Abstract
Mitogen-activated protein kinase kinase kinases (MAPKKKs), an important unit of MAPK cascade, play crucial roles in plant development and response to various stresses. However, little is known concerning the MAPKKK family in the important subtropical and tropical crop cassava. In this study, 62 MAPKKK genes were identified in the cassava genome, and were classified into 3 subfamilies based on phylogenetic analysis. Most of MAPKKKs in the same subfamily shared similar gene structures and conserved motifs. The comprehensive transcriptome analysis showed that MAPKKK genes participated in tissue development and response to drought stress. Comparative expression profiles revealed that many MAPKKK genes were activated in cultivated varieties SC124 and Arg7 and the function of MeMAPKKKs in drought resistance may be different between SC124/Arg7 and W14. Expression analyses of the 7 selected MeMAPKKK genes showed that most of them were significantly upregulated by osmotic, salt and ABA treatments, whereas slightly induced by H2O2 and cold stresses. Taken together, this study identified candidate MeMAPKKK genes for genetic improvement of abiotic stress resistance and provided new insights into MAPKKK -mediated cassava resistance to drought stress.
Introduction
To survival, plants have elaborated signaling network involved in sensing and transmitting signals. Among the stress-activated molecular pathways, mitogen-activated protein kinases (MAPKs) signaling cascade plays central roles in multiple biological processes1,2. The basic MAPK cascade is consist of three classes of protein kinases, including MAPK kinase kinase (MAPKKK), MAPK kinase (MAPKK) and MAPK, which are linked to upstream and downstream regulators by phosphorylation3.
In recent years, studies have suggested that MAPK cascade is involved in various biological processes in plants, including cell differentiation and growth4,5, plant ripening, hormone signaling6–8, plant immunity1,9, and respondent to the multiplicity of biotic and abiotic stress10–13. Arabidopsis MEKK1-MKK4/5-MPK3/6 cascade participates in innate immunity, which is the first identified MAPK signal pathway in plants1,14. Subsequently, a series of MAPK signal pathways are defined. Tobacco NPK1-MEK1-Ntf6 plays an important role in resistance to tobacco mosaic virus15. The MEKK1-MKK2-MPK4 cascade takes part in cold stress response16. Arabidopsis ANP3-MKK6-MPK4 and YDA-MKK4/5-MPK3/6 are involved in the regulation of cytokinesis and stomatal development, respectively17,18. Besides, some genes related to MAPK cascade pathway are identified to play a vital role in environmental stress responses. For example, MAP3Kd4 plays an important role in increasing salt stress resistance and ABA response by over-expression analysis11. Mutation of raf43–1 results in increased sensitivity to mannitol, NaCl and H2O2 19. Another Arabidopsis Raf-like MAPKKK gene, HT1, positively regulates response to CO2 20. A cotton Raf-like MAPKKK gene, GhRaf19, positively regulates resistance to cold stress, but negatively regulates resistance to drought and salt stresses using virus-induced gene silencing21. Overexpression of tobacco NPK1, belonging to the MAPKKK family, enhances maize resistance to drought22. Together, the above studies support that MAPK cascade pathway is a crucial regulator of plant development and stress response.
To date, many MAPKKK genes have been identified in several plant species based on genome sequencing, including Arabidopsis (80 members)23, Oryza sativa (75 members)24, Zea mays (71 members)25, Lycopersicon esculentum (89 members)26, Cucumis sativus (59 members)27, Glycine max (150 members)28, Medicago truncatula (73 members)29, Musa acuminate (77 members)30, and Vitis vinifera (45 members)31. Currently, there is little information concerning the MAPKKK family in cassava that is an important crop across the tropical and sub-tropical world32–36. Although cassava has the characteristic of drought resistance, the underlying mechanism is elusive because of the limitation of cassava cultivation zones. Thus, investigation of the gene families related to MAPK cascade is necessary to elucidate the biological processes of cassava development and stress responses.
In this study, we identified 62 MeMAPKKKs from the cassava genome, and further investigated their phylogenetic relationship, protein motifs, gene structure, expression patterns in diverse tissues, and the responses to drought stress in three cassava accessions. This comprehensive study will increase our understanding of MAPKKK associated with the developmental process and abiotic stress responses, and build a solid foundation for future studies aimed at MAPK signaling cascade-mediated genetic improvement of cassava.
Results
Identification and phylogenetic analysis of cassava MAPKKK family
By BLAST and Hidden Markov Model searches, a total of 62 MeMAPKKKs were obtained from the complete cassava genome with Arabidopsis and rice MAPKKK sequences as queries. The 62 predicted MAPKKK proteins ranged from 284 (MeMAPKKK17) to 1021 (MeMAPKKK33) amino acids (aa) in length with an average of 612.2 aa, the molecular weight (MW) varied from 32.5 kDa (MeMAPKKK17) to 111.8 kDa (MeMAPKKK33), and the theoretical isoelectric point (pI) ranged from 4.74 (MeMAPKKK61) to 9.5 (MeMAPKKK5) with 49 numbers pI <7 and the others pI <7 (Table S1).
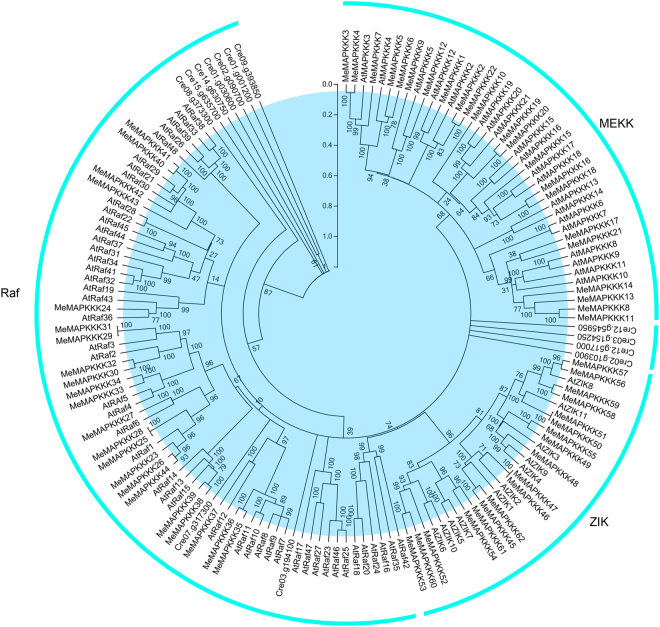
To investigate the evolutionary relationships of MAPKKK proteins between cassava and Arabidopsis/Chlamydomonas reinhardtii, a phylogenetic tree was constructed after alignment of amino acid sequences of all 155 MAPKKKs (62 in cassava, 80 in Arabidopsis, and 13 in Chlamydomonas reinhardtii) using Clustal X 2.0 and MEGA 5.0 softwares. As indicated in Fig. 1, 62 MeMAPKKK proteins were grouped into three clusters, namely Raf, ZIK and MEKK subfamily, in accordance with the categories of MeMAPKKKs in Arabidopsis. There were 22 MeMAPKKKs, 48 AtRAFs and 9 MAPKKKs from Chlamydomonas reinhardtii in the Raf subfamily, 18 MeMAPKKKs and 11 AtZIKs in the ZIK subfamily, and 22 MeMAPKKKs and 21 AtMAPKKKs in the MEKK subfamily. The additional 4 MAPKKKs in Chlamydomonas reinhardtii were not clustered, suggesting their independent evolution. Phylogenetic analysis also showed that there were some closely related orthologous MAPKKKs between cassava and Arabidopsis, suggesting that an ancestral set of MAPKKK genes existed before the divergence of cassava and Arabidopsis.
