Abstract
The ribosomal translocation involves both intersubunit rotations between the small 30S and large 50S subunits and the intrasubunit rotations of the 30S head relative to the 30S body. However, the detailed molecular mechanism on how the intersubunit and intrasubunit rotations are related to the translocation remains unclear. Here, based on available structural data a model is proposed for the ribosomal translocation, into which both the intersubunit and intrasubunit rotations are incorporated. With the model, we provide quantitative explanations of in vitro experimental data showing the biphasic character in the fluorescence change associated with the mRNA translocation and the character of a rapid increase that is followed by a slow single-exponential decrease in the fluorescence change associated with the 30S head rotation. The calculated translation rate is also consistent with the in vitro single-molecule experimental data.
Keywords: Ribosome, Translocation, Intrasubunit rotation, Intersubunit rotation, Hybrid state, Translation
Highlights
-
•
A model of mRNA translocation with inclusion of 30S head rotations is presented.
-
•
Experimental data on biphasic kinetics of translocation are quantitatively explained.
-
•
Experimental data on kinetics of 30S head rotation are quantitatively explained.
1. Introduction
One of the important and complex activities during translation elongation by the ribosome is the coupled translocation of transfer RNAs (tRNAs) and messenger RNA (mRNA) in the ribosome, which is catalyzed by elongation factor G (EF-G), hydrolyzing GTP. It has been well characterized that the translocation takes place generally via two steps. After the peptidyl transfer, the two tRNAs move relative to the large 50S ribosomal subunit from the classical non-rotated pretranslocation (A/A and P/P) state to the intermediate hybrid (A/P and P/E) state [1], [2], [3], [4], [5], [6]. The formation of the hybrid state can occur spontaneously and reversibly, without the need of EF-G, but the binding of EF-G stabilizes the hybrid state [1], [2], [3], [4], [5], [6]. This first step is correlated with the forward (counterclockwise, as viewed from the exterior of the 30S) and reverse (clockwise) intersubunit rotations of the small 30S ribosomal subunit relative to the 50S subunit. In the second step, the tRNAs coupled with the mRNA move relative to the 30S subunit from the hybrid state to the posttranslocation (P/P and E/E) state, placing the next codon of the mRNA in the A site. This step is catalyzed by EF-G.GTP and is correlated with the reverse intersubunit rotation [7], [8], [9].
Besides the intersubunit rotations, a lot of structural studies showed that the intrasubunit rotation of the 30S head relative to the body is also implicated in the translocation of the tRNA–mRNA complex in the 30S subunit [10], [11], [12], [13]. Moreover, Guo and Noller [14] observed in real time the rotation of the 30S head during the translocation using ensemble stopped-flow fluorescence resonance energy transfer (FRET) with ribosomes containing fluorescent probes attached to specific positions in the head and body of the 30S subunit. Nevertheless, the detailed molecular mechanism on how the intersubunit and intrasubunit rotations are related to the translocation remains unclear.
In our previous paper [15], we proposed a simplified model of mRNA translocation in the ribosome. In that model only intersubunit rotations were considered while the intrasubunit rotation of the 30S head during the translocation was not included, and thus only the experimental data on the biphasic character of the mRNA translocation can be explained [15]. In this paper, we extend the previous model by considering the effect of the intrasubunit rotation of the 30S head on the mRNA translocation, providing quantitative explanations of the experimental data both on the biphasic kinetics of the mRNA translocation and on the kinetics of the 30S head rotations observed recently by Guo and Noller [14].
2. Models
2.1. Translocation without inclusion of intra-subunit rotations
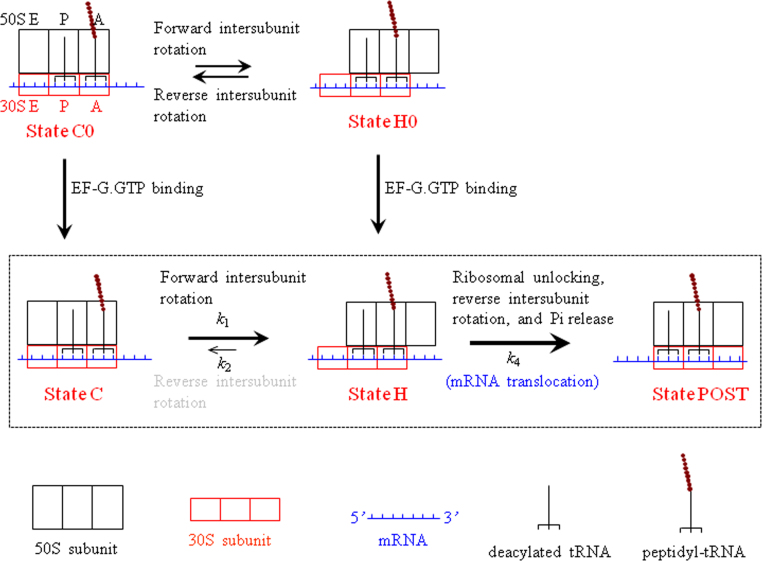
For convenience of reading, here we re-describe the model proposed before (Fig. 1) [15]. Before EF-G.GTP binding, the pretranslocation ribosomal complex transits spontaneously between the classical non-rotated pretranslocation state (State C0) and hybrid state (State H0), with the two states being in thermodynamic equilibrium with each other [1], [2], [3]. After adding EF-G.GTP into the solution containing the pretransolcation complex, EF-G.GTP can bind to both State C0 and State H0, as shown by structural [16], [17] and single-molecule [18], [19] studies. (i) If EF-G.GTP binds to State H0 (becoming State H), after rapid GTP hydrolysis the ribosomal unlocking occurs, widening the mRNA channel in the 30S subunit. The subsequent reverse intersubunit rotation couples with the translocation of the tRNA–mRNA complex in the 30S subunit, with State H transiting to State POST. This gives a fraction of mRNA translocation events having a larger translocation rate. (ii) If EF-G.GTP binds to State C0 (becoming State C), the EF-G-facilitated forward intersubunit rotation induces the transition of State C to State H. After the ribosomal unlocking in State H, the translocation of tRNA–mRNA complex in the 30S subunit occurs. The transition from State C to State H to State POST gives another fraction of mRNA translocation events having a smaller translocation rate. Thus, the kinetics of mRNA translocation would have the biphasic character, giving an explanation of the available experimental data [9], [20], [21], [22], [23].
