Abstract
Timely progression of living cells through the cell cycle is precisely regulated. This involves a series of phosphorylation events which are regulated by various cyclins, activated in coordination with the cell cycle progression. Phosphorylated proteins govern cell growth, division as well as duplication of the genetic material and transcriptional activation of genes involved in these processes. A subset of these tightly regulated genes, which depend on the MBF transcription factor and are mainly involved in DNA replication and cell division, is transiently activated at the transition from G1 to S phase. A Saccharomyces cerevisiae mutant in the Dpb2 non-catalytic subunit of DNA polymerase ε (Polε) demonstrates abnormalities in transcription of MBF-dependent genes even in normal growth conditions. It is, therefore, tempting to speculate that Dpb2 which, as described previously, participates in the early stages of DNA replication initiation, has an impact on the regulation of replication-related genes expression with possible implications for genomic stability.
Keywords: Dpb2, Polymerase ε, Cell cycle, MBF transcription factor
DNA replication is a tightly controlled process which occurs only once per cell cycle. Moreover, the duplication of the genetic material has to be coordinated with the cell growth and division. Thus, multiple mechanisms have evolved to ensure proper timing of all processes throughout all cell cycle phases. This includes the control of replisomes assembly at the origins of replication and replication arrest in response to perturbations in DNA synthesis (Hustedt et al. 2013; Gadaleta et al. 2016). In eukaryotic cells, high fidelity duplication of the genetic material is performed by three main multisubunit DNA polymerases: the polymerase α (Polα) synthesizes primers which are extended by Polε or Polδ, the two major replicative polymerases. Polε is postulated to be the leading strand polymerase, for review see (Lujan et al. 2016) and here we will focus on this polymerase.
Polε is composed of the catalytic subunit Pol2 and three non-catalytic subunits Dpb2, Dpb3 and Dpb4. Although the N-terminal part of Pol2 is dispensable, its C-terminal half is essential for cell survival (Dua et al. 1999; Isoz et al. 2012). This region of Pol2 interacts with the other essential subunit Dpb2, which in turn binds Psf1, a subunit of the GINS complex (Takayama et al. 2003; Sengupta et al. 2013). GINS is a multiprotein complex composed of Psf1, Psf2, Psf3 and Sld5 subunits (Takayama et al. 2003) which, together with Cdc45 and Mcm2-7, form the CMG helicase complex essential in both the initiation and elongation of DNA replication (Moyer et al. 2006). Therefore, Dpb2, which links the Polε with the CMG complex, is important for the assembly of the replisome and targeting of Polε to the leading strand (Langston et al. 2014; Grabowska et al. 2014). Mutations in DPB2 allels dpb2-100 or dpb2-103 affect the interaction of Dpb2 not only with Pol2 (Jaszczur et al. 2008) but also with Psf1 and Psf3 subunits of the GINS complex (Garbacz et al. 2015; Dmowski et al. 2017). Dpb2 dysfunction in dpb2-100 or dpb2-103 mutants results in DNA replication elongation defects, prolonged S phase and increased spontaneous mutagenesis (Jaszczur et al. 2008). Importantly, mutations in DPB2 or deletion of this gene do not influence the catalytic properties of Polε (Ganai et al. 2015; Garbacz et al. 2015).
