Abstract
MM-1α is a c-Myc-binding protein and acts as a transcriptional co-repressor in the nucleus. MM-1α is also PDF5, a subunit of prefoldin that is chaperon comprised of six subunits and prevents misfolding of newly synthesized nascent polypeptides. Prefoldin also plays a role in quality control against protein aggregation. It has been reported that mice harboring the missense mutation L110R of MM-1α/PFD5 exhibit neurodegeneration in the cerebellum and also male infertility, but the phenotype of infertility has not been fully characterized. In this study, we first analyzed morphology of the testis and epididymis of L110R of MM-1α mice. During differentiation of spermatogenesis, spermatogonia, spermatocytes and round spermatids were formed, but formation of elongated spermatids was compromised in L110R MM-1α mice. Furthermore, reduced number/concentration of sperm in the epididymis was observed. MM-1α was strongly expressed in the round spermatids and sperms with round spermatids, suggesting that MM-1α affects the differentiation and maturation of germ cells. Changes in expression levels of spermatogenesis-related genes in mice testes were then examined. The fatty-acid-binding protein (fabp4) gene was up-regulated and three genes, including sperm-associated glutamate (E)-rich protein 4d (speer-4d), phospholipase A2-Group 3 (pla2g3) and phospholipase A2-Group 10 (pla2g10), were down-regulated in L110R MM-1α mice. L110R MM-1α and wild-type MM-1α bound to regions of up-regulated and down-regulated genes, respectively. Since these gene products are known to play a role in maturation and motility of sperm, a defect of at least MM-1α transcriptional activity is thought to induce expressional changes of these genes, resulting in male infertility.
Keywords: Prefoldin, Spermatogenesis, Fatty acid synthesis
Highlights
-
•
Mice harboring MM-1α/prefoldin 5 mutation exhibit male infertility in addition to neurodegeneration.
-
•
Differentiation from the round spermatids to elongated spermatids was compromised.
-
•
Defect of at least MM-1α transcriptional activity compromises differentiation of spermatids.
1. Introduction
MM-1α has been identified by us as a c-Myc-binding protein [1]. MM-1α is a tumor suppressor protein and inhibits c-Myc function in various ways, resulting in suppression of cell cycle movement and cell transformation [2], [3], [4], [5]. As for transcriptional activity of MM-1α, MM-1α acts as a co-repressor through binding to DNA-binding transcriptional factors such c-Myc, p73 and Egr-1 to repress their target genes [1], [3], [4], [5], [6]. MM-1α is also PD5, a subunit of prefoldin. Prefoldin is a molecular chaperone comprised of six subunits containing two α-subunits (PFD3 and PFD5) and four β-subunits (PFD1, PFD2, PFD4 and PFD6) and holds newly synthesized proteins by binding to and transporting them to chaperonine Tric/CCT in cooperation with HSP70 and HSP40 [7], [8], [9], [10]. Prefoldin thus plays a role in maintaining homeostasis in cells through stimulation of proper folding of proteins. We have reported that prefoldin inhibited aggregate and inclusion formation of exogenously added pathogenic Huntingtin or α-synuclein in cells [11], [12] and that knockdown of prefoldin expression and mutation of PFD5 caused accumulation of ubiquitinated protein aggregates in cells and mice, resulting in reduced cell viability [13], suggesting that prefoldin plays a modifier role against the toxicity of misfolded proteins that cause neurodegenerative diseases. Like MM-1α/PDF5, each prefoldin subunit has its respective function. PFD3 is a pVHL-binding protein (VBP1) and stimulates protein degradation through the ubiquitin–proteasome system [14], [15]. PFD2 as a transcription factor regulates the expression of nutrition-related genes [16].
Homozygous C57BL/6-Pfdn5nmf5a/Pfdn5nmf5a mice harboring a missense mutation of the MM-1α/PFD5 gene that causes an amino acid substitution from leucine to arginine at amino acid number 110 (L110R) [17] are referred to here as MM-1α L110R mice. These mice, L110R MM-1α mice, exhibit phenotypes of neurodegeneration, including cerebellar atrophy with death of Purkinje cells and retinitis dystrophy, and of male infertility [17]. We have shown that polyubiquitinated proteins were increased in L110R MM-1α mice irrespective of formation of the prefoldin complex and that cell lines from L110R MM-1α mice were more susceptible to various stresses than were those from wild-type mice, suggesting that recognition activity of prefoldin in L110R MM-1α mice is decreased [13]. PFD1-knockout mice also exhibit cerebellar atrophy [18]. It is not known whether phenotypes of these mice are due to dysfunction of the prefoldin complex or of its subunits.
Regarding male infertility, atrophy of the testis of L110R MM-1α mice has been reported [17]. Characterization of male infertility in L110R MM-1α mice has, however, not been fully addressed.
In this study, we first characterized phenotypes of the testis and epididymis of wild-type and L110R MM-1α mice and found abnormal morphology of these tissues and germ cells in L110R MM-1α mice. We then examined expression changes of spermatogenesis-related genes in wild-type and L110R MM-1α mice. The results suggest that at least dysfunction of transcriptional of activity of MM-1α affects germ cell differentiation during spermatogenesis in L110R MM-1α mice.
2. Materials and methods
2.1. Cells and mice
Homozygous MM-1α L110R mice [17] were kindly provided by Dr. Patsy M. Nishina. C57BL/6 and MM-1α L110R mice were fed a normal diet (D12337, Research Diets, Inc. New Brunswick, NJ). Since homozygous MM-1α L110R mice were infertile, heterozygous MM-1α L110R mice were mated, genotypes of their offspring were determined by DNA sequencing of mouse tail DNA, and then homozygous MM-1α L110R mice were obtained. Male mice at 17 weeks of age were used in this study. The nucleotide sequence for a primer used for DNA sequencing is 5′-ATGTACGTCCCCGGGAAGCTACACG-3′.
Establishment of cell lines from wild-type and L110R MM-1α mice was described previously [13]. Briefly, primary cells from newborn mice were immortalized with T antigen of simian virus 40 and cultured in Dulbecco׳s modified Eagle׳s medium (DMEM) with 10% calf serum.
2.2. Real-time PCR and RT-PCR
Nucleotide sequences of the upper and lower strands and PCR conditions for real-time PCR and RT-PCR are shown in Table 1. The testis and epididymis were cut out from wild-type and MM-1α L110R mice and stored in an RNA later solution (Life Technologies, Carlsbad, CA) at 4 °C. Their RNAs were then extracted using ISOGEN (Nippon Gene, Toyama, Japan) according to manufacturer׳s protocol.
Table 1.