Figure 1.
Phylogenetic analysis of MAPKKKs from cassava, Arabidopsis, and Chlamydomonas reinhardtii using the complete protein sequences. The Neighbor-joining (NJ) tree was reconstructed using Clustal X 2.0 and MEGA 5.0 softwares with the pair-wise deletion option. 1000 bootstrap replicates were used to assess tree reliability.
Conserved motifs and gene structure of MeMAPKKKs
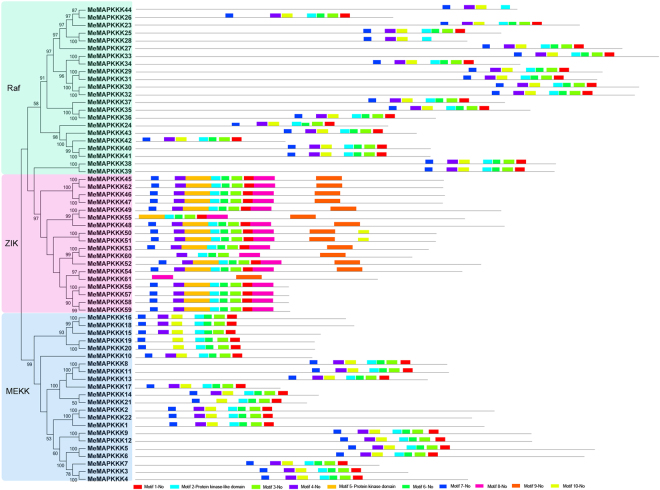
To further understand the structural features of MeMAPKKKs, 10 conserved motifs were identified using MEME software and further annotated by InterPro Scan 5 (Fig. 2). Motif 2 and Motif 5 were marked as protein kinase domain, while the remaining 8 motifs had no annotation. All the identified MeMAPKKKs had motif 2, and all the ZIK subfamily members specially showed motif 5, expect for MeMAPKKK61. Additionally, all the MeMAPKKKs, except for MeMAPKKK19, -20, -21, -28, -44, -60, and -61, contained motifs 1–4. Motif 8 only presented in ZIK subfamily, while motif 10 uniquely displayed in Raf and MEKK subfamily. These results suggest that each subfamily of MAPKKKs shares the similar protein structure and domain composition, suggesting that protein architecture is dramatically conserved within a specific subfamily of MAPKKKs.
Figure 2.
The conserved motifs of cassava MAPKKKs according to phylogenetic relationship. All motifs were identified by MEME database with the complete amino acid sequences of cassava MAPKKKs.
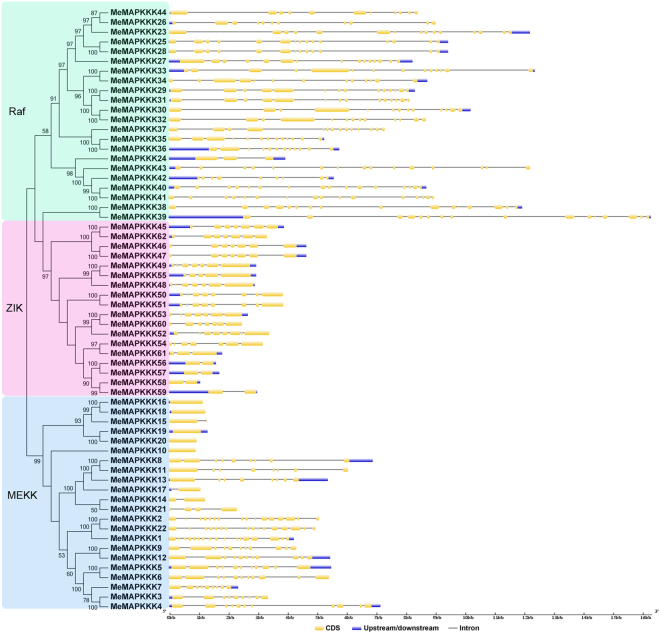
To reveal the structural features of MeMAPKKKs, intron-exon organizations were analyzed by Gene Structure Display Server (GSDS) v2.0. The number of intron in MeMAPKKKs varied from 0 (MeMAPKKK16, -18, -15, -19, -20, and -10) to 16 (MeMAPKKK1) (Fig. 3). Most of Raf subfamily genes contained more than 10 introns, while the intron in ZIK group genes varied from 1 to 7. Generally, MeMAPKKKs in the same cluster of the phylogenetic tree show similar exon-intron structures, indicating the link between evolutionary relationship and gene structure.
Figure 3.
Gene structure analyses of cassava MAPKKKs according to phylogenetic relationship. Exon-intron structure analyses were performed by GSDS database. The blue boxes, yellow boxes, and the black lines indicate upstream/downstream, exons, and introns, respectively.
Expression profiles of MeMAPKKKs in different tissues of two cassava genotypes
To detect the possible role of MeMAPKKKs in cassava growth and development, the stems, leaves and storage roots of wild subspecies (W14) and cultivated variety (Arg7) were collected for RNA-seq analyses. 80% (50/62) of MeMAPKKK genes were captured from the RNA-seq data (Fig. 4; Table S2). Generally, 13 out of 50 MeMAPKKK genes had high expression levels (value >5) in the tested tissues of the two genotypes, such as MeMAPKKK3, -4, -12, -13, -24, -25, -30, -36, -38, -43, -45, -46 and -59. In contrast, 8 MeMAPKKK genes (MeMAPKKK2, -7, -14, -15, -21, -22, -37, and -54) showed low transcript abundance (value <5) in the tissues of the two accessions.
