Abstract
Background
Although AQP10 is mainly expressed in the human GI tract, its physiological role is unclear. In fact, we previously reported that mouse AQP10 is a pseudogene. It is possible that AQP10 is also a pseudogene in other animals.
Methods
Genome databases were searched for AQP10 orthologs and the genomic DNA of each candidate pseudogene was sequenced to confirm its mutations. The expression of the AQP10 mRNA was examined by RT-PCR in the small intestine where human AQP10 is highly expressed.
Results
The genomic database of some mammals had insertions and deletions in the exons of the AQP10 gene, including cattle (Bos taurus), sheep (Ovis aries) and goats (Capra hircus). In the bovine AQP10 gene, exon 1 and 5 had deletions resulting in a frame-shift or a premature termination, respectively, which were confirmed by the direct exon sequencing of the genomic DNA. In the RT-PCR experiments, the PCR primer sets for exon 1/2 and exon 4/5 failed to detect the bands for AQP10 mRNA in the duodenum and jejunum. Similar AQP10 gene mutations were also confirmed in the genomic DNA from sheep and goats. Although these animals were derived from porcine ancestors, the exons of the swine (Sus scrofa) AQP10 gene were complete without mutations. Therefore, AQP10 gene might have turned to a pseudogene around 65 million years before when cattle evolved from porcine ancestors.
Conclusion
AQP10 of ruminantia which regurgitate and rechew their food may have lost its role possibly due to the redundant expression of other aquaglyceroporins.
Keywords: Aquaporin, Pseudogene, Bovine, Evolution, Frame shift, Genome project
Highlights
-
•
AQP10, mainly expressed in the GI tract, has turned to a pseudogene in the cow.
-
•
The other ruminantia such as sheep and goat also have the AQP10 pseudogene.
-
•
The presence of authentic AQP10 in the pig may indicate that AQP10 turned to a pseudogene 65 million years ago.
1. Introduction
Aquaporins (AQPs) are a family of small integral plasma membrane proteins that primarily transport water across the plasma membrane driven by the osmotic gradient reviewed in [1]. AQP10 is the fourth and last aquaglyceroporin discovered in mammals following AQP3, AQP7, and AQP9 [2], [3], [4]. Interestingly, human AQP10 has two isofoms: a fully-spliced form (isoform 1, 301 amino acids) [3] and a partially-unspliced form (isoform 2, 264 amino acids) [2]. The sequence of the isoform 2 in 236–301 amino acid residues, most of the C-terminus, is different from that of the isoform 1, leading to the loss of the 6th transmembrane, which will explain the poor function of the isoform 2 [2]. In fact, the isoform 1 was expressed predominantly in the GI tract [2]. Therefore, the isoform 1 should be the focus of the functional and physiological studies.
Unfortunately, one commercially available anti-AQP10 antibody will react specifically to the isoform 2 at the 17 amino acid C-terminal sequence (Alpha Diagnostics), while another will react specifically to the isoform 1 at the 50 amino acid C-terminal sequence (Sigma). Therefore, the results with the antibody from Alpha Diagnostics should reflect those of the isoform 2. The result that AQP10 was expressed at the apical membrane of epithelial cells in the small intestine by Mobasheri et al. was based on the isoform 2-specific antibody [5]. On the other hand, the isoform 1-specific antibodies localized AQP10 at the granular vesicles of enterochromaffin cells in the duodenum [6]. Thus, AQP10 may play a role in enterochromaffin cells to modulate its secretion and not in the absorption of water at the epithelium. Recently, human AQP10 has been found to be expressed in the adipose tissue by the isoform 2-specific antibody [7]. Whether the isoform 1 is also expressed or not remained to be clarified. Human AQP10 was also expressed in the skin with the antibody against the isoform 1 [8]. However, the expression level was very low compared with AQP3.