Nucleotide sequences of the upper and lower strands and PCR conditions for real-time PCR and RT-PCR
| Mouse gene | Sense/anti-sense | Nucleotide sequence | PCR condition |
|---|---|---|---|
| GAPDH | Sense | 5′-AACCTGGCATTGTGGAAGG-3′ | 95 °C 2 min, 95 °C 30 s, 55 °C 30 s, 33 cycles of 72 °C 1 min |
| Anti-sense | 5′-ACACATTGGGGGTAGGAACA-3′ | ||
| FABP4 | Sense | 5′-ATGCCTTGTGGGAACCTGGAAGC-3′ | 95 °C 2 min, 95 °C 30 s, 55 °C 30 s, 33 cycles of 72 °C 1 min |
| Anti-sense | 5′-GGCCATGCCACTTTCCTT-3′ | ||
| MM-1 | Sense | 5′-GGGAATTCATGGCGCAGTCGATT-3′ | 95 °C 2 min, 95 °C 30 s, 55 °C 30 s, 35 cycles of 72 °C 1 min |
| Anti-sense | 5′-GGCTCGAGTCAGGCTTTGACCGT-3′ | ||
| PLa2g3 | Sense | 5′-GGCTGAGGCCACCTCATATACTTC-3′ | 95 °C 2 min, 95 °C 30 s, 55 °C 30 s, 33 cycles of 72 °C 1 min |
| Anti-sense | 5′-TCCTTTGCCCTCAGCACAGTCAAG-3′ | ||
| PLa2g10 | Sense | 5′-GGATTGTGTTGGGCCTCGAT-3′ | 95 °C 2 min, 95 °C 30 s, 55 °C 30 s, 33 cycles of 72 °C 1 min |
| Anti-sense | 5′-TGGTAGCAGCACCAGTCAAT-3′ | ||
| Speer-4d | Sense | 5′-GCCTTCCACCTGAGAGTAATGAAGGA-3′ | 95 °C 2 min, 95 °C 30 s, 55 °C 30 s, 33 cycles of 72 °C 1 min |
| Anti-sense | 5′-AATCAGCCATGGCTTCCTAGTAAAGG-3′ |
2.3. Western blotting
The testis and epididymis were cut out from wild-type and MM-1α L110R mice and stored at −80 °C. Tissues were then homogenized. Proteins were extracted from tissue homogenates or from cultured cells after incubation of homogenates with a buffer containing 50 mM Tris–HCl (pH 7.4), 120 mM NaCl, 1 mM EDTA, 0.5% NP-40 and protease inhibitors (20 mg/ml leupeptin, 10 mg/ml pepstatin A, 10 mg/ml aprotinin, 2 mM phenylmethylsulfonyl fluoride) for 20 min at 4 °C. Proteins were then dissolved in the buffer containing 30 mM Tris–HCl (pH 6.8), 3% SDS, 6% β-mercaptoethanol and 30% glycerol, boiled for 5 min, and subjected to Western blot analysis with anti-MM-1 rabbit polyclonal, anti-FABP4 rabbit monoclonal (Cell Signaling, Danvers, MA) and anti-GAPDH rabbit polyclonal (Millipore, Billerica, MA) antibodies. Proteins were then reacted with an IRDye800 (Rockland, Philadelphia, PA) or Alexa Fluor 680 (Molecular Probes, Eugene, OR)-conjugated secondary antibody and visualized by using an infrared imaging system (Odyssey, LI-COR, Lincoln, NE). The anti-MM-1 rabbit polyclonal antibody was established by us by injection of GST-MM-1 into rabbits. The anti-MM-1 antibody was purified from rabbit serum using an affinity column containing recombinant GST-free MM-1.
2.4. Tissue preparation and immunohistochemistry
The testis and epididymis of male mice at 17 weeks of age were fixed in 10% formalin at neutral pH and embedded in paraffin. Five-μm-thick paraffin sections were then prepared using a microtome, treated with 3% H2O2 in methanol for 15 min at room temperature and reacted with anti-FABP4 (1:100 dilution) and anti-MM-1 (1:50 dilution) antibodies for 1 day at 4 °C. Sections were then stained with hematoxylin eosin (HE). After several washes, sections were subjected to DAB staining using a Histofine SAB-PO kit (Nicirei Bioscience, Tokyo, Japan). Stained images were then observed under a fluorescent microscope without fluorescence filters (Biorevo BZ-9000, Osaka, Japan).
Sperm was extracted with 50 μl of PBS (-) from the cauda epididymis at 37 °C and fixed with methanol for 1 h and then with 0.02% Triton X in PBS for 10 min and reacted with an anti-MM-1 (1:50 dilution) antibody for 4 days at 4 °C. After several washes, sperm was then reacted with an FITC-conjugated anti-rabbit IgG and stained images were obtained under a fluorescent microscope.
2.5. Histochemical analysis of tissues and sperm
Tissue sections cut into 5-μm-thick slices as described above were stained with HE and their images were then observed under a fluorescent microscope without fluorescence filters (Biorevo BZ-9000). Sperm was extracted as described above. The number of sperm was measured using a hemocytometer (0.100 mm at depth and 1/25 mm2 in area), and total 470 and 75 sperm in 100- and 50-diluted solution from wild-type and L110R mice, respectively, were counted. Sperm was also subjected to Diff-QuiK staining and stained images were then observed under a fluorescent microscope without fluorescence filters (Biorevo BZ-9000). Sperm motility was counted using Standard Count 2 Chamber Slide as a chamber (Leja Products, Nieuw-Vennep, Netherlands) at 37 °C. The replicated number of experiments is 2.
2.6. Microarray
RNA was extracted from whole testes of wild-type and L110R mice at 17 weeks of age. Sample labeling was carried out using Quick-Amp Labeling Kit (Agilent Technologies, Santa Clara, CA) following manufacturer׳s instructions. Amplified cRNAs were quantified using the Agilent 2100 bioanalyzer and RNA 6000 Nano kit (Agilent). Hybridization and scan were performed using Gene Expression Hybridization Kit (Agilent) and an Agilent G2505C scanner, respectively. Images were quantified using Agilent Feature Extraction Software (version 10.7.3.1).
2.7. Chromatin immunoprecipitation (ChIP) assay
ChIP assays using cultured wild-type and L110R MM-1α cells derived from wild-type and L110R cells were performed according to the protocol of the ChIP Assay Kit (Millipore, Billerica). Briefly, after proteins had been cross-linked with DNA, cell pellets were suspended in an SDS-lysis buffer and sonicated on ice using a sonicator (UR-20P, TOMY, Tokyo, Japan) 3 times for 20 s each time at 5 W. Genomic DNA was sheared to 300 to 5000 base pairs of length. Chromatin solution from 1×106 cells/dish was preincubated with salmon sperm DNA and Protein A-agarose and incubated with species-matched IgG or with an anti-MM-1 antibody as described previously [5] overnight at 4 °C. DNA fragments immunoprecipitated with the anti-MM-1 antibody were then used as templates for PCR with Ex taq (TaKaRa Bio, Kyoto, Japan) and reacted. Nucleotide sequences of primers and PCR conditions used for ChIP assays are shown in Table 2. PCR products were separated on a 2% agarose gel and stained with ethidium bromide.
Table 2.