Polymerase ε and the S phase checkpoint
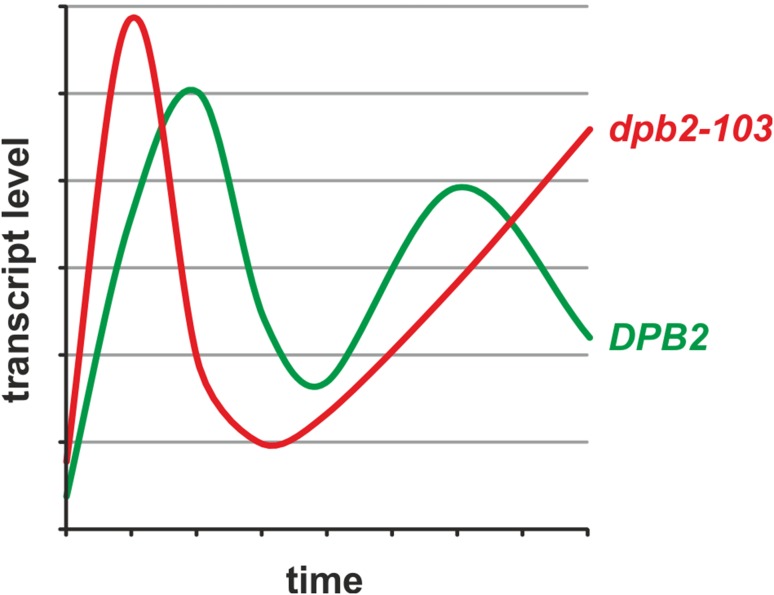
The S phase checkpoint is activated when DNA replication is threatened (Hustedt et al. 2013; Skoneczna et al. 2015; Palou et al. 2016) and it has been postulated that besides its role in DNA synthesis, Polε is also involved in this process. Early studies suggested that pol2-11 and pol2-12 mutants in the Pol2 C-terminus are impaired in correct response to the replication stress (Navas et al. 1995; Dua et al. 1998). However, the mechanism has not been identified because C-terminal mutations in the catalytic subunit Pol2 give very sick cells and their characterization is difficult. Recently, the involvement of the essential non-catalytic subunit Dpb2 in the response to the replication stress has been demonstrated by studies of the dpb2-103 mutant (Dmowski et al. 2017). Yeast cells, when challenged with the RNR inhibitor hydroxyurea, activate the Rad53 checkpoint kinase, which in turn activate the Dun1–Crt1 branch (mainly dNTP upregulation) and the MBF-Nrm1 branch, which stimulates the transcription of G1/S transition genes (explained below) (Bastos de Oliveira et al. 2012; Hustedt et al. 2013). The MBF-Nrm1-dependent genes analyzed in the dpb2-103 mutant are involved in environmental stresses response, sister chromatide cohesion, chromosome condensation or morphogenesis (Smolka et al. 2012). In contrast to the wild type cells, dpb2-103 cells treated with hydroxyurea fail to activate the MBF-Nrm1 branch of the replication checkpoint. Thus, in this mutant, G1/S MBF-dependent genes are repressed in the S phase regardless of the hydroxyurea treatment. Another interesting observation comes from the experiments performed during unperturbed growth. In dpb2-103 cells transcriptional activation of MBF-dependent G1/S transition was faster and, more importantly, prematurely switched off, when compared to the wild type cells (Fig. 1) (Dmowski et al. 2017). Thus these observations may be the starting point for investigations of the effect of mutations in the Dpb2 subunit on the transcription of G1/S transition genes and its consequences for the cell cycle progression and genome stability.
Fig. 1.
Simplified graph presenting the transcript levels of MBF-regulated G1/S transition genes in a wild type and dpb2-103 cells after the release from G1 block. Based on Fig. 6 from (Dmowski et al. 2017)
Cell cycle regulation by cyclin-dependent kinases
Expression of G1/S transition genes, like other cell cycle-related processes in eukaryotes, is tightly regulated by highly conserved cyclin-dependent kinases (CDKs) which are activated by cell cycle-specific cyclins. In Saccharomyces cerevisiae the Cdc28/Cdk1 kinase is controlled by nine periodically expressed cyclins: three G1 cyclins (Cln1-3) and six B-type cyclins (Clb1-6). This enables thigh control of cellular processes such as DNA replication, cell growth and division, as well as the transcriptional control of genes involved in these processes (Koch et al. 1996; Tanaka et al. 2007a; Benanti 2016; Deshmukh et al. 2016).