Figure 4.
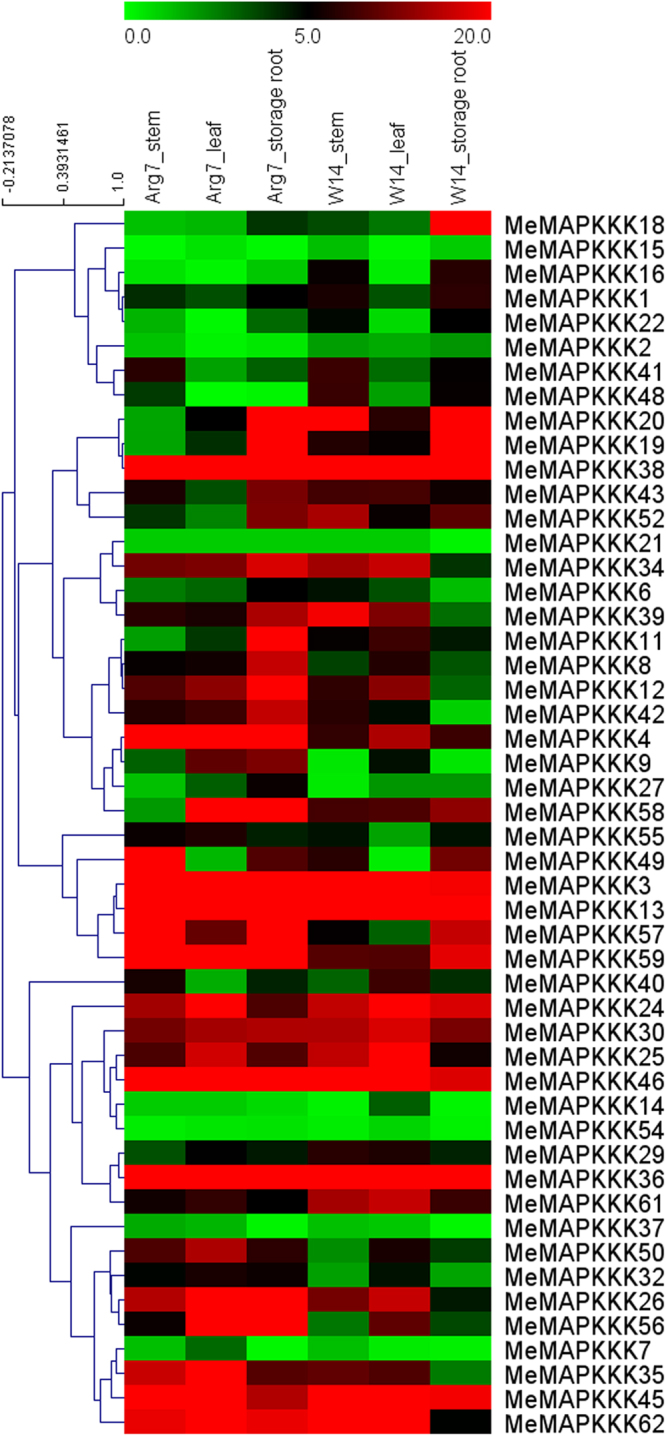
Expression profiles of cassava MAPKKKs in stems, leaves, and storage roots of Arg7 and W14. The heat map was constructed according to the FPKM value of cassava MAPKKKs. FPKM value is shown in color as the scale.
For Arg7 varieties, 26 of 50 MeMAPKKKs showed higher transcripts in storage roots, relative to that in stems or leaves; and 14 of 50 MeMAPKKKs displayed relatively higher transcripts in leaves, compared with that in stems or storage roots. For W14 subspecies, 12 of 50 MeMAPKKKs showed higher transcripts in storage roots, relative to that in stems and leaves; and 23 of 50 MeMAPKKKs displayed relatively higher transcripts in leaves, compared with that in stems or storage roots. Besides, some MAPKKK genes had differential expression patterns between the two accessions, such as MeMAPKKK-1, -11, -16, -19, -20, -29, -48, -52, -58 showing high transcript abundance (value >5) in W14 stems, whereas exhibiting low expression (value <5) in Arg7 stems, suggesting their differential roles in different organs of the two cassava accessions. Taken together, the tissue expression patterns of MAPKKK genes in different genotypes may provide valuable information for further study of organ development and function.
Expression profiles of MeMAPKKK genes after drought treatment in three cassava accessions
To seek insights into the clues of MeMAPKKKs in cassava response to drought, three accessions of cassava seedlings were exposed to drought conditions for 12 days, and then the leaves and roots were sampled to perform RNA-seq analyses (Fig. 5; Table S3). From the results, 56 MeMAPKKKs, except for MeMAPKKK10, -28, -31, -34, -47, -51, showed the corresponding transcriptomic data.
Figure 5.
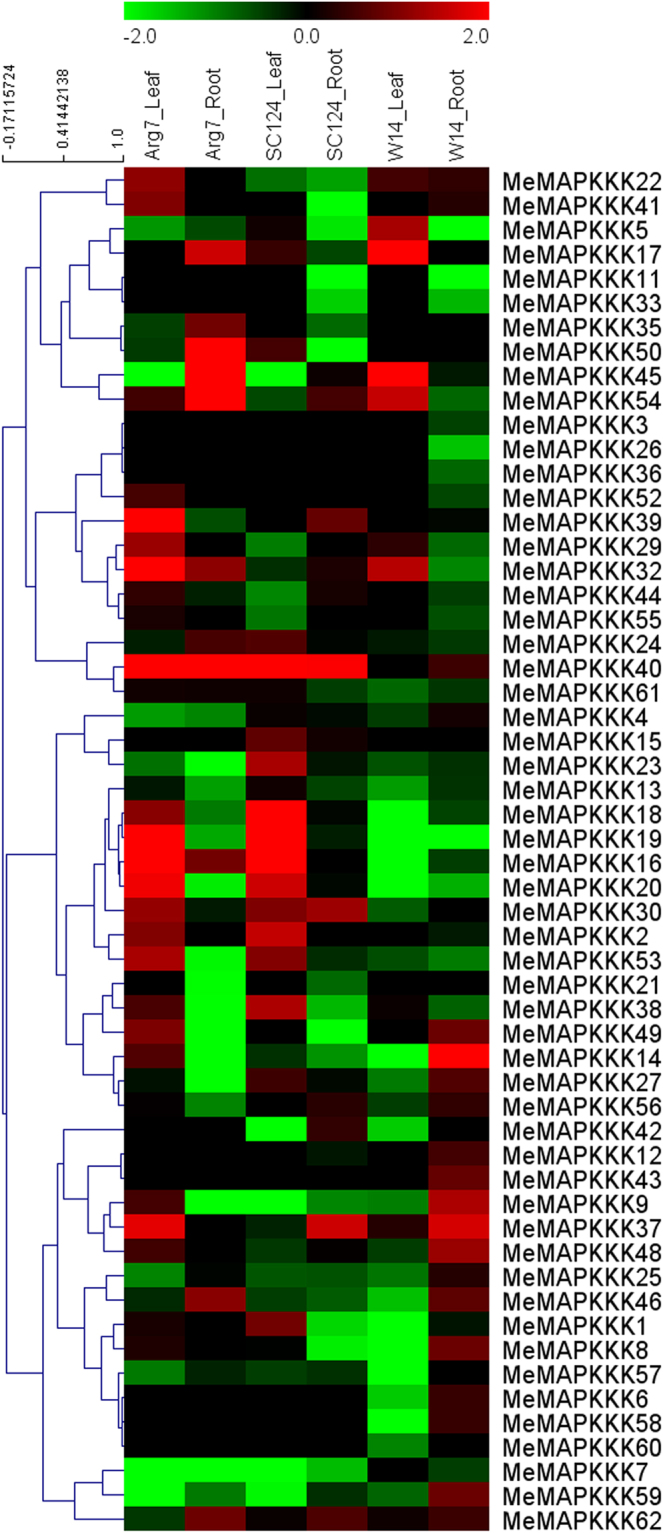
Expression profiles of cassava MAPKKKs in response to drought stress in leaves and roots of Arg7, SC124, and W14. Log2 based fold change was used to create the heat map. Fold changes in gene expression are shown in color as the scale.
