Abstract
The canonical Wnt pathway is critical for embryonic stem cell (ESC) pluripotency and aberrant control of β-catenin leads to failure of exit from pluripotency and lineage commitments. Hence, maintaining the appropriate level of β-catenin is important for the decision to commit to the appropriate lineage. However, how β-catenin links to core transcription factors in ESCs remains elusive. C-terminal-binding protein (CtBP) in Drosophila is essential for Wnt-mediated target gene expression. In addition, Ctbp acts as an antagonist of β-catenin/TCF activation in mammals. Recently, Ctbp2, a core Oct4-binding protein in ESCs, has been reported to play a key role in ESC pluripotency. However, the significance of the connection between Ctbp2 and β-catenin with regard to ESC pluripotency remains elusive. Here, we demonstrate that C-terminal-binding protein 2 (Ctbp2) associates with major components of the β-catenin destruction complex and limits the accessibility of β-catenin to core transcription factors in undifferentiated ESCs. Ctbp2 knockdown leads to stabilization of β-catenin, which then interacts with core pluripotency-maintaining factors that are occupied by Ctbp2, leading to incomplete exit from pluripotency. These findings suggest a suppressive function for Ctbp2 in reducing the protein level of β-catenin, along with priming its position on core pluripotency genes to hinder β-catenin deposition, which is central to commitment to the appropriate lineage.
Introduction
Embryonic stem cell (ESC) pluripotency is governed by intrinsic signaling pathways that include non-coding RNAs, chromatin regulators and transcriptional regulators.1, 2, 3, 4 In addition, extrinsic signaling pathways influence intrinsic signaling pathways that are important for ESC self-renewal and pluripotency.5, 6, 7, 8, 9, 10 Furthermore, ESC pluripotency is maintained by blocking differentiation-related signaling pathways using small-molecule inhibitors, which block the mitogen-activated protein kinase (Mek)/extracellular signal-related kinase (Erk1/2; Mapk3/1) pathway and the glycogen synthase kinase 3 (Gsk3) pathway.11 In particular, the Gsk3 signaling pathway negatively controls the canonical Wnt signaling pathway, which plays a crucial role in ESC pluripotency,12 via phosphorylation of β-catenin, resulting in its ubiquitylation and proteasomal degradation.13, 14
However, the requirement for Wnt in ESC pluripotency is controversial.15 Activation of Wnt signaling has been reported to maintain ESC pluripotency and to prevent exit from pluripotency. In addition, treatment with Leukemia inhibitory factor (LIF) plus Wnt produced new strained ESCs and synergistic effects on ESC maintenance and pluripotency.16, 17, 18, 19, 20 On the other hand, canonical Wnt signaling has been reported to induce mesendoderm progenitors,21, 22 indicating that Wnt signaling is involved in both ESC propagation and ESC differentiation, probably by context- and time-dependent regulation of Wnt signaling.15
β-Catenin is a key effector in the canonical Wnt signaling pathway, and it acts as a transactivator of target genes by cooperating with Tcf/Lef transcription factors.23, 24, 25 Deficiency in β-catenin did not influence ESC self-renewal26, 27 but produced defects in differentiation to form the mesendodermal germ layer formation and neuronal cell types,27 demonstrating that β-catenin is dispensable for ESC maintenance but indispensable for proper lineage commitment. However, upregulation or stabilization of β-catenin sustained ESC pluripotency upon induction of differentiation by nuclear translocation and subsequent de-repression of Tcf3 target genes26 or a subsequent increase in Oct4 activity in a Tcf3-independent manner.28 Thus, maintaining the appropriate level of β-catenin is crucial for decisions regarding the appropriate lineage commitment. However, how the level of β-catenin itself is controlled in ESCs and how nuclear β-catenin affects the activation of its target genes remains elusive.
The level of β-catenin is regulated by a destruction complex consisting of Ck1, Gsk-3β, axin1 and Apc.29, 30 Apart from Gsk3 inhibition, aberrant regulation of Apc also leads to β-catenin stabilization, which affects the exit from pluripotency,31, 32 indicating that proper control of β-catenin destruction components is essential for ESC pluripotency. β-Catenin-mediated transcription of the genes regulated by the destruction complex occurs in a cell-type- and context-dependent manner,24 suggesting that there are other components of the destruction complex in ESCs for fine-tuning the control of β-catenin with regard to ESC pluripotency.
C-terminal-binding protein-2 (Ctbp2)-knockout mice show early embryonic lethality.33 In addition, Ctbp2 was identified as a core Oct4-interacting protein in ESCs.34 Recently, Ctbp2 was revealed to control the epigenetic states of H3K27 in active ESC genes by interconnecting the NuRD complex and Oct4 in undifferentiated ESCs.35 Meanwhile, CtBP in Drosophila was revealed to be required for Wnt-mediated target gene expression during development.36 In addition, Ctbp acts as an antagonist of β-catenin activation through the assembly of the Apc/Ctbp/β-catenin complex, which lessens the amount of free nuclear β-catenin available for binding to TCF in mammals.37 However, the significance of the connection between Ctbp2 and β-catenin with regard to ESC pluripotency remains elusive.