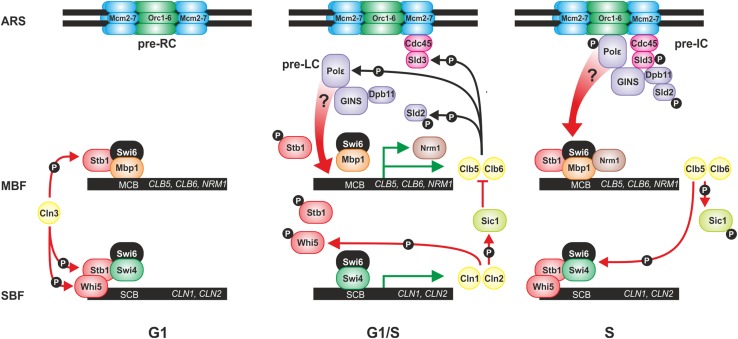
In early G1 phase transcription of G1-specific genes involved in transition to the S phase is inactive. These genes are controlled by two transcription factors, the SBF (SCB-binding factor) activator and the MBF (MCB-binding factor) repressor composed of DNA-binding subunits Swi4 and Mbp1, respectively, and a common regulatory subunit Swi6 (Iyer et al. 2001). The genes regulated by the SBF are mainly involved in cell cycle progression (e.g. CLN1 and CLN2) while MBF regulates the genes involved in DNA replication and repair (de Bruin et al. 2006). The MBF repressor with the Nrm1 corepressor interacting with Swi6, remains bound to specific promoter sequences throughout the cell cycle, while the transcription of SBF-dependent genes is inactivated by the transcriptional inhibitor Whi5 bound to SBF from M to late G1 phase (Fig. 2) (Koch et al. 1996; de Bruin et al. 2006). As the cell progress through G1 phase, Cdc28 is activated through binding of the Cln3 cyclin. The Cln3–Cdc28 complex phosphorylates the transcriptional inhibitor Whi5, to promote its dissociation from SBF and thus, the transcriptional activation of dozens of G1-specific genes involved in the transition to the S phase (Bertoli et al. 2013). SBF activation is reinforced by a positive feedback loop where Whi5 phosphorylation by Cdc28 is further activated by two SBF-dependent early-expressed Cln1 and Cln2 cyclines (Eser et al. 2011) whose expression peaks at G1-S transition (Harris et al. 2013). Additionally, both SBF- and MBF-dependent promoters are activated through Cln3–Cdc28-dependent phosphorylation of Stb1 (mediating early G1 repression) which then dissociates from MBF- and SBF-dependent promoters (Fig. 2) (de Bruin et al. 2008; Ferrezuelo et al. 2010). At this stage, cells are committed to pass START and enter the cell cycle (Doncic et al. 2011).
Fig. 2.
Cell cycle-related events at the origins of replication (ARS) and at promoter regions of G1/S transition genes regulated by MBF and SBF transcription factors. Thin arrows denote phosphorylation events which have inhibiting (red) and activating (black) effects. Green arrows denote active transcription. Details are given in the text
The cell entry into S phase requires Clb5 and Clb6 cyclins whose MBF-regulated expression peaks at G1-S transition. However, the activation of Clb5–Cdc28 and Clb6–Cdc28 complexes requires the ubiquitin-dependent degradation of Sic1, the inhibitor of these complexes. Degradation of Sic1 is promoted not only by Cln1,2–Cdc28 but also by Clb5–Cdc28 through a positive feedback (Nash et al. 2001; Kõivomägi et al. 2011). Clb5 and Clb6 cyclins activate Cdc28 to phosphorylate the Swi4–Swi6 complex which then dissociates from SBF-dependent promoters to inactivate theme (de Bruin et al. 2008). In parallel, MBF-dependent promoters are inactivated by the accumulating Nrm1 repressor (expressed in an MBF-dependent manner), presumably stabilized by Cdc28-dependent phosphorylation (Fig. 2) (de Bruin et al. 2006; Ostapenko and Solomon 2011). However, if the replication stress checkpoint is activated, MBF-dependent transcription is maintained during the S phase (Bastos de Oliveira et al. 2012; Travesa et al. 2012).
In parallel, Clb5,6–Cdc28 also activate the DNA replication. First, the origins of replication (ARS—autonomously replicating sequences) are bound by the ORC (origin recognition complex) composed of Orc1-6 proteins (Liang and Stillman 1997). Next, from late M to G1 phase, the ORC assisted by the Cdc6 ATPase and Cdt1 (the DNA-licensing factor) recruits the inactive Mcm2–7 (minichromosome maintenance) helicase complex to form the pre-RC (pre-replicative complex), reviewed in (Li and Araki 2013). Then, in the late G1 phase, at early-firing origins, DDK (Dbf4-dependent kinase) phosphorylates Mcm2–7, which then recruits Cdc45 together with Sld3 (Kamimura et al. 2001; Araki 2010; Sheu and Stillman 2010). Further steps of DNA replication initiation are regulated by Clb5- and Clb6-dependent Cdc28 (Tanaka et al. 2007a; Tanaka and Araki 2010). First, Clb5,6-dependent phosphorylation of Sld2 by Cdc28 promotes the association of the pre-LC (pre-loading complex) composed of Polε, GINS, Dpb11 and Sld2 (Masumoto et al. 2002). Next, Clb5,6-dependent phosphorylation of Sld3 recruits the pre-LC to the pre-RC to form the pre-IC (pre-initiation complex) (Tanaka et al. 2007b; Zegerman and Diffley 2007). Since Sld2 phosphorylation promotes its association with Dpb11 and Polε but not GINS, it has been suggested that Polε is necessary for the association of GINS with Sld2–Dpb11 (Araki 2010). Therefore, it has been postulated that Pol2–Dpb2 interaction is involved in the process of pre-LC assembly (Muramatsu et al. 2010). Moreover, Dpb2 has been shown to interact with Orc1 and Orc4 from the ORC complex (Krogan et al. 2006).
