Abstract
Hepatic steatosis is a common histological finding in obese patients. Even mild steatosis is associated with delayed hepatic regeneration and poor outcomes following liver resection or transplantation. We sought to identify and target molecular pathways that mediate this dysfunction. Lean mice and mice made obese through feeding of a high-fat, hypercaloric diet underwent 70 or 80% hepatectomy. After 70% resection, obese mice demonstrated 100% survival but experienced increased liver injury, reduced energy stores, reduced mitoses, increased necroapoptosis, and delayed recovery of liver mass. Increasing liver resection to 80% was associated with mortality of 40% in lean and 80% in obese mice (P < 0.05). Gene expression profiling showed decreased epidermal growth factor receptor (EGFR) in fatty liver. Meta-analysis of expression studies in mice, rats, and patients also demonstrated reduction of EGFR in fatty liver. In mice, both EGFR and phosphorylated EGFR decreased with increasing percent body fat. Hydrodynamic transfection of EGFR plasmids in mice corrected fatty liver regeneration, reducing liver injury, increasing proliferation, and improving survival after 80% resection. Loss of EGFR expression is rate limiting for liver regeneration in obesity. Therapies directed at increasing EGFR in steatosis might promote liver regeneration and survival following hepatic resection or transplantation.
Keywords: epidermal growth factor receptor, proliferation, hepatectomy, gene transfer, steatosis, microarray
steatosis is the most common abnormal liver histological finding among U.S. patients and currently affects one-third of American adults (26). All forms of fatty liver, including the most mild, increase rates of liver failure and death following liver resection (2, 12, 13). This limits the use of surgery for the treatment of many patients with liver tumors and renders many livers unsuitable for cadaveric or living-donor transplantation (20, 25, 30). Thus understanding the abnormal regenerative capacity of fatty liver and the propensity for hepatocellular necroapoptosis remains a critically important problem in need of novel therapies for patients with liver disease.
Using primarily the ob/ob genetic model of obesity, studies by Diehl and colleagues have demonstrated that obese mice with fatty liver following resection, ischemia reperfusion injury, and other models of injury undergo delayed hepatocyte proliferation and increased hepatocyte necroapoptosis (5, 32). To date, the molecular mechanisms responsible for these defects remain poorly understood.
We hypothesized that the delayed regenerative response and increased apoptosis observed might be due to abnormal mitogenic pathway(s) in fatty liver. We used a discovery approach in models of diet-induced obesity and liver regeneration. Our studies herein strongly implicate epidermal growth factor (EGF) signaling as an essential abnormality of fatty liver regeneration.
METHODS
Reagents, kits, and antibodies.
Reagents were purchased from Sigma (St. Louis, MO), including ATP, aspartate aminotransferase (AST), alanine aminotransferase (ALT), and bilirubin assay kits, unless otherwise specified. Other reagents were SuperSignal West Pico or Femto chemiluminescent substrate and Coomassie protein assay (Pierce, Chicago, IL); protease inhibitor cocktail and polyvinylidene difluoride membrane (Roche Diagnostics, Basel, Switzerland); antibodies to EGFR, GAPDH, Akt, phospho-Akt, STAT3, phospho-STAT3, p44/42 MAPK (ERK1/2), phospho-p44/42 MAPK (ERK1/2), peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α), and cyclin D1 (Cell Signaling Technology, Danvers, MA); apoptosis-inducing factor (AIF; Santa Cruz Biotechnology, Santa Cruz, CA); PCNA, β-actin, and 5-bromo-2′-deoxyuridine (BrdU; Sigma); cytochrome c (BD Biosciences, San Diego, CA); ImmunoPure peroxidase-conjugated secondary antibodies (Pierce); and diseased human liver tissue array (LV1201; US Biomax, Rockville, MD). Plasmid pEGFR-W (for wild-type) was constructed by inserting the full-length human EGFR cDNA into the vector pLNCX1. Control plasmid was pLNCX1 empty vector.
Mouse experiments.
All mouse experiments were approved by the Thomas Jefferson University and Indiana University School of Medicine Institutional Animal Care and Use Committees and are in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, publication no. 86-23. Male C57BL/6J control and diet-induced obese (DIO) mice (Jackson Laboratory) were fed a 10% fat diet (control) and a 60% fat diet (DIO) beginning at 5 wk. Mice were ordered at 16–18 wk and were acclimated to our colony for 2 wk before use, with continued feeding of control or high-fat diet. For hydrodynamic transfection, plasmid DNA was injected into the tail vein of 18–20-wk-old mice in a volume of saline equivalent to 7% of the body mass of the mouse in <5 s (4.7 µg DNA/g body wt). Seventy percent hepatectomy or sham surgery was performed as previously described (9, 10). In 80% hepatectomy the usual middle and left lateral liver lobes were resected along with the right inferior (caudate) lobe. Labeling of hepatocytes with BrdU (40 mg/kg ip 2 h before euthanasia) was done as previously described (9, 10). For all analyses, mice were euthanized under general anesthesia at reported time points or when moribund. Body composition was determined by EchoMRI in conscious mice.
Microarray analysis and gene expression profiling.
Total RNA was extracted from flash-frozen liver using TRIzol as previously described (35, 36). Total RNA was quantified with a Nanodrop 8000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE), and its quality was assessed with a Bioanalyzer 2100 using the RNA 6000 Nano Kit (Agilent, Santa Clara, CA). Microarray hybridization, detection, and data normalization were performed by the Oncogenomics Core Facility at the University of Miami Sylvester Cancer Center. Biotinylated cRNA was prepared using the Illumina TotalPrep RNA Amplification Kit (Ambion, Austin, TX) according to the manufacturer’s instructions, starting with 400-ng total liver RNA. Successful cRNA generation was checked using the Bioanalyzer 2100. Samples were added to the BeadChip using randomized block design to reduce batch effects. Hybridization to the MouseWG-6 v2.0 Expression BeadChips (Illumina, San Diego, CA), washing, and scanning were performed according to the Illumina BeadStation 500 manual (revision C). The resulting raw microarray data were generated using Illumina BeadStudio. GeneSpring GX 7.3 was used for data normalization, statistical analysis (ANOVA and t-test), and hierarchical clustering. Only genes that were detected as present (Illumina detection call P < 0.01) in at least one group were included in the analysis. NextBio Professional and GeneGo Metacore were used for gene and pathway analysis. All microarray data are Minimum Information About a Microarray Experiment (MIAME) compliant and have been deposited in the Gene Expression Omnibus (GEO) database (National Center for Biotechnology Information) as record GSE33296.
Meta-analysis of gene expression studies.
