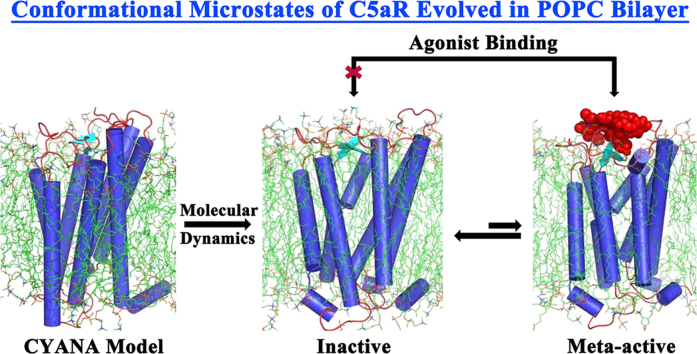
Abstract
C5a receptor (C5aR) is one of the major chemoattractant receptors of the druggable proteome that binds C5a, the proinflammatory polypeptide of complement cascade, triggering inflammation and SEPSIS. Here, we report the model structures of C5aR in both inactive and peptide agonist (YSFKPMPLaR; a=D-Ala) bound meta-active state. Assembled in CYANA and evolved over molecular dynamics (MD) in POPC bilayer, the inactive C5aR demonstrates a topologically unique compact heptahelical bundle topology harboring a β-hairpin in extracellular loop 2 (ECL2), derived from the atomistic folding simulations. The peptide agonist bound meta-active C5aR deciphers the “site2” at an atomistic resolution in the extracellular surface (ECS), in contrast to the previously hypothesized inter-helical crevice. With estimated Ki≈2.75 μM, the meta-active C5aR excellently rationalizes the IC50 (0.1–13 μM) and EC50 (0.01–6 μM) values, displayed by the peptide agonist in several signaling studies. Moreover, with Ki≈5.3×105 μM, the “site2” also illustrates selectivity, by discriminating the stereochemical mutant peptide (YSFkPMPLaR; k=D-Lys), known to be inert toward C5aR, up to 1 mM concentration. Topologically juxtaposed between the structures of rhodopsin and CXCR1, the C5aR models also display excellent structural correlations with the other G-protein coupled receptors (GPCRs). The models elaborated in the current study unravel many important structural insights previously not known for regulating the agonist binding and activation mechanism of C5aR.
Keywords: GPCR modeling, Peptide agonist, Docking, MD simulation, POPC bilayer, Activation mechanism
Graphical abstract
Highlights
-
•
Topologically unique inactive and meta-active atomistic models of C5aR.
-
•
Model demonstrates excellent structural correlation with the other reported GPCRs.
-
•
Model deciphers the “site2” in the ECS and also demonstrates agonist selectivity.
-
•
Agonist binding and activation requires “cation–π” interaction with F275 of C5aR.
-
•
Inactive to meta-active transition involves TM3–TM6 movements (ΔΘ≈+11.1°) in C5aR.
1. Introduction
C5aR is one of the two chemoattractant receptors [1] phylogenetically related to the rhodopsin subfamily of GPCRs found in human genome. C5aR interacts with C5a, the most potent proteolytic by-product of complement component 5 (C5) that triggers numerous proinflammatory and immunosuppressive disorders, including SEPSIS [2]. Thus, pharmaceutical industries have identified the C5aR as a potential target for development of future anti-inflammatory therapies [3].
Advances in protein engineering, including the membrane reconstitution techniques, have been phenomenal for structural studies of GPCRs [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23]. However, the growing list still lacks many rhodopsin family GPCRs including the C5aR, whose native structures both in inactive and agonist bound meta-active states are yet to be determined. Nevertheless, biochemical studies suggest that C5aR harbors two distinct binding sites for C5a, “site1” in N-terminus and “site2” in inter-helical crevice [24], obscured by the ECL2 in the inactive state [25]. For high affinity binding and signaling, the “site1” on C5aR engages the bulk of C5a, whereas “site2” engages the C-terminus of C5a. In agreement, it is evidenced that stereochemically engineered linear peptide agonist [26] (YSFKPMPLaR), carved out of the C-terminus of C5a [27] (ISHKDMQLGR, identical residues), potentially activates the C5aR signaling axis solo, albeit with weak potency. In addition, it is also noted that the proteolytic removal of the last C-terminal residue on C5a (des-R74-C5a) severely down regulates the C5aR signaling [28], [29], [30], [31], [32], [33], [34], which suggests that perhaps the interaction of the agonist at the “site2” plays a significant role over “site1” for triggering downstream signaling in C5aR. However, the precise intermolecular interactions between the C-terminus of C5a or the peptide agonist with C5aR, still remains a mystery, due to the unavailability of any such structural complexes, at an atomistic resolution. In the absence of structural data, several hypotheses have been postulated for C5a–C5aR interactions, purely based on the mutational studies performed on both C5a and C5aR. Interestingly, most of the functional data reported for C5a–C5aR or peptide agonist–C5aR has not been independently quantified with a full-fledged molecular model of C5aR. Even the coarse grain molecular model [35] proposed earlier for C5a–C5aR appears inadequate for precisely probing the interactions, respectively at the “site1” and “site2” of C5aR. As a result, the exact molecular mechanism underlying the agonist binding and selectivity in C5aR is still unclear, which greatly hinders the advancement of the prospective discovery and development of C5aR targeted therapeutics.
On the other hand, it is also not clear how C5aR switches from inactive to meta-active conformer that facilitates the agonist binding, followed by the interaction with G-proteins leading to the fully active state. The popular theory describes that activation of GPCRs involves a series of “molecular switches” [36] conserved across the “microdomains” [37] of the receptors. In particular, the GPCR activation involves the rotamer toggling on TM6 [38], [39] or the loss of the conserved ionic lock interaction between TM3 and TM6 [40], [41], facilitating the movement of TM3–TM6 at the intracellular face of the receptors, resulting in the subsequent binding and activation of G-proteins. Interestingly, C5aR does not feature the ionic lock, due to the lack of appropriate residues on TM3 and TM6 [42]. In fact the popular notion does not fit always, as nearly 80% of the rhodopsin family receptors do not have the right residues to support either the “rotamer toggling” or the “ionic lock” or sometimes the both [43]. So it is natural to wonder, whether this conformational transition from inactive to agonist ready meta-active conformer in C5aR requires the agonist binding (IF: induced fit) first or the agonist ready meta-active conformers are readily available due to the population dynamics of C5aR conformational microstates (CS: conformational selection) in tissue bilayers [44]. Surprisingly, no such information is currently available in the literature to clearly understand the activation mechanism of C5aR.
In the absence of a structure of C5aR, the present study aims to (1) generate a highly refined unique model structure of inactive C5aR, by recruiting the available structural data on GPCRs, (2) systematically decipher the meta-active conformer of C5aR with “site2”, (3) identify the residues in “site2” involved in peptide agonist binding and selectivity, in agreement with the reported binding and signaling studies [25], [28], [29], [30], [31], [32], [33], [34], [42], [45], [46], [47], [48], [49], [50], (4) test the stability of the C5aR–peptide agonist complex in POPC bilayer for further rationalizing the interaction specificity in light of C5a, and (5) understand the activation mechanism of C5aR triggered by the conformational transition from the inactive to meta-active state in light of other GPCRs.
2. Material and methods
2.1. Data sets and general computational methods
PDB coordinates of 20 rhodopsin family GPCRs (14 inactive and 6 active) [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23] were downloaded from www.rcsb.org. Primary sequence of C5aR was obtained from www.gpcr.org/7tm/. Visualization, analysis and presentation of model structure were performed respectively in PyMOL (The PyMOL Molecular Graphics System, Version 1.1r1, Schrödinger, LLC), MoLMoL [51], and Discovery studio (Accelrys). MoLMoL was implemented specifically for calculating the interhelical angles between the helix axes. Sequence alignments of receptors and of individual helices were achieved using ClustalX [52], which was further curated manually. Data were plotted in GraphPad Prism (version 6 for Windows, GraphPad Software, La Jolla California USA, www.graphpad.com). Solvent accessible surface area (SASA) of the receptors and of ligand binding sites were calculated with Naccess [53] with default van der Waals radii, by rolling a probe of radius 1.4 Å around the van der Waals surface. “Cation-π” angles were calculated as described [54] using the in house program. TM residues were numbered following Ballesteros–Weinstein system [37]. The two templates implemented for modeling each TMs of C5aR were selected from the matrix of 7×14 helices of inactive receptors, based on the highest sequence identity and similarity with TMs in C5aR. Homology models of the individual TMs in C5aR were achieved by recruiting two distinct templates in MODELLER [55], which were subsequently energy minimized and subjected to further analysis.
2.2. MD simulation in explicit water
The ECL2s of CXCR4 and β2AR were directly extracted from the crystal structures. The resulting peptides were end capped (COCH3 at N-terminus and CONH2 at C-terminus) and then subjected to careful energy minimization in a cubic box with periodic boundary, appropriately surrounded with simple point charge (SPC) water model, by recruiting the gromos-96 43a1 force field in GROMACS package [56]. Numerical integrations were performed in step size of 2 fs and coordinates were updated every 5 ps. Solvent density was set to the value corresponding to 1 atm at 300 K. Peptide and solvent were coupled independently to a modified Berendsen bath at 300 K, to the coupling time constant 0.1 ps. The appropriately equilibrated ECL2 structures were further subjected to MD studies at 300 K for 50 ns. The folding simulations for the ECL2 polypeptide of C5aR were initiated by implementing the above protocol, respectively validated against the ECL2s of CXCR4 and β2AR receptors. The end capped extended ECL2 polypeptide [Ac-Y174-RVVREEYFPPKVLCGVDYSHDKR-R198-NH2; ϕ, ψ=±120°, ϕ, ψ=−60°, 120° for proline] of C5aR was modeled using the in-house program PDBmake. Initially, the polypeptide was subjected to MD for 2 ns at 300 K. The resultant, partially collapsed random conformation of ECL2 was further subjected to folding studies, respectively, at 300 K and 310 K over 100 ns in explicit water. The 310 K trajectory was repeated again following the identical protocol. Analyses of the trajectories were achieved by recruiting the modules built into GROMACS.