Nucleotide sequences of primers and PCR conditions used for ChIP
| Gene | Nucleotide sequence | PCR condition | |
|---|---|---|---|
| mFabp4 | ChIP-1F | 5′-TTGGGACCCATTTTTGAAGGG-3′ | 96 °C 1 min, 96 °C 30 s, 60 °C 30 s, 72 °C 4 min×40 cycles, 72 °C 5 min |
| ChIP-1R | 5′-AACTTTGGTTCTCCCTGGCA-3′ | ||
| ChIP-2F | 5′-CAGTCACATGGTCAGGGCAT-3′ | 96 °C 1 min, 96 °C 30 s, 60 °C 30 s, 72 °C 4 min×40 cycles, 72 °C 5 min | |
| ChIP-2R | 5′-TACGGTTGCCAGTTCCACTC-3′ | ||
| ChIP-3F | 5′-GAGTGGAACTGGCAACCGTA-3′ | 96 °C 1 min, 96 °C 30 s, 60 °C 30 s, 72 °C 4 min×40 cycles, 72 °C 5 min | |
| ChIP-3R | 5′-TTCACCTTCCTGTCGTCTGC-3′ | ||
| ChIP-4F | 5′-CCCTGTAGGAGTGGGCTTTG-3′ | 96 °C 1 min, 96 °C 30 s, 60 °C 30 s, 72 °C 4 min×40 cycles, 72 °C 5 min | |
| ChIP-4R | 5′-GGCCTCTTCCTTTGGCTCAT-3′ | ||
| mPLa2g3 | ChIP-1F | 5′-ACATCAGCTTGGGGACATGG-3′ | 96 °C 1 min, 96 °C 30 s, 60 °C 30 s, 72 °C 4 min×40 cycles, 72 °C 5 min |
| ChIP-1R | 5′-TGCAAGGGCGAAATGGTTTG-3′ | ||
| ChIP-2F | 5′-ACAAACCATTTCGCCCTTGC-3′ | 96 °C 1 min, 96 °C 30 s, 60 °C 30 s, 72 °C 4 min×40 cycles, 72 °C 5 min | |
| ChIP-2R | 5′-CCAAACTTTGTCGGTGCCTG-3′ | ||
| ChIP-3F | 5′-AGTCTGGCACGTTTCCTGAG-3′ | 96 °C 1 min, 96 °C 30 s, 60 °C 30 s, 72 °C 4 min×40 cycles, 72 °C 5 min | |
| ChIP-3R | 5′-TTTCTTCTGTCCCCTGCGTC-3′ | ||
| ChIP-4F | 5′-TGCTGGTTTTGACCCCTTGT-3′ | 96 °C 1 min, 96 °C 30 s, 60 °C 30 s, 72 °C 4 min×40 cycles, 72 °C 5 min | |
| ChIP-4R | 5′-GCACCCCTGTGAGGTAGTTC-3′ | ||
| mPLa2g10 | ChIP-1F | 5′-GCTTCCCAGTTCTCAGACCC-3′ | 96 °C 1 min, 96 °C 30 s, 60 °C 30 s, 72 °C 4 min×40 cycles, 72 °C 5 min |
| ChIP-1R | 5′-ATCCCGAAGCCTTCCTACCT-3′ | ||
| ChIP-2F | 5′-AGGTAGGAAGGCTTCGGGAT-3′ | 96 °C 1 min, 96 °C 30 s, 60 °C 30 s, 72 °C 4 min×40 cycles, 72 °C 5 min | |
| ChIP-2R | 5′-TAGCATGTTTGAGGCCCAGG-3′ | ||
| ChIP-3F | 5′-CCTGGGCCTCAAACATGCTA-3′ | 96 °C 1 min, 96 °C 30 s, 60 °C 30 s, 72 °C 4 min×40 cycles, 72 °C 5 min | |
| ChIP-3R | 5′-AGAGAGGCAGGAAGGAGAGG-3′ | ||
| ChIP-4F | 5′-CCTCTCCTTCCTGCCTCTCT-3′ | 96 °C 1 min, 96 °C 30 s, 60 °C 30 s, 72 °C 4 min×40 cycles, 72°C 5 min | |
| ChIP-4R | 5′-CAGACAGACCCCAGAAGCTG-3′ | ||
| mSpeer-4d | ChIP-1F | 5′-GCTTAATGAGTGTGGCCCCT-3′ | 96 °C 1 min, 96 °C 30 s, 60 °C 30 s, 72 °C 4 min×40 cycles, 72 °C 5 min |
| ChIP-1R | 5′-CTCGGTCACGTAGATCAGGC-3′ | ||
| ChIP-2F | 5′-TGCTCCAAAACCTGTGGTGT-3′ | 96 °C 1 min, 96 °C 30 s, 60 °C 30 s, 72 °C 4 min×32 cycles, 72 °C 5 min | |
| ChIP-2R | 5′-AGGGAGGGCGTTCTATCCTT-3′ | ||
| ChIP-3F | 5′-AGGATAGAACGCCCTCCCTT-3′ | 96 °C 1 min, 96 °C 30 s, 60 °C 30 s, 72 °C 4 min×40 cycles, 72 °C 5 min | |
| ChIP-3R | 5′-GCTGCTCCTCTTGGACACTT-3′ | ||
2.8. Statistical analyses
Data are expressed as means±S.E. Statistical analyses were performed using Student׳s t-test.
2.9. Ethics statement
All animal experiments were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the protocols were approved by the Committee for Animal Research at Hokkaido University (permit numbers 06-0467 and 08-0467).
3. Results
3.1. Reduced weights of the testis and epididymis from L110R MM-1α mice
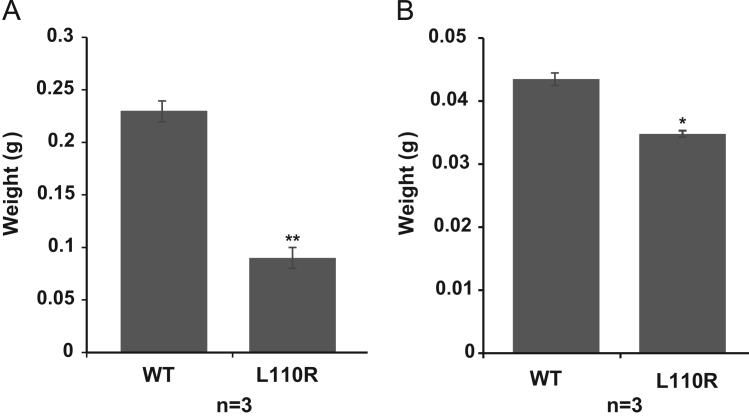
To precisely characterize MM-1α L110R mice, the testis weight and whole epididymis weight were then examined. As shown in Fig. 1, the testis weight and epididymis weigh in MM-1α L110R mice were reduced by 60% and 20% compared to those in wild-type mice. These results suggest that in addition to the testis, atrophy of the epididymis occurred in MM-1α L110R mice.
Fig. 1.
Reduced weights of the testis and epididymis in L110R MM-1α mice. Testes and epididymides were isolated from wild-type and L110R MM-1α mice at 17 weeks of age and their weights were measured. Values are means±S.E. n=3 experiments. Significance: *p<0.05, **p<0.01.
