela JL, Voigt K, Bibashi E, Walther G. Species recognition and clinical relevance of the zygomycetous genus Lichtheimia (syn. Absidia pro parte, Mycocladus) J Clin Microbiol. 2010;48:2154–2170. doi: 10.1128/JCM.01744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amore A, Giacobbe S, Faraco V. Regulation of cellulase and hemicellulase gene expression in fungi. Curr Genomics. 2013;14:230–249. doi: 10.2174/1389202911314040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André ALCM, Hoffmann K, Lima DX, de Oliveira RJV, Vieira HEE, Malosso E, Maia LC, da Silva GA. A new species of Lichtheimia (Mucoromycotina, Mucorales) isolated from Brazilian soil. Mycol Prog. 2014;13:343–352. doi: 10.1007/s11557-013-0920-8. [DOI] [Google Scholar]
- Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta Gen Subj. 1999;1473:4–8. doi: 10.1016/S0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- Beg QK, Bhushan B, Kapoor M, Hoondal GS. Enhanced production of a thermostable xylanase from Streptomyces sp. QG-11-3 and its application in biobleaching of eucalyptus kraft pulp. Enzyme Microb Technol. 2000;27:459–466. doi: 10.1016/S0141-0229(00)00231-3. [DOI] [PubMed] [Google Scholar]
- Beg QK, Kapoor M, Mahajan L, Hoondal GS. Microbial xylanases and their industrial applications: a review. Appl Microbiol Biotechnol. 2001;56:326–338. doi: 10.1007/s002530100704. [DOI] [PubMed] [Google Scholar]
- Caisová L, Marin B, Melkonian M. A close-up view on ITS2 evolution and speciation—a case study in the Ulvophyceae (Chlorophyta, Viridiplantae) BMC Evol Biol. 2011;11:262. doi: 10.1186/1471-2148-11-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman AW. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet. 2003;19:370–375. doi: 10.1016/S0168-9525(03)00118-5. [DOI] [PubMed] [Google Scholar]
- Collins T, Meuwis MA, Stals I, Claeyssens M, Feller G, Gerday C. A novel family 8 xylanase, functional and physicochemical characterization. J Biol Chem. 2002;277:35133–35139. doi: 10.1074/jbc.M204517200. [DOI] [PubMed] [Google Scholar]
- de Silva CA, Lacerda MPF, Leite RSR, Fonseca GG. Production of enzymes from Lichtheimia ramosa using Brazilian savannah fruit wastes as substrate on solid state bioprocessess. Electron J Biotechnol. 2013 [Google Scholar]
- Fawzi E. Highly thermostable xylanase purified from Rhizomucor miehei NRL 3169. Acta Biol Hung. 2011;62:85–94. doi: 10.1556/ABiol.61.2011.1.9. [DOI] [PubMed] [Google Scholar]
- Ferreira JA, Lennartsson PR, Edebo L, Taherzadeh MJ. Zygomycetes-based biorefinery: present status and future prospects. Bioresour Technol. 2013;135:523–532. doi: 10.1016/j.biortech.2012.09.064. [DOI] [PubMed] [Google Scholar]
- García-Huante Y, Cayetano-Cruz M, Santiago-Hernández A, Cano-Ramírez C, Marsch-Moreno R, Campos JE, Aguilar-Osorio G, Benítez-Cardoza CG, Trejo-Estrada S, Hidalgo-Lara ME. The thermophilic biomass-degrading fungus Thielavia terrestris Co3Bag1 produces a hyperthermophilic and thermostable β-1,4-xylanase with exo- and endo-activity. Extremophiles. 2017;21:175–186. doi: 10.1007/s00792-016-0893-z. [DOI] [PubMed] [Google Scholar]
- Goluguri BR, Thulluri C, Cherupally M, Nidadavolu N, Achuthananda D, Mangamuri LN, Addepally U. Potential of thermo and alkali stable xylanases from Thielaviopsis Basicola (MTCC-1467) in biobleaching of wood kraft pulp. Appl Biochem Biotechnol. 2012;167:2369–2380. doi: 10.1007/s12010-012-9765-x. [DOI] [PubMed] [Google Scholar]