Fig. 1.
The simplified model of mRNA translocation in the ribosome with the consideration of only intersubunit rotations (see text for detailed description).
Although the mRNA translocation has the biphasic character, the decay of the probability of the ribosome in the rotated conformation (State H) with time can be fit to a single exponential [15], which is also consistent with the experimental data on the kinetics of the reverse intersubunit rotation versus time [9]. In addition, Walker et al. [20] showed that although the mutation to the 50S E site alters significantly the heterogeneity of the pretranslocation ribosomal complex that can spontaneously fluctuate between the classical non-rotated and hybrid states, the mutant ribosome showed the similar relative amplitudes of the two phases to the wild-type case in the biphasic mRNA translocation. With the model that is modified from Fig. 1 these experimental data of Walker et al. [20] were also reasonably explained [15].
2.2. Translocation with inclusion of intra-subunit rotations
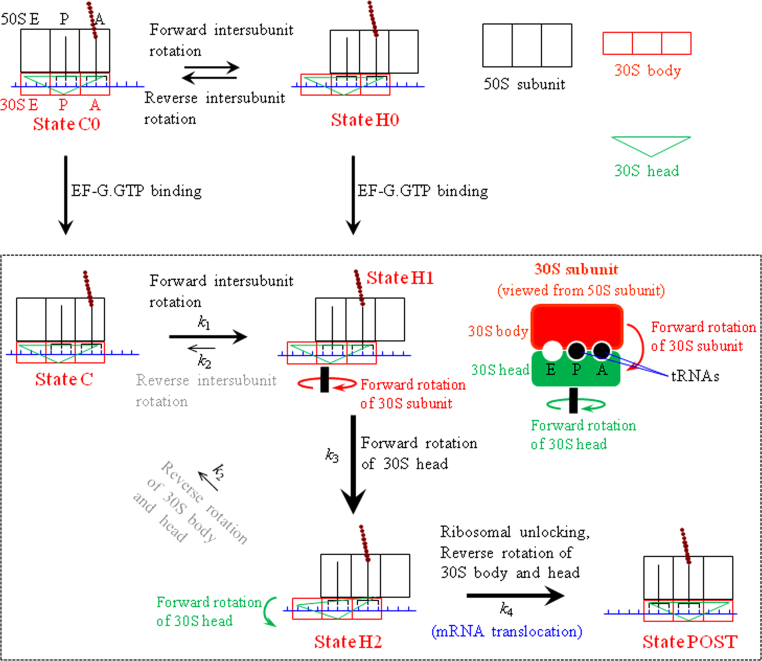
Here, we extend the model (Fig. 1) by considering the effect of the rotation of the 30S head on the mRNA translocation. Based on available structural data, we make three arguments. (i) Available structural data showed that although for ribosomes with no tRNA both the 30S subunit relative to the 50S subunit and the 30S head relative to the 30S body are not in the fixed orientations [24], [25], [26], [27], [28], [29], [30], the ribosomes complexed with tRNAs in the classical non-rotated state have no or nearly no 30S head rotation [31], [32], [33], [34], [35], [36], [37], [38], [39]. As during the translocation the ribosome is always bound with tRNAs, it is expected that during the translocation the ribosomal complex with no intersubunit rotation has the non-rotated 30S head. Thus, we argue that the transition from the rotated/hybrid state to the non-rotated state is accompanied or followed immediately by the reverse rotation of the 30S head if it is rotated. (ii) Inspired by cryo-EM data of Taylor et al. [10], we argue that in the hybrid state, after GTP hydrolysis to GDP.Pi or in GDP.Pi form small conformational changes in EF-G cause the tip (loops I and II) of domain IV to shift towards and interact with the decoding center in the 30S subunit, inducing the forward rotation of the 30S head.1 (iii) As it is implicated from the available structural studies [11], [13], we argue that the forward rotation of the 30S head leads to widening of the mRNA channel (termed ribosomal unlocking). From arguments (i)–(iii), it is noted that the ribosomal unlocking can only occur in the hybrid state with the 30S head rotation.