NextBio Professional (NextBio) was used to query wild-type mouse or human studies focused on high-fat diet, fatty liver, or steatosis and examined EGFR expression within each GEO data set and individual bioset. Null results from individual comparisons are not informative in the absence of any data that EGFR was detected; thus only studies with at least one reported result are presented. Studies found include GSE6089 (Expression patterns in liver after resveratrol treatment of mice on a high-calorie diet; 1), GSE11845 [Resveratrol diet aging (heart, liver, muscle and fat tissue); 17], GSE10493 (Novartis 12 strain diet sex survey; 4), GSE53131 (Expression data from livers of C57BL/6J mice fed a high-fat diet and coffee; 27), GSE51885 (Liver mRNA microarray study for mice treated with various diets; 21), GSE57425 (Gene expression data of livers from C57BL/6 fed a normal diet or high-fat-diet; 14), GSE45327 (High-fat diet induced changes to mouse liver mRNA; 6, 28), GSE39778 (Role of SIRT1 in diet-induced metabolic diseases; 19), GSE19102 (SRT1720 extends healthspan and lifespan in diet-induced obese mice; 15), GSE24031 (Adipose tissue dysfunction signals progression of hepatic steatosis towards nonalcoholic steatohepatitis in C57BL/6 mice; 7), and E-CBIL-24 (Transcription profiling of liver from B6 and 129 mice fed on low and high fat diets; 3). For human studies, GEO data sets are GSE33814 (Gene expression profiling unravels cancer-related hepatic molecular signatures in steatohepatitis but not in steatosis; 24) and GSE24807 (Increased hemoglobin expression in NASH livers; 34).
Western-blotting analysis.
Liver samples were homogenized in ice-cold modified radioimmunoprecipitation assay buffer (50 mmol/l Tris-HCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mmol/l NaCl, 1 mmol/l EDTA, 1 mmol/l phenylmethylsulfonyl fluoride, 1 mmol/l sodium orthovanadate, 1 mmol/l NaF, and protease inhibitor cocktail, pH 7.4). Fractionation of cytosol was done as previously described (9). Equivalent amounts of homogenate (50 µg per well), determined by Coomassie assay, were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose or polyvinylidene difluoride membranes, and detected by SuperSignal West Femto or Pico chemiluminescence. Densitometry and quantitation of protein expression were performed using ImageJ software (National Institutes of Health, Bethesda, MD).
Histologic examination and immunohistochemistry.
Liver tissues were isolated at designated time points and were placed in 10% neutral-buffered formalin, processed, sectioned, and stained with hematoxylin and eosin or periodic acid-Schiff (PAS). EGFR and BrdU immunohistochemical stainings were performed as previously described (9–11).
Statistical analyses.
Data analysis was done using GraphPad Prism version 5.0. All parametric data are presented as means ± SD. Data from two-group comparisons were analyzed for significance using an unpaired two-tailed t-test. Data from multiple-group comparisons were analyzed by one-way analysis of variance (ANOVA) with Fisher’s least significant difference (LSD) test. The log-rank test was applied to compare survival curves. Differences were considered statistically significant if P < 0.05.
RESULTS
DIO mice demonstrated increased injury after 70% hepatectomy.
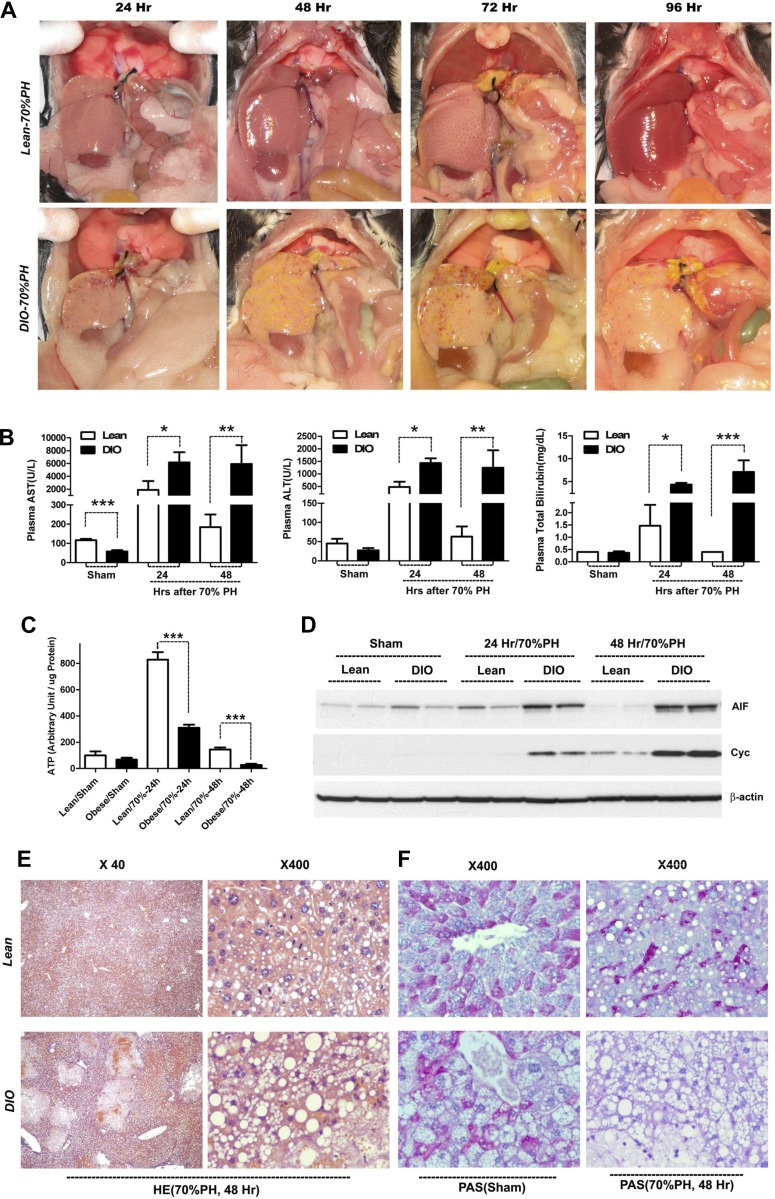
Eight-week-old C57BL/6J mice fed regular (control) or high-fat (DIO) diets were subjected to 70% partial hepatectomy (PH). Interval timed necropsies were performed in both groups. Whereas the regenerating liver appeared grossly normal in color and regained mass over time in the lean group, DIO mice had grossly abnormal livers at all time points examined after hepatectomy, characterized by smaller size, a pale yellow appearance, presence of multiple petechiae, and areas suggestive of hepatic parenchymal damage (Fig. 1A). Bilirubin staining was evident in multiple tissues of DIO mice following hepatectomy, indicating liver dysfunction. Indeed, plasma concentrations of AST and ALT were markedly increased in DIO mice at 24 and 48 h after hepatectomy (Fig. 1B). In addition, DIO mice demonstrated increased perioperative bilirubin serum concentrations consistent with impaired hepatic secretory function (Fig. 1B). Measurements of liver tissue ATP levels demonstrated energy depletion in DIO mice vs. lean mice at both 24 and 48 h following hepatectomy (Fig. 1C). Histologic sections of these animals demonstrated severe vacuolar changes and large areas of hepatocellular necrosis at 24–48 h after 70% PH in DIO mice predominantly in a zone I, periportal pattern (Fig. 1E) that may have arisen after biliary ductule injury. PAS staining of livers at 48 h showed a loss of pink staining consistent with near-complete glycogen depletion (Fig. 1F). Western blotting of cytoplasmic fractions of liver lysates for the normally mitochondrial resident proteins apoptosis-inducing factor (AIF) and cytochrome c (Cyc) also showed increased levels of these proteins in the cytoplasm, suggesting that DIO mice suffered mitochondrial disruption after hepatectomy (Fig. 1D). Taken together, these data demonstrate increased hepatocellular injury, evidence for mitochondrial damage, decreased cellular energy, and impaired liver function in DIO mice following 70% hepatectomy.