2.3. CYANA model and MD simulation in POPC bilayer
The initial model of C5aR was assembled in CYANA [57]. Distance restraints used as inputs for structural modeling of C5aR in CYANA were directly calculated from the individual secondary structures of TMs and ECL2. Utility tools in Gromacs provided the dihedral angles (ϕ, ψ) of the individual amino acids in TMs and in folded ECL2. This alternatively helped in setting up the appropriate range of angular constraints for individual amino acids, which was subsequently used as input in CYANA. Hydrogen bond constraints were derived from the individual TMs and ECL2. The above restraints, including the interhelical distance restraints derived from the CXCR1 based template helped the structural modeling of C5aR, by appropriately guiding the conformation of other I / ECLs in CYANA. The models were screened for violations in amide geometry and backbone dihedral angles at individual amino acids. The best CYANA model with minimal violations was subjected to further refinements in MD studies over 250 ns, at 300 K in POPC [1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine] bilayer. We preferred to use POPC bilayer in our studies, as it has been mostly used as a model lipid for GPCR simulations [18], [20], [58], [59]. Following the InflateGRO approach [60], the CYANA modeled C5aR was inserted into the pre-equilibrated POPC bilayer (Prof. Peter Tieleman, Univ. Calgary) having 64 POPC molecules, each in upper and lower leaflets. POPC was packed around the C5aR until the areas per lipid (A) values were little over its reported experimental value (A=68.3±1.5 Å2) [61], [62]. The final system with 123 POPC molecules was neutralized by randomly placing 13 chloride ions that contained a total of 4620 water molecules. The system was equilibrated twice, first for 500 ps under NVT, followed by 1 ns under NPT conditions prior to the MD studies. Given the phase transition of pure POPC at 270 K [62], [63], the MD studies were done at 300 K (modified Berendsen) with C5aR, POPC in one group and water and ions in another group. Numerical integrations were performed in step size of 2 fs and coordinates were updated every 5 ps. Bonds were constrained with LINCS with order 4. Non-bonded pair list cut-off was 1.2 nm with a grid function. PME was implemented for electrostatic calculations. The peptide agonist bound meta-active conformer of C5aR was also subjected to MD studies over 250 ns in POPC bilayer (system contained 123 POPC, 4373 water molecules, and 15 chloride ions) as described above. The stereochemical mutant peptide that displayed a weak binding affinity toward the meta-active conformer of C5aR was not subjected to MD studies in POPC bilayer. In order to maintain the homogeneity throughout our studies, we also used the gromos-96 43a1 force field for lipid bilayer simulation. We did not try any other force fields in GROMACS. The MD trajectories were analyzed using the modules built into GROMACS.
2.4. Conformational clustering
Conformational clustering was performed (every 5 ps) as described [64], to the RMSD cut-off≤3 Å for the folding trajectories of ECL2 in water, as both ECL2s in CXCR4 and β2AR displayed a backbone RMSD over 3 Å. However, cut-off was reduced to≤1.5 Å, while analyzing the trajectories of folded ECL2s in water. Clusters having≥100 conformers were considered as major microstates/clusters. On the other hand, conformational clustering of the C5aR in POPC bilayer was performed (every 10 ps) to the RMSD cut-off≤1.5 Å and the free energies of the microstates were derived from the relative populations of individual microstates, using the following equation [64]:
| (1) |
where κB is the Boltzmann constant, PA and PB are the relative probabilities of finding the system in states A and B respectively, at temperature T. PA and PB are taken as the number of structures in microstates A and B respectively. For C5aR, clusters with minimum 200 conformers were selected as major microstates. The clusters were ranked based on their free energies. Cluster with highest number of conformers represented the inactive state of C5aR.
2.5. Docking of the peptide agonist
The linear YSFKPMPLaR agonist was modeled with PDBmake using the 2D NMR data as described [26], which was further subjected to energy minimization prior to the docking studies. The stereochemical mutant YSFkPMPLaR of the peptide agonist was also modeled similarly and energy minimized. The flexible ligand docking (RMSD tolerance of 5 Å), followed by a rigid docking (RMSD tolerance of 2 Å), was performed with AutoDock4.2 [65] for a population size of 150, implementing the genetic search algorithm with maximum number of generation set to 27,000. Structurally distinct conformational clusters of the peptides were ranked in terms of increasing energy. The best interacting conformers, respectively of the peptide agonist and the C5aR, represented the agonist peptide bound meta-active state of C5aR. Similar protocols were also used for docking the stereochemical mutant peptide to the meta-active conformer of C5aR.
3. Results and discussion
3.1. Structural analysis of the GPCRs for modeling C5aR
In our quest for a model structure of C5aR, we analyzed 14 inactive and 6 active structures of GPCRs (Table S1) [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23]. The structural analysis revealed that GPCRs share a canonical hepta helical bundle topology with a short 8th helix at the intra cellular side of the C-terminus. In addition, it is also evidenced that barring ECL2 (Fig. S1), the longest polypeptide loop, no other intra- and extra-cellular loops (I/ECLs) in GPCRs demonstrate structural diversity. It is interesting to note that except the rhodopsin [4], a major part of the N- and C-terminus is structurally unresolved in most of the GPCRs, including several protein ligand binding chemokine receptors, such as CXCR1, CXCR4 and CCR5. Rhodopsin is an exception, as it harbors a covalently linked non-protein ligand within its helical bundle, which activates the receptor by absorbing light. However, similar to the chemokine receptors, the N-terminus of C5aR (T7-K28) is also unordered, as evidenced in complex with the immune evasion protein CHIPS [66], established by NMR structural studies. The structural analysis described here, including the previous mutational studies [42], prompted us to model a truncated structure of C5aR (D27-P316; lacks 26 residues on N-terminus and 34 residues on C-terminus), harboring a possible folded structure of ECL2. Despite the canonical nature of transmembrane regions, GPCRs are topologically distinguishable, which makes them structurally unique. In order to invoke that certain uniqueness into the C5aR, direct template based homology models were avoided in our study.
3.2. Prediction and homology models of the TMs in C5aR
Among the inactive GPCRs (Table S2), the chemokine receptors CXCR1 (27.4/52) [14] and CXCR4 (24.5/50) [7], including the μ-opioid (27/53.4) [13] receptor, displayed a relatively high % sequence identity/similarity with C5aR. Out of the two closest phylogenetic siblings, the ligand free inactive structure of CXCR1 was preferred over CXCR4 for predicting the length of TMs in C5aR. Further, the homology models (Fig. S2) of the predicted TMs (TM1: P36-F65, TM2: N71-H99, TM3: A107-V138, TM4: A150-L173, TM5: E199-R228, TM6: R236-V260, TM7: C272-A303, and TM8: Q305-L315) were respectively built in MODELLER [55], by recruiting the two distinct templates, screened out for each TMs, from the matrix of 7×14 helices (Table S3) of inactive GPCRs. Integrity of the models were tested thoroughly prior to the further studies.
3.3. Modeling the folded structure of the ECL2 in C5aR
In silico folding of extended polypeptides [67], [68], [69], [70], [71], recruiting the molecular dynamics (MD) techniques have been very successful in past for predicting the putative structures of small peptides and proteins. Thus, to predict the possible folded structure of ECL2 in C5aR, the extended ECL2 polypeptide was subjected to an all atom folding simulation in explicit water, by recruiting the GROMACS [56], as described in Section 2.2. First, the MD protocols were benchmarked, respectively against the ECL2s of CXCR4 and β2-adrenergic receptors [5], [7] over 50 ns at 300 K. Both the ECL2s lacked the restraint of the conserved disulfide bond, including the transmembrane helices over the period of MD. It is noteworthy that the disruption of the structurally conserved disulfide linkage between the ECL2 and the TM3/ECL1 is known to affect the overall expression, maturation and folding of GPCRs. However, it is not established either biophysically or structurally that lack of this conserved disulfide bond particularly affects the folding and the structural integrity of the ECL2 in GPCRs. The results summarized in Appendix A, Appendix A indicate that the absence of transmembrane helices, including the ECL2-TM3 conserved disulfide bond does not perturb the structural integrity of the ECL2s significantly, which further suggests that the ECL2s are also among the independently foldable structural motifs in GPCRs. However, in the absence of both the conserved disulfide bond and the connecting helices, the ECL2s display high conformational flexibility (backbone RMSD≈4 Å and 5.87 Å, respectively for CXCR4 and β2AR, Fig. S5), which is not surprising, as GPCRs are inherently dynamic and are known for their modularity in ligand recognition and functional diversity [72].
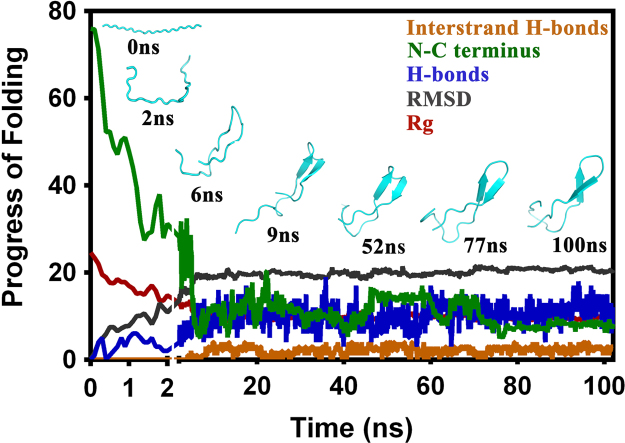
The benchmarked protocol was then implemented for evaluating the folded structure of the ECL2 in C5aR. The folding simulations were started, respectively, at 300 K and 310 K, from the partially collapsed random conformation (Fig. S6) obtained from the initial 2 ns MD study at 300 K. Compared to 310 K, the 300 K trajectory did not progress appreciably in folding pathways over 50 ns, as it got stuck in some sort of local minima, similar to a molten globule state (Fig. S7). Thus, the 300 K trajectory was not extended further. However, the 310 K trajectory displayed signatures of folding as early as 6 ns, which transformed into a well folded β-hairpin at around 9 ns (Fig. 1) and remained stable over 100 ns. A repeat run at 310 K also produced similar results (Fig. S8).
Fig. 1.
Folding trajectory of the extended ECL2 polypeptide in C5aR demonstrating the progress of folding over 100 ns at 310 K in explicit water.