Based on the above arguments, the extended model of the mRNA translocation is schematically shown in Fig. 2. As mentioned above, before EF-G.GTP binding the pretranslocation ribosomal complex transits spontaneously between State C0 and State H0, with the two states being in thermodynamic equilibrium with each other. After addition of EF-G.GTP into the solution, EF-G.GTP can bind to both State C0 and State H0. (i) If EF-G.GTP binds to the hybrid state, State H0 becomes State H1, which could be accompanied by little forward rotation of the 3S head [11], [13], [40].2 After rapid GTP hydrolysis to GDP.Pi, the 30S head makes a large forward rotation relative to the 30S body, with State H1 becoming State H2. Then, after the ribosomal unlocking takes place in State H2, widening the mRNA channel, the translocation of tRNA–mRNA complex in the 3S subunit is associated with the subsequent reverse intersubunit rotation that is accompanied or followed immediately by the reverse rotation of the 30S head, with State H2 transiting to State POST. As after the ribosomal unlocking the reverse intersubunit rotation occurs rapidly, the transition from State H2 to State POST is rate-limited by the ribosomal unlocking [41]. The ribosomal unlocking is also followed by rapid Pi release, which is independent of the reverse intersubunit rotation [42]. After transition to the non-rotated state (State POST), as the 30S head is not rotated the mRNA channel becomes tight again, as proposed by Frank and Agrawal [43]. (ii) If EF-G.GTP binds to the classical non-rotated pretranslocation state, State C0 becomes State C. After EF-G-facilitated transition of State C to State H1, the large forward rotation of the 30S head takes place, with State H1 becoming State H2. After the ribosomal unlocking in State H2, the translocation of tRNA–mRNA complex in the 30S subunit is associated with the subsequent reverse intersubunit rotation that is accompanied or followed immediately by the reverse rotation of the 30S head.
Fig. 2.
The model of mRNA translocation in the ribosome with the inclusion of both intersubunit rotations and intrasubunit rotations of the 30S head (see text for detailed description). The panel on the right hand of State H1 shows the 30S subunit viewed from the 50S subunit, where the ribosomal complex is in the hybrid state with two tRNAs bound to the 30S P and A sites. Green arrows indicate the direction of the forward rotation of the 30S head relative to the 30S body and red arrows indicate the direction of the forward rotation of the 30S subunit relative to the 50S subunit.
2.3. Pathway of the elongation cycle
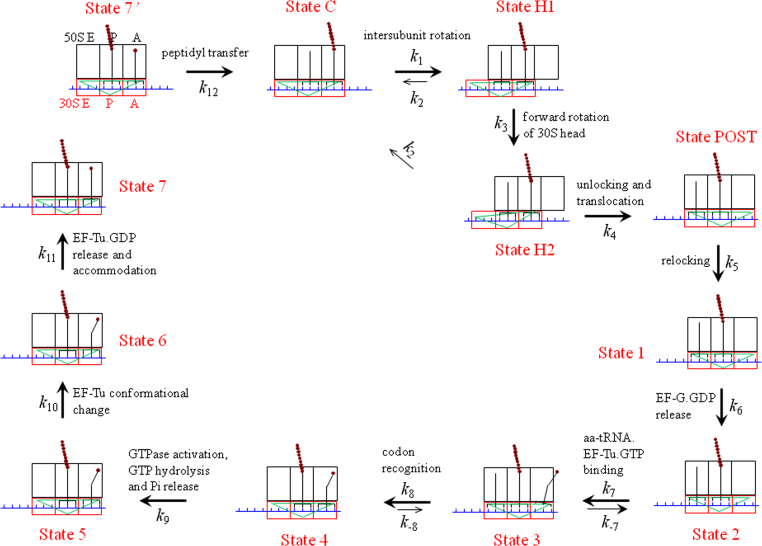
From the translocation model of Fig. 2, the pathway of an elongation cycle under saturating concentration of EF-G.GTP is shown in Fig. 3. We begin the elongation cycle with just after the peptidyl transfer and the ribosomal complex is in the classical non-rotated pretranslocation state, with deacylated tRNA being in the P/P site and the peptidyl-tRNA being in the A/A site. Then, EF-G.GTP binds immediately to the ribosomal complex (State C) before the spontaneous intersubunit rotation occurs. After EF-G-facilitated transition of State C to State H1, the forward rotation of the 30S head occurs, with State H1 becoming State H2. Then, after the ribosomal unlocking, the subsequent reverse intersubunit rotation couples with the translocation of tRNA–mRNA complex in the 30S subunit, with State H2 transiting to State POST. In State POST with the non-rotated conformation, the mRNA channel in the 30S subunit becomes tight again and then the ribosome becomes relocked (State 1), facilitating EF-G.GDP release (State 2). Then, the ternary complex composed of the cognate aminoacyl-tRNA, elongation factor EF-Tu and GTP binds to the ribosome in the partially bound “A/T” state (State 3).3 The subsequent codon recognition (State 4) triggers GTPase activation, GTP hydrolysis and Pi release (State 5) [44], resulting in a large-scale conformational change of EF-Tu to the GDP-bound form (State 6) [45], [46], [47]. EF-Tu.GDP is then released and the aminoacyl-tRNA is accommodated into its fully bound A/A state inside the ribosome (State 7 or State 7′). Then, the peptidyl transfer occurs, resulting in deacylated tRNA in the P site and the peptidyl-tRNA prolonged by one amino acid in the A site, from which the next elongation cycle will proceed.
