Abstract
The AML1/Runx1 transcription factor and its heterodimerization partner CBFβ are essential regulators of myeloid differentiation. The chromosomal translocation t(8;21), fusing the DNA binding domain of AML1 to the corepressor eight-twenty-one (ETO), is frequently associated with acute myeloid leukemia and generates the AML1/ETO (AE) fusion protein. AE represses target genes usually activated by AML1 and also affects the endogenous repressive function of ETO at Notch target genes. In order to analyze the contribution of CBFβ in AE-mediated leukemogenesis and deregulation of Notch target genes, we introduced two point mutations in a leukemia-initiating version of AE in mice, called AE9a, that disrupt the AML1/CBFβ interaction (AE9aNT). We report that the AE9a/CBFβ interaction is not required for the AE9a-mediated aberrant expression of AML1 target genes, while upregulation/derepression of Notch target genes does require the interaction with CBFβ. Using retroviral transduction to express AE9a in murine adult bone marrow-derived hematopoietic progenitors, we observed that both AE9a and AE9aNT lead to increased myeloproliferation in vivo. However, both development of leukemia and long-term replating capacity are only observed with AE9a but not with AE9aNT. Thus, deregulation of both AML1 and Notch target genes is required for the development of AE9a-driven leukemia.
Introduction
Acute myeloid leukemia (AML) requires the cooperation of proliferative signals in hematopoietic stem and progenitor cells and defects in myeloid differentiation.1, 2 AML is genetically heterogeneous and frequently characterized by chromosomal rearrangements that produce fusion proteins with aberrant transcriptional regulatory activities. The t(8;21)(q22;q22) translocation, present in 10–15% of AML cases, generates the AML1/ETO (AE) fusion protein. Expression of the AE fusion protein in vivo induces a pre-leukemic state,3, 4 but the underlying molecular and biological mechanisms used by AE for AML initiation still remain unclear.
The transcription factor AML1 (also known as Runx1) is an essential regulator of both fetal and adult hematopoiesis.5, 6 AML1 is required for the development of hematopoietic stem and progenitor cells.5 The DNA-binding Runt domain of AML1 heterodimerizes with the CBFβ protein, forming a complex called core binding factor. The consensus DNA-binding site of core binding factor has been identified as a 7 bp sequence ‘TGTGGTPy’.7, 8 Eight-twenty-one (ETO, also known as MTG8) protein is a member of the MTG family of transcriptional corepressors.9 Being the homolog of the Drosophila melanogaster protein Nervy,10 the ETO protein contains conserved nervy homology regions (NHR). NHR2 forms an amphipathic helix important for homodimerization, whereas NHR4 contains two putative zinc fingers, required for the interaction with nuclear receptor corepressor 1 and nuclear receptor corepressor 2 (also called SMRT (silencing mediator for retinoid or thyroid-hormone receptors))11, 12 linking ETO to histone deacetylases.12, 13
The t(8;21) translocation fuses the Runt domain of AML1 in frame with almost all of the ETO protein14, 15, 16, 17 (see also Figure 1a), leading to deletion of the carboxy-terminal transactivation domain of AML1. The resulting AE protein binds to AML1 binding sites,18 but, in contrast to AML1, represses its target genes.19, 20 The exact role for CBFβ in the transformation process driven by AE is still unclear.21, 22, 23
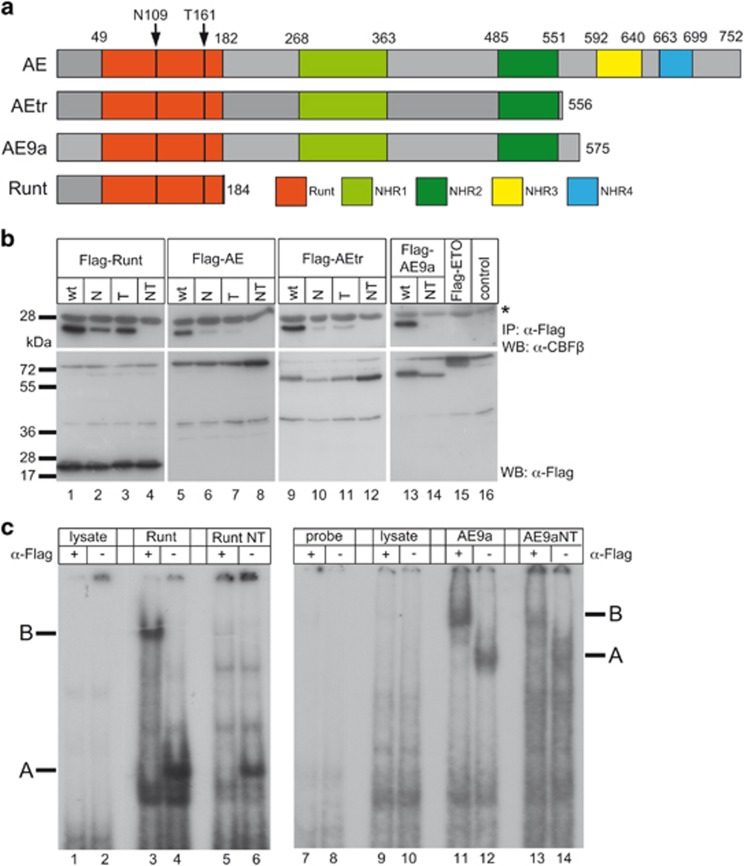
Figure 1.
AE but not AE9aNT forms a heterodimer with CBFβ but both proteins bind to DNA. (a) Representation of the proteins used in this study: Full-length AE, AEtr, splice-variant AE9a and DNA-binding Runt domain of AML1. The arrows indicate the location of the two mutations within the Runt domain, asparagine 109 (N109) and threonine 161 (T161) to alanines. (b) Coimmunoprecipitation experiments of endogenous CBFβ together with wt, single mutant (N or T) or double mutant (NT) of Runt, AE, AEtr or AE9a. The wt proteins (runt, AE, AEtr and AE9a) strongly interact with endogenous CBFβ (upper panels, lanes 1, 5, 9 and 13), whereas single N or T mutants interact only weakly (upper panels, lanes 2, 3, 6, 7, 10 and 11); the NT double mutant no longer binds to CBFβ (upper panels, lanes 4, 8, 12 and 14). Input controls are shown in the lower panels. Flag-ETO was used as negative control (lane 15). The asterisk denotes the light chain of the antibody used for immunoprecipitation. (c) Electrophoretic mobility shift assays showing that Runt and Runt NT (lanes 3 to 6) as well as AE9a and AE9aNT (lanes 11 to 14) bind to DNA (see label ‘A’). Addition of an anti-Flag antibody resulted in super shifted complexes (see label ‘B’). Flag-tagged protein expression was verified by western blot shown in Supplementary Figures S1d and e.
Different mouse models have been used to investigate the function of AE. The knock-in of AE in the endogenous AML1 locus inhibited hematopoiesis, resulting in lethality.24, 25 A conditional AE knock-in approach resulted in enhanced replating capacity of myeloid progenitors without blocking their differentiation or induction of leukemia.3, 26 Furthermore, AE requires additional mutations to induce leukemogenesis in a murine model in vivo.4 Two more recently identified distinct carboxy-terminal truncation variants of AE (AEtr and AE9a) though directly induce leukemia in mice.27, 28, 29 While AEtr is generated as consequence of a single-nucleotide insertion in mice,27 AE9a is a splice variant detected in AML patients and coexpressed with the full-length AE.28 Surprisingly, the repressive NHR3 and NHR4 domains are deleted in AE9a, indicating that, in contrast to previous suggestions, AE does not primarily act as a dominant negative regulator of AML1 target genes to induce leukemia.25, 30 We already demonstrated that ETO also represses, in a complex with the RBP-J binding protein SHARP (SMRT and HDAC associated repressor protein, also called Spen), Notch target genes, and this repressive function is impaired by AE.31 In order to further investigate the molecular mechanisms of AE9a-dependent transformation, here we dissected its dual role in the deregulation of the AML1-activating and the ETO-repressing gene regulatory role. Our data demonstrate that deregulation of both Notch and AML1 target genes is required for leukemia initiation by AE9a.