Involvement of Dpb2 in regulation of processes at the G1/S transition
In the G1 phase, the Dpb2 subunit of Polε is phosphorylated in a Cln–Cdc28-dependent manner, while in S phase through the activity of Clb5,6–Cdc28 (Kesti et al. 2004). Phosphorylation of Dpb2 is not only independent from Pol2 binding but also enhances its association with Pol2. Therefore, it has been suggested, that phosphorylation of Dpb2 in G1 phase is involved in Dpb2 interaction with Pol2. This is supported by the finding that DPB2 mutant in Cdc28 CDK sites demonstrate as a synthetic defect with a Pol2 mutant in the C-terminus (pol2-11) which interacts with Dpb2 (Kesti et al. 2004). Furthermore, the excessive dephosphorylation of Dpb2 in a Cdc14 mutant strain combined with mutations in CDK sites of Dpb2 severely impairs cell viability (Bloom and Cross 2007). Therefore, although there was no severe phenotypic effect of mutations in Dpb2 CDK sites, it cannot be excluded that the cell cycle-dependent phosphorylation of Dpb2 is involved in other aspects of DNA replication and the cell cycle progression.
Given that Dpb2 is involved in DNA replication initiation, is phosphorylated in a cell cycle-dependent manner and that the dpb2-103 mutant demonstrates a different pattern of G1/S transition genes transcription, it might be speculated that Dpb2 is also involved in the regulatory processes during replication initiation. How does it happen? Firstly, the Dpb2 protein, which is phosphorylated by Cdc28, may directly influence G1/S genes expression by yet unknown mechanisms. Secondly, Dpb2 may be involved in the communication between the assembling replisomes and MBF regulators. This would define a mechanism coordinating the pre-IC complexes assembly and G1/S gene expression prior to the transition to the S phase. Finally, the observed anomalies in transcriptional repression of G1/S transition genes may constitute a more indirect cellular response to DNA replication (initiation) perturbation that occurs in the DPB2 mutant (this hypothesis is not favored given that derepression of MBF-dependent genes in nrm1Δ cells suppresses, at least partially, some of dpb2-103 phenotypes)—(Fig. 5 in Dmowski et al. 2017). Strikingly, the premature exit of dpb2-103 cells from the G1 phase precedes a prolonged S phase observed in the analyses of DNA content (FACS) and G1/S-specific transcripts after the release from G1 block—(Fig. 6 in Dmowski et al. 2017). The premature entry into S phase (when not enough origins have been licensed) and the prolonged S phase have been observed as a result of the overexpression of Cln2 or deletion of the Sic1 inhibitor of Clb5-6 (Lengronne and Schwob 2002). This effect can be alleviated by the deletion of CLB5 and CLB6 (Lengronne and Schwob 2002). Because the MBF factor regulates various genes involved in DNA replication, their premature repression may also, at least partially, contribute to the observed S phase perturbations, the mutagenic effect of mutations in DPB2, and alleviation of dpb2-103 phenotypes by nrm1Δ. However, the elucidation of how the mutations in DPB2 affect the regulation of MBF-dependent genes and, as a consequence, genomic stability, needs further investigations.
Acknowledgements
This work was supported by Grant No. TEAM/2011-8/1 from the Foundation for Polish Science, co financed from the European Union-Regional Development Fund “New players involved in the maintenance of genomic stability”.