3.2. Abnormal morphology of the testis and epididymis and expression of MM-1α in MM-1α L110R mice
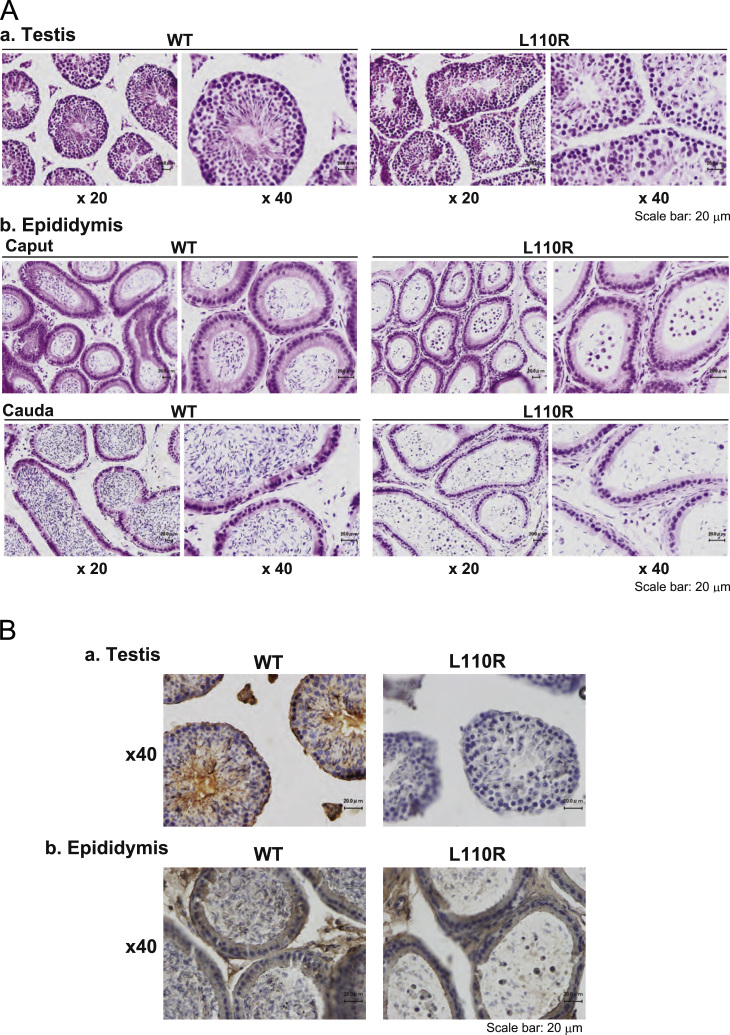
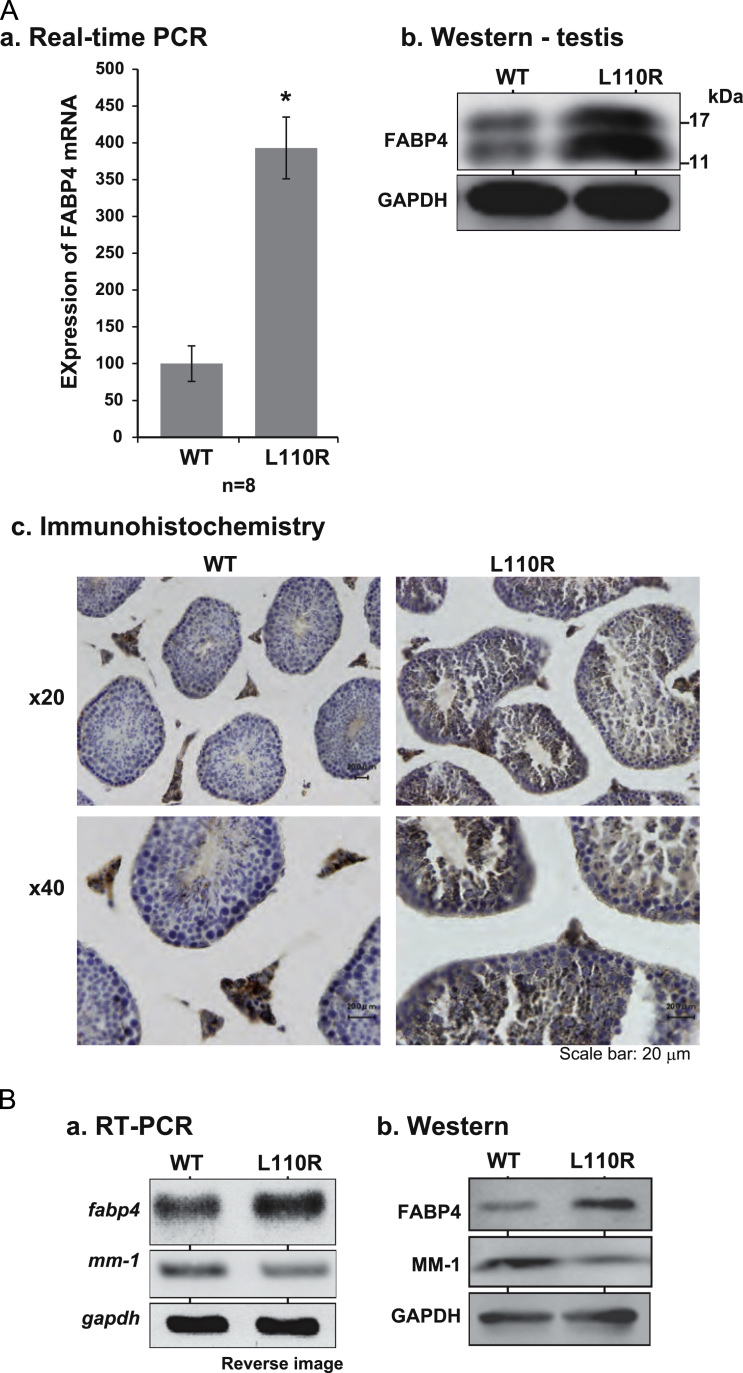
Tissue sections from the testis and epididymis of wild-type and MM-1α L110R mice were prepared and stained with hematoxylin eosin (HE). As shown in Fig. 2A-a, spermatogonia, spermatocytes and early spermatids (round spermatid) were morphologically normal but that few later spermatids (elongated spermatids) were observed in L110R mice, suggesting that formation of later spermatids (elongated spermatids) are compromised in L110R mice. Only a small number of sperm was observed in the caput and cauda of epididymis in MM-1α L110R mice compared to that in wild-type mice, and many round cells, probably round spermatids, were observed in both the caput and cauda of the epididymis in MM-1α L110R mice (Fig. 2A-b). The expression of MM-1α/prefoldin5 in the testis and epididymis was then examined after their sections had been stained with an anti-MM-1 antibody and with HE. First, intensity of stained MM-1α in the testis and epididymis of wild-type mice was stronger than that in the testis and epididymis of MM-1α L110R mice, which is consistent with the results of the expression level of MM-1α in wild-type and MM-1α L110R mice by Western blotting (see Fig. 4B-b). Spermatocytes, round cells, probably round spermatids, and elongated spermatids in the testis were stained with the anti-MM-1 antibody (Fig. 2B-a). As for the epididymis, on the other hand, MM-1α was strongly expressed in round cells in MM-1α L110R mice (Fig. 2B-b).
Fig. 2.
Histochemical analyses of the testis and epididymis and expression of MM-1α in L110R MM-1α mice. (A) The testis (a) and epididymis (b) were isolated from wild-type and L110R MM-1α mice at 17 weeks of age and tissue slices were prepared. Tissue sections were then subjected to HE staining as described in Section 2. Two figures of the same section with different magnification are shown in (A-a). (B) Tissue sections prepared as described in the legend for (A) were fixed with 3% H2O2 in methanol and reacted with an anti-MM-1 antibody followed by HE and DAB staining as described in Section 2. Two figures of the same section with different magnification are shown in (A-b).
Fig. 4.
Up-regulation of FABP4 in the testis of L110R MM-1α mice. (A-a) and (A-b) Total RNAs and proteins were extracted from testes of wild-type and L110R MM-1α mice, and RT-PCR (a) and Western blotting with anti-FABP4 and anti-GAPDH antibodies (b) were carried out as described in Section 2. Relative expression of fabp4 versus gapdh is shown in (a). Values are means±S.E. n=8 experiments. Significance: *p<0.05. (A-c) Testes were isolated from wild-type and L110R MM-1α mice and tissue sections were prepared. Tissue sections were then reacted with an anti-FABP4 antibody followed by HE staining, and their images were obtained as described in the legend for Fig. 2B. Two figures of the same section with different magnification are shown. (B) Total RNAs and proteins were extracted from cell lines derived from wild-type and L110R and MM-1α mice and subjected to real-time PCR and Western blotting with anti-FABP4, anti-MM-1 and anti-GAPDH antibodies as described in Section 2.