In the Arg7 variety, 7/56 and 14/56 MeMAPKKKs were induced (log2-based values >1) after drought stress in roots and leaves, respectively, with two genes (MeMAPKKK32 and MeMAPKKK40) showing upregulation in both roots and leaves. In the SC124 variety, 3/56 and 9/56 of MeMAPKKKs expression increased after drought stress in roots and leaves, respectively, with MeMAPKKK40 showing upregulation in both roots and leaves. In W14 subspecies, 4/56 and 5/56 of MeMAPKKKs genes showed induction after drought stress in roots and leaves, respectively. These results suggested that the number of MeMAPKKKs upregulated by drought was greater in Arg7 and SC124 than those in W14.
Besides, some MeMAPKKKs showed similar expression profiles in SC124 and Arg7, which is different from W14. Seven genes (MeMAPKKK2, -16, -18, -19, -20, -40, -53) were upregulated in leaves of Arg7 and SC124, whereas were downregulated or no response in leaves of W14 after drought treatment. We also found that two genes (MeMAPKKK9, -14) showed upregulation in roots of W14, but repression in roots of Arg7 and SC124. Thus, the MAPKKKs mediated mechanisms of drought response may be differentiated between wild subspecies and cultivated varieties.
Expression patterns of MeMAPKKK genes under various stress and related signalling treatments
According to the RNA-seq data, 7 MeMAPKKK genes (MeMAPKKK-16, -18, -19, -20, -30, -32 and -40) upregulated by drought were chosen to investigate their expression after salt, osmotic, cold, ABA and H2O2 treatments (Figs 6–8; Table S4). In response to osmotic, salt and ABA treatments, all the tested MeMAPKKKs were significantly up-regulated during the whole treated time points (Figs 6 and 8). Under cold stress condition, MeMAPKKK20 transcript was significantly induced during all the tested time points, and the other 6 MeMAPKKKs showed upregulation at several time points (Figs 7A and 8). After H2O2 treatment, MeMAPKKK40 displayed significant downregulation at all time points, whereas the other 6 genes were slightly induced at several time points (Figs 7B and 8). Together, these results indicated that most of the MeMAPKKKs could be significantly upregulated by osmotic, salt, and ABA treatments, whereas slightly affected by cold and H2O2 treatments (Fig. 8; Table S4), suggesting that MeMAPKKKs may participate in multiple signalling transduction pathways in cassava.
Figure 6.
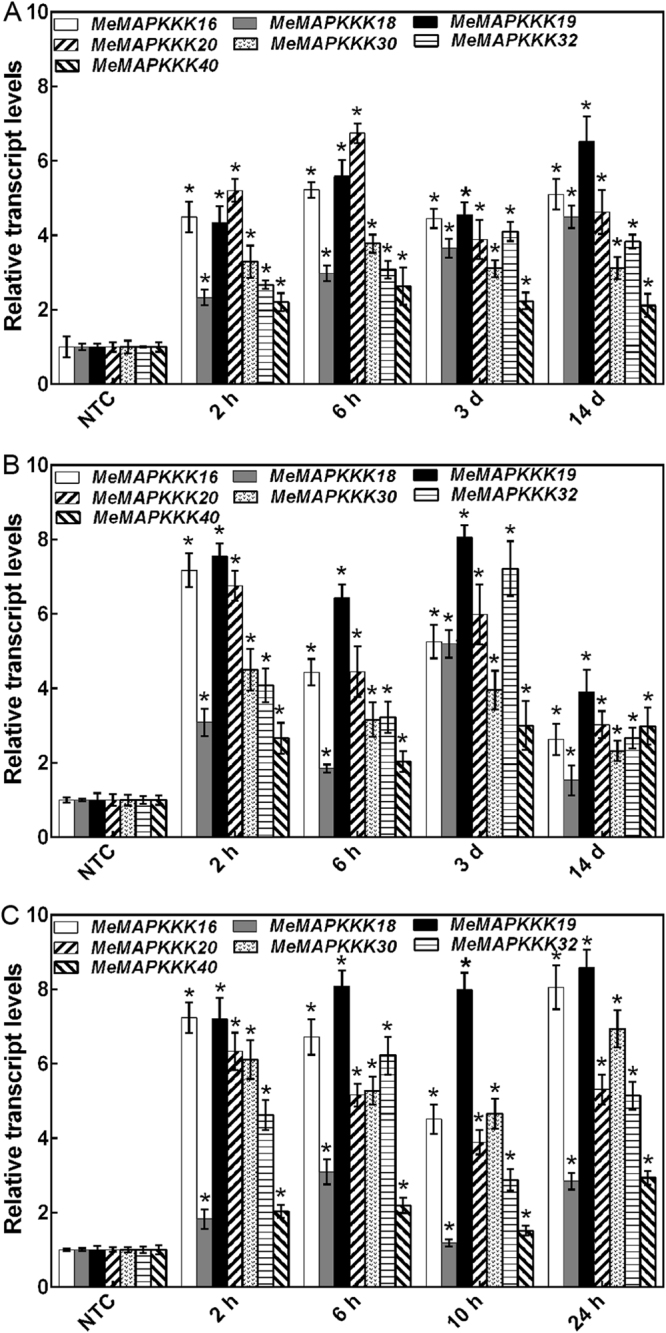
Expression patterns of selected MeMAPKKKs after osmotic (A), salt (B), and ABA (C) treatments in cassava. The mean fold changes of each gene between treated and control samples at each time point were used to calculate its relative expression levels. NTC indicates no treatment controls (mean value = 1). Data are means ± SD of n = 3 biological replicates.