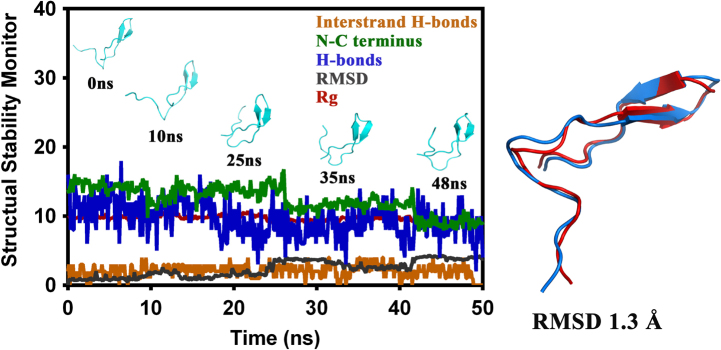
Cluster analysis (Fig. S9) suggested that the β-hairpin structure could be the major conformational microstate accessible to the ECL2 of C5aR. To probe it further, the central structure of the largest cluster was subjected to MD studies at 300 K over 50 ns. The results summarized in Fig. 2 suggest that indeed the ECL2 in C5aR harbors a topologically and structurally stable β-hairpin that spans from P184-H194 with C188-G189-V190 in loop conformations. Sequence analysis of the β-hairpin structures of the ECL2s (Table S4) across the GPCRs revealed that the ECL2 in C5aR shares the highest sequence identity/similarity (28%/48%) with the ECL2 of rhodopsin [4]. However, the folded ECL2 of C5aR demonstrates a distinctly different structure from the ECL2 of rhodopsin (backbone RMSD 6.8 Å, Fig. S10b). Further structural comparison indicated that while the conserved C188 adopted loop conformations in ECL2 of C5aR, the conserved C187 in rhodopsin adopted the β-sheet conformation (Fig. S10a). In contrast, the rhodopsin template based homology model of C5aR-ECL2 (Fig. S10c) displays a β-hairpin with C188 in the β-sheet region, as observed in rhodopsin (backbone RMSD 3 Å; 3CAP, www.gpcr.org/7tm/), spanning from E179-V190 with P183-P184-K185 in loop conformations. It is noteworthy that the ECL2 structure in C5aR obtained out of the folding simulation studies does not involve any such structural bias. Moreover, the β-hairpin loop in the folded ECL2 of C5aR features a glycine (C188-G189-V190), which is in agreement with ≥50% ECL2s in GPCRS with a β-hairpin structure. It is also interesting that similar to the folded ECL2 of C5aR, the ECL2 of rhodopsin also carries an additional unpaired cysteine (C185) in its hairpin loop (G182-M183-Q184), beside the conserved C187 on the β-sheet (Fig. S10b) linked to C1103.25 in TM3. This further assures that the folded structure obtained in the absence of the conserved disulfide bond for the ECL2 of C5aR is indeed of high quality.
Fig. 2.
Monitoring the structural stability of the β-hairpin obtained for the ECL2 of C5aR, over 50 ns of MD in explicit water at 300 K (left). Structural superposition of the folded β-hairpin at 0 ns (red) with the central conformer of the major microstate (blue) evolved over 50 ns, displaying backbone RMSD of 1.3 Å (right).
3.4. Structural assembly of C5aR in CYANA
CYANA [57] is an excellent tool that converts nuclear overhauser effect (NOE) intensities to distance restraints and further uses it as inputs for modeling the NMR structures of peptides and proteins. The hydrogen bond and the dihedral constraints used in CYANA are usually derived from the chemical shift index data, which provides information about the secondary structure of the proteins or peptides. In our studies, the very same principle of CYANA was harnessed for assembling the distance restraints based model structures of C5aR. In the absence of NOE data, the distance and other restraints used as inputs in CYANA were directly calculated from the independently modeled secondary structures of the TMs and ECL2, by recruiting the utility tools available in GROMACS or in other visualization softwares. Given the paucity of secondary structures in other I/ECLs, CYANA was allowed to model them appropriately, in the presence of 3981 distance restraints (includes interhelical restraints), 562 dihedral constraints, and 360 hydrogen bond constraints, derived from the individual TMs and ECL2. As noted in Table S3, 5 out of the 7 TMs in C5aR share over 50% sequence similarity with CXCR1 helices. Thus, the interhelical distance restraints used in CYANA, for modeling C5aR were derived from the CXCR1 template. The C188 being conserved in 90% of GPCRs [25] was assumed to form a disulfide bond with C1093.25 (TM3) in C5aR. Prior experimental studies have identified D27/S30 on N-terminus and S272 on third extracellular loop (ECL3) as important residues [42] on C5aR that upon mutation to cysteines, perhaps introduce a N-terminus-ECL3 disulfide bond, as observed in chemokine receptors CXCR4, CXCR1 and CCR5 [7], [14], [21]. Thus, structural modeling of C5aR in CYANA also involved an additional restraint between S30 and S272, which was removed subsequently during energy minimization steps.
3.5. Refinement of C5aR model structure
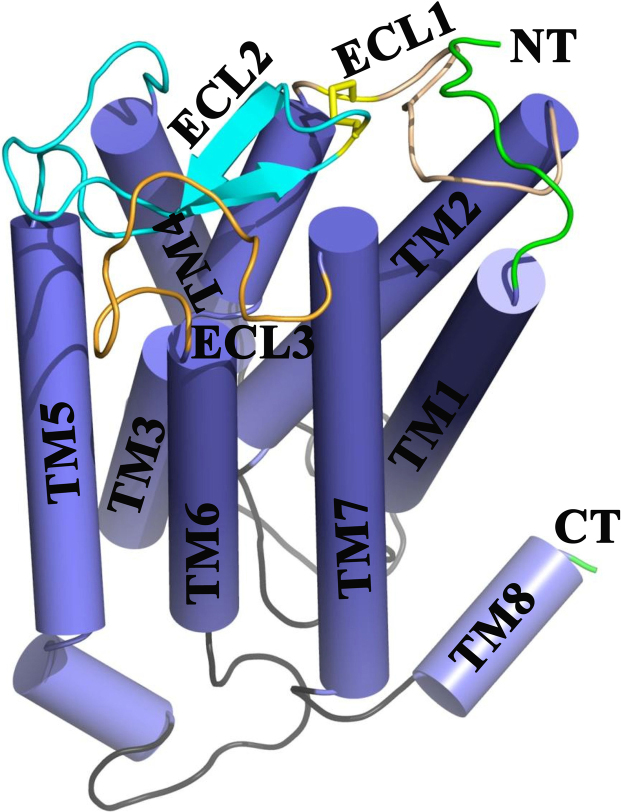
The resultant best model with≤5% violation was subjected to further refinements over 250 ns of MD simulations in POPC bilayer (Fig. S11), following the InflateGRO method, as described in Section 2.3. The presence of POPC bilayer helped the CYANA model to attain an equilibrated conformation over time. Given the slow conformational transition in bilayer, cluster analysis of the system provided only 7 major conformational microstates of C5aR (Fig. S12). With a backbone RMSD~4.68 Å from the CYANA model, the central structure of the microstate 1 (16,061 conformers) with lowest energy was chosen to represent the inactive state of C5aR (Fig. 3). The inactive C5aR with TM1: P36-F64, TM2: F75-Q98, TM3: I111-F139, TM4: W154-F172, TM5: E199-R228, TM6: L241-V260, TM7: F275-V302 and TM8: F307-L315 displayed minor deviation from the previously predicted TM lengths (Fig. S2). Overall, the inactive C5aR, presented in Fig. 3, displays a compact hepta helical bundle topology with a truncated N- and C-terminus, including three I/ECLs with a perfectly folded β-hairpin in ECL2 (L173-R198). The C188 on ECL2 form a disulfide bond with the C109 on TM3/ECL1, which covers the interhelical crevice in C5aR, as observed respectively in rhodopsin and CXCR1 receptors.
Fig. 3.
The unique heptahelical bundle topology of the inactive C5aR with truncated N- and C-terminus, featuring three I/ECLs, including the β-hairpin in ECL2 derived from the atomistic folding simulation studies. Topological features are labeled and highlighted. The conserved disulfide linkage is highlighted in yellow.
3.6. Structural comparison of C5aR with other GPCRs
The model structure of C5aR, presented in Fig. 3, is unique in nature, as no such refined model structure has been postulated for C5aR in the literature so far. To probe its topological uniqueness, the model C5aR was further validated against the inactive structures of other GPCRs. As noted in Table S5, the model displays excellent structural correlation with the other reported inactive GPCRs. As evidenced, most of the ICL3 engineered, ligand bound inactive GPCRs, shares ≈0.6–5 Å backbone RMSD with each other, whereas rhodopsin displays≈1.8–5.8 Å backbone RMSD across the inactive GPCRs (Table S5). In contrast, the C5aR (Fig. 3) displays≈4.6–6.5 Å backbone RMSD with other inactive GPCRs, including the CXCR1 (5.3 Å), which also shares≈5.8–7.9 Å backbone RMSD with other inactive GPCRs (Table S5). It is noteworthy that the structure of the inactive C5aR (Fig. 3) is perfectly juxtaposed between the structures of rhodopsin and CXCR1. It is interesting that the phylogenetically related receptors such as CXCR1 and CXCR4 also share a backbone RMSD over 6.5 Å, which makes them topologically distinct. On the other hand, a CXCR4 template (3rd best in homology, Table S2) based homology model of C5aR (3OE6, www.gpcr.org/7tm/) display only a backbone RMSD of 0.5 Å from CXCR4, making them topologically indistinguishable from each other. Interestingly, with backbone RMSD≈6 Å from CXCR4, the C5aR model structure presented in this study is both structurally and topologically unique.
3.7. Interhelical angle analysis of GPCRs compared with C5aR
With a common hepta helical bundle topology, GPCRs are incredibly smart in decoding messages carried out by a variety of ligands. It is certain that receptors undergo some kind of conformational arrangement to correctly decode the variety of signals. Given the fact that these receptors are packed inside the tissue bilayer, the degree of conformational freedom should be rather minimum. In such a scenario, the interhelical angle could be one of the important topological descriptors that can help in understanding the gradual conformational change within the helix bundle, associated with the transition from inactive to meta-active to fully active conformations.