Fig. 1.
Increased hepatic injury following partial hepatectomy (PH) in diet-induced obese (DIO) mice. A: gross evidence of liver injury after 70% hepatectomy. Timed necropsy at successive hours following 70% hepatectomy demonstrated intrahepatic hemorrhage and regions of bilious discoloration in DIO mice (bottom), in contrast to remnant hypertrophy and restoration of grossly normal liver appearance in lean mice (top). B: serum markers of liver injury after 70% hepatectomy. DIO mice had increased blood aspartate aminotransferase (AST), alanine aminotransferase (ALT), and bilirubin after 70% hepatectomy, indicating greater injury and reduced function relative to similarly treated lean mice (n = 3–8 per group, 1-way ANOVA and Fisher’s LSD). C: decreased hepatic ATP was measured following hepatectomy in DIO vs. lean mice (n = 4 per group, 1-way ANOVA and Fisher’s LSD). D: Western-blotting analysis of liver cytosolic fractions shows that DIO livers had increased cytosolic apoptosis-inducing factor (AIF) and cytochrome c (Cyc) after hepatectomy, consistent with increased mitochondrial injury vs. lean mice. E: histological changes [hematoxylin and eosin (HE) staining] demonstrate increased areas of necrosis with hemorrhage and more severe vacuolar fat accumulation in livers of DIO mice 48 h after hepatectomy. This injury pattern predominantly appears to involve zone 1 of the liver (periportal). F: periodic acid-Schiff (PAS) staining of DIO and control livers demonstrates a relative loss of PAS-positive staining in DIO livers, consistent with glycogen depletion. Bars are ±SD. *P < 0.05, **P < 0.01, ***P < 0.001.
DIO mice demonstrated delayed liver regeneration after 70% hepatectomy and increased mortality following 80% hepatectomy.
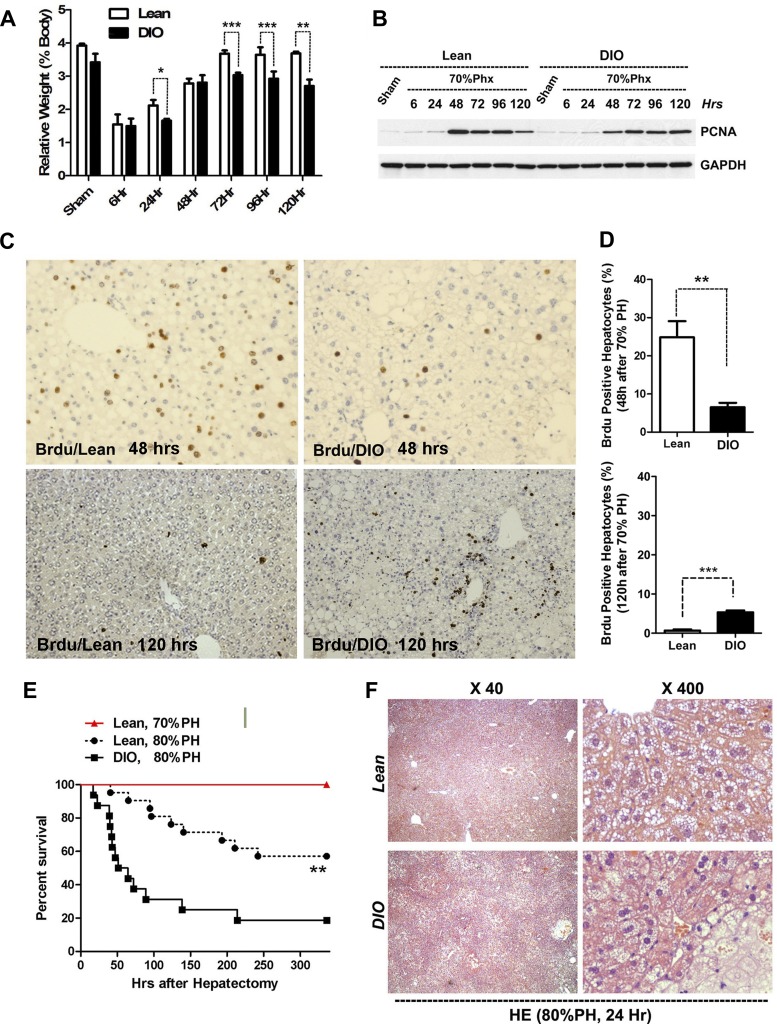
To determine whether there was an associated delay in reestablishing liver mass in DIO mice, we examined hepatocyte proliferation of lean and DIO mice following 70% hepatectomy. Return of liver mass in DIO animals lagged normal mice following 70% PH (Fig. 2A); regeneration, although somewhat slower, was nonetheless observed. A significant delay of 10–20% mass recovery was observed at 96–120 h in DIO mice. These differences, although significant, appear modest (Fig. 2A). By Western blot, PCNA expression peaked at 48 h in lean mice. In contrast, decreased PCNA expression was noted in DIO mice at 48 h vs. both lean controls at 48 h or at later time points obtained in DIO mice. Persistence of PCNA elevation was noted in DIO livers at 120 h, suggestive of a delayed but ongoing regenerative response that correlated with the delay in mass recovery (Fig. 2B). BrdU labeling of livers at the approximate peak of DNA synthesis in lean mice (48 h) demonstrated markedly reduced labeling fractions of hepatocytes in DIO mice (Fig. 2, C and D). BrdU staining of sections taken at 120 h after 70% hepatectomy, however, demonstrated increased hepatocyte-labeling rates in DIO mice. DIO mice demonstrated significantly increased BrdU incorporation relative to lean mice at 120 h as well, confirming that liver regeneration, although delayed, was occurring. Taken together, these data demonstrate that DIO mice have a modestly delayed and impaired hepatocyte proliferation and recovery of mass following 70% hepatectomy. This delay following 70% hepatectomy was not associated with increased mortality in either lean or DIO mice. We next examined whether the delayed regeneration observed in DIO mice following 70% hepatectomy could cause increased mortality in DIO mice following more extensive hepatectomy. Extent of hepatic resection is closely correlated with rate of liver failure and mortality; lean and DIO mice were next subjected to 80% hepatectomy. In 80% hepatectomy the usual middle and left lateral liver lobes are resected along with the right inferior (caudate) lobe. This resection leaves the animal with right superior and mammillary lobes and requires a considerably greater hepatic regenerative response. Of note, the 80% resection is not almost uniformly lethal as is 87–95% hepatectomy (9). Here 80% hepatectomy resulted in a 14-day survival rate of ~60% in lean mice (Fig. 2E). DIO mice subjected to 80% hepatectomy, however, showed only a 20% survival rate at 14 days (Fig. 2E; P < 0.05, n > 14 per group). Histological sections showed far more necrotic loci in DIO livers following 80% hepatectomy (Fig. 2F).