Figure 8.
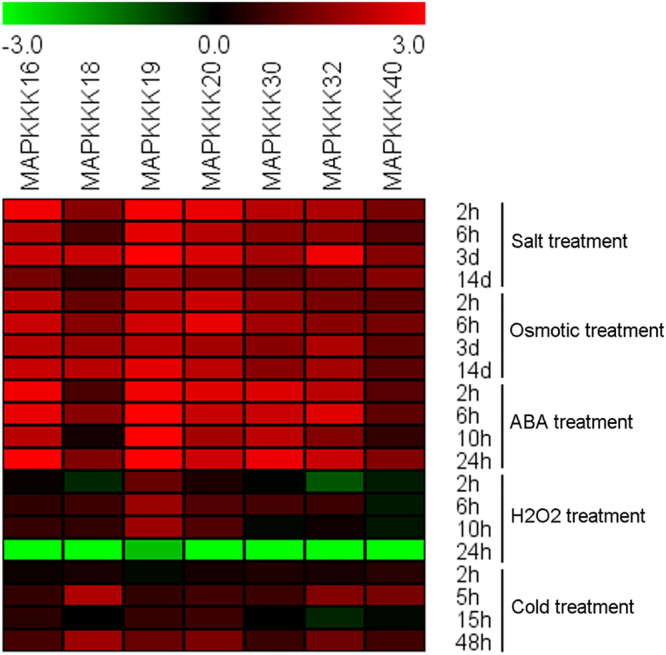
Expression of cassava selected MeMAPKKKs in response to various stimuli. The heatmap was constructed using Log2 based fold change from three biological replicates of qRT-PCR data. The scale represents the relative signal intensity.
Figure 7.
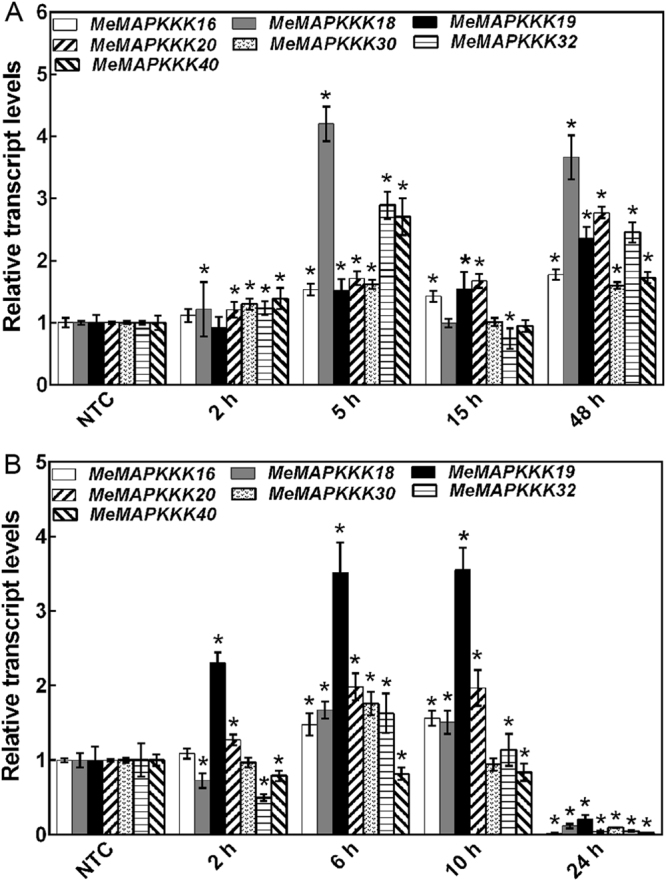
Expression patterns of selected MeMAPKKKs after cold (A) and H2O2 (B) treatments in cassava. The mean fold changes of each gene between treated and control samples at each time point were used to calculate its relative expression levels. NTC indicates no treatment controls (mean value = 1). Data are means ± SD of n = 3 biological replicates.
Discussion
Due to the central role of MAPK cascade in plant development and stress response and the characteristic of drought resistance of cassava, it is necessary to investigate the possible role of MAPKKKs in cassava. So far, there is no information for the MAPKKK family in cassava. In the present study, 62 MeMAPKKK genes were identified from cassava. Previously, genome-wide analyses have identified many MAPKKKs in various plant species, including 80 members in Arabidopsis 23, 75 in Oryza sativa 24, 71 in Zea mays 25, 89 in Lycopersicon esculentum 26, 59 in Cucumis sativus 27, 150 in Glycine max 28, 45 in Vitis vinifera 31, 73 in Medicago truncatula 29, and 77 in Musa acuminate 30. This indicated that MAPKKKs in cassava had expanded compared to that in Cucumis sativus and Vitis vinifera, while had shrunk compared to that in Arabidopsis, Oryza sativa, Zea mays, Lycopersicon esculentum, Glycine max, Medicago truncatula, and Musa acuminate. Cassava MAPKKKs were grouped into three subfamilies together with Arabidopsis MAPKKKs based on the evolutionary analysis (Fig. 1). This is in line with the previous evolutionary analysis of MeMAPKKKs from Arabidopsis, rice, tomato, and cucumber3,24,26,27. To better understand the evolution of MAPKKK family, 13 members from the ancestral Chlamydomonas reinhardtii were acquired using Phytozome V10.0 database. The evolutionary investigation indicated that most Chlamydomonas reinhardtii MAPKKKs (9 members) were clustered into Raf subfamily, implying that Raf subfamily may represent the original members of MAPKKK family (Fig. 1). We also observed that the Raf subfamily showed significantly more introns than the ZIK and MEKK subfamilies in cassava (Fig. 2). Previous report suggested that the rate of intron loss is faster than the rate of intron gain after segmental duplication37. This allows us to infer that Raf subfamily may contain the original genes, from which those in other subfamilies were derived. Intron-exon organization analyses showed that the number of intron in MeMAPKKKs was highly variable, varying from 0 to 16 (Fig. 2). The large-scale variation in structures of MeMAPKKKs implied that the cassava genome has significantly variable during its long evolutionary history. This phenomenon is also observed in maize (from 1 to 17) and grapevine (from 4 to 16)25,31. Conserved motif and gene structure analyses suggested that MeMAPKKKs in the same subfamily showed similar conserved motifs and exon-intron structures, further supporting the classification of MeMAPKKKs (Figs 2 and 3). Moreover, all the identified MeMAPKKKs had the typical protein kinase domain, which was also confirmed in other plants, including Arabidopsis, rice, cucumber, and tomato3,24,26,27.