3.3. Reduced number and motility of sperm in MM-1α L110R mice
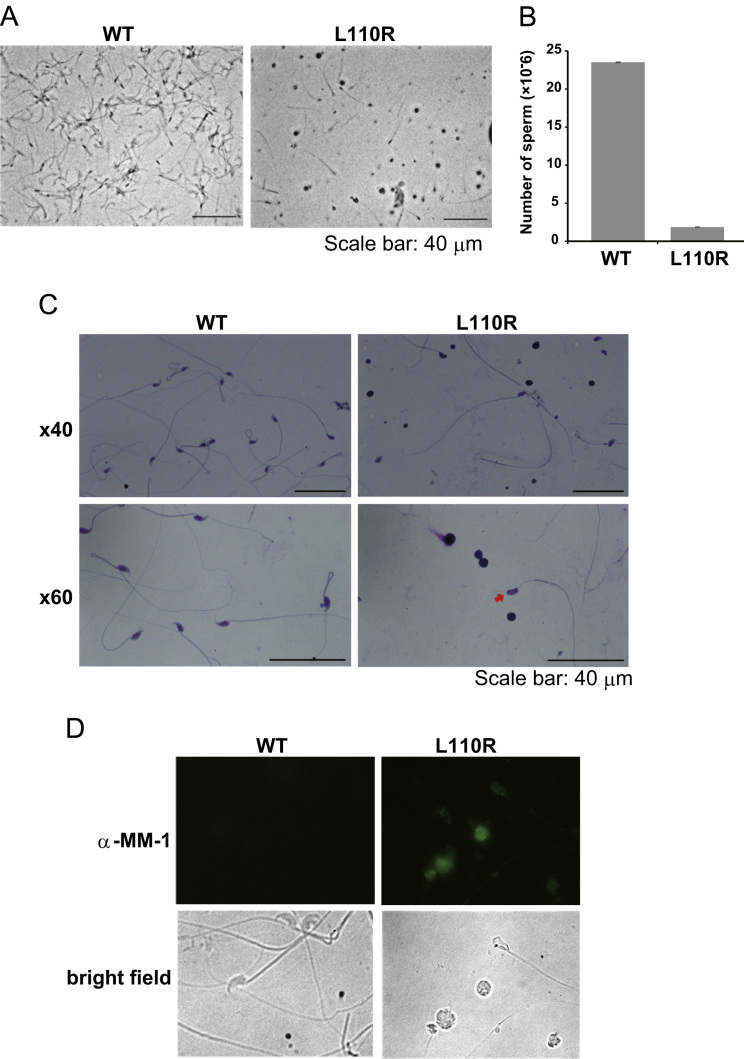
Sperm was extracted from the cauda epididymis of wild-type and MM-1α L110R mice, and the morphology and concentration of sperm were examined. As shown in Fig. 3A, the sperm number in MM-1α L110R mice was at a glance smaller than that in wild-type mice, and sperm with abnormal shape was observed in MM-1α L110R mice. The sperm concentration in the epididymis of MM-1α L110R mice was 3.743×107/ml (the number of sperm is 1.871×106), which was less than 10% of that in wild-type mice (4.704×108/ml (the number of sperm is 2.352×107)) (Fig. 3B). Sperm motility was then measured under a microscope. Although the ratios of sperm with forward movement, non-forward movement and immobile sperm were 46.1%, 9.4% and 44.5% (the exact numbers of sperm counted were 177, 36 and 171), respectively, in wild-type mice, almost all of the sperms (98.0%) were immobile in MM-1α L110R mice: the ratios of sperm with forward movement, non-forward movement and immobile sperm were 0.2%, 1.9% and 98.0% (the exact numbers of sperm counted were 1, 11 and 578), respectively. Sperm morphology was further examined by Diff-QuiK staining. As shown in Fig. 3C, sperms with round heads and round cells, which might be round spermatids that had not differentiated into elongated spermatids, were observed in the sperm solution from the epididymis of MM-1α L110R mice. Furthermore, when sperm samples were stained with an anti-MM-1 antibody, MM-1 was found to be present in round cells in MM-1α L110R mice (Fig. 3D). These results suggest that the reason for infertility of MM-1α L110R mice is both reduced number of mature sperm and defect of germ cell maturation and that MM-1α affects the differentiation of germ cell.
Fig. 3.
Reduced number of sperm and abnormal morphology of sperm in L110R MM-1α mice. (A) Sperms were extracted from the epididymides of wild-type and L110R MM-1α mice at 17 weeks of age and observed under a microscope. (B) The numbers of sperms extracted from the epididymides in wild-type and L110R MM-1α mice were counted using a hemocytometer. (C) Sperms were extracted from the epididymides of wild-type and L110R MM-1α mice. After the numbers of sperms from wild-type and L110R MM-1α mice had been adjusted to equal numbers, sperms were stained with a Diff-QuiK solution and their images were observed under a fluorescent microscope. An arrow indicates sperm with a spermatocyte-like cell. (D) Sperms from wild-type and L110R MM-1α mice were prepared as described in the legend for Fig. 3C, fixed with methanol and Triton X, and reacted with an anti-MM-1 antibody. After sperms had been reacted with an FITC-conjugated secondary antibody, their images were observed under a fluorescent microscope as described in Section 2.
3.4. Expression changes of and association of MM-1α with spermatogenesis-related genes
To examine expression changes of spermatogenesis-related genes in MM-1α L110R mice, total RNAs were extracted from testes of wild-type and MM-1α L110R mice and applied to microarray analysis using a 60 K microarray chip (Agilent Technologies). As shown in Table 3, expression levels of 23 genes and those of 17 genes were increase and decreased by more than 3 fold, respectively, in MM-1α L110R mice compared to those in wild-type mice. Since genes that are known to be related to spermatogenesis are fatty-acid-binding protein (fabp4), sperm-associated glutamate (E)-rich protein 4d (speer-4d), phospholipase A2-Group 3 (pla2g3) and phospholipase A2-Group 10 (pla2g10) and since expression levels of fabp4 and those of speer-4d, pla2g3 and pla2g10 were shown by the microarray analysis to be increased and decreased, respectively, in MM-1α L110R mice, expression levels of these genes in testes were examined.
Table 3.
Genes with changed expression levels in testes of MM-1α L110R mice.
| Gene name | Ratio |
|---|---|
| (A) Increased expression of genes in testes of L110R mice | |
| Fabp4 | 9.478 |
| Hp | 4.931 |
| Slc25a5 | 4.677 |
| Cdk1 | 4.297 |
| A_55_P1970536 | 4.216 |
| Snrpc | 4.208 |
| Cbx7 | 4.099 |
| Aurkb | 3.798 |
| NAP092947-001 | 3.746 |
| NAP115343-1 | 3.513 |
| Gm3146 | 3.476 |
| ENSMUST00000059110 | 3.318 |
| A_55_P2097072 | 3.247 |
| Gm9372 | 3.225 |
| Spink8 | 3.181 |
| Inmt | 3.180 |
| NAP094721-001 | 3.146 |
| Gm5067 | 3.139 |
| Mapkap1 | 3.135 |
| ENSMUST00000170969 | 3.080 |
| A_55_P2107682 | 3.056 |
| Olfr1500 | 3.052 |
| A_55_P2080902 | 3.049 |
| (B) Decreased expression of genes in testes of MM-1α L110R mice | |
| Gm5458 | 0.264 |
| Nprl3 | 0.307 |
| Csnk1g2 | 0.298 |
| Adc | 0.307 |
| Il20ra | 0.308 |
| Speer4d | 0.309 |
| ENSMUST00000143605 | 0.309 |
| Hk1 | 0.312 |
| Heatr8 | 0.314 |
| Gm17019 | 0.315 |
| Speer4b | 0.318 |
| Rnf38 | 0.325 |
| Nfe2 | 0.327 |
| Gm17019 | 0.328 |
| Kif17 | 0.332 |
| Lypla1 | 0.332 |
| Srcin1 | 0.333 |
| Omission | |
| Pla2g3 | 0.588 |
| Pla2g10 | 0.591 |
Real-time PCR and Western blot analyses showed that expression levels of FABP4 mRNA and protein in MM-1α L110R mice were increased by about 4 times and by 1.8 times, respectively, compared to those in wild-type mice (Fig. 4A-a and A-b). Two FABP4 bands with 15- and 17-kD were observed, and 17-kD FABP4 might be post-translationally modified 15-kD FABP4. Immunohistochemical analyses showed that FABP4 was strongly expressed in the testis, especially in spermatocytes and round cells, of MM-1α L110R mice (Fig. 4A-c). Furthermore, increased expression levels of FABP4 mRNA and protein in MM-1α L110R cells compared to those in wild-type cells were also observed (Fig. 4B).
