Abstract
Escherichia coli synthesizes three selenocysteine-dependent formate dehydrogenases (Fdh) that also have a molybdenum cofactor. Fdh-H couples formate oxidation with proton reduction in the formate hydrogenlyase (FHL) complex. The activity of Fdh-H in solution can be measured with artificial redox dyes but, unlike Fdh-O and Fdh-N, it has never been observed by chromogenic activity staining after non-denaturing polyacrylamide gel electrophoresis (PAGE). Here, we demonstrate that Fdh-H activity is present in extracts of cells from stationary phase cultures and forms a single, fast-migrating species. The activity is oxygen labile during electrophoresis explaining why it has not been previously observed as a discreet activity band. The appearance of Fdh-H activity was dependent on an active selenocysteine incorporation system, but was independent of the [NiFe]-hydrogenases (Hyd), 1, 2 or 3. We also identified new active complexes of Fdh-N and Fdh-O during fermentative growth. The findings of this study indicate that Fdh-H does not form a strong complex with other Fdh or Hyd enzymes, which is in line with it being able to deliver electrons to more than one redox-active enzyme complex.
Keywords: Formate dehydrogenase H, [NiFe]-hydrogenase, Chromogenic activity staining, Enzyme complexes, Stationary phase
Highlights
-
•
A chromogenic activity stain to identify formate dehydrogenase H was developed.
-
•
Fdh-H activity was identified in stationary phase fermenting cells.
-
•
Fdh-H activity was only observed if electrophoresis was performed anaerobically.
-
•
Fdh-H activity was independent of an active hydrogenase 3 enzyme.
-
•
New active forms of formate dehydrogenases O and N were identified.
1. Introduction
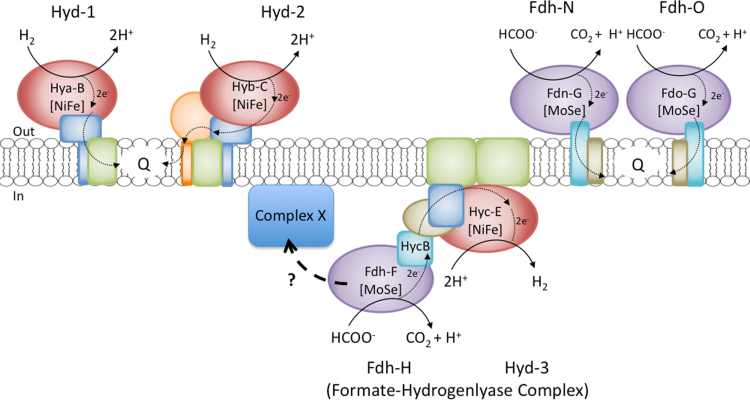
Formate and hydrogen are important electron donors, as well as key fermentation products, in the metabolism of numerous anaerobic bacteria and archaea [1]. During anaerobic growth, the model bacterium Escherichia coli synthesizes three formate dehydrogenases (Fdh) and up to four [NiFe]-hydrogenases (Hyd) [2]. The Fdh enzymes include Fdh-H (H for hydrogen, i.e. fermentation), Fdh-N (N for nitrate respiration) and Fdh-O (O for aerobic growth) to signify the conditions under which they were originally identified and that were primarily used for their characterization [2]. All three Fdh have a large subunit that contains both a selenocysteine residue and a molybdenum cofactor in their active site. Both of these cofactors are directly involved in catalysis [2]. This catalytic subunit also includes a [4Fe-4S] cluster that mediates electron transfer between the active site and [4Fe-4S] clusters of an electron-transferring small subunit. In contrast, the four Hyd enzymes include Hyd-1 and Hyd-2, which are respiratory enzymes that primarily couple hydrogen oxidation to quinone reduction, while Hyd-3, together with Fdh-H, forms part of the formate hydrogenlyase (FHL) complex, which catalyzes H2 production from formate [2]. Hyd-4 is also proposed to form a complex including Fdh-H, which is related to FHL [3]. All seven enzymes form key components of membrane-associated multi-protein complexes and these are depicted schematically in Fig. 1.
Fig. 1.
