Abstract
Background
Breast cancer is the most commonly diagnosed cancer in women, and has become the second leading cause of cancer death among women worldwide. Chemoresistance has become an important problem in breast cancer clinics. The identification of new mechanisms affecting chemosensitivity is of great clinical value for the treatment of breast cancer.
Methods
The expression levels of chemoresistance‐associated long non‐coding RNA (CRALA), a newly discovered long non‐coding RNA, were measured by quantitative real time‐PCR in 79 pre‐treatment biopsied primary breast cancer samples. Small interfering RNAs were used to knockdown CRALA expression. The effect of CRALA on chemosensitivity was evaluated using cell growth assay.
Results
Non‐responding tumors (poor response to chemotherapy, 32 samples) had fourfold higher CRALA expression than responding tumors (good response to chemotherapy, 47 samples). CRALA is upregulated in chemoresistant breast cancer cell lines compared to their parental lines. Silencing of CRALA in chemoresistant breast cancer cells resensitizes the cells to chemotherapy in vitro. Furthermore, univariate and multivariate analysis showed that higher CRALA expression was significantly associated with poor prognosis in 144 breast cancer patients.
Conclusion
The study findings indicate that CRALA expression may be an important biomarker for predicting the clinical response to chemotherapy and prognosis in breast cancer patients. It is possible to target CRALA to reverse chemoresistance in breast cancer patients.
Keywords: Chemoresistance, CRALA, long non‐coding RNA, primary breast cancer, response to chemotherapy
Introduction
Breast cancer is the most commonly diagnosed cancer in women and has become the second leading cause of cancer death among women worldwide.1, 2Because of the integrated application of preventive, early detection, and multimodality treatment, breast cancer outcomes continue to improve. Among all of the therapeutic strategies, chemotherapy remains the primary systematic adjuvant approach for most women with either human epidermal growth factor receptor 2 (HER2) positive or triple‐negative disease. In recent years, large randomized trials and meta‐analyses have shown that estrogen receptor (ER) positive patients also benefit from chemotherapy. Chemotherapy is effective in reducing tumor size, inhibiting recurrence and distant metastasis, and therefore, prolonging patient survival.3, 4, 5 Unfortunately, not all breast cancer patients are sensitive to chemotherapy, because of the innate or acquired chemoresistance induced by continuous drug application, which impedes the clinical cure of breast cancers.6 Thus, identifying patients who are sensitive to chemotherapy is of tremendous clinical significance in improving cost‐effectiveness and reducing the side effects from overtreatment.
It is well known that protein‐coding genes account for <2% of the total genome DNA, whereas the vast majority of genomes can be transcribed into non‐coding RNAs.7, 8 Among these are long non‐coding RNAs (lncRNAs), which are larger than 200 nucleotides in length but do not encode proteins.9, 10 In recent years, it was discovered that lncRNAs serve as pivotal molecules regulating gene expression at epigenetic, transcriptional, and posttranscriptional levels.8 Increasing evidence has indicated that lncRNAs play important roles in tumorigenesis and tumor progression.10, 11, 12 In breast cancer, lncRNAs are reported to regulate multiple tumor biological properties by diverse mechanisms of actions. NKILA was identified to repress NF‐κB signaling and cancer‐associated inflammation by inhibiting IκB phosphorylation and p65 nucleus translocation.13 LncRNA H19 functions as a competing endogenous RNA to sponge microRNA let‐7, leading to an increase of LIN28 and promoting breast cancer stem cell properties.14 Downregulated lncRNA‐ROR (also called lincRNA‐ST8SIA3) could inhibit the epithelial–mesenchymal transition (EMT) of breast cancer cells and enhance the sensitivity of breast cancer cells to tamoxifen by increasing miR‐205 expression and suppressing ZEB1/2.15 In addition, the dysregulation of lncRNAs has been associated with breast cancer survival and could serve as a biomarker to predict prognosis, such as NKILA, H19, and HOTAIR.13, 14, 16
Previous studies have not considered the role of lncRNAs in predicting chemotherapy response or serving as prognostic factors for chemosensitivity. Liu et al. analyzed a dataset in The Cancer Genome Atlas (TCGA) and found that the upregulation of four lncRNAs (LINC00657, LINC00346, LINC00654, and HCG11) was associated with poor overall survival (OS), while the upregulation of nine lncRNAs (LINC00705, LINC00310, LINC00704, LINC00574, FAM74A3, UMODL1‐AS1, ARRDC1‐AS1, HAR1A, and LINC00323) could predict tumor recurrence in breast cancer.16 We proposed that at least one of these upregulated lncRNAs might play an important role in regulating chemotherapy sensitivity in breast cancer patients. In this study, we found LINC00574 and HAR1A expression was increased in chemoresistant breast cancer cell lines; however, only LINC00574 was involved in regulating the chemotherapy response of breast cancer cells (data for HAR1A is not shown). Therefore, we identified LINC00574 as a chemoresistance‐associated lncRNA (CRALA). Furthermore, we analyzed clinical data from our breast cancer center and determined that higher CRALA expression was related to poor patient response to chemotherapy and shorter survival. Our findings provide a helpful marker to distinguish which patients will respond to chemotherapy in advance, so that effective treatment strategies can be applied.
