Abstract
Background
MicroRNAs are often abnormally expressed in human non‐small cell lung cancer (NSCLC) and are thought to play a critical role in the emergence or maintenance of NSCLC by binding to its target messenger RNA. We assessed the effects of miR‐155 on cell proliferation and invasion to elucidate the role played by miR‐155/PDCD 4 in NSCLC.
Methods
Quantitative reverse transcription‐PCR, Western blotting, and cell counting kit‐8, luciferase, and transwell invasion assays were conducted on a normal human bronchial epithelial cell line (BEAS‐2B) and three NSCLC cell lines (SPC‐A‐1, A549, and H2170).
Results
We confirmed that miR‐155 was upregulated, while PDCD 4 messenger RNA and protein levels were downregulated in NSCLC cell lines. miR‐155 negatively regulated PDCD 4 at both transcriptional and post‐transcriptional levels. Moreover, PDCD 4 was forecast as an assumed target of miR‐155 using bioinformatic methods and we demonstrated that PDCD 4 was a direct target of miR‐155 using luciferase reporter assays. Furthermore, PDCD 4 overexpression could restrain NSCLC proliferation and invasion induced by miR‐155.
Conclusion
Our results collectively demonstrate that miR‐155 exerts an oncogenic role in NSCLC by directly targeting PDCD 4.
Keywords: Invasion, miR‐155, NSCLC, PDCD4, proliferation
Introduction
Lung cancer is a leading cause of death globally. Non‐small cell lung cancer (NSCLC) is the most frequent type, accounting for approximately 80–85%.1, 2Squamous cell carcinoma and adenocarcinoma are the main types of NSCLC.3The five‐year overall survival (OS) rate associated with NSCLC is a dismal 11%, in spite of enormous breakthroughs in treatment techniques.4, 5 Therefore, there is an urgent need to find novel targets important to the progression and development of NSCLC.6
MicroRNAs (miRNAs) are a set of non‐coding RNA molecules that are small (<22 nt) yet significant in many biological processes.7 MiRNA usually exerts its function by means of base pairing with the 3′‐untranslated region (3′‐UTR) of corresponding genes.8 Accumulated evidence has shown that the alteration or dysfunction of miRNAs might play a vital role in the cell cycle, progression, apoptosis, autophagy, and migration and invasion.9, 10, 11 Moreover, abnormal miRNA expression has been observed in the progression and development of certain types of cancer and tumors, including esophageal and lung cancers, lymphocytic leukemia, and neuroblastoma.12, 13, 14, 15
The expression of the 64‐kDa protein PDCD4 is dramatically downregulated in numerous cancers, including colorectal, lung, gastric, and breast cancers, and is therefore generally recognized as a vital tumor suppressor.16, 17, 18, 19 To date, a set of miRNAs have been confirmed to target PDCD4, such as miR‐96 in glioma cancer, miR‐4262 in hepatocellular carcinoma, and miR‐499 in oropharyngeal cancer.20, 21, 22 Although miR‐155 and PDCD4 have been shown to play distinct roles in NSCLC, the exact mechanism of miR‐155/PDCD4 is not yet clear.16, 23 Therefore, we assessed the effects of miR‐155and PDCD4 on cell proliferation and invasion in NSCLC.
Methods
Cell culture
We cultured a normal human bronchial epithelial cell line (BEAS‐2B), as well as three NSCLC cell lines (SPC‐A‐1, A549, and H2170; Invitrogen, Carlsbad, CA, USA) in Dulbecco's modified Eagle medium containing 10% fetal bovine serum at 37°C with 5% CO2.
Tissue samples
Between 2015 and 2016, we collected a total of 26 primary NSCLC and adjacent non‐cancerous tissues from the Department of Cardiothoracic Surgery, the First Affiliated Hospital of Nanjing Medical University. All patients provided informed consent before surgery. All tissue specimens were snap frozen immediately in liquid nitrogen. The patients did not undergo chemotherapy or radiotherapy before surgery. Experienced pathologists histologically determined both cancerous and adjacent non‐tumor tissues. This project passed Nanjing Medical University ethical censorship.