- Gonçalves FA, Leite RSR, Rodrigues A, Argandoña EJS, Fonseca GG. Isolation, identification and characterization of a novel high level β-glucosidase-producing Lichtheimia ramosa strain. Biocatal Agric Biotechnol. 2013;2:377–384. [Google Scholar]
- Hatanaka K. Incorporation of fluorous glycosides to cell membrane and saccharide chain elongation by cellular enzymes. Top Curr Chem. 2012;308:294–306. doi: 10.1007/128_2011_276. [DOI] [PubMed] [Google Scholar]
- He J, Yu B, Zhang K, Ding X, Chen D. Expression of endo-1, 4-beta-xylanase from Trichoderma reesei in Pichia pastoris and functional characterization of the produced enzyme. BMC Biotechnol. 2009;9:56. doi: 10.1186/1472-6750-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann K. Identification of the genus Absidia (Mucorales, Zygomycetes): a comprehensive taxonomic revision. In: Gherbawy Y, Voigt K, editors. Molecular identification of fungi. Berlin: Springer; 2010. pp. 439–460. [Google Scholar]
- Hoffmann K, Discher S, Voigt K. Revision of the genus Absidia (Mucorales, Zygomycetes) based on physiological, phylogenetic, and morphological characters; thermotolerant Absidia spp. form a coherent group, Mycocladiaceae fam. nov. Mycol Res. 2007;111:1169–1183. doi: 10.1016/j.mycres.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Hrmová M, Biely P, Vršanská M. Cellulose-and xylan-degrading enzymes of Aspergillus terreus and Aspergillus niger. Enzyme Microb Technol. 1989;11:610–616. doi: 10.1016/0141-0229(89)90090-2. [DOI] [Google Scholar]
- Hrmová M, Petraková E, Biely P. Induction of cellulose-and xylan-degrading enzyme systems in Aspergillus terreus by homoand heterodisaccharides composed of glucose and xylose. J Gen Microbiol. 1991;137:541–547. doi: 10.1099/00221287-137-3-541. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Cong Q, Yan Q, Kumar N, Du X. Characterisation of a thermostable xylanase from Chaetomium sp. and its application in Chinese steamed bread. Food Chem. 2010;120:457–462. doi: 10.1016/j.foodchem.2009.10.038. [DOI] [Google Scholar]
- Juturu V, Wu JC. Microbial xylanases: engineering, production and industrial applications. Biotechnol Adv. 2012;30:1219–1227. doi: 10.1016/j.biotechadv.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Keller A, Schleicher T, Förster F, Ruderisch B, Dandekar T, Müller T, Wolf M. ITS2 data corroborate a monophyletic chlorophycean DO-group (Sphaeropleales) BMC Evol Biol. 2008;8:218. doi: 10.1186/1471-2148-8-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knob A, Carmona EC. Purification and characterization of two extracellular xylanases from Penicillium sclerotiorum: a novel acidophilic xylanase. Appl Biochem Biotechnol. 2010;162:429–443. doi: 10.1007/s12010-009-8731-8. [DOI] [PubMed] [Google Scholar]
- Krisana A, Rutchadaporn S, Jarupan G, Lily E, Sutipa T, Kanyawim K. Endo-1,4-β-xylanase B from Aspergillus cf. niger BCC14405 Isolated in Thailand: purification, characterization and gene isolation. J Biochem Mol Biol. 2005;38:17–23. doi: 10.5483/bmbrep.2005.38.1.017. [DOI] [PubMed] [Google Scholar]
- Kulkarni N, Shendye A, Rao M. Molecular and biotechnological aspects of xylanases. FEMS Microbiol Rev. 1999;23:411–456. doi: 10.1111/j.1574-6976.1999.tb00407.x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee SH, Lee YE. Cloning and characterization of a multidomain GH10 xylanase from Paenibacillus sp. DG-22. J Microbiol Biotechnol. 2014;24:1525–1535. doi: 10.4014/jmb.1407.07077. [DOI] [PubMed] [Google Scholar]