To our surprise, the murine AQP10 gene has turned to a non-processed pseudogene without the authentic gene [9]. A pseudogene is usually produced by gene duplications where the duplicated gene turns to a pseudogene so that the remaining original gene can secure the function. For example, human AQP7 has three pseudogenes in addition to the authentic gene [10]. Thus, the absence of authentic AQP10 gene in mice suggests that AQP10 is generally not essential or the other aquaglyceroporins may compensate for the loss of the functional AQP10 protein.
We speculated that some mammals other than mice may also have lost the functional AQP10 gene. The complete or near-complete genome sequences from many mammals in public databases have enabled us to search for the AQP10 gene ortholog. We found that the bovine AQP10 gene is also a pseudogene as well as its relatives, sheep and goats. They are subgrouped as ruminantia which regurgitate and rechew their food.
2. Materials and methods
2.1. Identification of the AQP10 ortholog in mammals
We blast-searched the NCBI database (http://www.ncbi.nlm.nih.gov/genome/) with human AQP10 cDNA (Genbank accession # AB066105.1), AQP1 (#AB451275.1), and AQP11 (AB028147.1) as queries. Each candidate gene was assigned to be the AQP ortholog based on the absence of other highly homologous genes. Furthermore, the completed genomic database was also searched for the annotated AQPs.
2.2. Genomic DNA isolation and the exon sequencing
The genomic DNA was isolated from the muscle of cattle, sheep and goat using DNeasy Blood & Tissue Kit (QIAGEN) following the manufacturer׳s instructions. The meats were obtained from local shops. The primers were set to amplify the exons which had deletions or insertions in the database. The amplified DNA was gel-purified and subcloned into a plasmid vector using TOPO TA cloning Kit (Invitrogen Japan K.K.). The sequencing reaction was performed with BigDye Terminator V3.1 Cycle Sequencing Kit (Applied Biosystems) with M13 Reverse Primer and T7 promoter primer to sequence both strands. The primers used to amplify each genomic DNA are shown in Table 1.
Table 1.
The primers used for PCR.
| Primers | Forward primer | Reverse primer |
|---|---|---|
| Bos AQP10 N-end | cagtctctggttgaagtcaagg | tgacctgagacattaccactcac |
| Bos AQP10 e4–5 | ttccatctttgccacctacc | gtcctggacagggttgattg |
| Bos GAPDH | tcaagaaggtggtgaagcag | gattctcagtgtggcggaga |
| Sheep AQP10 e5 | ggacaaagtcagatgccatgg | ggaacccaccaccaaccatcg |
| Goat AQP10 e5 | tgtcgactccaaggttctgg | ctgtcctgagatgccaagag |
2.3. Total RNA isolation and RT-PCR analysis for mRNA expression
We obtained bovine tissues from a supplier (Shibaurazouki Inc., Tokyo). Total RNA was extracted from each tissue using RNeasy Mini Kit (QIAGEN) following the manufacturer׳s instructions. Reverse transcription was performed with Takara RNA PCR Kit (AMV) Ver.3.0 (Takara Bio, Ootsu, Japan) following the manufacturer׳s instructions. Reverse transcription: 50 °C for 30 min, 95 °C for 5 min, 5 °C for 5 min followed by PCR using the primers spanning the intron (Table 1). PCR: 94 °C for 4 min, then 35 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and a final extension of 72 °C for 5 min. The sizes of the PCR products were analyzed by 1% agarose gel electrophoresis. RT-PCR was also performed using the primers for the bovine GAPDH gene spanning an intron (GenBank accession # NM_001034034) (Table 1).