Extraction of RNA and quantitative reverse transcription‐PCR
We isolated total RNA from human tissues and cell lines using Trizol (Invitrogen). We performed quantitative real‐time PCR (qRT‐PCR) experiments on an ABI 7500 version of a fast real‐time PCR system (Stratagene, La Jolla, CA, USA) with a high‐specificity miR‐155 qRT‐PCR detection kit (Stratagene), in accordance with the manufacturer's protocol. U6 small nuclear RNA was chosen as a built‐in control. A 2△CT method was applied to calculate the relative gene expression level.
Plasmid construction
We obtained miR‐155 mimics, anti‐miR‐155 mimics, and their negative control oligonucleotides (miR‐155 NC and anti‐miR‐155 NC) from RiboBio (Guangzhou, China). We then amplified 3′‐UTR of PDCD4 messenger RNA utilizing the follow primers: forward 5′‐GAATCTAGAATATAAGAACTCTTGCAGTC‐3′ and reverse 5′‐CTTCTAGAACCAGGTTCATTTTCC‐3′. The PCR products obtained after amplification were implanted into the pGL3 control vector (Promega, Madison, WI, USA). A fast mutation kit (NEB, Ipswich, Canada) was then used for mutation experiments. Finally, we inserted PDCD4 into the specific region of pcDNA 3.1and pcDNA3.1‐PDCD4 was ultimately constructed.
Cell transfection
We divided the transfected cells into seven groups in accordance with the treatment applied: (i) miR‐155, (ii) miR‐155 NC, (iii) anti‐miR‐155, (iv) anti‐miR‐155 NC, (v) PDCD4 3′‐UTR‐wild, (vi) PDCD4 3′‐UTR‐mut, and (vii) pcDNA3.1‐PDCD4. We performed all transfection experiments using Lipofectamin 2000 (Invitrogen). The selection of cells transfected with miR‐155 or PDCD4 was carried out using G418 and finally, we obtained the stable transfected cells.
Western blot
Total proteins were extracted from cultured cells using a RIPA Lysis Buffer (Beyotime, Shanghai, China). A dismembrator was used to homogenize 50–100 mg of tissues to extract proteins. Protein concentration was detected using a BCA Protein Assay Kit (BioRad, Hercules, CA, USA). Equal doses of protein were obtained from samples and were transfected onto polyvinylidene fluoride membrane after sodium dodecyl sulfate‐polyacrylamide gel electrophoresis integration. The membranes were immersed in tris‐buffered saline plus tween 20 with 5% non‐fat milk for two hours. The membranes were then incubated with diluted primary antibody against PDCD4 (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) at 1:400 overnight at 4°C and then with the secondary antibody for 1 hour at room temperature. The signals were visualized using ECL reagents (Pierce Biotechnology, Rockford, IL, USA).
Cell counting kit‐8 and tumor formation test
Cell proliferation was evaluated using cell counting kit‐8 assay (CCK‐8, Dojindo Molecular Technologies, Kumamoto, Japan). We seeded the cells into 96‐well plates at 2 × 103 cells per well. The proliferation rate was measured at zero, 24, 48, and 72 hours after cell transfection. A microplate reader (BioRad) was used to determine the absorbance at 450 nm. BALB/C nude mice aged five weeks were purchased from the Experiment Animal Center of Nanjing Medical University. The Institutional Animal Care and Treatment Committee of Nanjing Medical University approved all animal protocols. We gathered the A549 cells during the logarithmic phase. Cells were washed twice using phosphate buffered saline and resuspended at a concentration of 2 × 107 cells/mL. The female nude mice were then subcutaneously injected with 0.1 mL of the suspended cells at either side of their flank. The size of the tumor was measured twice a week.
Luciferase test
We seeded the cells into 24‐well plates with 2 × 105 cells per well. Cells were co‐transfected 24 hours later with PDCD4 3′‐UTR wild or PDCD4 3′‐UTR mut and anti‐miR‐155 or anti‐miR‐155 NC. We harvested the cells 48 hours after transfection and treated them using a Dual‐Luciferase Reporter Assay Kit (Promega) following the manufacturer's instructions.
Transwell invasion assay
Cell transfection was conducted with miR‐155, miR‐155 NC, and pcDNA3.1‐PDCD4, respectively. The transfected cells (2 × 105) were placed in transwell chambers precoated with 20 μg Matrigel. The medium containing 10% fetal bovine serum located in the lower chamber was treated as chemoattractant. Non‐invading cells above the membrane were scraped off after 24 hours of incubation. The invasive cells that were attached to the bottom were tinted with 0.05% crystal violet. We then counted the number of invaded cells under an inverted microscope at ×200 magnification. Each assay was carried out in triplicate.