- Lee JW, Park JY, Kwon M, Choi IG. Purification and characterization of a thermostable xylanase from the brown-rot fungus Laetiporus sulphureus. J Biosci Bioeng. 2009;107:33–37. doi: 10.1016/j.jbiosc.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Leyva A, Quintana A, Sánchez M, Rodríguez EN, Cremata J, Sánchez JC. Rapid and sensitive anthrone-sulfuric acid assay in microplate format to quantify carbohydrate in biopharmaceutical products: method development and validation. Biologicals. 2008;36:134–141. doi: 10.1016/j.biologicals.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Li X, She Y, Sun B, Song H, Zhu Y, Lv Y, Song H. Purification and characterization of a cellulase-free, thermostable xylanase from Streptomyces rameus L2001 and its biobleaching effect on wheat straw pulp. Biochem Eng J. 2010;52:71–78. doi: 10.1016/j.bej.2010.07.006. [DOI] [Google Scholar]
- Li H, Kankaanpää A, Xiong H, Hummel M, Sixta H, Ojamo H, Turunen O. Thermostabilization of extremophilic Dictyoglomus thermophilum GH11 xylanase by an N-terminal disulfide bridge and the effect of ionic liquid [emim]OAc on the enzymatic performance. Enzyme Microb Technol. 2013;53:414–419. doi: 10.1016/j.enzmictec.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Liab K, Azadi P, Collins R, Tolan J, Kim JS, Eriksson KEL. Relationships between activities of xylanases and xylan structures. Enzyme Microb Technol. 2000;27:89–94. doi: 10.1016/S0141-0229(00)00190-3. [DOI] [PubMed] [Google Scholar]
- Liao H, Sun S, Wang P, Bi W, Tan S, Wei Z, Mei X, Liu D, Raza W, Shen Q, Xu Y. A new acidophilic endo-β-1,4-xylanase from Penicillium oxalicum: cloning, purification, and insights into the influence of metal ions on xylanase activity. J Ind Microbiol Biotechnol. 2014;41:1071–1083. doi: 10.1007/s10295-014-1453-0. [DOI] [PubMed] [Google Scholar]
- Liao H, Zheng H, Li S, Wei Z, Mei X, Ma H, Shen Q, Xu Y. Functional diversity and properties of multiple xylanases from Penicillium oxalicum GZ-2. Sci Rep. 2015;5:12631. doi: 10.1038/srep12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Ndlovu LM, Singh S, Pillay B. Purification and biochemical characteristics of β-d-xylanase from a thermophilic fungus, Thermomyces lanuginosus-SSBP. Biotechnol Appl Biochem. 1999;30(Pt 1):73–79. [PubMed] [Google Scholar]
- Maalej I, Belhaj I, Masmoudi NF, Belghith H. Highly thermostable xylanase of the thermophilic fungus Talaromyces thermophilus: purification and characterization. Appl Biochem Biotechnol. 2009;158:200–212. doi: 10.1007/s12010-008-8317-x. [DOI] [PubMed] [Google Scholar]
- Mandels M, Sternberg D. Recent advances in cellular technology. J Ferment Technol. 1967;54:267–286. [Google Scholar]
- Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurelli L, Giovane A, Esposito A, Moracci M, Fiume I, Rossi M, Morana A. Evidence that the xylanase activity from Sulfolobus solfataricus Oα is encoded by the endoglucanase precursor gene (sso1354) and characterization of the associated cellulase activity. Extremophiles. 2008;12:689–700. doi: 10.1007/s00792-008-0175-5. [DOI] [PubMed] [Google Scholar]
- McPhillips K, Waters DM, Parlet C, Walsh DJ, Arendt EK, Murray PG. Purification and characterisation of a β-1,4-xylanase from Remersonia thermophila CBS 540.69 and its application in bread making. Appl Biochem Biotechnol. 2014;172:1747–1762. doi: 10.1007/s12010-013-0640-1. [DOI] [PubMed] [Google Scholar]