To further study the model structure of inactive C5aR, the interhelical angles (Θ, Fig. S13) between the helix axes were analyzed in 14 inactive and 6 active GPCRs. (Table S6). The result summarized in Table S6 suggests that the common canonical topology also share a common mean Θ value across the GPCRs (SD of ±3–8° for inactive and SD of±2–8° for meta-active GPCRs), irrespective of the % sequence homology, location of binding sites and the type of the bound ligands. On comparison, the inactive C5aRs also displayed similar Θ values (Table S7), matching significantly with the chemokine receptors CXCR1, CXCR4, and CCR5 (Table S8) [7], [14], [21]. The observed similarity with the reported structures further suggests that the topological uniqueness in C5aR is not a result of unusual interhelical packing and indeed the models are of high quality.
3.8. Conformational scanning for a probable “site2” on C5aR
Evaluation of the ligand free inactive state of CXCR1 and antagonist bound inactive state of CXCR4 provides a contrasting snapshot of ECS and its modularity to present an active site on exposure to ligands (Fig. S14). While the inactive CXCR1 lacks an active site, the CXCR4 demonstrate an active site on the ECS. Due to the limited structural information, it cannot be clearly established, whether the availability of an active site in the ECS of CXCR4 is primarily due to CS or IF mechanism [44] is recruited post exposure to the ligand. However, the modularity in ECS could be unique to native peptide/protein binding receptors, as the ligand is generally big in size, compared to the many small molecule (non-protein) binding rhodopsin family GPCRs [5], [9], [10], [11], [13], [15], [19], [21], where ligand binding sites are usually located little deeper into the interhelical crevices.
We desired to investigate whether such modularity to accommodate a polypeptide ligand exists in the ECS of C5aR. For a possible answer, the 7 major microstates of C5aR obtained over 250 ns of MD simulation in POPC bilayer were further energetically ranked as described (Fig. S15a). Among the 3 top high energy microstates of C5aR, microstate 6 with 178 conformers displayed highest number of conformational transition (Fig. S15b) and thus, subjected to further structural and surface area analysis. Surprisingly, several conformers in microstate 6 displayed features of an apparent active “site2” on ECS, in contrast to the interhelical crevice, which was visibly absent on the conformers of microstate 1, representing the inactive state of C5aR (Figs. 3 and S14). With no trace of a ligand, the availability of an apparent active “site2” on ECS of C5aR is surely a result of conformational transition within the bilayer.
3.9. Deciphering the affinity of the YSFKPMPLaR agonist toward the “site2” on C5aR
To decipher the best meta-active conformer of C5aR from the pool of conformers in microstate 6, we recruited AutoDock [65] to scan the apparent affinity of the “site2” toward the linear decapeptide agonist YSFKPMPLaR (Fig. S16a) [26]. The peptide agonist is known to target the “site2” of C5aR with relatively high affinity (Table S9) and thus was chosen as the preferred molecular probe over the wild type C-terminus peptide of C5a. It is also evidenced that the peptide agonist triggers C5aR signaling in several mammalian tissues (EC50≈0.01–6 μM), including the engineered yeast [42], albeit with weak potency (IC50≈0.1–13 μM) compared to C5a [73], [74], [75], [76], [77].
Surprisingly, in agreement with the experiments (Table S9), one of the conformer from the microstate 6 displayed a strong binding affinity (Ki≈2.75 μM, −7.59 kcal/mole) for the peptide agonist, and thus was qualified as the meta-active conformer of C5aR (Fig. 4a). In addition, the meta-active conformer of C5aR also displayed selectivity by discriminating (Ki≈5.3×105 μM, −0.38 kcal/mole) the stereochemical mutant of the peptide agonist YSFkPMPLaR [78] that replaces the l-Lys on the peptide agonist with a d-Lys (Fig. S16b). The result is also in agreement with the previous functional data [78], where it is demonstrated that the mutant peptide is inert toward C5aR up to 1 mM concentration. This further justifies the worthiness of the “site2” on the meta-active conformer of C5aR (Fig. 4b).
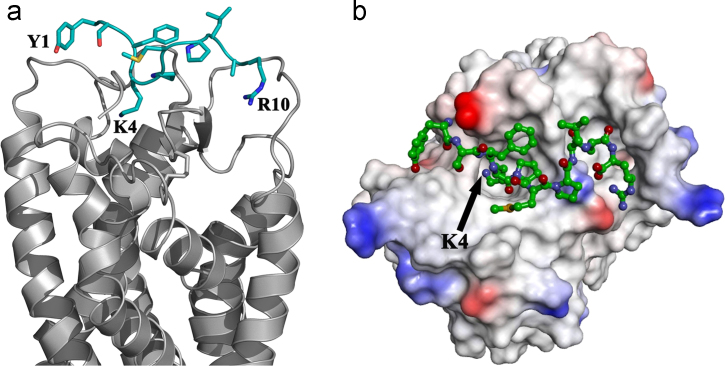
Fig. 4.
The peptide agonist bound meta-active state of C5aR. (a) The meta-active conformer of C5aR in complex with the peptide agonist illustrated in ribbon diagram. (b) C5aR surface rendered in ionizability mode highlights the YSFKPMPLaR (a=d-Ala) agonist (green) at the “site2”. The crucial pocket on the ECS occupied by the K4 of the agonist is also highlighted.
3.10. Mechanism of the YSFKPMPLaR agonist binding and selectivity at the “site2” of C5aR
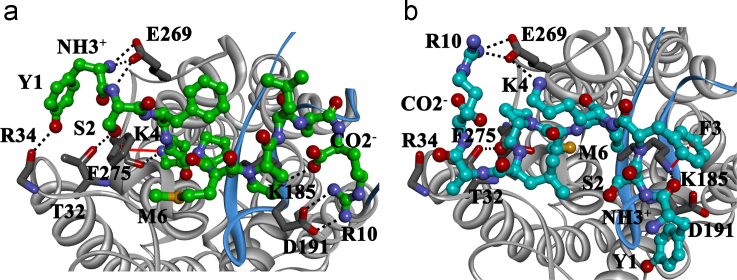
The YSFKPMPLaR agonist displays a significant conformational change with a backbone RMSD of ≈2.9 Å, post binding to the ECS of C5aR (Fig. S17a). The specific interactions driving the conformational change are detailed in Fig. 5a (Table S10, Fig. S17b). While Y1-R34, S2-T32 and R10-D191, including the termini (NH3+-E269 and COO−-K185) help in anchoring the peptide agonist to C5aR, the K4 of the agonist occupies the crucial pocket on the ECS (Figs. 4b, 5a and S17b) of the C5aR. This favors a strong “cation-π” [79] as well as the hydrogen bond interaction between the K4 of the agonist and the F2757.28 (beginning of TM7) of C5aR, triggering the binding affinity and perhaps the signaling in C5aR [46].
Fig. 5.
(a) Specific interactions driving the selective binding of the YSFKPMPLaR agonist (green) at the “site2” of C5aR. (b) Nonspecific intermolecular interactions displayed by the mutant peptide (cyan) at the “site2” of C5aR. Interacting residues of C5aR (gray) are respectively shown in sticks, highlighting the cation–π (solid red) and salt bridge/hydrogen bond (dotted black) interactions. Snapshots of the respective binding pockets are also illustrated in Fig. S17.
On the other hand, the stereochemical mutation [78] (l-Lys/d-Lys) not only alters the conformation of K4 side chain, but also affects the overall backbone conformation of the mutant peptide with a backbone RMSD≈3.1 Å (Fig. S17c). This potentially disrupts the specific “cation-π”, as well as the hydrogen bond interactions between the K4 and F2757.28. As a result, M6 occupies the crucial pocket on the ECS (Fig. S17d), forcing an unfavorable anchoring of the mutant peptide, driven by the non-specific K185-F3 “cation-π” interaction (Fig. 5b). This explains the known inactivity of YSFkPMPLaR toward C5aR [78].
3.11. Comparison of the “site2” on C5aR with CXCR4
The interaction of SDF-1 with CXCR4 is also believed to involve a similar two site binding interaction [7], as hypothesized for C5a and C5aR [24]. Thus, the peptide agonist bound “site2” on C5aR was compared against the cyclic peptide antagonist bound “site2” on CXCR4. As illustrated in Fig. 4b, the peptide agonist bound C5aR displays an extended and open active “site2” on ECS, similar to the “site2” observed in CXCR4, but in sharp contrast with the “site2”, previously hypothesized for C5aR [24]. As evidenced in Table 1, total 23 residues (6 N-terminus, 8 ECL2, 5 ECL3 and 4 TM7 residues) contribute toward the “site2” on C5aR that present a solvent accessible surface area (SASA) of 899 Å2 to the linear peptide agonist, roughly comparable to the SASA of the “site2” bound to the cyclic peptide CVX15 on CXCR4 (20 residues, SASA≈606 Å2). The ECLs and N-terminus residues are the major contributors toward the “site2” on C5aR that includes only 4 residues from the extracellular stems of TM7, whereas the “site2” on CXCR4 has significant contribution from the extracellular part of the helices 3, 4, 5 and 7. In fact 14 out of the 20 residues in “site2” on CXCR4 are from the transmembrane helices. This is not surprising, as the ICL3 engineered antagonist bound inactive CXCR4 has a more open topology than the peptide agonist bound C5aR. As evidenced from the SASA calculation (Table 1) the inactive C5aR has a relatively closed topology, similar to the inactive CXCR1. Thus, it is not surprising that agonist binding at the “site2” on meta-active conformer of C5aR involves more number of residues from the N-terminus and loops than from the extracellular parts of the transmembrane helices. Indeed, literature evidences that mutation of many residues [25], [45], [46], [47], [80] (T29, D191, P270, S272, F275, N279, F182 and P184) identified on “site2” of C5aR (Table 1) affect C5aR signaling. In addition, the residues that are sequentially close to “site2” but appear apparently farther from the “site2” [28], [80] (D27, L277, and D282) in our model are also known to affect the C5aR signaling in response to C5a. It is quite possible that such residues may be structurally important to C5aR or other sections of C5a might be engaging these residues for tighter binding and signaling of C5aR. This further confirms the exceptional quality of the meta-active conformer of the C5aR.
Table 1.