References
- Araki H. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr Opin Cell Biol. 2010;22:766–771. doi: 10.1016/j.ceb.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Bastos de Oliveira FM, Harris MR, Brazauskas P, et al. Linking DNA replication checkpoint to MBF cell-cycle transcription reveals a distinct class of G1/S genes. EMBO J. 2012;31:1798–1810. doi: 10.1038/emboj.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benanti JA. Create, activate, destroy, repeat: Cdk1 controls proliferation by limiting transcription factor activity. Curr Genet. 2016;62:271–276. doi: 10.1007/s00294-015-0535-5. [DOI] [PubMed] [Google Scholar]
- Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14:518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J, Cross FR. Novel role for Cdc14 sequestration: Cdc14 dephosphorylates factors that promote DNA replication. Mol Cell Biol. 2007;27:842–853. doi: 10.1128/MCB.01069-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin RAM, Kalashnikova TI, Chahwan C, et al. Constraining G1-Specific Transcription to late G1 phase: the MBF-associated corepressor Nrm1 Acts via negative feedback. Mol Cell. 2006;23:483–496. doi: 10.1016/j.molcel.2006.06.025. [DOI] [PubMed] [Google Scholar]
- de Bruin RAM, Kalashnikova TI, Wittenberg C. Stb1 collaborates with other regulators to modulate the G1-specific transcriptional circuit. Mol Cell Biol. 2008;28:6919–6928. doi: 10.1128/MCB.00211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh AS, Agarwal M, Dhar SK. Regulation of DNA replication proteins in parasitic protozoans: possible role of CDK-like kinases. Curr Genet. 2016;62:481–486. doi: 10.1007/s00294-015-0562-2. [DOI] [PubMed] [Google Scholar]
- Dmowski M, Rudzka J, Campbell JL, et al. Mutations in the non-catalytic subunit Dpb2 of DNA polymerase epsilon affect the Nrm1 branch of the DNA replication checkpoint. PLoS Genet. 2017;13(1):e1006572. doi: 10.1371/journal.pgen.1006572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncic A, Falleur-Fettig M, Skotheim JM. Distinct interactions select and maintain a specific cell fate. Mol Cell. 2011;43:528–539. doi: 10.1016/j.molcel.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua R, Levy DL, Campbell JL. Role of the putative zinc finger domain of Saccharomyces cerevisiae DNA polymerase epsilon in DNA replication and the S/M checkpoint pathway. J Biol Chem. 1998;273:30046–30055. doi: 10.1074/jbc.273.45.30046. [DOI] [PubMed] [Google Scholar]
- Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol ε and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274:22283–22288. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- Eser U, Falleur-Fettig M, Johnson A, Skotheim JM. Commitment to a cellular transition precedes genome-wide transcriptional change. Mol Cell. 2011;43:515–527. doi: 10.1016/j.molcel.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrezuelo F, Colomina N, Futcher B, Aldea M. The transcriptional network activated by Cln3 cyclin at the G1-to-S transition of the yeast cell cycle. Genome Biol. 2010;11:R67. doi: 10.1186/gb-2010-11-6-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadaleta MC, González-Medina A, Noguchi E. Timeless protection of telomeres. Curr Genet. 2016;62:725–730. doi: 10.1007/s00294-016-0599-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganai RA, Osterman P, Johansson E. Yeast DNA polymerase ε catalytic core and holoenzyme have comparable catalytic rates. J Biol Chem. 2015;290:3825–3835. doi: 10.1074/jbc.M114.615278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbacz M, Araki H, Flis K, et al. Fidelity consequences of the impaired interaction between DNA polymerase epsilon and the GINS complex. DNA Repair (Amst) 2015;29:23–35. doi: 10.1016/j.dnarep.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Grabowska E, Wronska U, Denkiewicz M, et al. Proper functioning of the GINS complex is important for the fidelity of DNA replication in yeast. Mol Microbiol. 2014;92:659–680. doi: 10.1111/mmi.12580. [DOI] [PubMed] [Google Scholar]