Fig. 3.
Pathway of translation elongation. Here we draw that the dissociation of the deacylated tRNA from the ribosome occurs after the codon recognition. In fact, before the binding of the ternary complex, the dissociation of the deacylated tRNA can also occurs [61], [62].
3. Equations
First, we derive equations for the kinetics of mRNA translocation and 30S head rotations at saturating concentration of EF-G.GTP using model of Fig. 2 (inside the box). We denote by P1, P2, P3 and P4 the probabilities of State C, State H1, State H2 and State POST, respectively. Since whether before or after the release of EF-G.GDP from State POST, the ribosome is kept unchanged relative to the mRNA, implying that the pyrene that is labeled at the 3′ end of mRNA in the experiments [9], [14], [20], [21], [22], [23] has the same fluorescence intensity. Thus, we can denote by P4 the probability of the posttranslocation states both before and after EF-G.GDP release. Then, the temporal evolutions of the four probabilities are described by following equations:
| (1) |
| (2) |
| (3) |
| (4) |
For the procedure as used in the experiment [9], [14], [20], [21], [22], [23], the initial conditions at t=0 are as follows: P1(0)=1–a, P2(0)=a, and P3(0)=P4(0)=0, where time t=0 represents the moment when the pretranslocation complex is mixed with EF-G.GTP of saturating concentration and a denotes the probability of the ribosomal complex being in the hybrid state before the mixing of EF-G.GTP.
Available experimental data indicated that the binding of EF-G.GTP shifts the equilibrium toward the hybrid state, increasing the forward transition rate k1 and greatly decreasing the backward transition rate k2 [2]. Thus, we have k2≪k1. For the approximation of k2=0, we can obtain the analytical solutions of Eqs. (1), (2), (4), which have following forms:
| (5) |
| (6) |
Based on the model of Fig. 2, P3(t) corresponds to the fluorescence change associated with the rotation of the 30S head in the experiments of Guo and Noller [14] and P4(t) to the fluorescence change associated with the mRNA translocation.
Then, we derive the equation of translation rate in the elongation cycle at saturating concentrations of EF-G.GTP and the ternary complex. Based on Fig. 3, we can easily derive the mean time, , for transition from State C to State POST as
| (7) |
Under k3≫k1, k2 and k4, Eq. (7) becomes
| (8) |
It is noted that from the scheme of Fig. 1 (inside box) we can obtain the same equation of as Eq. (8), implying that when k3≫k1, k2 and k4, the model of Fig. 2 becomes the same as that of Fig. 1. The mean time for transition from State POST to State C (Fig. 3) at saturating concentration of the ternary complex is calculated by
| (9) |
With Eqs. (7), (9), the translation rate at saturating concentrations of EF-G.GTP and the ternary complex is calculated by
| (10) |
4. Results
The experimental data [9], [14] on the kinetics of mRNA translocation catalyzed by EF-G with GTP can be described by following function:
| (11) |
where Ppost is the probability of the posttranslocation state (State POST, Fig. 1) and A1+A2=1. The experimental data in Ermolenko and Noller [9] gave A1=0.57, A2=0.43, =6.9 and =0.7 for the wild-type ribosome. In the previous paper [15], using model of Fig. 1 we showed that by fixing value of k2 and adjusting values of parameters a, k1 and k4, the theoretical data on the biphasic kinetics of mRNA translocation (characterized by A1, A2, and ) are identical to the corresponding experimental data [9]. In more detail, using model of Fig. 1 we showed that with the following values of parameters we obtained the above results for A1, A2, and : (i) for fixed k2=0, we have a=0.614, k1=0.7 and k4=6.9 ; (ii) for fixed k2=0.1 , we have a=0.624, k1=0.715 and k4=6.79 ; (iii) for fixed k2=0.2 , we have a=0.634, k1=0.724 and k4=6.675 ; (iv) for fixed k2=0.4 , we have a=0.656, k1=0.748 and k4=6.452 ; or (v) for fixed k2=0.6 , we have a=0.68, k1=0.776 and k4=6.225 [15]. Moreover, we showed that with the above parameter values the obtained theoretical data on the single-exponential kinetics of the reverse intersubunit rotation versus time [15] are also in quantitative agreement with the experimental data [9].