In this study, we demonstrate that Ctbp2 associates with the major β-catenin destruction complex in ESCs. Ablation of Ctbp2 results in upregulation of β-catenin, which is directed to core ESC factors that were occupied by Ctbp2, such as Oct4 and Nanog, thereby leading to incomplete exit from pluripotency during ESC differentiation. This is a novel mechanism in which Ctbp2 controls β-catenin by reducing the expression level of β-catenin as a component of the β-catenin destruction complex, as well as by priming its position in core ESC genes to hinder β-catenin deposition, which is central to appropriate lineage commitment.
Materials and methods
Cell culture
ESCs were cultured and passaged on 0.1% gelatinized (Sigma-Aldrich, St Louis, MO, USA) plates as reported previously.38 Briefly, the mouse ESC medium was composed of KNOCKOUT DMEM (Gibco-Invitrogen, Waltham, MA, USA) and 15% (v/v) fetal bovine serum (FBS; Gibco), supplemented with 2 mM L-glutamine, 55 μM β-mercaptoethanol, 1% (v/v) non-essential amino acids, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (all from Gibco), and 1000 U ml−1 ESGRO (Millipore, Billerica, MA, USA). ESC differentiation was induced by LIF withdrawal from ESC medium in monolayer cultures.
Self-renewal assay
The self-renewal assay (colony-forming assay) was performed as described previously.39 Briefly, ESCs were trypsinized and re-plated at 600 cells per six-well plate. After incubation for 7 days with/without LIF, plates were stained for alkaline phosphatase and scored by differentiation status.
Lentiviral shRNA-mediated Ctbp2-knockdown
PLKO-puro constructs expressing shRNAs were purchased from Sigma. The shRNA constructs targeting Ctbp2 are TRCN0000109335 and TRCN0000307495. Lentivirus production was performed in 293FT cells by co-transfection of 1 μg each of pMD2.G, pMDLg/pRRE, pRSV-rev and pLKO-shRNA using Lipofectamine 2000 (Invitrogen, Waltham, MA, USA). At 48 h after transfection, virus-containing medium was collected and filtered using 0.45 μm filters. Polybrene (10 μg ml−1) was added to target cells immediately before infection, and infection was performed for 5 h. Puromycin selection (2 μg ml−1) was performed at 48 h after infection and continued for 2 days. As a control, we used pLKO.1 puro vector.
Lentiviral shRNA-mediated β-catenin-knockdown in Ctbp2-knockdown ESCs
shRNAs were purchased from Sigma. The shRNA construct targeting β-catenin is TRCN0000012691. We substituted blasticidine for puromycin in a shCtnnb1.puro vector (shCtnnb1.bla). To generate β-catenin-knockdown ESCs in Ctbp2-knockdown ESCs, Ctbp2-knockdown ESCs were infected with lentivirus containing shCtnnb1.bla, and 8 μg ml−1 blasticidine selection was performed after 48 h. Blasticidine-resistant cells were expanded and tested for β-catenin expression by western blotting.
Reporter gene assay
ESCs were cultured and transfected with the reporter plasmids TOP-FLASH and FOP-FLASH (Addgene, Cambridge, MA, USA) using Lipofectamine 2000 (Invitrogen). After 48 h, reporter gene activity was measured using Magellan software (Tecan, Switzerland).
Immunoprecipitation, western blot and immunofluorescence
Immunoprecipitation from Flag-tagged CTBP2-expressing ESCs and wild-type ESCs, western blotting, and immunofluorescence were performed as described previously.38 Anti-Ctbp1 (612042) and anti-Ctbp2 (612044) antibodies were purchased from BD Transduction Laboratories (Franklin Lakes, NJ, USA); anti-Nanog (ab14959) antibody was from Abcam (Cambridge, MA, USA); anti-Oct4 (sc-5279), Gsk-3β (sc-9166) and β-catenin (sc-7199) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, USA); anti-Apc (A300-981A) antibody was from Bethyl Laboratories (Montgomery, TX, USA); and anti-phospho Gsk-3β (S9) (9336s) antibody was from Cell Signaling (Danvers, MA, USA).
Real-time qPCR
RT-qPCR was performed as previously described.38 Briefly, total RNA was extracted using Trizol (Invitrogen), and cDNA was synthesized according to the manufacturer’s protocol (AMV Reverse Transcriptase, Takara (Mountain View, CA, USA)). Quantitative real-time-PCR was performed using the SYBR Green qPCR Kit (Finnzymes, Waltham, MA, USA, F-410L) with the iQ5 Real-time PCR Detection System (Bio-Rad, Hercules, CA, USA). We analyzed relative mRNA expression levels using the 2-ΔΔCT calculation method. Sequences of the primers for real-time PCR are listed in Supplementary Table 1.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed as described.38 Briefly, cells were cross-linked with 1% formaldehyde and lysed and sonicated. Then, 10-fold-diluted lysates in IP buffer were incubated overnight at 4 °C with protein A/G PLUS agarose and 1–2 μg of the appropriate antibodies.