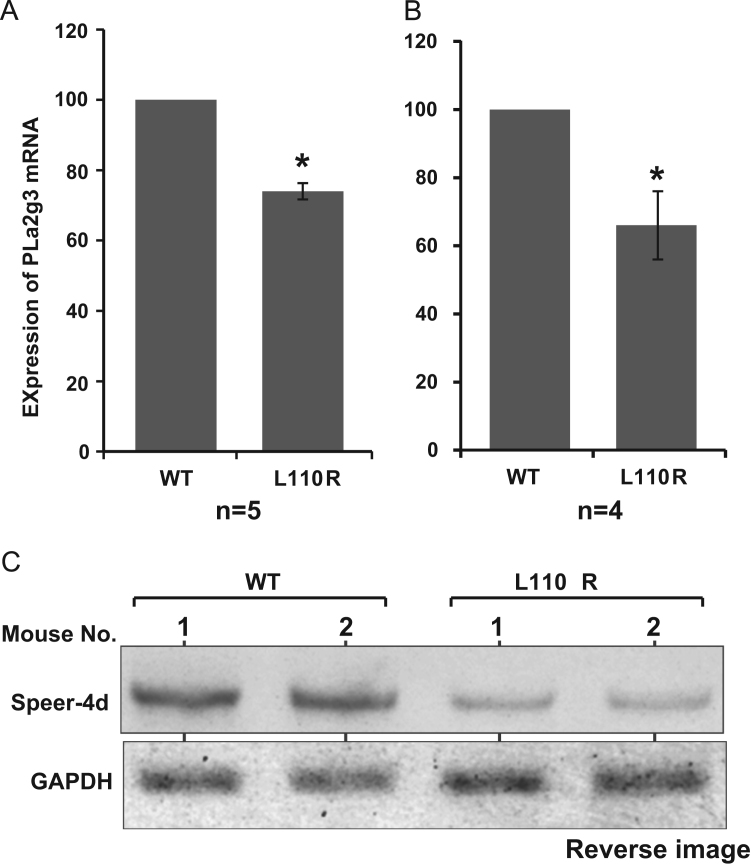
Expression levels of pla2g3 and pla2g10 in mice testes were examined by real-time PCR, and decreased levels of their expression in MM-1α L110R mice were confirmed (Fig. 5A and B). Since there are various isoforms of speer-4d mRNA, we could not design real-time PCR primers specific to the speer-4d gene, and semi-quantitative RT-PCR was therefore carried out to examine the expression level of speed-4d. As shown in Fig. 5C, reduced expression level of speer-4d in MM-1α L110R mice compared to that in wild-type mice was confirmed.
Fig. 5.
Down-regulation of PLa2g3, PLa2g10 and Speer-4d in the testis of L110R MM-1α mice. Total RNAs were extracted from testes of wild-type and L110R MM-1α mice, and real-time PCR (A and B) and RT-PCR (C) were carried out as described in Section 2. Relative expression of PLa2g3 and PLa2g10 versus gapdh is shown in (A) and (B), respectively. Values are means±S.E. n=4–5 experiments. Significance: *p<0.05. Reverse images of stained patterns in RT-PCR are shown (C).
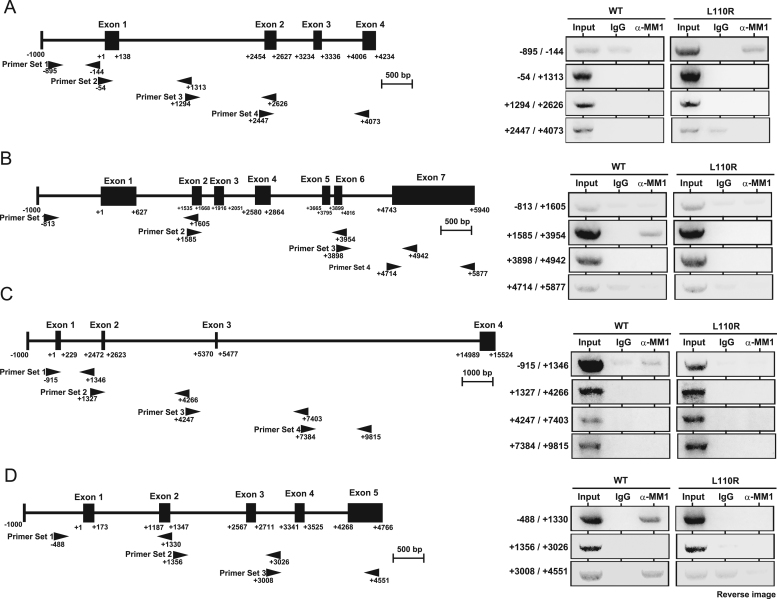
To examine the molecular mechanism of MM-1α toward expression changes of spermatogenesis-related genes, chromatin immunoprecipitation (ChIP) assays were carried out using the chromatin-protein complex from wild-type and MM-1α L110R cells and an anti-MM-1 antibody or IgG. Primers were set to amplify approximately 800–1000 base pairs in each gene. As shown in Fig. 6, L100R MM-1α but not wild-type MM-1α bound to the region spanning −895 to −144 in the Fabp4 gene, which was up-regulated in MM-1α L100R mice, suggesting that gain-of-function by mutation of MM-1α results in transcriptional activation of the Fabp4 gene. Wild-type MM-1α but not L100R MM-1α, on the other hand, bound to regions in Pla2g3, Pla2g10 and Speer-4d genes, which were down-regulated in MM-1α L100R mice, suggesting that wild-type MM-1α negatively regulates these genes at the transcriptional level.
Fig. 6.
Association of MM-1α with spermatogenesis-related genes. Chromatin immunoprecipitation assays were carried out using chromatin prepared from wild-type and L110R MM-1α cells. Chromatin was immunoprecipitated with an anti-MM-1α antibody or non-specific IgG. After extraction of DNA from precipitated chromatin, regions in Fabp4, PLa2g3, PLa2g10 and Speer-4d genes were amplified by PCR with specific primers and with amplified DNA (left figures) and were separated on agarose gels (right figures) as described in Section 2. n=3.
4. Discussion
Germ cells are differentiated from spermatogonia, spermatocytes and then spermatids in the testis and are secreted into the epididymis, in which mature sperms are present. In this study, we examined mice with a missense mutation of prefoldin5/MM-1α that introduces a substitution mutation in MM-1 containing L110R, and these MM-1α L110R mice exhibit male infertility. We first found that the weights of the testis and whole epididymis in MM-1α L110R mice were reduced compared to those in wild-type mice and that few elongated spermatids were observed in MM-1α L110R mice. Reduced number/concentration of sperm in the testis and epididymis and the presence of round cells, presumably round spermatids, in the epididymis, was observed in MM-1α L110R mice. Sperms from the epididymis of MM-1α L110R mice had lost their motility and were not differentiated. These results suggest that differentiation and maturation steps of germ cells are inhibited in MM-1α L110R mice, leading to male infertility.
The sperm concentration in the epididymis of MM-1α L110R mice was less than 10% of that in wild-type mice, and 98% of the sperms in MM-1α L110R mice were immobile. Indeed, the number of the offspring after mating between heterozygous MM-1α L110R mice was less than that after mating between wild-type mice, reflecting the reduced number and motility of sperms in MM-1α L110R mice.
MM-1α was expressed in spermatocytes and round cells, presumably round spermatids, in the testis in wild-type and MM-1α L110R mice, respectively. Furthermore, MM-1α was strongly expressed in round cells in the epididymis and in sperms with round cells in MM-1α L110R mice. Since the epididymis contains mature germ cells but not undifferentiated germ cells such as spermatocytes and spermatids, these results suggest that MM-1α affects differentiation of spermatids and maturation of germ cells and that its activity is reduced by L110R mutation into MM-1α, resulting in accumulation of undifferentiated round spermatids in the epididymis and of round cell-containing sperms in MM-1α L110R mice. This is the first report showing the possibility that MM-1α participates in spermatogenesis.