Methods
Patients and tissue specimens
A total of 176 primary breast cancer patients from Breast Tumor Center, Sun Yat‐Sen Memorial Hospital, Sun Yat‐sen University, from January 2014 to November 2015, were enrolled in this study. Snap‐frozen tissues from core needle biopsy were collected from 79 patients before neoadjuvant chemotherapy. Another 97 specimens were obtained from core needle biopsy or surgically removed tumors. None of the patients had received any therapy prior to the biopsy. The Research Ethics Board of Sun Yat‐Sen Memorial Hospital approved this retrospective study and waived the need for written informed patient consent. The patients tissues and clinical data were anonymized throughout the study.
Seventy‐nine patients received neoadjuvant chemotherapy every 21 days according to National Comprehensive Cancer Network guidelines. The chemotherapy regimen consisted of four 21‐day cycles of an epirubicin‐cyclophosphamide (EC) regimen (epirubicin 100 mg/m2 and cyclophosphamide 600 mg/m2 on day 1), followed by four 21‐day cycles of a docetaxel (100 mg/m2 on day 1) or epirubicin‐cyclophosphamide‐docetaxel (ECT) regimen (epirubicin 100 mg/m2, cyclophosphamide 500 mg/m2 and docetaxel 75 mg/m2 on day 1). Whether trastuzumab was used or not depended on HER2 status. The patients then underwent mastectomy or conserving breast surgery four weeks after the last cycle of chemotherapy. The response to chemotherapy was clinically evaluated after every two cycles by measuring the change in tumor size according to Response Evaluation Criteria in Solid Tumors (RECIST).17 RECIST is defined as follows: (i) complete response (CR), disappearance of all known disease, and reduction of any pathological lymph nodes in short axis to <10 mm; (ii) partial response (PR), at least a 30% decrease in tumor size; (iii) stable disease (SD), a less than 30% decrease or a less than 20% increase in tumor size; and (iv) progressive disease (PD), at least a 20% increase and a 5 mm absolute increase in tumor size or the appearance of new lesions. In this study, we classified SD and PD as non‐responders, and CR and PR as responders.
Quantitative real time‐PCR
Total RNA from tissue samples and cell lines was extracted using Trizol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Complementary DNA was obtained using reverse transcription of total RNA with the PrimeScriptRT reagent Kit (TaKaRa, DaLian, China). Amplification and analysis were performed using the Roche LightCycler480 Real‐time PCR System (Roche Diagnostics, Basel, Switzerland). The gene‐specific primers were as follows: CRALA (forward: CTCACTCCTCTGCCGATGCT; reverse: CCACACCAGGACCATTCTCTTG); glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) (forward: ATCACCATCTTCCAGGAGCGA; reverse: CCTTCTCCATGGTGGTGAAGAC).
Cell culture and treatment
MDA‐MB‐231 breast cancer cell line was obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). MDA‐MB‐231‐Paclitaxel‐resistant cell line (MDA‐MB‐231P) and MDA‐MB‐231‐http://www.baidu.com/link?url=8KP7tJHph40SJW8LifxzQynrX1SvBzSqis54H-7SFbXT6t2wZEcxfw2CtSHfmF50SR4YuosiRIPMbLuTQnxEtHZw5-seWboCVOrZOJCPFgfO_B1k5ZladBMvmbJqwHRz‐resistant cell line (MDA‐MB‐231Cis) were established by gradual administration of increasing concentrations of chemotherapy drugs into MDA‐MB‐231 cells up to 50 nm and 30 μm for paclitaxel and cisplatin, respectively. Cells were maintained in Dulbecco's modified Eagle medium with 10% fetal bovine serum (Life Technologies, Carlsbad, CA, USA) in a humidified atmosphere containing 5% CO2 at 37°C.
Transfection of the cells with small interfering RNA (siRNA) was performed using Lipofectamine 3000 (Life Technologies) according to the manufacturer's protocol. The sequences for specific siRNA against CRALA were as follows:
Negative control (NC): sense, UUCUCCGAACGUGUCACGUTT,antisense, ACGUGACACGUUCGGAGAATT.
SiRNA‐1 (Si‐1): sense, GGAAGAUGGUUAAUUCCAUTT,antisense, AUGGAAUUAACCAUCUUCCTT.