Cross-linking was reversed by adding 20 μl of 5 M NaCl and incubating overnight at 65 °C. DNA was precipitated with ethanol and kept at −20 °C for later use. Sequences of the ChIP primers for real-time PCR are listed in Supplementary Table 1.
ChIP-sequencing data analysis
Mapping, identification and annotation of enriched regions
Reads after sequencing were mapped against the mouse genome (NCBI build 37/mm9) using Bowtie v.0.12.740 with the default parameters. The BAM format outputs were sorted by genomic coordinates (samtools sort), and reliable reads based on mapping scores were used in subsequent processes. GO annotation was performed using GREATv2.0.2.41 The ChIP-seq raw data are available under the accession number E-MTAB-2002.
Motif and co-bound factor analysis
HOMER was used for motif and co-bound factor analysis. De novo motif and known motif discovery algorithms were applied with default settings. We considered two factors to be co-bound when their peak centers were located within 200 bp of each other.
Results
Ctbp2 associates with the β-catenin destruction complex in ESCs
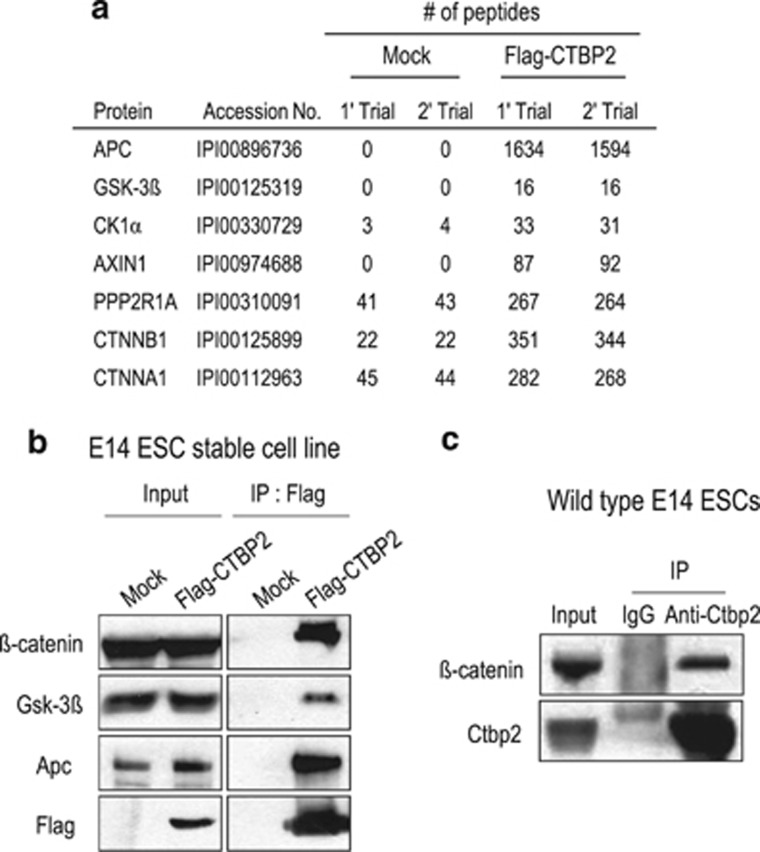
We previously isolated Ctbp2-binding proteins in ESCs by mass spectrometry and revealed Ctbp2-interacting proteins related to transcription factors and chromatin regulators that linked to stem cell pluripotency and differentiation (Supplementary Table 2).35 In addition, we identified Ctnnb1 (β-catenin) and major components of its destruction complex, such as Apc, Gsk3β, Ck1α, Axin1, Ppp2r1a and Ctnna1 (α-catenin), which play a key role in regulating the transcriptional activity and stability of β-catenin (Figure 1a).24, 25 Among these proteins, we confirmed that Ctbp2 interacted with β-catenin and major components of the β-catenin destruction complex, such as Apc and Gsk-3β, by immunoprecipitation using Flag-CTBP2-expressing ESCs that had characteristics similar to an undifferentiated state35 (Figure 1b). In addition, we observed that Ctbp2 interacted with β-catenin endogenously in wild-type ESCs (Figure 1c).
Figure 1.
Ctbp2 associates with the β-catenin destruction complex in ESCs. (a) Using mass spectrometry,35 we identified Ctbp2-interacting proteins that were β-catenin and core β-catenin destruction components in ESCs with reasonable peptide counts. (b) Using immunoprecipitation with Flag antibodies from ESCs, showing that Flag-tagged CTBP2 was stably incorporated, and endogenous β-catenin, Gsk-3β and Apc were detected with the indicated antibodies. (c) Using immunoprecipitation with Ctbp2 antibodies from wild-type ESCs, endogenous β-catenin was detected with the indicated antibodies.