3. Results
3.1. Screening of the genome database
We screened the completed genome sequences of 14 primates, 12 rodents, 14 even-toed ungulates and whales (Cetartiodactyla), and 30 other mammals for AQP1, AQP10 and AQP11 orthlogs. Both AQP1 and AQP11 had orthologs in their genomes with perfect open reading frames. However, some AQP10 orthlogs were frame-shifted due to insertions and deletions. These included Mus musculus (house mouse: NG_021121.1 as reported in [9]), Microtus ochrogaster (prairie vole: XM_005367307.1: two bases inserted in one codon), Peromyscus maniculatus bairdii (Prairie Deer Mouse: XM_006976464.1: three bases inserted in 2 codons; five bases deleted in three codons; one base substituted to be a stop codon), Bos taurus (cattle: XM_003581937.2: two bases inserted in one codon), Bos mutus (XM_005898757.1: two bases inserted in one codon; one base substituted to be a stop codon), Bubalus bubalis (water buffalo: XM_006044200.1: two bases inserted in one codon; one base substituted to be a stop codon), Vicugna pacos (alpaca: XM_006220151.1: one base inserted in one codon; one base deleted in one codon), Capra hircus (goat: XM_005677552.1: one base inserted in one codon; two bases deleted in one codon), Ovis aries (sheep: XM_004003657.1: one base deleted in one codon), Pantholops hodgsonii (chiru: XM_005975129.1: one base inserted in one codon), Balaenoptera acutorostrata scammoni (whale: XM_007198353.1: one base substituted to be a stop codon), Orcinus orca (killer whale: XM_004284659.1: one base inserted in one codon; one base deleted in one codon; two bases substituted to be two stop codons), Physeter catodon (sperm whale: XM_007130325.1: one base inserted in one codon; one base deleted in one codon), Tursiops truncatus (bottlenosed dolphin: XM_004330549.1: one base inserted in one codon; one base deleted in one codon), Lipotes vexillifer (Yangtze River dolphin: XM_007457995.1: one base inserted in one codon; three bases deleted in two codons; two bases substituted to be two stop codons), and Camelus ferus (Wild Bactrian camel: XM_006195679.1: two bases inserted in two codons; one base deleted in one codon).
Each was assigned to be the AQP10 ortholog based on the absence of other highly homologous genes to AQP10. We also referred to the AQP10 orthologs from annotation pipeline available at “ortholog_gene_89872” in NCBI from 103 organisms.
It should be noted that Sus scrofa (pig), the ancestor of cattle, has a perfect AQP10 gene without deletions or insertions (NCBI Reference Sequence: NC_010446.4).
3.2. The genomic AQP10 gene of ruminantia
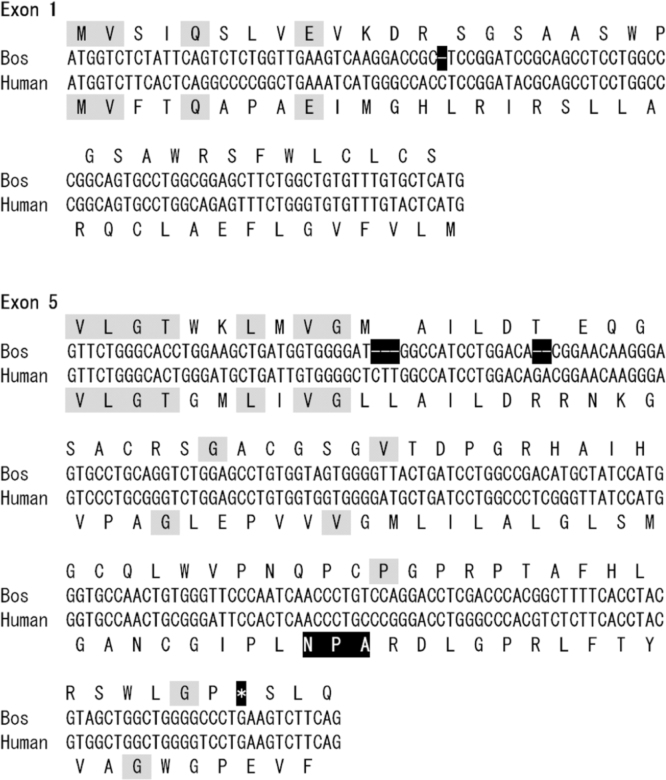
In the genome of B. Taurus, we identified the AQP10 ortholog (NCBI Reference Sequence: AC_000160.1). The exon nucleotide sequences of the bovine AQP10 gene were highly homologous to those of the human AQP10 gene. However, the open reading frame for the bovine AQP10 gene had two deletions leading to frame-shifts at exon 1 and exon 5 (exon 1; C and exon 5; CTT, GA)(Fig. 1, Fig. 2). These frame-shifts will prevent the proper translation of the full AQP10 protein, especially at exon 5 which encodes the second NPA box, essential for the function of water transport. Exon 5 also had a premature stop codon which is present at exon 6 in the human AQP10 gene.