Schematic representation of the membrane-bound formate dehydrogenases and hydrogenases of E. coli. Fdh-H, Fdh-N and Fdh-O, along with Hyd-1, Hyd-2 and Hyd-3, the latter of which forms a key component of the formate-hydrogenlyase (FHL) complex, are shown. With the exception of HycB, the electron-transferring small subunit of Fdh-H, only the catalytic subunits of the enzymes are shown with their respective names. The periplasmic (out) and cytoplasmic (in) sides of the membrane are indicated, while the abbreviations NiFe, MoSe and Q signify the [NiFe] cofactor, the molybdenum cofactor/selenocysteine and quinone pool, respectively. Electron flow within the enzyme complexes and to the quinone pool are indicated by thin arrows in the enzyme complexes, while the thick dashed arrow with the question mark signifies that the Fdh-H also forms a complex with another complex (complex X) or complexes. Complex X could represent the second FHL complex comprising the Hyf components of Hyd-4 [3]; however, whether this complex is membrane-associated remains to be determined.
Biochemical, and particularly physiological, analysis of individual Fdh enzymes has been hampered by the fact that activity can only efficaciously be determined using artificial redox dyes. Nevertheless, use of dyes such as benzyl viologen (BV) and phenazine methosulfate/2,6-dichlorophenolindophenol (PMS/DCPIP) for enzyme assays in solution, along with selenopolypeptide analyses, demonstrated that Fdh-H and Fdh-N are indeed distinct enzymes [4], [5]. Subsequent biochemical [6], [7] and molecular biological studies [8] showed that Fdh-O (encoded by fdoGHI), although highly similar to Fdh-N (encoded by fdnGHI), is distinct from it and revealed that Fdh-O probably represents the aerobic formate-oxidizing activity originally identified in the early 1950s [9].
Development of an effective and facile in-gel activity-staining procedure has proved useful in the study of the physiology of the Hyd enzymes, particularly with regard to what governs their synthesis and activity [10]. Although the activities of both Fdh-O and Fdh-N have been determined with PMS/nitroblue tetrazolium (NBT) [10], to date, however, no evidence has ever been provided that the activity of the highly labile Fdh-H enzyme [4], encoded by the fdhF gene [2], withstands polyacrylamide gel electrophoretic (PAGE) separation from other anaerobic enzyme complexes. Moreover, no systematic in-gel activity analysis of the Fdh enzymes of E. coli during fermentative growth has been conducted. In this study we identify conditions that reveal Fdh-H enzyme activity after non-denaturing PAGE. The Fdh-H activity is very oxygen-labile, perhaps explaining why previous studies have failed to identify it after gel electrophoresis. Furthermore, these studies identify previously unobserved active enzyme complexes of Fdh-O and Fdh-N during fermentative growth of E. coli.
2. Materials and methods
2.1. Strains and growth conditions
The strains used in this study are listed in Table 1. E. coli strains were routinely grown at 37 °C on LB-agar plates or with shaking in LB-broth [11]. Anaerobic growths for the preparation of extracts to study enzyme activity were performed at 37 °C as standing liquid cultures [12] in M9-minimal medium containing 1× M9 salts [11], 2 mM MgSO4, 0.1 mM CaCl2, 3 µM thiamine hydrochloride, trace element solution SL-A [13], and 0.8% (w/v) glucose. Kanamycin, when required, was used at the final concentration of 50 μg/ml. Cells were harvested anaerobically by centrifugation at 15,000g for 15 min and at 4 °C after the cultures had reached an OD600 nm of either 0.6–0.8 (exponential phase) or 1.2 (stationary phase). Cell pellets were used immediately or stored at −20 °C until use.
Table 1.
E. coli strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| MC4100 | F-, araD139, Δ(argF-lac)U169, λ-, rpsL150, relA1 deoC1, flhD5301, Δ(fruK-yeiR)725(fruA25), rbsR22, Δ(fimB-fimE)632(: IS1) | [28] |
| FTD147 | MC4100 ΔhyaB ΔhybC ΔhycE | [29] |
| FM460 | MC4100 ΔselC (KanR) | [30] |
| CP585 | MC4100 ΔfdhF | This study |
| CP734 | MC4100 ΔhyaB hybC | [31] |
| CP1002 | CP734 ΔhycB (KanR) | This study |
| CP1010 | CP734 ΔfdhF (KanR) | This study |
| HD705 | MC4100 ΔhycE | [32] |
| JW3862 | BW25113 ΔfdhE (KanR) | [14] |
| SH1 | FTD147 ΔselC (KanR) | This study |
| SH173 | FTD147 ΔfdnG | This study |
| SH174 | FTD147 ΔfdoG | This study |
| SH175 | SH173 ΔfdoG (KanR) | This study |
| SH196 | SH175 with KanR-cassette removed | This study |
| SH200 | SH196 ΔselC (KanR) | This study |
2.2. Strain construction
Strains were constructed using P1kc-mediated phage transduction [11] to introduce the respective defined deletion mutation from the appropriate strains obtained from the Keio collection [14]. When required, the plasmid pCP20 was used to remove the kanamycin antibiotic resistance cassette as described [15].