Solvent accessible surface area (SASA)a of the ECS in C5aR, respectively compared with the CXCR1 and CXCR4. The SASA of the “site2” in C5aR is also compared with the “site2” observed in CXCR4.
| GPCRs (PDB) |
SASA (Å2) |
Active site residues | |
|---|---|---|---|
| ECS (No. of residues) | Site2 (No. of residues) | ||
| CXCR1 (2LNL) | 2286 (67) | ||
| CXCR4 (3OE0); Bound to cyclic peptide antagonist CVX15 | 3238 (60) | 606 (20) | [H1133.29, Y1163.32, T1173.33, D1714.60, D187, R188, F189, Y190, P191, D193, V1965.35, F1995.38, Q2005.39, D2626.58, L2666.62, E2777.28, H2817.32, I2847.35, S2857.36, E2887.39] |
| C5aR (Inactive) | 2739 (70) | ||
| C5aR (Meta-active); Bound to linear peptide agonist | 2660 (70) | 899 (23) | [T29, S30, N31, T32, R34, V35, Y174, R175, F182, P184, K185, V186, V190, D191, L268, E269, P270, S271, S272, F2757.28, L2767.29, L2787.31, N2797.32] |
a=SASA calculations are done using the default van der Waals radii with Ala–X–Ala template as reference in Naccess; TM residues shown in bold are numbered following Ballesteros–Weinstein system.
3.12. Rationalizing the YSFKPMPLaR agonist binding to C5aR in light of C5a
It is important to note that a full agonist like C5a can potentially access the “site1” and “site2”, whereas the linear peptide agonist can only access the “site2” on C5aR (Figs. 4b and 5a). Thus, unlike the mutant peptide [78] (Fig. 5b, Table S10), mutation of K68 to E68/M68 in C5a does not induce complete inactivity [33], rather decreases the ED50 value, respectively by 2.5 and 6.8 fold toward C5aR. Similarly, the des-R74-C5a with intact stereochemistry at K68 displays relatively weak binding and signaling [33], [34], [48] compared to C5a (Table S11), which abrogates completely by further mutation of K68/E68 [33], suggesting the functional importance of K68 over R74 in C5a for C5aR signaling. Further, the loss of signaling (~100 fold) in D191G-C5aR [25] in response to W5Cha (Me-F-K-P-dCha-Cha-dR; dR=d-Arg), clearly justifies the role of D191 in C5aR for anchoring the agonist by involving the dR6 on W5Cha. In agreement, the R10 on YSFKPMPLaR agonist also demonstrates interaction with D191 (Fig. 5a) in our studies, which could also be the case for R74 on C5a. This brings back the attention to D282 of C5aR, previously suggested to interact with R74 of C5a or other related peptides [28], [31]. However, a closer analysis of the binding and signaling data of D282 mutants suggests an alternative mechanism. As evidenced, both wild type and D282A-C5aR bind to C5a with identical affinity (IC50~14 nM), which clearly indicates the negligible role of D282 in anchoring R74 of C5a to C5aR. Moreover, compared to wild type C5aR (EC50~5.82 nM), the D282A-C5aR demonstrates~3 fold weaker signaling (IC50~14.6 nM) in response to C5a. But, surprisingly D282A-C5aR demonstrates better signaling (91% degranulation) than the wild type C5aR (79% degranulation) in response to des-R74-C5a. Further, R74D-C5a does not bind to D282R-C5aR, but demonstrates weaker binding to wild type C5aR (IC50~3790 nM). It is noteworthy that both C5a (EC50~325 nm, IC50~57 nm) and R74D-C5a (EC50~365 nM, IC50~not known) trigger comparable signaling activity in D282R-C5aR. In summary, the reported functional data on D282 mutants does not favorably support or establish a direct interaction between D282 of C5aR with R74 of C5a. This is further in support of the C5aR–peptide agonist complex (Fig. 5a), where it is demonstrated that the R10 of the YSFKPMPLaR agonist interacts with the D191 at the “site2” of C5aR, instead of the D282, which is positioned little deeper into the transmembrane region in the model. GPCRs are conformationally dynamic and are known to demonstrate allosterism [81]. Thus, it is quite possible that mutations of structurally important residues such as D282 in C5aR can induce allosterism, which can modulate the overall pharmacology of C5aR in response to C5a. Though 3D models provide important insights, only future structural studies can unequivocally establish the functional importance of many such residues and their contribution toward ligand binding and allosterism in C5aR.
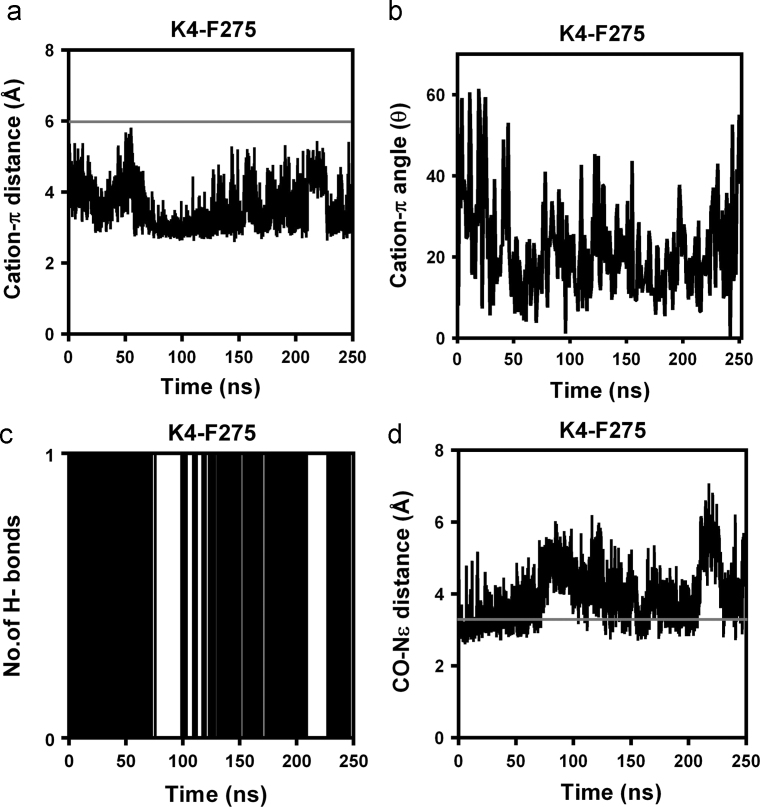
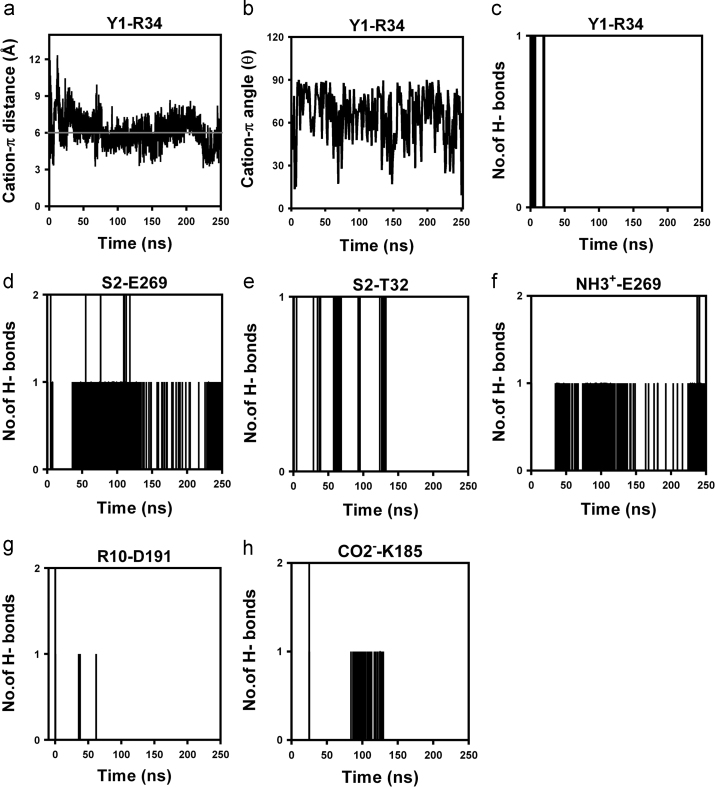
Nevertheless, the specificity of the intermolecular interactions illustrated for the C5aR–peptide complex (Fig. 5a) was maintained over 250 ns in POPC bilayer (Fig. 6, Fig. 7, S18, S19 and S20). Besides the general hydrophobic and hydrogen bonding interactions, the moderate “cation-π” interaction evolved between the Y1-R34 side chains (Figs. 7a and 7b) by the disappearance of the weak hydrogen bonding between the side chain of Y1 and backbone carbonyl of R34 (Fig. 7c) during the MD simulation also contribute toward the overall stability of the complex. Moreover, the strong K4-F2757.28 “cation-π” (d≤6 Å; Θ<90°) and hydrogen bonding interactions between the side chain of K4 and backbone carbonyl of F2757.28 (Fig. 6), including the transient R10-D191 salt bridge interaction (Fig. 7g), immensely strengthen the specific binding mode demonstrated by the peptide agonist in C5aR complex (Figs. 4 and 5a).
Fig. 6.
The stability of the specific intermolecular interactions monitored between the K4 of the YSFKPMPLaR agonist with the F2757.28 of C5aR, over 250 ns of MD at 300 K in POPC bilayer. The distance (a) and the interaction angle (b) monitored between the K4 head group and aromatic ring of F2757.28 indicates a strong K4-F2757.28 “cation–π” interaction. (c) The strong hydrogen bond interaction between the K4 head group and the backbone carbonyl of F2757.28. (d) The distance between the heavy atoms participating in the hydrogen bonding.
Fig. 7.
The other intermolecular interactions monitored between the YSFKPMPLaR agonist and the C5aR, over 250 ns of MD at 300 K in POPC bilayer. NH3+ and CO2− respectively represent the N and C-terminus of the peptide agonist. Y1, S2, and R10, respectively represent the interacting residues on the peptide. T32, R34, K185, D191, and E269, respectively represent the interacting residues on the C5aR. Demonstration of the distance (a) and angle (b) of the moderate “cation–π” interaction observed between the Y1-R34 side chains, by depletion of the weak hydrogen bonding (c) between the Y1 (HH)-R34 (CO). (d) The strong hydrogen bonding demonstrated between the back bone NH of S2 and the side chain of E269. (e) The strong hydrogen bonding between the side chains of S2 and T32 is maintained modestly over the 50% of the time. (f) The presence of hydrogen bond between the N-terminus and the side chain of E269 implicates the existence of a strong salt bridge interaction between NH3+-E269. (g) The transient nature of hydrogen bonding between the side chain of the R10 and D191 suggest the presence of a salt bridge interaction between R10-D191. (h) The strong hydrogen bond interaction between the C-terminus of the peptide agonist and the side chain of K185 is modestly maintained over 20% of the time.