Materials and methods
Cell culture and preparation of cell extracts
Cell lines HEK-293 (ATCC CRL 1573), HeLa (ATCC CCL 2) and 3T3 (ATCC CRL 1658), as well as the Phoenix packaging cells (Orbigen, Inc., San Diego, CA, USA), were grown in Dulbecco’s Modified Eagle Medium (Thermo Fisher Scientific/Gibco, Waltham, MA, USA) supplemented with 10% fetal calf serum, and penicillin/streptomycin. HoxB4 cells (kindly provided by Drs Simona Saccani and Dominic van Essen and described in Stump et al.32) were grown in Iscove’s Modified Dulbecco Medium (Thermo Fisher Scientific/Gibco) supplemented with 2% fetal calf serum, 1% interleukin 3, 0.5% interleukin 6, 20 ng/ml stem cell factor, 0.3 mg/l peptone, 5 mg/l insulin, nonessential amino acids, β-mercaptoethanol and penicillin/streptomycin. Cells were grown at 37 °C with 5% CO2. For western blot and coimmunoprecipitation experiments, whole-cell lysates were prepared as previously described.31 Protein concentrations were determined using the Bradford assay method (Bio-Rad, Hercules, CA, USA). Details on the retroviral infection of HoxB4 cells are provided in Supplementary Information.
DNA transfection
HEK-293 and HeLa cells were transfected using the Nanofectin transfection reagent (PAA, Pasching, Austria). For transfection of Phoenix cells, the Calcium Phosphate Transfection Kit (Thermo Fisher Scientific/Invitrogen, Waltham, MA, USA) was used. All transfections were performed according to the manufacturer’s instructions.
Plasmids
The following plasmids were previously described: pGa981/6, pcDNA3-Flag-SPOC, pcDNA3-RBP;33 pcDNA3-Flag-SPOC pcDNA3-F3, pcDNA3-F3-AML1-ETO, pcDNA3-AML1-ETO(tr)-GFP;31 pcDNA3-Flag-SPOC (Y3602A), pcDNA3-Flag-SPOC (K3516A), pcDNA3-Flag-SPOC (R3552A/R3554A), pcDNA3-RBP-SPOC pcDNA3-RBP-SPOC (Y3602A), pcDNA3-RBP-SPOC (K3516A), pcDNA3-RBP-SPOC (R3552A/R3554A).34 The plasmid pMYs-IRES-GFP was commercially achieved from Cell Biolabs, San Diego, CA, USA. Sequence information of all additional vectors used in this study is available on request.
Coimmunoprecipitation experiments
The coimmunoprecipitation experiments were carried out essentially as previously described.35 Briefly, cells were lysed 24 h after transfection with 600 μl CHAPS lysis buffer consisting of 10 mM CHAPS, 50 mM Tris-HCl (pH 7.8), 150 mM NaCl, 5 mM NaF, 1 mM DTT, 0.5 mM PMSF and 40 μl/ml 'Complete Mix' protease inhibitor cocktail (Roche, Basel, Switzerland). Extracts were incubated overnight with 40 μl agarose-conjugated anti-Flag antibody (M2, Sigma-Aldrich, St Louis, MO, USA) at 4 °C. Precipitates were washed 6 to 8 times with CHAPS lysis buffer and finally resuspended in SDS-polyacrylamide gel loading buffer. Proteins were resolved via SDS-PAGE and analyzed via western blotting.
GST pulldown assays
GST fusion proteins were expressed in Escherichia coli strain BL21 (Stratagene, San Diego, CA, USA) and stored as whole bacterial lysates at −80 °C. For GST pulldown assays, proteins were translated in vitro in the presence of [35S]-Methionine using the TNT-assay (#L4610) from Promega (Madison, WI, USA) according to the manufacturer’s instructions. Pulldown experiments were carried out essentially as previously described.31
Electromobility shift assay
Transfected HEK-293 cells were lysed using a freeze–thaw method in a lysis buffer containing 0.2 mM EDTA, 20 mM HEPES (pH 7.9), 1.5 mM MgCl2, 420 mM NaCl and 25% glycerol. Six micrograms of protein lysate was incubated with 1.5 μg poly(dI-dC) (GE Healthcare, Chalfont St Giles, UK) and approximately 0.5 ng of [32P]-dCTP-labeled oligonucleotides in a binding buffer consisting of 0.5 mM DTT, 5 mM EDTA, 5 mM MgCl2, 250 mM NaCl, 50 mM Tris-HCl (pH 7.5) and 25% glycerol. The sequence of the double-stranded oligonucleotide Runt2X (5′-CTAGAGAGTGGTGTGGTAGTGCGGTCGGGGT-3′) corresponds to the AE9a-binding sites published in Okumura et al.36 Super shifting of complexes was achieved by adding 1 μg of anti-Flag antibody (M5, Sigma-Aldrich, #F4042). The reaction products were resolved at room temperature in a 7.5% PAGE with 0.5 × Tris-Borate-EDTA. Gels were dried and exposed to X-ray films (GE Healthcare).
RNA extraction, RT-PCR and qPCR
Total RNA was purified using Trizol reagent (Thermo Fisher Scientific/Ambion, Waltham, MA, USA, #15596018) according to the manufacturer’s instructions and 1 mg of RNA was retro-transcribed in cDNA using random hexamers and M-MuLV reverse transcriptase (NEB, Ipswich, MA, USA). qPCR reactions were performed with Absolute QPCR ROX Mix (Thermo Fisher Scientific, Waltham, MA, USA) #AB-1139, gene-specific oligonucleotides and double-dye probes (listed in Supplementary Table S1) and analyzed using the 7300 ABI PRISM sequence detector system (Thermo Fisher Scientific/Applied Biosystems, Waltham, MA, USA). Data were normalized to either the housekeeping gene TATA Box Binding Protein or Glyceraldehyde-3-Phosphate-Dehydrogenase.
Retroviral particle production
The pMY vectors (IRES-GFP, AE9a IRES-GFP- and AE9aNT IRES-GFP) were transiently transfected into Phoenix packaging cells. At 24, 36 and 60 h after transfection, retrovirus containing supernatants were collected, filtered (Whatman, #10462100) and stored at −80 °C. Viral supernatants were titrated on 3T3 cells.
Animals
C57BL/6 and B6.SJL-Ptprca Pep3b/BoyJ mice were either obtained from in-house colonies of the animal facility of Ulm or purchased from Janvier. All animal experiments were carried out in cooperation with the animal facility at the University of Ulm in accordance with the ‘Tierschutzgesetz §8, Abs. 1 und 3’. Experiments were approved by the ‘Regierungspräsidium Tübingen’.
Retroviral gene transfer and bone marrow (BM) transplantation
Retroviral gene transfer and BM transplantation were carried out basically as described in Vas et al.37 A detailed protocol is provided in Supplementary Information.
Flow cytometry analysis of peripheral blood (PB), BM and spleen samples
Chimerism and engraftment in PB was determined in recipient mice every 6 weeks post transplantation. For end-point in-depth analysis, mice were killed and BM and spleen cells were isolated. Chimerism and engraftment in BM and spleen cells were analyzed as described above. After red blood cell lysis with standard NH4Cl buffer, cells were analyzed on a LSRII flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). Detailed staining protocols and antibodies are provided in Supplementary Information.
Colony-forming cell assays
GFP-positive hematopoietic progenitors from retroviral transduced BM cells were fluorescence-activated cell scanner sorted and 400 cells were seeded on methylcellulose-based semi-solid media supplemented with cytokine mixtures (Stem Cell Technologies, Vancouver, BC, Canada). Colony-forming progenitors were scored every 7 days and replated in new methylcellulose medium at a concentration of 1 × 104 cells/well (six-well plates). Experiments were performed in triplicates.