2.3. Preparation of crude cell extracts
Wet cell paste was re-suspended at a ratio of 1 g/3 ml in 50 mM MOPS pH 7 including 5 μg DNase ml−1 and 0.2 mM phenylmethylsulfonyl fluoride. Cells were disrupted aerobically by sonication (30 W power for 5 min with 0.5 s pulses) and then placed immediately under a N2 atmosphere. Unbroken cells and debris were removed by centrifugation for 30 min at 50,000g at 4 °C. The resultant supernatant was termed the crude extract and was used for all studies reported herein.
2.4. Polyacrylamide gel electrophoresis, activity staining, and protein determination
Unless stated otherwise, non-denaturing polyacrylamide gel electrophoresis (PAGE) was carried out under anaerobic conditions in a Coy™ chamber in an atmosphere of 95% N2:5% H2. Non-denaturing PAGE was performed using 6% (w/v) polyacrylamide and gels were maintained under anaerobic conditions prior to staining. Hydrogenase activity-staining was done in 50 mM MOPS buffer pH 7.0, as described [16], and included 0.5 mM benzyl viologen (BV) and 1 mM 2,3,5-triphenyltetrazolium chloride (TTC). Gels were incubated at RT under an atmosphere of 100% highly pure hydrogen gas [10]. Alternatively, staining was done in a 100% hydrogen atmosphere using 0.3 mM phenazine methosulfate (PMS) as mediator and 0.2 mM nitroblue tetrazolium (NBT) as redox dyes [10]. Unless otherwise stated, formate (50 mM final concentration) was added to the activity staining buffer to visualize formate dehydrogenase activity [17], [18]. Determination of protein concentration was done as described [19].
3. Results and discussion
3.1. Active formate dehydrogenase H is present in stationary phase cells
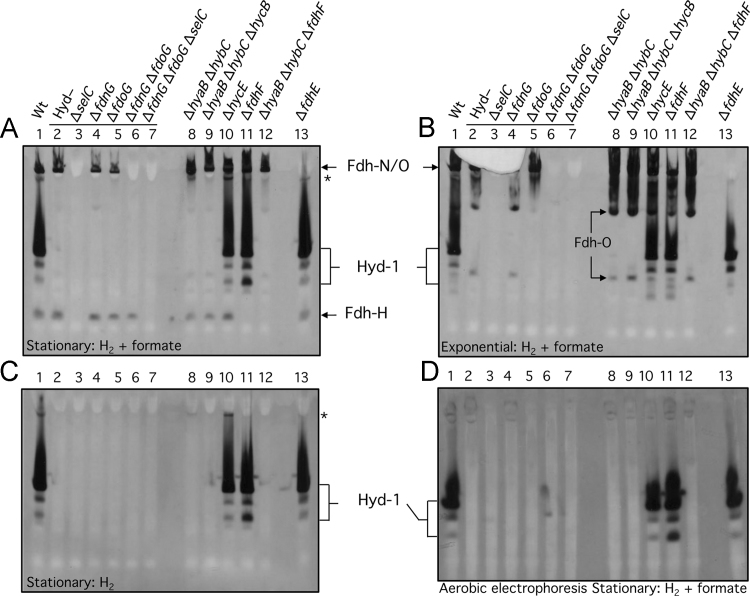
Fdh-H enzyme activity is oxygen-labile [4] and has so far never been observed after non-denaturing PAGE. A previously unobserved, fast-migrating and formate-dependent enzyme activity was revealed after separation of enzyme complexes in extracts derived from the parental strain MC4100 by non-denaturing PAGE (Fig. 1A, lane 1). The gel was run under anaerobic conditions and the enzyme activity was stained with PMS/NBT using a combination of 50 mM formate and a H2 atmosphere for the activity staining procedure. The enzyme activity appeared within seconds of placing the gel in the formate-containing activity stain, without the necessity for bubbling with a 100% H2 atmosphere. This enzyme activity was observed in extracts derived from stationary phase cells only and not in extracts derived from cells harvested in the exponential phase (compare lane 1 in Fig. 2A and B). Moreover, no activity band could be observed when formate was omitted from the staining procedure (Fig. 2C). Both of these findings are consistent with this activity being due to the fdhF gene product, Fdh-H; expression of fdhF occurs preferentially in the stationary phase of growth [2]. To prove that this enzyme activity was indeed due to Fdh-H, an extract derived from an E. coli selC mutant, which cannot insert selenocysteine into any of the Fdh enzymes [2], was shown to be devoid of the activity band (Fig. 2A, lane 3). Notably, the activity bands corresponding to Fdh-N and Fdh-O were also absent from strain FM460 (ΔselC). Introduction of a ΔfdhF allele into the parental strain MC4100, delivering strain CP585 (ΔfdhF), as well as into strain CP734 (ΔhyaB, ΔhybC), delivering strain CP1010 (ΔhyaB, ΔhybC, ΔfdhF), also prevented synthesis of the enzyme activity (Fig. 2A, lanes 11 and 12), again consistent with this species being due to Fdh-H.