Moreover, previous studies have also shown that K4/E4 mutation in the peptide agonist dampens the C5aR signaling, which is further rescued to a great extent by R4 with a cationic side chain [78]. In addition, F275A mutation has also been suggested to have a deleterious effect on C5aR function [46]. It is worth mentioning that similar to the des-R74-C5a, the removal of R10 from the peptide agonist reduces the estimated affinity by~30 fold toward the meta-active conformer of C5aR (data not shown). Thus, it is implied that while R10/R74 helps in docking the agonist, the K4/K68 plays an important role in triggering the binding and signaling at the “site2” involving F2757.28 in C5aR. Moreover, comparison of the bound peptide agonist with the unbound C-terminus of C5a [27] (Fig. S21) reveals an incredible similarity between the side chain conformations of R10/R74, K4/K68, S2/S66 and M6/M70 residues. Indeed, the specificity of the binding demonstrated by the peptide agonist at the “site2” of C5aR is also reflected by the native C-terminus peptide (NISHKDMQLGR) of C5a (data not shown).
3.13. Activation mechanism in C5aR compared to other GPCRs
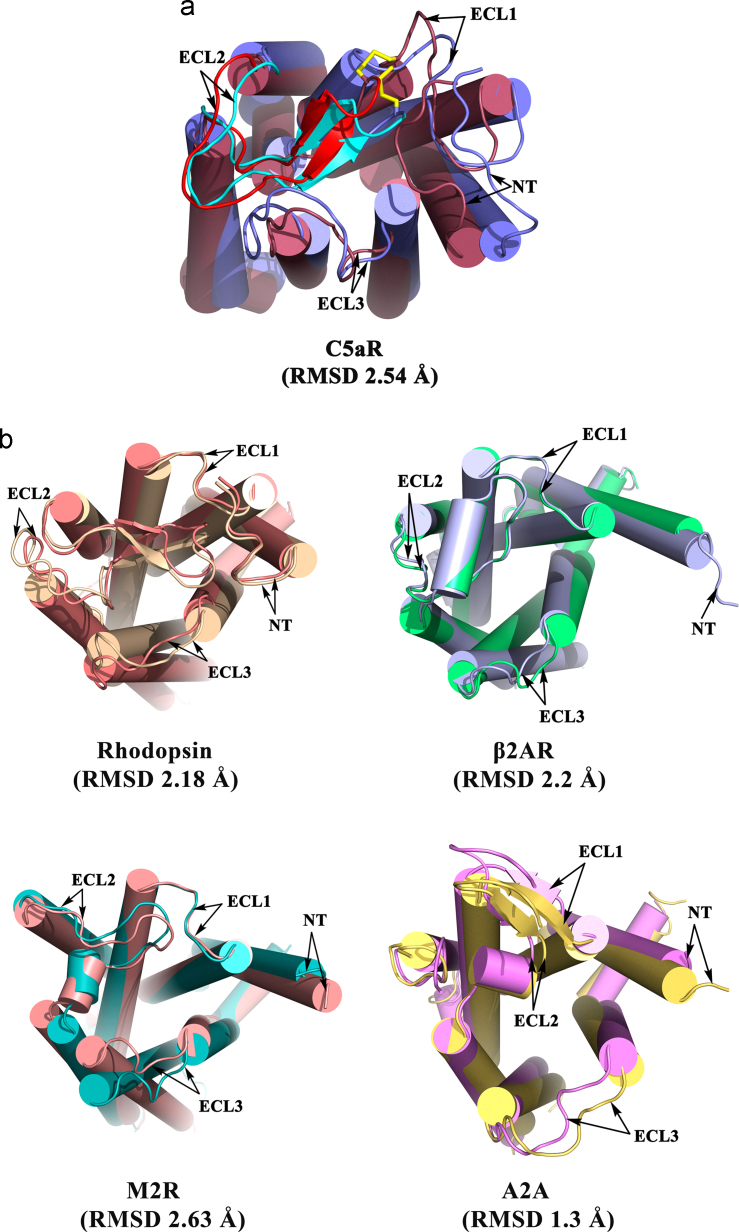
Random saturation mutagenesis studies on the ECL2 of C5aR have suggested that the ECL2 works as a gatekeeper and significant movement in ECL2 is associated with auto-activation of C5aR [25]. In addition, single or double mutation in TM3 has also been shown to auto-activate C5aR [42], [82]. As noted in Fig. 8a, with a backbone RMSD of 2.54 Å, the transition from the inactive to meta-active conformer in C5aR not only displays the movement of the ECL2, but also displays significant movements in N-terminus and other ECLs. Interestingly, a backbone RMSD of ≈1.3–2.6 Å, coupled with significant movements of helices and loops, is also noted across the known inactive and agonist bound meta-active conformers of GPCRs (Fig. 8b). The conformational dynamics in POPC bilayer provided both the inactive and the meta-active conformer of the C5aR, with no direct influence from the peptide agonist. Thus, we hypothesize that perhaps agonist binding in C5aR follows a conformational selection approach than an induced fit approach [44]. This appears plausible as many GPCRs in native and non-native tissues display some amount of basal activity, which further enhances in response to the ligand or to an activating point mutation [83]. However, whether ligand binding in GPCR is universally conformational selection based is a matter of challenging structural studies of active GPCRs, both in ligand free and in bound states.
Fig. 8.
Illustration of the conformational changes (movements in TMs, N-terminus and ECLs) highlighting the backbone RMSDs involved in transition of inactive to agonist bound meta-active conformer in C5aR in light of other GPCRs. (a) the meta-active (raspberry, ECL2 in red) and inactive (slate, ECL2 in cyan) conformers of the model C5aRs, and (b) the meta-active (3DQB, deep salmon) and inactive (1F88, wheat) conformers of rhodopsin (top left), the meta-active (3P0G, light blue) and inactive (2RH1, lime green) conformers of β2 AR (top right), the meta-active (4MQS, salmon) and inactive (3UON, cyan) conformers of M2R (bottom right), the meta-active (3QAK, yellow orange) and inactive (3REY, violet) conformers of A2A (bottom left).
Analysis of the change in interhelical angles (ΔΘ=Θinactive−ΘActive) of known GPCR pairs, summarized in Table S12 indicate that inactive-to-meta-active transition involves the movement of almost all the helices across the GPCR pairs, including the model C5aRs. However, the highest number of significant movements (ΔΘ≥±4°) are noted for the helix6 in opsin [6], helix1, helix6 in β2AR [5], [10], helix7 in A2A [9], [11], and helix5 in M2R [15], [19], compared to TM3, and TMs 5–7 in C5aR. In agreement with the popular activation mechanism [84], the helix3–helix6 pair displays significantly higher magnitude of change in ΔΘ (ΔΘ≈−7.9° for rhodopsin, −8.5° for A2A and −9.4° for β2AR). However, it is noted that the ΔΘ related to helix3–helix6 pair is not consistently highest across the GPCR pairs. Surprisingly, only M2R (ΔΘ≈-22.2°) display the highest magnitude of change for helix3–helix6 pair, whereas the highest is noted for the helix5–helix6 in opsin (ΔΘ≈−13.5°), helix6–helix7 in β2AR (ΔΘ≈+17.2°), and helix6–helix7 pair (ΔΘ≈+16.7°) in A2A. Interestingly, the TM3–TM6 pair displays the highest magnitude of change (ΔΘ≈+11.1°) in C5aR, which does not feature the “ionic lock” mostly attributed for the activation of the above mentioned receptors. This analysis suggests, perhaps there is no uniform mechanism that operates across the GPCRs and also highlights the complexity involved in GPCR activation. Thus, it can be implied that the nature and the type of the agonist play an important role in regulating the population of GPCR microstates, which in turn chooses an activation mechanism [85] suitable for the tissues expressing the GPCRs.
4. Conclusions
Highly refined model structures of C5aR are reported for the first time in excellent correlation with experiments, both in inactive state and in complex with a stereochemically engineered linear YSFKPMPLaR agonist [26], carved out of the C-terminus of C5a [27]. The meta-active conformer of C5aR with an atomistic resolution of “site2” on ECS provides important structural insights not appreciated before for understanding the selectivity of agonist binding and signaling. The conformational transition from inactive to meta-active state in C5aR, which facilitates the agonist binding and activation not only involves the movements of TM3–TM6 (ΔΘ≈+11.1°) but also involves the movement of N-terminus, ECL2 and other ECLs. The study highlights F2757.28 and D191 as major residues on C5aR for anchoring the K4 and R10 of the peptide agonist and also suggests a similar binding mechanism between C5a and C5aR.
Acknowledgments
We thank Prof. S. Durani, IIT Bombay, for giving access to CYANA. We also thank Prof. T. J. Baranski, Washington University School of Medicine in St. Louis, for critically reading the manuscript. Use of BRAF facility at CDAC, Pune, is highly appreciated. This research is partly supported by the SEED Grant IIT Bhubaneswar.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.03.002.
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.03.002.