Fig. 1.
The nucleic acid and the three reading-frames of translated amino acid sequences from exon 1 and exon 5 in the bovine AQP10 gene. The identical amino acids with human AQP10 were shaded. The open reading frame of the bovine AQP10 gene was frame-shifts at exon 1 and exon 5 due to the deletions as shown in the next figure.
Fig. 2.
The nucleic acid and the translated amino acid sequences from exon 1 and exon 5 in the bovine and human AQP10 genes. The gene deletions of the bovine AQP10 gene in exon 1 and exon 5 were corrected with dashed inserts to improve the nucleic acid alignments. The highly conserved second NPA box in human AQP10 was boxed which was absent in bovine AQP10. A premature stop site (*) of bovine AQP10 was also boxed.
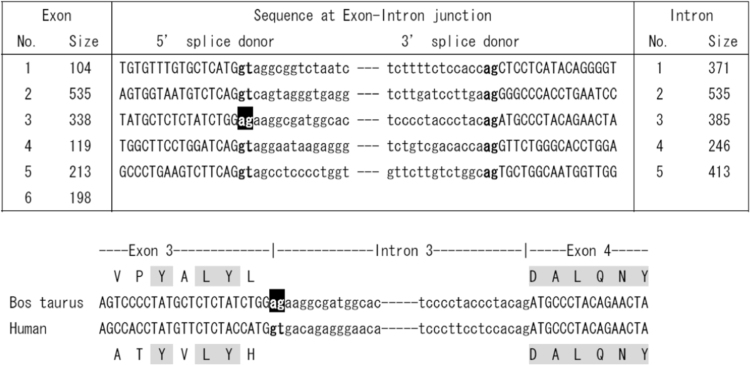
The analysis of the nucleotide sequences at exon/intron boundaries revealed that the all followed GT/AG rules except for exon 3/intron 3, which will inhibit a proper splicing (Fig. 3).
Fig. 3.
The analysis of nucleotide sequences at the exon/intron boundaries of the bovine AQP10 gene. All exon/intron boundaries except the exon 3/intron 3 boundary followed the GT/AG rule for splicing.
The 5׳ upper stream promoter region had consensus sequences for the binding of transcription factors such as AP-1 and -4, C/EBP, GATA-1 and -2 as well as the transcription initiation site, TATA box (Fig. 4). Thus, it is possible that the bovine AQP10 gene is transcribed even with a frame-shifted and truncated open reading frame.
Fig. 4.
The 5′ upstream region of the bovine AQP10 gene. The consensus sequences for the binding of general transcription factors such as AP-1,-4, C/EBP, GATA-1,-2 as well as the transcription initiation site, TATA box, were marked by an underline. The translation initiation codon ATG for methionine was boxed.
As cattle belong to ruminantia which regurgitate and rechew their food, we then examined other ruminantia, goats and sheep in the database in more detail.
The goat AQP10 (XM_005677552.1) had a deletion at exon 5: ATCCAGGTG-CCCAGAAC, which leads to a reading-frame shift (Fig. 5). The sheep AQP10 (XM_004003657.1) had a stop codon at exon 5: ACCTGAATG, and a deletion at exon 5: CTGAGCTCCCAATC-AACCTGCCCA, which leads to a reading-frame shift (Fig. 6). Therefore, both goat and sheep AQP10 genes were frame shifted and will be pseudogenes.
Fig. 5.
The translated amino acid sequences in the first and third reading-frames of the goat AQP10 gene. The sequences in bold type indicated the homologous amino acid sequence to that of human AQP10, which spanned the different reading-frames. The two underlines indicate NPA boxes which are the signature sequences of the aquaporin family.
Fig. 6.