Ctbp2 destabilizes β-catenin levels in ESCs
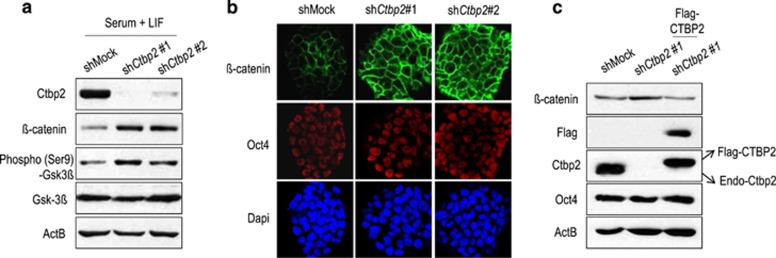
Since the β-catenin destruction complex regulates β-catenin stability,29, 30 we hypothesized that Ctbp2 is involved in β-catenin stability in ESCs. To determine whether Ctbp2 controls β-catenin stability, we produced two shRNA-mediated Ctbp2-knockdown ESC lines, which had no effect on ESC maintenance in the undifferentiated state35 (Figure 2a). We observed that the expression level of β-catenin was higher in Ctbp2-knockdown ESCs than in wild-type ESCs (Figure 2a). In addition, the level of Gsk3β that was phosphorylated at serine 9, an inactivated form of Gsk3β, was elevated upon Ctbp2-knockdown, suggesting that β-catenin destruction complex is inefficiently assembled. Using immunostaining, we confirmed that the level of β-catenin in Ctbp2-knockdown ESCs was higher than in wild-type ESCs (Figure 2b). Furthermore, rescue of CTBP2 in Ctbp2-knockdown ESCs (rescued ESCs) restored β-catenin expression (Figure 2c). Taken together, these results showed that Ctbp2 regulates the protein level of β-catenin in ESCs.
Figure 2.
Ctbp2 is required for β-catenin stability in ESCs. (a) Western blot analysis of Ctbp2, β-catenin, phospho-Gsk3β (Ser9) and Gsk3β in wild-type ESCs and two shRNA-mediated Ctbp2-knockdown ESCs with the indicated antibodies. ActB was used as an internal control. The expression levels of β-catenin and phosphor-Gsk3β (Ser9), the inactive form of Gsk3β, were upregulated in Ctbp2-knockdown ESCs compared to wild-type ESCs. (b) Immunostaining of β-catenin and Oct4 in wild-type ESCs and two shRNA-mediated Ctbp2-knockdown ESCs. (c) Western blot analysis of β-catenin with the indicated antibodies. The increased expression level of β-catenin upon Ctbp2-knockdown was rescued by Flag-tagged CTBP2 overexpression in Ctbp2-knockdown ESCs. Oct4 level was not affected by Ctbp2. ActB was used as an internal control.
Upregulated β-catenin in Ctbp2-knockdown ESCs directly targets active ESC genes
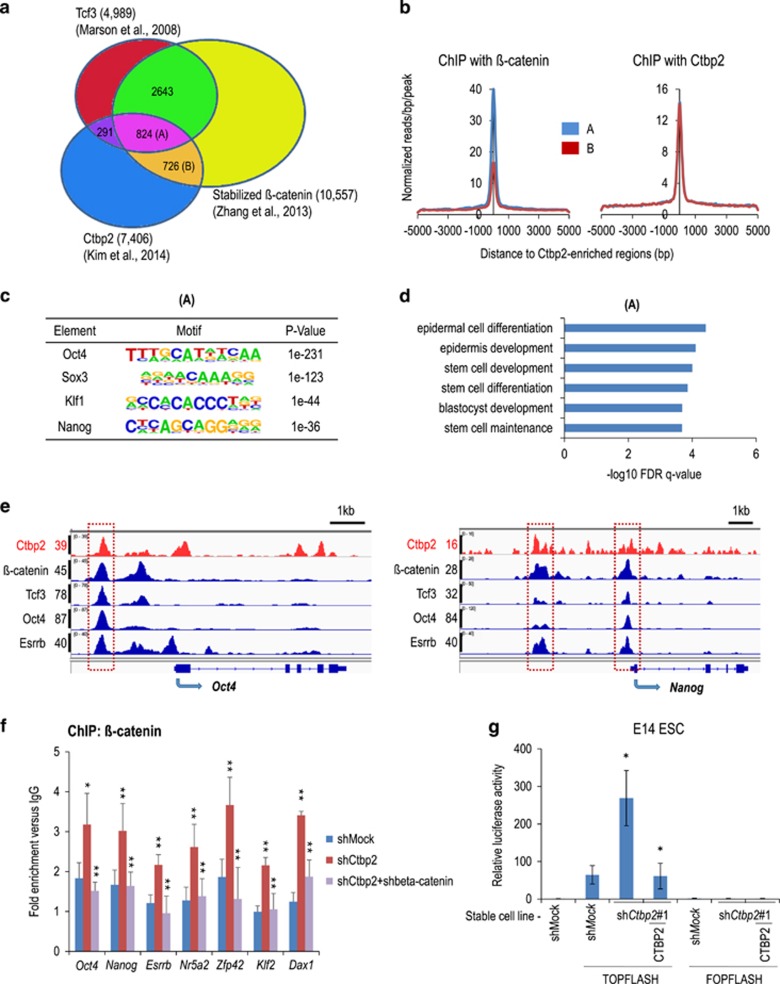
To investigate the role of the upregulation of β-catenin upon Ctbp2-knockdown in chromatin, we compared the genome-wide occupancy of Ctbp2 (Supplementary Table 3),35 Tcf342 and stabilized β-catenin43 using published ChIP-seq data to identify their target genes. We identified 824 regions that were co-occupied by Ctbp2, Tcf3 and β-catenin (A regions; Supplementary Table 4) and 726 regions that were co-occupied by Ctbp2 and β-catenin but not by Tcf3 (B regions; Supplementary Table 5) (Figure 3a). Intriguingly, we found that β-catenin occupancy was more enriched in A regions than B regions, and even Ctbp2 occupancy was comparable between them, indicating that among Ctbp2-occupied regions, stabilized β-catenin tended to be enriched in Tcf3-occupied regions compared to regions without Tcf-3 binding (Figure 3b; Supplementary Figure 1a).