Luciferase assay
HeLa cells (5 × 104) were transfected in 48-well plates with 0.5 μg of reporter plasmid DNA and different amounts of the desired expression plasmids. Luciferase activity was determined from at least three independent transfections using 10 μl of cleared lysate in a LB 9501 luminometer (Berthold Technologies, Bad Wildbad, Germany) by using the luciferase assay system from Promega.
Mouse survival and statistical analyses
The Kaplan–Meier survival curve and statistical analyses were performed using GraphPad Prism4 software (GraphPad Software, San Diego, CA, USA).
Results
Mutant AE9aNT lacks CBFβ-interaction ability but still binds to DNA
Based on the published structure of the AML1/CBFβ complex,38 we generated mutants of AE9a where asparagine 109 and threonine 161 were mutated to alanines (N109A and T161A, respectively) (indicated by arrows in Figure 1a) and tested their influence on the AE9a/CBFβ interaction (Figure 1b). Coimmunoprecipitation of Flag-tagged Runt domain of AML1, AE, AEtr and AE9a, each with either a single N109A (N) or T161A (T) mutation or N109A/T161A double point mutations (NT), revealed that Runt, AE, AEtr and AE9a wild-type (wt) proteins bind strongly to endogenous CBFβ (Figure 1b, upper panels, lanes 1, 5, 9 and 13). In contrast, single mutations in N or T decreased the AE/CBFβ interaction to a low but still detectable level (Figure 1b, upper panels, lanes 2, 3, 6, 7, 10 and 11), whereas the NT double point mutations completely abrogated the interaction (Figure 1b, upper panels, lanes 4, 8, 12 and 14). Of note, the ETO protein did not interact with CBFβ (Figure 1b, upper panel, lane 15). The AE9a splice variant is detectable in AML patients, as RNA-Seq of t(8;21) AML (n=10) as well of 23 patients with either inv(16) or a cytogenetically normal karyotype with a NPM1 mutation revealed the expression of the alternative exon E9a to be mutually exclusive in t(8;21) cases, accounting for approx. 24% of the fusion transcript (Supplementary Figures S1a–c). Therefore, we focused the study on AE9a.
Considering that the NT mutations lie within the DNA-binding Runt-domain of AML1,6 we next evaluated the DNA-binding ability of AE9aNT. Electromobility shift assays revealed that the NT mutated Runt domain (Runt NT, Figure 1c, lanes 5 and 6) as well as AE9aNT (Figure 1c, lanes 13 and 14) still possess DNA-binding capacity, but to a lower extent compared to the wt Runt domain (Figure 1c, lanes 3 and 4) and AE9a (Figure 1c, lanes 11 and 12), respectively. Western blot analysis showed almost equal levels of protein expression (Supplementary Figures S1d and e), thereby ruling out that the observed differences might be simply due to different protein levels. Altogether, these data suggest that the NT mutation within the Runt domain of AE9a reduces, but does not abrogate, its DNA-binding ability.
AE9a and AE9aNT show different subnuclear localization
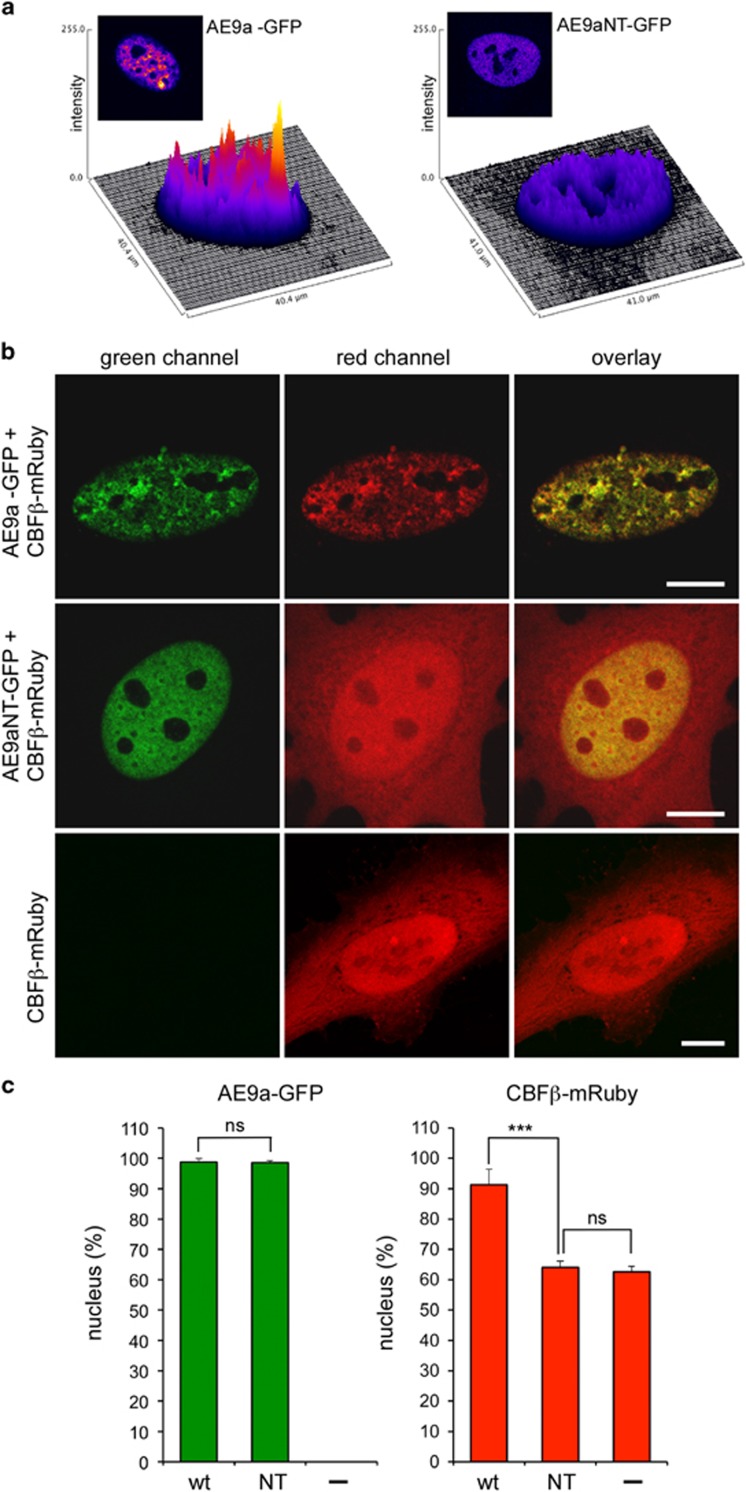
To determine whether AE9a and AE9aNT localize distinctly, cells transiently transfected with GFP-tagged AE9a or AE9aNT alone (AE9a-GFP or AE9aNT-GFP) or together with mRuby-tagged CBFβ (CBFβ-mRuby) were analyzed by fluorescence microscopy (Supplementary Figure S2) and confocal laser scanning microscopy (Figure 2). Both AE9a and AE9aNT localized to the nucleus (Supplementary Figure S2 and Figures 2a–c). A closer inspection by confocal laser scanning microscopy revealed that AE9a appeared punctuated and highly structured in a speckled pattern (Figures 2a and b), consistent with previous reports suggesting that AE is associated with the nuclear matrix.39, 40 In contrast, AE9aNT presented with a homogenous localization throughout the nucleus (Figures 2a and b). In addition, in the presence of AE9a-GFP, CBFβ-mRuby was detected in the nucleus with virtually no signal in the cytoplasm (Figure 2b, upper panel, middle column and Figure 2c) while, in the presence of AE9aNT-GFP, the distribution of CBFβ-mRuby was, as expected, not restricted to the nucleus (Figure 2b, middle panel, middle column and Figure 2c) and very similar to the distribution in cells transfected with CBFβ-mRuby alone (Figure 2b, lower panel, middle column, Figure 2c). AE9aNT thus failed to retain CBFβ in the nucleus. Altogether, these data suggest that the AE9a/CBFβ interaction has a dual function: while CBFβ stabilizes the DNA binding of AE9a, as previously described in the case of AML1,41 AE9a supports the nuclear localization of CBFβ.