The translated amino acid sequences in the first and third reading-frames of the sheep AQP10 gene. The sequences in bold type indicated the homologous amino acid sequences to that of human AQP10, which spanned the different reading-frames. The two underlines indicate NPA boxes.
3.3. The exon sequencing of the genomic DNA
As the above mutations in the ruminantia AQP10 genes could be caused by sequence errors, we then sequenced the genomic AQP10 genes by ourselves. The mutated exons were amplified by PCR and sequenced in the both strands. The sequencing confirmed their frame-shifting mutations in the database as described above (Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6).
Therefore, the AQP10 gene of cattle, sheep and goats should be a pseudogene as each cannot produce a functional protein even if it is translated.
3.4. AQP10 mRNA expression in the bovine duodenum
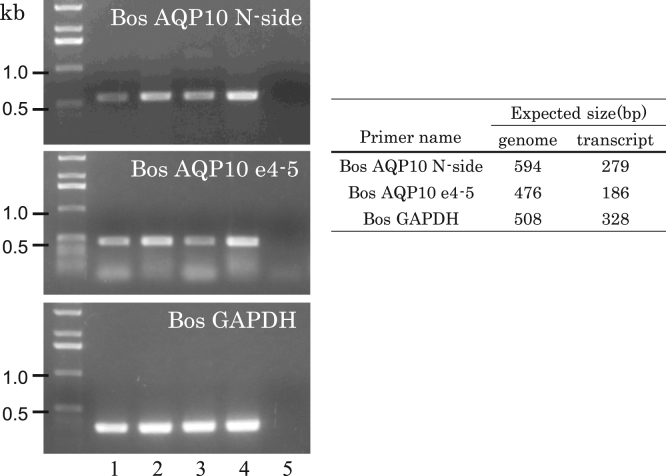
We then examined the expression of bovine AQP10 transcript in the duodenum and jejunum where AQP10 is highly expressed in human. We employed a more sensitive RT-PCR technique than Northern blot analyses. The primer sets spanning an intron were used to distinguish the expected bands for mRNA by RT-PCR from the bands from the contaminated genomic DNA. The primer sets were derived from exon 1–exon 2 and exon 4–exon 5 spanning an intron (Table 1).
The expected bands for the bovine AQP10 mRNA were not detected although the expected bands for the contaminated genomic DNA were detected (Fig. 7). The integrity of mRNA was confirmed by RT-PCR for GAPDH mRNA (Fig. 7). One RT-PCR product derived from B. taurus genomic DNA was sequenced and found to have the deletion of cytosine at exon 1 with intron 1 as expected by the database.
Fig. 7.
The absence of the bovine AQP10 mRNA expression in the GI tract. Lanes 1–3 were from the mRNA of the duodenum, Lane 4 from the mRNA of the jejunum, and Lane 5 from water (a negative control). The primers were designed to span an intron to distinguish the mRNA bands from the genomic DNA bands. The primers were from exon 1/exon 2 (N-side), and exon 4/exon 5 (e4–5). Only the bands from the contaminated genomic DNA were detected, which indicated the absence of the expression of the bovine mRNA. The bands for the GAPDH mRNA were detected in all tissues (a positive control).
Furthermore, the blast search for AQP10 cDNA clones in bovine ESTs libraries at NCBI failed to detect any clones. On the other hand, a porcine AQP10 EST clone, GenBank: EW289993.2 from small intestine and jejunum from KVL (Royal Veterinary and Agricultural University) was identified by the blast search. Another porcine AQP10 EST clone, GenBank: DY418303.1 from the library made with RNA pooled from multiple tissues including brain, liver, muscle, placenta/endometrium, ovary, testes, and bone marrow was also found. Therefore, bovine AQP10 gene may not be transcribed or degraded for its unstable mRNA.