Figure 3.
Upregulated β-catenin in Ctbp2-knockdown ESCs directly targets pluripotency genes. (a) Comparison of co-occupancy regions for Tcf3, stabilized β-catenin and Ctbp2 using published ChIP-seq data.35, 42, 43 In total, 824 regions were co-occupied by Tcf3, stabilized β-catenin and Ctbp2 (A regions), and 726 regions were co-occupied by stabilized β-catenin and Ctbp2 but not Tcf3 (B regions). (b) The mean enrichment levels of β-catenin and Ctbp2 in A regions versus B regions. The mean enrichment levels of β-catenin were higher in A regions than B regions, but the levels of Ctbp2 were similar in both types of regions. (c) Using motif discovery algorithms, Oct4, Sox3, Klf1 and Nanog motifs were predicted with high scores in A regions. (d) The functional analysis of A regions using GREAT showed that enrichment regions were related to epidermis development and stem cell development. (e) Integrated genomics viewer (IGV) represents Ctbp2, stabilized β-catenin, Tcf3, Oct4 and Esrrb in Oct4 and Nanog loci in ESCs. (f) ChIP-qPCR analysis of β-catenin in Ctbp2-occupied Oct4, Nanog, Esrrb, Rex1, Klf2 and NrOb1 loci regions between wild-type and Ctbp2-knockdown ESCs. Values represent the mean±standard deviation (n=3). (g) The TOP-FLASH and FOP-FLASH reporter activity were measured in wild-type, Ctbp2-knockdown and rescued ESCs. Increased β-catenin activity in Ctbp2-knockdown ESCs compared to wild-type ESCs was decreased in rescued ESCs. Value represent mean±standard deviation (n=3). *P⩽0.05, **P⩽0.01.
Based on motif discovery algorithms,44 the Oct4 motif was most significantly enriched in A regions, while the Esrrb motif was most enriched in B regions, even though its P-value was lower in B regions (P=1e−74) than in A regions (P=1e−231) (Figure 3c; Supplementary Figure 1b). In addition, based on gene ontology analysis using GREAT,41 A regions were linked to epidermis development and stem cell development (Figure 3d), while B regions were linked to chromosome and chromatin organization (Supplementary Figure 1c).
Next, we examined whether the upregulation of β-catenin upon Ctbp2-knockdown leads to the activation of ESC genes. β-Catenin was enriched at Oct4, Nanog, Esrrb, Nr5a2, Rex1, Klf2 and Dax1 loci, also sites of Ctbp2 occupation, in Ctbp2-knockdown ESCs compared to wild-type ESCs (Figure 3f), which is consistent with the ChIP-seq data analysis in which stabilized β-catenin localizes to Ctbp2-occupied loci (Figure 3e). Moreover, we found that transcriptional activity of β-catenin was significantly increased in Ctbp2-knockdown ESCs compared to wild-type ESCs. Further, such increased activity disappeared in rescued ESCs in the TOP-FLASH reporter assay, while reporter activity was not detected in the FOP-FLASH reporter assay (Figure 3g).
Collectively, these results demonstrate that Ctbp2 controls β-catenin in chromatin, which directs to core ESC genes.