Statistical analysis
All statistical analyses were performed using SPSS version 21.0 (IBM Corp., Armonk, NY, USA). Mean values ± standard deviations were presented in all data. A student's t‐test was applied to assess differences. A P value of < 0.05 was considered statistically significant.
Results
MiR‐155 was upregulated in non‐small cell lung cancer (NSCLC) cell lines, while PDCD 4 messenger RNA and protein levels were downregulated
We tested messenger RNA (mRNA) and protein in PDCD4 and miR‐155 expression in four cell lines: a normal human bronchial epithelial cell line (BEAS‐2B) and three NSCLC cell lines (A549, H2170, and SPC‐A‐1). The level of miR‐155 expression in normal cells was lower than in NSCLC cell lines (Fig 1a). Inversely, the PDCD4 mRNA level was higher (Fig 1b). Western blot results demonstrated a higher level of PDCD4 protein in normal cells than in cancer cell lines (Fig 1c,d). These results indicated notable upregulation of miR‐155 expression in NSCLC cell lines and downregulation of PDCD4 mRNA and protein levels.
Figure 1.
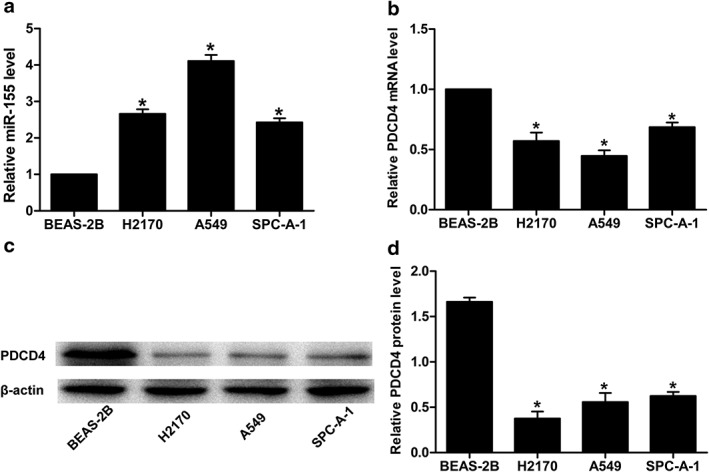
Relative (a) miR‐155 expression, (b) PDCD 4 messenger RNA (mRNA) and (c) PDCD 4 protein levels in three non‐small cell lung cancer cell lines (A549, H2170, and SPC‐A‐1) and a normal human bronchial epithelial cell line (BEAS‐2B). U6 small nuclear RNA, glyceraldehyde 3‐phosphate dehydrogenase, and β‐actin were used as internal controls, respectively. (d) Relative grayscale values of Western blotting were calculated to analyze the PDCD 4 protein levels in the four cell lines (*P < 0.05).
Downregulation of PDCD 4 and negative correlation with miR‐155 expression in NSCLC tissues
To further prove that miR‐155 negatively regulates PDCD4 not only at a transcriptional level but also at a post‐transcriptional level, we performed qRT‐PCR and Western blotting to analyze PDCD4 mRNA, PDCD4 protein, and miR‐155 expression in resected tumors and adjacent normal tissues in 26 NSCLC patients. The level of PDCD4 protein was notably lower in the tumor tissues than in the corresponding normal tissues (Fig 2a,b). Analogously, low levels of PDCD4 mRNA were observed in the tumor tissues compared to the adjacent normal tissues (Fig 2d). In contrast, we detected that miR‐155 was upregulated in NSCLC tissues compared to normal tissues (Fig 2c). This in vivo data further indicated that miR‐155 negatively regulates PDCD4 at both transcriptional and post‐transcriptional levels.
Figure 2.

PDCD4 protein levels in representative examples of eight out of 26 matched tumor tissues/normal (T/N) were analyzed via (a) Western blotting and (b) by calculating the grayscale Western blot values; β‐actin was used as an internal control. (c) Relative miR‐155 expression and (d) average level of PDCD 4 messenger RNA (mRNA) in non‐small cell lung cancer (NSCLC) (n = 26) and adjacent non‐tumor tissues (n = 26) assessed by quantitative reverse transcription (qRT)‐PCR. U6 small nuclear RNA and glyceraldehyde 3‐phosphate dehydrogenase were used as endogenous controls, respectively (*P < 0.05).