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- Moreira AC, Ferreira D, De Almeida FG, Rodrigues-Filho E, Fernandes JB, Silva MFGF, Vieira PC, Pagnocca FC, Souza DHF. Molecular and kinetic characterization of two extracellular xylanases isolated from Leucoagaricus gongylophorus. Appl Biochem Biotechnol. 2014;173:694–704. doi: 10.1007/s12010-014-0872-8. [DOI] [PubMed] [Google Scholar]
- Neves MLC, Da Silva MF, Souza-Motta CM, Spier MR, Soccol CR, Porto TS, Moreira KA, Porto ALF. Lichtheimia blakesleeana as a new potencial producer of phytase and xylanase. Molecules. 2011;16:4807–4817. doi: 10.3390/molecules16064807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson KH. Intraspecific ITS variability in the Kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol Bioinforma. 2008;2008:193–201. doi: 10.4137/ebo.s653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Lutzoni FM, Ward TJ, Benny GL. Evolutionary relationships among mucoralean fungi (Zygomycota): evidence for family polyphyly on a large scale. Mycologia. 2001;93:286–296. doi: 10.2307/3761650. [DOI] [Google Scholar]
- Pavón-Orozco P, Santiago-Hernández A, Rosengren A, Hidalgo-Lara ME, Stålbrand H. The family II carbohydrate-binding module of xylanase CflXyn11A from Cellulomonas flavigena increases the synergy with cellulase TrCel7B from Trichoderma reesei during the hydrolysis of sugar cane bagasse. Bioresour Technol. 2012;104:622–630. doi: 10.1016/j.biortech.2011.11.068. [DOI] [PubMed] [Google Scholar]
- Pawłowska J, Walther G, Wilk M, De Hoog S, Wrzosek M. The use of compensatory base change analysis of ITS2 as a tool in the phylogeny of Mucorales, illustrated by the Mucor circinelloides complex. Org Divers Evol. 2013;13:497–502. doi: 10.1007/s13127-013-0139-1. [DOI] [Google Scholar]
- Pellerin P, Gosselin M, Lepoutre JP, Samain E, Debeire P. Enzymic production of oligosaccharides from corncob xylan. Enzyme Microb Technol. 1991;13:617–621. doi: 10.1016/0141-0229(91)90074-K. [DOI] [Google Scholar]
- Poczai P, Varga I, Hyvönen J. Internal transcribed spacer (ITS) evolution in populations of the hyperparasitic European mistletoe pathogen fungus, Sphaeropsis visci (Botryosphaeriaceae): the utility of ITS2 secondary structures. Gene. 2015;558:54–64. doi: 10.1016/j.gene.2014.12.042. [DOI] [PubMed] [Google Scholar]
- Polizeli ML, Riazzatti AC, Monti R, Terenzi HF, Jorge JA, Amorim DS. Xylanases from fungi: properties and industrial applications. Appl Microbiol Biotechnol. 2005;67:577–591. doi: 10.1007/s00253-005-1904-7. [DOI] [PubMed] [Google Scholar]
- Quintero JA, Cardona CA. Ethanol dehydration by adsorption with starchy and cellulosic materials. Ind Eng Chem Res. 2009;48:6783–6788. doi: 10.1021/ie8015736. [DOI] [Google Scholar]
- Royer JC, Nakas JP. Simple, sensitive zymogram technique for detection of xylanase activity in polyacrylamide gels. Appl Environ Microbiol. 1990;56:1516–1517. doi: 10.1128/aem.56.6.1516-1517.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Maisel S, Gerlach D, Müller T, Wolf M. A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA. 2005;11:361–364. doi: 10.1261/rna.7204505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P, Du Y, Yang H, Huang H, Zhang X, Wang Y, Yao B. Molecular characterization of a new alkaline-tolerant xylanase from Humicola insolens Y1. Biomed Res Int. 2015 doi: 10.1155/2015/149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Madlala AM, Prior BA. Thermomyces lanuginosus: properties of strains and their hemicellulases. FEMS Microbiol Rev. 2003;27:3–16. doi: 10.1016/S0168-6445(03)00018-4. [DOI] [PubMed] [Google Scholar]