Figure 2.
Subcellular localization of CBFβ after coexpression of AE9a or AE9aNT. Confocal fluorescent microscopy after cotransfection of HeLa cells with GFP-tagged AE9a or AE9aNT (AE9a-GFP and AE9aNT-GFP, respectively) together with mRuby-tagged CBFβ (CBFβ-mRuby). (a) AE9a but not AE9aNT shows a speckled pattern in the nucleus. Representation of green fluorescence intensity as three-dimensional plots (fluorescence intensities are false color coded). (b) Green channel (AE9a or AE9aNT), red channel (CBFβ) and their overlay in cotransfected HeLa cells. Scale bar, 10 μm. (c) Fluorescence quantification within the nucleus. Intensities of the green (AE9a and AE9aNT) and red (CBFβ) signals in the nucleus and the cytoplasm were analyzed. Graphs show percentages of signal intensities in the nucleus. Cells were fixed 24 h after transfection and analyzed by confocal microscopy. Nuclear and cytoplasmic distributions of green and red fluorescence intensities were measured in 40 cells, respectively. Mean values ± s.d. (error bars) are shown (nsNot significant, ***P<0.001, unpaired Student’s t-test).
AE9a but not AE9aNT interferes with RBP-J/SHARP-mediated repression of Notch target genes
We have previously demonstrated that AE interferes with the endogenous SHARP/ETO-dependent repression of Notch target genes.31 SHARP, a component of the RBP-J corepressor complex, physically interacts and co-localizes with ETO as well as AE via its highly conserved SPOC domain.31 RBP-J is the key transcription factor that modulates expression of Notch target genes.42, 43 We therefore investigated if the NT mutation in AE9a influences its physical/functional binding to SHARP. GST pulldown experiments using a bacterially purified GST-tagged SPOC domain (amino acids 3477–3664) and in vitro transcribed/translated AE9a, AE9aNT or ETO as positive control revealed that GST-SPOC interacts with both AE9a and AE9aNT in cell-free assays (Figure 3a, right panel). The interaction was further validated by coimmunoprecipitation experiments using Flag-tagged SPOC and GFP-tagged AE9a or AE9aNT (Supplementary Figure S3a). We recently reported that an RBP-SPOC fusion protein acts as a dominant repressor of Notch-dependent transcription.34 Luciferase assays in HeLa cells using an RBP-dependent reporter construct and either RBP44 or RBP-SPOC alone or in combination with ETO, AE9a or AE9aNT revealed that AE9a, but not AE9aNT or ETO, possesses derepressing activity (Figure 3b). Interestingly, AE9a-dependent derepression was reduced in response to expression of the Runt domain but not of the mutated Runt NT domain defective for CBFβ binding (Supplementary Figure S3b). Since CBFβ is expressed in HeLa cells (Supplementary Figure S3c), these data suggest that the AE9a/CBFβ interaction is a prerequisite for its derepressing activity. To characterize the AE9a/SPOC interaction in more detail we used specific point mutations (Figure 3c) within the SPOC domain of SHARP.45
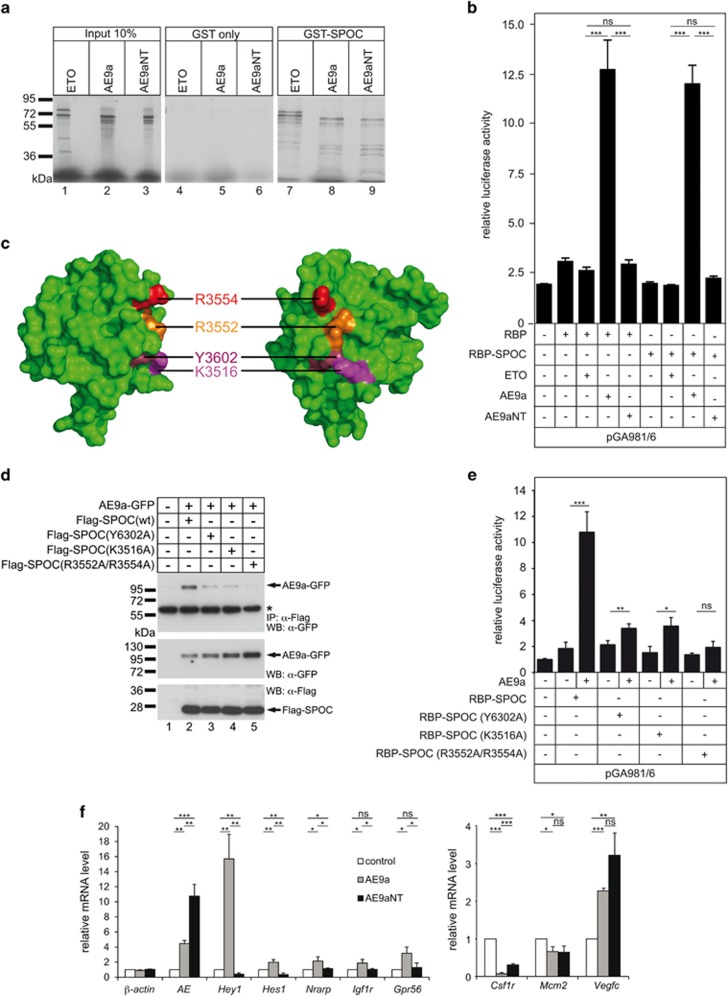
Figure 3.
Both AE9a and AE9aNT interact with the Notch-corepressor SHARP but only AE9a derepresses Notch target genes. (a) AE9a and AE9aNT interact with the SPOC domain of SHARP. GST pulldown assay was performed by using bacterially purified GST-SPOC or GST only as control and cell free synthesized AE9a, AE9aNT or ETO as positive control. (b) AE9a but not AE9aNT derepresses RBP-J-dependent transcription. Activity of AE9a and AE9aNT was tested in HeLa cells by luciferase assays using the RBP-dependent reporter construct pGA891/6 and RBP or RBP-SPOC fusion proteins together with ETO, AE9a or AE9aNT. Mean values ± SD (error bars) based on at least three independent experiments are shown (nsNot significant, ***P<0.001, unpaired Student’s t-test). (c) Surface view of the human SPOC domain of SHARP (PDB1OW1). The four amino acids R3554 (red), R3552 (orange), Y3602 (dark red) and K3516 (magenta), which were mutated to alanines are shown. Approximately 90° views are shown. (d) SPOC domain mutations show decreased interaction with AE9a (upper panel, lanes 3, 4 and 5). HEK-293 cells were transfected with the indicated expression constructs for Flag-tagged wt SPOC domain [Flag-SPOC(wt)] or mutant SPOC domains [Flag-SPOC (Y3602A), Flag-SPOC(K3516A) and Flag-SPOC(R3552A/R3554A)] together with an AE9a-GFP expression construct. Expression was verified by western blotting for AE9a (middle panel, lanes 2, 3, 4, 5), SPOC(wt) (lower panel, lane 2) and SPOC mutants (lower panel, lanes 3, 4 and 5). (e) Mutations of the SPOC domain of SHARP compromise the derepressing activity of AE9a. Activity of AE9a was tested in luciferase assays using the RBPJ-dependent reporter construct pGA891/6 and RBPJ-SPOC fusion proteins together with AE9a. The RBP-SPOC mutants show decreased AE9a mediated derepression of transcription. Mean values ± SD (error bars) based on four independent experiments are shown (nsNot significant, *P<0.05, **P<0.01, ***P<0.001, unpaired Student’s t-test). (f) The AE9a/CBFβ interaction is required to dysregulate Notch target genes (left panel) but not AML1 (right panel) target genes in HoxB4 cells. Total RNA from HoxB4 cells transfected with empty vector (control), AE9a or AE9aNT was reverse transcribed in cDNA and analyzed via qPCR. Data were normalized to the housekeeping gene TATA-binding protein. The mean ± SD of triplicate experiments is shown (nsNot significant, *P<0.05, **P<0.01, ***P<0.001, unpaired Student’s t-test).