Fig. 2.
Identification of conditions revealing the activity of the Fdh-H enzyme. Crude extracts (30 µg of protein) derived from the anaerobically grown E. coli strains indicated were subjected to 6% (w/v polyacrylamide) non-denaturing PAGE and subsequently stained for Fdh and Hyd enzyme activities (A, B, D) or only Hyd activity (C) using the PMS-NBT staining procedure (see Materials and methods). The gels shown in panels A, B, and C were run anaerobically, while that shown in panel D was run aerobically. Cells were harvested when the cultures reached the exponential phase (B) or the stationary phase (A, C, D) of growth. The phase of growth and the electron donors used for staining are indicated at the bottom of each panel. The strains included: Lane 1, MC4100 (wild type); lane 2, FTD147 (ΔhyaB, ΔhybC, ΔhycE); lane 3, SH1 (ΔhyaB, ΔhybC, ΔhycE, ΔselC); lane 4, SH173 (ΔhyaB, ΔhybC, ΔhycE, ΔfdnG); lane 5, SH174, (ΔhyaB, ΔhybC, ΔhycE, ΔfdoG); lane 6, SH175 (ΔhyaB, ΔhybC, ΔhycE, ΔfdnG, ΔfdoG); lane 7, SH200 (ΔhyaB, ΔhybC, ΔhycE, ΔfdnG, ΔfdoG, ΔselC); lane 8, CP734 (ΔhyaB, ΔhybC); lane 9, CP1002 (ΔhyaB, ΔhybC, ΔhycB); lane 10, HD705 (ΔhycE); lane 11, CP585 (ΔfdhF); lane 12, CP1010 (ΔhyaB, ΔhybC, ΔfdhF); lane 13, JW3862 (ΔfdhE). The phenotype or genotype of the respective strain is indicated above the lanes. Note that the horizontal line above lanes 2–7 in A and B indicates that all strains lacked active Hyd-1, -2, and -3. The activity band(s) due to the respective enzyme complexes are indicated beside the panels and the asterisk denotes an unidentified H2-dependent enzyme complex. The migration of two faster-migrating Fdh-O-dependent enzyme complexes observed in exponentially growing cells (B) is indicated within the figure.
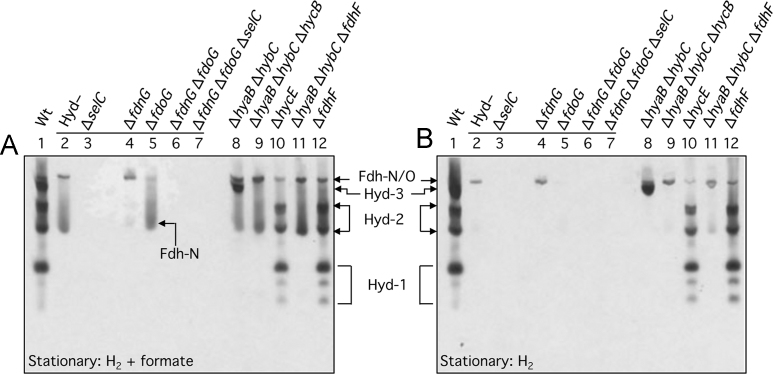
Next, we examined whether Fdh-H activity was reliant on the presence of either Fdh-O or Fdh-N. Deletion of the fdoG gene or the fdnG gene, encoding the catalytic subunit of Fdh-O or Fdh-N, respectively, or indeed both genes together, did not affect the appearance of the Fdh-H activity (Fig. 2A, compare lanes, 4, 5 and 6). Introduction of the ΔselC allele into the Fdh-N−/ Fdh-O− double null strain, delivering strain SH200 (see Table 1), abolished Fdh-H activity, as anticipated (Fig. 2A, lane 7). Finally, analysis of a mutant lacking the fdhE gene, which encodes an iron-sulfur protein with an essential function in the maturation of the periplasmically-oriented Fdh-O and Fdh-N, but not that of Fdh-H [20], [21], exhibited a clear activity band due to Fdh-H, but completely lacked enzyme activity for either Fdh-N or Fdh-O (Fig. 2A, lane 13). Together, these data identify the fast-migrating formate-dependent enzyme activity as Fdh-H and demonstrate that activity of the enzyme is high in the stationary phase of growth and is independent of the other two Fdh enzymes. The latter observation is consistent with earlier findings [2], [4].