SiRNA‐1 (Si‐2): sense, GCACUCUGUCCAUUUCAUATT, antisense, UAUGAAAUGGACAGAGUGCTT.
Cell growth assay
Cell growth in the presence or absence of chemotherapy drugs was assayed by cell counting. Briefly, the cells were seeded at appropriate densities (2.0 × 105 cells per well) in six‐well plates in triplicate and treated with interest drugs (0–120 μm cisplatin, 0–1000 nm paclitaxel) for indicated times. Beckman Coulter Z1 (Fullerton, CA, USA) was used to count the cells.
Apoptosis assay
Cell apoptosis was performed using fluorescein isothiocyanate‐labeled Annexin V and propidium iodide staining (BD Biosciences, San Jose, CA, USA), followed by flow cytometry according to the manufacturer's protocol. BD Accuri C6 (BD Biosciences) was used to count the cells.
Statistics analysis
Statistical analysis was performed using SPSS version 20.0 (IBM Corp., Armonk, NY, USA). Student's t‐tests were used to compare differences between the groups. The Mann–Whitney U test was used to compare the CRALA expression levels between responding and non‐responding tumors. A receiver operating characteristic (ROC) curve was used to evaluate CRALA sensitivity and specificity of CRALA. P < 0.05 was considered statistically significant.
Results
CRALA is an indicator for predicting response to chemotherapy in breast cancer
To evaluate the association between CRALA expression and response to chemotherapy in breast cancer, we detected CRALA expression in 79 snap‐frozen pretreatment core needle biopsies from primary breast cancer patients by quantitative real time‐PCR. We then analyzed the association between CRALA levels and chemotherapeutic response. Seven patients achieved a CR, 40 achieved a PR, 29 exhibited SD, and three showed PD. SD and PD were classified as non‐responders, and CR and PR were classified as responders. The average level of CRALA expression in the non‐responders was 4.09‐fold higher than that in responders (P < 0.001) (Fig 1a).We then constructed ROC curves to evaluate the average sensitivity and specificity of CRALA for predicting response to chemotherapy in breast cancer. The ROC curve analysis showed that CRALA performed well in predicting a response to chemotherapy (area under the curve 0.735, 95% confidence interval [CI] 0.626–0.843; P < 0.001) (Fig 1b). These data suggested that CRALA was negatively related to the response of breast cancer patients to chemotherapy.
Figure 1.
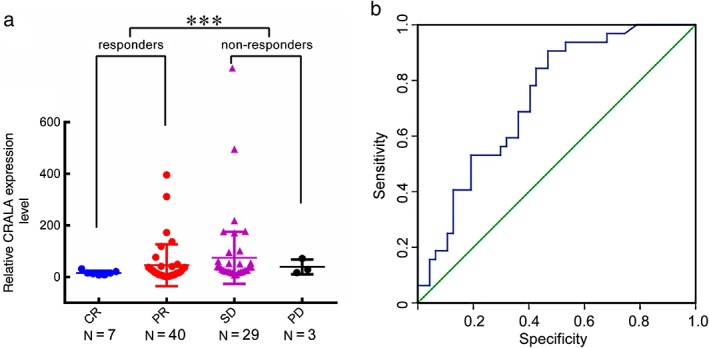
High CRALA expression is associated with chemoresistance in primary breast cancer. (a) Scatter plot of CRALA expression levels in 79 primary breast cancer tissues. (b) Receiver operating characteristic curves were conducted to evaluate the average sensitivity and specificity of CRALA for predicting response to chemotherapy in breast cancer. ***P < 0.001. CR, complete response; PD, progressive disease; PR, partial response; SD, stable disease.
CRALA is upregulated in chemoresistant breast cancer cell lines
As our clinical data indicated that CRALA expression levels correlated with chemotherapy resistance, we further examined the CRALA level in chemotherapy drug resistant cell lines. Similar to the CRALA expression pattern in clinical samples, we found its expression was much higher in MDA‐MB‐231P and MDA‐MB‐231Cis cells than in their parental cells, 8.89‐fold and 4.13‐fold, respectively (Fig 2a).
Figure 2.