We observed that mutations within the SPOC domain of SHARP (Flag-SPOC (Y3602A), Flag-SPOC(K3516A), Flag-SPOC(R3552A/R3554A)) strongly reduce the AE9a/SPOC interaction in both biochemical (Figure 3d) and functional (Figure 3e) analyses. Altogether, these data support that the SPOC/AE9a/CBFβ interaction is required for derepressing promoter elements bound by RBP-J, further implying that AE9a is not just a dominant negative form of AML1.
Differential requirement of the AE9a/CBFβ interaction in deregulating gene expression
AE9a, but not AE9aNT, when expressed in hematopoietic progenitor cells immortalized by overexpression of HoxB4 (HoxB4 cells32) leads to derepression of several Notch target genes, such as Hey1, Hes1, Nrarp, Igf1r and Gpr56 (Figure 3f, left). These effects are direct, as RBP-J is bound to the Notch-responsive elements of Hey1, Hes1 and Nrarp in HoxB4 cells (Supplementary Figure S3d and schematic representation in Supplementary Figure S3e). In contrast, both AE9a and AE9aNT are able to repress AML1 target genes Csf1r and Mcm2 and activate Vegfc (Figure 3f, right). Altogether, these data suggest that the AE9a/CBFβ interaction is required to derepress Notch target genes but not to deregulate AML1 target genes.
The AE9a/CBFβ interaction is dispensable to increase the self-renewal potential of hematopoietic progenitor cells
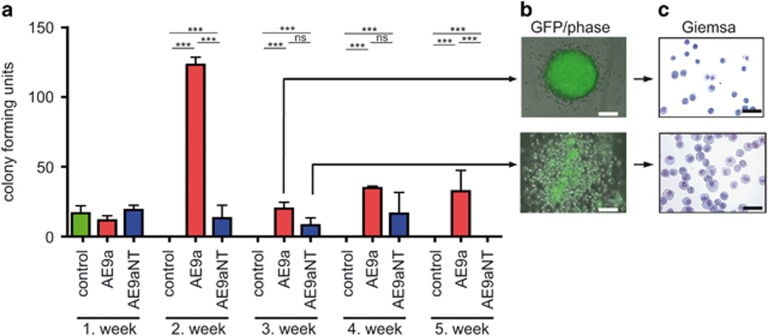
To test the ability of AE9a and AE9aNT to sustain hematopoietic self-renewal upon serial rounds of replating, colony-forming cell assays with retrovirally transduced cells were performed (overview in Supplementary Figure S4a, expression control in Supplementary Figure S4b). While, as expected, self-renewing ability was absent in IRES-GFP control cells (Figure 4a, no colonies upon replating), both AE9a-IRES-GFP (AE9a) or AE9aNT-IRES-GFP (AE9aNT) sustained significant self-renewing ability, as multiple rounds of replating still resulted in colonies (Figure 4a) indicating a strong differentiation block conferred by both AE9a28 and AE9aNT. Colonies from AE9a transduced cells though presented with a more immature morphology, whereas AE9aNT colonies contained a higher frequency of more differentiated mononuclear cells (Figures 4b and c), consistent with the lower AE9aNT replating activity compared to AE9a. Altogether these data suggest that both AE9a and AE9aNT induce self-renewal of progenitor cells but the differentiation block conferred by AE9aNT might be distinct from the one induced by AE9a.
Figure 4.
Colony forming potential of transduced progenitor populations. See overview in Supplementary Figure S4a. (a) AE9a displays higher replating efficiency compared to GFP control and CBFβ-binding deficient mutant AE9aNT. Colony-forming potential was measured by serial replating of sorted BM cells expressing IRES-GFP (control), AE9a-IRES-GFP (AE9a) and AE9aNT-IRES-GFP (AE9aNT) on semi-solid methylcellulose medium in seven days intervals over 5 weeks. Mean values and standard deviation (error bars) based on at least three independent experiments are shown. (nsNot significant, ***P<0.001). (b) Images of representative colonies of AE9a (upper panel) and AE9aNT (lower panel). (c) Analysis of cells in the colonies of AE9a (upper panel) and AE9aNT (lower panel) via Giemsa staining. While AE9a colonies consist of cells with immature morphology, AE9aNT colonies contain a higher number of mononuclear cells with a massive cytoplasmatic incorporation of methylcellulose (bars, 100 μm). Protein expression of AE9a and AE9aNT in cells isolated from methylcellulose was verified by western blot shown in Supplementary Figure S4b.
The AE9a/CBFβ interaction is required for leukemogenesis
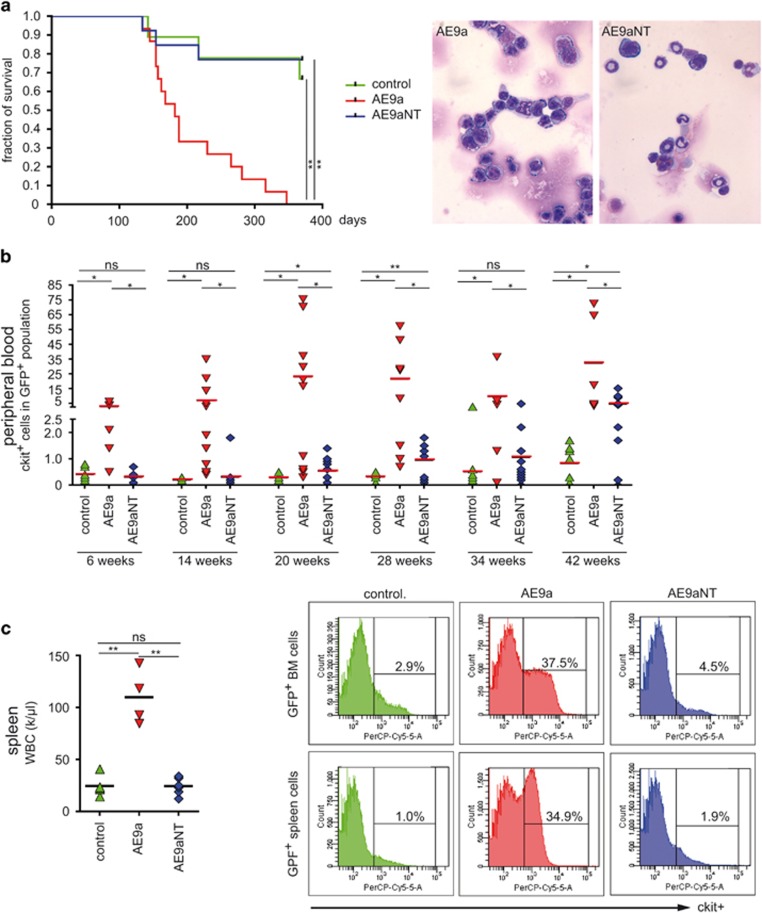
BM transplantation experiments were performed to determine the in vivo leukemic potential of AE9a and AE9aNT (Supplementary Figure S4a). BM cells from C57BL/6 Ly5.2 mice were retrovirally transduced either with IRES-GFP (control), AE9a-IRES-GFP (AE9a) or AE9aNT-IRES-GFP (AE9aNT) prior to transplantation into recipient C57BL/6 Ly5.1 (BoyJ) mice. Overall engraftment (Supplementary Figure S5a) and frequency of GFP+ cells (Supplementary Figure S5b) was constantly monitored in PB and revealed stable values. In the transplanted control group (IRES-GFP), three out of nine animals died within the observation period of 380 days due to solid tumors or wound infections from skin lesions (Figure 5a, left, green line). In the AE9aNT group, 3 out of 13 mice died within the observation period (Figure 5a, left, blue line) but leukemia was excluded as cause of death (data not shown). In the AE9a group, mice showed first signs of leukemia at 100 days post transplantation and animals died of leukemia between day 150 and 200 post transplantation (Figure 5a, left, red line). At 7 months post transplantation, BM cells from AE9a mice showed a homogenous population of immature blasts, whereas cells from AE9aNT mice were more heterogeneous and differentiated (Figure 5a, right). Analysis of white blood cell (WBC) counts (Supplementary Figure S6a) and expression of the immature blast marker c-kit (Figure 5b) further confirmed that AE9a mice presented an AML-like phenotype. Recipients of AE9a-transduced BM cells showed elevated WBC as early as 20 weeks post transplantation (Supplementary Figure S6), paralleled by increased frequency of c-kit+ cells in the PB (Figure 5b). Mice transplanted with either control or AE9aNT-transduced BM cells displayed baseline levels of WBC and low frequencies of c-kit+cells in PB (Supplementary Figures S6a and 5b, respectively). AE9a-transplanted mice further presented with splenomegaly (data not shown), high WBC in the spleen (Figure 5c, left) and high frequency of c-kit+ cells in BM and spleen (Figure 5c, right). In addition, BM cells isolated at day 214 post transplantation showed derepression of Notch targets Hey1, Hes1, Nrarp and Gpr56, but not Igf1r in AE9a-transduced mice, while BM cells from IRES-GFP (control) or AE9aNT-transduced mice did not (Supplementary Figure S6b). Taken together, these data imply that the AE9a/CBFβ interaction and derepression of Notch target genes via the interaction with the corepressor SHARP are necessary for initiation of AML by AE9a.