3.2. Barriers to determining Fdh-H in-gel activity
The oxygen-labile nature of the Fdh-H enzyme activity during purification has been well documented [4], [22], [23]. To determine whether the Fdh-H activity detected after anaerobic non-denaturing PAGE could also be visualized after performing the electrophoresis under aerobic conditions, the same samples as those shown in Fig. 2A were subjected to aerobic non-denaturing PAGE (Fig. 2D). No Fdh-H enzyme activity could be observed. Despite the cells being briefly exposed to O2 during disruption, Fdh-H activity was retained in the crude extract. It appears that separation of the enzyme from the rest of the FHL complex during electrophoresis renders it more sensitive to the deleterious effects of oxygen [4], [22] (R.G. Sawers unpublished data).
When the same crude extracts from stationary phase cells were separated by anaerobic non-denaturing PAGE and the gel was incubated in a H2 atmosphere with 50 mM formate as electron donor and BV/TTC as electron acceptors, no Fdh-H enzyme activity band could be observed, despite the detection of the well-characterized bands of the three hydrogenases Hyd-1, Hyd-2 and Hyd-3 and weak activities due to Fdh-N and Fdh-O (Fig. 3A). The redox potential of H2-saturated, formate buffer used for the BV/TTC activity stain (E′=−320 mV) was significantly lower than that of the PMS/NBT stain (E′=−230 mV), suggesting that the Fdh-H enzyme catalyzes electron transfer more efficiently to the buffer with the more positive redox potential.
Fig. 3.
The Fdh-H enzyme lacks in-gel formate: BV-TTC oxidoreductase activity. Crude extracts (30 µg of protein) derived from the same E. coli strains shown in Fig. 1 were stained for Fdh and Hyd enzyme activities (A) or only Hyd activity (A) using the BV-TTC staining procedure (see Materials and methods). Cells were harvested when the cultures reached the stationary phase of growth. The strains included: Lane 1, MC4100 (wild type); lane 2, FTD147 (ΔhyaB, ΔhybC, ΔhycE); lane 3, SH1 (ΔhyaB, ΔhybC, ΔhycE, ΔselC); lane 4, SH173 (ΔhyaB, ΔhybC, ΔhycE, ΔfdnG); lane 5, SH174, (ΔhyaB, ΔhybC, ΔhycE, ΔfdoG); lane 6, SH175 (ΔhyaB, ΔhybC, ΔhycE, ΔfdnG, ΔfdoG); lane 7, SH200 (ΔhyaB, ΔhybC, ΔhycE, ΔfdnG, ΔfdoG, ΔselC); lane 8, CP734 (ΔhyaB, ΔhybC); lane 9, CP1002 (ΔhyaB, ΔhybC, ΔhycB); lane 10, HD705 (ΔhycE); lane 11, CP1010 (ΔhyaB, ΔhybC, ΔfdhF); lane 12, CP585 (ΔfdhF). The activity band(s) due to the respective enzyme complexes are indicated beside the panels. The migration position of a fast-migrating Fdh-N-dependent enzyme complex observed after activity staining in the presence of formate (A) is indicated within the figure.