PDCD 4 is a direct target of miR‐155
We presented the 3′‐UTR of PDCD4 and its possible binding site with miR‐155 based on the bioinformatics yielded by TargetScan and miRanda (Fig 3a). We then conducted Western blotting and a luciferase test. Western blotting demonstrated that PDCD4 was distinctly upregulated in H2170 and A549 cells after transfection with anti‐miR‐155 mimic (Fig 3b). Moreover, we further conducted a luciferase reporter test to verify whether the 3′‐UTR of PDCD4 was a functional target of miR‐155 in NSCLC. pGL3‐PDCD4 3′‐UTR wild and pGL3‐PDCD4 3′‐UTR mut were established (Fig 3a). When co‐transfected with anti‐miR‐155, significantly increased luciferase activity containing PDCD4 3′‐UTR was observed in H2170 and A549 cells (Fig 3c,d). Thus, we concluded that miR‐155 combined with site of the 3′‐UTR sequence of PDCD4 regulates the PDCD4 level and PDCD4 serves as a direct target of miR‐155.
Figure 3.
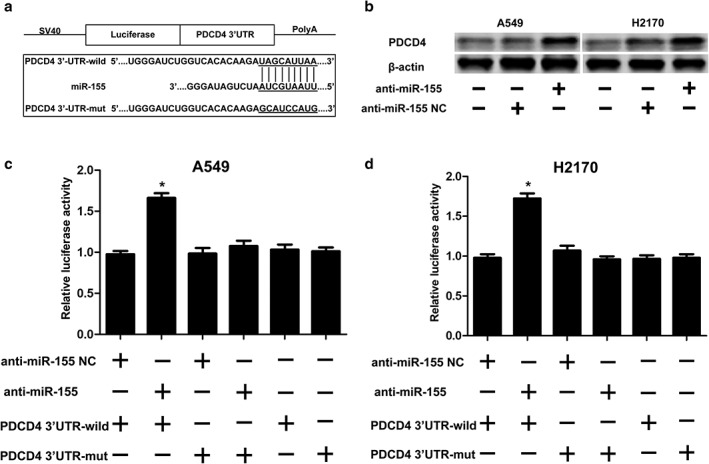
PDCD 4 was identified as a direct target of miR‐155 in non‐small cell lung cancer cell lines. (a) The predicted PDCD 4 3′‐untranslated region (UTR)‐wild and PDCD4 3′‐UTR‐mut binding sequences in miR‐155. (b) PDCD 4 protein levels were analyzed by Western blot. β‐actin was used as an internal control. After co‐transfection with miR‐155 NC or miR‐155 and PDCD 4 3′‐UTR‐wild or PDCD 4 3′‐UTR‐mut (reporter vectors), the luciferase activity in (c) A549 and (d) H2170 cells was analyzed. The measured luciferase activity was normalized to renilla luciferase activity (*P < 0.05).
Upregulated expression of PDCD 4 could restrain the tumor promoting effect of miR‐155 in NSCLC
Western blotting, CCK‐8 testing, tumor formation in nude mouse models, and transwell invasion assay were performed in order to further explore the correlation between PDCD4 and miR‐155. After miR‐155 NC or miR‐155 were co‐transfected with pcDNA3.1‐PDCD4, the H2170 and A549 cells all displayed high PDCD4 protein levels (Fig 4a,b). After CCK‐8 testing and transfection with pcDNA3.1‐PDCD4, the absorbance value in H2170 and A549 cells was reduced, but after co‐transfection with miR‐155 we did not observe any growth (Fig 4c,d). Furthermore, we set up four types of nude mouse models by injecting A549 cells into either side of the flank of female nude mice. Our results demonstrated that the tumor was reduced after transfection with pcDNA3.1‐PDCD4, but co‐transfection with miR‐155 did not reverse this phenomenon (Fig 4e). Regarding transwell invasion assay, after transfection with miR‐155, the average number of H2170 or A549 cells penetrating the transwell membrane increased. However, the average number of transwell cells after co‐transfection with pcDNA3.1‐PDCD4 and miR‐155 reduced, which meant that the invasion effect of miR‐155 in NSCLC was abrogated (Fig 4f,g). Overall, these results imply that overexpression of PDCD4 restrained the tumor‐promoting effect of miR‐155 in NSCLC.