Ctbp2 is indispensable for exit from pluripotency, partially through β-catenin destruction
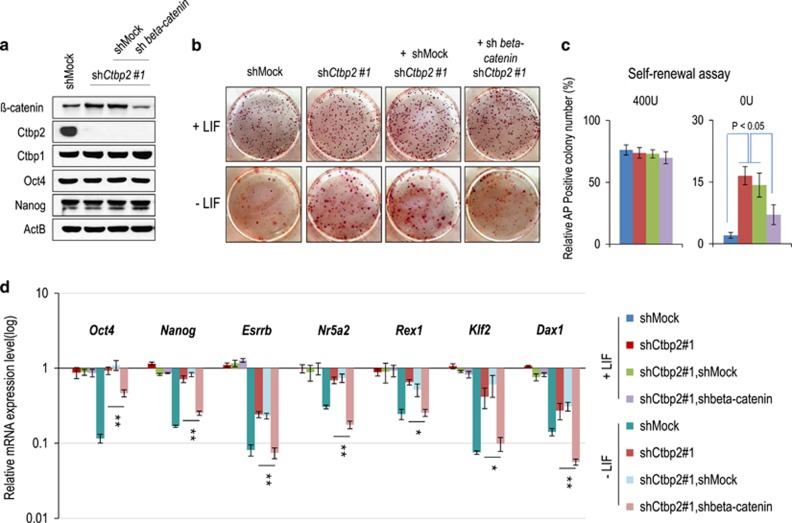
Next, we determined whether a physiological association between Ctbp2 and β-catenin affects ESC maintenance or exit from pluripotency. We have previously shown that ESCs failed to exit from pluripotency upon Ctbp2 knockdown.35 To elucidate whether this phenomenon is dependent on the up-regulation of β-catenin by Ctbp2, we produced shRNA-mediated β-catenin-knockdown ESCs in Ctbp2-knockdown ESCs, which did not affect ESC maintenance as shown by comparable expression levels of Oct4 and Nanog and similar activity of alkaline phosphatase (AP) (Figure 4a and b). The population prevented from exiting pluripotency in Ctbp2-knockdown ESCs was partially rescued upon β-catenin knockdown during ESC differentiation, as shown by the colony-formation assay (Figure 4b and c). In addition, insufficient downregulation of pluripotency-associated genes, such as Oct4, Nanog, Esrrb, Nr5a2, Rex1, Klf2 and Dax1, in Ctbp2-knockdown ESCs versus wild-type ESCs was rescued upon β-catenin knockdown in Ctbp2-knockdown ESCs during differentiation (Figure 4d). Furthermore, based on the colony-formation assay and qRT-PCR analysis of pluripotent-associated genes during ESC differentiation, administration of IWP2, which is known as a β-catenin inhibitor,45 to Ctbp2-knockdown ESCs partially restored the ability to exit from pluripotency (Supplementary Figure 2), which is consistent with the results of the knockdown of both β-catenin and Ctbp2 in ESCs (Figure 4). Taken together, we demonstrated that Ctbp2 is indispensable for exit from pluripotency in some contexts of β-catenin regulation in ESCs (Figure 5).
Figure 4.
Prevention of the exit from pluripotency in Ctbp2-knockdown ESCs was partially restored upon β-catenin knockdown in Ctbp2-knockdown ESCs. (a) Western blot analysis of β-catenin after shRNA-mediated knockdown in Ctbp2-knockdown ESCs. The increased expression level of β-catenin upon Ctbp2-knockdown was decreased upon β-catenin knockdown in Ctbp2-knockdown ESCs. Ctbp1, Oct4 and Nanog levels were not affected by either Ctbp2 or β-catenin in ESCs. ActB was used as an internal control. (b, c) Self-renewal assay and alkaline phosphatase (AP) staining in wild-type, Ctbp2-knockdown and β-catenin-knockdown in Ctbp2-knockdown ESCs. Incomplete exit from pluripotency in Ctbp2-knockdown ESCs was partially rescued upon β-catenin knockdown. (d) mRNA expression of pluripotent-associated genes of wild-type, Ctbp2-knockdown and β-catenin knockdown in Ctbp2-knockdown ESCs in an undifferentiated state (+LIF) and a differentiated state (−LIF, 6 days). Values represent the mean±standard deviation (n=3). *P⩽0.05, **P⩽0.01.
Figure 5.
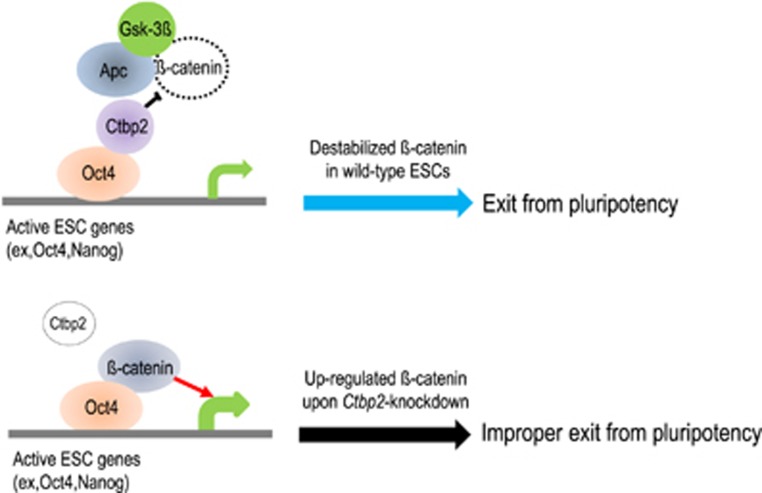
A schematic model for Ctbp2-mediated β-catenin regulation to pluripotency exit. In undifferentiated ESCs, Ctbp2, together with the β-catenin destruction complexes, decreases the protein level of β-catenin and localizes on core pluripotency genes, such as Oct4 and Nanog loci, to hinder β-catenin deposition. Upon Ctbp2-knockdown, upregulated β-catenin localizes to Ctbp2-occupied pluripotency genes and leads to improper exit from pluripotency.