Although cassava has strong resistance to drought stress, the underlying mechanism is less known. Previous studies demonstrated the positive role of MAPKKKs in plants resistant to abiotic stress, including drought, salt, and cold11,19,21,22. In the present study, we found that many MeMAPKKK genes showed induction after drought treatment in three cassava accessions, indicating their possible role in the regulation of cassava resistance to drought stress. By comparing the expression profiles of MeMAPKKK genes in different cassava accessions, we found that the number of MeMAPKKKs upregulated by drought was greater in leaves of Arg7 (14/56) and SC124 (9/56) than those in W14 (5/56) (Fig. 5). W14 is a drought resistant genotype, which is more resistant than Arg7 and SC12433–36. Some studies have revealed the positive role of MAPKKKs in drought response in Arabidopsis, rice, and maize by biochemical and genetic approaches19,22,38,39. Based on the above results, we speculated that the cultivated varieties (SC124 and Arg7) need to activate more MAPKKK genes in response to drought stress, thus well-fitting the drought environment, compared to wild subspecies W14. Notably, some MeMAPKKKs showed different expression profiles between SC124/Arg7 and W14, indicating the MAPKKK mediated drought resistance mechanism may be differentiated between wild subspecies and cultivated varieties.
As the core regulation components of MAPK cascade, MAPKKK widely participates in plants response to abiotic stress11,19,21,22. Based on the transcriptomic data, some MeMAPKKK genes could respond to drought stress. Therefore, it is essential to investigate the possible role of MeMAPKKK genes under various abiotic stress and stress-related signal molecule treatments. Interestingly, all the tested 7 MeMAPKKK genes (MeMAPKKK-16, -18, -19, -20, -30, -32 and -40) were significantly upregulated after osmotic, salt and ABA treatments during the whole treated time points, indicating that these genes may be the cross points of multiple stress signaling (Figs 6 and 8). In banana, 13.2% and 2.6% MAPKKKs were significantly induced by osmotic and salt stress in BX variety, and 15.8% and 5.3% MAPKKKs were upregulated under each stress in FJ variety30. In grapevine, most of the MAPKKK genes showed induction after 8 d and 12 d drought treatment31. In tomato, most of the MAPKKK genes were upregulated by drought, salt, cold, and heat stress, among which SlMAPKKK51, SlMAPKKK53, and SlMAPKKK55 were upregulated by more than 100-fold after heat or drought treatment26. Further biochemical and genetic studies revealed that some MAPKKK members, including GhRaf19, AtMAPKKK18, AtRaf43, and DSM1 (a rice MAPKKK) were involved in plants resistance to abiotic stress21,38–40. These results suggested that MAPKKK family genes could widely participate in abiotic stress response. Moreover, MAPK cascade is considered to be crucial components of the ABA signaling transduction network39–42. Recently, the MAP3K17/18-MKK3-MPK1/2/7/14 module was found to be under the control of the ABA core signalling pathway at transcriptional and post-translational levels42. Due to the crucial role of ABA signalling in plants response to abiotic stress, these MeMAPKKK genes may be involved in ABA mediated abiotic stress response. In contrast, most of the tested MeMAPKKK genes were slightly affected by cold treatments (Fig. 7A), which could be explained by the warm habitat and cold-sensitive characteristic of cassava43,44.
In conclusion, this study identified 62 MAPKKKs from cassava and investigated their classification, protein motif, and gene structure. Organ expression analysis indicated the different expression profiles of MeMAPKKK genes in two different accessions. Transcriptomic analysis revealed the possible role of MeMAPKKK genes against drought stress in different cassava genotypes. Finally, several MeMAPKKK genes were identified as important candidates for improving crop resistances to multiple stresses. Together, these results will advance the understanding of MeMAPKKK-mediated abiotic stress response and provide candidate MeMAPKKK genes used for genetic improvement of crop resistance to abiotic stress.
Methods
Plant materials and treatments
The characteristic of W14, SC124 and Arg7 was described in previous studies45,46. Segments of cassava stems were taken from mother plants, and cultured in pots in a growth room with a 16 h/35 °C day and 8 h/20 °C night regime, and a relative humidity of 70%. Then, 90-day-old stems, 90-day-old leaves, and 150-day-old tuberous roots were acquired from W14 and Arg7 under normal conditions to study the expression levels of MeMAPKKKs in distinct organs. To detect the transcriptional changes of MeMAPKKKs in response to drought, roots and leaves were collected from W14, SC124, and Arg7, respectively, under drought conditions for 12 d. For osmotic, cold, salt, ABA and H2O2 treatments, two-month-old seedlings of Arg7 were suffered from 200 mM mannitol for 14 d, 300 mM NaCl for 14 d, 100 µM abscisic acid (ABA) for 24 h, 10% H2O2 for 24 h and low temperature (4 °C) for 48 h, respectively.
Identification and evolutionary analyses
Protein sequences of AtMAPKKKs and OsMAPKKKs were downloaded from RGAP and UniPort databases, respectively47,48. The candidate MeMAPKKKs were firstly acquired with local hidden Markov Model-based search and BLAST search from cassava genome database using Arabidopsis and rice MAPKKKs as queries49,50. Subsequently, the candidate MeMAPKKKs were further checked with PFAM and CDD dataases51,52. Then, the protein sequences of 62 MeMAPKKKs and 80 AtMAPKKKs, and 13 MAPKKKs from Chlamydomonas reinhardtii were used to construct a neighbor-joining (NJ) phylogenetic tree using Clustal X2.0 and MEGA 5.0 software with 1000 bootstrap replicates53,54.
Protein properties and sequence analyses
ExPASy proteomics server was used to predict the molecular weight (MW) and isoelectric points (pI) of cassava MAPKKK family proteins55. MEME program was used to identify the conserved proteins motifs56. Furthermore, all identified motifs were annotated according to InterProScan57. The gene structures were identified using GSDS database58.