Figure 5.
AE9a but not AE9aNT causes myeloid leukemia and a typical leukemic differentiation pattern. Upon 5-FU injection, BM progenitor cells, transduced with either IRES-GFP (control), AE9a-IRES-GFP (AE9a) or AE9aNT-IRES-GFP (AE9aNT), were transplanted into lethally irradiated (11Gy) C57BL/6 Ly5.1 mice (see overview in Supplementary Figure S4a). (a, left) Kaplan–Meier plot showing that the majority of AE9a transplanted mice died between day 150 and 200 post transplantation whereas AE9aNT mice and control animals did not succumb to leukemia. (a, right) Representative images of BM cells stained with Giemsa from AE9a or AE9aNT mice analyzed 214 days post-transplantation. While AE9a mice presented with homogenous population of immature blasts, AE9aNT BM cells contained heterogenous and differentiated populations. (b) c-kit expression in GFP+ cells in PB was monitored at 6–8 weeks intervals via flow cytometry. AE9a transplanted mice displayed significantly elevated c-kit+ cell numbers compared to GFP or AE9aNT transplanted mice over the entire observation period. (nsNot significant, *P<0.05, **P<0.01, unpaired Student’s t-test). (c) Analysis of spleen and BM cells 214 days post-transplantation. (c, left) WBC counts from the spleen of mice transplanted with AE9a (red) are significantly elevated compared to mice transplanted with GFP (control, green) and AE9aNT (blue) (nsNot significant, **P<0.01, unpaired Student’s t-test, k=103). Mean values based on the number of animals. (c, right) Flow cytometric analysis of c-kit expression in BM and spleen of representative GFP (control, green), AE9a (red) and AE9aNT (blue) transplanted mice indicate a higher percentage of c-kit+ cells in BM and spleen of AE9a transplanted mice.
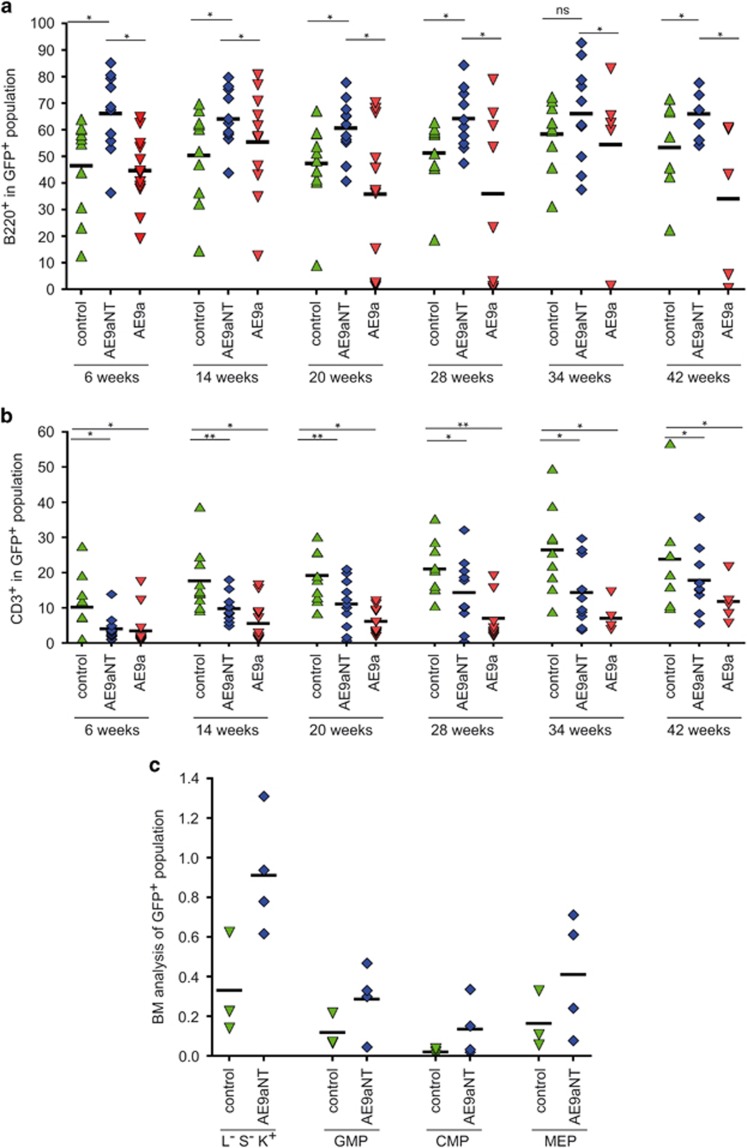
AE9aNT regulates lymphocyte differentiation and hematopoietic stem and progenitor cells in vivo
Compared to animals transplanted with empty vector control (IRES-GFP), AE9aNT-transplanted mice displayed increased frequencies of B220+ B-cells in PB (Figure 6a), which was mirrored by a reduced frequency of CD3+ T-cells (Figure 6b). AE9aNT might thus skew the differentiation of common lymphoid progenitor cells towards the B-cell lineage. Finally, compared to controls, we observed an elevated frequency of hematopoietic progenitor lineage−/sca-1−/c-kit+ (L−S−K+) cells, granulocyte/macrophage progenitors (GMP), common myeloid progenitors (CMP), and megakaryocyte-erythrocyte progenitors (MEP) in AE9aNT BM 2 years post transplantation (Figure 6c), implying that AE9aNT might also support enhanced self-renewal properties in vivo, similar to its enhanced replating ability in vitro (Figure 4a).
Figure 6.
Analysis of PB composition of AE9a and AE9aNT mice at indicated time points post transplant. (a) Levels of B220+ cells within the GFP positive/Ly5.1− cell population. (b) CD3+ cells within the GFP+ /Ly5.1− cell population in PB over time (nsNot significant, *P<0.05, **P<0.01, unpaired Student’s t-test). Mean values based on the number of animals. (c) Flow cytometric measurement of myeloid progenitor cells (lineage−/Sca-1−/c-kit+ [L−S−K+]), granulocyte/macrophage progenitors (GMP); (CD16/32+, CD34+), CMP (common myeloid progenitors; CD16/32−, CD34+) and megakaryocyte-erythrocyte progenitors (MEP); (CD16/32−, CD34−) in the GFP+ BM cell population of control (green) and AE9aNT (blue) transplanted mice after approximately 2 years post transplantation. Mean values based on the numbers of animals.