3.3. Fdh-H is active in the absence of a functional Hyd-3
The Fdh-H enzyme has, so far, only been observed to be associated with the FHL complex [24], although it has been suggested that it might also form a complex with the hyf gene products, encoding Hyd-4 [3]. The fdhF gene, which encodes Fdh-H, is located separately, and in isolation, from the hyf genes and from those encoding Hyd-3 and the FHL complex, which would be in accord with this proposal. This is also in line with our finding here that Fdh-H activity, after separation in non-denaturing PAGE, does not co-migrate with Hyd-3 activity (Fig. 2A). A recent study has demonstrated that if the fdhF gene is deleted, the in-gel activity of Hyd-3 is still measureable but is significantly reduced in intensity [10]. This result was confirmed in the current study (Fig. 3, compare lanes 1, 8, 11 and 12). To test whether the in-gel Fdh-H activity is influenced by the absence of Hyd-3, an extract from HD705 (ΔhycE) was prepared and after non-denaturing PAGE the gel was stained for Fdh-H activity (Fig. 2A, lane 10). The result clearly demonstrates that Fdh-H activity is independent of the presence of Hyd-3 activity in stationary phase E. coli cells. Moreover, Fdh-H activity could also be visualized in a mutant lacking the HycB protein (Fig. 2A, lane 9), which has been proposed to act as the small subunit channeling electrons from Fdh-H to the FHL complex [2], [24]. It should be noted that the fdhF gene product has an iron-sulfur cluster and when purified anaerobically the protein is able to transfer electrons to redox dyes [4]. Together, these results indicate that Fdh-H retains activity in the absence of the catalytic subunit HycE of the Hyd-3 enzyme or in strains that lack HycB, the protein that is presumed to act as the docking site for Fdh-H on the FHL complex [2], [24].
3.4. Active Fdh-O and Fdh-N sub-complexes in exponential phase cells
While the activities of both Fdh-N and Fdh-O are detectable in extracts of exponential and stationary phase cells, activity of both enzymes is highest in exponential phase cells (Fig. 2B). This contrasts with the activity of Fdh-H, which is more active in stationary phase cells (see Fig. 2A). It was noted that two new, faster migrating sub-complexes that were dependent on the presence of the fdoG gene, encoding the catalytic subunit of Fdh-O, were observed in extracts derived from exponential phase cells after activity staining with PMS/ NBT (Fig. 2B, lanes 8–12). These two activity bands were observed in extracts of a strain lacking Fdh-N (Fig. 2B, lane 4), but not in extracts of strains lacking Fdh-O (Fig. 2B, lane 5), strongly suggesting that they are additional complexes of Fdh-O. Consistent with this suggestion, these new activity bands were absent in extracts of the selC mutant FM460 (Fig. 2B, lane 3). It is currently unclear whether these active species are sub-complexes of a presumptive trimer-of-trimers (3 x FdnGHI), as has been observed for Fdh-N [25], or whether they are complexes of Fdh-O interacting with other redox enzymes.
A rather diffuse additional, faster-migrating activity band that was dependent on Fdh-N was also observed when extracts from stationary phase cells were subjected to the activity staining procedure using BV/TTC as electron acceptor (Fig. 3A, lanes, 2, 5, 8, 9 and 11). This additional activity band was almost completely absent in extracts derived from strain SH173 (Fig. 3A, lane 4), which, in addition to lacking genes encoding the catalytic subunits of Hyd-1 through -3, is also devoid of fdnG (Table 1). Notably, the activity was dependent upon formate (Fig. 3B). Thus, despite the Fdh-O and Fdh-N enzymes sharing high levels of amino acid sequence similarity, e.g. the catalytic subunits of both enzymes share 76% identity, the fact that the newly identified Fdh-O and Fdh-N complexes specifically reduced PMS/NBT and BV/TTC, respectively, indicates that they differ in the substrates to which they deliver electrons. In the case of Fdh-O this might be related to the fact that the enzyme is synthesized under aerobic as well as anaerobic conditions [7], and that it forms ‘super-complexes’ with cytochrome o and cytochrome d oxidases [26]. Consequently, Fdh-O possibly uses ubiquinone rather than menaquinone as physiological electron donor, as has been previously suggested [27], and hence its preferential use of the redox dye combination of PMS/ NBT, which has a more positive redox potential than BV/TTC. Nevertheless, both enzymes can also oxidize H2 when BV/TTC are the electron acceptors (Fig. 3, compare A and B), which is in agreement with the findings of earlier work [18].
Finally, a further, previously unobserved, H2-dependent activity band was identified in extracts of the parental strain MC4100 and in HD750 (ΔhycE) after staining with PMS/NBT buffer (Fig. 2C, see lanes 1 and 8). This activity was not observed when electrophoretically separated extracts were stained using BV/TTC buffer (Fig. 3). The activity band observed with PMS/NBT was independent of active Hyd-3, but showed a clear reliance on Fdh-H, and was absent if Hyd-1, -2 and -3 activities were absent. If only Hyd-1 and Hyd-2 were absent, then the activity was present but was weak (Fig. 2A and C, lane 8). The nature of this enzyme activity remains to be elucidated.