Fig. 2.
Delayed hepatocyte proliferation after 70% hepatectomy and increased mortality after 80% hepatectomy in DIO mice. A: recovery of liver mass (expressed as liver mass/total body mass) was delayed in DIO mice after 70% hepatectomy (n = 4 per group, 1-way ANOVA and Fisher’s LSD). B–D: evidence for delayed proliferation in DIO mice. B: Western-blotting analysis shows reduced hepatic PCNA expression in DIO after 70% hepatectomy (Phx). Peak DNA synthesis is ~48 h in lean mice following 70% hepatectomy; marked decreased PCNA expression is observed at 48 h in DIO mice. C and D: reduced 48-h hepatic cell proliferation in DIO mice measured by BrdU incorporation, immunostaining, and quantification (n = 4 per group, unpaired 2-sided t-test) demonstrates reduced BrdU incorporation at 48 h. Conversely, increased BrdU incorporation is observed at 120 h in DIO mice suggesting delayed but persistent hepatocyte proliferation. E: DIO and lean mice demonstrate equivalent, 100% survival, following 70% partial hepatectomy. In contrast, DIO mice show increased mortality after 80% hepatectomy relative to lean controls (n = 12 lean 70% mice and n = 21 lean or DIO mice in the 80% group, log-rank test). F: hematoxylin and eosin staining demonstrates increased regions of hepatocyte necrosis in DIO mice following 80% hepatectomy. Bars are ±SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Expression profiling revealed clustering of gene expression by metabolic status and loss of EGFR expression.
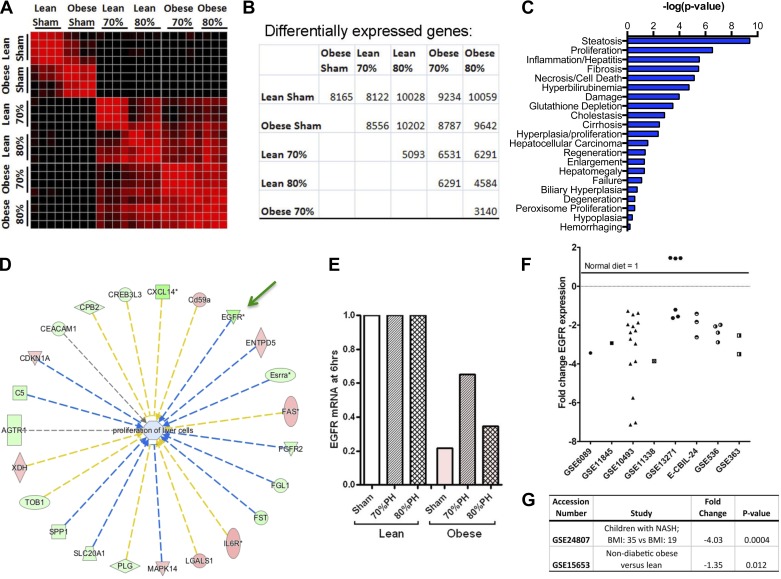
We posited that the increased injury and delayed liver regeneration observed in DIO mice following hepatectomy might be due to failure of a necessary mitogenic signal that is otherwise activated in nonobese, control mice. Certainly, it is well established that the addition of hepatic mitogens is associated with accelerated entry into the cell cycle and resistance to necroapoptotic signals (8, 9). To uncover these molecular defects, we performed expression profiling of control and regenerating livers. Diet-induced obese and lean C57BL/6J mice were subjected to 70%, 80%, or sham hepatectomy and euthanized 6 h later. Six hours was chosen to identify the most proximal transcriptional events in initiating liver regeneration. Individual samples clustered well within groups. Livers from sham-treated lean and obese mice were separable from each other and from all resected samples. Within the resected groups, samples clustered by nutritional/obesity status, more than degree of hepatectomy. Heat map analysis showing relatedness of gene expression signatures between individual samples demonstrated clustering by obesity status within the sham and hepatectomy group and numbers of differentially regulated genes (Fig. 3, A and B). Ingenuity pathway analysis (IPA) of genes differentially expressed after hepatectomy between obese and lean mice demonstrated that proliferation was the second most significant gene category of expression (Fig. 3C).
Fig. 3.
Discovery of genes differentially regulated in obesity and resection. A: heat map showing relatedness of gene expression signatures between individual samples shows clustering by obesity status within the sham and hepatectomy group. Note that obese samples cluster with each other more than with the same extent hepatectomy in lean mice. Black, 0% correlation; red, 99% correlation. B: table showing numbers of differentially expressed probe sets between the indicated groups. Within these comparisons, EGFR was the sixth-ranked gene across obese vs. matched lean comparisons. C: ingenuity pathway analysis (IPA) of genes differentially expressed after hepatectomy between obese and lean mice. D: hub-and-spoke rendering of upstream effectors of the proliferation of liver cell IPA pathways in C. Genes increased are shown in red, and those decreased are shown in green. Those predicted to enhance the pathway have spokes shown in yellow, and those predicted to inhibit the pathway have spokes in blue. Thus the reduction of EGFR is predicted to inhibit proliferation. E: microarray data showed that EGFR mRNA is reduced in obese mice at baseline and after 70 and 80% hepatectomy (PH). P << 0.001 for all obese vs. lean comparisons and for obese 70% vs. obese sham and obese 80%. Meta-analysis of independent studies of mice of different sex, age, institution, diet supplier, duration of diet, and microarray platform deposited in the Gene Expression Omnibus (GEO) microarray data showed that EGFR mRNA is reduced in obese mice at baseline and after 70 and 80% hepatectomy (PH). F: meta-analysis of independent studies of mice of different sex, age, institution, diet supplier, duration of diet, and microarray platform deposited in GEO demonstrated that EGFR expression was reproducibly reduced in liver of mice with high-fat diet treatment vs. normal diet in many experimental comparisons. G: analysis of the two human steatosis studies in the GEO database demonstrated that EGFR expression was reduced 1.35-fold in livers from obese subjects without diabetes (n = 4) relative to lean subjects (n = 5, P = 0.013; 18) and reduced 4.03-fold in obese adolescents with nonalcoholic steatohepatitis (n = 7) vs. those without (n = 4, P = 0.004).