Figure 4.
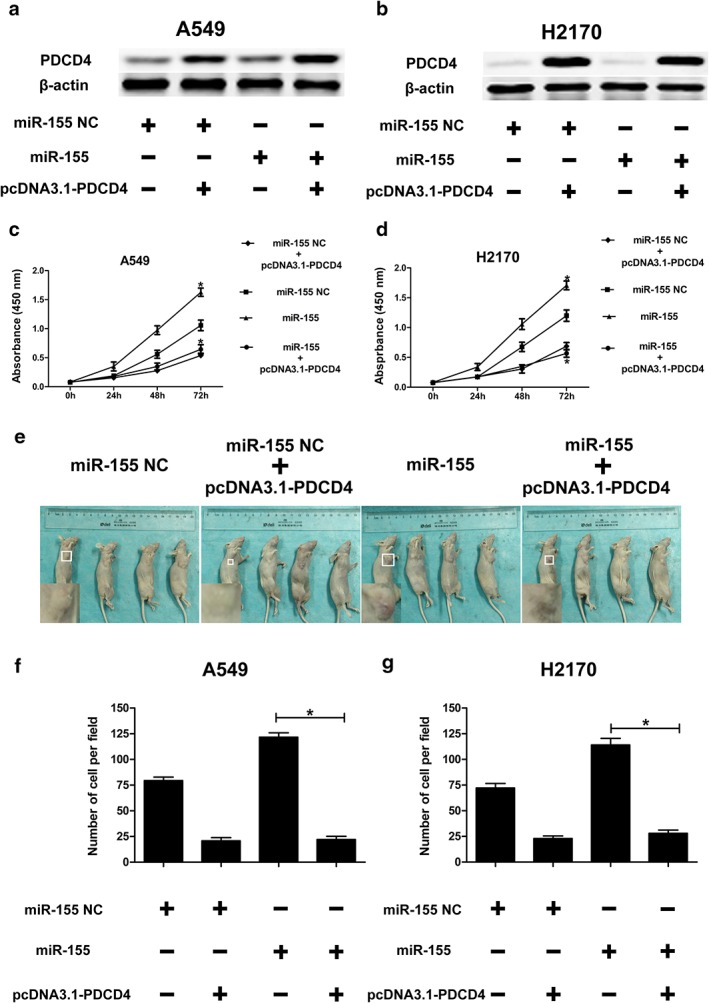
PDCD 4 overexpression restrains proliferation and invasion induced by miR‐155. After co‐transfection with miR‐155 NC or miR‐155 and pcDNA3.1‐PDCD 4, the PDCD 4 protein level was analyzed by Western blot in (a) A549 and (b) H2170 cells. β‐actin was used as an internal control. Cell growth activity after co‐transfection with miR‐155 NC or miR‐155 and pcDNA3.1‐PDCD 4 was assessed using CCK‐8 assays in (c) A549 and (d) H2170 cells (*P < 0.05). (e) Six weeks after injecting the mice with miR‐155 NC, miR‐155 NC + pcDNA3.1‐PDCD 4, miR‐155, or miR‐155 + pcDNA3.1‐PDCD 4 group cells (2 × 106 A549 or H2170 cells), the nude mice were all sacrificed to compare the tumor volume in the four different groups (*P < 0.05). The average numbers of (f) A549 and (g) H2170 cells penetrating the transwell membrane were analyzed using transwell invasion assay (*P < 0.05).
Discussion
Accumulating evidence suggests that miRNAs function as either tumor suppressors or oncogenes by regulating various biological processes of cancer cells, such as cell proliferation, apoptosis, migration, and invasion.24, 25 Numerous studies have described the effect of miR‐155 in regulating the progression of varied tumors.26, 27There has been a trend to define miR‐155 as a marker of solid and hematological malignancies for diagnosis and prognosis;28however, the exact role miR‐155 plays in the occurrence and progression of NSCLC remains unknown. Therefore, illuminating the molecular mechanism in the occurrence and prognosis of NSCLC is of great importance.