We then examined expression changes of genes in wild-type and MM-1α L110R mice by using a DNA microarray, and expression changes of four spermatogenesis-related genes were evaluated by real-time PCR or RT-PCR. Fatty acid-binding protein 4 (FABP4) is a member of a multigene family encoding approximately 15-kDa proteins. Proteins of the FABP family, comprised of more than 10 members, bind to fatty acids and other hydrophobic biomolecules in an internal cavity, and they act for solubilization and metabolic trafficking of fatty acid [19]. Each FABP is expressed in distinct tissues and cells, and FABP4 was originally identified as an adipose-specific protein [20] but was later found to be expressed in various cells and tissues, including macrophages [21], [22], [23], the lymphatic system [24] and the testis and epididymis [25], and during embryogenesis [26]. FABP4 functions as an adipokine in regulating macrophage and adipocyte interactions during inflammation, and dysregulation of FABP4 is associated with obesity and nonalcoholic fatty liver disease (see a recent review, [27]). In this study, expression levels of FABP4 were found to be increased in the testis and in cell lines derived from MM-1α L110R mice at mRNA and protein levels, and strong expression of FABP4 was observed in spermatocyte-like cells. Mammalian spermatozoa membranes are rich in polyunsaturated fatty acids. Metabolism of fatty acids plays an important role in spermatogenesis from spermatocytes to spermatids (see a review, [28]). It is therefore thought that overexpression of FABP4 in the testis of MM-1α L110R mice induces dysregulation of trafficking and/or intake of fatty acids, compromising differentiation of sperm. Lipid metabolism or lipid content of sperm is important for sperm motility, and phospholipases A2 participate in lipid rearrangements. The phospholipase A2 superfamily comprises intracellular and secreted enzymes (see review, [29]), and phospholipase A2-Group 3 (PLa2g3) and phospholipase A2-Group 10 (PLa2g10) proteins are expressed in male reproductive organs and/or in sperms. Reduced sperm motility is observed in PLa2g3-knockout mice, suggesting that phospholipase A2-Group 3 proteins play a role in sperm maturation and fertility through rearrangement of lipid and fatty acids [30], [31], [32]. Sperm-associated glutamate (E)-rich protein 4d (Speer-4d) is expressed specifically in the testis, epididymis and sperm and is thought to be important for sperm maturation [33]. In this study, expression levels of all of the above three genes were reduced in MM-1α L110R mice compared to those in wild-type mice, suggesting that reduced expression of these genes in MM-1α L110R mice induces decreased sperm number and motility. ChIP assays showed that L100R MM-1α but not wild-type MM-1α binds to a region in the Fabp4 gene, and wild-type MM-1α but not L100R MM-1α binds to regions of PLa2g3, PLa2g10 and Speer-4d genes, suggesting that gain-of-function and loss-of-function of MM-1α affect positive and negative regulation, respectively, of these genes at the transcriptional levels
Although defect of at least transcriptional activity of MM-1α affects male infertility of MM-1α L110R mice, it is not clear at present whether male infertility of MM-1α L110R mice is also due to dysfunction of MM-1α/PFDF5 mutation-containing prefoldin. We have reported that prefoldin was properly formed between MM-1α/PFDF5 mutant and the other five prefoldin subunits but that polyubiquitinated proteins were increased in L110R MM-1α mice and that cell lines from L110R MM-1α mice were more susceptible to various stresses than were those from wild-type mice, suggesting that substrate specificity of the prefoldin complex is reduced by MM-1α/PFDF5 mutation [13]. In addition to a subunit of prefoldin, MM-1α alone has various functions, including transcriptional regulation [1], [3], [5], [6], [34] and activation of protein degradation through the ubiquitin–proteasome system [35], [36]. It is important to address these points to clarify the effect of a single amino acid change of MM-1α on male infertility in future study.
Acknowledgments
We thank Patsy M. Nishina for C57BL/6-Pfdn5nmf5a/Pfdn5nmf5a mice. We also thank Tsutomu Osanai and Takae Koshiyama for their technical assistance of mice. This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (No. 24390013).
Footnotes
Transparency document associated with this article can be found in the online version at 10.1016/j.bbrep.2015.03.005.
Appendix A. Transparency document
Transparency docoment
References
- 1.Mori K., Maeda Y., Kitaura H., Taira T., Iguchi-Ariga S.M., Ariga H. MM-1, a novel c-Myc-associating protein that represses transcriptional activity of c-Myc. J. Biol. Chem. 1998;273:29794–29800. doi: 10.1074/jbc.273.45.29794. [DOI] [PubMed] [Google Scholar]
- 2.Fujioka Y., Taira T., Maeda Y., Tanaka S., Nishihara H., Iguchi-Ariga S.M., Nagashima K., Ariga H. MM-1, a c-Myc-binding protein, is a candidate for a tumor suppressor in leukemia/lymphoma and tongue cancer. J. Biol. Chem. 2001;276:45137–45144. doi: 10.1074/jbc.M106127200. [DOI] [PubMed] [Google Scholar]
- 3.Satou A., Taira T., Iguchi-Ariga S.M., Ariga H. A novel transrepression pathway of c-Myc. Recruitment of a transcriptional corepressor complex to c-Myc by MM-1, a c-Myc-binding protein. J. Biol. Chem. 2001;276:46562–46567. doi: 10.1074/jbc.M104937200. [DOI] [PubMed] [Google Scholar]
- 4.Satou A., Hagio Y., Taira T., Iguchi-Ariga S.M., Ariga H. Repression of the c-fms gene in fibroblast cells by c-Myc-MM-1-TIF1beta complex. FEBS Lett. 2004;572:211–215. doi: 10.1016/j.febslet.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida T., Kitaura H., Hagio Y., Sato T., Iguchi-Ariga S.M., Ariga H. Negative regulation of the Wnt signal by MM-1 through inhibiting expression of the wnt4 gene. Exp. Cell Res. 2008;314:1217–1228. doi: 10.1016/j.yexcr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe K., Ozaki T., Nakagawa T., Miyazaki K., Takahashi M., Hosoda M., Hayashi S., Todo S., Nakagawara A. Physical interaction of p73 with c-Myc and MM1, a c-Myc-binding protein, and modulation of the p73 function. J. Biol. Chem. 2002;277:15113–15123. doi: 10.1074/jbc.M111281200. [DOI] [PubMed] [Google Scholar]
- 7.Siegers K., Waldmann T., Leroux M.R., Grein K., Shevchenko A., Schiebel E., Hartl F.U. Compartmentation of protein folding in vivo: sequestration of non-native polypeptide by the chaperonin-GimC system. EMBO J. 1999;18:75–84. doi: 10.1093/emboj/18.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vainberg I.E., Lewis S.A., Rommelaere H., Ampe C., Vandekerckhove J., Klein H.L., Cowan N.J. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- 9.Geissler S., Siegers K., Schiebel E. A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J. 1998;17:952–966. doi: 10.1093/emboj/17.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartl F.U., Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 11.Tashiro E., Zako T., Muto H., Itoo Y., Sörgjerd K., Terada N., Abe A., Miyazawa M., Kitamura A., Kitaura H., Kubota H., Maeda M., Momoi T., Iguchi-Ariga S.M., Kinjo M., Ariga H. Prefoldin protects neuronal cells from polyglutamine toxicity by preventing aggregation formation. J. Biol. Chem. 2013;288:19958–19972. doi: 10.1074/jbc.M113.477984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takano M., Tashiro E., Kitamura A., Maita H., Iguchi-Ariga S.M., Kinjo M., Ariga H. Prefoldin prevents aggregation of α-synuclein. Brain Res. 2014;1542:186–194. [PubMed] [Google Scholar]