Appendix A. Supplementary materials
Supplementary data
Appendix B. Transparency document
Transparency document
References
- 1.Gerard N.P., Gerard C. The chemotactic receptor for human C5a anaphylatoxin. Nature. 1991;349:614–617. doi: 10.1038/349614a0. [DOI] [PubMed] [Google Scholar]
- 2.Guo R.F., Ward P.A. Role of C5a in inflammatory responses. Annu. Rev. Immunol. 2005;23:821–852. doi: 10.1146/annurev.immunol.23.021704.115835. [DOI] [PubMed] [Google Scholar]
- 3.Ricklin D., Lambris J.D. Complement-targeted therapeutics. Nat. Biotechnol. 2007;25:1265–1275. doi: 10.1038/nbt1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palczewski K., Kumasaka T., Hori T., Behnke C.A., Motoshima H., Fox B.A., Le Trong I., Teller D.C., Okada T., Stenkamp R.E., Yamamoto M., Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 5.Cherezov V., Rosenbaum D.M., Hanson M.A., Rasmussen S.G., Thian F.S., Kobilka T.S., Choi H.J., Kuhn P., Weis W.I., Kobilka B.K., Stevens R.C. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheerer P., Park J.H., Hildebrand P.W., Kim Y.J., Krauss N., Choe H.W., Hofmann K.P., Ernst O.P. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 7.Wu B., Chien E.Y., Mol C.D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F.C., Hamel D.J., Kuhn P., Handel T.M., Cherezov V., Stevens R.C. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chien E.Y., Liu W., Zhao Q., Katritch V., Han G.W., Hanson M.A., Shi L., Newman A.H., Javitch J.A., Cherezov V., Stevens R.C. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu F., Wu H., Katritch V., Han G.W., Jacobson K.A., Gao Z.G., Cherezov V., Stevens R.C. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasmussen S.G., Choi H.J., Fung J.J., Pardon E., Casarosa P., Chae P.S., Devree B.T., Rosenbaum D.M., Thian F.S., Kobilka T.S., Schnapp A., Konetzki I., Sunahara R.K., Gellman S.H., Pautsch A., Steyaert J., Weis W.I., Kobilka B.K. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dore A.S., Robertson N., Errey J.C., Ng I., Hollenstein K., Tehan B., Hurrell E., Bennett K., Congreve M., Magnani F., Tate C.G., Weir M., Marshall F.H. Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure. 2011;19:1283–1293. doi: 10.1016/j.str.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimamura T., Shiroishi M., Weyand S., Tsujimoto H., Winter G., Katritch V., Abagyan R., Cherezov V., Liu W., Han G.W., Kobayashi T., Stevens R.C., Iwata S. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manglik A., Kruse A.C., Kobilka T.S., Thian F.S., Mathiesen J.M., Sunahara R.K., Pardo L., Weis W.I., Kobilka B.K., Granier S. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park S.H., Das B.B., Casagrande F., Tian Y., Nothnagel H.J., Chu M., Kiefer H., Maier K., De Angelis A.A., Marassi F.M., Opella S.J. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 2012;491:779–783. doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haga K., Kruse A.C., Asada H., Yurugi-Kobayashi T., Shiroishi M., Zhang C., Weis W.I., Okada T., Kobilka B.K., Haga T., Kobayashi T. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granier S., Manglik A., Kruse A.C., Kobilka T.S., Thian F.S., Weis W.I., Kobilka B.K. Structure of the delta-opioid receptor bound to naltrindole. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanson M.A., Roth C.B., Jo E., Griffith M.T., Scott F.L., Reinhart G., Desale H., Clemons B., Cahalan S.M., Schuerer S.C., Sanna M.G., Han G.W., Kuhn P., Rosen H., Stevens R.C. Crystal structure of a lipid G protein-coupled receptor. Science. 2012;335:851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruse A.C., Hu J., Pan A.C., Arlow D.H., Rosenbaum D.M., Rosemond E., Green H.F., Liu T., Chae P.S., Dror R.O., Shaw D.E., Weis W.I., Wess J., Kobilka B.K. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruse A.C., Ring A.M., Manglik A., Hu J., Hu K., Eitel K., Hubner H., Pardon E., Valant C., Sexton P.M., Christopoulos A., Felder C.C., Gmeiner P., Steyaert J., Weis W.I., Garcia K.C., Wess J., Kobilka B.K. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C., Srinivasan Y., Arlow D.H., Fung J.J., Palmer D., Zheng Y., Green H.F., Pandey A., Dror R.O., Shaw D.E., Weis W.I., Coughlin S.R., Kobilka B.K. High-resolution crystal structure of human protease-activated receptor 1. Nature. 2012;492:387–392. doi: 10.1038/nature11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan Q., Zhu Y., Li J., Chen Z., Han G.W., Kufareva I., Li T., Ma L., Fenalti G., Li J., Zhang W., Xie X., Yang H., Jiang H., Cherezov V., Liu H., Stevens R.C., Zhao Q., Wu B. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science. 2013;341:1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srivastava A., Yano J., Hirozane Y., Kefala G., Gruswitz F., Snell G., Lane W., Ivetac A., Aertgeerts K., Nguyen J., Jennings A., Okada K. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature. 2014;513:124–127. doi: 10.1038/nature13494. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J., Zhang K., Gao Z.G., Paoletta S., Zhang D., Han G.W., Li T., Ma L., Zhang W., Muller C.E., Yang H., Jiang H., Cherezov V., Katritch V., Jacobson K.A., Stevens R.C., Wu B., Zhao Q. Agonist-bound structure of the human P2Y12 receptor. Nature. 2014;509:119–122. doi: 10.1038/nature13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siciliano S.J., Rollins T.E., DeMartino J., Konteatis Z., Malkowitz L., Van Riper G., Bondy S., Rosen H., Springer M.S. Two-site binding of C5a by its receptor: an alternative binding paradigm for G protein-coupled receptors. Proc. Natl. Acad. Sci. USA. 1994;91:1214–1218. doi: 10.1073/pnas.91.4.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klco J.M., Wiegand C.B., Narzinski K., Baranski T.J. Essential role for the second extracellular loop in C5a receptor activation. Nat. Struct. Mol. Biol. 2005;12:320–326. doi: 10.1038/nsmb913. [DOI] [PubMed] [Google Scholar]
- 26.Vogen S.M., Prakash O., Kirnarsky L., Sanderson S.D., Sherman S.A. Determination of structural elements related to the biological activities of a potent decapeptide agonist of human C5a anaphylatoxin. J. Pept. Res. 1999;54:74–84. doi: 10.1034/j.1399-3011.1999.00087.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X., Boyar W., Toth M.J., Wennogle L., Gonnella N.C. Structural definition of the C5a C terminus by two-dimensional nuclear magnetic resonance spectroscopy. Proteins. 1997;28:261–267. doi: 10.1002/(sici)1097-0134(199706)28:2<261::aid-prot13>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 28.Higginbottom A., Cain S.A., Woodruff T.M., Proctor L.M., Madala P.K., Tyndall J.D., Taylor S.M., Fairlie D.P., Monk P.N. Comparative agonist/antagonist responses in mutant human C5a receptors define the ligand binding site. J. Biol. Chem. 2005;280:17831–17840. doi: 10.1074/jbc.M410797200. [DOI] [PubMed] [Google Scholar]
- 29.Kalant D., Cain S.A., Maslowska M., Sniderman A.D., Cianflone K., Monk P.N. The chemoattractant receptor-like protein C5L2 binds the C3a des-Arg77/acylation-stimulating protein. J. Biol. Chem. 2003;278:11123–11129. doi: 10.1074/jbc.M206169200. [DOI] [PubMed] [Google Scholar]
- 30.Schatz-Jakobsen J.A., Yatime L., Larsen C., Petersen S.V., Klos A., Andersen G.R. Structural and functional characterization of human and murine C5a anaphylatoxins. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2014;70:1704–1717. doi: 10.1107/S139900471400844X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cain S.A., Coughlan T., Monk P.N. Mapping the ligand-binding site on the C5a receptor: arginine74 of C5a contacts aspartate282 of the C5a receptor. Biochemistry. 2001;40:14047–14052. doi: 10.1021/bi011055w. [DOI] [PubMed] [Google Scholar]
- 32.Scola A.M., Higginbottom A., Partridge L.J., Reid R.C., Woodruff T., Taylor S.M., Fairlie D.P., Monk P.N. The role of the N-terminal domain of the complement fragment receptor C5L2 in ligand binding. J. Biol. Chem. 2007;282:3664–3671. doi: 10.1074/jbc.M609178200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crass T., Bautsch W., Cain S.A., Pease J.E., Monk P.N. Receptor activation by human C5a des Arg74 but not intact C5a is dependent on an interaction between Glu199 of the receptor and Lys68 of the ligand. Biochemistry. 1999;38:9712–9717. doi: 10.1021/bi990139q. [DOI] [PubMed] [Google Scholar]
- 34.Cain S.A., Monk P.N. The orphan receptor C5L2 has high affinity binding sites for complement fragments C5a and C5a des-Arg(74) J. Biol. Chem. 2002;277:7165–7169. doi: 10.1074/jbc.C100714200. [DOI] [PubMed] [Google Scholar]
- 35.Nikiforovich G.V., Marshall G.R., Baranski T.J. Modeling molecular mechanisms of binding of the anaphylatoxin C5a to the C5a receptor. Biochemistry. 2008;47:3117–3130. doi: 10.1021/bi702321a. [DOI] [PubMed] [Google Scholar]
- 36.Nygaard R., Frimurer T.M., Holst B., Rosenkilde M.M., Schwartz T.W. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol. Sci. 2009;30:249–259. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Visiers I., Ballesteros J.A., Weinstein H. Three-dimensional representations of G protein-coupled receptor structures and mechanisms. Methods Enzymol. 2002;343:329–371. doi: 10.1016/s0076-6879(02)43145-x. [DOI] [PubMed] [Google Scholar]
- 38.Shi L., Liapakis G., Xu R., Guarnieri F., Ballesteros J.A., Javitch J.A. Beta2 adrenergic receptor activation. Modulation of the proline kink in transmembrane 6 by a rotamer toggle switch. J. Biol. Chem. 2002;277:40989–40996. doi: 10.1074/jbc.M206801200. [DOI] [PubMed] [Google Scholar]
- 39.Ahuja S., Crocker E., Eilers M., Hornak V., Hirshfeld A., Ziliox M., Syrett N., Reeves P.J., Khorana H.G., Sheves M., Smith S.O. Location of the retinal chromophore in the activated state of rhodopsin*. J. Biol. Chem. 2009;284:10190–10201. doi: 10.1074/jbc.M805725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahalingam M., Martinez-Mayorga K., Brown M.F., Vogel R. Two protonation switches control rhodopsin activation in membranes. Proc. Natl. Acad. Sci. USA. 2008;105:17795–17800. doi: 10.1073/pnas.0804541105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ballesteros J.A., Jensen A.D., Liapakis G., Rasmussen S.G., Shi L., Gether U., Javitch J.A. Activation of the beta 2-adrenergic receptor involves disruption of an ionic lock between the cytoplasmic ends of transmembrane segments 3 and 6. J. Biol. Chem. 2001;276:29171–29177. doi: 10.1074/jbc.M103747200. [DOI] [PubMed] [Google Scholar]