- Stricker AR, Mach RL, De Graaff LH. Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei) Appl Microbiol Biotechnol. 2008;78:211–220. doi: 10.1007/s00253-007-1322-0. [DOI] [PubMed] [Google Scholar]
- Sukhumsirichart W, Deesukon W, Kawakami T, Matsumoto S, Seesom W, Sakamoto T. Expression and characterization of recombinant GH11 xylanase from thermotolerant Streptomyces sp. SWU10. Appl Biochem Biotechnol. 2014;172:436–446. doi: 10.1007/s12010-013-0508-4. [DOI] [PubMed] [Google Scholar]
- Sun J, Tian C, Diamond S, Glass NL. Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in Neurospora crassa. Eukaryot Cell. 2012 doi: 10.1128/EC.05327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunna A, Antranikian G. Xylanolytic Enzymes from Fungi and Bacteria. Crit Rev Biotechnol. 1997;17:39–67. doi: 10.3109/07388559709146606. [DOI] [PubMed] [Google Scholar]
- Tien M, Kirk TK. Lignin peroxidase of Phanerochaete chrysosporium. Methods Enzymol. 1988;161:238–249. doi: 10.1016/0076-6879(88)61025-1. [DOI] [Google Scholar]
- Viikari L, Alapuranen M, Puranen T, Vehmaanperä J, Siika-Aho M. Thermostable enzymes in lignocellulose hydrolysis. Adv Biochem Eng Biotechnol. 2007;108:121–145. doi: 10.1007/10_2007_065. [DOI] [PubMed] [Google Scholar]
- Voigt K, Cigelnik E, Kerry O, Donnell KO. Phylogeny and PCR identification of clinically important Zygomycetes based on nuclear ribosomal-dna sequence data. J Clin Microbiol. 1999;37:3957–3964. doi: 10.1128/jcm.37.12.3957-3964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther G, Pawłowska J, Alastruey-Izquierdo A, Wrzosek M, Rodriguez-Tudela JL, Dolatabadi S, Chakrabarti A, de Hoog GS. DNA barcoding in Mucorales: an inventory of biodiversity. Persoonia Mol Phylogeny Evol Fungi. 2013;30:11–47. doi: 10.3767/003158513X665070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Luo H, Wang Y, Huang H, Shi P, Yang P, Meng K, Bai Y, Yao B. A novel cold-active xylanase gene from the environmental DNA of goat rumen contents: direct cloning, expression and enzyme characterization. Bioresour Technol. 2011;102:3330–3336. doi: 10.1016/j.biortech.2010.11.004. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns S, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc A Guid Methods Appl. 1990;18:315–322. [Google Scholar]
- Wolf M, Achtziger M, Schultz J, Dandekar T, Müller T. Homology modeling revealed more than 20,000 rRNA internal transcribed spacer 2 (ITS2) secondary structures. RNA. 2005;11:1616–1623. doi: 10.1261/rna.2144205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Song J, Liu C, Luo K, Han J, Li Y, Pang X, Xu H, Zhu Y, Xiao P, Chen S. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS ONE. 2010 doi: 10.1371/journal.pone.0013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Siika-aho M, Puranen T, Tang M, Tenkanen M, Viikari L. Thermostable recombinant xylanases from Nonomuraea flexuosa and Thermoascus aurantiacus show distinct properties in the hydrolysis of xylans and pretreated wheat straw. Biotechnol Biofuels. 2011;4:12. doi: 10.1186/1754-6834-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Bai J, Deng S, Wang J, Zhu J, Wu M, Wang W. Cloning of a xylanase gene from Aspergillus usamii and its expression in Escherichia coli. Bioresour Technol. 2008;99:831–838. doi: 10.1016/j.biortech.2007.01.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets were presented in the main paper.