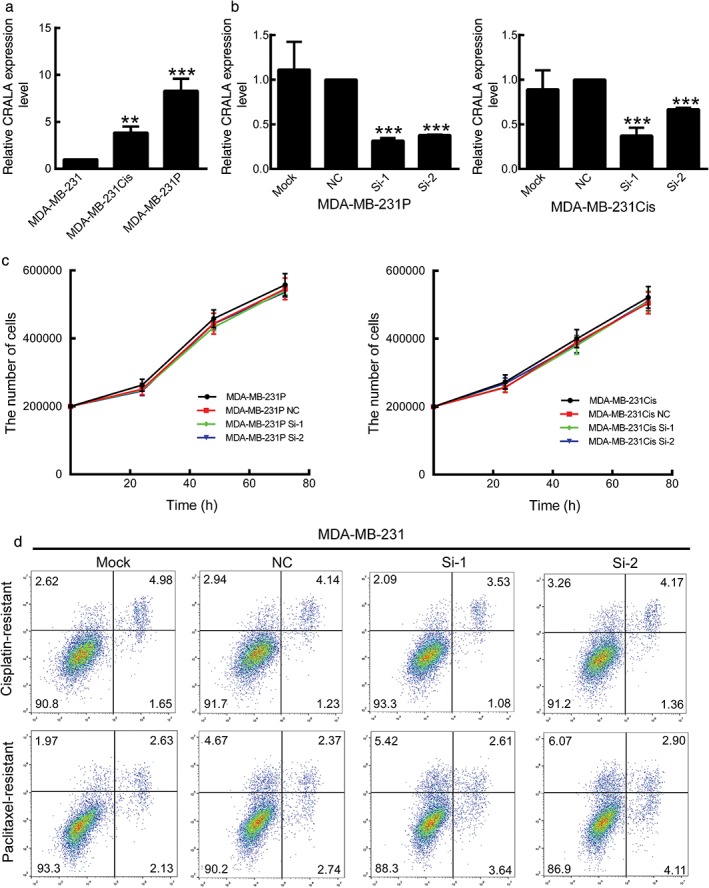
CRALA is not required for chemoresistant cell survival. (a) CRALA expression was determined using quantitative real time (qRT)‐PCR in parental MDA‐MB‐231 and chemoresistant breast cancer cell lines. (b) The knockdown efficiency of two specific small interfering RNAs (siRNAs) against CRALA was examined by qRT‐PCR in chemoresistant breast cancer cell lines. (c) The chemoresistant cells MDA‐MB‐231P and MDA‐MB‐231Cis were transfected with specific siRNA against CRALA or negative control (NC) for 24, 48, and 72 hours in the absence of any chemotherapeutic drug. (d) Apoptosis was determined by flow cytometry after transfection of siRNAs for 48 hours. **P < 0.01, ***P < 0.001.
Silencing of CRALA in chemoresistant breast cancer cells resensitizes cells to chemotherapy
We further investigated whether CRALA upregulation in chemoresistant cells contributes to drug resistance. CRALA siRNAs markedly decreased CRALA expression in MDA‐MB‐231P and MDA‐MB‐231Cis cells (Fig 2b). Without exposure to chemotherapeutic drugs, cell proliferation (Fig 2c) and apoptosis (Fig 2d) in MDA‐MB‐231P and MDA‐MB‐231Cis cells were not significantly altered after 72 hours of CRALA silencing, suggesting that CRALA dysregulation is not required for chemoresistant cell survival.
However, when chemoresistant cells were treated with corresponding chemotherapeutic drugs, a decrease in CRALA expression obviously slowed down the proliferation of MDA‐MB‐231Cis and MDA‐MB‐231P cells in a time‐dependent manner (Fig 3a). We then treated cells with increasing concentrations of chemotherapeutic drugs for 48 hours, and as demonstrated in Figure 3b, MDA‐MB‐231Cis cells were more resistant to cisplatin than parental cells, as the concentration response curves of cisplatin shifted to the right, with inhibitory concentration (IC)50 increasing from 5.63 ± 1.04 μm to 89.22 ± 3.45 μm (P < 0.001). CRALA silencing with si‐1 and si‐2 enhanced the suppressive effect of cisplatin on cell proliferation with inhibition curves shifting back to the left of MDA‐MB‐231Cis and MDA‐MB‐231Cis NC cells, and a decreased IC50 value from 85.90 ± 1.79 μm to 43.42 ± 1.40 μm (P < 0.001) and 49.03 ± 3.95 μm (P < 0.001), respectively (Fig 3b,c right).
Figure 3.