We hypothesized that differentially expressed genes in obese vs. lean groups, across the stresses of sham, 70%, and 80% hepatectomy, would be important. Using NextBio for analysis of those 3 groups, the sixth gene of 4,869 significantly changed genes in a rank-ordered list by fold change and P value was EGFR. Microarray data showed that EGFR mRNA is reduced in obese mice at baseline and after 70 and 80% PH (Fig. 3E). Within this list, we did not observe dysregulation of other essential mitogenic and antiapoptotic ligands and their receptors, particularly hepatocyte growth factor (HGF), insulin, insulin-like growth factor (IGF), or IL-6. (A more detailed analysis of these data detracts from this study and will thus be published elsewhere.) A hub-and-spoke rendering of upstream effectors of the proliferation of liver cell IPA pathways was also undertaken that illustrates the likely role of EGFR loss in the abnormal regeneration of fatty liver (Fig. 3D). Genes increased are shown in red, and those decreased are shown in green. Those predicted to enhance the pathway are shown in yellow, to inhibit the pathway in blue. Thus the reduction of EGFR is predicted to inhibit proliferation.
This observation of reduced EGFR in fatty liver is further supported by a large number of independent observations. Meta-analysis of independent studies of mice of different sex, age, institution, diet supplier, duration of diet, and microarray platform deposited in GEO demonstrated that EGFR expression was reproducibly reduced in liver of mice on high-fat diets vs. normal diet in many experimental comparisons (Fig. 3F) whereas none of the other top five genes identified from our experiment were similarly changed (data not shown). Moreover, analysis of the two human steatosis studies in the GEO database demonstrated that EGFR expression was reduced 1.35-fold in livers from obese subjects without diabetes (n = 4) relative to lean subjects (n = 5, P = 0.013; 18) and reduced 4.03-fold in obese adolescents with nonalcoholic steatohepatitis (n = 7) vs. those without (n = 4, P = 0.004; Fig. 3G).
Gene expression data regarding EGFR levels were supported by protein data. Western-blotting analysis demonstrated that EGFR protein levels were reduced in DIO mice (Fig. 4A). Furthermore, linear regression analysis of Western-blotting results vs. body composition showed a significant relationship between reduced EGFR and pY1068-EGFR and increased percent body fat in DIO, but not in lean mice (Fig. 4B). Less EGFR staining was also observed in sections of human fatty liver by immunostaining (Fig. 4C). These results indicate that low EGFR is apparently a common feature of fatty liver, both in mice and in human patients.
Fig. 4.
Reduced EGFR expression with increasing obesity in mice. A: Western blotting shows reduced EGFR and pY1068-EGFR in normal, freely fed lean (10% fat diet) and diet-induced obese (DIO) mice (60% fat diet). Each lane represents an individual mouse. Representative result of duplicate experiments. B: linear regression analysis of EGFR or pY1068-EGFR levels, normalized to GAPDH, vs. percent body fat determined by EchoMRI in the conscious mouse. No significant (N.S.) relationship was seen for lean mice or for DIO mice with EGFR/pEGFR vs. total body weight or epididymal fat pad mass. C: EGFR immunohistochemical staining of human fatty liver, normal human liver, and hepatocellular carcinoma tissue sections shows lowest staining in fatty liver. Note lack of discernible localization of signal to the cell membrane in fatty liver.
Replenishment of EGFR improved liver regeneration in DIO mice.
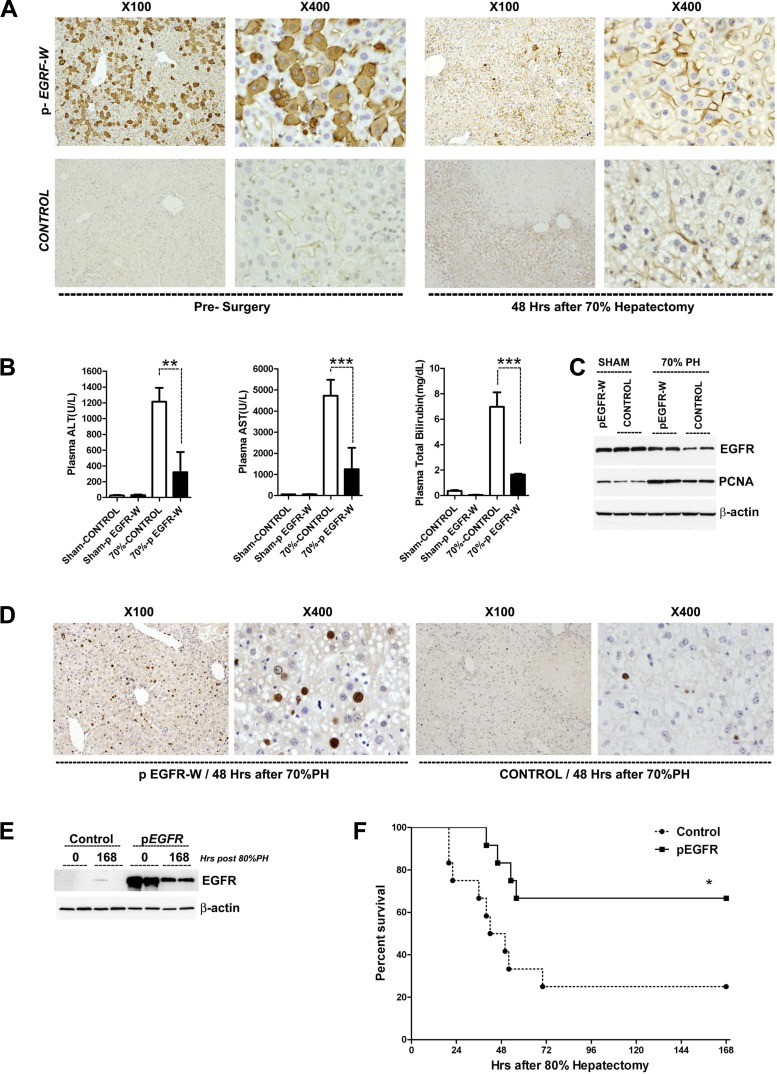
Given that low EGFR was linked to defective regeneration, we sought to restore normal regeneration via EGFR gene transfer. Plasmids for expression of full-length EGFR (pEGFR-W) or empty vector/no exogenous protein (control) were delivered to the liver of DIO mice via hydrodynamic transfection. Transfection rates of ~30–50% were observed by staining for the carboxyl portion of the EGFR after pEGFR-W transfection (Fig. 5A, left) or by direct fluorescence imaging after similar transfection using a vector expressing green fluorescent protein (data not shown). Twenty-four hours after transfection, mice were subjected to 70 or 80% hepatectomy (time = 0 h), when EGFR levels were highly elevated in pEGFR-transfected mice vs. controls (Fig. 5E). EGFR protein elevation persisted but was localized to cell membranes 48 h after hepatectomy (Fig. 5A, right). EGFR transgenesis was protective in DIO liver regeneration. Markedly reduced levels of serum ALT, AST, and bilirubin were observed after 70% hepatectomy in DIO mice transfected with the full-length EGFR vs. control plasmid (Fig. 5B). Increased PCNA expression and increased BrdU incorporation at 48 h after hepatectomy in DIO mice demonstrated that exogenous EGFR replenishment repaired the defect regenerative capacity of steatotic hepatocytes (Fig. 5, C and D). Next we assayed after 80% hepatectomy in DIO mice transfected with either the control or EGFR plasmids. Mice transfected with pEGFR demonstrated a markedly improved survival vs. control-transfected mice, with survival rates similar to that observed in lean mice (Figs. 2E vs. 5F). On the basis of these results, we conclude that replenishment of EGFR corrected the abnormal liver regeneration observed in fatty liver. Furthermore, these data suggest that EGFR downregulation appears to play a critical role in the abnormal liver regeneration observed in DIO mice.