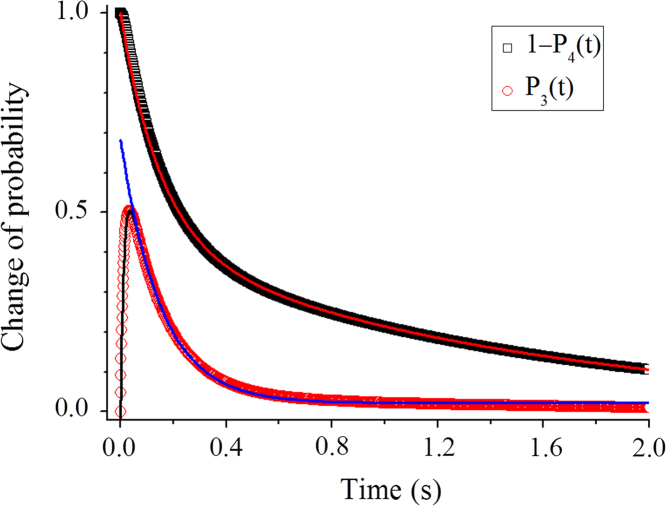
Here, we still use above values of parameters a, k1, k2 and k4 to study the kinetics of mRNA translocation and 30S head rotations with the extended model (Fig. 2). In addition, to be consistent with the experimental data [14], we take k3=80 . For the case of fixed k20, we solve Eqs. (1), (2), (4) numerically by using Runge–Kutta method to obtain the temporal evolutions of the state probabilities. For the case of fixed k2=0, we use both the analytical solutions described by Eqs. (5), (6) and the numerical method to calculate temporal evolutions of P3(t) and P4(t) (the two methods give the same results), which correspond respectively to the fluorescence change associated with the rotation of the 30S head and to the fluorescence change associated with the mRNA translocation in the experiments of Guo and Noller [14]. In Fig. 4 we show results of P3(t) and versus time for the case of fixed k2=0 (dots). It is seen that the results of versus time can be fit to the two-exponential function, , with A1=0.57, =6.6 and =0.7 (red line). The results of P3(t) versus time show a rapid increase in P3(t), followed by a slow decrease. The slow phase can be fit to a single-exponential function, (blue line), where B1, B2 and are constants, with =6.6 that is identical to =6.6 . These features are in good agreement with the experimental data of Guo and Noller [14] showing that within the experimental errors the rate of the reverse 30S head rotation is equal to that of the fast mRNA translocation phase. Note that the results of P3(t) versus time can be fit to the function, , where C1, C2, C3, and are constants, with =6.6 and =80 (black line). For the case of fixed k20, the corresponding results are given in Fig. S1 (see Supplementary material), where it is seen that the features in Fig. S1 are the same as those in Fig. 4.
Fig. 4.
Temporal evolutions of P3(t) (open circles) and (open squares) for the case of fixed k2=0. P3(t) corresponds to the fluorescence change associated with the rotation of the 30S head and to the fluorescence change associated with the mRNA translocation. The data of P3(t) as a function of t are fit to the function, , where C1=0.66, C2=0.65, C3=0.02, =6.6 and =80 (black line). The data on the slow phase of P3(t) as a function of t are fit to the single exponential, , where B1=0.66, B2=0.02 and = 6.6 (blue line). The data of are fit to the two-exponential function, , with A1+A2=1, A1=0.57, =6.6 and =0.7 (red line).
With above values of rate constants, e.g., k1=0.7 , k2=0, k3=80 , k4=6.9 (see above), and values of other rate constants k5–k12 (Fig. 3) given in Table 1, which are taken from the available in vitro biochemical data [48], [49], [50], [51], [52], [53], using Eqs. (7), (9), (10) we calculate the translation rate to be 0.45 codons/s during the elongation cycle. This value of translation rate is consistent with the single-molecule experimental data on the mean rate (0.4–0.5 codons/s) of translation through the single-stranded mRNA with a homogeneous codon sequence obtained by Noller and his colleagues [54].
Table 1.
In vitro values of rate constants for Escherichia coli ribosome, which are taken from the available biochemical data [48], [49], [50], [51], [52], [53].
| Rate constants (s−1) | Values |
|---|---|
| k5 | 5 |
| k6 | 20 |
| k8 | 100 |
| k−8 | 0.2 |
| k9 | 260 |
| k10 | 60 |
| k11 | 3 |
| k12 | 50 |
5. Discussion
In the previous paper [15], a simplified model of mRNA translocation (Fig. 1) has been presented, where only the intersubunit rotations were considered. Here, an extended model is proposed (Fig. 2), where besides the intersubunit rotations the intrasubunit rotations of the 30S head are also included.