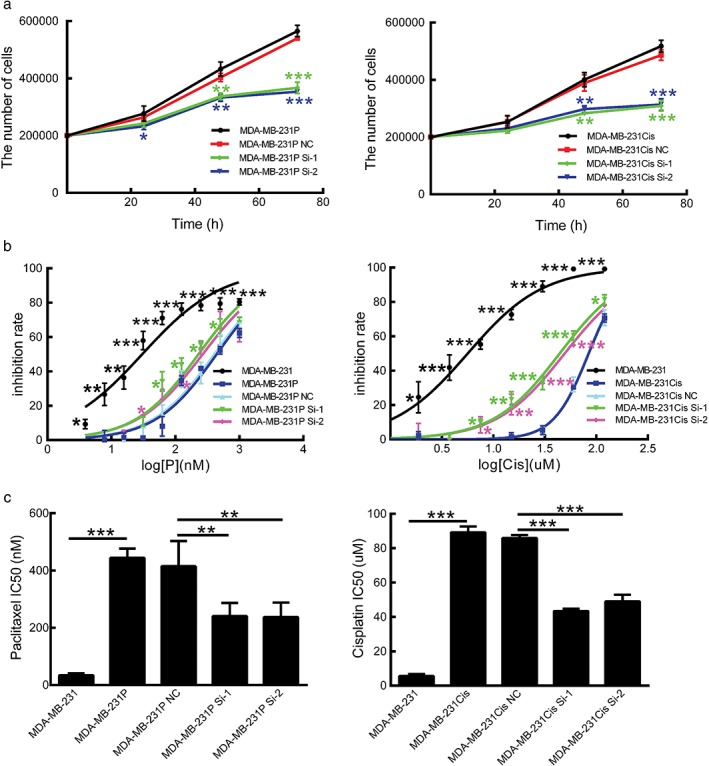
CRALA silencing in chemoresistant breast cancer cells resensitizes cells to chemotherapy. (a) Cell proliferation of chemoresistant breast cancer cells was measured by cell counting after transfection with small interfering RNAs (siRNAs) for 24, 48, and 72 hours in the presence of 30 μm cisplatin or 50 nM paclitaxel. (b) CRALA siRNAs enhanced the inhibitory effects of chemotherapeutic drugs on chemoresistant cells and shifted the concentration response curves of chemotherapeutic drugs to the left of their negative control (NC). (c) Inhibitory concentration (IC)50 values were calculated based on the concentration response curves. *P < 0.05, **P < 0.01, ***P < 0.001.
Similarly, paclitaxel had less inhibitory effects on MDA‐MB‐231P cells than parental cells with IC50 of 444.46 ± 32.09 nm and 34.32 ± 6.35 nm (P < 0.001), respectively. CRALA Si‐1 and CRALA Si‐2 significantly increased the sensitivity of MDA‐MB‐231P cells to the different concentrations of paclitaxel, with inhibition ratio curves shifting to the left of MDA‐MB‐231P and MDA‐MB‐231P NC cells. The IC50 decreased from 414.65 ± 88.34 nm (for MDA‐MB‐231P NC cells) to 241.08 ± 45.71 nm (P = 0.002) and 237.22 ± 50.92 nm (P = 0.002), respectively (Fig 3b,c left).
These data indicate that CRALA was upregulated in chemoresistant breast cancer cells, and that CRALA silencing could restore chemosensitivity.
High CRALA expression predicts poor prognosis in patients with breast cancer
To further understand the roles of CRALA in breast cancer, we analyzed the correlation between CRALA expression and clinicopathological status of 176 breast cancer patients. As shown in Table 1, elevated CRALA expression in breast cancer was significantly associated with larger tumor size (P < 0.001), lymph node metastasis (P < 0.001), advanced tumor node metastasis (TNM) stage (P = 0.001), high Ki67 expression (P < 0.001), negative estrogen receptor (P = 0.005), negative progestogen receptor (P < 0.001), molecular subtype (P = 0.006), the status of local and regional recurrence (P < 0.001), and distant metastasis (P < 0.001). However, statistical analysis showed that CRALA expression levels had no significant correlation with age and HER2 status.
Table 1.
Correlation between CRALA expression level and clinicopathological characteristics
| Characteristic | Low CRALA expression | High CRALA expression | P |
|---|---|---|---|
| Age | 0.063 | ||
| <45 | 18 | 34 | |
| ≥45 | 71 | 53 | |
| Tumor size | <0.001 | ||
| T1 | 46 | 13 | |
| ≥T2 | 43 | 74 | |
| Nodal status | |||
| N0 | 56 | 34 | <0.001 |
| ≥ N1 | 33 | 53 | |
| Metastasis | <0.001 | ||
| Yes | 2 | 26 | |
| No | 87 | 61 | |
| Relapse | <0.001 | ||
| Yes | 0 | 10 | |
| No | 89 | 77 | |
| Ki67 | <0.001 | ||
| <14% | 35 | 4 | |
| ≥14% | 54 | 83 | |
| ER | 0.005 | ||
| Negative | 20 | 44 | |
| Positive | 69 | 43 | |
| PR | <0.001 | ||
| Negative | 29 | 62 | |
| Positive | 60 | 25 | |
| HER2 | 0.098 | ||
| Negative | 60 | 55 | |
| Positive | 29 | 32 | |
| TNM stage | 0.001 | ||
| I–II | 76 | 55 | |
| III–IV | 13 | 32 | |
| Molecular subtype | 0.006 | ||
| Basal‐like | 9 | 24 | |
| ERBB2+ | 13 | 20 | |
| Luminal A | 19 | 2 | |
| Luminal B | 48 | 41 | |
Bold text indicates that P < 0.05 was considered significant. CRALA, chemoresistance‐associated long non‐coding RNA; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor; TNM, tumor node metastasis.