Discussion
Extrinsic signaling cues and their subsequent activation of intrinsic signaling pathways determine either ESC propagation or lineage commitment. Among extrinsic signals, the canonical Wnt pathway is critical for ESC pluripotency according to the reports that aberrant control of β-catenin, a key effective protein of canonical Wnt pathway, results in failure of exit from pluripotency and lineage commitment.26, 27, 28 Hence, proper expression control of β-catenin is indispensable for cell fate decisions.
In this study, we demonstrated that Ctbp2 associates with the β-catenin destruction complex (Figure 1; Supplementary Table 2) and regulates the expression level of β-catenin (Figure 2). Upon Ctbp2 knockdown, upregulated β-catenin directly targets pluripotency-maintenance genes that were occupied by Ctbp2, and its transcriptional activity is increased (Figure 3), resulting in incomplete exit from pluripotency (Figure 4; Supplementary Figure 2).
We previously reported that Ctbp2 associates with the NuRD complex in active ESC genes for proper H3K27 acetylation, which is pivotal to commitment toward a differentiated lineage.35 Based on these results, knockdown of β-catenin appeared insufficient for the exit from pluripotency in Ctbp2-knockdown ESCs, even though the β-catenin level was similar to that in wild-type ESCs (Figure 4). This may be because Ctbp2-NuRD-mediated deacetylation in active ESC genes is critical to the exit from pluripotency upon the induction of a lineage commitment. However, the finding that Ctbp2 binds major NuRD- and Wnt-related components in ESCs (Supplementary Table 2)35 merits further study of the Ctbp2-mediated cooperative relationship between NuRD and Wnt in ESC pluripotency.
By comparing ChIP-seq data with Tcf3, stabilized β-catenin and Ctbp2, we observed that the mean peak density of β-catenin was higher at 824 regions (Supplementary Figure 4; A regions) that were co-occupied by β-catenin, Ctbp2 and Tcf3 than at 726 regions (Supplementary Figure 5; B regions) that were co-occupied by β-catenin and Ctbp2 but not by Tcf3 (Figure 3a and b; Supplementary Figure 1a). This suggests that although β-catenin could strengthen ESC pluripotency with increased Oct4 activity through a Tcf-independent mechanism,28 the effect of β-catenin on exit from pluripotency is dependent on Tcf3 in the context of Ctbp2-mediated β-catenin regulation. Consistent with this, XCtBP is known to be an XTcf3 co-repressor during Xenopus development.46
The significance of this work lies in the finding that Ctbp2 links a canonical Wnt-effector, β-catenin, with the exit from pluripotency. Based on our observations, Ctbp2 is a fine-tuning regulator of β-catenin and acts by controlling the expression level of β-catenin and by priming its position on pluripotency-maintenance genes to hinder the enrichment of β-catenin in ESCs, which is essential for successful lineage commitment. CtBP-mediated repression of Wnt target genes via Apc in normal cells was abolished in colorectal cancer cells,47 suggesting that CtBP is important for normal life span via fine-tuning the regulation of Wnt-mediated gene expression.
In conclusion, we demonstrated that Ctbp2, together with the β-catenin destruction complex, plays a central role in ESCs’ exit from pluripotency by regulating the protein level of β-catenin. We also showed that Ctbp2 on chromatin is indispensable for the exit from pluripotency by priming positions in core pluripotency genes to hinder β-catenin deposition. Finally, we showed that Ctbp2 knockdown caused β-catenin upregulation, which was directed to core pluripotency genes and ultimately led to improper exit from pluripotency, suggesting that Ctbp2 is a key factor for normal lineage commitment in some contexts of β-catenin regulation.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grants, funded by the Korean government (MSIP) (National Creative Research Laboratory Program (2012R1A3A2048767 to H-DY)).
Author contributions
TWK, SK, JS, BHK, SEL, MYS, JHK, IYH, JHL and JC: collection and assembly of data and data analysis and interpretation; EJC: data analysis and interpretation; TWK, SK and H-DY: conception, design and manuscript writing; H-DY: final approval of manuscript.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
The authors declare no conflict of interest.