In the extended model (Fig. 2), the forward rotation of the 30S head is purposed to widen the mRNA channel and the subsequent reverse intersubunit rotation couples with the mRNA translocation in the 30S subunit. The model also proposes that the reverse intersubunit rotation is accompanied or followed immediately by the reverse 30S head rotation. With the proposal, the available structural data of trapped intermediate states with the reverse rotation of the 30S body from about 8° to about 2.5° and the forward rotation of the 30S head to about 18° in the presence of antibiotic fusidic acid [11], [12], [13] can be readily explained. The altered conformation (particularly the altered orientation of domain IV) of EF-G caused by fusidic acid bound to it [29] would inhibit the 30S head from rotating backward, which can be noted from the structural data showing that domain IV of EF-G makes contact with helices h34 and h35 in the 30S head [29] and the two helices are critical for the 30S head rotation [55]. As the backward 30S head rotation accompanies or follows immediately the reverse intersubunit rotation, the 30S head that is trapped in the forward rotation would thus resist the 30S body from rotating backwards completely to the non-rotated conformation, making the 30S body stay in the intermediate rotation of about 2.5°. It is important to note here that the proposal is only applicable to the case when the ribosome is bound with tRNA, as available structural data showed [31], [32], [33], [34], [35], [36], [37], [38], [39]. However, when the ribosome is not bound with tRNA, both the 30S subunit relative to the 50S subunit and the 30S head relative to the 30S body are not in the fixed orientations [24], [25], [26], [27], [28], [29], [30]. Thus, the structural data of the ribosome without tRNA but with release factor RF3, which has close structural similarities to EF-G but completely lacks domain IV, having a large head rotation [56] are also understandable.
With the extended model (Fig. 2), the experimental data both on the biphasic kinetics of mRNA translocation and on the kinetics of 30S head rotations [14] are quantitatively explained. By comparison, we consider the model for the order of dynamic structural events during mRNA translocation proposed by Guo and Noller [14] (see Fig. S2a in Supplementary material). It is shown that the analytical solutions based on Fig. S2a are not consistent with the experimental data [14] (see Text S1 and Fig. S2b in Supplementary material).
Based on our explanations, the rate of the forward intersubunit rotation after the binding of EF-G.GTP is about k1=0.7 , which is consistent with the available single-molecule FRET (smFRET) data obtained by Noller and his colleagues [2], as seen below. The smFRET data showed that for the ribosomal complex with peptidyl-tRNA analog N-Ac-Phe-tRNAPhe bound to the 30S A site and deacylated tRNAfMet bound to the 30S P site, the rate of transition from State C0 to State H0 in the absence of EF-G is about k01=0.270.08 [2]. The smFRET data also showed that when EF-G.GDPNP is bound to the ribosomal complex with deacylated tRNAfMet bound to the 30S P site, the rate of transition from classical non-rotated to hybrid states is increased by about 2.33-fold [2]. Thus, for the ribosomal complex with two tRNAs bound to the 30S A and P sites, the rate of transition from State C to State H1 in the presence of EF-G.GDPNP or EF-G.GTP is estimated to be =0.630.16 , which is consistent with the value of about k1=0.7 determined here. In the rotated/hybrid state after rapid GTP hydrolysis to GDP.Pi, the rate of the forward rotation of the 30S head is about k3=80 . Then the ribosomal unlocking occurs, which is followed by mRNA translocation, with the rate constant of about k4=6.9 . Moreover, with above values of rate constants and values of other rate constants in the elongation cycle which are taken from the available in vitro biochemical data, the calculated translation rate is also consistent with the in vitro single-molecule experimental data obtained by Noller and his colleagues [54]. However, it is noted that the translation rate observed in vivo is more than one order of magnitude larger than the in vitro single-molecule experimental data [54], which could be due to the fact that the in vivo rate constants of the transitions in the elongation cycle are much larger than the corresponding in vitro ones [57]. In addition, it is noted that since the rate constants of state transitions during the translocation are dependent on the 70S preparation and buffer condition, in vitro experiments by other research groups [20], [23], [58] gave values of the rate constant such as k1 being much larger than 0.7 that is obtained here by fitting to the experimental data of Guo and Noller [14], as explained before [15], [59].
In conclusion, based on the available structural data we present a model of ribosomal translocation which is coupled with both the intrasubunit and intersubunit rotations. The model gives quantitative explanations of the in vitro biochemical data on both the biphasic character in the fluorescence change associated with the mRNA translocation and the character of a rapid increase that is followed by a slow single-exponential decrease in the fluorescence change associated with the 30S head rotation.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant no. 11374352).
Footnotes
Structural data indicated that the binding of EF-G with the nonhydrolyzable GTP analog GDPNP or GMPPNP can also induce the large forward rotation of the 30S head [11], [13]. It is argued here that EF-G in GDPNP or GMPPNP form is much less efficient than in GDP.Pi form to induce the large forward 30S head rotation.