To determine the relationship between CRALA expression and breast cancer prognosis, we evaluated the correlation between CRALA expression and progression‐free survival (PFS) and OS by Kaplan–Meier analysis. Among the 176 patients, 144 patients were followed up for over two years. We analyzed the prognostic value of CRALA expression in these 144 patients and found that breast cancer patients with high CRALA expression exhibited significantly inferior clinical outcomes (P < 0.001 for PFS, Fig 4a; P = 0.002 for OS, Fig 4b). According to our results, TNM stage, tumor size, Ki67, ER, progesterone receptor, and molecular subtype also showed significant prognostic effects on PFS (Table 2). To further assess whether CRALA expression can be identified as an independent prognostic predictor for breast cancer patients, we performed multivariate analysis. According to our results, the CRALA expression level showed significant prognostic effects on PFS (P < 0.001, hazard ratio 44.272, 95% confidence interval 5.968–328.403), independent of various clinical variables (Table 2). These results indicate that CRALA serves as an independent prognostic factor for PFS in breast cancer patients.
Figure 4.
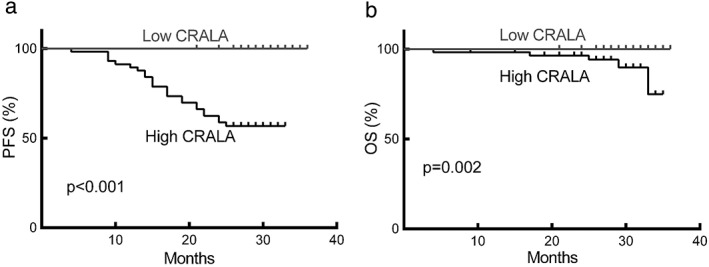
High CRALA expression is significantly correlated with poor prognosis in breast cancer. Survival curves according to CRALA expression in breast cancer for (a) progression‐free survival (PFS) and (b) overall survival (OS).
Table 2.
Univariate and multivariate analysis of different prognostic variables in primary breast cancer patients
| PFS | OS | ||||||
|---|---|---|---|---|---|---|---|
| Variables | N | Univariate | Multivariate | Univariate | Multivariate | ||
| P | P | HR (95% CI) | P | P | HR (95% CI) | ||
| Age | 0.363 | NS | N/A | NS | NS | N/A | |
| <45 | 40 | ||||||
| ≥5 | 104 | ||||||
| Tumor size | 0.016 | NS | N/A | NS | NS | N/A | |
| T1 | 58 | ||||||
| ≥T2 | 86 | ||||||
| Nodal status | 0.122 | NS | N/A | NS | NS | N/A | |
| N0 | 80 | ||||||
| ≥N1 | 64 | ||||||
| Ki67 | 0.020 | NS | N/A | NS | NS | N/A | |
| <14% | 38 | ||||||
| ≥4% | 106 | ||||||
| ER | 0.007 | NS | N/A | NS | NS | N/A | |
| Negative | 50 | ||||||
| Positive | 94 | ||||||
| PR | 0.001 | NS | N/A | NS | NS | N/A | |
| Negative | 68 | ||||||
| Positive | 76 | ||||||
| HER2 | 0.937 | NS | N/A | NS | NS | N/A | |
| Negative | 99 | ||||||
| Positive | 45 | ||||||
| TNM stage | 0.001 | 0.036 | 2.311 (1.056–5.056) | NS | NS | N/A | |
| I–II | 114 | ||||||
| III–IV | 30 | ||||||
| Molecular subtype | 0.006 | NS | N/A | NS | NS | N/A | |
| Basal‐like | 29 | ||||||
| ERBB2+ | 25 | ||||||
| Luminal A | 20 | ||||||
| Luminal B | 70 | ||||||
| CRALA level | < 0.001 | < 0.001 | 44.272 (5.968–328.403) | 0.002 | NS | N/A | |
| Low | 86 | ||||||
| High | 58 | ||||||
CI, confidence interval; CRALA, chemoresistance‐associated long non‐coding RNA; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; HR, hazard ratio; N/A, not available; NS, non‐significant; OS, overall survival; PFS, progression‐free survival; PR, progesterone receptor; TNM, tumor node metastasis.
Discussion
Breast cancer is the most commonly diagnosed cancer in women and has become the second leading cause of cancer death in women worldwide.1 At present, chemotherapy is the common treatment strategy and plays an important role in the clinical cure of breast cancer; however, some patients have a poor response to chemotherapy and their prognosis is unfavorable. Chemoresistance has become a significant challenge in the clinical setting; therefore, further investigation needs to be conducted in order to be able to predict the patients who will respond well to chemotherapy. Such knowledge will assist in optimizing the clinical decision‐making process and treatment program to provide minimum cost and maximum benefit for breast cancer patients.