Transcriptomic analysis
Total RNA of each sample was extracted with plant RNA extraction kit (TIANGEN, China) and used for cDNA library construction. The sequencing was performed with an Illumina GAII following manufacturer’s instructions. Adapter sequences were removed with FASTX-toolkit. Clean reads were generated by removing low quality sequences using FastQC. Tophat v.2.0.10 was used to map the clean reads to the cassava genome. Using cufflinks, the transcriptome data was assembled59. Genes were scored as not expressed if the corresponding RNA-seq reads could not align to the genome. Gene expression levels were calculated as Reads Per Kilobase of exon model per Million mapped reads (FPKM). DEGseq was used to identify differentially expressed genes in response to drought60. The transcriptiomic data was submitted to NCBI and the accession number was listed in Supplementary Table S5.
Quantitative RT-PCR analysis
The leaf samples of Arg7 in response to salt, osmotic, cold, ABA and H2O2 were collected to isolate total RNA. Then, the expression of MeMAPKKKs was detected by qRT-PCR analysis using TransStart Green qPCR SuperMix (TRANS, Beijing, Chian) chemistry on a Stratagene Mx3000 P (Stratagene, CA, USA) instrument. Agarose gel electrophoresis, melting curve, and sequencing analyses were performed to confirm the specificities of primer pairs. The primers of target genes were listed in Supplementary Tables S6. For each target gene, expression data were normalized with expression levels of β-tubulin gene (TUB) and elongation factors 1α gene (EF1) and calculated by the formula 2−ΔΔCt method61,62.
Electronic supplementary material
Acknowledgements
This work was supported by the Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (1630052016005, 1630052016006), Central Public-interest Scientific Institution Basal Research Fund for Innovative Research Team Program of CATAS (17CXTD-28), and the earmarked fund for Modern Agro-industry Technology Research System (CARS-11).
Author Contributions
K.L., H.Z., W.H., and J.Y. conceived the study. H.Z., H.Y., H.S., Y.W., W.T., Z.D., Y.Y., Y.L., Z.X., W.W., and M.P. performed the experiments and carried out the analysis. W.H., and J.Y. designed the experiments and wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Jianqiu Ye, Hai Yang and Haitao Shi contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-13988-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kaimian Li, Email: likaimian@itbb.org.cn.
He Zhang, Email: atzzhef@163.com.
Wei Hu, Email: huwei2010916@126.com.
References
- 1.Asai T, et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 2.Rohila JS, Yang Y. Rice mitogen-activated protein kinase gene family and its role in biotic and abiotic stress response. J. Integr. plant biol. 2007;49:751–759. doi: 10.1111/j.1744-7909.2007.00501.x. [DOI] [Google Scholar]
- 3.Jonaka C, Ökrészb L, Bögrec L, Hirta H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 2002;5:415–424. doi: 10.1016/S1369-5266(02)00285-6. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi Y, Soyano T, Kosetsu K, Sasabe M, Machida Y. HINKEL kinesin, ANP MAPKKKs and MKK6/ANQ MAPKK, which phosphorylates and activates MPK4 MAPK, constitute a pathway that is required for cytokinesis in Arabidopsis thaliana. Plant Cell Physiol. 2010;51:1766–1776. doi: 10.1093/pcp/pcq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao FY, et al. MAPKs regulate root growth by influencing auxin signaling and cell cycle-related gene expression in cadmium-stressed rice. Environ. Sci. Pollut. Res. Int. 2013;20:5449–5460. doi: 10.1007/s11356-013-1559-3. [DOI] [PubMed] [Google Scholar]
- 6.Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-B. [DOI] [PubMed] [Google Scholar]
- 7.Yue H, Li Z, Xing D. Roles of Arabidopsis bax inhibitor-1in delaying methyl jasmonate-induced leaf senescence. Plant Signal. Behav. 2012;7:1488–1489. doi: 10.4161/psb.21776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang G, et al. Genome-wide identification and analysis of mitogen activated protein kinase kinase kinase gene family in grapevine (Vitis vinifera) BMC Plant Biol. 2014;14:219. doi: 10.1186/s12870-014-0219-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genot, B., Lang, J., Berriri, S., Garmier, M. & Gilard, F. Constitutively active Arabidopsis MAP Kinase 3 triggers defense responses involving salicylic acid and SUMM2 resistance protein. Plant Physiol., doi: 10.1104/pp.17.00378 (2017). [DOI] [PMC free article] [PubMed]
- 10.Kumar K, Sinha AK. Overexpression of constitutively active mitogen activated protein kinase kinase 6 enhances tolerance to salt stress in rice. Rice (NY). 2013;6:25. doi: 10.1186/1939-8433-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shitamichi N, Matsuoka D, Sasayama D, Furuya T, Nanmori T. Over-expression of MAP3Kδ4, an ABA-inducible Raf-like MAP3K that confers salt tolerance in Arabidopsis. Plant Biotechnol. 2013;30:111–118. doi: 10.5511/plantbiotechnology.13.0108a. [DOI] [Google Scholar]
- 12.Jiang M, et al. Genome-wide exploration of the molecular evolution and regulatory network of mitogen-activated protein kinase cascades upon multiple stresses in Brachypodium distachyon. BMC Genomics. 2015;16:228. doi: 10.1186/s12864-015-1452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Çakır B, Kılıçkaya O. Mitogen-activated protein kinase cascades in Vitis vinifera. Front Plant Sci. 2015;6:556. doi: 10.3389/fpls.2015.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pitzschke A, Djamei A, Bitton F, Hirt H. A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Mol. Plant. 2009;2:120–137. doi: 10.1093/mp/ssn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Schiff M, Dinesh-Kumar SP. Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J. 2004;38:800–809. doi: 10.1111/j.1365-313X.2004.02085.x. [DOI] [PubMed] [Google Scholar]
- 16.Furuya T, Matsuoka D, Nanmori T. Membrane rigidification functions upstream of the MEKK1-MKK2-MPK4 cascade during cold acclimation in Arabidopsis thaliana. FEBS Lett. 2014;588:2025–2030. doi: 10.1016/j.febslet.2014.04.032. [DOI] [PubMed] [Google Scholar]
- 17.Zeng Q, Chen JG, Ellis BE. AtMPK4 is required for male-specific meiotic cytokinesis in Arabidopsis. Plant J. 2011;67:895–906. doi: 10.1111/j.1365-313X.2011.04642.x. [DOI] [PubMed] [Google Scholar]
- 18.Eckardt NA. A complete MAPK signaling cascade that functions in stomatal development and patterning in Arabidopsis. Plant Cell. 2007;19:7. doi: 10.1105/tpc.107.190110. [DOI] [Google Scholar]