Structural data indicated that the binding of EF-G.GTP to the hybrid state could induce the forward rotation of the 30S head with a small angle of about 3–5°, [11], [13], [40], reducing mildly the interaction of the 30S subunit with the tRNA–mRNA complex, which is consistent with the biochemical data showing that the binding of EF-G.GDPNP promotes mRNA back-slippage [60]. The further large forward 30S head rotation of about 18° [11], [13], which is induced by GTP hydrolysis, induces ribosomal unlocking, widening completely the mRNA channel.
For simplicity of analysis, we consider here the translation of the mRNA with a homogeneous codon sequence and thus, we do not consider the effect of the near- and non-cognate ternary complexes.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.05.004.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Blanchard S.C., Kim H.D., Gonzalez R.L., Jr., Puglisi J.D., Chu. S. tRNA dynamics on the ribosome during translation. Proc. Natl Acad. Sci. USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornish P.V., Ermolenko D.N., Noller H.F., Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fei J., Kosuri P., MacDougall D.D., Gonzalez R.L., Jr. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Moazed D., Noller H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 5.Valle M., Zavialov A., Sengupta J., Rawat U., Ehrenberg M., Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 6.Zavialov A.V., Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- 7.Rodnina M.V., Savelsbergh A., Katunin V.I., Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 8.Savelsbergh A., Rodnina M.V., Wintermeyer W. Distinct functions of elongation factor G in ribosome recycling and translocation. RNA. 2009;15:772–780. doi: 10.1261/rna.1592509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ermolenko D.N., Noller H.F. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat. Struct. Mol. Biol. 2011;18:457–463. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor D.J., Nilsson J., Merrill A.D., Andersen G.R., Nissen P., Frank J. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ratje A.H., Loerke J., Mikolajka A. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramrath D.J., Lancaster L., Sprink T., Mielke T., Loerke J., Noller H.F., Spahn C.M.T. Proc. Natl. Acad. Sci. USA. 2013;110:20964–20969. doi: 10.1073/pnas.1320387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J., Lancaster L., Donohue J.P., Noller H.F. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340:1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Z., Noller H.F. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc. Natl. Acad. Sci. USA. 2012;109:20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie P. An explanation of biphasic characters of mRNA translocation in the ribosome. Bio Systems. 2014;118:1–7. doi: 10.1016/j.biosystems.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Whitford P.C., Ahmed A., Yu Y., Hennelly S.P., Tama F., Spahn C.M.T., Onuchic J.N., Sanbonmatsu K.Y. Excited states of ribosome translocation revealed through integrative molecular modeling. Proc. Natl. Acad. Sci. USA. 2011;108:18943–18948. doi: 10.1073/pnas.1108363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J., Gagnon M.G., Bulkley D., Steitz T.A. Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell. 2015;160:219–227. doi: 10.1016/j.cell.2014.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C., Stevens B., Kaur J., Cabral D., Liu H., Wang Y., Zhang H., Rosenblum G., Smilansky Z., Goldman Y.E., Cooperman B. Single-molecule fluorescence measurements of ribosomal translocation dynamics. Mol. Cell. 2011;42:367–377. doi: 10.1016/j.molcel.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J., Petrov A., Tsai A., O’Leary S.E., Puglisi J.D. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat. Struct. Mol. Biol. 2013;20:718–727. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker S.E., Shoji S., Pan D., Cooperman B.S., Fredrick K. Role of hybrid tRNA-binding states in ribosomal translocation. Proc. Natl. Acad. Sci. USA. 2008;105:9192–9197. doi: 10.1073/pnas.0710146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peske F., Savelsbergh A., Katunin V.I., Rodnina M.V., Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA–mRNA translocation. J. Mol. Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 22.Shi X., Chiu K., Ghosh S., Joseph S. Bases in 16S rRNA important for subunit association, tRNA binding, and translocation. Biochemistry. 2009;48:6772–6782. doi: 10.1021/bi900472a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q., Fredrick K. Contribution of intersubunit bridges to the energy barrier of ribosomal translocation. Nucleic Acids Res. 2013;41:565–574. doi: 10.1093/nar/gks1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pulk A., Cate J.H.D. Control of ribosomal subunit rotation by elongation factor G. Science. 2013;340:1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W., Dunkle J.A., Cate J.H.D. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borovinskaya M.A., Shoji S., Holton J.M., Fredrick K., Cate J.H.D. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem. Biol. 2007;2:545–552. doi: 10.1021/cb700100n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borovinskaya M.A., Shoji S., Fredrick K., Cate J.H. Structural basis for hygromycin B inhibition of protein biosynthesis. RNA. 2008;14:1590–1599. doi: 10.1261/rna.1076908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H.D. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 29.Schuwirth B.S., Day J.M., Hau C.W., Janssen G.R., Dahlberg A.E., Cate J.H.D., Vila-Sanjurjo A. Structural analysis of kasugamycin inhibition of translation. Nat. Struct. Mol. Biol. 2006;13:879–886. doi: 10.1038/nsmb1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ben-Shem A., de Loubresse N.G., Melnikov S., Jenner L., Yusupova G., Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 31.Selmer M., Dunham C.M., Murphy F.V., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 32.Dunkle J.A., Wang L., Feldman M.B. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenner L.B., Demeshkina N., Yusupova G., Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 34.Stanley R.E., Blaha G., Grodzicki R.L., Strickler M.D., Steitz T.A. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat. Struct. Mol. Biol. 2010;17:289–293. doi: 10.1038/nsmb.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng S., Chen Y., Gao Y.-G. Crystal structure of 70S ribosome with both cognate tRNAs in the E and P sites representing an authentic elongation complex. PLoS ONE. 2013;8:e58829. doi: 10.1371/journal.pone.0058829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao Y.-G., Selmer M., Dunham C.M., Weixlbaumer A., Kelley A.C., Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H.D., Noller H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 38.Laurberg M., Asahara H., Korostelev A., Zhu J.Y., Trakhanov S., Noller H.F. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 39.Korostelev A., Asahara H., Lancaster L. Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl. Acad. Sci. USA. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tourigny D.S., Fernández I.S., Kelley A.C., Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie P. Dynamics of forward and backward translocation of mRNA in the ribosome. PLoS ONE. 2013;8:e70789. doi: 10.1371/journal.pone.0070789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savelsbergh A., Katunin V.I., Mohr D., Peske F., Rodnina M.V., Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA–mRNA translocation. Mol. Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 43.Frank J., Agrawal R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 44.Rodnina M.V., Fricke R., Kuhn L., Wintermeyer W. Codondependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. EMBO J. 1995;14:2613–2619. doi: 10.1002/j.1460-2075.1995.tb07259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dell V.A., Miller D.L., Johnson A.E. Effects of nucleotide- and aurodox-induced changes in elongation factor Tu conformation upon its interactions with aminoacyl tranfer RNA. A fluorescence study. Biochemistry. 1990;29:1757–1763. doi: 10.1021/bi00459a014. [DOI] [PubMed] [Google Scholar]
- 46.Abel K., Yoder M.D., Hilgenfeld R., Jurnak F. An to β conformational switch in EF-Tu. Structure. 1996;4:1153–1159. doi: 10.1016/s0969-2126(96)00123-2. [DOI] [PubMed] [Google Scholar]
- 47.Polekhina G., Thirup S., Kjeldgaard M., Nissen P., Lippmann C., Nyborg J. Helix unwinding in the effector region of elongation factor EF-Tu–GDP. Structure. 1996;4:1141–1151. doi: 10.1016/s0969-2126(96)00122-0. [DOI] [PubMed] [Google Scholar]
- 48.Pape T., Wintermeyer W., Rodnina M.V. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pape T., Wintermeyer W., Rodnina M.V. Induced fit in initial selection and proofreading of aminocyl-tRNA on the ribosome. EMBO J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stark H., Rodnina M.V., Wieden H.J., Zemlin F., Wintermeyer W., van Heel M. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat. Struct. Biol. 2002;9:849–854. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- 51.Wintermeyer W., Peske F., Beringer M., Gromadski K.B., Savelsbergh A., Rodnina M.V. Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem. Soc. Trans. 2004;32:733–737. doi: 10.1042/BST0320733. [DOI] [PubMed] [Google Scholar]
- 52.Wilden B., Savelsbergh A., Rodnina M.V., Wintermeyer W. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc. Natl Acad. Sci. USA. 2006;103:13670–13675. doi: 10.1073/pnas.0606099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodnina M.V., Beringer M., Wintermeyer W. How ribosomes make peptide bonds. Trends Biochem. Sci. 2007;32:20–26. doi: 10.1016/j.tibs.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 54.Qu X., Wen J.-D., Lancaster L., Noller H.F., Bustamante C., Tinoco I., Jr. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mohan S., Donohue J.P., Noller H.F. Molecular mechanics of 30S subunit head rotation. Proc. Natl. Acad. Sci. USA. 2014;111:13325–13330. doi: 10.1073/pnas.1413731111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou J., Lancaster L., Trakhanov S., Noller H.F. Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. RNA. 2012;18:230–240. doi: 10.1261/rna.031187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rudorf S., Thommen M., Rodnina M.V., Lipowsky R. Deducing the kinetics of protein synthesis in vivo from the transition rates measured in vitro. PLoS Comput. Biol. 2014;10:e1003909. doi: 10.1371/journal.pcbi.1003909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Munro J.B., Altman R.B., O_Connor N., Blanchard S.C. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie P. Biphasic character of ribosomal translocation and non-Michaelis–Menten kinetics of translation. Phys. Rev. E. 2014;90:062703. doi: 10.1103/PhysRevE.90.062703. [DOI] [PubMed] [Google Scholar]
- 60.Spiegel P.C., Ermolenko D.N., Noller H.F. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uemura S., Aitken C.E., Korlach J., Flusberg B.A., Turner S.W., Puglisi J.D. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie P. Dynamics of tRNA occupancy and dissociation during translation by the ribosome. J. Theor. Biol. 2013;316:49–60. doi: 10.1016/j.jtbi.2012.09.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material