LncRNAs are non‐protein coding transcripts that are longer than 200 nucleotides.18, 19 Increasing evidence has revealed that LncRNAs may function as oncogenes or tumor suppressors and play important roles in regulating tumor cell biological properties, including chemoresistance.20 HOTAIR is upregulated in breast and colorectal cancers and hepatocellular carcinoma,21, 22, 23, 24 and can reportedly regulate the cisplatin‐resistant ability of human lung adenocarcinoma cells by affecting p21 expression, thus regulating apoptosis and cell cycle distribution.25 LncRNA H19 has been proven to function as an oncogene.26 H19‐induces P‐glycoprotein expression and MDR1‐associated drug resistance in liver cancer cells by regulating MDR1 promoter methylation.27 In addition, p95‐overexpressing multidrug‐resistant cell lines of human lung carcinoma NCI‐H1688 and breast carcinoma MCF‐7 displayed higher levels of H19 messenger RNA.28 LncRNAs also participate in chemoresistant regulation of breast cancer. For instance, higher PANDA expression contributes to doxorubicin resistance in breast cancer cells.26 lncRNA adriamycin‐resistance associated (ARA) inhibition reverses drug resistance in adriamycin‐resistant cells in breast cancer cell lines by regulating multiple cellular processes and signaling pathways.6 However, no previous research has proposed lncRNAs as a prognostic factor for chemosensitivity.
CRALA is a recently discovered long intergenic non‐protein coding RNA encoded by a gene at the chromosome 6q27 region.29 We performed a retrospective analysis based on patients who received neoadjuvant chemotherapy to evaluate the chemotherapeutic response of CRALA. We collected tumor samples before the patients received any treatment, therefore the predictive value of lncRNA to neoadjuvant chemotherapy was independent of other treatment. We found that non‐responding tumors expressed higher levels of lncRNA CRALA, and ROC curve analysis showed good sensitivity and specificity of CRALA to predict the response of breast cancer patients to chemotherapy. We also attempted to analyze the ROC curve for tumor size, ER, progesterone receptor, and Ki67 to compare the prognostic accuracy of these variables with CRALA. Unfortunately, it was not possible to develop a curve for ER, progesterone receptor, or Ki67 (data not shown). This may have been a result of treatment selection bias in that patients with certain expression levels of these biomarkers are prone to undergo neoadjuvant chemotherapy. Another reason is related to the limited sample size. There are no other widely accepted markers to predict chemotherapy efficacy, therefore we can only evaluate the accuracy of CRALA by AUC and P value, which showed that CRALA performed well in predicting a response to chemotherapy.
Our in vitro studies showed that CRALA expression was increased in chemoresistant breast cancer cell lines. CRALA knockdown resulted reversed chemoresistance. In addition, CRALA was expressed at significantly higher levels in chemoresistant breast cancer samples than that in chemosensitive samples. Moreover, our results showed that higher CRALA expression was significantly correlated with poor clinical outcomes, assayed by PFS and OS. More importantly, CRALA served as an independent prognostic factor for PFS in breast cancer patients. Our results strongly suggest that CRALA acts as an oncogene to regulate breast cancer chemosensitivity and may potentially be used as a therapeutic target for chemoresistant breast cancer patients.
There are some shortcomings to our study. First, the precise mechanism of how CRALA contributes to chemoresistance is yet to be defined. Second, because only in vitro experiments were performed, current CRALA research is incomplete. Whether CRALA plays a similar role in an in vivo setting is unclear. Third, our clinical results were based on retrospective analysis, and only 79 breast cancer tissues were sampled for the study. We need to validate our results with a prospective multicenter clinical trial. In addition, because of inadequate follow‐up and the limited sample size, we could not determine an association between TNM stage, molecular subtype, and lymph node status and their impact on OS. We will address these shortcomings in future research.
In summary, our results suggest that CRALA may emerge as an important biomarker for predicting the clinical response to chemotherapy and as an independent prognostic factor for PFS in breast cancer patients. As a newly discovered lncRNA, further research is required to explore its potential. It is of great clinical value to develop effective therapeutic strategies by targeting CRALA to reverse chemoresistance in breast cancer patients.
Disclosure
No authors report any conflict of interest.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (81672622), and Sun Yat‐sen University Training Project(14ykpy20).