- 13.Abe A., Takahashi-Niki K., Takekoshi Y., Shimizu T., Kitaura H., Maita H., Iguchi-Ariga S.M., Ariga H. Prefoldin plays a role as a clearance factor in preventing proteasome inhibitor-induced protein aggregation. J. Biol. Chem. 2013;288:27764–27776. doi: 10.1074/jbc.M113.476358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsuchiya H., Iseda T., Hino O. Identification of a novel protein (VBP-1) binding to the von Hippel-Lindau (VHL) tumor suppressor gene product. Cancer Res. 1996;56:2881–2885. [PubMed] [Google Scholar]
- 15.Mousnier A., Kubat N., Massias-Simon A., Ségéral E., Rain J.C., Benarous R., Emiliani S., Dargemont C. von Hippel Lindau binding protein 1-mediated degradation of integrase affects HIV-1 gene expression at a postintegration step. Proc. Natl. Acad. Sci. USA. 2007;104:13615–13620. doi: 10.1073/pnas.0705162104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gstaiger M., Luke B., Hess D., Oakeley E.J., Wirbelauer C., Blondel M., Vigneron M., Peter M., Krek W. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science. 2003;302:1208–1212. doi: 10.1126/science.1088401. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y., Smith R.S., Jordan W., King B.L., Won J., Valpuesta J.M., Naggert J.K., Nishina P.M. Prefoldin 5 is required for normal sensory and neuronal development in a murine model. J. Biol. Chem. 2011;286:726–736. doi: 10.1074/jbc.M110.177352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao S., Carlesso G., Osipovich A.B., Llanes J., Lin Q., Hoek K.L., Khan W.N., Ruley H.E. Subunit 1 of the prefoldin chaperone complex is required for lymphocyte development and function. J. Immunol. 2008;181:476–484. doi: 10.4049/jimmunol.181.1.476. [DOI] [PubMed] [Google Scholar]
- 19.Hertzel A.V., Bernlohr D.A. The mammalian fatty acid-binding protein multigene family: molecular and genetic insights into function. Trends Endocrinol. Metab. 2000;11:175–180. doi: 10.1016/s1043-2760(00)00257-5. [DOI] [PubMed] [Google Scholar]
- 20.Burczynski F.J., Zhang M.N., Pavletic P., Wang G.Q. Role of fatty acid binding protein on hepatic palmitate uptake. Can. J. Physiol. Pharmacol. 1997;75:1350–1355. [PubMed] [Google Scholar]
- 21.Carlsson L., Nilsson I., Oscarsson J. Hormonal regulation of liver fatty acid-binding protein in vivo and in vitro: effects of growth hormone and insulin. Endocrinology. 1998;139:2699–2709. doi: 10.1210/endo.139.6.6041. [DOI] [PubMed] [Google Scholar]
- 22.Tataranni P.A., Baier L.J., Paolisso G., Howard B.V., Ravussin E. Role of lipids in development of noninsulin-dependent diabetes mellitus: lessons learned from Pima Indians. Lipids. 1996;31(Suppl.):S267–S270. doi: 10.1007/BF02637088. [DOI] [PubMed] [Google Scholar]
- 23.Baier L.J., Bogardus C., Sacchettini J.C. A polymorphism in the human intestinal fatty acid binding protein alters fatty acid transport across Caco-2 cells. J. Biol. Chem. 1996;271:10892–10896. doi: 10.1074/jbc.271.18.10892. [DOI] [PubMed] [Google Scholar]
- 24.Yamada K., Yuan X., Ishiyama S., Koyama K., Ichikawa F., Koyanagi A., Koyama W., Nonaka K. Association between Ala54Thr substitution of the fatty acid-binding protein 2 gene with insulin resistance and intra-abdominal fat thickness in Japanese men. Diabetologia. 1997;40:706–710. doi: 10.1007/s001250050737. [DOI] [PubMed] [Google Scholar]
- 25.Liu R.Z., Li X., Godbout R. A novel fatty acid-binding protein (FABP) gene resulting from tandem gene duplication in mammals: transcription in rat retina and testis. Genomics. 2008;92:436–445. doi: 10.1016/j.ygeno.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Ito K., Nakatani K., Fujii M., Katsuki A., Tsuchihashi K., Murata K., Goto H., Yano Y., Gabazza E.C., Sumida Y., Adachi Y. Codon 54 polymorphism of the fatty acid binding protein gene and insulin resistance in the Japanese population. Diabetic Med. 1999;16:119–124. doi: 10.1046/j.1464-5491.1999.00034.x. [DOI] [PubMed] [Google Scholar]
- 27.Thumser A.E., Moore J.B., Plant N.J. Fatty acid binding proteins: tissue-specific functions in health and disease. Curr. Opin. Clin. Nutr. Metab. Care. 2014;17:124–129. doi: 10.1097/MCO.0000000000000031. [DOI] [PubMed] [Google Scholar]
- 28.Wang C., Huang X. Lipid metabolism and Drosophila sperm development. Sci. China Life Sci. 2012;55:35–40. doi: 10.1007/s11427-012-4274-2. [DOI] [PubMed] [Google Scholar]
- 29.Schaloske R.H., Dennis E.A. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta. 2006;1761:1246–1259. doi: 10.1016/j.bbalip.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Sato H., Taketomi Y., Isogai Y., Miki Y., Yamamoto K., Masuda S., Hosono T., Arata S., Ishikawa Y., Ishii T., Kobayashi T., Nakanishi H., Ikeda K., Taguchi R., Hara S., Kudo I., Murakami M. Group III secreted phospholipase A2 regulates epididymal sperm maturation and fertility in mice. J. Clin. Investig. 2010;120:1400–1414. doi: 10.1172/JCI40493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Escoffier J., Jemel I., Tanemoto A., Taketomi Y., Payre C., Coatrieux C., Sato H., Yamamoto K., Masuda S., Pernet-Gallay K., Pierre V., Hara S., Murakami M., De Waard M., Lambeau G., Arnoult C. Group X phospholipase A2 is released during sperm acrosome reaction and controls fertility outcome in mice. J. Clin. Investig. 2010;120:1415–1428. doi: 10.1172/JCI40494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Escoffier J., Pierre V.J., Jemel I., Munch L., Boudhraa Z., Ray P.F., De Waard M., Lambeau G., Arnoult C. Group X secreted phospholipase A2 specifically decreases sperm motility in mice. J. Cell Physiol. 2011;226:2601–2609. doi: 10.1002/jcp.22606. [DOI] [PubMed] [Google Scholar]
- 33.Spiess A.N., Walther N., Müller N., Balvers M., Hansis C., Ivell R. SPEER – a new family of testis-specific genes from the mouse. Biol. Reprod. 2003;68:2044–2054. doi: 10.1095/biolreprod.102.011593. [DOI] [PubMed] [Google Scholar]
- 34.Ma H.C., Lin T.W., Li H., Iguchi-Ariga S.M., Ariga H., Chuang Y.L., Ou J.H., Lo S.Y. Hepatitis C virus ARFP/F protein interacts with cellular MM-1 protein and enhances the gene trans-activation activity of c-Myc. J. Biomed. Sci. 2008;15:417–425. doi: 10.1007/s11373-008-9248-9. [DOI] [PubMed] [Google Scholar]
- 35.Kimura Y., Nagao A., Fujioka Y., Satou A., Taira T., Iguchi-Ariga S.M., Ariga H. MM-1 facilitates degradation of c-Myc by recruiting proteasome and a novel ubiquitin E3 ligase. Int. J. Oncol. 2007;31:829–836. [PubMed] [Google Scholar]
- 36.Narita R., Kitaura H., Torii A., Tashiro E., Miyazawa M., Ariga H., Iguchi-Ariga S.M. Rabring7 degrades c-Myc through complex formation with MM-1. PLoS One. 2012;7:e41891. doi: 10.1371/journal.pone.0041891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency docoment