Supplementary Material
References
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 2008; 132: 567–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell 2011; 144: 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell 2011; 145: 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008; 133: 1106–1117. [DOI] [PubMed] [Google Scholar]
- Pera MF, Tam PP. Extrinsic regulation of pluripotent stem cells. Nature 2010; 465: 713–720. [DOI] [PubMed] [Google Scholar]
- Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP et al. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 1988; 336: 684–687. [DOI] [PubMed] [Google Scholar]
- Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M et al. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 1988; 336: 688–690. [DOI] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev 1998; 12: 2048–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T et al. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J 1999; 18: 4261–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 2009; 460: 118–122. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J et al. The ground state of embryonic stem cell self-renewal. Nature 2008; 453: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med 2004; 10: 55–63. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly. Science 1996; 272: 1023–1026. [DOI] [PubMed] [Google Scholar]
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J 1997; 16: 3797–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol SY. Maintaining embryonic stem cell pluripotency with Wnt signaling. Development 2011; 138: 4341–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Li TG, Qi X, Zhao DF, Zhao GQ. WNT/beta-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Dev Biol 2006; 290: 81–91. [DOI] [PubMed] [Google Scholar]
- Miyabayashi T, Teo JL, Yamamoto M, McMillan M, Nguyen C, Kahn M. Wnt/beta-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc Natl Acad Sci USA 2007; 104: 5668–5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla DK, Schneider DJ, LeWinter MM, Sobel BE. wnt3a but not wnt11 supports self-renewal of embryonic stem cells. Biochem Biophys Res Commun 2006; 345: 789–795. [DOI] [PubMed] [Google Scholar]
- ten Berge D, Kurek D, Blauwkamp T, Koole W, Maas A, Eroglu E et al. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol 2011; 13: 1070–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Nishinakamura R, Iwamatsu Y, Shimosato D, Niwa H. Synergistic action of Wnt and LIF in maintaining pluripotency of mouse ES cells. Biochem Biophys Res Commun 2006; 343: 159–166. [DOI] [PubMed] [Google Scholar]
- Lindsley RC, Gill JG, Kyba M, Murphy TL, Murphy KM. Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development 2006; 133: 3787–3796. [DOI] [PubMed] [Google Scholar]
- Bakre MM, Hoi A, Mong JC, Koh YY, Wong KY, Stanton LW. Generation of multipotential mesendodermal progenitors from mouse embryonic stem cells via sustained Wnt pathway activation. J Biol Chem 2007; 282: 31703–31712. [DOI] [PubMed] [Google Scholar]
- Wray J, Hartmann C. WNTing embryonic stem cells. Trends Cell Biol 2012; 22: 159–168. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell 2006; 127: 469–480. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 2009; 17: 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R et al. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol 2011; 13: 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyashenko N, Winter M, Migliorini D, Biechele T, Moon RT, Hartmann C. Differential requirement for the dual functions of beta-catenin in embryonic stem cell self-renewal and germ layer formation. Nat Cell Biol 2011; 13: 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly KF, Ng DY, Jayakumaran G, Wood GA, Koide H, Doble BW. beta-catenin enhances Oct-4 activity and reinforces pluripotency through a TCF-independent mechanism. Cell Stem Cell 2011; 8: 214–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman D, Xu W. beta-catenin destruction complex: insights and questions from a structural perspective. Oncogene 2006; 25: 7482–7491. [DOI] [PubMed] [Google Scholar]
- Stamos JL, Weis WI. The beta-catenin destruction complex. Cold Spring Harb Perspect Biol 2013; 5: a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielman MF, Rindapää M, Gaspar C, van Poppel N, Breukel C, van Leeuwen S et al. Apc modulates embryonic stem-cell differentiation by controlling the dosage of beta-catenin signaling. Nat Genet 2002; 32: 594–605. [DOI] [PubMed] [Google Scholar]
- Atlasi Y, Noori R, Gaspar C, Franken P, Sacchetti A, Rafati H et al. Wnt signaling regulates the lineage differentiation potential of mouse embryonic stem cells through Tcf3 down-regulation. PLoS Genet 2013; 9: e1003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand JD, Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol Cell Biol 2002; 22: 5296–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch D, Vahokoski J, Groves MR, Pogenberg V, Cojocaru V, Vom Bruch H et al. A unique Oct4 interface is crucial for reprogramming to pluripotency. Nat Cell Biol 2013; 15: 295–301. [DOI] [PubMed] [Google Scholar]
- Kim TW, Kang BH, Jang H, Kwak S, Shin J, Kim H et al. Ctbp2 modulates the acetylated states of H3K27 in active ESC genes for exit from pluripotency. Stem Cells 2015; 33: 2442–2455. [DOI] [PubMed] [Google Scholar]
- Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, Cadigan KM. C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. EMBO J 2006; 25: 2735–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada F, Bienz M. The APC tumor suppressor binds to C-terminal binding protein to divert nuclear beta-catenin from TCF. Dev Cell 2004; 7: 677–685. [DOI] [PubMed] [Google Scholar]
- Jang H, Kim TW, Yoon S, Choi SY, Kang TW, Kim SY et al. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell 2012; 11: 62–74. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M et al. Nanog safeguards pluripotency and mediates germline development. Nature 2007; 450: 1230–1234. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010; 28: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 2008; 134: 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Peterson KA, Liu XS, McMahon AP, Ohba S. Gene regulatory networks mediating canonical Wnt signal-directed control of pluripotency and differentiation in embryo stem cells. Stem Cells 2013; 31: 2667–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol 2009; 5: 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannon M, Brown JD, Bates R, Kimelman D, Moon RT. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development 1999; 126: 3159–3170. [DOI] [PubMed] [Google Scholar]
- Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev 2006; 20: 586–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.