4. Conclusions
The findings of this study identify for the first time the activity of the oxygen-labile Fdh-H in extracts of stationary phase fermenting E. coli using chromogenic activity staining. Visualization of the enzyme activity required that the non-denaturing polyacrylamide gel electrophoresis had to be performed anaerobically. Surprisingly, the activity of the enzyme could only be observed using the redox dye combination of PMS and NBT, presumably because under the assay conditions used it has a more positive redox potential than the combination of BV and TTC. The latter combination functions optimally for the determination of hydrogenase activity. Fdh-H activity was shown to depend on formate and the machinery for incorporation of selenocysteine. The enzyme activity was independent of the other two Fdh enzymes and of a functional Hyd-3 enzyme. The latter finding correlates with the fact that Fdh-H might associate with different enzyme complexes, as depicted in the model presented in Fig. 1, to supply electrons derived from formate [2].
Acknowledgment
This work was supported by the Deutsche Forschungsgemeinschaft (FOR 1530 and SA 494/3-2).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.03.006.
Appendix A. Supplementary materials
Supplementary data
Supplementary data
Supplementary data
References
- 1.Sieber J.R., McInerney M.J., Gunsalus R.P. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Ann. Rev. Microbiol. 2012;66:429–452. doi: 10.1146/annurev-micro-090110-102844. [DOI] [PubMed] [Google Scholar]
- 2.R.G. Sawers, M. Blokesch, A. Böck, Anaerobic formate and hydrogen metabolism, in: R. Curtiss III (Ed. in Chief), EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology, ASM Press, Washington, D.C. [Online] 〈http://www.ecosal.org〉, September 2004, Chapter 3.5.4.
- 3.Trchounian K., Poladyan A., Vassilian A., Trchounian A. Multiple and reversible hydrogenases for hydrogen production by Escherichia coli: dependence on fermentation substrate, pH and FOF1-ATPase. Crit. Rev. Biochem. Mol. Biol. 2012;47:236–249. doi: 10.3109/10409238.2012.655375. [DOI] [PubMed] [Google Scholar]
- 4.Axley M.J., Grahame D.A., Stadtman T.C. Escherichia coli formate-hydrogen lyase: purification and properties of the selenium-dependent formate dehydrogenase component. J. Biol. Chem. 1990;265:18213–18218. [PubMed] [Google Scholar]
- 5.Cox J.C., Edwards E.S., DeMoss J.A. Resolution of distinct selenium-containing formate dehydrogenases from Escherichia coli. J. Bacteriol. 1981;145:1317–1324. doi: 10.1128/jb.145.3.1317-1324.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pommier J., Mandrand M.A., Holt S.E., Boxer D.H., Giordano G. A second phenazine methosulphate-linked formate dehydrogenase isoenzyme in Escherichia coli. Biochim. Biophys. Acta. 1992;1107:305–313. doi: 10.1016/0005-2736(92)90417-k. [DOI] [PubMed] [Google Scholar]
- 7.Sawers G., Heider J., Böck A. Expression and operon structure of the sel genes of Escherichia coli and identification of a third selenium-containing formate dehydrogenase isoenzyme. J. Bacteriol. 1991;173:4983–4993. doi: 10.1128/jb.173.16.4983-4993.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abaibou H., Pommier J., Benoit S., Giordano G., Mandrand-Berthelot M.A. Expression and characterization of the Escherichia coli fdo locus and a possible physiological role for aerobic formate dehydrogenase. J. Bacteriol. 1995;177:7141–7149. doi: 10.1128/jb.177.24.7141-7149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinsent J. The need for selenite and molybdate in the formation of formic dehydrogenase by members of the coli-aerogenes group of bacteria. Biochem. J. 1953;57:10–16. doi: 10.1042/bj0570010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinske C., Jaroschinsky M., Sargent F., Sawers G. Zymographic differentiation of [NiFe]-hydrogenases 1, 2 and 3 of Escherichia coli K-12. BMC Microbiol. 2012;12:134. doi: 10.1186/1471-2180-12-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller J. Cold Spring Harbor Laboratories; 1972. Experiments in Molecular Genetics. [Google Scholar]
- 12.Pinske C., Jaroschinsky M., Linek S., Kelly C.L., Sargent F., Sawers R.G. Physiology and bioenergetics of [NiFe]-hydrogenase 2-catalyzed H2-consuming and H2-producing reactions in Escherichia coli. J. Bacteriol. 2015;197:296–306. doi: 10.1128/JB.02335-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hormann K., Andreesen J. Reductive cleavage of sarcosine and betaine by Eubacterium acidaminophilum via enzyme systems different from glycine reductase. Arch. Microbiol. 1989;153:50–59. [Google Scholar]