Discussion
The molecular mechanisms of the contribution of AE to AML initiation are still not fully understood.5, 25, 46 Yan et al.27 showed that only a truncated version of AE (AEtr) but not the full-length fusion protein was able to induce leukemia in a murine transduction/transplantation model. They further identified a shorter, spliced isoform of AE (AE9a) in AML patients that is structurally similar to the truncated AE product and also strongly induces leukemia in the transduction/transplantation model28 (and see Figure 5). Given that AML patients coexpress both the full-length AE and the AE9a variant at ratios that are very distinct28 (and Supplementary Figures S1a–c) the ‘personal’ AE/AE9a ratio might have a significant impact on the aggressiveness of the leukemia. In line with that, high levels of AE9a in patients indicate a poor disease outcome.47 However, the underlying mechanisms of AE9a-mediated leukemogenesis remain to be elucidated. The structure of AE9a though argues against a model in which AE simply converts the AML1 activator into an AE repressor, since AE9a is a weaker repressor compared to AE and the specific disruption of the NHR4 region of AE that is also deleted in AE9a (see also Figure 1a) reduces the interaction with corepressors nuclear receptor corepressor 1 and SMRT.12, 11
Our study for the first time demonstrates that deregulation of both AML1 target genes as well as Notch target genes by AE9a is required for AML initiation. Notch1-activating mutations were previously shown to either act as an oncogene48, 49 or in other instances as a tumor suppressor.50 Previous studies postulate a negative role of Notch signaling in myeloid leukemia.51 In an animal model of AML, Notch gain of function induced rapid cell cycle arrest, differentiation and apoptosis of AML-initiating cells.52 However, our data support the notion that derepression of Notch target genes, due to interference with the RBP-J corepressor complex, facilitates the development of leukemia. This is in agreement with a previous study showing that AE expression leads to upregulation of Notch target gene.53 In addition to our ChIP data, we have also analyzed anti-AE ChIP-Seq data previously published by the Stunnenberg lab54 and observed that AE localizes at the Notch-responsive elements of Hes1 and Hey1 (Supplementary Figure S7).
Moreover, we show that binding of AE9a to the coactivator CBFβ is necessary for leukemia initiation (Figure 5). Two previous studies already analyzed the role of CBFβ in AE’s activity by mutating Y113A/T161A23 or M106V22 within the Runt domain of AE. Whereas Kwok et al.22 observed no need for interaction with CBFβ the study from Roudaia et al.23 showed absolute requirement for leukemogenesis. Based on structural analysis, we decided not to use the same mutations but instead the double-mutant AE9a-N109A/T161A (AE9aNT) that did not show any major effects in protein stability compared to the wt AE9a protein. In regard to expression of AML1 target genes, our data show no differences between AE9a and the CBFβ-binding defective mutant AE9aNT, agreeing with Kwok et al. On the other hand, there are clear differences observed between AE9a and AE9aNT when expression of Notch target genes is analyzed, in agreement with the study by Roudaia et al. Regarding leukemogenesis, we do observe that overexpression of AE9a but not AE9aNT results in leukemia, agreeing with Roudaia et al. However, we do observe features of myeloproliferation not only in AE9a but also in AE9aNT. Furthermore, the functionality of both AE9a and AE9aNT is also observed in the T-cell lineage given that levels of peripheral T-cells are reduced in animals transplanted with either AE9a or AE9aNT. These data further support that AE9aNT is a functional protein. To note, at the same time, AE9a, and even more AE9aNT, leads to increased B-cell frequencies, suggesting wide hematopoietic defects conferred by these fusion proteins and furthermore suggesting a distinct functionality of AE9aNT. It remains to be established whether this observation might serve as a novel diagnostic tool to better characterize and identify distinct forms of AML.
In summary, we show that AE9a but not the CBFβ-binding deficient mutant AE9aNT mediates leukemogenesis in vivo. However, both AE9a and AE9aNT are active in replating assays indicating functional progenitor self-renewal. At the molecular level, interaction of AE9a with CBFβ is a pre-requisite for AE9a to dysregulate Notch target genes but not AML1 target genes (see Table 1 for summary). Therefore, our data demonstrate a significant role for aberrant regulation of Notch signaling in leukemia initiation by AE9a.
Table 1. Comparison of biochemical and hematological properties of AE9a versus AE9aNT as revealed in the study.
| AE9a | AE9aNT | Figure | |
|---|---|---|---|
| CBFβ interaction | Yes | No | 1b/2 |
| SHARP-SPOC interaction | Yes | Yes | 3a/3d/S3a |
| Transactivation capacitya | Yes | No | 3b/S3b |
| AML target genes | Affected | Affected | 3f |
| Notch target genes | Strongly affected (derepressed)b | Less affected (repressed)b | 3f/S6c |
| Serial replating capacity | Increased | Slightly increased | 4a |
| cKit+ | Elevated | Less elevated | 5b/5c, right |
| WBC | Elevated | As controls | 5c, left/S6a |
| Survival | Reduced | As controls | 5a, left |
| Disease initiation | Yes | No | 5 |
| Stem cell and progenitor pool | N. a.c | Increased | 6c |
| B-cells (B220+) | Reduced | Elevated | 6a |
| T-cells (CD3+) | Reduced | Reduced | 6b |
Abbreviation: WBC, white blood cell count.
Reporter gene assay.
Hes1, Hey1.
Not applicable, no mice survived.
Acknowledgments
We thank S Schirmer and R Rittelmann for excellent technical assistance. This work was supported by the DFG through a collaborative research grant to FO and HG (SFB 1074/A3) and by the BMBF (Federal Ministry of Education and Research, research nucleus SyStAR) to FO and HG The study was also supported in part by the DFG (SFB 1074/B3) to KD and LB and Heisenberg-Professur (BU 1339/8-1) to LB Further support was provided by the collaborative research grant TRR81 and the Heisenberg program (BO 1639/5-1) by the DFG (German Research Foundation) and the Max Planck Society to TB.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
The authors declare no conflict of interest.