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016; 66: 7–30. [DOI] [PubMed] [Google Scholar]
- 2. American Cancer Society . How Common Is Breast Cancer? 2017. [Cited 21 Jul 2017.] Available from URL: https://www.cancer.org/cancer/breast-cancer/about/how-common-is-breast-cancer.html
- 3. Cancer Research UK . Breast cancer: Treatment [Cited 24 Jul 2017.] Available from URL: http://www.cancerresearchuk.org/about-cancer/breast-cancer/treatment
- 4. Breastcancer.org . Chemotherapy [Cited 24 Jul 2017.] Available from URL: http://www.breastcancer.org/treatment/chemotherapy
- 5. WebMD . Breast Cancer: Chemotherapy [Cited 24 Jul 2017.] Available from URL: http://www.webmd.com/breast‐cancer/breast‐cancer‐chemotherapy.
- 6. Jiang M, Huang O, Xie Z et al A novel long non‐coding RNA‐ARA: Adriamycin resistance‐associated. Biochem Pharmacol 2014; 87: 254–83. [DOI] [PubMed] [Google Scholar]
- 7. Thomas CA Jr. The genetic organization of chromosomes. Annu Rev Genet 1971; 5: 237–56. [DOI] [PubMed] [Google Scholar]
- 8. Kung JT, Colognori D, Lee JT. Long noncoding RNAs: Past, present, and future. Genetics 2013; 193: 651–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mendell JT. Targeting a long noncoding RNA in breast cancer. N Engl J Med 2016; 374: 2287–9. [DOI] [PubMed] [Google Scholar]
- 10. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011; 43: 904–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non‐coding RNAs and cancer: A new frontier of translational research? Oncogene 2012; 31: 4577–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol 2011; 21: 354–61. [DOI] [PubMed] [Google Scholar]
- 13. Liu B, Sun L, Liu Q et al A cytoplasmic NF‐kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015; 27: 370–81. [DOI] [PubMed] [Google Scholar]
- 14. Peng F, Li TT, Wang KL et al H19/let‐7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis 2017; 8: e2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang HY, Liang F, Zhang JW, Wang F, Wang L, Kang XG. Effects of long noncoding RNA‐ROR on tamoxifen resistance of breast cancer cells by regulating microRNA‐205. Cancer Chemother Pharmacol 2017; 79: 327–37. [DOI] [PubMed] [Google Scholar]
- 16. Liu H, Li J, Koirala P et al Long non‐coding RNAs as prognostic markers in human breast cancer. Oncotarget 2016; 7: 20584–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eisenhauer EA, Therasse P, Bogaerts J et al New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45: 228–47. [DOI] [PubMed] [Google Scholar]
- 18. Li L, Gu M, You B et al Long non‐coding RNA ROR promotes proliferation, migration and chemoresistance of nasopharyngeal carcinoma. Cancer Sci 2016; 107: 1215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao W, Luo J, Jiao S. Comprehensive characterization of cancer subtype associated long non‐coding RNAs and their clinical implications. Sci Rep 2014; 4: 6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Silva A, Bullock M, Calin G. The clinical relevance of long non‐coding RNAs in cancer. Cancers (Basel) 2015; 7: 2169–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non‐coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci 2013; 104: 458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gupta RA, Shah N, Wang KC et al Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang Z, Zhou L, Wu LM et al Overexpression of long non‐coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 2011; 18: 1243–50. [DOI] [PubMed] [Google Scholar]
- 24. Kogo R, Shimamura T, Mimori K et al Long noncoding RNA HOTAIR regulates polycomb‐dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011; 71: 6320–6. [DOI] [PubMed] [Google Scholar]
- 25. Liu Z, Sun M, Lu K et al The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregulation of p21(WAF1/CIP1) expression. PLoS ONE 2013; 8: e77293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pan JJ, Xie XJ, Li X, Chen W. Long non‐coding RNAs and drug resistance. Asian Pac J Cancer Prev 2015; 16: 8067–73. [DOI] [PubMed] [Google Scholar]
- 27. Tsang WP, Kwok TT. Riboregulator H19 induction of MDR1‐associated drug resistance in human hepatocellular carcinoma cells. Oncogene 2007; 26: 4877–81. [DOI] [PubMed] [Google Scholar]
- 28. Doyle LA, Yang W, Rishi AK, Gao Y, Ross DD. H19 gene overexpression in atypical multidrug‐resistant cells associated with expression of a 95‐kilodalton membrane glycoprotein. Cancer Res 1996; 56: 2904–7. [PubMed] [Google Scholar]
- 29. University of California Santa Cruz . Human Gene LINC00574: Description and Page Index [Cited 24 Jul 2017.] Available from URL: http://genome.ucsc.edu/cgi‐bin/hgGene?hgg_gene=uc032yzj.2&hgg_prot=ENST00000625483.1&hgg_chrom=chr6_KI270797v1_alt&hgg_start=65522&hgg_end=78075&hgg_type=knownGene&db=hg38&hgsid=577008195_yOXAgd8xzkdDy7ayTOj7lsIvmIor