We demonstrated that miR‐155 is upregulated in NSCLC cell lines, while PDCD4 mRNA and protein levels are downregulated. Furthermore, our results showed that PDCD4 was downregulated in NSCLC tumor tissues and was negatively correlated with miR‐155 expression. Luciferase reporter testing revealed that miR‐155 exerts its tumor‐promoting effect by directly targeting PDCD4. PDCD4 overexpression reversed the malignant phenotypes of NSCLC, thus it may be a potential target for NSCLC treatment. In accordance with the results of previous studies, we confirmed that PDCD4 is a tumor suppressor.29, 30 PDCD4 suppresses cell invasion in colon cancer by inhibiting the expression of mitogen‐activated protein kinase. It has also been reported that PDCD4 could inhibit cell invasion in breast cancer by suppressing metalloproteinase 2.31, 32 These results indicate that emphasis should be placed on an exploration of the relationship between PDCD4 and miR‐155, which may cast light on the pathogenesis of NSCLC.
In conclusion, our results demonstrate the effects of miR‐155 on multiplication and invasion by targeting PDCD4 in NSCLC. miR‐155 plays a crucial role in NSCLC tumorigenesis and could represent a potential NSCLC treatment strategy. miR‐155 and PDCD4 (protein and mRNA) have contrasting expression levels in NSCLC tissues and cell lines. PDCD4 is a functional target for miR‐155 and regulates proliferation or invasion by targeting PDCD4 in NSCLC. Therefore, our data indicate that downregulating miR‐155 or upregulating PDCD4 could be potential treatments strategies for NSCLC.
Disclosure
No authors report any conflict of interest.
References
- 1. Rivera MP. Multimodality therapy in the treatment of lung cancer. Semin Respir Crit Care Med 2004; 25 (Suppl. 1): 3–10. [DOI] [PubMed] [Google Scholar]
- 2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- 3. Shtivelman E, Hensing T, Simon GR et al Molecular pathways and therapeutic targets in lung cancer. Oncotarget 2014; 5: 1392–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scagliotti GV, Novello S. Adjuvant therapy in completely resected non‐small‐cell lung cancer. Curr Oncol Rep 2003; 5: 318–25. [DOI] [PubMed] [Google Scholar]
- 5. Stinchcombe TE, Fried D, Morris DE, Socinski MA. Combined modality therapy for stage III non‐small cell lung cancer. (Published erratum appears in Oncologist 2006; 11: 958.) Oncologist 2006; 11: 809–23. [DOI] [PubMed] [Google Scholar]
- 6. Socinski MA. Seeking new options for the treatment of small‐cell lung cancer. Lung Cancer 2005; 50 (Suppl. 1): S25–6. [DOI] [PubMed] [Google Scholar]
- 7. Jiang YW, Chen LA. microRNAs as tumor inhibitors, oncogenes, biomarkers for drug efficacy and outcome predictors in lung cancer (review). Mol Med Rep 2012; 5: 890–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iorio MV, Casalini P, Piovan C, Braccioli L, Tagliabue E. Breast cancer and microRNAs: Therapeutic impact. Breast 2011; 20 (Suppl. 3): S63–70. [DOI] [PubMed] [Google Scholar]
- 9. Piovan C, Palmieri D, Di Leva G et al Oncosuppressive role of p53‐induced miR‐205 in triple negative breast cancer. Mol Oncol 2012; 6: 458–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang D, Liu D, Gao J et al TRAIL‐induced miR‐146a expression suppresses CXCR4‐mediated human breast cancer migration. FEBS J 2013; 280: 3340–53. [DOI] [PubMed] [Google Scholar]
- 11. Yang S, Li Y, Gao J et al MicroRNA‐34 suppresses breast cancer invasion and metastasis by directly targeting Fra‐1. Oncogene 2013; 32: 4294–303. [DOI] [PubMed] [Google Scholar]
- 12. Iorio MV, Ferracin M, Liu CG et al MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65: 7065–70. [DOI] [PubMed] [Google Scholar]
- 13. Iorio MV, Visone R, Di Leva G et al MicroRNA signatures in human ovarian cancer. Cancer Res 2007; 67: 8699–707. [DOI] [PubMed] [Google Scholar]