4. Discussion
The present study has expanded the scope of pseudogenes for AQP10 in mammals. So far, only the murine AQP10 gene was identified as a pseudogene [9]. Here, we showed that the bovine AQP10 gene is also a pseudogene. The genome sequences of bovine AQP10 have multiple structural defects including frame-shifts and a splice junction mutation. The degree of mutations in the bovine AQP10 gene seems to be less than that of the murine AQP10 gene and both seemed to be unrelated. For example, the murine AQP10 gene has a deletion of a guanine (G) at the ATG translation-initiation site, which will strongly inhibit the translation. However, this is not the case with the bovine AQP10 gene. The murine AQP10 gene has an insertion and a deletion at exon 2 and exon 5, respectively, while the bovine AQP10 gene has deletions at exon 1 and exon 5. As the frame-shifts are differently located in mice and cattle, the mutations in the AQP10 genes may not be related. Thus both may have independently turned to a pseudogene.
As the transcript of the bovine AQP10 gene was not detected in the duodenum where human AQP10 is highly expressed [2], [3], this pseudogene may have a transcription problem although the 5′ upstream non-coding region seemed to have functional consensus sequences including a TATA box as is the case with murine AQP10 gene [9]. Even if the bovine AQP10 gene should be transcribed in tissues other than the small intestine, it will not play a role in antisense interference or generation of small interfering RNAs (siRNA) as no authentic AQP10 gene is present [11].
Interestingly, the AQP10 genes of sheep and goats, the relatives of cattle, have also turned to a pseudogene. These animals belong to ruminantia, which regurgitate and rechew their food. Furthermore, whales may have AQP10 pseudogenes if the genomic database is correct. In that case, the extent of AQP10 pseudogenes will be large and beyond ruminantia. As there seemed to be shared mutations among these animals, AQP10 may have turned to a pseudogene at the time of the advent of artiodactyla (even-toed ungulates) with the exception of swine. Since pseudogenes do not have functional constraint, they usually accumulate more mutations than functional genes. Further analysis in the phylogeny will be necessary to examine the time course and extent of the AQP10 pseudogene formation in mammals as well as in the lower animals [12].
Pseudogenes are usually produced by gene duplications. The duplicated gene turns to a pseudogene so that the authentic gene can preserve the original function, whereas the pseudogene could acquire a new function to survive in a hazardous environment [11]. However, the absence of the authentic AQP10 in the bovine genome suggests that the function of AQP10 is not essential for cattle. Alternatively, the function of AQP10 may be compensated by the other aquaglyceroporins. As the knockout mice of AQP3 [13], AQP7 [14], [15], [16] and AQP9 [17] did not suffer from GI tract symptoms such as diarrhea, such compensations may be absent or difficult to be detected.
As human AQP10 is expressed intracellularly at the enterochromaffin cells [6], AQP10 may be needed for the hormone excretion as suggested by the enhanced expression in cholera patients [18]. Some reports indicated that AQP10 permeates ammonia, erythritol (a nutrient) and metalloids including toxic inorganic arsenic as well as glycerol [19]. Therefore, AQP10 may play more extensive unknown roles than water transport. Recent reports on the expression of human AQP10 in the skin stratum corneum and in the adipocyte suggest that AQP10 may be expressed widely [7], [8]. However, their western blot indicated several bands of AQP10 without monomer bands in contrast to the clear expression of AQP3 in the skin, suggesting a minimum expression of AQP10 [8]. The localization of the AQP10 isoform 1 in the adipocyte still remains to be clarified as the isoform 2-specific antibody was used in the study [7].
Why AQP10 gene become a pseudogene in some animals and not in other animals is an intriguing question. Speculatively, AQP10 protein could be a pathogen receptor which was originally expressed at the plasma membrane of the intestinal epithelium and later shifted to the enterochromaffin cell even intracellularly. If it was the case, the defective AQP10 may be advantageous with the lessened susceptibility to pathogen toxins such as arsenites [20]. Such an example was found in Bombyx mori, where a mutant of an amino acid transporter in the midgut membrane caused the resistance to B. mori densovirus type 2 (BmDNV-2) as this membrane protein is a functional receptor for BmDNV-2 [21].
In summary, the AQP10 gene is a pseudogene in cattle as is the case with mice. The current genome database suggests a much wider distribution of AQP10 pseudogenes in many members in even-toed ungulates including whales [22].