Supplementary Material
References
- Gilliland DG. Targeted therapies in myeloid leukemias. Ann Hematol 2004; 83 (Suppl 1): S75–S76. [DOI] [PubMed] [Google Scholar]
- Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med 1999; 341: 1051–1062. [DOI] [PubMed] [Google Scholar]
- Higuchi M, O'Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell 2002; 1: 63–74. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Zhou L, Miyamoto T, Iwasaki H, Harakawa N, Hetherington CJ et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci USA 2001; 98: 10398–10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 1996; 84: 321–330. [DOI] [PubMed] [Google Scholar]
- Lam K, Zhang DE. RUNX1 and RUNX1-ETO: roles in hematopoiesis and leukemogenesis. Front Biosci 2012; 17: 1120–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova IN, Crute BE, Wang S, Speck NA. Sequence specificity of the core-binding factor. J Virol 1993; 67: 2408–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijssen MR, Cvejic A, Joshi A, Hannah RL, Ferreira R, Forrai A et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev Cell 2011; 20: 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi F, Mitani S, Mitsuhashi N, Kitabayashi I, Takahashi E, Suzuki M et al. Structure and expression pattern of a human MTG8/ETO family gene, MTGR1. Gene 2000; 241: 287–295. [DOI] [PubMed] [Google Scholar]
- Feinstein PG, Kornfeld K, Hogness DS, Mann RS. Identification of homeotic target genes in Drosophila melanogaster including nervy, a proto-oncogene homologue. Genetics 1995; 140: 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lausen J, Cho S, Liu S, Werner MH. The nuclear receptor co-repressor (N-CoR) utilizes repression domains I and III for interaction and co-repression with ETO. J Biol Chem 2004; 279: 49281–49288. [DOI] [PubMed] [Google Scholar]
- Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol 1998; 18: 7176–7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci PG, Lazar MA. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol 1998; 18: 7185–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S et al. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood 1992; 80: 1825–1831. [PubMed] [Google Scholar]
- Nisson PE, Watkins PC, Sacchi N. Transcriptionally active chimeric gene derived from the fusion of the AML1 gene and a novel gene on chromosome 8 in t(8;21) leukemic cells. Cancer Genet Cytogenet 1992; 63: 81–88. [DOI] [PubMed] [Google Scholar]
- Kozu T, Miyoshi H, Shimizu K, Maseki N, Kaneko Y, Asou H et al. Junctions of the AML1/MTG8(ETO) fusion are constant in t(8;21) acute myeloid leukemia detected by reverse transcription polymerase chain reaction. Blood 1993; 82: 1270–1276. [PubMed] [Google Scholar]
- Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y et al. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J 1993; 12: 2715–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardini A, Cesaroni M, Luzi L, Okumura AJ, Biggs JR, Minardi SP et al. AML1/ETO oncoprotein is directed to AML1 binding regions and co-localizes with AML1 and HEB on its targets. PLoS Genet 2008; 4: e1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank R, Zhang J, Uchida H, Meyers S, Hiebert SW, Nimer SD. The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B. Oncogene 1995; 11: 2667–2674. [PubMed] [Google Scholar]
- Liu Y, Chen W, Gaudet J, Cheney MD, Roudaia L, Cierpicki T et al. Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO's activity. Cancer Cell 2007; 11: 483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorczynski MJ, Grembecka J, Zhou Y, Kong Y, Roudaia L, Douvas MG et al. Allosteric inhibition of the protein-protein interaction between the leukemia-associated proteins Runx1 and CBFbeta. Chem Biol 2007; 14: 1186–1197. [DOI] [PubMed] [Google Scholar]
- Kwok C, Zeisig BB, Qiu J, Dong S, So CW. Transforming activity of AML1-ETO is independent of CBFbeta and ETO interaction but requires formation of homo-oligomeric complexes. Proc Natl Acad Sci USA 2009; 106: 2853–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudaia L, Cheney MD, Manuylova E, Chen W, Morrow M, Park S et al. CBFbeta is critical for AML1-ETO and TEL-AML1 activity. Blood 2009; 113: 3070–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T, Cai Z, Yang S, Lenny N, Lyu CJ, van Deursen JM et al. Expression of a knocked-in AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood 1998; 91: 3134–3143. [PubMed] [Google Scholar]
- Yergeau DA, Hetherington CJ, Wang Q, Zhang P, Sharpe AH, Binder M et al. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet 1997; 15: 303–306. [DOI] [PubMed] [Google Scholar]
- Fenske TS, Pengue G, Mathews V, Hanson PT, Hamm SE, Riaz N et al. Stem cell expression of the AML1/ETO fusion protein induces a myeloproliferative disorder in mice. Proc Natl Acad Sci USA 2004; 101: 15184–15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Burel SA, Peterson LF, Kanbe E, Iwasaki H, Boyapati A et al. Deletion of an AML1-ETO C-terminal NcoR/SMRT-interacting region strongly induces leukemia development. Proc Natl Acad Sci USA 2004; 101: 17186–17191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Kanbe E, Peterson LF, Boyapati A, Miao Y, Wang Y et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med 2006; 12: 945–949. [DOI] [PubMed] [Google Scholar]
- Yan M, Ahn EY, Hiebert SW, Zhang DE. RUNX1/AML1 DNA-binding domain and ETO/MTG8 NHR2-dimerization domain are critical to AML1-ETO9a leukemogenesis. Blood 2009; 113: 883–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers S, Lenny N, Hiebert SW. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol Cell Biol 1995; 15: 1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salat D, Liefke R, Wiedenmann J, Borggrefe T, Oswald F. ETO, but not leukemogenic fusion protein AML1/ETO, augments RBP-Jkappa/SHARP-mediated repression of notch target genes. Mol Cell Biol 2008; 28: 3502–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf M, Yue X, Schmitz S, Luche H, Reddy JK, Borggrefe T. Specific erythroid-lineage defect in mice conditionally deficient for Mediator subunit Med1. Proc Natl Acad Sci USA 2010; 107: 21541–21546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Kostezka U, Astrahantseff K, Bourteele S, Dillinger K, Zechner U et al. SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway. EMBO J 2002; 21: 5417–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Rodriguez P, Giaimo BD, Antonello ZA, Mira L, Mittler G et al. A phospho-dependent mechanism involving NCoR and KMT2D controls a permissive chromatin state at Notch target genes. Nucleic Acids Res 2016; 44: 4703–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker SA, Alvarado C, von Wichert G, Knippschild U, Wiedenmann J, Clauss K et al. RITA, a novel modulator of Notch signalling, acts via nuclear export of RBP-J. EMBO J 2011; 30: 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura AJ, Peterson LF, Okumura F, Boyapati A, Zhang DE. t(8;21)(q22;q22) Fusion proteins preferentially bind to duplicated AML1/RUNX1 DNA-binding sequences to differentially regulate gene expression. Blood 2008; 112: 1392–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vas V, Wandhoff C, Dorr K, Niebel A, Geiger H. Contribution of an aged microenvironment to aging-associated myeloproliferative disease. PLoS One 2012; 7: e31523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YY, Shi J, Zhang L, Davis A, Bravo J, Warren AJ et al. Energetic and functional contribution of residues in the core binding factor beta (CBFbeta) subunit to heterodimerization with CBFalpha. J Biol Chem 2000; 275: 39579–39588. [DOI] [PubMed] [Google Scholar]
- McNeil S, Zeng C, Harrington KS, Hiebert S, Lian JB, Stein JL et al. The t(8;21) chromosomal translocation in acute myelogenous leukemia modifies intranuclear targeting of the AML1/CBFalpha2 transcription factor. Proc Natl Acad Sci USA 1999; 96: 14882–14887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn EY, Yan M, Malakhova OA, Lo MC, Boyapati A, Ommen HB et al. Disruption of the NHR4 domain structure in AML1-ETO abrogates SON binding and promotes leukemogenesis. Proc Natl Acad Sci USA 2008; 105: 17103–17108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TL, Goetz TL, Graves BJ, Speck NA. Auto-inhibition and partner proteins, core-binding factor beta (CBFbeta) and Ets-1, modulate DNA binding by CBFalpha2 (AML1). Mol Cell Biol 2000; 20: 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci 2009; 66: 1631–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaimo BD, Oswald F, Borggrefe T. Dynamic chromatin regulation at Notch target genes. Transcription 2016;; 27: e1265702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou S, Zeng X, Cortes P, Erdjument-Bromage H, Tempst P, Honjo T et al. The recombination signal sequence-binding protein RBP-2N functions as a transcriptional repressor. Mol Cell Biol 1994; 14: 3310–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyoshi M, Schwabe JW. A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev 2003; 17: 1909–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Guzman CG, Warren AJ, Zhang Z, Gartland L, Erickson P, Drabkin H et al. Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1-ETO translocation. Mol Cell Biol 2002; 22: 5506–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao B, Wu CF, Liang Y, Chen HM, Xiong SM, Chen B et al. AML1-ETO9a is correlated with C-KIT overexpression/mutations and indicates poor disease outcome in t(8;21) acute myeloid leukemia-M2. Leukemia 2009; 23: 1598–1604. [DOI] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, JPt Morris, Silverman LB, Sanchez-Irizarry C et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271. [DOI] [PubMed] [Google Scholar]
- Chiang MY, Radojcic V, Maillard I. Oncogenic Notch signaling in T-cell and B-cell lymphoproliferative disorders. Curr Opin Hematol 2016; 23: 362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch U, Radtke F. Notch signaling in solid tumors. Curr Top Dev Biol 2010; 92: 411–455. [DOI] [PubMed] [Google Scholar]
- Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature 2011; 473: 230–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry C, Ntziachristos P, Ndiaye-Lobry D, Oh P, Cimmino L, Zhu N et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J Exp Med 2013; 210: 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcalay M, Meani N, Gelmetti V, Fantozzi A, Fagioli M, Orleth A et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J Clin Invest 2003; 112: 1751–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JH, Mandoli A, Simmer F, Wierenga BJ, Saeed S, Singh AA et al. ERG and FLI1 binding sites demarcate targets for aberrant epigenetic regulation by AML1-ETO in acute myeloid leukemia. Blood 2012; 120: 4038–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.