- 14. Schetter AJ, Leung SY, Sohn JJ et al MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008; 299: 425–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yanaihara N, Caplen N, Bowman E et al Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006; 9: 189–98. [DOI] [PubMed] [Google Scholar]
- 16. Chen Y, Knösel T, Kristiansen G et al Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J Pathol 2003; 200: 640–6. [DOI] [PubMed] [Google Scholar]
- 17. González‐Villasana V, Nieves‐Alicea R, McMurtry V, Gutiérrez‐Puente Y, Tari AM. Programmed cell death 4 inhibits leptin‐induced breast cancer cell invasion. Oncol Rep 2012; 27: 861–6. [DOI] [PubMed] [Google Scholar]
- 18. Liu Y, Tan X, Wang Z et al Down‐regulation of tumor suppressor PDCD4 expression in endometrium of adenomyosis patients. Curr Res Transl Med 2016; 64: 123–8. [DOI] [PubMed] [Google Scholar]
- 19. Yang HS, Matthews CP, Clair T et al Tumorigenesis suppressor Pdcd4 down‐regulates mitogen‐activated protein kinase kinase kinase kinase 1 expression to suppress colon carcinoma cell invasion. Mol Cell Biol 2006; 26: 1297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu S, Wu J, Gao Y, Han G, Ding W, Huang X. MicroRNA‐4262 activates the NF‐kappaB and enhances the proliferation of hepatocellular carcinoma cells. Int J Biol Macromol 2016; 86: 43–9. [DOI] [PubMed] [Google Scholar]
- 21. Ma QQ, Huang JT, Xiong YG, Yang XY, Han R, Zhu WW. MicroRNA‐96 regulates apoptosis by targeting PDCD4 in human glioma cells. Technol Cancer Res Treat 2017; 16: 92–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang X, Gee H, Rose B et al Regulation of the tumour suppressor PDCD4 by miR‐499 and miR‐21 in oropharyngeal cancers. BMC Cancer 2016; 16: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hou L, Chen J, Zheng Y, Wu C. Critical role of miR‐155/FoxO1/ROS axis in the regulation of non‐small cell lung carcinomas. Tumour Biol 2016; 37: 5185–92. [DOI] [PubMed] [Google Scholar]
- 24. Teoh SL, Das S. The role of microRNAs in diagnosis, prognosis, metastasis and resistant cases in breast cancer. Curr Pharm Des 2017; 23: 1845–59. [DOI] [PubMed] [Google Scholar]
- 25. Takahashi RU, Prieto‐Vila M, Hironaka A, Ochiya T. The role of extracellular vesicle microRNAs in cancer biology. Clin Chem Lab Med 2017; 55: 648–56. [DOI] [PubMed] [Google Scholar]
- 26. Lawrie CH. MicroRNAs and lymphomagenesis: A functional review. Br J Haematol 2013; 160: 571–81. [DOI] [PubMed] [Google Scholar]
- 27. Yu DD, Lv MM, Chen WX et al Role of miR‐155 in drug resistance of breast cancer. Tumour Biol 2015; 36: 1395–401. [DOI] [PubMed] [Google Scholar]
- 28. Liang H, Dong Z, Liu JF, Chuang W, Gao LZ, Ren YG. Targeting miR‐155 suppresses proliferation and induces apoptosis of HL‐60 cells by targeting Slug/PUMA signal. Histol Histopathol 2017; 32: 899–907. [DOI] [PubMed] [Google Scholar]
- 29. Leupold JH, Yang HS, Colburn NH, Asangani I, Post S, Allgayer H. Tumor suppressor Pdcd4 inhibits invasion/intravasation and regulates urokinase receptor (u‐PAR) gene expression via Sp‐transcription factors. Oncogene 2007; 26: 4550–62. [DOI] [PubMed] [Google Scholar]
- 30. Schmid T, Jansen AP, Baker AR, Hegamyer G, Hagan JP, Colburn NH. Translation inhibitor Pdcd4 is targeted for degradation during tumor promotion. Cancer Res 2008; 68: 1254–60. [DOI] [PubMed] [Google Scholar]
- 31. Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR‐21 in breast cancer cells. J Biol Chem 2008; 283: 1026–33. [DOI] [PubMed] [Google Scholar]
- 32. Matsuhashi S, Hamajima H, Xia J et al Control of a tumor suppressor PDCD4: Degradation mechanisms of the protein in hepatocellular carcinoma cells. Cell Signal 2014; 26: 603–10. [DOI] [PubMed] [Google Scholar]