Acknowledgments
This study was supported by grants from the Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to K.I. (Nos. 24591243 and 21590242).
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.03.009.
Appendix A. Transparency document
STransparency document
References
- 1.Ishibashi K., Kondo S., Hara S., Morishita Y. The evolutionary aspects of aquaporin family. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011;300:R566–R576. doi: 10.1152/ajpregu.90464.2008. [DOI] [PubMed] [Google Scholar]
- 2.Hatakeyama S., Yoshida Y., Tani T., Koyama Y., Nihei K., Ohshiro K., Kamiie J.I., Yaoita E., Suda T., Hatakeyama K., Yamamoto T. Cloning of a new aquaporin (AQP10) abundantly expressed in duodenum and jejunum. Biochem. Biophys. Res. Commun. 2001;287:814–819. doi: 10.1006/bbrc.2001.5661. [DOI] [PubMed] [Google Scholar]
- 3.Ishibashi K., Morinaga T., Kuwahara M, Sasaki S., Imai M. Cloning and identification of a new member of water channel (AQP10) as an aquaglyceroporin. Biochim. Biophys. Acta. 2002;1576:335–340. doi: 10.1016/s0167-4781(02)00393-7. [DOI] [PubMed] [Google Scholar]
- 4.Ishibashi K. New members of mammalian aquaporins: AQP10–AQP12. Handb. Exp. Pharmacol. 2009;190:251–262. doi: 10.1007/978-3-540-79885-9_13. [DOI] [PubMed] [Google Scholar]
- 5.Mobasheri A., Shakibaei S., Marples D. Immunohistochemical localization of aquaporin 10 in the apical membranes of the human ileum: a potential pathway for luminal water and small solute absorption. Histochem. Cell Biol. 2004;121:463–471. doi: 10.1007/s00418-004-0657-1. [DOI] [PubMed] [Google Scholar]
- 6.Li H., Kamiie J., Morishita Y., Yoshida Y., Yaoita E., Ishibashi K., Yamamoto T. Expression and localization of two isoforms of AQP10 in human small intestine. Biol. Cell. 2005;97:823–829. doi: 10.1042/BC20040091. [DOI] [PubMed] [Google Scholar]
- 7.Laforenza U., Scaffino M.F., Gastaldi G. Aquaporin-10 represents an alternative pathway for glycerol efflux from human adipocytes. PLoS One. 2013;8:e54474. doi: 10.1371/journal.pone.0054474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jungersted J.M., Bomholt J., Bajraktari N, Hansen J.S., Klærke D.A., Pedersen P.A., Hedfalk K., Nielsen K.H., Agner T., Hélix-Nielsen C. In vivo studies of aquaporins 3 and 10 in human stratum corneum. Arch. Dermatol. Res. 2013;305:699–704. doi: 10.1007/s00403-013-1365-2. [DOI] [PubMed] [Google Scholar]
- 9.Morinaga T., Nakakoshi M, Hirao A., Imai M, Ishibashi K. Mouse aquaporin 10 gene (AQP10) is a pseudogene. Biochem. Biophys. Res. Commun. 2002;294:630–634. doi: 10.1016/S0006-291X(02)00536-3. [DOI] [PubMed] [Google Scholar]
- 10.Kondo H., Shimomura I, Kishida K., Kuriyama H., Makino Y., Nishizawa H., Matsuda M, Maeda N, Nagaretani H., Kihara S., Kurachi Y., Nakamura T., Funahashi T., Matsuzawa Y. Human aquaporin adipose (AQPap) gene. Genomic structure, promoter analysis and functional mutation. Eur. J. Biochem. 2002;269:1814–1826. doi: 10.1046/j.1432-1033.2002.02821.x. [DOI] [PubMed] [Google Scholar]