- Harris MR, Lee D, Farmer S, et al. Binding specificity of the G1/S transcriptional regulators in budding yeast. PLoS ONE. 2013;8:1–7. doi: 10.1371/annotation/d0132de3-56ca-4258-9119-bdab0ceb2cff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustedt N, Gasser SM, Shimada K. Replication checkpoint: tuning and coordination of replication forks in s phase. Genes (Basel) 2013;4:388–434. doi: 10.3390/genes4030388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoz I, Persson U, Volkov K, Johansson E. The C-terminus of Dpb2 is required for interaction with Pol2 and for cell viability. Nucleic Acids Res. 2012;40:11545–11553. doi: 10.1093/nar/gks880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, et al. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Jaszczur M, Flis K, Rudzka J, et al. Dpb2p, a noncatalytic subunit of DNA polymerase ε, contributes to the fidelity of DNA replication in Saccharomyces cerevisiae. Genetics. 2008;178:633–647. doi: 10.1534/genetics.107.082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, Tak YS, Sugino A, Araki H. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 2001;20:2097–2107. doi: 10.1093/emboj/20.8.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti T, McDonald WH, Yates JR, Wittenberg C. Cell cycle-dependent phosphorylation of the DNA polymerase epsilon subunit, Dpb2, by the Cdc28 cyclin-dependent protein kinase. J Biol Chem. 2004;279:14245–14255. doi: 10.1074/jbc.M313289200. [DOI] [PubMed] [Google Scholar]
- Koch C, Schleiffer A, Ammerer G. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at Start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 1996;10:129–141. doi: 10.1101/gad.10.2.129. [DOI] [PubMed] [Google Scholar]
- Kõivomägi M, Valk E, Venta R, et al. Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature. 2011;480:128–131. doi: 10.1038/nature10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Langston LD, Zhang D, Yurieva O, et al. CMG helicase and DNA polymerase ε form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc Natl Acad Sci. 2014;111:15390–15395. doi: 10.1073/pnas.1418334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Schwob E. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G1. Mol Cell. 2002;9:1067–1076. doi: 10.1016/S1097-2765(02)00513-0. [DOI] [PubMed] [Google Scholar]
- Li Y, Araki H. Loading and activation of DNA replicative helicases: the key step of initiation of DNA replication. Genes Cells. 2013;18:266–277. doi: 10.1111/gtc.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan SA, Williams JS, Kunkel TA. DNA polymerases divide the labor of genome replication. Trends Cell Biol. 2016;26:640–654. doi: 10.1016/j.tcb.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Muramatsu S, Kamimura Y, Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–655. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S, Hirai K, Tak YS, et al. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol ε, and GINS in budding yeast. Genes Dev. 2010;24:602–612. doi: 10.1101/gad.1883410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Navas TA, Zhou Z, Elledge SJ. DNA polymerase ε links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- Ostapenko D, Solomon MJ. Anaphase promoting complex-dependent degradation of transcriptional repressors Nrm1 and Yhp1 in Saccharomyces cerevisiae. Mol Biol Cell. 2011;22:2175–2184. doi: 10.1091/mbc.E11-01-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palou R, Palou G, Quintana DG. A role for the spindle assembly checkpoint in the DNA damage response. Curr Genet. 2016;63:1–6. doi: 10.1007/s00294-016-0634-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Van Deursen F, De Piccoli G, Labib K. Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr Biol. 2013;23:543–552. doi: 10.1016/j.cub.2013.02.011. [DOI] [PubMed] [Google Scholar]
- Sheu Y-J, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463:113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoneczna A, Kaniak A, Skoneczny M. Genetic instability in budding and fission yeast—sources and mechanisms. FEMS Microbiol Rev. 2015;39:917–967. doi: 10.1093/femsre/fuv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka MB, Bastos De Oliveira FM, Harris MR, De Bruin RAM. The checkpoint transcriptional response: make sure to turn it off once you are satisfied. Cell Cycle. 2012;11:3166–3174. doi: 10.4161/cc.21197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y, Kamimura Y, Okawa M, et al. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003;17:1153–1165. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Araki H. Regulation of the initiation step of DNA replication by cyclin-dependent kinases. Chromosoma. 2010;119:565–574. doi: 10.1007/s00412-010-0291-8. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Tak YS, Araki H. The role of CDK in the initiation step of DNA replication in eukaryotes. Cell Div. 2007;2:16. doi: 10.1186/1747-1028-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, et al. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Travesa A, Kuo D, de Bruin RA, et al. DNA replication stress differentially regulates G1/S genes via Rad53-dependent inactivation of Nrm1. EMBO J. 2012;31:1811–1822. doi: 10.1038/emboj.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JFX. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]