Fig. 5.
EGFR gene transfer reduces hepatic injury and accelerates hepatocyte proliferation in DIO mice following hepatectomy. A: EGFR immunostaining following hydrodynamic gene transfer of EGFR-expressing plasmids into DIO mice results in gene expression in ~30–50% of hepatocytes at 24 h after tail vein injection of plasmids (presurgery) and enhanced EGFR expression at 48 h after 70% hepatectomy (72 h after tail vein injection). Control samples are obtained from DIO mice that received a nonexpressing plasmid injection. B: serum markers of liver injury and dysfunction after hepatectomy are dramatically reduced by EGFR gene transfer in DIO mice (n = 4 mice per group). C: Western blotting shows that EGFR gene transfer increased PCNA expression with enhanced EGFR in DIO fatty livers 48 h after hepatectomy. D: BrdU immunostaining demonstrating EGFR gene transfer increased hepatocyte proliferation in DIO fatty livers at 48 h after 70% hepatectomy. E: Western blotting demonstrates that enhanced EGFR expression by hydrodynamic gene transfer persists 7 days after 80% hepatectomy in DIO mice. F: Kaplan-Meier survival plots after 80% hepatectomy for DIO mice injected 24 h before surgery via tail vein with EGFR-expressing plasmid (pEGFR) or control plasmid. Survival was significantly increased by EGFR gene therapy. *P < 0.05, **P < 0.01, ***P < 0.001.
DISCUSSION
Herein, we have identified EGFR downregulation as a common finding in steatosis and increased total body adiposity. This downregulation of EGFR expression is associated with the delayed hepatic regeneration seen in fatty liver. Increasing EGFR expression is also demonstrated to accelerate the delayed return of liver mass seen in liver resection and dramatically improve survival in the setting of obesity. Thus this study identifies EGFR loss as likely a mechanistically significant abnormality in the delayed regenerative response of fatty liver. It also identifies EGFR replenishment therapy as a potentially effective therapy to correct or improve the abnormal liver regeneration seen in obese patients undergoing liver injury or resection.
Patients who have fatty liver are well documented to have an abnormal hepatic regenerative response with a significantly increased risk of serious perioperative complications and death (2). Studies in the genetically obese ob/ob mouse have similarly identified increased hepatocyte cell death and delayed proliferation as essential elements of this abnormal process (32). Herein, we examined the liver regenerative response in a diet-induced model of obesity, similar to that associated with a high-fat Western diet. As has been noted in the ob/ob mouse, we observed delayed regeneration and increased hepatocellular injury in DIO mice vs. normal lean animals. Of note, the phenotype of abnormal liver regeneration following 70% hepatectomy is much milder with less dramatic regenerative abnormalities and less hepatocellular injury than that observed in ob/ob mice following hepatectomy. Previously, we have demonstrated that addition of the potent antiapoptotic factor and potential hepatic mitogen IL-6 prevented hepatocyte necroapoptosis and accelerated normal hepatic regeneration following partial hepatectomy (10, 37, 38). Administration of IL-6 to normal lean control mice facilitated recovery from an otherwise lethal hepatic resection (87% hepatectomy; 9). Thus we postulated that the increased necroapoptosis and delayed regeneration might be due to loss or dysregulation of an otherwise normal antiapoptotic and mitogenic signal such as that associated with IL-6 or other known hepatic mitogens (31).
To identify potential mitogenic pathways that might be disregulated in fatty liver, we undertook a discovery approach using gene chip arrays, which revealed defective EGFR expression in fatty liver. Strikingly, we did not observe dysregulation of other known essential mitogenic and antiapoptotic pathways such as that associated with HGF, insulin, IGF, or IL-6 receptor signaling. EGFR in lean animals has been shown to be not absolutely essential for liver regeneration, but loss of this pathway is associated with impaired regeneration and evidence for increased necroapoptosis (16, 22, 33). This observation is apparently robust and highly reproducible across experimental conditions and laboratories, on the basis of our further analysis of publicly available data from GEO. The finding of EGFR downregulation was also previously reported by Shirai et al. in genetically obese, leptin receptor-deficient db/db mice following 70% partial hepatectomy (23). Collin et al. have also identified EGFR downregulation in steatosis observed in ob/ob mice, in mice fed a choline-deficient diet, and in steatotic human liver. Our data confirm these findings but also suggest that EGFR expression closely correlates with total adiposity (6). Nonetheless, not all authors have identified decreased EGFR expression in fatty liver. Wang et al., examining gene arrays from human samples, have reported increased EGFR expression in the fatty liver samples they analyzed (29). Further studies will be needed to explain this apparently discordant finding.
After our discovery-based approach identified a decrease in EGFR expression in steatosis and obesity, we examined the potential benefits of replenishing the expression of EGFR protein. Using hydrodynamic transfection, we achieved a high rate of plasmid incorporation into hepatocytes and generated a considerable induction of EGFR protein in fatty livers that persisted for over a week following transfection. Administration of full-length EGFR receptor dramatically improved the regenerative response and significantly improved survival in DIO mice after 80% hepatectomy. Taken together, these data strongly support the hypothesis that dysregulation of the EGFR signaling pathway, a known mitogenic and antiapoptotic pathway, is a critical component of the abnormal liver regeneration seen in fatty liver. Nonetheless, these observations do not prove that EGFR downregulation mediates this process as replenishment of EGFR may have been supraphysiologic and treatment with other hepatic growth factors such as IL-6 will also improve the regenerative response (10, 37).