- 19.Virk N, et al. Arabidopsis Raf-like mitogen-activated protein kinase kinase kinase gene Raf43 is required for tolerance to multiple abiotic stresses. PLoS One. 2015;10:e0133975. doi: 10.1371/journal.pone.0133975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashimoto-Sugimoto M, et al. Dominant and recessive mutations in the Raf-like kinase HT1 gene completely disrupt stomatal responses to CO2 in Arabidopsis. J. Exp. Bot. 2016;67:3251–3261. doi: 10.1093/jxb/erw134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia H, et al. A Raf-like MAPKKK gene, GhRaf19, negatively regulates tolerance to drought and salt and positively regulates resistance to cold stress by modulating reactive oxygen species in cotton. Plant Sci. 2016;252:267–281. doi: 10.1016/j.plantsci.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 22.Shou H, Bordallo P, Wang K. Expression of the Nicotiana protein kinase (NPK1) enhanced drought tolerance in transgenic maize. J. Exp. Bot. 2004;55:1013–1019. doi: 10.1093/jxb/erh129. [DOI] [PubMed] [Google Scholar]
- 23.Kong Q, et al. The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell. 2012;24:2225–2236. doi: 10.1105/tpc.112.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao KP, Richa T, Kumar K, Raghuram B, Sinha AK. In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA Res. 2010;17:139–153. doi: 10.1093/dnares/dsq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, et al. RNA-seq analysis reveals MAPKKK family members related to drought tolerance in Maize. PloS One. 2015;10:e0143128. doi: 10.1371/journal.pone.0143128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J, et al. Genome-wide identification of MAPKK and MAPKKK gene families in tomato and transcriptional profiling analysis during development and stress response. PloS One. 2014;9:e103032. doi: 10.1371/journal.pone.0103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, et al. Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genomics. 2015;16:1. doi: 10.1186/1471-2164-16-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neupane A, et al. Identification, nomenclature, and evolutionary relationships of mitogen-activated protein kinase (MAPK) genes in soybean. Evol. Bioinform. 2013;9:363–386. doi: 10.6026/97320630009363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, et al. Bioinformatics Analysis of MAPKKK family genes in Medicago truncatula. Genes (Basel). 2016;7:E13. doi: 10.3390/genes7040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu W, et al. Genome-wide analyses of the bZIP family reveal their involvement in the development, ripening and abiotic stress response in banana. Sci Rep. 2016;6:30203. doi: 10.1038/srep30203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G, et al. Genome-wide identification and analysis of mitogen activated protein kinase kinase kinase gene family in grapevine (Vitis vinifera) BMC Plant Biol. 2014;14:219. doi: 10.1186/s12870-014-0219-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliveira EJ, Santana FA, Oliveira LA, Santos VS. Genetic parameters and prediction of genotypic values for root quality traits in cassava using REML/BLUP. Genet. Mol. Res. 2014;13:6683–6700. doi: 10.4238/2014.August.28.13. [DOI] [PubMed] [Google Scholar]
- 33.Wei Y, et al. Genome-wide identification and Eexpression analysis of the WRKY gene family in cassava. Front Plant Sci. 2016;7:25. doi: 10.3389/fpls.2016.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan W, et al. The ERF transcription factor family in cassava: genome-wide characterization and expression analyses against drought stress. Sci. Rep. 2016;6:37379. doi: 10.1038/srep37379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan Y, et al. Genome-wide identification and expression analysis of the mitogen-activated protein kinase gene family in cassava. Front Plant Sci. 2016;7:1294. doi: 10.3389/fpls.2016.01294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu W, et al. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 2016;6:22783. doi: 10.1038/srep22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nuruzzaman M, et al. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 38.Ning J, Li X, Hicks LM, Xiong L. A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 2010;152:876–890. doi: 10.1104/pp.109.149856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, et al. Arabidopsis MAPKKK18 positively regulates drought stress resistance via downstream MAPKK3. Biochem. Biophys. Res. Commun. 2017;484:292–297. doi: 10.1016/j.bbrc.2017.01.104. [DOI] [PubMed] [Google Scholar]
- 40.Virk N, et al. Arabidopsis Raf-Like Mitogen-Activated Protein Kinase Kinase Kinase Gene Raf43 Is Required for Tolerance to Multiple Abiotic Stresses. PLoS One. 2015;10:e0133975. doi: 10.1371/journal.pone.0133975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuoka D, Yasufuku T, Furuya T, Nanmori T. An abscisic acid inducible Arabidopsis MAPKKK, MAPKKK18 regulates leaf senescence via its kinase activity. Plant Mol. Biol. 2015;87:565–575. doi: 10.1007/s11103-015-0295-0. [DOI] [PubMed] [Google Scholar]
- 42.de Zelicourt A, Colcombet J, Hirt H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016;21:677–685. doi: 10.1016/j.tplants.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 43.Huang L, Ye Z, Bell RW, Dell B. Boron nutrition and chilling tolerance of warm climate crop species. Ann. Bot. 2005;96:755–767. doi: 10.1093/aob/mci228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.An D, Yang J, Zhang P. Transcriptome profling of low temperature-treated cassava apical shoots dynamic responses of tropical plant to cold stress. BMC Genomics. 2012;13:64. doi: 10.1186/1471-2164-13-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu W, et al. Genome-wide gene phylogeny of CIPK family in cassava and expression analysis of partial drought-induced genes. Front Plant Sci. 2015;6:914. doi: 10.3389/fpls.2015.00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W, et al. Cassava genome from a wild ancestor to cultivated varieties. Nat. Commun. 2014;5:5110. doi: 10.1038/ncomms6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The UniProt Consortium UniProt: A hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawahara Y, et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6:4. doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prochnik S, et al. The Cassava genome: current progress, future directions. Trop Plant Biol. 2012;5:88–94. doi: 10.1007/s12042-011-9088-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 54.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gasteiger E, et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown P, et al. MEME-LaB: Motif analysis in clusters. Bioinformatics. 2013;29:1696–1697. doi: 10.1093/bioinformatics/btt248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mulder N, Apweiler R. InterPro and InterProScan: Tools for protein sequence classification and comparison. Methods Mol. Biol. 2007;396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- 58.Hu B, et al. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trapnell, et al. TopHat and Cufflinks-Protocol. Nature Protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 61.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time Quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 62.Salcedo A, Zambrana C, Siritunga D. Comparative expression analysis of reference genes in field-grown cassava. Trop. Plant Biol. 2014;7:53–64. doi: 10.1007/s12042-014-9137-5. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.