- 14.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K., Tomita M., Wanner B., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cherepanov P., Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 16.Ballantine S.P., Boxer D.H. Nickel-containing hydrogenase isoenzymes from anaerobically grown Escherichia coli K-12. J. Bacteriol. 1985;163:454–459. doi: 10.1128/jb.163.2.454-459.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enoch H.G., Lester R.L. The purification and properties of formate dehydrogenase and nitrate reductase from Escherichia coli. J. Biol. Chem. 1975;250:6693–6705. [PubMed] [Google Scholar]
- 18.Soboh B., Pinske C., Kuhns M., Waclawek M., Ihling C., Trchounian K., Trchounian A., Sinz A., Sawers R.G. The respiratory molybdo-selenoprotein formate dehydrogenases of Escherichia coli have hydrogen: benzyl viologen oxidoreductase activity. BMC Microbiol. 2011;11:173. doi: 10.1186/1471-2180-11-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowry O., Rosebrough N., Farr A., Randall R. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 20.Schlindwein C., Giordano G., Santini C.L., Mandrand M.A. Identification and expression of the Escherichia coli fdhD and fdhE genes, which are involved in the formation of respiratory formate dehydrogenase. J. Bacteriol. 1990;172:6112–6121. doi: 10.1128/jb.172.10.6112-6121.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu¨ke I., Butland G., Moore K., Buchanan G., Lyall V., Fairhurst S.A., Greenblatt J.F., Emili A., Palmer T., Sargent F. Biosynthesis of the respiratory formate dehydrogenases from Escherichia coli: characterization of the FdhE protein. Arch. Microbiol. 2008;190:685–696. doi: 10.1007/s00203-008-0420-4. [DOI] [PubMed] [Google Scholar]
- 22.Sawers R.G., Ballantine S.P., Boxer D.H. Differential expression of hydrogenase isoenzymes in Escherichia coli K-12: evidence for a third isoenzyme. J. Bacteriol. 1985;164:1324–1331. doi: 10.1128/jb.164.3.1324-1331.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyington J.C., Gladyshev V.N., Khangulov S.V., Stadtman T.C., Sun P.D. Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science. 1997;275:1305–1308. doi: 10.1126/science.275.5304.1305. [DOI] [PubMed] [Google Scholar]
- 24.McDowall J.S., Murphy B.J., Haumann M., Palmer T., Armstrong F.A., Sargent F. Bacterial formate hydrogenlyase complex. Proc. Natl. Acad. Sci. USA. 2014;111:E3948–E3956. doi: 10.1073/pnas.1407927111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jormakka M., Törnroth S., Byrne B., Iwata S. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science. 2002;295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- 26.Sousa P.M.F., Videira M.A.M., Melo A.M.P. The formate:oxygen oxidoreductase supercomplex of the Escherichia coli aerobic respiratory chain. FEBS Lett. 2013;587:2559–2564. doi: 10.1016/j.febslet.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 27.Giordano G., Grillet L., Rosset R., Dou J.H., Azoulay E., Haddock B.A. Characterization of an Escherichia coli K12 mutant that is sensitive to chlorate when grown aerobically. Biochem. J. 1978;176:553–561. doi: 10.1042/bj1760553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casadaban M.J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 29.Redwood M., Mikheenko I., Sargent F., Macaskie L. Dissecting the roles of Escherichia coli hydrogenases in biohydrogen production. FEMS Microbiol. Lett. 2007;278:48–55. doi: 10.1111/j.1574-6968.2007.00966.x. [DOI] [PubMed] [Google Scholar]
- 30.Leinfelder W., Forchhammer K., Zinoni F., Sawers G., Mandrand-Berthelot M.A., Böck A. Escherichia coli genes whose products are involved in selenium metabolism. J. Bacteriol. 1988;170:540–546. doi: 10.1128/jb.170.2.540-546.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinske C., Kru¨ger S., Soboh B., Ihling C., Kuhns M., Braussemann M., Jaroschinsky M., Sauer C., Sargent F., Sinz A., Sawers R.G. Efficient electron transfer from hydrogen to benzyl viologen by the [NiFe]-hydrogenases of Escherichia coli is dependent on the coexpression of the iron-sulfur cluster-containing small subunit. Arch. Microbiol. 2011;193:893–903. doi: 10.1007/s00203-011-0726-5. [DOI] [PubMed] [Google Scholar]
- 32.Sauter M., Böhm R., Böck A. Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol. Microbiol. 1992;6:1523–1532. doi: 10.1111/j.1365-2958.1992.tb00873.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data