In summary, this report identifies EGFR downregulation as an apparent critical component of the dysfunctional regeneration seen in diet-induced fatty liver. Increasing EGFR expression in the setting of adiposity may have implications in the management of liver failure after resection, in transplantation requiring hepatic regeneration, and in other settings where the hepatic regenerative response is impaired.
GRANTS
This work was supported by the Lilly Endowment Incorporated Physician Scientist Initiative and National Institutes of Health Grants GM-6360301 (L. G. Koniaris), CA-122596 (T. A. Zimmers), and GM-092758 (T. A. Zimmers).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
T.A.Z., X.J., Y.J., and L.G.K. conceived and designed research; T.A.Z., X.J., Z.Z., and Y.J. performed experiments; T.A.Z., X.J., Y.J., and L.G.K. analyzed data; T.A.Z., X.J., Y.J., and L.G.K. interpreted results of experiments; T.A.Z., X.J., Z.Z., Y.J., and L.G.K. prepared figures; T.A.Z. and L.G.K. drafted manuscript; T.A.Z., X.J., Y.J., and L.G.K. edited and revised manuscript; T.A.Z., X.J., Z.Z., Y.J., and L.G.K. approved final version of manuscript.
REFERENCES
- 1.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444: 337–342, 2006. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behrns KE, Tsiotos GG, DeSouza NF, Krishna MK, Ludwig J, Nagorney DM. Hepatic steatosis as a potential risk factor for major hepatic resection. J Gastrointest Surg 2: 292–298, 1998. doi: 10.1016/S1091-255X(98)80025-5. [DOI] [PubMed] [Google Scholar]
- 3.Biddinger SB, Almind K, Miyazaki M, Kokkotou E, Ntambi JM, Kahn CR. Effects of diet and genetic background on sterol regulatory element-binding protein-1c, stearoyl-CoA desaturase 1, and the development of the metabolic syndrome. Diabetes 54: 1314–1323, 2005. doi: 10.2337/diabetes.54.5.1314. [DOI] [PubMed] [Google Scholar]
- 4.Burgess-Herbert SL, Cox A, Tsaih SW, Paigen B. Practical applications of the bioinformatics toolbox for narrowing quantitative trait loci. Genetics 180: 2227–2235, 2008. doi: 10.1534/genetics.108.090175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi SS, Diehl AM. Epithelial-to-mesenchymal transitions in the liver. Hepatology 50: 2007–2013, 2009. doi: 10.1002/hep.23196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collin de l’Hortet A, Zerrad-Saadi A, Prip-Buus C, Fauveau V, Helmy N, Ziol M, Vons C, Billot K, Baud V, Gilgenkrantz H, Guidotti JE. GH administration rescues fatty liver regeneration impairment by restoring GH/EGFR pathway deficiency. Endocrinology 155: 2545–2554, 2014. doi: 10.1210/en.2014-1010. [DOI] [PubMed] [Google Scholar]
- 7.Duval C, Thissen U, Keshtkar S, Accart B, Stienstra R, Boekschoten MV, Roskams T, Kersten S, Müller M. Adipose tissue dysfunction signals progression of hepatic steatosis towards nonalcoholic steatohepatitis in C57BL/6 mice. Diabetes 59: 3181–3191, 2010. doi: 10.2337/db10-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galun E, Zeira E, Pappo O, Peters M, Rose-John S. Liver regeneration induced by a designer human IL-6/sIL-6R fusion protein reverses severe hepatocellular injury. FASEB J 14: 1979–1987, 2000. doi: 10.1096/fj.99-0913com. [DOI] [PubMed] [Google Scholar]
- 9.Jin X, Zhang Z, Beer-Stolz D, Zimmers TA, Koniaris LG. Interleukin-6 inhibits oxidative injury and necrosis after extreme liver resection. Hepatology 46: 802–812, 2007. doi: 10.1002/hep.21728. [DOI] [PubMed] [Google Scholar]
- 10.Jin X, Zimmers TA, Perez EA, Pierce RH, Zhang Z, Koniaris LG. Paradoxical effects of short- and long-term interleukin-6 exposure on liver injury and repair. Hepatology 43: 474–484, 2006. doi: 10.1002/hep.21087. [DOI] [PubMed] [Google Scholar]
- 11.Jin X, Zimmers TA, Zhang Z, Pierce RH, Koniaris LG. Interleukin-6 is an important in vivo inhibitor of intestinal epithelial cell death in mice. Gut 59: 186–196, 2010. doi: 10.1136/gut.2008.151175. [DOI] [PubMed] [Google Scholar]
- 12.Koniaris LG, Levi DM, Pedroso FE, Franceschi D, Tzakis AG, Santamaria-Barria JA, Tang J, Anderson M, Misra S, Solomon NL, Jin X, DiPasco PJ, Byrne MM, Zimmers TA. Is surgical resection superior to transplantation in the treatment of hepatocellular carcinoma? Ann Surg 254: 527–537, 2011. doi: 10.1097/SLA.0b013e31822ca66f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koniaris LG, McKillop IH, Schwartz SI, Zimmers TA. Liver regeneration. J Am Coll Surg 197: 634–659, 2003. doi: 10.1016/S1072-7515(03)00374-0. [DOI] [PubMed] [Google Scholar]
- 14.Lu Y, Liu X, Jiao Y, Xiong X, Wang E, Wang X, Zhang Z, Zhang H, Pan L, Guan Y, Cai D, Ning G, Li X. Periostin promotes liver steatosis and hypertriglyceridemia through downregulation of PPARα. J Clin Invest 124: 3501–3513, 2014. doi: 10.1172/JCI74438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minor RK, Baur JA, Gomes AP, Ward TM, Csiszar A, Mercken EM, Abdelmohsen K, Shin YK, Canto C, Scheibye-Knudsen M, Krawczyk M, Irusta PM, Martín-Montalvo A, Hubbard BP, Zhang Y, Lehrmann E, White AA, Price NL, Swindell WR, Pearson KJ, Becker KG, Bohr VA, Gorospe M, Egan JM, Talan MI, Auwerx J, Westphal CH, Ellis JL, Ungvari Z, Vlasuk GP, Elliott PJ, Sinclair DA, de Cabo R. SRT1720 improves survival and healthspan of obese mice. Sci Rep 1: 70, 2011. doi: 10.1038/srep00070. [Corrigendum. Sci Rep 3(Jan.): 1131, 2013. doi:.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Natarajan A, Wagner B, Sibilia M. The EGF receptor is required for efficient liver regeneration. Proc Natl Acad Sci USA 104: 17081–17086, 2007. doi: 10.1073/pnas.0704126104. [Erratum. Proc Natl Acad Sci USA 104(49): 19656, 2007. doi:.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, Labinskyy N, Swindell WR, Kamara D, Minor RK, Perez E, Jamieson HA, Zhang Y, Dunn SR, Sharma K, Pleshko N, Woollett LA, Csiszar A, Ikeno Y, Le Couteur D, Elliott PJ, Becker KG, Navas P, Ingram DK, Wolf NS, Ungvari Z, Sinclair DA, de Cabo R. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab 8: 157–168, 2008. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pihlajamäki J, Boes T, Kim EY, Dearie F, Kim BW, Schroeder J, Mun E, Nasser I, Park PJ, Bianco AC, Goldfine AB, Patti ME. Thyroid hormone-related regulation of gene expression in human fatty liver. J Clin Endocrinol Metab 94: 3521–3529, 2009. doi: 10.1210/jc.2009-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purushotham A, Xu Q, Li X. Systemic SIRT1 insufficiency results in disruption of energy homeostasis and steroid hormone metabolism upon high-fat-diet feeding. FASEB J 26: 656–667, 2012. doi: 10.1096/fj.11-195172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redaelli CA, Wagner M, Krähenbühl L, Gloor B, Schilling MK, Dufour JF, Büchler MW. Liver surgery in the era of tissue-preserving resections: early and late outcome in patients with primary and secondary hepatic tumors. World J Surg 26: 1126–1132, 2002. doi: 10.1007/s00268-002-6321-9. [DOI] [PubMed] [Google Scholar]
- 21.Renaud HJ, Cui JY, Lu H, Klaassen CD. Effect of diet on expression of genes involved in lipid metabolism, oxidative stress, and inflammation in mouse liver-insights into mechanisms of hepatic steatosis. PLoS One 9: e88584, 2014. doi: 10.1371/journal.pone.0088584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seki E, Kondo Y, Iimuro Y, Naka T, Son G, Kishimoto T, Fujimoto J, Tsutsui H, Nakanishi K. Demonstration of cooperative contribution of MET- and EGFR-mediated STAT3 phosphorylation to liver regeneration by exogenous suppressor of cytokine signalings. J Hepatol 48: 237–245, 2008. doi: 10.1016/j.jhep.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 23.Shirai M, Yamauchi H, Nakayama H, Doi K, Uetsuka K. Expression of epidermal growth factor receptor protein in the liver of db/db mice after partial hepatectomy. Exp Toxicol Pathol 59: 157–162, 2007. doi: 10.1016/j.etp.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Starmann J, Fälth M, Spindelböck W, Lanz KL, Lackner C, Zatloukal K, Trauner M, Sültmann H. Gene expression profiling unravels cancer-related hepatic molecular signatures in steatohepatitis but not in steatosis. PLoS One 7: e46584, 2012. doi: 10.1371/journal.pone.0046584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strasberg SM, Howard TK, Molmenti EP, Hertl M. Selecting the donor liver: risk factors for poor function after orthotopic liver transplantation. Hepatology 20: 829–838, 1994. doi: 10.1002/hep.1840200410. [DOI] [PubMed] [Google Scholar]
- 26.Syn WK, Choi SS, Diehl AM. Apoptosis and cytokines in non-alcoholic steatohepatitis. Clin Liver Dis 13: 565–580, 2009. doi: 10.1016/j.cld.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi S, Saito K, Jia H, Kato H. An integrated multi-omics study revealed metabolic alterations underlying the effects of coffee consumption. PLoS One 9: e91134, 2014. doi: 10.1371/journal.pone.0091134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vickers KC, Remaley AT. HDL and cholesterol: life after the divorce? J Lipid Res 55: 4–12, 2014. doi: 10.1194/jlr.R035964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W, Smail N, Wang P, Chaudry IH. Increased gut permeability after hemorrhage is associated with upregulation of local and systemic IL-6. J Surg Res 79: 39–46, 1998. doi: 10.1006/jsre.1998.5385. [DOI] [PubMed] [Google Scholar]
- 30.Wu CC, Ho WL, Lin MC, Yeh DC, Wu HS, Hwang CJ, Liu TJ, P’eng FK. Hepatic resection for bilobar multicentric hepatocellular carcinoma: is it justified? Surgery 123: 270–277, 1998. doi: 10.1016/S0039-6060(98)70179-5. [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi K, Itoh Y, Yokomizo C, Nishimura T, Niimi T, Umemura A, Fujii H, Okanoue T, Yoshikawa T. Blockade of IL-6 signaling exacerbates liver injury and suppresses antiapoptotic gene expression in methionine choline-deficient diet-fed db/db mice. Lab Invest 91: 609–618, 2011. doi: 10.1038/labinvest.2011.2. [DOI] [PubMed] [Google Scholar]
- 32.Yang SQ, Lin HZ, Mandal AK, Huang J, Diehl AM. Disrupted signaling and inhibited regeneration in obese mice with fatty livers: implications for nonalcoholic fatty liver disease pathophysiology. Hepatology 34: 694–706, 2001. doi: 10.1053/jhep.2001.28054. [DOI] [PubMed] [Google Scholar]
- 33.Zerrad-Saadi A, Lambert-Blot M, Mitchell C, Bretes H, Collin de l’Hortet A, Baud V, Chereau F, Sotiropoulos A, Kopchick JJ, Liao L, Xu J, Gilgenkrantz H, Guidotti JE. GH receptor plays a major role in liver regeneration through the control of EGFR and ERK1/2 activation. Endocrinology 152: 2731–2741, 2011. doi: 10.1210/en.2010-1193. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Baker SS, Baker RD, Zhu R, Zhu L. Systematic analysis of the gene expression in the livers of nonalcoholic steatohepatitis: implications on potential biomarkers and molecular pathological mechanism. PLoS One 7: e51131, 2012. doi: 10.1371/journal.pone.0051131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmers TA, Jin X, Hsiao EC, McGrath SA, Esquela AF, Koniaris LG. Growth differentiation factor-15/macrophage inhibitory cytokine-1 induction after kidney and lung injury. Shock 23: 543–548, 2005. [PubMed] [Google Scholar]
- 36.Zimmers TA, Jin X, Hsiao EC, Perez EA, Pierce RH, Chavin KD, Koniaris LG. Growth differentiation factor-15: induction in liver injury through p53 and tumor necrosis factor-independent mechanisms. J Surg Res 130: 45–51, 2006. doi: 10.1016/j.jss.2005.07.036. [DOI] [PubMed] [Google Scholar]
- 37.Zimmers TA, McKillop IH, Pierce RH, Yoo JY, Koniaris LG. Massive liver growth in mice induced by systemic interleukin 6 administration. Hepatology 38: 326–334, 2003. doi: 10.1053/jhep.2003.50318. [DOI] [PubMed] [Google Scholar]
- 38.Zimmers TA, Pierce RH, McKillop IH, Koniaris LG. Resolving the role of IL-6 in liver regeneration. Hepatology 38: 1590–1591, 2003. doi: 10.1016/j.hep.2003.09.021. [DOI] [PubMed] [Google Scholar]