- 11.Pink R.C., Wicks K., Caley D.P., Punch E.K., Jacobs L., Carter D.R. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA. 2011;17:792–798. doi: 10.1261/rna.2658311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tingaud-Sequeira A., Calusinska M, Finn R.N., Chauvigné F, Lozano J., Cerdà J. The zebrafish genome encodes the largest vertebrate repertoire of functional aquaporins with dual paralogy and substrate specificities similar to mammals. BMC Evol. Biol. 2010;10:38. doi: 10.1186/1471-2148-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma T., Song Y., Yang B., Gillespie A., Carlson E.J., Epstein C.J., Verkman A.S. Nephrogenic diabetes insipidus in mice lacking aquaporin-3 water channels. Proc. Natl. Acad. Sci. USA. 2000;97:4386–4391. doi: 10.1073/pnas.080499597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara-Chikuma M, Sohara E., Rai T., Ikawa M, Okabe M, Sasaki S., Uchida S., Verkman A.S. Progressive adipocyte hypertrophy in aquaporin-7-deficient mice: adipocyte glycerol permeability as a novel regulator of fat accumulation. J. Biol. Chem. 2005;280:15493–15496. doi: 10.1074/jbc.C500028200. [DOI] [PubMed] [Google Scholar]
- 15.Hibuse T., Maeda N, Funahashi T., Yamamoto K., Nagasawa A., Mizunoya W., Kishida K., Inoue K., Kuriyama H., Nakamura T., Fushiki T., Kihara S., Shimomura I. Aquaporin 7 deficiency is associated with development of obesity through activation of adipose glycerol kinase. Proc. Natl. Acad. Sci. USA. 2005;102:10993–10998. doi: 10.1073/pnas.0503291102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumura K., Chang B.H., Fujimiya M, Chen W., Kulkarni R.N., Eguchi Y., Kimura H., Kojima H., Chan L. Aquaporin 7 is a beta-cell protein and regulator of intraislet glycerol content and glycerol kinase activity, beta-cell mass, and insulin production and secretion. Mol. Cell. Biol. 2007;27:6026–6037. doi: 10.1128/MCB.00384-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y., Promeneur D., Rojek A., Kumar N, Frøkiaer J., Nielsen S., King LS, Agre P., Carbrey J.M. Aquaporin 9 is the major pathway for glycerol uptake by mouse erythrocytes, with implications for malarial virulence. Proc. Natl. Acad. Sci. USA. 2007;104:12560–12564. doi: 10.1073/pnas.0705313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flach C.F., Qadri F, Bhuiyan T.R., Alam N.H., Jennische E., Holmgren J., Lönnroth I. Differential expression of intestinal membrane transporters in cholera patients. FEBS Lett. 2007;581:3183–3188. doi: 10.1016/j.febslet.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Litman T., Søgaard R., Zeuthen T. Ammonia and urea permeability of mammalian aquaporins. Handb. Exp. Pharmacol. 2009;190:327–358. doi: 10.1007/978-3-540-79885-9_17. [DOI] [PubMed] [Google Scholar]
- 20.Carbrey J.M., Song L., Zhou Y., Yoshinaga M., Rojek A., Wang Y., Liu H.L., Lujan Y., DiCarlo S.E., Nielsen S., Rosen B.P., Agre P., Mukhopadhyay R. Reduced arsenic clearance and increased toxicity in aquaglyceroporin-9-null mice. Proc. Natl. Acad. Sci. USA. 2009;106:15956–15960. doi: 10.1073/pnas.0908108106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito K., Kidokoro K., Sezutsu H., Nohata J., Yamamoto K., Kobayashi I., Uchino K., Kalyebi A., Eguchi R., Hara W., Tamura T., Katsuma S., Shimada T., Mita K., Kadono-Okuda K. Deletion of a gene encoding an amino acid transporter in the midgut membrane causes resistance to a Bombyx parvo-like virus. Proc. Natl. Acad. Sci. USA. 2008;105:7523–7527. doi: 10.1073/pnas.0711841105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gatesy J., Geisler J.H., Chang J., Buell C., Berta A., Meredith R.W., Springer M.S., McGowen M.R. A phylogenetic blueprint for a modern whale. Mol. Phylogenet. Evol. 2013;66:479–506. doi: 10.1016/j.ympev.2012.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
STransparency document