- 42.Rana S., Baranski T.J. Third extracellular loop (EC3)-N terminus interaction is important for seven-transmembrane domain receptor function: implications for an activation microswitch region. J. Biol. Chem. 2010;285:31472–31483. doi: 10.1074/jbc.M110.129213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sum C.S., Tikhonova I.G., Costanzi S., Gershengorn M.C. Two arginine-glutamate ionic locks near the extracellular surface of FFAR1 gate receptor activation. J. Biol. Chem. 2009;284:3529–3536. doi: 10.1074/jbc.M806987200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greives N., Zhou H.X. Both protein dynamics and ligand concentration can shift the binding mechanism between conformational selection and induced fit. Proc. Natl. Acad. Sci. USA. 2014;111:10197–10202. doi: 10.1073/pnas.1407545111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagemann I.S., Miller D.L., Klco J.M., Nikiforovich G.V., Baranski T.J. Structure of the complement factor 5a receptor–ligand complex studied by disulfide trapping and molecular modeling. J. Biol. Chem. 2008;283:7763–7775. doi: 10.1074/jbc.M709467200. [DOI] [PubMed] [Google Scholar]
- 46.Klco J.M., Nikiforovich G.V., Baranski T.J. Genetic analysis of the first and third extracellular loops of the C5a receptor reveals an essential WXFG motif in the first loop. J. Biol. Chem. 2006;281:12010–12019. doi: 10.1074/jbc.M600548200. [DOI] [PubMed] [Google Scholar]
- 47.Kolakowski L.F., Jr., Lu B., Gerard C., Gerard N.P. Probing the message:address sites for chemoattractant binding to the C5a receptor. Mutagenesis of hydrophilic and proline residues within the transmembrane segments. J. Biol. Chem. 1995;270:18077–18082. doi: 10.1074/jbc.270.30.18077. [DOI] [PubMed] [Google Scholar]
- 48.Okinaga S., Slattery D., Humbles A., Zsengeller Z., Morteau O., Kinrade M.B., Brodbeck R.M., Krause J.E., Choe H.R., Gerard N.P., Gerard C. C5L2, a nonsignaling C5A binding protein. Biochemistry. 2003;42:9406–9415. doi: 10.1021/bi034489v. [DOI] [PubMed] [Google Scholar]
- 49.Marder S.R., Chenoweth D.E., Goldstein I.M., Perez H.D. Chemotactic responses of human peripheral blood monocytes to the complement-derived peptides C5a and C5a des Arg. J. Immunol. 1985;134:3325–3331. [PubMed] [Google Scholar]
- 50.Gerard C., Chenoweth D.E., Hugli T.E. Response of human neutrophils to C5a: a role for the oligosaccharide moiety of human C5ades Arg-74 but not of C5a in biologic activity. J. Immunol. 1981;127:1978–1982. [PubMed] [Google Scholar]
- 51.Koradi R., Billeter M., Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 51-55. 1996;1996;14:29–32. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 52.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 53.Hubbard S.J., Thornton J.M. Department of Biochemistry and Molecular Biology, University College London; 1993. NACCESS, Computer Program. [Google Scholar]
- 54.Marshall M.S., Steele R.P., Thanthiriwatte K.S., Sherrill C.D. Potential energy curves for cation−π interactions: off-axis configurations are also attractive. J. Phys. Chem. A. 2009;113:13628–13632. doi: 10.1021/jp906086x. [DOI] [PubMed] [Google Scholar]
- 55.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 56.Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 57.Guntert P., Mumenthaler C., Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 58.Treptow W., Klein M.L. The membrane-bound state of K2P potassium channels. J. Am. Chem. Soc. 2010;132:8145–8151. doi: 10.1021/ja102191s. [DOI] [PubMed] [Google Scholar]
- 59.Wang T., Duan Y. Chromophore channeling in the G-protein coupled receptor rhodopsin. J. Am. Chem. Soc. 2007;129:6970–6971. doi: 10.1021/ja0691977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kandt C., Ash W.L., Tieleman D.P. Setting up and running molecular dynamics simulations of membrane proteins. Methods. 2007;41:475–488. doi: 10.1016/j.ymeth.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 61.Kucerka N., Tristram-Nagle S., Nagle J.F. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J. Membr. Biol. 2005;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 62.Tieleman D.P., Forrest L.R., Sansom M.S., Berendsen H.J. Lipid properties and the orientation of aromatic residues in OmpF, influenza M2, and alamethicin systems: molecular dynamics simulations. Biochemistry. 1998;37:17554–17561. doi: 10.1021/bi981802y. [DOI] [PubMed] [Google Scholar]
- 63.Leekumjorn S., Sum A.K. Molecular characterization of gel and liquid-crystalline structures of fully hydrated POPC and POPE bilayers. J. Phys. Chem. B. 2007;111:6026–6033. doi: 10.1021/jp0686339. [DOI] [PubMed] [Google Scholar]
- 64.Daura X., Gademann K., Jaun B., Seebach D., van Gunsteren W.F., Mark A.E. Peptide folding: when simulation meets experiment. Angew. Chem. Int. Ed. 1999;38:236–240. [Google Scholar]
- 65.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ippel J.H., de Haas C.J., Bunschoten A., van Strijp J.A., Kruijtzer J.A., Liskamp R.M., Kemmink J. Structure of the tyrosine-sulfated C5a receptor N terminus in complex with chemotaxis inhibitory protein of Staphylococcus aureus. J. Biol. Chem. 2009;284:12363–12372. doi: 10.1074/jbc.M808179200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thukral L., Smith J.C., Daidone I. Common folding mechanism of a beta-hairpin peptide via non-native turn formation revealed by unbiased molecular dynamics simulations. J. Am. Chem. Soc. 2009;131:18147–18152. doi: 10.1021/ja9064365. [DOI] [PubMed] [Google Scholar]
- 68.Wu X., Wang S., Brooks B.R. Direct observation of the folding and unfolding of a beta-hairpin in explicit water through computer simulation. J. Am. Chem. Soc. 2002;124:5282–5283. doi: 10.1021/ja0257321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou R., Berne B.J., Germain R. The free energy landscape for beta hairpin folding in explicit water. Proc. Natl. Acad. Sci. USA. 2001;98:14931–14936. doi: 10.1073/pnas.201543998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pande V.S., Rokhsar D.S. Molecular dynamics simulations of unfolding and refolding of a beta-hairpin fragment of protein G. Proc. Natl. Acad. Sci. USA. 1999;96:9062–9067. doi: 10.1073/pnas.96.16.9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferrara P., Caflisch A. Folding simulations of a three-stranded antiparallel beta-sheet peptide. Proc. Natl. Acad. Sci. USA. 2000;97:10780–10785. doi: 10.1073/pnas.190324897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Katritch V., Cherezov V., Stevens R.C. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol. Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.March D.R., Proctor L.M., Stoermer M.J., Sbaglia R., Abbenante G., Reid R.C., Woodruff T.M., Wadi K., Paczkowski N., Tyndall J.D., Taylor S.M., Fairlie D.P. Potent cyclic antagonists of the complement C5a receptor on human polymorphonuclear leukocytes. Relationships between structures and activity. Mol. Pharmacol. 2004;65:868–879. doi: 10.1124/mol.65.4.868. [DOI] [PubMed] [Google Scholar]
- 74.Ulrich J.T., Cieplak W., Paczkowski N.J., Taylor S.M., Sanderson S.D. Induction of an antigen-specific CTL response by a conformationally biased agonist of human C5a anaphylatoxin as a molecular adjuvant. J. Immunol. 2000;164:5492–5498. doi: 10.4049/jimmunol.164.10.5492. [DOI] [PubMed] [Google Scholar]
- 75.Finch A.M., Vogen S.M., Sherman S.A., Kirnarsky L., Taylor S.M., Sanderson S.D. Biologically active conformer of the effector region of human C5a and modulatory effects of N-terminal receptor binding determinants on activity. J. Med. Chem. 1997;40:877–884. doi: 10.1021/jm960727r. [DOI] [PubMed] [Google Scholar]
- 76.Short A.J., Paczkowski N.J., Vogen S.M., Sanderson S.D., Taylor S.M. Response-selective C5a agonists: differential effects on neutropenia and hypotension in the rat. Br. J. Pharmacol. 1999;128:511–514. doi: 10.1038/sj.bjp.0702847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vogen S.M., Paczkowski N.J., Kirnarsky L., Short A., Whitmore J.B., Sherman S.A., Taylor S.M., Sanderson S.D. Differential activities of decapeptide agonists of human C5a: the conformational effects of backbone N-methylation. Int. Immunopharmacol. 2001;1:2151–2162. doi: 10.1016/s1567-5769(01)00141-2. [DOI] [PubMed] [Google Scholar]
- 78.Vogen S.M., Finch A.M., Wadi S.K., Thatcher J., Monk P.N., Taylor S.M., Sanderson S.D. The influence of Lys68 in decepeptide agonists of C5a on C5a receptor binding, activation and selectivity. J. Pept. Res. 1999;53:8–17. doi: 10.1111/j.1399-3011.1999.tb01612.x. [DOI] [PubMed] [Google Scholar]
- 79.Ma J.C., Dougherty D.A. The Cation−π Interaction. Chem. Rev. 1997;97:1303–1324. doi: 10.1021/cr9603744. [DOI] [PubMed] [Google Scholar]
- 80.Mery L., Boulay F. The NH2-terminal region of C5aR but not that of FPR is critical for both protein transport and ligand binding. J. Biol. Chem. 1994;269:3457–3463. [PubMed] [Google Scholar]
- 81.Smith N.J., Milligan G. Allostery at G protein-coupled receptor homo- and heteromers: uncharted pharmacological landscapes. Pharmacol. Rev. 2010;62:701–725. doi: 10.1124/pr.110.002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baranski T.J., Herzmark P., Lichtarge O., Gerber B.O., Trueheart J., Meng E.C., Iiri T., Sheikh S.P., Bourne H.R. C5a receptor activation. Genetic identification of critical residues in four transmembrane helices. J. Biol. Chem. 1999;274:15757–15765. doi: 10.1074/jbc.274.22.15757. [DOI] [PubMed] [Google Scholar]
- 83.Kobilka B.K., Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol. Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 84.Hofmann K.P., Scheerer P., Hildebrand P.W., Choe H.W., Park J.H., Heck M., Ernst O.P. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem. Sci. 2009;34:540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 85.Liu J.J., Horst R., Katritch V., Stevens R.C., Wuthrich K. Biased signaling pathways in beta2-adrenergic receptor characterized by 19F-NMR. Science. 2012;335:1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Transparency document